Abstract
Clinical metagenomic next-generation sequencing (mNGS), the comprehensive analysis of microbial and host genetic material (DNA and RNA) in samples from patients, is rapidly moving from research to clinical laboratories. This emerging approach is changing how physicians diagnose and treat infectious disease, with applications spanning a wide range of areas, including antimicrobial resistance, the microbiome, human host gene expression (transcriptomics) and oncology. Here, we focus on the challenges of implementing mNGS in the clinical laboratory and address potential solutions for maximizing its impact on patient care and public health.
Subject terms: Infectious diseases, Microbial genetics, Next-generation sequencing, Metagenomics
Clinical metagenomic next-generation sequencing (mNGS) is rapidly moving from bench to bedside. This Review discusses the clinical applications of mNGS, including infectious disease diagnostics, microbiome analyses, host response analyses and oncology applications. Moreover, the authors review the challenges that need to be overcome for mNGS to be successfully implemented in the clinical laboratory and propose solutions to maximize the benefits of clinical mNGS for patients.
Introduction
The field of clinical microbiology comprises both diagnostic microbiology, the identification of pathogens from clinical samples to guide management and treatment strategies for patients with infection, and public health microbiology, the surveillance and monitoring of infectious disease outbreaks in the community. Traditional diagnostic techniques in the microbiology laboratory include growth and isolation of microorganisms in culture, detection of pathogen-specific antibodies (serology) or antigens and molecular identification of microbial nucleic acids (DNA or RNA), most commonly via PCR. While most molecular assays target only a limited number of pathogens using specific primers or probes, metagenomic approaches characterize all DNA or RNA present in a sample, enabling analysis of the entire microbiome as well as the human host genome or transcriptome in patient samples. Metagenomic approaches have been applied for decades to characterize various niches, ranging from marine environments1 to toxic soils2 to arthropod disease vectors3,4 to the human microbiome5,6. These tools have also been used to identify infections in ancient remains7, discover novel viral pathogens8 and characterize the human virome in both healthy and diseased states9–11 and for forensic applications12.
The capacity to detect all potential pathogens — bacteria, viruses, fungi and parasites — in a sample and simultaneously interrogate host responses has great potential utility in the diagnosis of infectious disease. Metagenomics for clinical applications derives its roots from the use of microarrays in the early 2000s13,14. Some early successes using this technology include the discovery of the SARS coronavirus15, gene profiling of mutations in cancer16 and in-depth microbiome analysis of different sites in the human body17. However, it was the advent of next-generation sequencing (NGS) technologies in 2005 that jump-started the metagenomics field18. For the first time, millions to billions of reads could be generated in a single run, permitting analysis of the entire genetic content of a clinical or environmental sample. The proliferation of available sequencing instruments and exponential decreases in sequencing costs over the ensuing decade drove the rapid adoption of NGS technology.
To date, several studies have provided a glimpse into the promise of NGS in clinical and public health settings. For example, NGS was used for the clinical diagnosis of neuroleptospirosis in a 14-year-old critically ill boy with meningoencephalitis19; this case was the first to demonstrate the utility of metagenomic NGS (mNGS) in providing clinically actionable information, as successful diagnosis prompted appropriate targeted antibiotic treatment and eventual recovery of the patient. Examples in public health microbiology include the use of NGS, in combination with transmission network analysis20, to investigate outbreaks of the Escherichia coli strain O104:H4 (ref.21) and for surveillance of antimicrobial resistance in the food supply by bacterial whole-genome sequencing22. Increasingly, big data provided by mNGS is being leveraged for clinical purposes, including characterization of antibiotic resistance directly from clinical samples23 and analysis of human host response (transcriptomic) data to predict causes of infection and evaluate disease risk24,25. Thus, mNGS can be a key driver for precision diagnosis of infectious diseases, advancing precision medicine efforts to personalize patient care in this field.
Despite the potential and recent successes of metagenomics, clinical diagnostic applications have lagged behind research advances owing to a number of factors. A complex interplay of microbial and host factors influences human health, as exemplified by the role of the microbiome in modulating host immune responses26, and it is often unclear whether a detected microorganism is a contaminant, colonizer or bona fide pathogen. Additionally, universal reference standards and proven approaches to demonstrate test validation, reproducibility and quality assurance for clinical metagenomic assays are lacking. Considerations of cost, reimbursement, turnaround time, regulatory considerations and, perhaps most importantly, clinical utility also remain major hurdles for the routine implementation of clinical mNGS in patient care settings27.
We review here the various applications of mNGS currently being exploited in clinical and public health settings. We discuss the challenges involved in the adoption of mNGS in the clinical laboratory, including validation and regulatory considerations that extend beyond its initial development in research laboratories, and propose steps to overcome these challenges. Finally, we envisage future directions for the field of clinical metagenomics and anticipate what will be achievable in the next 5 years.
Applications of clinical metagenomics
To date, applications of clinical metagenomics have included infectious disease diagnostics for a variety of syndromes and sample types, microbiome analyses in both diseased and healthy states, characterization of the human host response to infection by transcriptomics and the identification of tumour-associated viruses and their genomic integration sites (Fig. 1; Table 1). Aside from infectious disease diagnostics, adoption of mNGS in clinical laboratories has been slow, and most applications have yet to be incorporated into routine clinical practice. Nonetheless, the breadth and potential clinical utility of these applications are likely to transform the field of diagnostic microbiology in the near future.
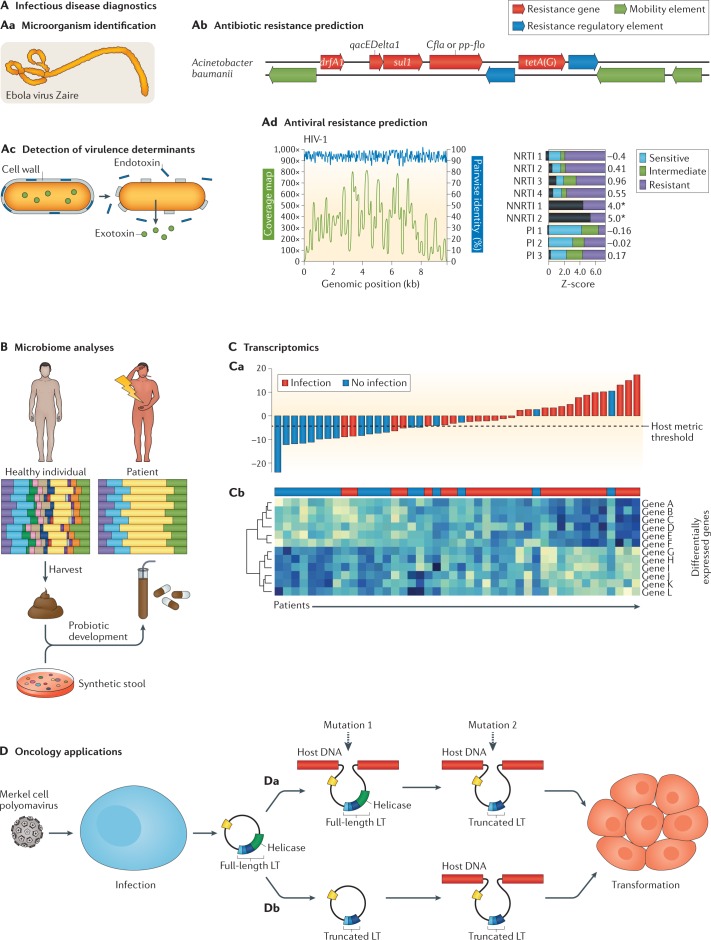
Fig. 1. Clinical applications of metagenomic sequencing.
A | Applications in infectious disease diagnostics include direct identification of microorganisms from primary clinical samples (part Aa); antimicrobial resistance prediction by characterization of resistance genes (part Ab); detection of species-level or strain-level virulence determinants, such as secretion of specific endotoxins or exotoxins (part Ac); and antiviral resistance prediction (part Ad). As shown for HIV-1, recovery of the complete viral genome from a patient sample by metagenomic next-generation sequencing (mNGS) (part Ad, graph) facilitates sequence analysis to predict susceptibility or resistance to antiretroviral drugs (part Ad, bar plot); the susceptibility profile for the analysed strain (black bars) predicts resistance to the non-nucleoside reverse transcriptase inhibitor (NNRTI) class of drugs (denoted by an asterisk), as opposed to nucleoside reverse transcriptase inhibitors (NRTIs) or protease inhibitors (PIs). B | Microbiome analyses can inform disease prognosis in acute and chronic disease states and underlie the development of probiotic therapies. Coloured bars represent individual microbiota species. A reduction in species diversity is seen in dysbiosis (an unhealthy state), such as present in patients with Clostridium difficile-associated disease. Stool from healthy individuals can be harvested to treat patients with C. difficile infection by faecal stool transplantation or as orally administered encapsulated faecal pills. Alternatively, synthetic stool generated from microbiota species observed in healthy individuals can be used as probiotics to treat patients. In addition to C. difficile infection, chronic diseases such as obesity, inflammatory bowel disease and diabetes mellitus are potential targets for probiotic therapy. C | RNA-sequencing-based transcriptomics can improve the diagnosis of infectious and non-infectious conditions on the basis of the human host response. Host transcriptomic profiling by NGS can enable the construction of a classifier metric to discriminate between patients with infection (red bars) from uninfected patients (blue bars) with high accuracy (part Ca). Metric scores above the dotted line indicate infection, whereas scores below the dotted line indicate absence of infection; the overall accuracy of the classifier metric shown is 83%. Cluster heat map analysis identifies individual, differentially expressed host genes associated with infection (genes A–F) versus those associated with no infection (genes G–L) (part Cb). D | Sequencing of viral tumours or liquid biopsy analyses in oncology can be used for simultaneous pathogen detection and characterization of host genetic mutations. mNGS can be used to detect Merkel cell polyomavirus, the virus associated with the development of Merkel cell carcinoma. Simultaneous sequencing of host DNA can identify mutations that arise from integration of the viral genome containing the full-length large T antigen (LT) followed by subsequent truncation of the LT antigen (part Da) or truncation of the LT antigen before viral genome integration (part Db). Both of these two mutations lead to cellular transformation that drives tumour proliferation. Although promising, many of these sequencing-based applications have yet to be incorporated into routine clinical practice. Part C is adapted from ref.25, CC BY-NC-ND 4.0 (https://creativecommons.org/licenses/by-nc-nd/4.0/). Part D is adapted from ref.134, CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/).
Table 1.
Clinical microbiology approaches using next-generation sequencing
| Sequencing method | Clinical sample type | Potential clinical indications | Clinical test available? | Refs |
|---|---|---|---|---|
| Infectious disease diagnosis — targeted analyses | ||||
| Amplicon sequencing (universal bacterial, fungal or parasitic rRNA sequencing) | Multiple body fluids and tissues | Multiplexed pathogen detection | Yesa | 39 |
| Amplicon sequencing (multiplexed primer panels) | Multiple body fluids and tissues | Multiplexed pathogen detection | No | 135 |
| Capture probe enrichment | Multiple body fluids and tissues | Viral genome recovery for infection control, epidemiology and public health | No | 43, 44, 46, 47 |
| Capture probe enrichment | Multiple body fluids and tissues | Multiplexed pathogen detection | No | 49– 52 |
| Capture probe enrichment | Multiple body fluids and tissues | Antibiotic resistance characterization | No | 23, 136 |
| Infectious disease diagnosis — untargeted analyses | ||||
| Metagenomic sequencing | Blood (plasma) | Culture-negative sepsis, endocarditis, febrile neutropenia, fever of unknown origin or monitoring of immunocompromised patients | Yesb | 33, 57 |
| Metagenomic sequencing | Respiratory secretions | Culture-negative and/or PCR-negative pneumonia | Yesc | 25, 37, 58, 137, 138 |
| Metagenomic sequencing | Cerebrospinal fluid | Undiagnosed meningitis, encephalitis or myelitis | Yesd | 36, 37 |
| Metagenomic sequencing | Stool | Severe diarrhoea | No | 139 |
| Metagenomic sequencing | Infected tissue or other body fluid | Culture-negative infection | No | 118, 140 |
| Microbiome analyses | ||||
| Metagenomic sequencing | Stool | Consumer-based microbiome testinge | Noe | No reference |
| Metagenomic sequencing | Stool | Guiding management and treatment of Clostridium difficile infection | No | 141 |
| Metagenomic sequencing | Stool | Chronic illnesses | No | 64 |
| Metagenomic sequencing | Respiratory secretions | Aiding in diagnosis of acute respiratory infection | No | 137 |
| Human host response analyses | ||||
| RNA sequencing | Multiple sample types; whole blood or PBMC most common | Aiding diagnosis or characterization of infections such as bacterial sepsis or pneumonia; disease prognosis | No | 24, 25, 68 |
| Oncological analyses | ||||
| Whole-genome tumour sequencing | Tumour | Identification of viruses associated with cancer | No | 142 |
| Liquid biopsy sequencing | Cell-free body fluids | Simultaneous cancer and infectious disease testing | No | 57, 143 |
Infectious disease diagnosis
The traditional clinical paradigm for diagnosis of infectious disease in patients, applied for more than a century, involves a physician formulating a differential diagnosis and then ordering a series of tests (generally ‘one bug, one test’) in an attempt to identify the causative agent. The spectrum of conventional testing for pathogens in clinical samples ranges from the identification of microorganisms growing in culture (for example, by biochemical phenotype testing or matrix-assisted laser desorption/ionization (MALDI) time-of-flight mass spectrometry), the detection of organism-specific biomarkers (such as antigen testing by latex agglutination or antibody testing by enzyme-linked immunosorbent assay (ELISA)) or nucleic acid testing by PCR for single agents to multiplexed PCR testing using syndromic panels. These panels generally include the most common pathogens associated with a defined clinical syndrome, such as meningitis and encephalitis, acute respiratory infection, sepsis or diarrhoeal disease28–31.
Molecular diagnostic assays provide a fairly cost-effective and rapid (generally <2 hours of turnaround time) means to diagnose the most common infections. However, nearly all conventional microbiological tests in current use detect only one or a limited panel of pathogens at a time or require that a microorganism be successfully cultured from a clinical sample. By contrast, while NGS assays in current use cannot compare with conventional tests with respect to speed — the sequencing run alone on a standard Illumina instrument takes >18 hours — mNGS enables a broad range of pathogens — viruses, bacteria, fungi and/or parasites — to be identified from culture or directly from clinical samples on the basis of uniquely identifiable DNA and/or RNA sequences32. Another key advantage of NGS approaches is that the sequencing data can potentially be leveraged for additional analyses beyond the mere identification of a causative pathogen, such as microbiome characterization and parallel analyses of human host responses through transcriptome profiling by RNA sequencing (RNA-seq). Thus, the clinical utility of NGS in diagnosis may be in the most difficult-to-diagnose cases or for immunocompromised patients, in whom the spectrum of potential pathogens is greater. Eventually, mNGS may become cost competitive with multiplexed assays or used as an upfront ‘rule out’ assay to exclude infectious aetiologies. Of course, detection of nucleic acids, either by multiplex PCR panels or NGS, does not by itself prove that an identified microorganism is the cause of the illness, and findings have to be interpreted in the clinical context. In particular, discovery of an atypical or novel infectious agent in clinical samples should be followed up with confirmatory investigations such as orthogonal testing of tissue biopsy samples and demonstration of seroconversion or via the use of cell culture or animal models, as appropriate8, to ascertain its true pathogenic potential.
NGS of clinical samples as performed in either research or clinical laboratories involves a number of steps, including nucleic acid extraction, enrichment for DNA and/or RNA, library preparation, PCR amplification (if needed), sequencing and bioinformatics analysis (Fig. 2). Any body fluid or tissue yielding sufficient nucleic acid is amenable to NGS analysis, which can either be targeted, that is, enriching individual genes or genomic regions, or untargeted, as is the case for metagenomic ‘shotgun’ approaches (Fig. 2). The details for the specific steps vary by laboratory and are described extensively elsewhere33–37.
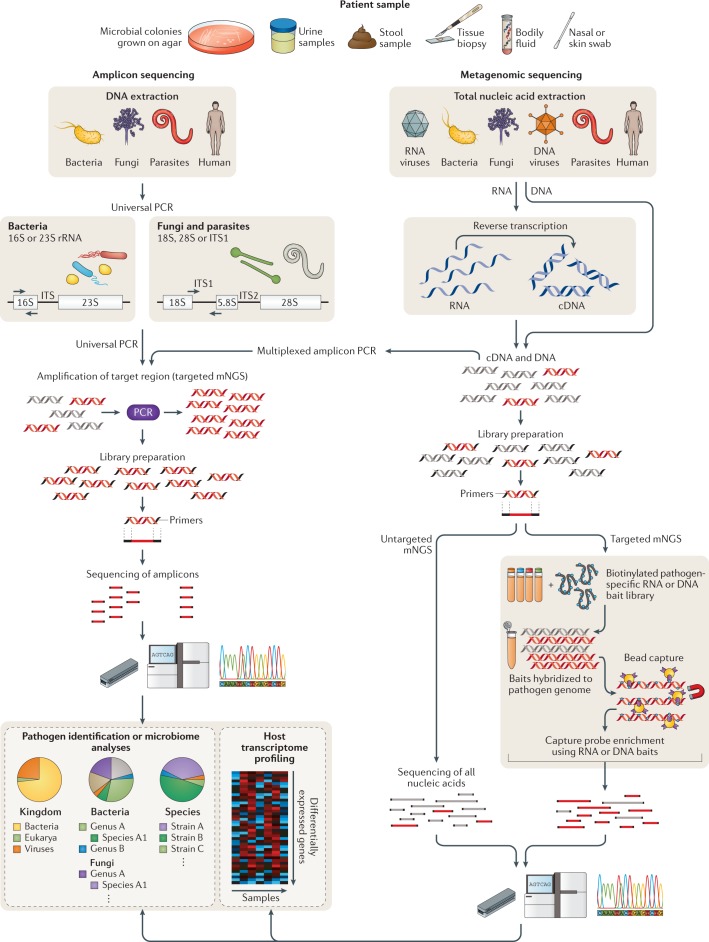
Fig. 2. Targeted versus untargeted shotgun metagenomic next-generation sequencing approaches.
A variety of patient samples, as well as cultured microbial colonies, can be analysed using targeted or untargeted metagenomic next-generation sequencing (mNGS) methods for pathogen identification, microbiome analyses and/or host transcriptome profiling. Universal PCR (left) is a targeted mNGS approach that uses primers designed from conserved regions such as the ribosomal RNA (rRNA) genes that are universally conserved among bacteria (16S or 23S rRNA) or fungi and parasites (18S rRNA, 28S rRNA or internal transcribed spacer (ITS)). Other sets of primers can be designed to target a defined set of pathogens and/or genes and used for multiplex reverse transcription PCR or PCR (multiplexed amplicon PCR). NGS library preparation and sequencing of the resultant amplicons enable pathogen identification down to the genus or species level. Metagenomic sequencing (right) entails unbiased shotgun sequencing of all microbial and host nucleic acids present in a clinical sample. Separate DNA and RNA libraries are constructed; the DNA library is used for identification of bacteria, fungi, DNA viruses and parasites, whereas the RNA library is used for identification of RNA viruses and RNA sequencing-based human host transcriptome profiling (heat map, bottom right). As no primers or probes are used in unbiased mNGS, the vast majority of reads corresponds to the human host and, thus, detection of pathogens from metagenomic libraries is a ‘needle-in-a-haystack’ endeavour. An optional capture probe enrichment step using magnetic beads enables targeted mNGS of pathogens and/or genes from metagenomic libraries. All these methods are compatible with sequencing on traditional benchtop instruments such as the Illumina HiSeq and portable nanopore sequencers such as the Oxford Nanopore Technologies MinION.
Targeted NGS analyses
Targeted approaches have the benefit of increasing the number and proportion of pathogen reads in the sequence data. This step can increase the detection sensitivity for microorganisms being targeted, although it limits the breadth of potential pathogens that can be identified. An example of a targeted approach is the use of highly conserved primers for universal PCR amplification and detection of all microorganisms corresponding to a specific type from clinical samples, such as 16S ribosomal RNA (rRNA) gene amplification for bacteria38,39 and 18S rRNA and internal transcribed spacer (ITS) gene amplification for fungi40 (Fig. 2). Previously, such approaches were followed by Sanger sequencing of the resulting PCR amplicon to identify the pathogen and make a diagnosis; now, this step is commonly accomplished using NGS. Universal PCR for detection of bacteria and fungi has now been adopted in many hospital laboratories and has increased the number and proportion of infectious diagnoses39,41, although the technique is limited by the breadth of detection (that is, bacteria or fungi only or even a more limited range of targets, such as mycobacteria only, depending on the primer sets used) and by concerns regarding sensitivity42.
Another example of a targeted NGS approach is the design of primers tiled across the genome to facilitate PCR amplification and amplicon NGS for recovery of viral genomes directly from clinical samples43. This method has been used to track the evolution and spread of Zika virus (ZIKV) in the Americas44–46 and of Ebola virus in West Africa47, with some demonstrations of real-time monitoring having an impact on public health interventions.
Another targeted approach is capture probe enrichment, whereby metagenomic libraries are subjected to hybridization using capture ‘bait’ probes48. These probes are generally 30–120 bp in length, and the number of probes can vary from less than 50 to more than 2 million49–52. Although this enrichment method has been shown to increase the sensitivity of metagenomic detection in research settings, especially for viruses, it has yet to be used routinely for clinical diagnosis. A promising application of this approach may be the enrichment of clinical samples for characterization of antibiotic resistance23, a considerable problem in hospitals and the primary focus of the US National Action Plan for Combating Antibiotic-Resistant Bacteria53. However, drawbacks of capture probe enrichment, compared with untargeted approaches for infectious disease diagnosis, include a bias towards targeted microorganisms, added steps, increased costs and long hybridization times (24–48 hours) as a result of the additional processing needed for maximal efficiency.
Untargeted metagenomic NGS analyses
Untargeted shotgun mNGS analyses forego the use of specific primers or probes54. Instead, the entirety of the DNA and/or RNA (after reverse transcription to cDNA) is sequenced. With pure cultures of bacteria or fungi, mNGS reads can be assembled into partial or complete genomes. These genome sequences are then used for subtyping and/or monitoring hospital outbreaks in support of infection control and/or public health surveillance efforts. For example, a seminal study described the use of whole-genome sequencing of multidrug-resistant, carbapenemase-producing Klebsiella pneumoniae to track the origin and evolution of a hospital outbreak55. This study demonstrated for the first time the high-resolution mapping of likely transmission events in a hospital, some of which were unexpected on the basis of initial epidemiological data, and also identified putative resistance mutations in emerging resistant strains. The integration of genomic and epidemiological data yielded actionable insights that would have been useful for curbing transmission.
Untargeted mNGS of clinical samples is perhaps the most promising approach for the comprehensive diagnosis of infections. In principle, nearly all pathogens, including viruses, bacteria, fungi and parasites, can be identified in a single assay56. mNGS is a needle-in-a-haystack endeavour, as only a small proportion (typically <1%) of reads are non-human, of which only a subset may correspond to potential pathogens. A limitation of mNGS is that the sensitivity of the approach is critically dependent on the level of background. Tissues, for example, have increased human host background relative to cell-free body fluids, resulting in a reduced number and proportion of microbial reads and hence a decrease in mNGS sensitivity33,36,37. Moreover, defining specific microbial profiles that are diagnostic or predictive of disease development can be difficult, especially from nonsterile sites that harbour a complex microbiome, such as respiratory secretions or stool6. Nevertheless, several groups have successfully validated mNGS in Clinical Laboratory Improvement Amendments (CLIA)-certified clinical laboratories for the diagnosis of infections, including meningitis or encephalitis36,37, sepsis33,57 and pneumonia58, and these assays are now available for clinical reference testing of patients.
Clinical microbiome analyses
Many researchers now use mNGS instead of targeted sequencing of the 16S rRNA gene for in-depth characterization of the microbiome59. There is growing public awareness of the microbiome and its likely involvement in both acute and chronic disease states60. However, no microbiome-based tests have been clinically validated for the diagnosis or treatment of disease, in part owing to an incomplete understanding of the complexity of the microbiome and its role in disease pathogenesis.
One future clinical application of microbiome analysis may be in the management and treatment of Clostridium difficile-associated disease. C. difficile is an opportunistic bacterium that can infect the gut, resulting in the production of toxins that can cause diarrhoea, dehydration, sepsis and death. C. difficile infection occurs only in the setting of a microbiome that is altered by factors such as exposure to broad-spectrum antibiotics or recent gastrointestinal surgery61. The importance of the microbiome in C. difficile infection is underscored by the 80–90% effectiveness of faecal stool transplantation in treating and potentially curing the disease62,63. The use of mNGS to characterize the microbiome in multiple studies has facilitated the development of bacterial probiotic mixtures that can be administered as pills for prophylaxis or treatment of C. difficile-associated disease (Fig. 1B).
Another potential application of the microbiome is in the analysis of bacterial diversity, which can provide clues as to whether a patient’s illness is infectious or non-infectious. For example, a study of mNGS for the identification of respiratory pathogens in patients with pneumonia found that individuals with culture-proven infection had significantly less diversity in their respiratory microbiome25. Alterations of the microbiome, known as dysbiosis, have also been shown to be related to obesity, diabetes mellitus and inflammatory bowel disease64, and manipulation of the microbiome may be a pathway to treating these pathological conditions.
Human host response analyses
Clinical mNGS typically focuses on microbial reads; however, there is a complementary role for the analysis of gene expression in studying human host responses to infection65 (Fig. 1c). mNGS of RNA libraries used for the detection of pathogens such as RNA viruses in clinical samples incidentally produces host gene expression data for transcriptome (RNA-seq) analyses66. Although RNA-seq analyses are commonly performed on whole blood or peripheral blood mononuclear cell (PBMC) samples, any body fluid or tissue type is potentially amenable to these analyses. Classification of genes by expression profiling using RNA-seq has been used to characterize several infections, including staphylococcal bacteraemia67, Lyme disease68, candidiasis69, tuberculosis (discriminating between latent and active disease risk)70–72 and influenza73–75. Machine-learning-based analyses of RNA-seq data have been used for cancer classification76, and translation of these approaches may be promising for infectious diseases. Panels containing a limited number of host biomarkers are being developed as diagnostic assays for influenza77, tuberculosis70 and bacterial sepsis78.
Although no RNA-seq-based assay has been clinically validated to date for use in patients, the potential clinical impact of RNA-seq analyses is high. Interrogation of RNA reads from microorganisms corresponding to active microbial gene expression might enable the discrimination between infection versus colonization25 and live (viable) versus dead organisms79. Moreover, RNA-seq analyses of the human host can be used to identify novel or underappreciated host–microbial interactions directly from clinical samples, as previously shown for patients with Lyme disease68, dengue80 or malaria81. RNA-seq may be particularly useful in clinical cases in which the causative pathogen is only transiently present (such as early Lyme disease82 or arboviral infections, including West Nile virus83 or ZIKV84); analogous to serologic testing, indirect diagnosis of infections may be possible on the basis of a pathogen-specific human host response. Analysis of pathogen-specific host responses may also be useful in discriminating the bona fide causative pathogen or pathogens in a complex clinical metagenomic sample, such as a polymicrobial abscess or respiratory fluid25. Yet another promising application of RNA-seq is in discriminating infectious versus non-infectious causes of acute illness25. If an illness is judged more likely to be non-infectious (for example, an autoimmune disease) on the basis of the host response, for example, clinicians may be more willing to discontinue antibiotics and treat the patient aggressively with steroids and other immunosuppressive medications. As large-scale sequencing data continue to be generated, perhaps driven by routine clinical mNGS testing, secondary mining of human reads might improve the accuracy of clinical diagnoses by incorporating both microbial and host gene expression data.
Applications in oncology
In oncology, whole-genome or directed NGS approaches to identify mutated genes can be used to simultaneously uncover viruses associated with cancer (that is, herpesviruses, papillomaviruses and polyomaviruses) and/or to gather data on virus–host interactions85. For example, mNGS was critical in the discovery of Merkel cell polyomavirus (Fig. 1d), now believed to be the cause of Merkel cell carcinoma, a rare skin cancer seen most commonly in elderly patients86. To date, the US Food and Drug Administration (FDA) has approved the clinical use of two NGS panels testing for actionable genomic aberrations in tumour samples87. Detection of reads corresponding to both integrated and exogenous viruses in these samples would be possible with the addition of specific viral probes to the panel or accomplished incidentally while sequencing the whole tumour genome or exome.
Additional knowledge of integrated or active viral infections in cancers and their involvement in signalling pathways may inform preventive and therapeutic interventions with targeted antiviral and/or chemotherapeutic drugs88, as evidenced by the decreased risk of hepatitis C virus-associated hepatocellular carcinoma after treatment with direct-acting antiviral agents89. In the future, mNGS of cell-free DNA from liquid biopsy samples (for example, plasma) might be leveraged for the simultaneous identification of early cancer and diagnosis of infection in immunocompromised patients (Box 1).
Box 1 Where is the signal — cellular or cell-free DNA?
Metagenomic sequencing for clinical diagnostic purposes typically uses a shotgun approach by sequencing all of the DNA and/or RNA in a clinical sample. Clinical samples can vary significantly in their cellularity, ranging from cell-free fluids (that is, plasma, bronchoalveolar lavage fluid or centrifuged cerebrospinal fluid) to tissues. In the next-generation sequencing (NGS) field, there is great interest in the use of liquid biopsies from cell-free DNA (cfDNA) extracted from body fluids, such as plasma, to identify chromosomal or other genetic mutations and thus diagnose malignancies in the presymptomatic phase123. Similarly, cfDNA analysis has been useful for non-invasive prenatal testing applications, such as for the identification of trisomy 21 (ref.124). One study has described the potential utility of cfDNA analysis in diagnosing invasive fungal infection in cases where biopsy is not possible57. Another advantage to cfDNA analysis is the higher sensitivity of metagenomic sequencing owing to less cellular background from the human host. However, limitations of cfDNA analysis may include decreased sensitivity for detection of predominantly intracellular pathogens, such as human T cell lymphotropic virus, Rickettsia spp. and Pneumocystis jirovecii, and loss of the ability to interrogate cellular human host responses with RNA sequencing.
Clinical implementation of metagenomic NGS
Implementation of mNGS in the clinical laboratory is a complex endeavour that requires customization of research protocols using a quality management approach consistent with regulatory standards90. Library preparation reagents, sequencing instrumentation and bioinformatics tools are constantly changing in the research environment. However, in the clinical laboratory, assays need to be implemented following standardized (locked-down) protocols. Changes made to any component of the assay need to be validated and shown to have acceptable performance before testing in patients. Periodic updates and repeat validation studies are performed as deemed necessary to incorporate interim technological advances in NGS reagents, protocols and instrumentation.
Metagenomic methods for pathogen detection present a particularly challenging scenario for clinical validation (Fig. 3), as it is not practical to test an essentially unlimited number of different organisms for the assay to be considered validated. Although the FDA has provided general guidelines for clinical validation of NGS infectious disease testing91, there are no definitive recommendations for the clinical implementation of mNGS testing, nor is there mention of specific requirements. However, a best-practice approach can be taken that includes failure-mode analysis and evaluations of performance characteristics using representative organisms with ongoing assay monitoring and independent confirmation of unexpected results.
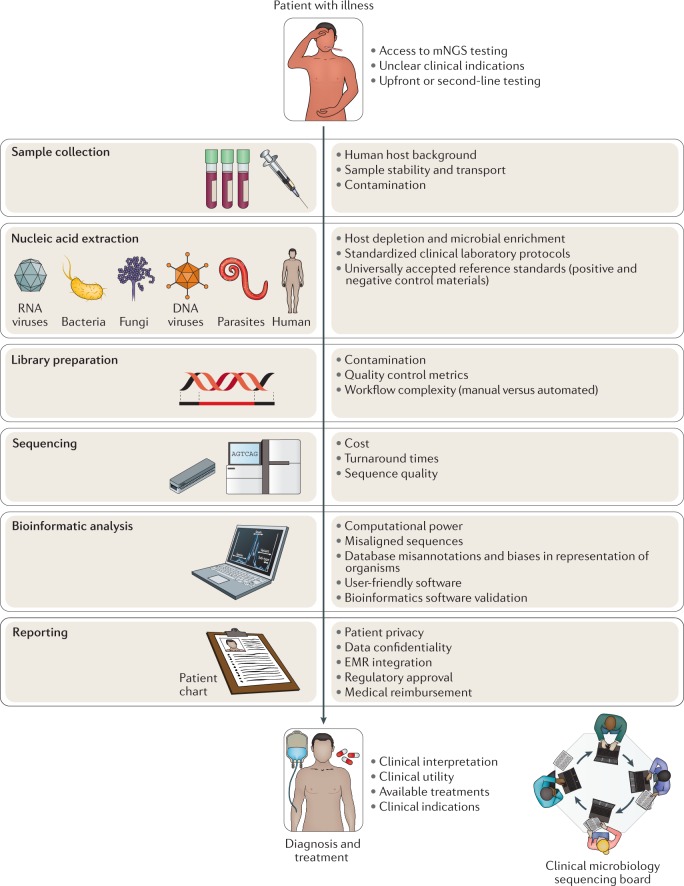
Fig. 3. Challenges to routine deployment of metagenomic sequencing in the clinical setting.
At each step in the process, multiple factors (bullet points) must be taken into account when implementing a clinical metagenomic pipeline for diagnosis of infections to maximize accuracy and clinical relevance. In particular, it is often useful to interpret and discuss the results of metagenomic next-generation-sequencing (mNGS) testing in a clinical context as part of a clinical microbial sequencing board, akin to a tumour board in oncology. EMR, electronic medical record.
Sensitivity and enrichment or depletion methods
A key limitation of mNGS is its decreased sensitivity with high background, either predominantly from the human host (for example, in tissue biopsies) or the microbiome (for example, in stool). The background can be clinically relevant as the pathogen load in infections, such as Shigella flexneri in stool from patients with diarrhoea92 or ZIKV in plasma from patients with vector-borne febrile illness93, can be very low (<103 copies per ml).
Host depletion methods for RNA libraries have been developed and shown to be effective, including DNase I treatment after extraction to remove residual human background DNA94; the use of RNA probes followed by RNase H treatment95; antibodies against human and mitochondrial rRNA (the most abundant host RNA types in clinical samples)96; and/or CRISPR–Cas9-based approaches, such as depletion of abundant sequences by hybridization97.
Unfortunately, there are no comparably effective parallel methods for DNA libraries. Limited enrichment in the 3–5 times range can be achieved with the use of antibodies against methylated human host DNA98, which enriches microbial reads owing to the lack of methylated DNA in most pathogen genomes. Differential lysis of human cells followed by degradation of background DNA with DNase I — thus retaining and enriching for nucleic acid from organisms with cell walls, which include some bacteria and fungi — has been shown to provide substantial microbial enrichment of up to 1,000 times94,99,100. However, the performance of differential lysis methods can be limited by a number of factors. These limitations include potential decreased sensitivity for microorganisms without cell walls, such as Mycoplasma spp. or parasites; a possible paradoxical increase in exogenous background contamination by use of additional reagents101; and the inability to detect free nucleic acid from dead organisms that are lysed in vivo by human host immune cells or antibiotic treatment. The importance of retaining the ability for cell-free DNA detection from culture-negative samples from dead organisms is also why incorporation of a propidium monoazide treatment step to select for DNA from live organisms may not be clinically useful as an enrichment method for mNGS102. In general, both the differential lysis and propidium monoazide approaches would also be cumbersome to implement in a highly reproducible fashion, which is needed for clinical laboratory validation.
To some extent, the human host background limitation may be overcome with brute force, made possible by the increasing capacities of available sequencers. For instance, an astrovirus was detected in a child with encephalitis by ultradeep sequencing of brain tissue, yielding only 1,612 reads out of ~134 million (0.0012%) sequences103. Yet another approach to improve sensitivity is to leverage a hybrid method for enrichment, such as metagenomic sequencing with spiked primers46. Combining targeted with untargeted sequencing, the method uses variably sized panels (100–10,000) of short primers that are added (‘spiked’) into reaction mixtures to enrich for specific target organisms while retaining the breadth of metagenomic sequencing for off-target organisms. When spiked at the reverse transcription step, a panel of ZIKV-specific primers was found to increase the number of ZIKV reads by more than tenfold without appreciably decreasing broad metagenomic sensitivity for other pathogens, enabling whole-genome viral sequencing to characterize ZIKV spread from Brazil into Central America and Mexico46.
Laboratory workflow considerations
The complexity of mNGS analysis requires highly trained personnel and extreme care in sample handling to avoid errors and cross-contamination. Even miniscule amounts of exogenous DNA or RNA introduced during sample collection, aliquoting, nucleic acid extraction, library preparation or pooling can yield a detectable signal from contaminating reads. In addition, laboratory surfaces, consumables and reagents are not DNA free. A database of background microorganisms commonly detected in mNGS data and arising from normal flora or laboratory contamination101,104 typically needs to be maintained for accurate mNGS analyses. Microorganisms on this list are either not reported or will require higher thresholds for reporting if they are clinically significant organisms.
Clinical laboratory operations are characterized by a defined workflow with scheduled staffing levels and are less amenable to on-demand testing than those of research laboratories. As samples are typically handled in batches, the frequency of batch analysis is a major determinant of overall turnaround time. Unless fully automated sample-handling systems are readily available, wet lab manipulations for mNGS require considerable hands-on time to perform, as well as clinical staff who are highly trained in molecular biology procedures. There are ergonomic concerns with repetitive tasks such as pipetting, as well as potential for inadvertent sample mix-up or omission of critical steps in the workflow. Maintaining high quality during complex mNGS procedures can be stressful to staff, as slight deviations in sample handling can lead to major changes in the results generated. Separating the assay workflow into multiple discrete steps to be performed by rotating shifts can be helpful to avoid laboratory errors.
Reference standards
Well-characterized reference standards and controls are needed to ensure mNGS assay quality and stability over time. Most available metagenomic reference materials are highly customized to specific applications (for example, ZymoBIOMICS Microbial Community Standard for microbiome analyses and bacterial and fungal metagenomics105) and/or focused on a more limited spectrum of organisms (for example, the National Institute of Standards and Technology (NIST) reference materials for mixed microbial DNA detection, which contain only bacteria106). Thus, these materials may not be applicable to untargeted mNGS analyses.
Custom mixtures consisting of a pool of microorganisms (mock microbial communities) or their nucleic acids can be developed as external controls to establish limits of detection for mNGS testing. Internal spike-in control standards are available for other NGS applications such as transcriptome analysis by RNA-seq, with External RNA Controls Consortium (ERCC) RNA standards composed of synthetic RNA oligonucleotides spanning a range of nucleotide lengths and concentrations107. The complete set or a portion of the ERCC RNA standards (or their DNA equivalents) can be used as spike-in internal controls to control for assay inhibition and to quantify titres of detected pathogens by standard curve analysis108. Nonetheless, the lack of universally accepted reference standards for mNGS makes it difficult to compare assay performances between different laboratories. There is a critical need for standardized reference organisms and genomic materials to facilitate such comparisons and to define optimal analysis methods.
Bioinformatics challenges
User-friendly bioinformatics software for analysis of mNGS data is not currently available. Thus, customized bioinformatics pipelines for analysis of clinical mNGS data56,109–111 still require highly trained programming staff to develop, validate and maintain the pipeline for clinical use. The laboratory can either host computational servers locally or move the bioinformatics analysis and data storage to cloud platforms. In either case, hardware and software setups can be complex, and adequate measures must be in place to protect confidential patient sequence data and information, especially in the cloud environment. Storage requirements for sequencing data can quickly become quite large, and the clinical laboratory must decide on the quantity, location and duration of data storage.
Bioinformatics pipelines for mNGS analysis use a number of different algorithms, usually developed for the research setting and constantly updated by software developers. As for wet lab procedures, it is usually necessary to make custom modifications to the pipeline software and then lock down both the software and reference databases for the purposes of clinical validation112. A typical bioinformatics pipeline consists of a series of analysis steps from raw input FASTQ files including quality and low-complexity filtering, adaptor trimming, human host subtraction, microorganism identification by alignment to reference databases, optional sequence assembly and taxonomic classification of individual reads and/or contiguous sequences (contigs) at levels such as family, genus and species (Fig. 4). Each step in the pipeline must be carefully assessed for accuracy and completeness of data processing, with consideration for propagation of errors. Sensitivity analyses should be performed with the inclusion of both in silico data and data generated from clinical samples. Customized data sets can be prepared to mimic input sequence data and expand the range of microorganisms detected through in silico analysis37. The use of standardized reference materials and NGS data sets is also helpful in comparative evaluation of different bioinformatics pipelines105.
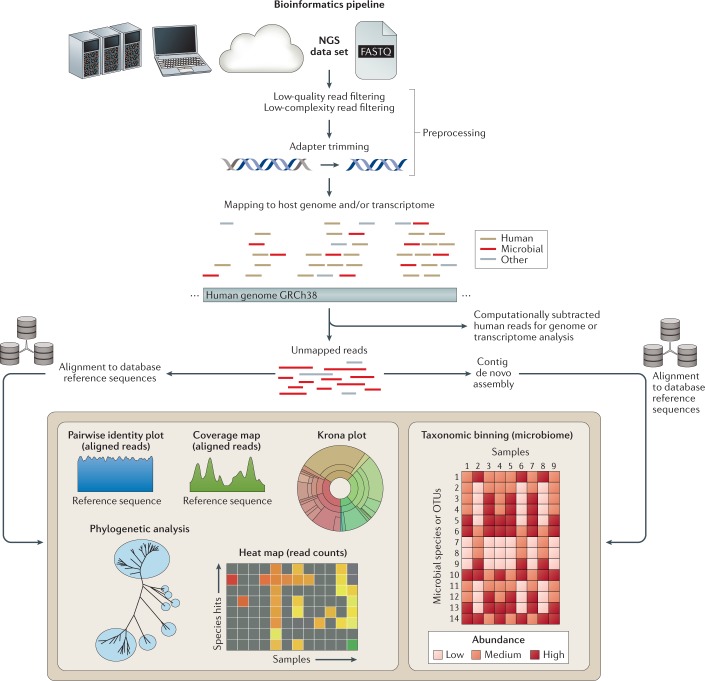
Fig. 4. A typical metagenomic next-generation sequencing bioinformatics pipeline.
A next-generation sequencing (NGS) data set, generally in FASTQ or sequence alignment map (SAM) format, is analysed on a computational server, portable laptop or desktop computer or on the cloud. An initial preprocessing step consists of low-quality filtering, low-complexity filtering and adaptor trimming. Computational host subtraction is performed by mapping reads to the host (for example, human) genome and setting aside host reads for subsequent transcriptome (RNA) or genome (DNA) analysis. The remaining unmapped reads are directly aligned to large reference databases, such as the National Center for Biotechnology Information (NCBI) GenBank database or microbial reference sequence or genome collections, or are first assembled de novo into longer contiguous sequences (contigs) followed by alignment to reference databases. After taxonomic classification, in which individual reads or contigs are assigned into specific taxa (for example, species, genus and family), the data can be analysed and visualized in a number of different formats. These include coverage map and pairwise identity plots to determine how much of the microbial genome has been recovered and its similarity to reference genomes in the database; Krona plots to visualize taxonomic diversity in the metagenomic library; phylogenetic analysis to compare assembled genes, gene regions or genomes to reference sequences; and heat maps to show microorganisms that were detected in the clinical samples. OTU, operational taxonomic unit.
Additionally, public databases for microbial reference genomes are being continuously updated, and laboratories need to keep track of the exact versions used in addition to dealing with potential misannotations and other database errors. Larger and more complete databases containing publicly deposited sequences such as the National Center for Biotechnology Information (NCBI) Nucleotide database are more comprehensive but also contain more errors than curated, more limited databases such as FDA-ARGOS91,113 or the FDA Reference Viral Database (RVDB)114. A combined approach that incorporates annotated sequences from multiple databases may enable greater confidence in the sensitivity and specificity of microorganism identification.
Performance validation and verification for bioinformatics analysis constitute a time-consuming endeavour and include analysis of control and patient data sets and comparisons, with orthogonal clinical testing to determine the accuracy of the final result36. Establishing thresholds enables separation of true-positive matches from the background, and these thresholds can incorporate metrics such as the number of sequence reads aligning to the detected microorganism, normalized to reads per million, external no-template control samples or internal spike-in material; the number of nonoverlapping genomic regions covered; and the read abundance in clinical samples relative to negative control samples (to avoid reporting of contaminant organisms). Receiver–operator curve (ROC) analysis is a useful tool to determine optimal threshold values for a training set of clinical samples with known results, with verification of pre-established thresholds using an independent validation set36.
As in the wet lab workflow, analysis software and reference databases should ideally be locked down before validation and clinical use. Many laboratories maintain both production and up-to-date development versions of the clinical reference database (for example, the NCBI nucleotide database is updated every 2 weeks), with the production database being updated at regular, prespecified intervals. Standardized data sets should be used to verify the database after any update and to ensure that assay results are accurate and reproducible, as errors can be introduced from newly deposited sequences and clinical metadata.
Cost considerations
Although there have been substantial cost reductions in the generation of sequence data, the overall per-sample reagent cost for sequencing remains fairly high. Most laboratories lack the robotic equipment and established automated protocols to multiplex large numbers of patient samples in a single run. Thus, the majority of library preparation methods for mNGS are performed manually and hence incur considerable staff time. The additional resources needed to run and maintain a bioinformatics analysis pipeline are also considerable, and steps taken to ensure regulatory oversight can add notably to costs as well. This leads to an overall cost of several hundreds to thousands of dollars per sample analysed, which is higher than that for many other clinical tests.
Technical improvements in hardware are needed for mNGS sample processing to increase throughput and to reduce costs. As NGS procedures become more standardized, there has been a drive towards increasing automation with the use of liquid-handling biorobots115. Typically, two biorobots are needed for clinical mNGS for both the pre-amplification and post-amplification steps to avoid PCR amplicon cross-contamination. Increased multiplexing is also possible with the greatly enhanced output from the latest generation of sequencers, such as the Illumina NovaSeq instruments. However, a potential limitation with running larger numbers of samples per run is longer overall turnaround times for clinical use owing to the requirement for batch processing as well as sample workflow and computational analysis considerations. Additionally, high-throughput processing of clinical samples for NGS may only be possible in reference laboratories. The development of microfluidic devices for NGS sample library preparation, such as VolTRAX116, could eventually enable clinicians to use mNGS more widely in hospital laboratories or point-of-care settings.
Regulatory considerations
Clinical laboratories are highly regulated, and general laboratory and testing requirements apply to all molecular diagnostic assays reported for patient care90. Quality control is paramount, and methods must be developed to ensure analytic accuracy throughout the assay workflow. Important quality control steps can include initial sample quality checks, library parameters (concentration and size distribution), sequence data generation (cluster density and Q-score), recovery of internal controls and performance of external controls. Validation data generated during assay development and implementation should be recorded and made available to laboratory inspectors (for laboratory-developed tests) or submitted to regulatory agencies, such as the FDA in the USA or the European Medicines Agency (EMA) in Europe, for approval.
Ongoing monitoring is particularly important for mNGS assays to verify acceptable performance over time and to investigate atypical findings36. Monitoring is accomplished using sample internal controls, intra-run control samples, swipe tests for contamination and periodic proficiency testing. Unexpected or unusual results are further investigated by reviewing patients’ clinical charts or by confirmatory laboratory testing using orthogonal methods. Identification of microorganisms that have not been identified before in the laboratory should be independently confirmed, usually through clinical reference or public health laboratory testing. Atypical or novel organisms should be assessed for their clinical significance, and these findings should be reported and discussed with health-care providers, with consideration for their potential pathogenicity and for further testing and treatment options. Clinical microbial sequencing boards, modelled after tumour boards in oncology, can be convened via real-time teleconferencing to discuss mNGS results with treatment providers in clinical context (Fig. 3). Detection of microorganisms with public health implications such as Sin Nombre hantavirus or Ebola virus should be reported, as appropriate, to the relevant public health agencies.
Conclusions and future perspectives
Technological advancements in library preparation methods, sequence generation and computational bioinformatics are enabling quicker and more comprehensive metagenomic analyses at lower cost. Sequencing technologies and their applications continue to evolve. Real-time sequencing in particular may be a game-changing technology for point-of-care applications in clinical medicine and public health, as laboratories have begun to apply these tools to diagnose atypical infections and track pathogen outbreaks, as demonstrated by the recent deployment of real-time nanopore sequencing for remote epidemiological surveillance of Ebola44 and ZIKV44,45, and even for use aboard the International Space Station117 (Box 2).
Nonetheless, formidable challenges remain when implementing mNGS for routine patient care. In particular, sensitivity for pathogen detection is decreased in clinical samples with a high nucleic acid background or with exceedingly low pathogen titres; this concern is only partially mitigated by increasing sequencing depth per sample as costs continue to drop. As a comprehensive direct detection method, mNGS may eventually replace culture, antigen detection and PCR methods in clinical microbiology, but indirect approaches such as viral serological testing will continue to play a key part in the diagnostic work-up for infections27, and functional assays such as culture and phenotypic susceptibility testing will likely always be useful for research studies. In summary, while current limitations suggest that mNGS is unlikely to replace conventional diagnostics in the short term, it can be a complementary, and perhaps essential, test in certain clinical situations.
Although the use of mNGS for informing clinical care has been demonstrated in multiple case reports and small case series118, nearly all studies have been retrospective, and clinical utility has yet to be established in a large-scale prospective clinical trial. Prospective clinical studies will be critical to understand when to perform mNGS and how the diagnostic yield compares with that of other methods. For example, the mNGS transcriptomic approach might enable effective treatment triage, whereby antimicrobials are only needed for patients showing an ‘infectious profile’ of gene expression and those with a ‘non-infectious profile’ can be treated for other causes. In particular, prospective clinical trial and economic data showing the cost-effectiveness of these relatively expensive tests in improving patient outcomes are needed to justify their use. These data will also support a pathway towards regulatory approval and clinical reimbursement. High-quality evidence that clinical metagenomic assays are effective in guiding patient management will require protocols that minimize potential assay and patient selection bias and compare relevant health outcomes using data sets generated from large patient cohorts119,120.
We predict that, over the next 5 years, prospective clinical trial data evaluating the clinical utility and cost-effectiveness of mNGS will become available; overall costs and turnaround time for mNGS will continue to drop; other aspects of mNGS beyond mere identification, such as incorporation of human host response and microbiome data, will prove clinically useful; robotic sample handling and microfluidic devices will be developed for push-button operation; computational analysis platforms will be more widely available, both locally and on the cloud, obviating the need for dedicated bioinformatics expertise; and at least a few mNGS-based diagnostic assays for infectious diseases will attain regulatory approval with clinical reimbursement. We will witness the widespread democratization of mNGS as genomic analyses become widely accessible not only to physicians and researchers but also to patients and the public via crowdsourcing initiatives121,122. Furthermore, in a world with constantly emerging pathogens, we envisage that mNGS-based testing will have a pivotal role in monitoring and tracking new disease outbreaks. As surveillance networks and rapid diagnostic platforms such as nanopore sequencing are deployed globally, it will be possible to detect and contain infectious outbreaks at a much earlier stage, saving lives and lowering costs. In the near future, mNGS will not be a luxury but a necessity in the clinician’s armamentarium as we engage in the perpetual fight against infectious diseases.
Box 2 Nanopore sequencing.
Nanopore sequencing is an emerging next-generation sequencing (NGS) technology that enables real-time analysis of sequencing data125. As such, it is particularly applicable to metagenomic NGS (mNGS) approaches because time is of the essence when treating patients with acute infectious diseases. To date, the only commercially available instruments for nanopore sequencing are from Oxford Nanopore Technologies and include the MinION (1 flow cell), GridION (5 flow cell capacity) and PromethION (48 flow cell capacity). In a published research study126, mNGS-based detection of Ebola and chikungunya virus infections on a nanopore sequencer was possible in <10 minutes of sequencing time and in <6 hours of sample-to-answer turnaround time overall. Research studies have also demonstrated the clinical potential of nanopore sequencing in targeted universal 16S ribosomal RNA (rRNA) bacterial detection127, microbiome analyses128, whole-genome sequencing of bacteria129 and outbreak viruses44,45,47, RNA sequencing (RNA-seq) using standardized controls130 and diagnosis of prosthetic joint131 and lower respiratory infections99. Untargeted approaches such as mNGS or whole-transcriptome RNA-seq, however, may be limited by the lower throughput of nanopore sequencing relative to short-read sequencing such as with an Illumina instrument.
Currently, no NGS-based clinical test for pathogens has been validated on a nanopore sequencing platform. The clinical adoption of these devices has been limited by the rapid pace of improvements to the platform, which can hinder clinical validation efforts requiring standardized instruments and locked-down protocols, and by ongoing issues regarding sequencing quality and yield. Nonetheless, there is enormous potential for nanopore sequencing in point-of-care clinical sequencing applications, such as mNGS testing done at a patient’s bedside or in an emergency room, local clinic or in the field132. Importantly, selective sequencing of pathogen reads has been demonstrated on the nanopore platform by early termination of the sequencing of the human reads as they are identified in real time133. Although attractive for purposes of protecting patient privacy and confidentiality, as human reads are depleted as part of the sequencing run, this approach is not currently scalable owing to the limited throughput of the nanopore sequencer to date (up to 10 million mNGS reads per run on the MinION nanopore sequencer as of 2019) and the need to computationally match reads to reference sequences in real time.
Acknowledgments
Reviewer information
Nature Reviews Genetics thanks J. C. Lagier, A. Nitsche and J. Dekker for their contribution to the peer review of this work.
Glossary
- Microbiome
The entirety of organisms that colonize individual sites in the human body.
- Microarrays
Commonly referred to as ‘chips’, these platforms consist of spots of DNA fragments, antibodies or proteins printed onto surfaces, enabling massive multiplexing of hundreds to thousands of targets.
- Reads
In DNA sequencing, reads are inferred sequences of base pairs corresponding to part of or all of a single DNA fragment.
- Metagenomic NGS
(mNGS). A shotgun sequencing approach in which all genomic content (DNA and/or RNA) of a clinical or environmental sample is sequenced.
- Transmission network analysis
The integration of epidemiological, laboratory and genomic data to track patterns of transmission and to infer origin and dates of infection during an outbreak.
- Precision medicine
An approach to medical care by which disease treatment and prevention take into account genetic information obtained by genomic or molecular profiling of clinical samples.
- Reference standards
In laboratory test development, well-characterized, standardized and validated reference materials or databases that enable measurement of performance characteristics of an assay, including sensitivity, specificity and accuracy.
- Latex agglutination
A clinical laboratory test for detection of a specific antibody in which the corresponding antigen is adsorbed on spherical polystyrene latex particles that undergo agglutination in the presence of the antibody.
- Seroconversion
The development of detectable antibodies in the blood that are directed against an infectious agent, such as HIV-1, after which the infectious disease can be detected by serological testing for the antibody.
- Library
In DNA sequencing, a collection of DNA fragments with known adapter sequences at one or both ends that is derived from a single clinical or environmental sample.
- Sanger sequencing
A classical method of DNA sequencing based on selective incorporation of chain-terminating dideoxynucleotides developed by Frederick Sanger and colleagues in 1977; now largely supplanted by next-generation sequencing.
- Subtyping
In microbiology, refers to the identification of a specific genetic variant or strain of a microorganism (for example, virus, bacterium or fungus), usually by sequencing all or part of the genome.
- Liquid biopsy
The detection of molecular biomarkers from minimally invasive sampling of clinical body fluids, such as DNA sequences in blood, for the purpose of diagnosing disease.
- Spike-in
In laboratory test development, refers to the use of a nucleic acid fragment or positive control microorganism that is added to a negative sample matrix (for example, plasma from blood donors) or clinical samples and that serves as an internal control for the assay.
- No-template control
In PCR or sequencing reactions, a negative control sample in which the DNA or cDNA is left out, thus monitoring for contamination that could produce false-positive results.
- Biorobots
The automated instrumentation in the clinical laboratory that enables parallel processing of many samples at a time.
- Point-of-care
Refers to diagnostic testing or other medical procedures that are done near the time and place of patient care (for example, at the bedside, in an emergency department or in a developing-world field laboratory).
- Cluster density
On Illumina sequencing systems, a quality control metric that refers to the density of the clonal clusters that are produced, with each cluster corresponding to a single read. An optimal cluster density is needed to maximize the number and accuracy of reads generated from a sequencing run.
- Q-score
A quality control metric for DNA sequencing that is logarithmically related to the base calling error probabilities and serves as a measurement of read accuracy.
- Proficiency testing
A method for evaluating the performance of individual laboratories for specific laboratory tests using a standard set of unknown samples that permits interlaboratory comparisons.
- Nanopore sequencing
A sequencing method in which DNA or RNA molecules are transported through miniature pores by electrophoresis. Sequencing reads are generated by measurement of transient changes in ionic current as the molecule passes through the pore.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
C.Y.C. is the director of the UCSF–Abbott Viral Diagnostics and Discovery Center (VDDC) and receives research support from Abbott Laboratories. C.Y.C. and S.A.M. are inventors on a patent application on algorithms related to SURPI+ software titled ‘Pathogen Detection using Next-Generation Sequencing’ (PCT/US/16/52912).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
External RNA Controls Consortium (ERCC): http://jimb.stanford.edu/ercc/
FDA-ARGOS: https://www.ncbi.nlm.nih.gov/bioproject/231221
FDA Reference Viral Database (RVDB): https://hive.biochemistry.gwu.edu/rvdb
National Center for Biotechnology Information (NCBI) Nucleotide database: https://www.ncbi.nlm.nih.gov/nucleotide/
References
- 1.Zhao F, Bajic VB. The value and significance of metagenomics of marine environments. Genomics Proteomics Bioinformatics. 2015;13:271–274. doi: 10.1016/j.gpb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ufarte L, Laville E, Duquesne S, Potocki-Veronese G. Metagenomics for the discovery of pollutant degrading enzymes. Biotechnol. Adv. 2015;33:1845–1854. doi: 10.1016/j.biotechadv.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Greay TL, et al. Recent insights into the tick microbiome gained through next-generation sequencing. Parasit. Vectors. 2018;11:12. doi: 10.1186/s13071-017-2550-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guegan M, et al. The mosquito holobiont: fresh insight into mosquito-microbiota interactions. Microbiome. 2018;6:49. doi: 10.1186/s40168-018-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pallen MJ. Diagnostic metagenomics: potential applications to bacterial, viral and parasitic infections. Parasitology. 2014;141:1856–1862. doi: 10.1017/S0031182014000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan JZ, et al. Metagenomic analysis of tuberculosis in a mummy. N. Engl. J. Med. 2013;369:289–290. doi: 10.1056/NEJMc1302295. [DOI] [PubMed] [Google Scholar]
- 8.Chiu CY. Viral pathogen discovery. Curr. Opin. Microbiol. 2013;16:468–478. doi: 10.1016/j.mib.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moustafa A, et al. The blood DNA virome in 8,000 humans. PLOS Pathog. 2017;13:e1006292. doi: 10.1371/journal.ppat.1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rascovan N, Duraisamy R, Desnues C. Metagenomics and the human virome in asymptomatic individuals. Annu. Rev. Microbiol. 2016;70:125–141. doi: 10.1146/annurev-micro-102215-095431. [DOI] [PubMed] [Google Scholar]
- 11.Somasekar S, et al. Viral surveillance in serum samples from patients with acute liver failure by metagenomic next-generation sequencing. Clin. Infect. Dis. 2017;65:1477–1485. doi: 10.1093/cid/cix596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hampton-Marcell JT, Lopez JV, Gilbert JA. The human microbiome: an emerging tool in forensics. Microb. Biotechnol. 2017;10:228–230. doi: 10.1111/1751-7915.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller MB, Tang YW. Basic concepts of microarrays and potential applications in clinical microbiology. Clin. Microbiol. Rev. 2009;22:611–633. doi: 10.1128/CMR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Streit WR, Schmitz RA. Metagenomics—the key to the uncultured microbes. Curr. Opin. Microbiol. 2004;7:492–498. doi: 10.1016/j.mib.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Rota PA, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 16.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 17.Palmer C, et al. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 2006;34:e5. doi: 10.1093/nar/gnj007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin. Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 19.Wilson MR, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nutman A, Marchaim D. ‘How to do it’-molecular investigation of a hospital outbreak. Clin. Microbiol. Infect. 2018 doi: 10.1016/j.cmi.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Loman NJ, et al. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4. JAMA. 2013;309:1502–1510. doi: 10.1001/jama.2013.3231. [DOI] [PubMed] [Google Scholar]
- 22.Oniciuc EA, et al. Genes (Basel) 2018. The present and future of whole genome sequencing (WGS) and whole metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain; p. E268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefan C, Koehler J, Minogue T. Targeted next-generation sequencing for the detection of ciprofloxacin resistance markers using molecular inversion probes. Sci. Rep. 2016;6:25904. doi: 10.1038/srep25904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gliddon HD, Herberg JA, Levin M, Kaforou M. Genome-wide host RNA signatures of infectious diseases: discovery and clinical translation. Immunology. 2018;153:171–178. doi: 10.1111/imm.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langelier C, et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl Acad. Sci. USA. 2018;115:E12353–E12362. doi: 10.1073/pnas.1809700115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017;18:2. doi: 10.1186/s12865-016-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greninger A. The challenge of diagnostic metagenomics. Expert Rev. Mol. Diagn. 2018;18:605–615. doi: 10.1080/14737159.2018.1487292. [DOI] [PubMed] [Google Scholar]
- 28.Khare R, et al. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J. Clin. Microbiol. 2014;52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leber AL, et al. Multicenter evaluation of BioFire FilmArray meningitis/encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J. Clin. Microbiol. 2016;54:2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruggiero P, McMillen T, Tang YW, Babady NE. Evaluation of the BioFire FilmArray respiratory panel and the GenMark eSensor respiratory viral panel on lower respiratory tract specimens. J. Clin. Microbiol. 2014;52:288–290. doi: 10.1128/JCM.02787-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang YW, et al. Clinical evaluation of the Luminex NxTAG respiratory pathogen panel. J. Clin. Microbiol. 2016;54:1912–1914. doi: 10.1128/JCM.00482-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. Next-generation sequencing for infectious disease diagnosis and management: a report of the association for molecular pathology. J. Mol. Diagn. 2015;17:623–634. doi: 10.1016/j.jmoldx.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Blauwkamp TA, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 2019 doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 34.Deurenberg RH, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol. 2017;243:16–24. doi: 10.1016/j.jbiotec.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 35.Gargis AS, Kalman L, Lubin IM. Assuring the quality of next-generation sequencing in clinical microbiology and public health laboratories. J. Clin. Microbiol. 2016;54:2857–2865. doi: 10.1128/JCM.00949-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller S, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Preprint at bioRxiv. 2019 doi: 10.1101/330381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlaberg R, et al. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab Med. 2017;141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 38.Rampini SK, et al. Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin. Infect. Dis. 2011;53:1245–1251. doi: 10.1093/cid/cir692. [DOI] [PubMed] [Google Scholar]
- 39.Salipante SJ, et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLOS ONE. 2013;8:e65226. doi: 10.1371/journal.pone.0065226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner K, Springer B, Pires VP, Keller PM. Molecular detection of fungal pathogens in clinical specimens by 18S rDNA high-throughput screening in comparison to ITS PCR and culture. Sci. Rep. 2018;8:6964. doi: 10.1038/s41598-018-25129-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basein T, et al. Clinical utility of universal PCR and its real-world impact on patient management. Open Forum Infect. Dis. 2017;4:S627. doi: 10.1093/ofid/ofx163.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corless CE, et al. Contamination and sensitivity issues with a real-time universal 16 S rRNA PCR. J. Clin. Microbiol. 2000;38:1747–1752. doi: 10.1128/jcm.38.5.1747-1752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quick J, et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faria NR, et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. 2017;546:406–410. doi: 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grubaugh N, et al. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature. 2017;546:401–405. doi: 10.1038/nature22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thézé J, et al. Genomic epidemiology reconstructs the introduction and spread of Zika virus in central America and Mexico. Cell Host Microbe. 2018;23:855–864. doi: 10.1016/j.chom.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Garcia G, et al. Assessment of the latest NGS enrichment capture methods in clinical context. Sci. Rep. 2016;6:20948. doi: 10.1038/srep20948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Briese T, et al. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio. 2015;6:e01491–15. doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Metsky HC, et al. Capturing sequence diversity in metagenomes with comprehensive and scalable probe design. Nat. Biotechnol. 2019;37:160–168. doi: 10.1038/s41587-018-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naccache S, et al. Distinct Zika virus lineage in Salvador, Bahia, Brazil. Emerg. Infect. Dis. 2016;22:1788–1792. doi: 10.3201/eid2210.160663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wylie TN, Wylie KM, Herter BN, Storch GA. Enhanced virome sequencing using targeted sequence capture. Genome Res. 2015;25:1910–1920. doi: 10.1101/gr.191049.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Presidential Council. National action plan for combating antibiotic-resistant bacteria (The White House, Washington, 2015).
- 54.Quince C, Walker A, Simpson J, Loman N, Segata N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 55.Snitkin E, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl Med. 2012;4:148ra116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naccache S, et al. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24:1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong D, et al. Liquid biopsy for infectious diseases: sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn. Microbiol. Infect. Dis. 2018;92:210–213. doi: 10.1016/j.diagmicrobio.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 58.Schlaberg R, et al. Viral pathogen detection by metagenomics and pan-viral group polymerase chain reaction in children with pneumonia lacking identifiable etiology. J. Infect. Dis. 2017;215:1407–1415. doi: 10.1093/infdis/jix148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jovel J, et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 2016;7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young V. The role of the microbiome in human health and disease: an introduction for clinicians. BMJ. 2017;356:j831. doi: 10.1136/bmj.j831. [DOI] [PubMed] [Google Scholar]
- 61.Samarkos M, Mastrogianni E, Kampouropoulou O. The role of gut microbiota in Clostridium difficile infection. Eur. J. Intern. Med. 2018;50:28–32. doi: 10.1016/j.ejim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Shogbesan O, et al. A Systematic review of the efficacy and safety of fecal microbiota transplant for Clostridium difficile infection in immunocompromised patients. Can. J. Gastroenterol. Hepatol. 2018;2018:1394379. doi: 10.1155/2018/1394379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 64.Boulangé C, Neves A, Chilloux J, Nicholson J, Dumas M. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kukurba K, Montgomery S. RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015;2015:951–969. doi: 10.1101/pdb.top084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahn S, et al. Gene expression-based classifiers identify Staphylococcus aureus infection in mice and humans. PLOS ONE. 2013;8:e48979. doi: 10.1371/journal.pone.0048979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bouquet J, et al. Longitudinal transcriptome analysis reveals a sustained differential gene expression signature in patients treated for acute Lyme disease. mBio. 2016;7:e00100–00116. doi: 10.1128/mBio.00100-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zaas A, Aziz H, Lucas J, Perfect J, Ginsburg G. Blood gene expression signatures predict invasive candidiasis. Sci. Transl Med. 2010;2:21ra17. doi: 10.1126/scitranslmed.3000715. [DOI] [PubMed] [Google Scholar]
- 70.Anderson S, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N. Engl. J. Med. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singhania A, et al. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat. Commun. 2018;9:2308. doi: 10.1038/s41467-018-04579-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zak DE, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.HIPC-CHI Signatures Project Team & HIPC-I Consortium Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci. Immunol. 2017;2:eaal4656. doi: 10.1126/sciimmunol.aal4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woods C, et al. A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2. PLOS ONE. 2013;8:e52198. doi: 10.1371/journal.pone.0052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaas A, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, et al. Identifying and analyzing different cancer subtypes using RNA-seq data of blood platelets. Oncotarget. 2017;8:87494–87511. doi: 10.18632/oncotarget.20903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McClain M, et al. A Genomic signature of influenza infection shows potential for presymptomatic detection, guiding early therapy, and monitoring clinical responses. Open Forum Infect. Dis. 2016;3:ofw007. doi: 10.1093/ofid/ofw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sweeney T, Wong H, Khatri P. Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci. Transl Med. 2016;8:346ra391. doi: 10.1126/scitranslmed.aaf7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emerson JB, et al. Schrodinger’s microbes: tools for distinguishing the living from the dead in microbial ecosystems. Microbiome. 2017;5:86. doi: 10.1186/s40168-017-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Banerjee A, et al. RNA-seq analysis of peripheral blood mononuclear cells reveals unique transcriptional signatures associated with disease progression in dengue patients. Transl Res. 2017;186:62–78. doi: 10.1016/j.trsl.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 81.Lee HJ, et al. Integrated pathogen load and dual transcriptome analysis of systemic host-pathogen interactions in severe malaria. Sci. Transl Med. 2018;10:eaar3619. doi: 10.1126/scitranslmed.aar3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marques A. Laboratory diagnosis of Lyme disease: advances and challenges. Infect. Dis. Clin. North Am. 2015;29:295–307. doi: 10.1016/j.idc.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Debiasi R, Tyler K. Molecular methods for diagnosis of viral encephalitis. Clin. Microbiol. Rev. 2004;17:903–925. doi: 10.1128/CMR.17.4.903-925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Landry M, St George K. Laboratory diagnosis of Zika virus infection. Arch. Pathol. Lab Med. 2017;141:60–67. doi: 10.5858/arpa.2016-0406-SA. [DOI] [PubMed] [Google Scholar]
- 85.Nakagawa H, Fujita M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018;109:513–522. doi: 10.1111/cas.13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng H, Shuda M, Chang Y, Moore P. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Allegretti M, et al. Tearing down the walls: FDA approves next generation sequencing (NGS) assays for actionable cancer genomic aberrations. J. Exp. Clin. Cancer Res. 2018;37:47. doi: 10.1186/s13046-018-0702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saha A, Kaul R, Murakami M, Robertson ES. Tumor viruses and cancer biology: modulating signaling pathways for therapeutic intervention. Cancer Biol. Ther. 2010;10:961–978. doi: 10.4161/cbt.10.10.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanwal F, et al. Risk of hepatocellular cancer in HCV patients treated with direct-acting antiviral agents. Gastroenterology. 2017;153:996–1005. doi: 10.1053/j.gastro.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 90.Burd E. Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev. 2010;23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Food and Drug Administration. Infectious disease next generation sequencing based diagnostic devices: microbial identification and detection of antimicrobial resistance and virulence markers (FDA, 2016). This draft guidance from the FDA covers considerations for validation and approval of sequencing-based diagnostic devices for infectious diseases.
- 92.DuPont HL, Levine MM, Hornick RB, Formal SB. Inoculum size in shigellosis and implications for expected mode of transmission. J. Infect. Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 93.Corman VM, et al. Assay optimization for molecular detection of Zika virus. Bull. World Health Organ. 2016;94:880–892. doi: 10.2471/BLT.16.175950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hasan M, et al. Depletion of human DNA in spiked clinical specimens for improvement of sensitivity of pathogen detection by next-generation sequencing. J. Clin. Microbiol. 2016;54:919–927. doi: 10.1128/JCM.03050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matranga C, et al. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol. 2014;15:519. doi: 10.1186/s13059-014-0519-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Neil D, Glowatz H, Schlumpberger M. Ribosomal RNA depletion for efficient use of RNA-seq capacity. Curr. Protoc. Mol. Biol. 2013;103:4.19.1–4.19.8. doi: 10.1002/0471142727.mb0419s103. [DOI] [PubMed] [Google Scholar]
- 97.Gu W, et al. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:41. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feehery G, et al. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLOS ONE. 2013;8:e76096. doi: 10.1371/journal.pone.0076096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Charalampous T, et al. Rapid diagnosis of lower respiratory infection using nanopore-based clinical metagenomics. Preprint at bioRxiv. 2018 doi: 10.1101/387548. [DOI] [Google Scholar]
- 100.Thoendel M, et al. Comparison of microbial DNA enrichment tools for metagenomic whole genome sequencing. J. Microbiol. Methods. 2016;127:141–145. doi: 10.1016/j.mimet.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Salter S, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li R, et al. Comparison of DNA-, PMA-, and RNA-based 16S rRNA Illumina sequencing for detection of live bacteria in water. Sci. Rep. 2017;7:5752. doi: 10.1038/s41598-017-02516-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Naccache S, et al. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin. Infect. Dis. 2015;60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Strong M, et al. Microbial contamination in next generation sequencing: implications for sequence-based analysis of clinical samples. PLOS Pathog. 2014;10:e1004437. doi: 10.1371/journal.ppat.1004437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McIntyre A, et al. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol. 2017;18:182. doi: 10.1186/s13059-017-1299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jackson, S. A., Kralj, J. G. & Lin, N. J. Report on the NIST/DHS/FDA workshop: standards for pathogen detection for biosurveillance and clinical applications (National Institute for Standards and Technology, 2018).
- 107.Pine P, et al. Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design. BMC Biotechnol. 2016;16:54. doi: 10.1186/s12896-016-0281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Avraham R, et al. A highly multiplexed and sensitive RNA-seq protocol for simultaneous analysis of host and pathogen transcriptomes. Nat. Protoc. 2016;11:1477–1491. doi: 10.1038/nprot.2016.090. [DOI] [PubMed] [Google Scholar]
- 109.Flygare S, et al. Taxonomer: an interactive metagenomics analysis portal for universal pathogen detection and host mRNA expression profiling. Genome Biol. 2016;17:111. doi: 10.1186/s13059-016-0969-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim D, Song L, Breitwieser F, Salzberg S. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wood D, Salzberg S. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roy S, et al. Standards and guidelines for validating next-generation sequencing bioinformatics pipelines: a joint recommendation of the Association for Molecular Pathology and the College of American Pathologists. J. Mol. Diagn. 2018;20:4–27. doi: 10.1016/j.jmoldx.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 113.Goldberg B, Sichtig H, Geyer C, Ledeboer N, Weinstock G. Making the leap from research laboratory to clinic: challenges and opportunities for next-generation sequencing in infectious disease diagnostics. mBio. 2015;6:e01888–15. doi: 10.1128/mBio.01888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goodacre N, Aljanahi A, Nandakumar S, Mikailov M, Khan AS. A reference viral database (RVDB) to enhance bioinformatics analysis of high-throughput sequencing for novel virus detection. mSphere. 2018;3:e00069–18. doi: 10.1128/mSphereDirect.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.May M. Automated sample preparation. Science. 2016;351:300–302. doi: 10.1126/science.351.6270.300. [DOI] [Google Scholar]
- 116.Levy SE, Myers RM. Advancements in next-generation sequencing. Annu. Rev. Genomics Hum. Genet. 2016;17:95–115. doi: 10.1146/annurev-genom-083115-022413. [DOI] [PubMed] [Google Scholar]
- 117.Castro-Wallace SL, et al. Nanopore DNA sequencing and genome assembly on the International Space Station. Sci. Rep. 2017;7:18022. doi: 10.1038/s41598-017-18364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simner PJ, Miller S, Carroll KC. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin. Infect. Dis. 2018;66:778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Afshinnekoo E, Ahsanuddin S, Mason CE. Globalizing and crowdsourcing biomedical research. Br. Med. Bull. 2016;120:27–33. doi: 10.1093/bmb/ldw044. [DOI] [PubMed] [Google Scholar]
- 120.Brooks JP, et al. The truth about metagenomics: quantifying and counteracting bias in 16 S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boja E, et al. Right data for right patient-a precisionFDA NCI-CPTAC multi-omics mislabeling challenge. Nat. Med. 2018;24:1301–1302. doi: 10.1038/s41591-018-0180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McDonald D, et al. American gut: an open platform for citizen science microbiome research. mSystems. 2018;3:e00031–18. doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Babayan A, Pantel K. Advances in liquid biopsy approaches for early detection and monitoring of cancer. Genome Med. 2018;10:21. doi: 10.1186/s13073-018-0533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Norton ME, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N. Engl. J. Med. 2015;372:1589–1597. doi: 10.1056/NEJMoa1407349. [DOI] [PubMed] [Google Scholar]
- 125.Jain M, Olsen H, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Greninger A, et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mitsuhashi S, et al. A portable system for rapid bacterial composition analysis using a nanopore-based sequencer and laptop computer. Sci. Rep. 2017;7:5657. doi: 10.1038/s41598-017-05772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kerkhof L, Dillon K, Häggblom M, McGuinness L. Profiling bacterial communities by MinION sequencing of ribosomal operons. Microbiome. 2017;5:116. doi: 10.1186/s40168-017-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tyler A, et al. Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci. Rep. 2018;8:10931. doi: 10.1038/s41598-018-29334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oikonomopoulos S, Wang Y, Djambazian H, Badescu D, Ragoussis J. Benchmarking of the Oxford Nanopore MinION sequencing for quantitative and qualitative assessment of cDNA populations. Sci. Rep. 2016;6:31602. doi: 10.1038/srep31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Street T, et al. Molecular diagnosis of orthopedic-device-related infection directly from sonication fluid by metagenomic sequencing. J. Clin. Microbiol. 2017;55:2334–2347. doi: 10.1128/JCM.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gardy J, Loman N. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Loose M, Malla S, Stout M. Real-time selective sequencing using nanopore technology. Nat. Methods. 2016;13:751–754. doi: 10.1038/nmeth.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stakaityte G, et al. Cancers (Basel) 2014. Merkel cell polyomavirus: molecular insights into the most recently discovered human tumour virus; pp. 1267–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brinkmann A, et al. Development and preliminary evaluation of a multiplexed amplification and next generation sequencing method for viral hemorrhagic fever diagnostics. PLOS Negl. Trop. Dis. 2017;11:e0006075. doi: 10.1371/journal.pntd.0006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Quan J, et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Preprint at bioRxiv. 2018 doi: 10.1101/426338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Langelier C, et al. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am. J. Respir. Crit. Care Med. 2018;197:524–528. doi: 10.1164/rccm.201706-1097LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zinter MS, et al. Pulmonary metagenomic sequencing suggests missed infections in immunocompromised children. Clin. Infect. Dis. 2018 doi: 10.1093/cid/ciy802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou Y, et al. Metagenomic approach for identification of the pathogens associated with diarrhea in stool specimens. J. Clin. Microbiol. 2016;54:368–375. doi: 10.1128/JCM.01965-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ivy MI, et al. Direct detection and identification of prosthetic joint infection pathogens in synovial fluid by metagenomic shotgun sequencing. J. Clin. Microbiol. 2018;56:e00402–18. doi: 10.1128/JCM.00402-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Milani C, et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci. Rep. 2016;6:25945. doi: 10.1038/srep25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tang KW, Larsson E. Tumour virology in the era of high-throughput genomics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:20160265. doi: 10.1098/rstb.2016.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aravanis AM, Lee M, Klausner RD. Next-generation sequencing of circulating tumor DNA for early cancer detection. Cell. 2017;168:571–574. doi: 10.1016/j.cell.2017.01.030. [DOI] [PubMed] [Google Scholar]