Abstract
BiP is a major ER chaperone and suggested to act as primary sensor in the activation of the unfolded protein response (UPR). How BiP operates as a molecular chaperone and as an ER stress sensor is unknown. Here, by reconstituting components of human UPR, ER stress and BiP chaperone systems, we discover that the interaction of BiP with the luminal domains (LD) of UPR proteins, IRE1 and PERK, switch BiP from its chaperone cycle into an ER stress sensor cycle by preventing the binding of its cochaperones, with loss of ATPase stimulation. Furthermore, misfolded protein-dependent dissociation of BiP from IRE1 is primed by ATP but not ADP. Our data elucidate a previously unidentified mechanistic cycle of BiP function that explains its ability to act as a Hsp70 chaperone and ER stress sensor.
Introduction
The unfolded protein response (UPR) is a signaling system that detects misfolded proteins within the endoplasmic reticulum (ER) and coordinates a cellular response that aims to restore protein homeostasis. There are three key UPR signal activators, IRE1, PERK and ATF6 that give rise to separate branches of the response1–4. The activator proteins traverse the ER membrane presenting a luminal domain (LD) that is involved in signal detection; they also possess cytosolic effector domains that propagate the signal3,4. The LD of UPR proteins participate in detecting misfolded protein leading to UPR activation. IRE1 LD and PERK LD share a high degree of structural similarity5–7, suggesting that they operate the same mechanism for detecting misfolded proteins. The cytosolic portions of both IRE1 and PERK contain a kinase domain that auto-phosphorylates in trans8–11. Activated PERK then phosphorylates eIF2a, which ephemerally leads to global mRNA translation attenuation11. IRE1 phosphorylation results in stimulation of endoribonuclease activity which splices mRNA of XBP1 to form a potent transcriptional activator12–16. This acts to upregulate UPR target genes such as chaperones, which then help to alleviate the burden of misfolded proteins within the ER1–3.
The chaperone, BiP, is one of the most abundant proteins within the ER17. It operates a typical Hsp70 chaperone-substrate mechanism that involves recruitment of misfolded proteins to BiP substrate-binding domain (SBD) by J-domain cochaperones18–20. This greatly stimulates ATPase activity within the nucleotide binding domain (NBD), enabling BiP to adopt an ADP-bound (low Kon and Koff) closed conformation which traps misfolded protein substrate. The exchange of ADP to ATP by nucleotide exchange factors (NEF) permits transition to an open conformation (high Kon and Koff) that releases the bound protein substrate18,19.
Besides its role as a major ER chaperone, BiP has been suggested to be a direct ER stress sensor leading to UPR activation4. BiP binds to the UPR activator proteins, IRE1 and PERK via its NBD and it’s release is dependent upon misfolded protein binding to BiP SBD21,22. This is possibly followed by binding of misfolded protein to LD to further enhance UPR activation23 leading to IRE1 association with ribosomes which may affect ER protein folding load24. BiP may also bind to IRE1 via SBD to repress UPR signaling25. However, the precise mechanistic details of UPR signal activation are unclear.
The ability of BiP to function as an ER stress sensor21,22, suggests that it acts independently of its well characterized Hsp70 chaperone-substrate regulated cycle in as yet an unknown method. To shed light on the mechanistic events that determine how BiP operates as a molecular chaperone and as an ER stress sensor, we reconstitute components of UPR, ER stress, and BiP chaperone complex systems and assess the relationship between them using biochemical methods to inform us of molecular mechanism. We discover a previously unknown BiP ER stress sensor cycle that explains its ability to operate both as a Hsp70 chaperone in complex with its cochaperone proteins and as a sensor of ER stress in the UPR.
Results
IRE1 and PERK inhibit BiP ATPase stimulation
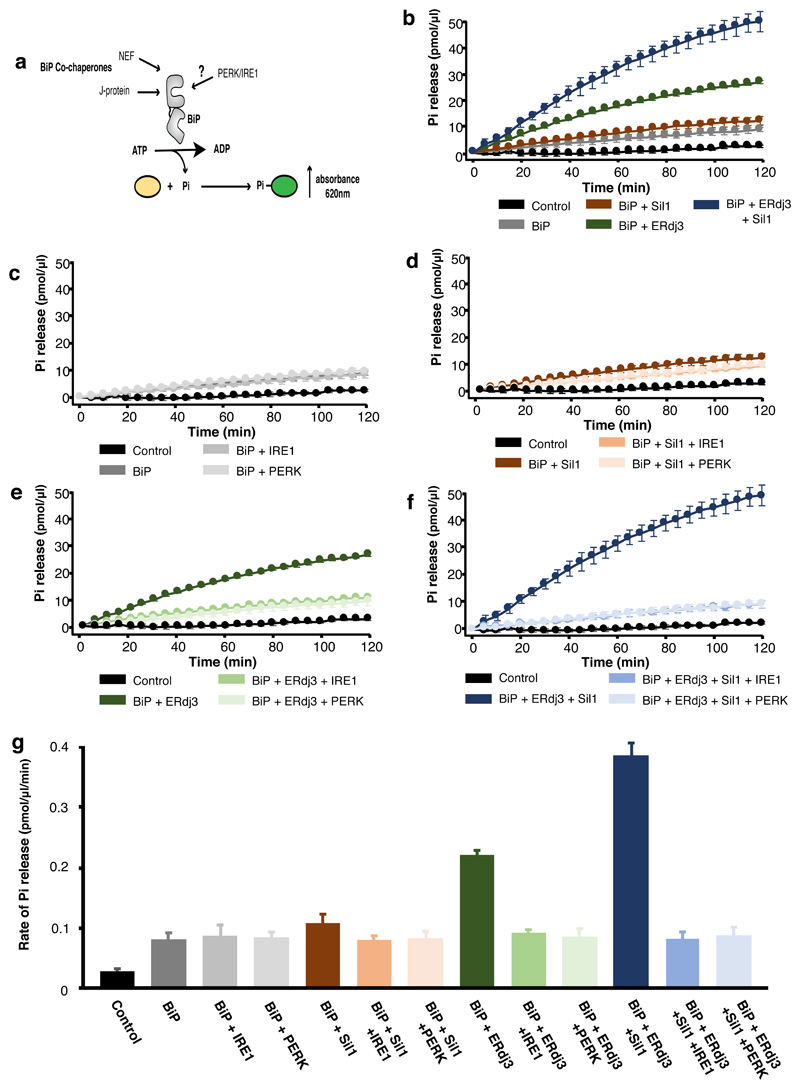
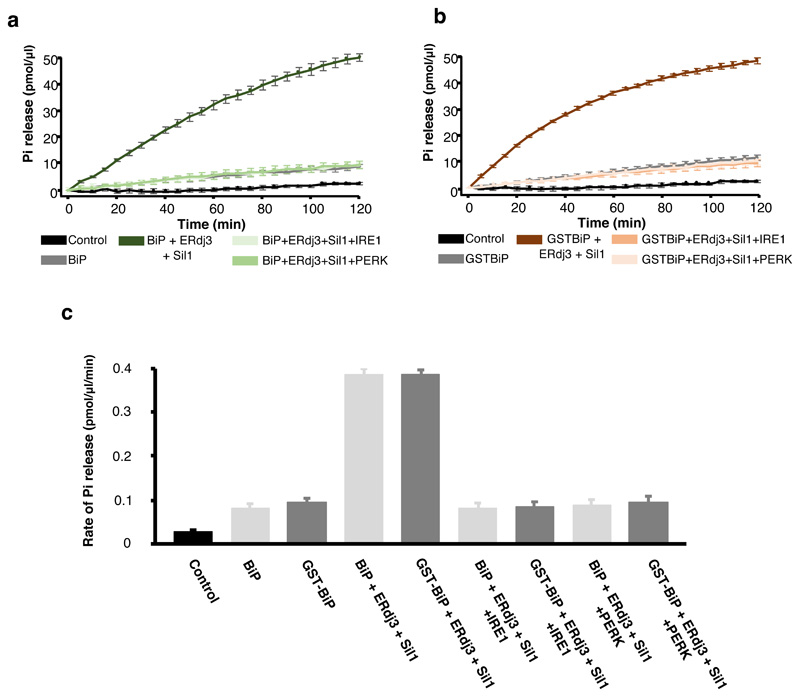
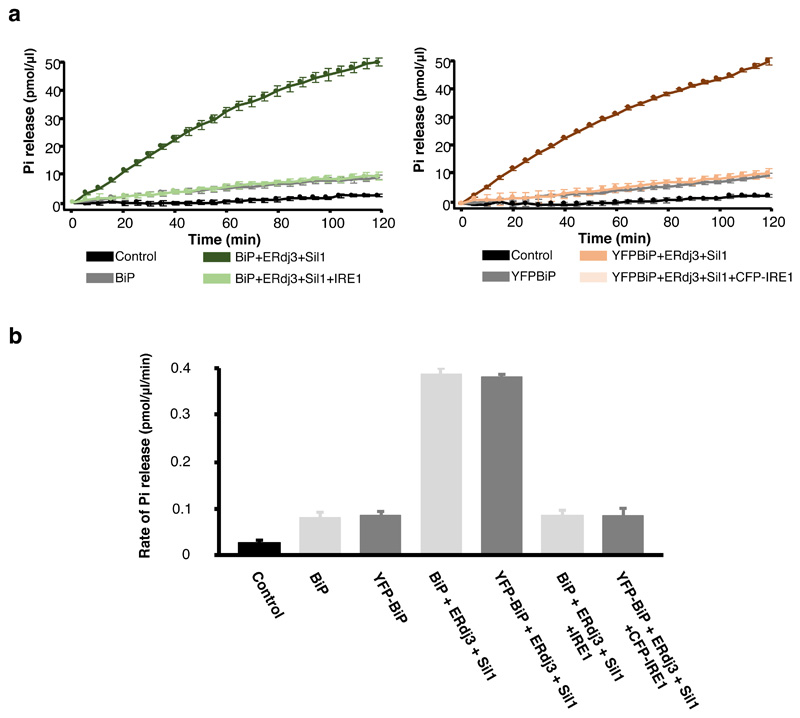
The ATPase activity of a Hsp70 chaperone is an integral part of its chaperone mechanism18,19. To determine what effect IRE1 and PERK has upon BiP ATPase activity, we recombinantly expressed and purified components of human BiP chaperone complex system including its J-protein containing cochaperone, ERdj3, and NEF, Sil1, along with the LD of human UPR proteins, IRE1 and PERK. The ATPase activity was followed colorimetrically at 620 nm wavelength as a result of free orthophosphate binding to a molybdate moiety causing a colour change upon ATP hydrolysis (Fig. 1). BiP had a low inherent ATPase activity (rate of Pi release = 0.080 pmol/μl/min) that was stimulated almost 5-fold on addition of ERdj3 and Sil1 (rate= 0.386 pmol/μl/min) (Fig. 1b and g, Supplementary Table 1). The characteristic stimulation of ATPase activity by J-protein and NEF was consistent with that of a typical Hsp70 type chaperone. When IRE1 LD and PERK LD were added to BiP without its cochaperones, there was no significant difference in the ATPase activity (Fig. 1c, g), with BiP retaining its low inherent ATPase level, indicating that IRE1 LD and PERK LD had no direct effect on BiP ATPase activity (Fig. 1 and Supplementary Table 1). Next, we assessed the ATPase activity when adding either individual or both cochaperones, ERdj3 and Sil1, together with IRE1 LD or PERK LD (Fig. 1d-g and Supplementary Table 1). We observed a reduction in BiP ATPase activity on addition of IRE1 LD and PERK LD to basal BiP levels in the presence of either single cochaperone or the whole chaperone complex encompassing both ERdj3 and Sil1 (Fig. 1d-g and Supplementary Table 1). This indicates that the addition of IRE1 LD and PERK LD caused loss of BiP ATPase stimulation by its J-domain cochaperone and NEF but retained its low inherent ATPase activity, which was unaffected by IRE1 LD and PERK LD.
Figure 1. IRE1 and PERK LD cause loss of BiP ATPase stimulation by cochaperone and NEF effectors, but BiP retains its inherent activity.
a, Schematic representation of assay used for measuring ATPase activity. b-f, Graphs displaying phosphate (Pi) release over time as a result of BiP ATPase activity. Data shown are mean ± sd, from nine independent experiments. Source data available online. BiP displayed low basal ATPase activity; the addition of Sil1 slightly increased activity, whereas ERdj3 or both cochaperone Sil1 and ERdj3 together greatly stimulated BiP activity (b). Addition of IRE1 LD or PERK LD did not affect BiP basal ATPase activity (c) but reversed the stimulation by Sil1 (d), by ERdj3 (e), and by both cochaperones ERdj3 and Sil1 together (f). g, Bar graph showing the rates of BiP ATPase activity upon addition of effector proteins. Data are mean ± sd, from nine independent experiments. ATPase rates are in Supplementary Table 1.
Folded or misfolded IRE1 LD affect BiP ATPase activity differently
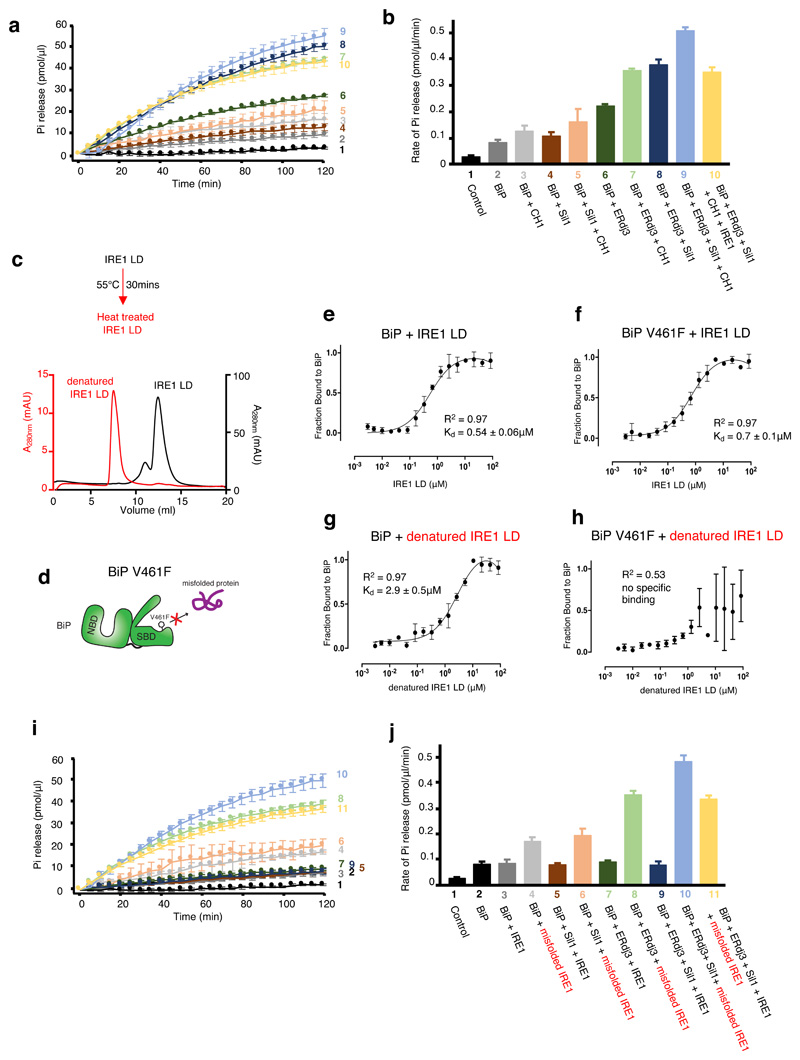
Thus far, the impact of UPR proteins and cochaperones on BiP ATPase activity were measured in the absence of misfolded protein. We repeated the BiP ATPase measurements, but added CH121,26, an immunoglobulin fragment that is inherently misfolded in the absence of its binding partner CL. The addition of CH1 stimulated BiP ATPase activity. Stimulation by misfolded protein was further observed in each sample in which either or both Sil1 and ERdj3 cochaperones were added, at a higher level than without CH1 (Fig. 2a,b and Supplementary Table 2). However, the addition of IRE1 LD had an inhibitory effect, with the ATPase rate showing a reduction compared to samples that lacked IRE1 LD. Thus, misfolded proteins stimulate BiP ATPase activity, whereas IRE1 LD has an inhibitory effect on BiP ATPase stimulation even in the presence of misfolded protein.
Figure 2. Folded and misfolded IRE1 LD have different effects on BiP interaction and activity.
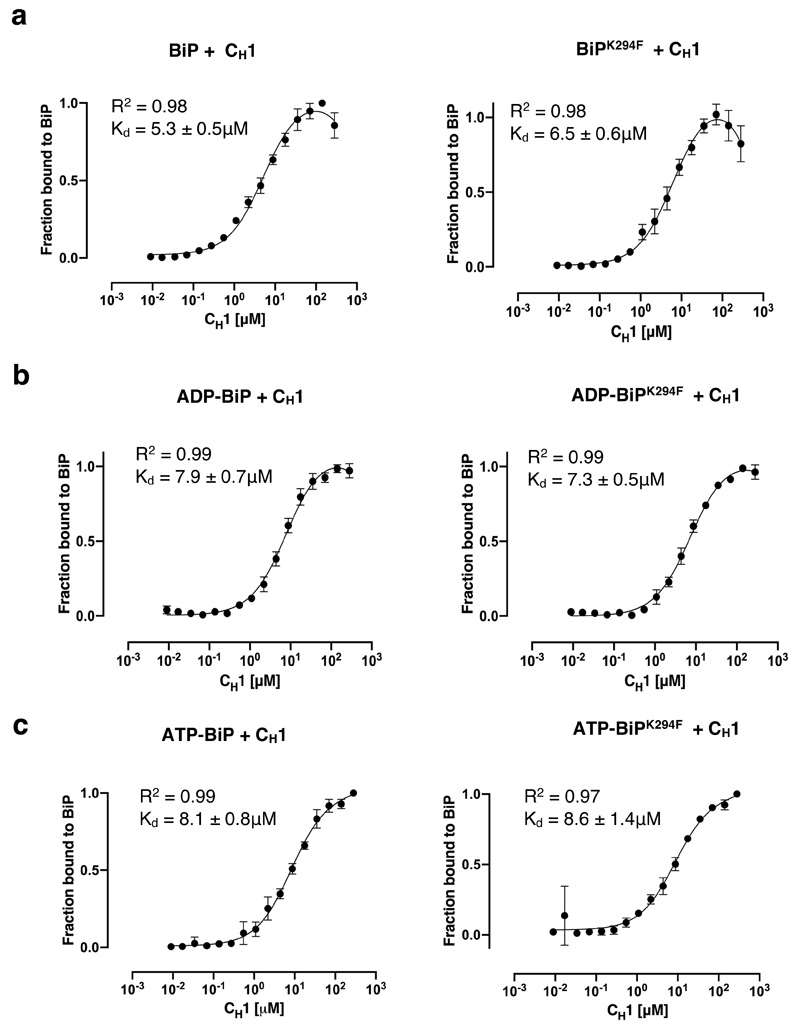
a-b, ATPase assay that measured BiP activity on addition of CH1 misfolded protein with cochaperones and IRE1 LD. Pi release over time (a) and rate of BiP ATPase activity (b) deduced from a. Data are mean ± sd from nine independent experiments. c, Size exclusion chromatography of IRE1 LD heat treated (55°C for 30 mins, red) or untreated control (black). The heat-treated sample eluted in the void volume indicating protein aggregation. d, A graphical representation of BiP V461F mutant. The SBD mutant prevents the engagement of misfolded chaperone-substrate protein to BiP28,29. e-f, Microscale thermophoresis (MST) profile of folded IRE1 LD interaction with BiPWT (e) or BiPV461F(f). The binding affinities are comparable and suggest folded IRE1 LD does not engage the SBD as a chaperone substrate. g-h, MST profile of misfolded IRE1 LD interaction with BiPWT (g), and BiPV461F(h). Data points in e-h are mean ± sd from three independent experiments. i, BiP ATPase assay comparing the effects of folded and misfolded IRE1 on BiP activity in the presence of cochaperones. j, rate of BiP ATP activity deduced from i (mean ± sd, n=9 independent experiments). Source data for graphs are available online. ATPase rate values for b and j are listed in Supplementary Tables 2 and 4. Binding affinities are in Supplementary Table 3.
Misfolded proteins associate with BiP to form a chaperone substrate interaction via BiP SBD; in contrast, IRE1 LD binds to BiP NBD21,22,27, although a UPR functional association that represses signaling has also been reported with SBD25 (see discussion). The binding of both CH1 and IRE1 LD to BiP are mutually exclusive21. We considered whether we could reverse the inhibitory effect of IRE1 LD by inducing its denaturing to mimic authentic misfolded protein, CH1. In this scenario, IRE1 LD would possibly change the way it associates to BiP from interacting with NBD to SBD as a chaperone substrate upon denaturation. Exposure to elevated temperatures is a typical method to induce protein misfolding. We heated IRE1 LD to 55ºC for 30 mins and then observed its elution profile by size exclusion chromatography and compared it to before heat treatment. IRE1 LD protein eluted, for the most part, as a dimer with small fraction forming a tetramer. However, heat treated IRE1 LD eluted exclusively in the void volume of the column indicating that the protein was aggregated and denatured as a result of thermal treatment (Fig. 2c).
To assess differences in binding between folded IRE1 LD and misfolded IRE1 LD to BiP, we measured their binding affinities to both wild type BiP (BiPWT) and a BiP SBD mutant, V461F (BiPV461F) 28,29, using microscale thermophoresis (MST). This mutation prevents misfolded protein engagement to BiP SBD as a chaperone-substrate (Fig. 2d). Folded IRE1 LD interacted with both BiPWT and BiPV461F with similar affinity (Fig. 2e,f and Supplementary Table 3). This suggests that folded IRE1 LD does not interact with BiP via its SBD as a chaperone-substrate. By contrast, misfolded IRE1 LD interacted with BiPWT but failed to associate with BiPV461F (Fig. 2g,h and Supplementary Table 3), indicating that misfolded IRE1 LD binds to BiP SBD. Thus, folded and misfolded IRE1 LD act in contrasting fashion by binding to different domains of BiP.
We then assessed the impact of misfolded IRE1 LD on BiP ATPase activity in the presence of its cochaperones. Misfolded IRE1 LD stimulated BiP ATPase activity in the presence of either or both Sil1 and ERdj3 to similar levels observed with CH1. This is the opposite effect seen with folded IRE1 LD, which had an inhibitory impact on BiP ATPase activity (Fig. 2i,j and Supplementary Table 4). Therefore, we were able to reverse the inhibitory effects of folded IRE1 LD on BiP ATPase stimulation by inducing its misfolding, which in turn, stimulated BiP ATPase activity similar to CH1. This indicates differences in the way BiP functions when interacting with folded IRE1 LD and misfolded IRE1 LD, consistent with its dual role as a chaperone and an ER stress sensor.
Binding of UPR proteins and cochaperones to BiP is mutually exclusive
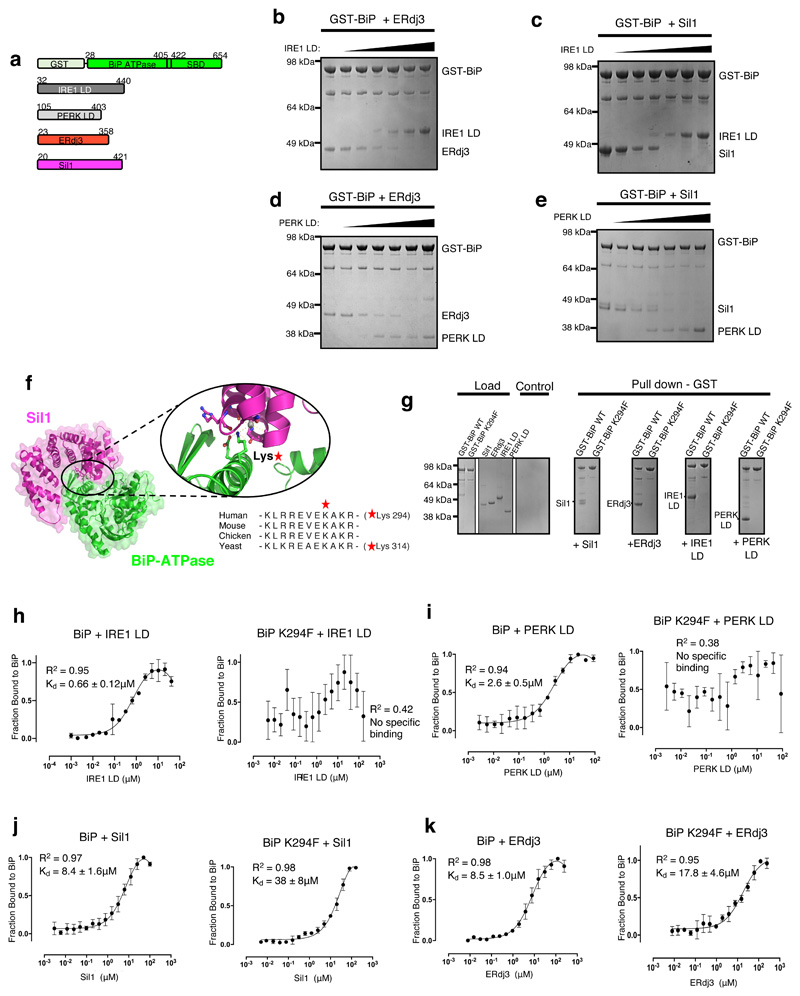
The observation that IRE1 LD and PERK LD inhibit stimulation of BiP ATPase, but not its inherent ATPase activity, suggests that IRE1 LD and PERK LD prevent the association of ERdj3 and Sil1 to BiP by competing for the same binding site on BiP NBD. To test this notion, we conducted a series of competitive pull-down assays, in which GST tagged BiP was incubated with either ERdj3 or Sil1 (Fig. 3a; Extended Data Fig. 1), and then increasing concentrations of IRE1 LD and PERK LD were added. The experiments indicate that when BiP formed a chaperone complex with ERdj3 or with Sil1, increasing concentrations of IRE1 LD (Fig. 3b,c) and PERK LD (Fig. 3d,e) outcompeted Sil1 and ERdj3 for binding to BiP. Thus, the data demonstrate that binding to BiP by UPR proteins and BiP cochaperones are mutually exclusive.
Figure 3. UPR proteins and cochaperones have mutually exclusive binding sites on BiP NBD that are impacted by BiPK294F mutant.
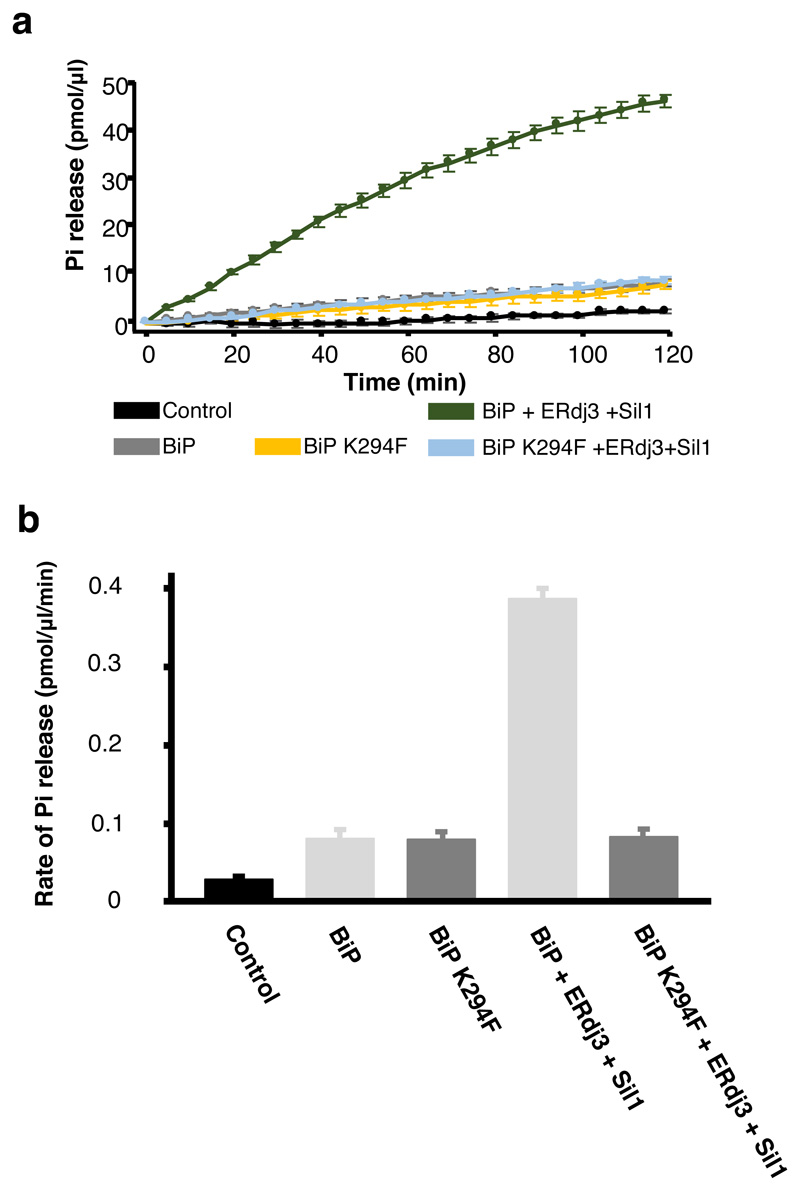
a, Schematic representation of the various constructs used for the pull-down assay, with the numbers denoting the residue number of the protein. b, Competitive pull-down assay in which increasing concentrations of IRE1 LD were added to GST-BiP – ERdj3 chaperone complex. IRE1 LD outcompeted ERdj3 for binding to BiP, suggesting mutually exclusive binding. c, Competitive pull-down assay where GST-BiP - Sil1 complex was immobilized to beads and increasing concentrations of IRE1 LD were added. d, Same as (b), but with PERK LD. e, Same as (c), but with PERK LD. f, Crystal structure of BiP ATPase – Sil1 complex (PDB 3QML), highlighting the interface between interacting molecules. BiP NBD domain residue Lys314 (corresponding to Lys294 in human) mediates key polar contacts between the two molecules. Sequence alignment (right) indicated that this residue is highly conserved across BiP species. g, Pull-down assay to assess interaction of GST- BiPWT or BiPK294F mutant with cochaperones and UPR proteins IRE1 LD and PERK LD. h-k, MST experiments comparing the binding affinity of GST tagged BiPWT and BiPK294F to cochaperones and UPR proteins. Data are mean ± sd from 3 independent experiments. Source Data are available online. BiPK294F mutant disrupted binding to IRE1 LD (h) and PERK LD (i). The BiPK294F mutant displayed greatly reduced binding affinity for Sil1 compared to BiPWT (j). The BiPK294F mutant displayed reduced, but still substantial binding for ERdj3 indicating that ERdj3 may have other contact sites with BiP. Uncropped gel images are available as Source Data. Binding affinities are in Supplementary Table 5.
To identify a mutation that affects LD association with BiP and to obtain further evidence that LD and Sil1 have overlapping binding sites on BiP, we examined the crystal structure of yeast BiP NBD in complex with Sil130 (Fig. 3f). At the interface, a Lysine residue (Lys314) of BiP mediates a number of polar contacts with Sil130; moreover, this residue and surrounding ones are well conserved across all BiP species. We reasoned that mutation of that Lys to a bulky Phe would deleteriously impact binding, by disrupting the polar interactions at the interface and by sterically blocking association with binding partner(s). We mutated the equivalent Lys residue in human BiP (Lys294) in GST tagged BiP (BiPK294F) (Extended Data Fig. 2 and 3) and assessed binding to cochaperones and UPR proteins by pull-down assay and MST analysis. The pull-down assays suggested that BiPK294F disrupted the interaction between BiP and its cochaperones, Sil1 and ERdj3; but more importantly, it inhibited the binding of IRE1 LD and PERK LD to BiP (Fig. 3g). Analysis of binding affinities by MST suggested a loss of specific binding between BiPK294F and IRE1 LD or PERK LD (Fig. 3h,i and Supplementary Table 5). The binding profiles did indicate small fluctuations in fluorescence, but the data did not fit a binding saturation curve. For Sil1, there was a considerable reduction in strength of binding to BiPK294F (Figure 3j and Supplementary Table 5). Similarly, ERdj3 also displayed a reduced affinity for BiPK294F although the reduction in binding strength was less than that observed for Sil1 (Fig. 3k and Supplementary Table 5). This suggests that the interaction between BiP NBD and its cochaperones, particularly ERdj3, may involve other contact sites. Indeed, ERdj3 possesses a J-domain, which is known to bind to a pocket between the NBD and the SBD of Hsp70s, contacting the interdomain linker31.
Thus, we identify a mutation in BiP that inhibits binding to IRE1 LD and PERK LD. The mutation reinforces the evidence that the interaction site for IRE1 LD and PERK LD is in the NBD of BiP, consistent with other studies21,22,27. BiP cochaperones also interact with this site but may have other substantial contacts with BiP31. The mutational analyses, along with the competitive pull-down experiments, suggest that BiP interaction with UPR proteins and cochaperones are mutually exclusive. By inhibiting the binding of cochaperones to BiP, the UPR proteins prevent BiP from functioning effectively as a chaperone; however, BiP still retains its ability to bind and detect misfolded proteins via its SBD.
Effect of nucleotides on BiP interactions
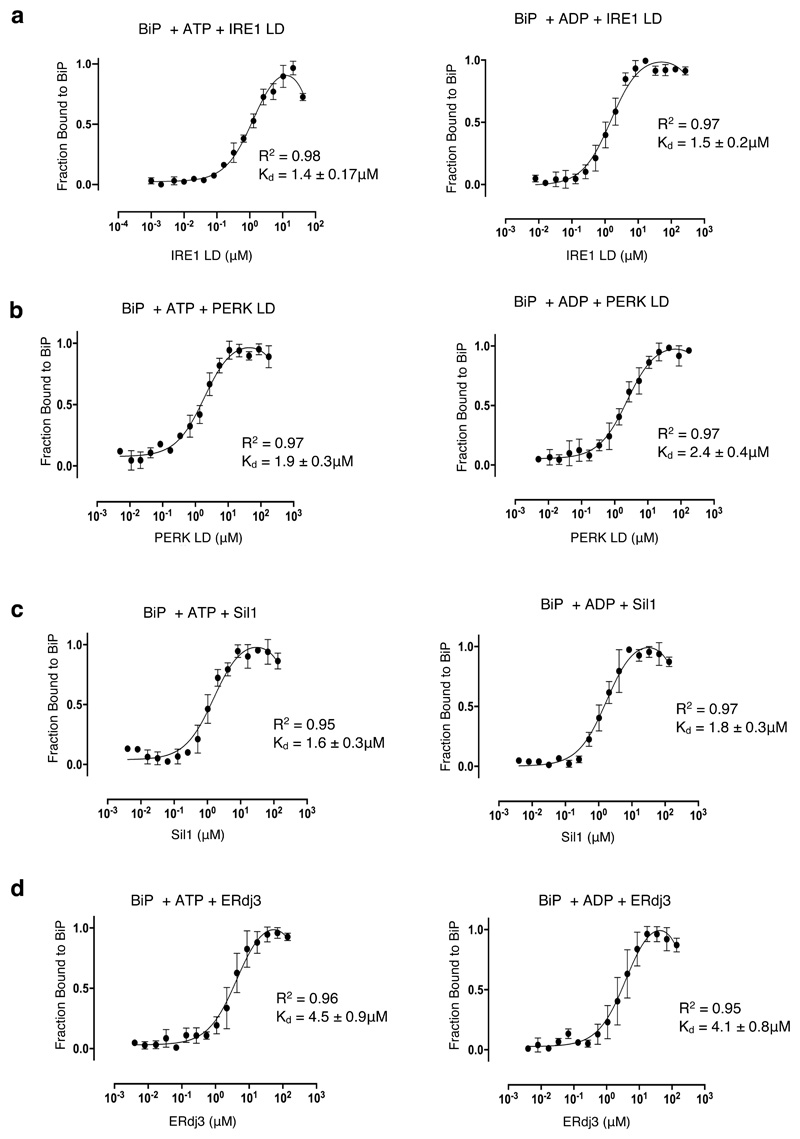
We have previously demonstrated that the interaction between LD and BiP is independent of nucleotides21,22. To support those previous observations, we measured the interaction between IRE1 LD and PERK LD to BiP in the presence of ATP and ADP using MST and extended our measurements to analyze the influence of nucleotides on BiP interaction with its cochaperones. The binding affinities for BiP interaction with IRE1 LD and PERK LD in the absence and in the presence of nucleotides were similar (Fig. 3h,i; Fig. 4a,b and Supplementary Table 6), indicating that association between BiP and LD is largely unaffected by nucleotides. The interaction between BiP and its cochaperones, ERdj3 and Sil1, increased in strength on addition of nucleotides when compared to BiP (Fig. 3j,k; Fig 4c,d and Supplementary Table 6). These results show that binding between LD and BiP is not influenced by nucleotides.
Figure 4. The effect of nucleotides on BiP binding to UPR proteins and cochaperones.
MST profiles measuring the binding between GST tagged BiP with UPR proteins and with cochaperones in the presence of ATP and ADP. a, The binding affinities for BiP association to IRE1 LD in the presence of ATP and ADP are similar to the binding affinity without nucleotides (Figure 3h). b, PERK LD incubated with ATP and ADP are similar to the binding affinity without nucleotides (Figure 3i). c-d, Sil1 (c) and ERdj3 (d) both show stronger binding to BiP with nucleotide present than without nucleotides (Figure 3 j-k). Data are mean ± sd from 3 independent experiments. Source Data are available online. Binding affinities are in Supplementary Table 6.
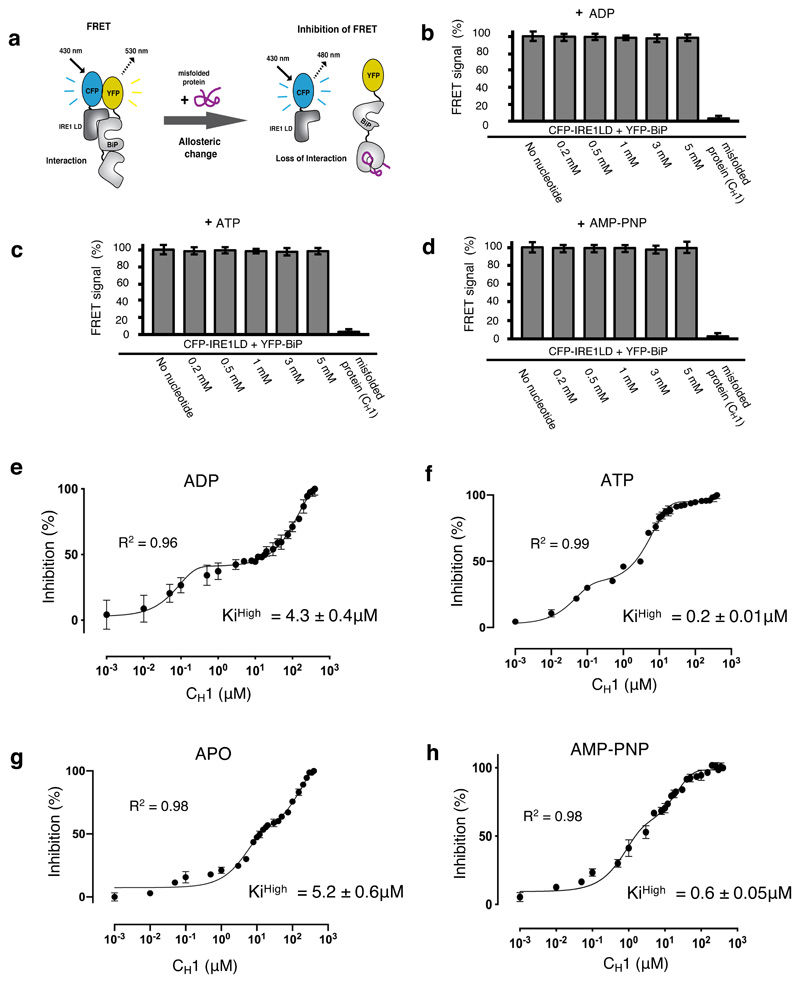
Given that the interaction between BiP NBD and IRE1 LD is independent of nucleotide influence, the dissociation of BiP from IRE1 LD would be solely dependent upon misfolded protein binding to BiP SBD. We considered whether nucleotides could sensitize or prime BiP SBD to detect misfolded proteins, whilst bound to IRE1 LD, thereby facilitating its release. To determine the effect of nucleotides on the ability of BiP to detect misfolded proteins whilst in complex with IRE1 LD, we used our in vitro FRET based assay22,32. This assay measures an interaction between IRE1 LD and BiP NBD via N-terminally tagged FRET CFP and YFP fluorophores (Fig. 5a). The binding of misfolded proteins to BiP SBD induces a conformational change that mediates BiP dissociation from IRE1 LD with corresponding loss of FRET signal from which we can deduce the IC50 value and the inhibition constant (Ki).
Figure 5. ATP – but not ADP – primes and facilitates the release of BiP from IRE1 LD.
a, Experimental design for three-component in vitro UPR FRET assay22,32 used to measure the inhibition of CFP-IRE1 – YFP-BiP complex by misfolded protein in the presence and absence of nucleotides. b-d, Bar graph showing FRET signal observed upon addition of ADP (b), ATP (c) or AMP-PNP (d). In each case, FRET inhibition due to dissociation of the complex was solely dependent on binding of misfolded protein CH1. Data are mean ± s.d., n=9 independent experiments. e-h, FRET signal inhibition (%) as a function of the CH1 concentration in the absence and presence of different nucleotides: ADP (e), ATP (f), APO (g) and AMP-PNP (h). Data were fitted to exponential curve (two phase association) and the IC50 (equivalent to half-life) for each phase was deduced. The IC50 measurement was then used to calculate the Ki32. The ATP bound BiP-IRE1 LD complex required 21-fold less misfolded protein to facilitate full dissociation of complex than the ADP bound state, suggesting a priming effect towards misfolded protein on ATP addition. Data are mean ± s.d., n = 9 independent experiments. Source Data are available online. The IC50 and inhibition constant values for both low and high concentration phases in the absence and presence of different nucleotides are in Supplementary Table 7.
We first assessed whether ADP, ATP or AMP-PNP, a non-hydrolysable analogue of ATP, caused any dissociation of YFP-BiP and CFP-IRE1 LD complex by measuring loss of FRET signal. Consistent with our MST experiments (Fig. 4) and our previous in vitro protein interaction analysis21,22, we found that addition of nucleotides at all concentrations tested did not cause any loss in the FRET signal, indicating that there was no dissociation of BiP-IRE1 LD complex (Fig. 5b, 5c and 5d and Extended Data Fig. 4). However, upon addition of CH1 we observed complete loss of FRET signal, thus demonstrating that dissociation of BiP-IRE1 LD complex is dependent upon misfolded protein association. Next, we incubated CFP-IRE1 LD and YFP-BiP in the absence and in the presence of either ATP, ADP, AMP-PNP, and then measured the FRET signal on addition of varying concentration of misfolded protein, CH1. The inhibition profile of FRET signal displayed a two-phase response for all samples tested including APO, ATP, AMP-PNP and ADP (Fig. 5e-h and Supplementary Table 7). The inhibition constant for the low concentration phase (Kilow) were similar for both ADP and ATP but differed for AMP-PNP and greatly differed for APO state. The inhibition constant for the high concentration phase (KiHigh) is a more significant measurement than the Kilow, as this is the concentration required for full dissociation of FRET complex (YFP-BiP and CFP-IRE1 LD). Interestingly, the KiHigh for FRET complex incubated with ADP (KiHigh = 4.3 ± 0.4μM) was similar to APO FRET complex (KiHigh = 5.2 ± 0.6μM) but significantly greater for the FRET complex when enriched with ATP (KiHigh = 0.2 ± 0.01μM). The KiHigh for FRET complex when bound to AMP-PNP (KiHigh = 0.6 ± 0.05μM) was similar to addition of ATP, consistent with AMP-PNP being an ATP mimic. The ATP bound BiP-IRE1 LD complex requires 21-fold less CH1 to inhibit the FRET signal and facilitate dissociation than the ADP bound FRET complex. The addition of ATP increases the sensitivity of BiP-IRE1 LD complex for CH1, whereas ADP abrogates the sensitivity and priming effect. Thus, ATP – but not ADP – primes IRE1-BiP SBD to sense misfolded protein, the binding of which facilitates BiP dissociation via NBD from UPR proteins.
Discussion
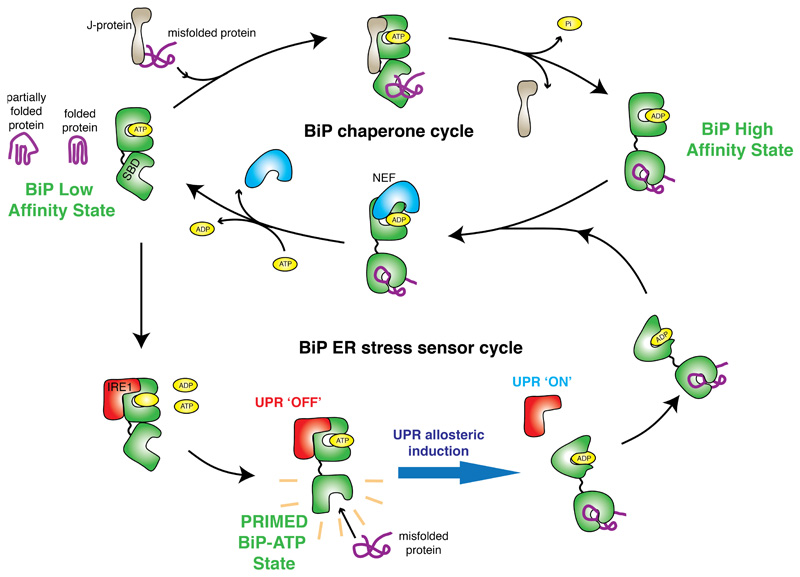
In the current study, we reconstitute components of UPR, ER stress, and BiP chaperone complex systems and assess their relationships. We identify a previously unknown ER stress sensor cycle (Fig. 6) that explains how BiP functions as a Hsp70 chaperone and as an ER stress sensor.
Figure 6. Mechanism of BiP function encompassing its chaperone and ER stress sensor cycles.
Interaction with UPR proteins switch BiP from chaperone cycle to ER stress sensor cycle by preventing binding to cochaperone and NEF proteins, with loss of BiP ATPase stimulation. The interaction between IRE1 LD is mediated via BiP NBD, whilst dissociation of complex is dependent on misfolded protein binding to BiP SBD. The BiP-IRE1 complex in the ATP bound state, has a greater sensitivity for misfolded protein, thereby facilitating release. By contrast the complex is desensitized to misfolded protein when bound to ADP. The release of BiP from UPR proteins via conformational change21,22 – is coupled to UPR activation34, enabling BiP to interact with cochaperones and revert to its chaperone cycle with misfolded protein attached.
The binding of UPR proteins inhibit BiP ATPase stimulation by preventing the association of cochaperones. The inhibitory effect of UPR proteins can be reversed by denaturing IRE1 LD, resulting in BiP stimulation similar to CH1. Our data suggest that BiP can interact with IRE1 in one of two ways. Firstly, as a UPR signaling partner protein that binds via NBD and inhibits BiP ATPase stimulation. Secondly, as a chaperone that binds via BiP SBD that stimulates BiP ATPase activity. We have previously reported that IRE1 LD interacts with BiP NBD, using a combination of techniques including: pull-down assays, MST, chemical cross-linking21, and in vitro FRET measurements22. This interaction has also been observed in another study using genetic analysis27. Furthermore, overexpression of NBD was shown to reduce IRE1 phosphorylation in cells21. Here, we utilized the Sil1-BiP crystal structure to identify a mutation within the NBD that inhibits binding between LD and BiP.
The inhibition of ATPase stimulation by IRE1 prevents BiP from functioning effectively as a Hsp70 type chaperone. In this bound state, however, BiP still retains the ability to engage misfolded proteins and act as a stress sensor. These two modes of binding and action highlight the different roles that BiP plays, both as an ER stress sensor and a Hsp70 chaperone. It is the association of UPR proteins that causes BiP to switch from its chaperone role to that of an ER stress sensor.
The interaction between BiP NBD and IRE1 LD is independent of nucleotides. The dissociation of the complex is solely dependent on misfolded protein binding to BiP SBD. However, nucleotides do influence the ability of BiP SBD to engage misfolded proteins. BiP that is bound to IRE1 LD has a greater sensitivity towards misfolded protein in the presence of ATP. In contrast, the priming of BiP to sense misfolded proteins is reversed or desensitized when BiP is bound to ADP, with over 21-fold increase in misfolded protein concentration required to facilitate the same level of dissociation as the ATP bound BiP-IRE1 LD complex on addition of CH1.
Interestingly, the dissociation of BiP from IRE1 occurs in two phases comprising of a low and high concentration phase irrespective of whether BiP is bound with various nucleotides or not. Although we do not know what these phases are, we speculate that the first phase could be a breakdown in oligomer species to form a single heterodimer complex, with the second phase being the dissociation of heterodimer. Alternatively, it may represent two distinct conformations of BiP that are required to enable full dissociation.
The discriminatory effects of different nucleotides on BiP ability to engage misfolded protein whilst bound to IRE1, is analogous to protein translocation across the inner mitochondrial membrane, where mitochondrial Hsp70 bound to a component of the inner membrane translocon, Tim44, engages the emerging translocating polypeptide chain in an ATP primed state33 but not ADP bound form.
In the primed state, IRE1 and PERK prevent the association of BiP cochaperones and nucleotide exchange factors to ensure that ATP turnover and exchange is halted so that BiP is geared towards sensing misfolded proteins. Once BiP that is bound to IRE1, engages misfolded protein via its SBD, it engenders an allosteric change that alters its affinity for IRE1 causing its releases – a process that is suggested to be coupled to UPR activation21,34. This enables BiP to interact with its cochaperones again and revert to its chaperone cycle with misfolded substrate in tow.
Our results also suggest a possible explanation for observations that indicate ATP triggers the dissociation of BiP-IRE1 co-immune complexes from cell lysates34. Present within the cell lysate are numerous peptides and proteins that have exposed hydrophobic patches and are partially misfolded. The addition of ATP would prime BiP SBD to engage misfolded protein without affecting IRE1-BiP NBD interaction, leading to enhanced misfolded protein dependent dissociation of the complex.
Analysis of binding affinities indicate that in the absence of ER stress, IRE1 will be bound with BiP. This association favours BiP acting as a stress sensor. In this state, BiP is unable to conduct its normal chaperone function as it cannot bind to its cochaperones. IRE1 is a scarce protein with 500—5000 copies in HeLa cells35. By contrast, BiP is one of the most abundant proteins in the ER with estimates suggesting around 2 x 107 as a baseline number17. Assuming all cellular IRE1 is occupied with BiP in the absence of UPR, it would have minimal effect on the total number of free BiP that could participate in protein folding and chaperone activity.
BiP detects its client substrate protein to activate UPR in a ratiometric fashion17. BiP that is bound to IRE1 would bind misfolded protein via its SBD and compete with chaperone productive free BiP to engage its client proteins. The huge difference in concentration between BiP that is bound to IRE1 and chaperone productive BiP, would protect against UPR activation at low misfolded protein levels, but would facilitate a graded response, with saturation of chaperone productive BiP, along with IRE1 bound BiP, leading to full UPR activation. In our model, IRE1 does not participate in competition with misfolded protein for association to BiP, as its interaction to BiP is mediated via NBD4.
After UPR activation and a reduction in misfolded protein levels, the vast disparity in concentration between free BiP and IRE1, would favour an equilibrium that drives BiP to re-associate with IRE1 leading to UPR repression. The inhibited state with BiP bound to IRE1 would represent the default ground state, in agreement with their cellular concentrations17,35 and binding affinity analysis.
Besides our model in which BiP acts as an ER stress sensor, there are two further established models that seek to explain how UPR activation occurs. An alternative BiP dependent model suggests that BiP interacts with IRE1 via SBD to inhibit UPR signaling analogous to heat shock factor 1 repression by Hsp7036. There is also a direct association model that demonstrates binding of misfolded protein to IRE1 LD to activate UPR23. One speculative scenario that can reconcile certain aspects of the different models suggest that upon ER stress, BiP detects misfolded protein via its SBD leading to reduced affinity of BiP NBD for IRE1 and subsequent dissociation. This triggers IRE1 dimerization and low level UPR signaling. The departure of BiP reveals a binding site on IRE1 (MHC-like peptide groove) that can now engage misfolded protein resulting in oligomerization and full scale UPR activation. To remove the bound misfolded protein from the peptide groove, BiP could re-associate with IRE1 but this time via its SBD - in cooperation with J-protein - to capture the misfolded protein from IRE1. However, exactly how these different mechanisms are interconnected is not clear and further work is required to understand this process in better detail.
In summary, the binding of IRE1 and PERK switch BiP into stress sensor cycle where ATP primes BiP to engage misfolded proteins, thereby facilitating dissociation of complex. Our study rationalizes how BiP can act as the major ER Hsp70 chaperone and as a primary detector of ER stress.
Methods
Expression and Purification
Human BiP (28-654), IRE1 LD (32-440), PERK LD (105-403) proteins were cloned and expressed with a N-terminal His6- tag or a N-terminal GST-tag followed by a PreScission Protease cleavage site. Human ERdj3 (23-358aa) and yeast Sil1 (20-421aa) were cloned into pGEX 6p1 vector containing a GST-tag and PreScission protease site. Fluorescent fusion constructs were designed with fluorescent proteins, CFP and YFP, attached N-terminally to human IRE1 LD and BiP via a short 15aa linker (GGAGGAGGAGGAGGA) (Kopp et al. 2018)22 that were cloned into pGEX 6p1 vector. All constructs were expressed in Escherichia coli BL21 (DE3) cells (Agilent, CA), and were grown in autoinduction medium with trace elements (Formedia, UK) at 37°C for 3 hrs and then overnight at 22°C with the exception of ERdj3 which was grown for 1 hr at 37°C then overnight at 18°C. His6-tag proteins were purified by Co2+-NTA affinity using TALON metal affinity resin (Clontech, CA,) in buffer A (50 mM HEPES pH 8.0, 400 mM NaCl) and eluted in the presence of 250 mM imidazole. GST-tagged proteins were purified using Agarose Glutathione Affinity resin (ThermoFisher, MA). GST- proteins were bound to resin and washed three times in buffer B (50 mM HEPES pH 7.3, 400 mM NaCl, 1 mM DTT), and once in a high salt buffer containing 1M NaCl. Proteins were eluted with buffer C (50 mM HEPES pH 8.0, 400 mM NaCl,1mM DTT, 10mM reduced glutathione). Cell lysis (sonication) and the initial BiP purification step were supplemented with 5 mM ATP and 10 mM MgCl2. The His6-tag or GST-tag were removed by overnight incubation with PreScission Protease at 4°C followed by an additional affinity purification step to remove uncleaved protein. All proteins were further purified by anion-exchange using a HiTrap Q HP column (GE Healthcare, UK) and size-exclusion chromatography on a HiLoad 16/60 Superdex 200 column in buffer D (50 mM HEPES pH 8.0, 200 mM NaCl, 1 mM DTT). For experiments, proteins were diluted into buffer E (50 mM HEPES pH 8.0, 50 mM NaCl, 10 mM MgCl2, 10 mM KCl). The misfolded protein CH1 was expressed and purified following the previously described protocol (Feige et al 2009)26. For thermal treatment of IRE1 LD, protein samples were kept at 55°C for 30 mins and then passed through a Superdex 200 10/300 Increase size-exclusion column. Estimation of multimeric states and void volume were based on comparison with known protein standards.
ATPase Assay
The Malachite Green Phosphate Kit solutions were used for experiments (Abnova, TW). A standard curve created from a serial dilution of K2PO4 was used to determine phosphate concentrations during experiments. Reaction samples were prepared for each 5-minute time point measured between 0-120 minutes. BiP at 5 μM concentration was used in all experiments, to which 5 μM of each component (IRE1 LD, PERK LD, ERdj3, or Sil1) were added in various combinations, making sure that stimulatory proteins (ERdj3 and Sil1) were in equimolar concentration with inhibitory proteins (IRE1 LD or PERK LD). The reaction mixtures were incubated with 1 mM ATP at room temperature (RT) for the specified time point, after which samples were quenched with 12 μL of the acid solution and left at RT for 10 min, then 34 μL of the Malachite Green solution was added to each sample and incubated for 20 min, according to manufactures protocol. Samples were then transferred to a 384 well microplate (Greiner Bio-one, AT) and absorbance was measured at 620 nM wavelength on a CLARIOstar® microplate reader (BMG Labtech, DE).
Microscale Thermophoresis
MST experiments were performed using a Monolith NT.115 instrument (Nanotemper Technologies, DE). BiPWT, BiPV461F and BiPK294F were labelled using the Monolith NT Protein Labelling Kit Red-NHS. Then, ~100 nM labelled BiP was mixed with 1 mM nucleotide where indicated and incubated on ice for 30 min in MST buffer (50 mM HEPES pH 7.8, 50 mM NaCl, 10 mM KCl, 10 mM MgCl2, 0.1% TWEEN-20). This mixture was then combined with equal volumes of a sixteen two-fold serial dilution of the unlabeled binding partner. Experiments were carried out in hydrophobic treated capillaries with 20% LED power and 80% IR-laser at 25 °C. Data were analyzed on Graphpad Prism 8 (Graphpad Software, CA, USA) and Kd was determined using one site binding saturation curve fit.
Single Mutant and Competition Pull-down assay
All pull-down experiments were conducted in 2 mL gravity flow columns. Glutathione resin pre-equilibrated with buffer E was incubated with 40 μM of the GST-tagged BiPWT or BiPK294F for 1 hr at RT. The resin was washed with 500 μL of buffer E to remove the unbound BiP. Then 50 μL of 120 μM IRE1, PERK, ERdj3, or Sil1 as indicated was added to the column and incubated for 1 hr at RT. The resin was washed with 500 μL of buffer E in 125 μL increments and resuspended in 75 μL of buffer E. Samples of the re-suspended resin were analyzed on a 4-12% gradient SDS-PAGE gel.
For the competition pull-downs, 75 μL of Glutathione resin pre-equilibrated with buffer E was incubated with 20 μM GST tagged BiP (GST-BiP). The resin was washed with 500 μL of buffer E to remove the unbound BiP. Then 50 μL of 60 μM ERdj3 or Sil1 was added to each column and incubated for 1 hr at RT. The resin was then washed with 500 μL of buffer E in 125 μL increments. IRE1 LD or PERK LD was added to each column at increasing concentrations (0 μM, 15 μM, 30 μM, 60 μM, 75 μM, 90 μM, and 120 μM) and incubated for 1 hr at RT. Resin was washed again as mentioned earlier and re-suspended with 75 μL of buffer E. Samples of the re-suspended resin were analyzed on a 4-12% gradient SDS-PAGE gel.
FRET Assay
FRET assay experiments were carried out using a CLARIOstar® microplate reader (BMG Labtech, DE) following the protocol for measuring a three-way protein-protein interaction using FRET from Martin et al (2008)32 and Kopp et al (2018)22 for apo protein. In brief, a mixture containing 30 μM of each fluorescent protein (CFP-IRE1 LD and YFP-BiP), and increasing concentrations of CH1 were made into a volume of 160 μL in buffer E. Next, 50 μL was transferred to a 384 well microplate (Greiner Bio-one, AT). The protocol was adapted for nucleotide experiments to include a 30 min incubation at RT with 1mM nucleotide in the reaction mixture. Calculation of Ki was based on the protocol from Martin et al (2008)32. The data were plotted using percentage inhibition as a function of the CH1 concentration and fitted to an exponential curve (two phase association), from which 50% dissociation value or IC50 (equivalent to half-life) for each phase were measured in GraphPad prism 8. These values were then used to calculate the Ki for each phase.
Statistics and reproducibility
Sample sizes and statistical tests where applicable for experiments are denoted in the figure legends. Analysis and graphs were generated in GraphPad prism 8 and excel.
Extended Data
Extended Data Fig. 1. Comparison of BiP and GST-BiP stimulation and inhibition.
(a) BiP ATPase activity showing stimulation by co-chaperones and inhibition by IRE1 and PERK LD. (b) same as (a), but using GST tagged BiP. (c) A comparison of the rate of ATPase activity for BiP and GST tagged BiP. Both tagged and untagged BiP are stimulated by co-chaperones and inhibited by IRE1 and PERK LD to the same level, indicating that the attachment of GST to BiP had no effect on BiP ATPase stimulation or inhibition. Statistics as in Figure 1, source data available online.
Extended Data Fig. 2. Mutant BiPK294F has same basal ATPase activity as BiPWT.
(a) BiPK294F ATPase activity on addition of co-chaperones. (b) A comparison of the rate of ATPase activity for BiP and BiPK294F. The K294F mutation based within the BiP NBD, had no effect on inherent BiP ATPase, but prevents ATPase stimulation consistent with-it inhibiting co-chaperone binding. Statistics as in Figure 1, source data available online.
Extended Data Fig. 3. Assessment of BiPK294F affinity for CH1 in presence of nucleotides.
BiPK294F has same binding affinity for CH1, in different nucleotide bound states, as BiPWT. (a) MST profile measuring the binding affinity between BiP and BiPK294F for CH1. (b) same as (a), but in the presence of ADP. (C) same as (a), but with ATP. The experiments indicate that the mutation had no effect on BiP interaction with misfolded substrate protein or affected BiP nucleotide bound conformations. Statistics as in Figure 4, source data available online.
Extended Data Fig. 4. Attachment of YFP had no effect on BiP functionality.
(a) ATPase activity of BiP and YFP tagged BiP. (b) Comparison of the rate of ATPase activity for BiP and YFP- tagged BiP on addition of co-chaperones and CFP-IRE1 LD. The attachment of YFP had no effect on BiP basal activity, or stimulation by co-chaperones, or inhibition by CFP tagged IRE1 LD. Statistics as in Figure 1, source data available online.
Supplementary Material
Acknowledgements
We thank Piotr Nowak for assistance with protein purifications. We thank A Mouskidis for assistance with FRET ADP measurements. Also, we thank the Freemont and Zhang labs, along with Jon Wilson for use of MST equipment. This work was funded by a cancer research UK senior research fellowship awarded to MMUA (C33269/A20752) and (C33269/A23215).
Footnotes
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Source data for Figures 1-5 and Extended Data Figures are provided with the paper.
Author contributions
M.C.K. designed experiments, expressed and purified proteins, conducted experiments including ATPase assays, competitive pull-down assays, MST and FRET assays, analyzed results, presented data, and contributed to figure preparation and manuscript. N.L. expressed and purified proteins, including mutant BiP, conducted MST experiments, analyzed data, contributed to figure preparation and manuscript. V.D. did some initial protein preps and ATPase assay measurements. C.J.A. assisted in expression of CH1 and MST experiments, and contributed to manuscript. M.M.U.A. conceptualized and designed experiments, analyzed results, prepared figures and wrote manuscript, supervised project and obtained funding.
Competing interests
The authors declare no competing interests.
References
- 1.Hetz C, Papa FR. The Unfolded Protein Response and Cell Fate Control. Mol Cell. 2017 doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- 3.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 4.Adams CJ, Kopp MC, Larburu N, Nowak PR, Ali MMU. Structure and Molecular Mechanism of ER Stress Signaling by the Unfolded Protein Response Signal Activator IRE1. Front Mol Biosci. 2019;6:11. doi: 10.3389/fmolb.2019.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J, et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrara M, Prischi F, Nowak PR, Ali MMU. Crystal structures reveal transient PERK luminal domain tetramerization in endoplasmic reticulum stress signaling. EMBO J. 2015;34:1589–1600. doi: 10.15252/embj.201489183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 9.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ali MMU, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 12.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 13.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 15.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–6. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 16.Prischi F, Nowak PR, Carrara M, Ali MMU. Phosphoregulation of Ire1 RNase splicing activity. Nat Commun. 2014;5 doi: 10.1038/ncomms4554. 3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakunts A, et al. Ratiometric sensing of BiP-client versus BiP levels by the unfolded protein response determines its signaling amplitude. Elife. 2017;6 doi: 10.7554/eLife.27518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 19.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–92. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behnke J, Feige MJ, Hendershot LM. BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions. J Mol Biol. 2015;427:1589–1608. doi: 10.1016/j.jmb.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carrara M, Prischi F, Nowak PR, Kopp MC, Ali MMU. Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. Elife. 2015;4 doi: 10.7554/eLife.03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopp MC, Nowak PR, Larburu N, Adams CJ, Ali MMU. In vitro FRET analysis of IRE1 and BiP association and dissociation upon endoplasmic reticulum stress. Elife. 2018;7 doi: 10.7554/eLife.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagoz GE, Acosta-Alvear D, Walter P. The Unfolded Protein Response: Detecting and Responding to Fluctuations in the Protein-Folding Capacity of the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol. 2019 doi: 10.1101/cshperspect.a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acosta-Alvear D, et al. The unfolded protein response and endoplasmic reticulum protein targeting machineries converge on the stress sensor IRE1. Elife. 2018;7 doi: 10.7554/eLife.43036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amin-Wetzel N, et al. A J-Protein Co-chaperone Recruits BiP to Monomerize IRE1 and Repress the Unfolded Protein Response. Cell. 2017;171:1625–1637.e13. doi: 10.1016/j.cell.2017.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feige MJ, et al. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell. 2009;34:569–579. doi: 10.1016/j.molcel.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todd-Corlett A, Jones E, Seghers C, Gething M-J. Lobe IB of the ATPase domain of Kar2p/BiP interacts with Ire1p to negatively regulate the unfolded protein response in Saccharomyces cerevisiae. J Mol Biol. 2007;367:770–787. doi: 10.1016/j.jmb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laufen T, et al. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc Natl Acad Sci USA. 1999;96:5452–5457. doi: 10.1073/pnas.96.10.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan M, Li J, Sha B. Structural analysis of the Sil1-Bip complex reveals the mechanism for Sil1 to function as a nucleotide-exchange factor. Biochem J. 2011;438:447–455. doi: 10.1042/BJ20110500. [DOI] [PubMed] [Google Scholar]
- 31.Kityk R, Kopp J, Mayer MP. Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones. Mol Cell. 2018;69:227–237 e4. doi: 10.1016/j.molcel.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Martin SF, Tatham MH, Hay RT, Samuel IDW. Protein Sci. Vol. 17. Cold Spring Harbor Laboratory Press; 2008. Quantitative analysis of multi-protein interactions using FRET: application to the SUMO pathway; pp. 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig EA. Hsp70 at the membrane: driving protein translocation. BMC Biol. 2018;16:11. doi: 10.1186/s12915-017-0474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 35.Boisvert FM, et al. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.011429. M111 011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preissler S, Ron D. Early Events in the Endoplasmic Reticulum Unfolded Protein Response. Cold Spring Harb Perspect Biol. 2018 doi: 10.1101/cshperspect.a033894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.