Abstract
Modification of specific Ser and Thr residues of nucleocytoplasmic proteins with O-GlcNAc, catalyzed by O-GlcNAc transferase (OGT), is an abundant post-translational event essential for proper animal development and dysregulated in various diseases. Due to the rapid concurrent removal by the single O-GlcNAcase (OGA), precise functional dissection of site-specific O-GlcNAc modification in vivo is currently not possible without affecting the entire O-GlcNAc proteome. Exploiting the fortuitous promiscuity of OGT, we show that S-GlcNAc is a hydrolytically stable and accurate structural mimic of O-GlcNAc that can be encoded in mammalian systems with CRISPR-Cas9 in an otherwise unperturbed O-GlcNAcome. Using this novel approach, we target an elusive Ser405 O-GlcNAc site on OGA, showing that this site-specific modification affects OGA stability.
Introduction
Protein O-GlcNAcylation is a ubiquitous and dynamic nucleocytoplasmic post-translational modification of serines and threonines with O-linked β-N-acetylglucosamine (O-GlcNAc)1. Thousands of protein substrates are modified by O-GlcNAc transferase (OGT)2 while a single enzyme, O-GlcNAc hydrolase (OGA)3, removes the modification. Importantly, OGT is required for proper development in mice and fruit flies4,5. On the other hand, disruption of O-GlcNAc cycling on specific proteins is associated with cancer6, diabetes7, neurodegeneration8 and a recently described intellectual disability syndrome9.
Functional dissection of individual O-GlcNAc sites has so far relied on alanine mutagenesis or modulation of OGA or OGT activity through inhibition or knockdown. However, these approaches are confounded by undesirable effects on the entire O-GlcNAcome. Methods for introducing site-specific stoichiometric O-GlcNAc modification are limited to in vitro chemical biology techniques such as expressed protein ligation. Yet, only a few O-GlcNAcylated proteins have been produced using this method, including tau and α-synuclein10,11. Another in vitro method, relying on the chemical conversion of a cysteine (introduced by site-directed mutagenesis) to dehydroalanine and subsequent reaction with a GlcNAc derivative, is limited to proteins that do not contain (many) native cysteine residues and is not stereo-specific12. Site-specific incorporation of O-GlcNAc in the context of a living biological system has not been achieved yet.
Given that the O-GlcNAc modification is prone to hydrolysis by OGA, site-specific O-GlcNAcylation in cells, even if achieved, would not be permanent. Interestingly, OGA itself belongs to a plethora of OGT substrates and the modification site (Ser405) is conserved among vertebrates (Fig. 1a)13,14. Previously, this O-GlcNAc modification was shown to be inducible by OGA inhibition, while being minimal at basal conditions15,16. However, studying this phenomenon presents a paradox – even if stoichiometric O-GlcNAcylation of catalytically active OGA could be achieved, its own hydrolase activity would remove it. Therefore, the investigation of the regulatory functions of O-GlcNAc modification on OGA, and other OGT substrates, would require site-specific installation of an OGA-resistant functional mimic of O-GlcNAc.
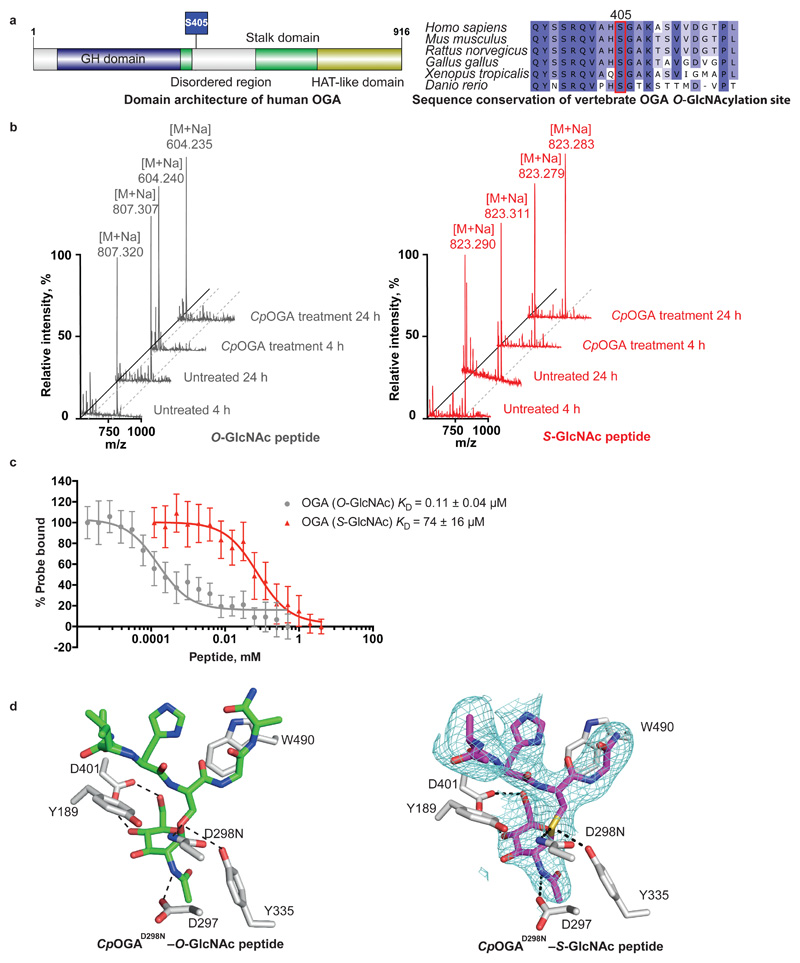
Figure 1. S-GlcNAc mimics O-GlcNAc in the context of an OGA peptide.
(a) Schematic representation of the human OGA sequence (including the O-GlcNAcylation site Ser405) and sequence conservation of Ser405 in vertebrates. GH domain – glycoside hydrolase domain. HAT-like domain – histone acetyltransferase-like domain.
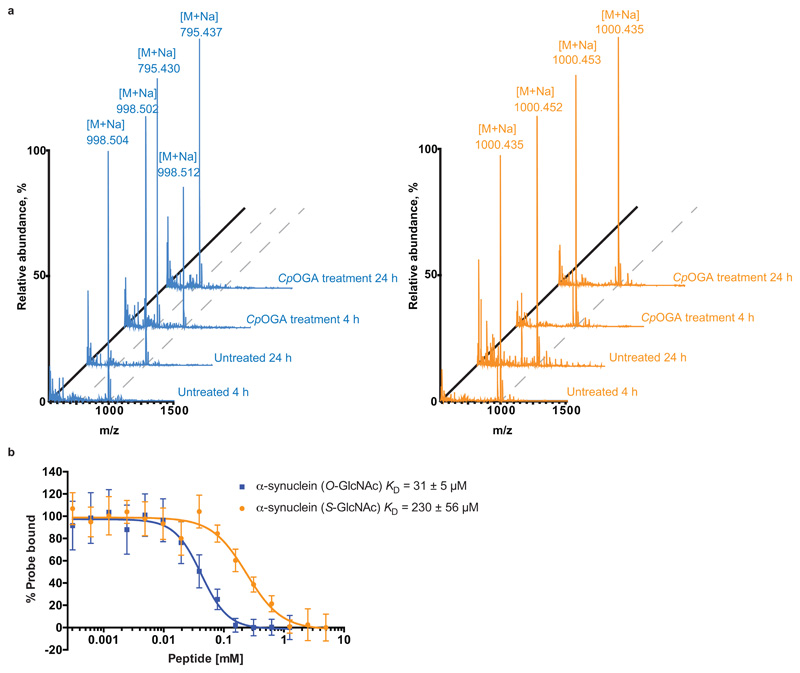
(b) MALDI-TOF mass spectra of synthetic O-/S-GlcNAc-modified peptides derived from hOGA with or without CpOGA treatment for 4 and 24 h. Loss of GlcNAc residue is characterized by the reduction in m/z by 203 Da. M – m/z, [M + Na] – m/z with a sodium adduct (23 Da), m/z – mass/charge ratio.
(c) Fluorescence polarization assay dose-response curves showing the displacement of a fixed concentration of fluorescent probe (15 nM GlcNAcstatin B-FITC) from CpOGAD298N by increasing concentrations of O- or S-GlcNAcylated peptides derived from hOGA. Highest amount of probe bound to CpOGAD298N in the absence of competing O- or S-GlcNAcylated peptides was arbitrarily set as 100%. Data points were fitted to a four-parameter equation for dose-dependent inhibition using Prism (GraphPad). Data are shown as mean ± s.e.m. of n = 3 (O-GlcNAc peptide) and n = 4 (S-GlcNAc peptide) independent experiments. KD values were calculated as described previously23.
(d) Previously published O-GlcNAcylated hOGA-derived peptide (depicted as green sticks) in complex with CpOGAD298N (PDB: 2YDQ) and a crystal structure of S-GlcNAcylated hOGA-derived peptide (purple sticks) in complex with CpOGAD298N (deposited in the PDB as 6RHE). The unbiased |Fo-Fc| map before inclusion of any S-GlcNAc peptide model is shown as a blue mesh contoured at 2.5σ. CpOGAD298N active site residues are shown as grey sticks. Hydrogen bonds are shown as dashed lines.
Source data for panel c are available with the paper online.
A thio-linked GlcNAc (S-GlcNAc) has potential to resist OGA-mediated hydrolysis. It was initially shown that the pseudo-substrate peptide Ala-Cys-(S-GlcNAc)-Ala inhibits a highly active bacterial OGA orthologue from Clostridium perfringens (CpOGA) at low micromolar concentrations, implying that S-GlcNAc may be a hydrolytically stable mimic of O-GlcNAc in the context of a peptide backbone17. The non-hydrolysable S-GlcNAc modification has been successfully applied as an O-GlcNAcylation mimic in vitro in the context of several recombinant proteins, such as casein kinase 2 (CK2), histone H2B, α-synuclein and tau18–21. Therefore, S-GlcNAc, if site-specifically incorporated, presents an attractive candidate for studying the effects of permanent O-GlcNAcylation. Nonetheless, production of such S-GlcNAcylated proteins has thus far been limited to in vitro approaches. Here we establish a straightforward method for site-specific installation of S-GlcNAc mimic in vivo, compatible with CRISPR-Cas9 genome editing. We show that the O-GlcNAc site on the O-GlcNAcase regulates its half-life, adding to the growing body of evidence that suggests a link between O-GlcNAcylation and protein stability.
Results
S-GlcNAc is a non-hydrolysable O-GlcNAc analogue
A prerequisite for the incorporation of S-GlcNAc as a stable O-GlcNAc mimic is resistance to the action of OGA. Although some studies support this17,20, we wondered whether S-GlcNAcylation would be resistant to OGA hydrolysis in the context of human OGA (hOGA). To investigate this, short synthetic peptides (bearing the S- or O-GlcNAc modification) derived from hOGA were subjected to treatment with CpOGA, a promiscuous, bacterial orthologue of the human enzyme with elevated activity17. Analysis of the products by MALDI-TOF mass spectrometry revealed loss of modification on the O-GlcNAcylated peptide after just 4 h of CpOGA treatment (Fig. 1b), whereas the S-GlcNAcylated peptide remained intact for at least 24 h (Fig. 1b).
Next we asked whether the S-GlcNAc modification is an adequate structural mimic of the O-GlcNAc modification in the context of proteins that recognize the O-GlcNAc modification, such as OGA and recently discovered O-GlcNAc readers22. We employed a fluorescence polarimetry competition assay23 to compare the binding affinities of Ser-O-GlcNAc and Cys-S-GlcNAc in the context of a peptide, using a catalytically incompetent mutant of CpOGA (CpOGAD298N) as a model O-GlcNAc ‘’reader”24. Encouragingly, CpOGAD298N did bind the hOGA S-GlcNAcylated peptide with a KD value in the μM range, albeit two orders of magnitude lower than the O-GlcNAcylated equivalent (Fig. 1c). These data suggest that S-GlcNAc will likely be recognized by a range of O-GlcNAc binding proteins, and is an adequate mimic of the O-GlcNAc modification.
Although Cys-S-GlcNAc is expected to most accurately mimic Ser-O-GlcNAc, we wondered whether Thr-O-GlcNAc could also be replaced with a non-hydrolysable S-GlcNAc. To investigate this, we synthesized O- and S-GlcNAcylated peptides derived from the Thr72 O-GlcNAcylation site of α-synuclein11 and subjected them to treatment with active CpOGA. The latter completely removed the modification on Thr-O-GlcNAc peptide after 24 h, while the Cys-S-GlcNAcylated counterpart remained stable (Extended Data Fig. 1a). Interestingly in a binding assay, the affinity of inactive CpOGA for Cys-S-GlcNAcylated peptide was just one order of magnitude weaker (Extended Data Fig. 1b). Thus, Cys-S-GlcNAc could potentially be used to mimic Thr-O-GlcNAc.
To further corroborate the structural mimicry, we investigated whether the conformation of Cys-S-GlcNAc is similar to Ser-O-GlcNAc in the context of a binding protein. We have previously reported crystal structures of O-GlcNAcylated peptides in complex with CpOGAD298N, including a complex with a peptide encompassing Ser405 of hOGA25. We soaked CpOGAD298N crystals with the S-GlcNAcylated hOGA peptide and obtained synchrotron diffraction data (Table 1). Molecular replacement with the previously reported CpOGA structure revealed well-defined difference density covering the S-GlcNAc moiety and the peptide backbone (Fig. 1d). Further refinement resulted in a final model with R/Rfree = 0.19/0.23 (Table 1). Comparison of the positions and conformations of the O- and S-GlcNAcylated peptides reveals them to bind in a similar fashion (sugar atomic positional shifts < 0.3 Å, Fig. 1d) with conservation of the hydrogen bonds of the sugar moiety with Asp401, Tyr335, Asn298 and Asp297 (Fig. 1d).
Table 1. Data collection and refinement statistics.
| CpOGA–hOGA (S-GlcNAc) peptide (PDB 6RHE) | |
|---|---|
| Data collection | |
| Space group | P61 |
| Cell dimensions | |
| a, b, c (Å) | 117.7, 117.7, 147.9 |
| α, β, γ (°) | 90º, 90º, 120º |
| Resolution (Å) | 48.17 - 3.1 (3.21 - 3.1)a |
| Rmerge | 0.1077 (1.123) |
| I / σI | 9.13 (1.10) |
| CC1/2 | 0.994 (0.379) |
| Completeness (%) | 93.85 (95.67) |
| Redundancy | 3.2 (3.3) |
| Refinement | |
| Resolution (Å) | 48.17-3.1 |
| No. reflections | 19790 (1990) |
| Rwork / Rfree | 0.1949/0.2270 |
| No. atoms | |
| Protein | 4594 |
| S-GlcNAc peptide | 43 |
| Cadmium ion | 25 |
| Water | 28 |
| B-factors | |
| Protein | 84.34 |
| S-GlcNAc peptide | 112.10 |
| Cadmium | 138.30 |
| Water | 69.83 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.61 |
The dataset was collected from a single crystal.
Values in parentheses are for highest-resolution shell.
S-GlcNAc can be genetically incorporated in the context of O-GlcNAc sites
Having confirmed that S-GlcNAc appears to be a non-hydrolysable functional mimic of O-GlcNAc, we sought a straightforward approach for site-specific incorporation of this modification in vivo. Interestingly, the existence of S-GlcNAc on mammalian proteins was demonstrated recently26. Moreover, using an in vitro assay it was shown that cysteine residues in certain peptide sequences could be S-GlcNAcylated by OGT26. Inspired by this report, we hypothesized that OGT could modify cysteine residues in the context of O-GlcNAcylation sites in proteins, such as a well-characterized single O-GlcNAcylation site (Ser395) on the innate immunity signaling node TGF-β activated kinase binding protein 1 (TAB1)27.
To test whether S395C mutation on TAB1 would result in S-GlcNAcylation, the recombinant protein was incubated with OGT and UDP-GlcNAc followed by MS/MS analysis using Electron-Transfer/Higher-Energy Collision Dissociation (EThcD). In line with our hypothesis, cysteine-S-GlcNAcylation was unambiguously detected on TAB1S395C (Fig. 2a), confirming the reported S-GlcNAc transferase activity of OGT26.
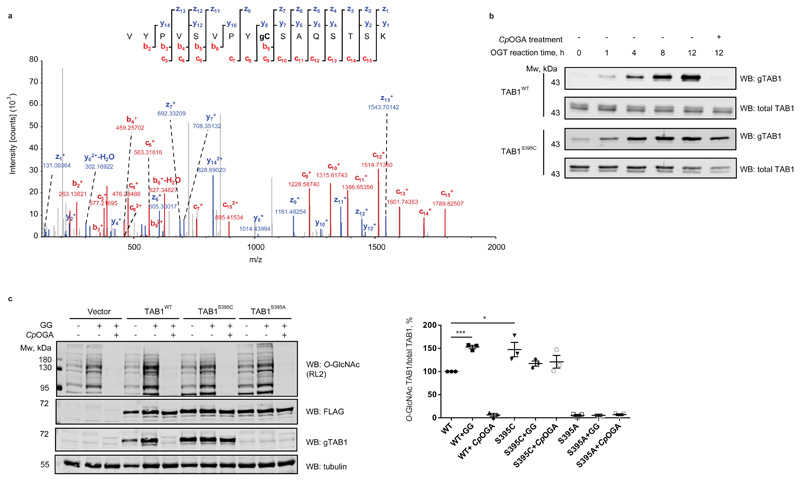
Figure 2. OGT catalyzes S-GlcNAcylation of TAB1S395C in vitro and in cells.
(a) LC-MS/MS EthCD S-GlcNAc site mapping on S395C TAB1 mutant. In vitro S-GlcNAcylated S395C TAB1 mutant (7-409) was digested with trypsin and subjected to LC–MS. The tryptic peptide VYPVSVPYCSAQSTSK (Mwcalc = 1918 Da) containing a HexNAc (+203.1 Da) was detected. The observed fragment ions (z, y, c and b ion series) are indicated in the fragmentation diagram. Mass differences between y7 to y8 and c8 to c9 allow confirmation of C395 as the site of S-GlcNAc modification. The c82+ ion corresponds to m/z 461.75528.
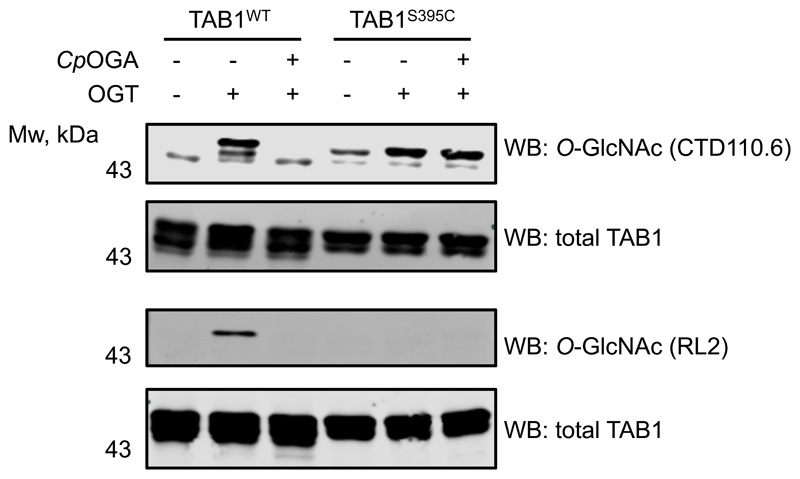
(b) Time-course of TAB1WT and TAB1S395C in vitro OGT reaction. After 12 h incubation of TAB1WT and TAB1S395C with OGT, the reactions were treated with CpOGA. O- and S-GlcNAc modifications were detected by Western blot using an anti O-GlcNAc TAB1 (gTAB1) antibody.
(c) OGT catalyzes S-GlcNAcylation of TAB1S395C in cells. IL-1R Hek293 cells were transfected with N-terminally FLAG-tagged TAB1 constructs that were wild type or carried S395C or S395A mutations of the O-GlcNAc modification site. The cells were treated with OGA inhibitor GlcNAcstatin G (GG) for to elevate O-GlcNAc levels. The lysate was then split in half and one half was treated with CpOGA to remove O-GlcNAc (as monitored by Western blot using an anti-O-GlcNAc antibody RL2). Overexpressed TAB1 levels were assessed with an anti-FLAG antibody. Glycosylation levels of TAB1 were monitored with a site-specific O-GlcNAc TAB1 antibody (gTAB1). Tubulin was used as a loading control. Normalization was performed by dividing the gTAB1 antibody signal (O-GlcNAc TAB1) by the FLAG signal (total TAB1). “*” denotes P = 0.0377, “***” denotes P = 0.0001, calculated by Student’s t-test (two-tailed, unpaired) Data are shown as mean ± s.e.m. of n = 3 biological replicates. Uncropped Western blot images and source data for panel 2c are available with the paper online.
We have previously generated a site-specific O-GlcNAc TAB1 Ser395 antibody (gTAB1)27 and used this on Cys395 S-GlcNAcylated TAB1 to further investigate structural mimicry. In a control experiment, in vitro OGT assay revealed a time-dependent increase of O-GlcNAcylation on wild type TAB1 using the gTAB1 antibody, while incubation with CpOGA completely removed the modification (Fig. 2b). Likewise, a time-dependent increase in TAB1S395C GlcNAcylation was observed, indicating that S-GlcNAcylation represents a suitable structural mimic recognizable by the gTAB1 antibody (Fig. 2b). Gratifyingly, TAB1S395C S-GlcNAcylation was resistant to CpOGA treatment.
To investigate whether protein S-GlcNAc transferase activity of OGT could occur in a different substrate sequence context, we generated a recombinant OGT linear fusion with a peptide derived from CK2 using a previously described approach28, in which the O-GlcNAcylation site was mutated to a Cys. In absence of a site-specific antibody for CK2 O-GlcNAc site (Ser347), we used the pan-specific O-GlcNAc antibody CTD110.6, which recognized both O-GlcNAcylated and S-GlcNAcylated CK2 fusion proteins (Extended Data Fig. 2). The latter was resistant against CpOGA hydrolysis, confirming our previous findings. Notably, S-GlcNAcylated TAB1 was also detectable with CTD110.6 (Extended Data Fig. 3), suggesting that this widely used antibody may be used as a tool to detect S-GlcNAcylation. These results are not entirely surprising, since CTD110.6 can cross-react with N-glycans and GlcNAcylated O-mannose, as well as terminal β-GlcNAc on complex N-glycans of cell surface glycoproteins29–31. On the other hand, another pan-specific O-GlcNAc antibody RL2 does not suffer from such poor specificity towards the linkage type32 and reacted exclusively with O-GlcNAcylated TAB1 (Extended Data Fig. 3).
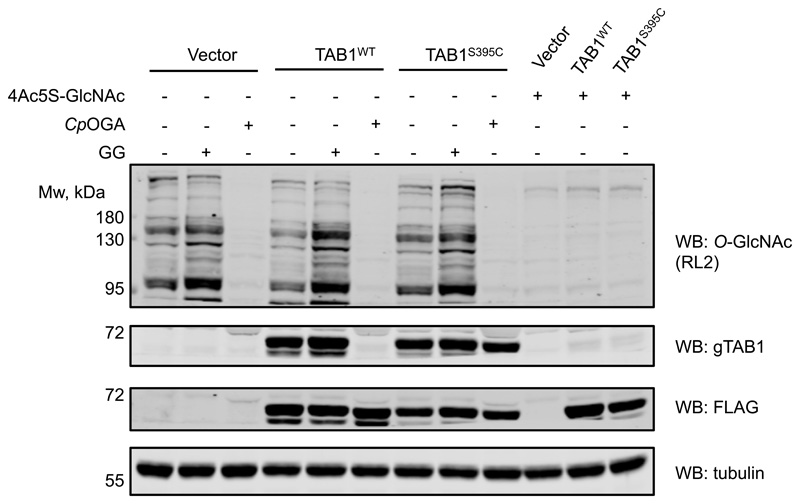
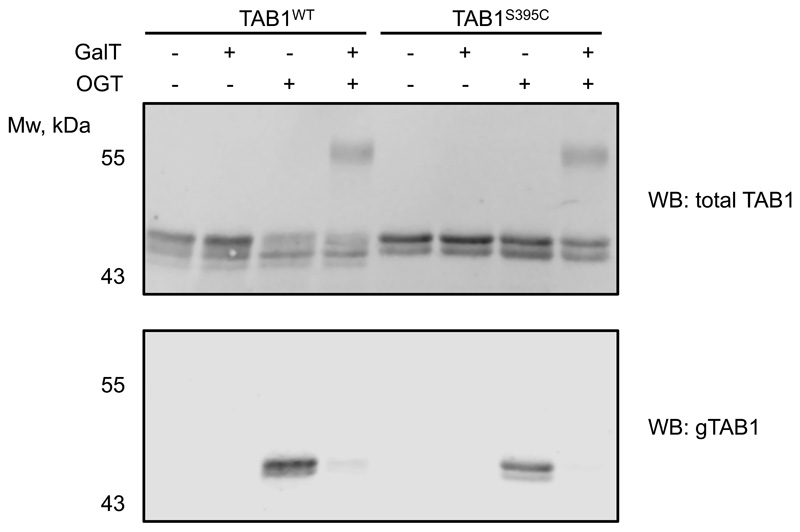
The crucial advantage of the OGT-assisted S-GlcNAcylation mimicry of protein O-GlcNAcylation is the ability to encode it genetically in living systems by engineering targeted, single Ser or Thr to Cys mutations. Having shown the application of our approach in vitro, we next investigated whether this could be extended to cultured mammalian cells. FLAG-tagged wild type and Ser395Cys TAB1 were expressed in IL-1R Hek293 cells27 that were optionally treated with GlcNAcstatin G33 to inhibit intracellular OGA. Additionally, lysates were treated with CpOGA to remove O-GlcNAc. As expected, O-GlcNAc-modified wild type TAB1 was detected, with the modification being sensitive to CpOGA treatment (Fig. 2c). Strikingly, we also detected GlcNAc modification on the Ser395Cys mutant, which was resistant to CpOGA hydrolysis (Fig. 2c). In a separate experiment, treatment of cells overexpressing WT or S395C TAB1 with 4Ac5S-GlcNAc34, a metabolic precursor of the OGT inhibitor UDP-5S-GlcNAc, in both cases resulted in a disappearance of signal detected by the gTAB1 antibody, suggesting that in cells the S-GlcNAc modification is enzymatic (Extended Data Fig. 4).
Genetically directed S-GlcNAcylation allows dissection of O-GlcNAc function on OGA
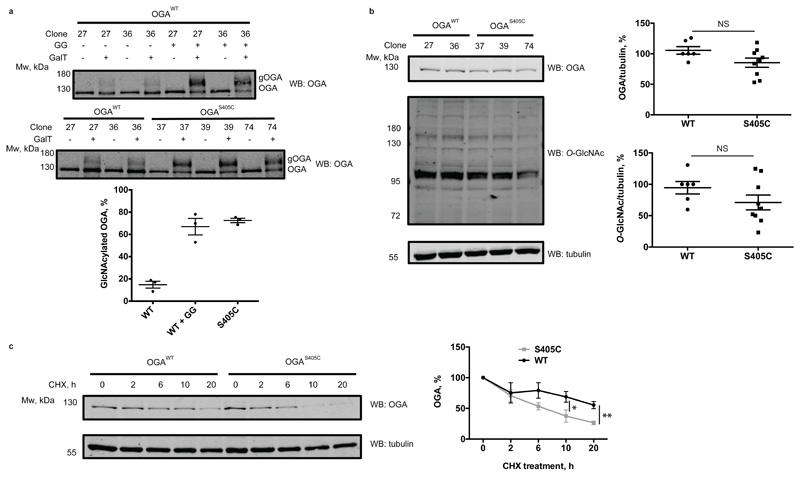
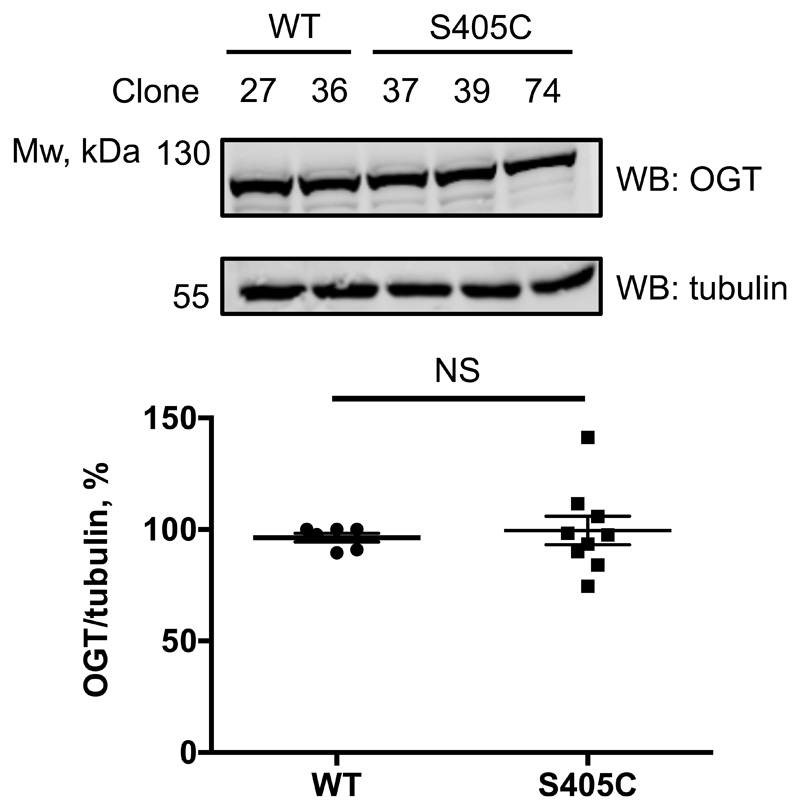
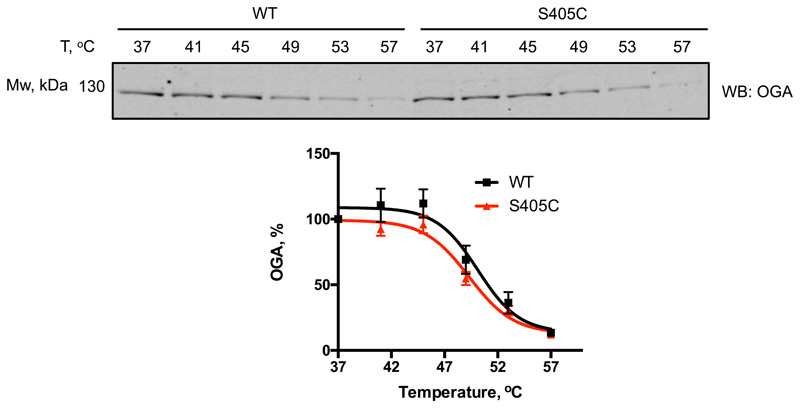
Finally, we aimed to extend this approach to investigate the role of a single O-GlcNAc site on O-GlcNAcase in cell culture. We pursued this goal by introducing the OGA Ser405Cys mutation in mouse embryonic stem cells (mESCs) with CRISPR-Cas9. Potential knock-in mutants were screened and several mutant lines were verified by sequencing (Supplementary Fig. 1-4). Due to the absence of a site-specific O-GlcNAc OGA antibody as well as a lack of a proven immunoprecipitation-grade OGA antibody, we set out to quantify the stoichiometry of the GlcNAc modification on OGA in cell lysates, by performing GalTY289L labelling with GalNAz followed by click-chemistry reaction with an alkyne-labelled PEG 5000 to allow separation of glycosylated and unmodified OGA by SDS-PAGE16. Although the galactosyltransferase mutant GalTY289L is expected to label terminal GlcNAc residues, irrespective of the linkage type35, this approach has been mostly used for O-GlcNAc and terminal N-GlcNAc16,36. To validate this method for S-GlcNAc, we first performed OGT reactions with WT and S395C recombinant TAB1, followed by a reaction with GalTY289L and subsequent PEGylation. Since the site-specific O-GlcNAc TAB1 antibody appears to recognize both O- and S-GlcNAcylated TAB1 with similar sensitivity, we used it as a control to monitor the successful OGT in vitro reaction as well as galactosyltransferase labelling efficiency based on the disappearance of GlcNAc signal detected by a gTAB1 antibody (Extended Data Fig. 5). Western blot analysis revealed complete labelling of S-GlcNAcylated TAB1, and comparable GlcNAcylation stoichiometry, indicating similar efficiency of both O- and S-GlcNAc transferase activities of OGT (Extended Data Fig. 5). Encouraged by these results, we applied this GlcNAc mass-tagging approach to OGA in mESC lysates. Quantification of GlcNAcylation stoichiometry revealed a rather low O-GlcNAcylation of OGA at a basal level (consistent with previously published data15,16), while the S405C mutation resulted in a GlcNAcylation stoichiometry of approximately 70%, similar to that induced by GlcNAcstatin G treatment on the wild type protein (Fig. 3a). It must be noted that due to the enzymatic nature of GalT labelling, the stoichiometry measured using this approach may be underestimated. Hyper-GlcNAcylation did not significantly affect OGA and OGT levels, while the O-GlcNAcase activity was preserved (Fig. 3b and Extended Data Fig. 6). Recent evidence has emerged suggesting that O-GlcNAcylation may directly affect protein stability37–39. This prompted us to investigate whether modification of OGA with GlcNAc could have an effect on its stability by monitoring the protein half-life (Fig. 3c). Interestingly, in a cycloheximide assay hyper-GlcNAcylated OGAS405C mutant had a significantly reduced half-life (approximately 6 h) compared to wild type OGA (approximately 20 h). To investigate whether this was through a direct effect on OGA stability, we performed a cell lysate thermal shift assay that revealed no significant differences between wild type and S405C mutant OGA (Extended Data Fig. 7). Due to the fact that at basal conditions, OGA protein level appears to be unchanged, we speculate that owing to a compensation mechanism to maintain O-GlcNAc homeostasis, OGAS405C may be synthesized at a faster rate to compensate for accelerated degradation of a permanently S-GlcNAcylated protein. This suggests the presence of additional factors that affect OGA stability in a GlcNAcylation dependent mechanism.
Figure 3. OGT catalyzes hydrolytically stable cysteine-S-GlcNAcylation of OGA in cells, which reduces OGA stability.
(a) Quantification of O- and S-GlcNAcylation stoichiometry on OGA was performed by chemoenzymatic labelling with GalNAz using a galactosyltransferase Y289L mutant (GalT) and subsequent click-chemistry reaction with a 5 kDa PEG-alkyne. Labelling was performed on cell lysates from cells expressing wild type OGA (cells were either untreated or treated with 1 ∝M GlcNAcstatin G (GG) as a positive control) and OGAS405C mutant. Reactions were assessed by Western blot using an anti-OGA antibody. Upward molecular weight shift corresponds to GlcNAc modified OGA (gOGA). Plotted are the GlcNAcylation stoichiometries of wild type (undetectable) and S405C mutant OGA. Data are shown as mean ± s.e.m. of n = 3 biological replicates.
(b) Quantification of OGA and total O-GlcNAc levels in wild type OGA and OGAS405C mutant mES cells by Western blot analysis. α-tubulin was used as a loading control. Data are shown as mean ± s.e.m. of n = 6 (WT OGA) and n = 9 (OGAS405C) biological replicates. NS – no significant difference (analyzed by two-tailed Student’s t-test).
(c) OGA stability was measured by treating cells with cycloheximide at a final concentration of 20 μg/ml and harvesting cells at indicated time points for Western blot analysis using an anti-OGA antibody. Normalization of band intensities was performed by dividing OGA signal by α-tubulin signal. Untreated normalized sample was assigned as 100% OGA. “*” denotes P = 0.0399, “**” denotes P = 0.00115, calculated by Student’s t-test (two-tailed, unpaired). Data are shown as mean ± s.e.m. of n = 6 biological replicates (using three independent replicates for clones 27, 36 and two independent replicates for clones 37, 39, 74). CHX – cycloheximide. Uncropped Western blot images and source data for panels a-c are available with the paper online.
Discussion
In summary, we describe a simple and effective method for the rapid site-specific incorporation of a stable O-GlcNAc analogue in vitro and in cells by harnessing the unexpected cysteine S-GlcNAc transferase activity of OGT in combination with genome editing methods. We demonstrated that S-GlcNAc is a bona fide O-GlcNAc mimic by determining the crystal structure of a synthetic S-GlcNAcylated hOGA-derived peptide in complex with a model O-GlcNAc “reader” protein. We also showed that, despite micromolar affinity, S-GlcNAcylation is stable against OGA hydrolysis in the context of the hOGA- and α-synuclein-derived peptides. By mutating the corresponding O-GlcNAc site to a cysteine, we validated the OGT-mediated S-GlcNAcylation approach on TAB1 and CK2 in vitro and for the former – in cells. Utilizing CRISPR-Cas9 technology, we then directed OGT activity to the single specific site on OGA via genetic encoding of a S405C mutation in mESCs and demonstrated quantitative S-GlcNAcylation of OGA, while preserving its hydrolase activity. Intriguingly, this technique allowed us to provide the first insights into the importance of O-GlcNAc removal from Ser405 of OGA for the maintenance of its stability, suggesting the presence of a feedback regulation mechanism through as yet unknown additional factors. We anticipate that site-specific incorporation of S-GlcNAc would be useful for studying O-GlcNAc modifications that possess low stoichiometry due to the particularly rapid OGA-mediated hydrolysis. The S-GlcNAc genetic recoding method is likely applicable to proteins with multiple O-GlcNAc sites. By sequential mutagenesis of these sites to cysteines, it will be possible to dissect the roles of individual modifications. The added advantage of this method in vitro, is the ability to remove the hydrolysis-prone O-GlcNAc modifications through the action of OGA (CpOGA), leaving only S-GlcNAc modification on sites of interest intact. In addition, we show that some of the detection tools available for O-GlcNAc modification (such as O-GlcNAc-specific antibodies and galactosyltransferase labelling) can also be applied to S-GlcNAc. The key advantage of our approach in cell studies is direct targeting of the residue of interest (a single atom substitution in case of Ser to Cys mutation) without the need to express any artificial enzymatic machinery or treat cells with chemical compounds. Cys mutagenesis can be easily performed by common molecular biology techniques to study the modification in vitro and in an overexpression system. Moreover, application of CRISPR-Cas9 gene editing technology now allows functional dissection of individual O-GlcNAc sites on endogenous proteins in cells as well as model animals.
Materials & Methods
Glycopeptide synthesis
Peptides VAHS(O-GlcNAc)GA, VAHC(S-GlcNAc)GA, GAVVT(O-GlcNAc)GVTA and GAVVC(S-GlcNAc)GVTA were synthesized as described previously25, using 3,4,6-triacetyl-O-GlcNAc-Fmoc-Ser-OH and 3,4,6-triacetyl-S-GlcNAc-Fmoc-Cys-OH as building blocks.
Synthesis of Cys-S-GlcNAc building block
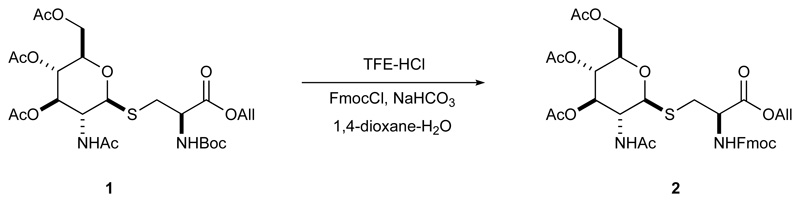
Details of instrumentation and analytical procedures were reported elsewhere40. Fmoc-GlcNAc-S-Cys-OAll was prepared from Boc-GlcNAc-S-Cys-OAll41.
To a stirred solution of 1 (1.98 g, 3.35 mmol) in trifluoroethanol (TFE; 20 mL) concentrated HCl (1.5 mL) was added at RT. The reaction mixture was further stirred for 45 min; tlc [PE–DCM]–Me2CO 30% showed complete consumption of the starting material and formation of a more polar product. The reaction was diluted with CHCl3 and toluene, concentrated, and briefly dried in vacuum. The residue was dissolved in 1,4-dioxan-water 3:1 mixture (40 mL) and treated sequentially with solid NaHCO3 (0.76 g, 9 mmol) and FmocCl (1 g, 4 mmol). The clear solution with some solid deposit was stirred at RT for 1 h; tlc [PE˗DCM]˗Me2CO 40% showed formation of a less polar new product. The residue was purified by flash-column chromatography [PE−DCM 4:1]−Me2CO 10→40% to give 2.35 g (3.33 mmol, quant) of the target product as amorphous solid.
1H NMR (500 MHz, DMSO-d6) δ 7.94 (d, J = 9.4 Hz, 1H), 7.90 (d, J = 7.6 Hz, 2H), 7.75 (d, J = 8.1 Hz, 1H), 7.72 (d, J = 7.5 Hz, 2H), 7.43 (t, J = 7.4 Hz, 2H), 7.37 – 7.31 (m, 2H), 5.91 (ddt, J = 17.2, 10.5, 5.2 Hz, 1H), 5.34 (dq, J = 17.3, 1.7 Hz, 1H), 5.21 (dq, J = 10.5, 1.6 Hz, 1H), 5.10 (t, J = 9.8 Hz, 1H), 4.88 (t, J = 9.7 Hz, 1H), 4.78 (d, J = 10.4 Hz, 1H), 4.61 (dt, J = 5.1, 1.6 Hz, 2H), 4.38 (td, J = 8.8, 4.9 Hz, 1H), 4.35 – 4.28 (m, 2H), 4.25 (t, J = 6.9 Hz, 1H), 4.14 (dd, J = 12.3, 5.1 Hz, 1H), 4.03 (dd, J = 12.2, 2.4 Hz, 1H), 3.92 (q, J = 10.3 Hz, 1H), 3.86 (ddd, J = 10.1, 5.0, 2.5 Hz, 1H), 3.15 (dd, J = 13.8, 4.8 Hz, 1H), 2.85 (dd, J = 13.8, 9.4 Hz, 1H), 1.99 (s, 3H), 1.99 (s, 3H), 1.93 (s, 3H), 1.76 (s, 3H) (Supplementary Fig. 5). m/z (ESI-TOF) found: 713.2344 expected for C35H40N2O12S (M+H+), 713.2380
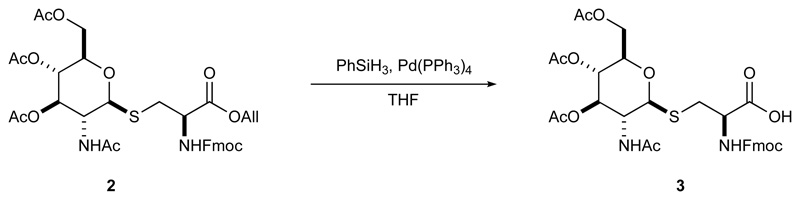
To a cold (ice-bath) solution of 2 (0.355 g, 0.5 mmol) in THF (2.5 mL, 0.2 M) phenyl silane (PhSiH3; 0.092 mL, 0.75 mmol) was added followed by tetrakistriphenylphosphine palladium (Pd(PPh3)4; 0.007 g, 0.00625 mmol). The reaction was removed from the cooling bath and stirred for 20 min; tlc [PE‒DCM 4:1]‒EA 30% revealed the reaction was complete. The reaction was concentrated. The crude acid 3 was dried in vacuum and used in the peptide synthesis without purification.
Analysis of O- and S-GlcNAc hydrolysis
hOGA- and α-synuclein-derived O- and S-GlcNAc peptides were diluted to 0.5 mM in 50 μl TBS buffer. To initiate the reaction, CpOGA was added to a final concentration of 0.3 μM. Hydrolysis reactions were performed at 37 °C for 4 or 24 h. Samples not treated with CpOGA were used as negative controls. To separate the peptides from CpOGA, all samples were passed through a 10 kDa molecular weight cut-off spin column. For MALDI-TOF MS analysis, samples were diluted in 0.1% TFA and 2 pmol was used for loading. 2,5 dihydroxy benzoic acid was used as a matrix. The samples were run in a rapifleX MALDI-TOF MS system.
Fluorescence polarization assay
Experiments were performed in black 384-well plates as described previously23. Reaction mixtures contained (added in order): O- or S-GlcNAc-modified peptide derived from hOGA (VAHS/C(GlcNAc)GA) or α-synuclein (GAVVT/C(GlcNAc)GVTA) at varying concentrations, 16 nM CpOGAD298N inactive mutant and 5 nM fluorescent probe (GlcNAcstatin-B-FITC) in TBS buffer pH 7.5 containing 1% DMSO. Fluorescence polarization (in millipolarization units) was measured using a Pherastar FS plate reader at excitation and emission wavelengths of 485 nm and 530 nm, respectively. Background subtraction was performed using reactions that did not contain CpOGAD298N and data were fitted with GraphPad Prism as described previously23. Assays using O-GlcNAcylated peptides were performed with three technical replicates in three independent experiments. Assays using S-GlcNAcylated peptides were performed with a single technical replicate in three (α-synuclein peptide) or four (hOGA peptide) independent experiments.
Crystallography
CpOGAD298N was purified and crystallized as previously described25. Soaking with S-modified peptide was performed by transferring crystals into a 0.5 μl drop of fresh crystallization buffer containing 10 mM of the peptide for 4 h at 20 °C. The crystals were mounted in nylon loops, cryo-protected in 0.175 M CdSO4, 0.6 M NaAc buffer containing 20% glycerol and flash-frozen in liquid nitrogen prior to data collection. Diffraction datasets were collected at the ID29 beam line (wavelength 0.9686 Å) of the European Synchrotron Radiation Facility (ESRF, Grenoble, France). Datasets were indexed and integrated with XDS42 and further reduced and scaled with Aimless. The CpOGA structure in complex with an S-GlcNAc hOGA peptide was solved by molecular replacement using published CpOGA-(O-GlcNAc hOGA peptide) complex structure (PDB: 2YDQ25) without the ligand as a phase donor in MOLREP43. The resulting model was submitted to several cycles of refinement with REFMAC544 followed by manual modelling with COOT45. Statistics of the data collection and model refinement are summarized in Table 1. The Ramachandran statistics are as follows: 96.55% favored, 3.45% additionally allowed. Figures of the structures were generated with Pymol46.
Cloning
The gene coding for full length TAB1 was cloned as a BamHI-NotI fragment into the pCMV-FLAG vector (obtained from DSTT, School of Life Sciences, Dundee, UK) for expression of N-terminal FLAG tag. The products were gel extracted and digested with BamHI and NotI restriction enzyme. The products were gel extracted and digested before being ligated into pCMV-FLAG cut with BamHI and NotI. The inserts were confirmed by DNA sequencing. Wild type pGEX6P1 TAB1 7-409 construct was produced as described previously28. The S395C mutation was introduced by site-directed mutagenesis based on the QuikChange protocol by Stratagene but using KOD Polymerase. All inserts were confirmed by DNA sequencing. Primers used for cloning and sequencing are listed in Supplementary Table 1.
In vitro OGT reactions and OGT-CK2 linear fusion
In vitro OGT reaction (100 μl) contained 10 μM TAB1 (7-409 construct), 50 nM full length human OGT in TBS buffer pH 7.5 with 0.1 mg/ml bovine serum albumin (BSA). The reaction was initiated by addition of UDP-GlcNAc to a final concentration of 100 μM. Reactions were performed at 25 °C. After incubation with OGT, the reactions were treated with 3 μM CpOGA for 2 h at 37 °C. 5 μl of the reaction was taken at different time points, run on the SDS PAGE gel and analyzed by Western blotting using O-GlcNAc TAB1 and total TAB1 antibodies. The OGT-CK2 peptide linear fusion was produced as described previously28.
Mass spectrometry
TAB1S395C OGT in vitro reaction was run on SDS-PAGE, the corresponding TAB1 band was excised and processed by in-gel digestion. The gel slice was washed with water, shrunk with 100 μl of acetonitrile (ACN) for 5 min at room temperature and reswollen with 50 μl of 50 mM Tris HCl pH 8.0 twice. Reduction and alkylation were performed in gel using 50 μl of 5 mM DTT in 50 mM Tris HCl pH 8.0 (shaken for 20 min at 65 °C) and 50 μl of 50 mM iodoacetamide in 50 mM Tris HCl pH 8.0 (shaken for 20 min at room temperature). The gel slice was shrunk using 500 μl ACN for 5 min at room temperature and 50 μl of 50 mM triethylammonium bicarbonate was added to reswell the gel slice. 50-100 μl of mass spectrometry grade trypsin in 50 mM triethylammonium bicarbonate, containing 5 μg/ml of trypsin protease (in 50 mM acetic acid) was added and the sample was shaken at 30 °C overnight. 100 μl of ACN was added to completely shrink the gel. The supernatant was transferred to a fresh tube. The gel piece was re-swollen with 50 μl of 0.1% trifluoroacetic acid (TFA). Digest was extracted twice with 100 μl of ACN. All extracts were combined, lyophilized in a speed-vac and stored at -20 °C. Mass spectrometric analysis was performed by LC-MS/MS on an Orbitrap Fusion tribrid mass spectrometer (Thermo Scientific) coupled to a U3000 RSLC HPLC (Thermo Scientific). Peptides were trapped on a nanoViper Trap column, 2 cm x 100 μm, C18 5 μm 100 Å (Thermo Scientific, 164564) and then separated on a 50 cm EASY-Spray column (Thermo Scientific, ES803) equilibrated with a flow of 300 nl/min of 3% Solvent B (Solvent A: 2% Acetonitrile, 0.1% formic acid; Solvent B: 80% acetonitrile, 0.08% formic acid). The elution gradient was as follows: time (min): solvent B (%); 0:3, 5:3, 55:25, 74:40, 79:99, 80:3, 90:3. The instrument was operated with the internal mass calibrant (EASY IC) option to improve the mass accuracy of precursor ions and data were acquired in the data-dependent mode. MS1 spectra (m/z 400-1600) were acquired in the Orbitrap with resolution 120,000. The method used was a ‘top speed’ method. The precursors were isolated using quadrupole with an isolation width of 1.6 Da. The activation type was HCD (Higher-energy Collisional Dissociation) with collision energy at 30% and the fragments were detected in ion trap mass analyzer. The AGC (Automatic Gain Control) target set was 100 and the maximum injection time was set at 250 ms. If an ion in this analysis shows the presence of certain ions, in this case any of HexNAc (204.1), HexNAc fragment (138.1) or HexNAcHex (366.1) then EThcD/Orbitrap analysis of the parent ion was triggered. The ions were isolated using quadrupole and fragmentation method used was ETD (Electron Transfer Dissociation) with HCD supplemental activation (EThcD). The Orbitrap resolution for this step was set at 30000. The AGC target was 30000 and the maximum injection time was 150 ms. Data files were analyzed by Proteome Discoverer 2.0 (Thermo Scientific), using Mascot 2.4.1 (www.matrixscience.com), and searching an in-house database containing the relevant sequences. Scaffold (www.ProteomeSoftware.com) was also used to examine the Mascot result files. Allowance was made for the following variable modifications Oxidation (M), HexNAc (S), HexNAc (C). Error tolerances were 10 ppm for MS1, 0.6 Da for HCD MS2 and 20 mmu for EThcD/Orbitrap spectra
Cell culture and transfection
IL-1R Hek293 cells27 were grown in DMEM medium supplemented with 10% FBS, 2 mM L-glutamine and 1% penicillin/streptomycin (100 U/ml and 100 μg/ml respectively) at 37 °C and 5% CO2. Hek293 cells were transfected with FLAG-tagged TAB1 using Lipofectamine 3000 reagent (Invitrogen) at a ratio 1:2 (μg:μl) according to manufacturer’s instructions. Male mouse embryonic stem cells (AW2 line from MRC Centre for Regenerative Medicine, Institute for Stem Cell Research, University of Edinburgh) were cultured in 0.1% (w/v) gelatin-coated plates in GMEM BHK-21 medium supplemented with 10% fetal bovine serum, 50 μM β-mercaptoethanol, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate and 100 U/ml leukemia inhibitory factor at 37 °C and 5% CO2. Cell lines tested negative for mycoplasma. GlcNAcstatin G treatment was performed at a final concentration of 1 μM for 24 h. 4Ac5S-GlcNAc treatment was performed at a final concentration of 200 μM for 24 h.
Generation of OGA S405C CRISPR-Cas9 knock-in mouse embryonic stem cells
Paired gRNAs were selected using the website http://www.sanger.ac.uk/htgt/wge/find_crisprs. Annealing oligonucleotides were designed with the appropriate overhangs for cloning into BpiI cut pX335 (Cas9 D10A) and pBABED puro U6. A 3 μM mix of each annealing oligonucleotide pair were combined in a 100 μl volume and treated with 1 μl polynucleotide kinase (Fermentas) in T4 Ligase buffer at 37 °C for 20 min followed by 10 min incubation at 75 °C and finally placed in a heating block at 95 °C. The metal block was removed from the heat source and allowed to cool gradually to room temperature. 1 μl of a 1/30 dilution of this mixture was added to a ligation reaction containing 20 ng BpiI cut, dephosphorylated destination vector and 1 μl DNA ligase (Fermentas) in T4 ligase buffer in a 20 μl final volume. After 25 min at RT a 1 μl aliquot of the reaction was used to transform DH5-alpha competent cells. Inserts were confirmed by DNA sequencing. A 2 kb PCR product was obtained from 46c mouse genomic DNA and cloned as a BamHI-NotI fragment into pGEX6P1. The insert was confirmed to be wild type by DNA sequencing. A 249 bp geneblock containing the desired mutation and silent mutations to eliminate the gRNA recognition sites was ordered from Integrated DNA Technologies. The geneblock was amplified by PCR, the product was gel extracted and introduced into the middle of the 2 kb insert by restrictionless cloning47. The changes were confirmed by DNA sequencing.
Male mouse embryonic stem cells were co-transfected with pBABED, pX335 and the repair template plasmids using Lipofectamine 3000 (Invitrogen) according to manufacturer’s instructions. 24 h post transfection, G-MEM medium (supplemented with 10% fetal bovine serum, β-mercapthoethanol, non-essential amino acids, sodium pyruvate and leukemia inhibitory factor) was substituted with fresh medium containing 1 μg/ml puromycin. Cells that survived after 24 h of puromycin treatment were dissociated with accutase, and plated onto several 10 cm plates. Colonies were grown for 1-2 weeks and then screened by restriction digestion of a 622 bp PCR product (covering the mutation site) with DraI enzyme. Wild type restriction digestion produced two 231 and 391 bp bands on a 1.3% agarose gel. The S405C mutant produced a single 622 bp band, which was verified by sequencing (Supplementary Fig. 1 and 2). To eliminate the possibility of a false positive due to random integration of the repair template into the genome and to detect the presence of hemizygous genomic deletions, a larger PCR product was obtained. This PCR product encompassed an area outside the repair template and sequencing confirmed the change in the correct location (Supplementary Fig. 3). This confirmed the desired homologous recombination had taken place. Primers used for knock-in generation and sequencing are listed in Supplementary Table 1.
In addition to DNA based analysis of mutated cell lines, RNA was extracted from candidate clones containing the S405C mutation. This RNA was subjected to one-step RT-PCR (using Takara Primescript High Fidelity RT-PCR Kit) with primers specific to the next exon outside the area covered by the repair template in both directions. This resulted in a 951 bp PCR product. Sequencing of the resultant product from both ends confirmed the precision of the alteration of the mRNA expressed in these cells (Supplementary Fig. 4). Primers and geneblock sequence are listed in Supplementary Table 1.
Western blotting
Cells were scraped in lysis buffer (Cell Signaling Technology, #9803) supplemented with 1 mM PMSF, sonicated and lysates were spun down at 17000 g for 10 min. Supernatants were transferred into fresh tubes and protein concentration was quantified using Bradford assay. Protein samples were prepared using LDS buffer containing 5% β-mercaptoethanol. Samples were run on a 10% SDS PAGE gel. Proteins were then transferred onto nitrocellulose membrane at 100 V for 1 h. Membranes were stained with Ponceau-S to check for successful transfer, washed with 0.2% TBS-Tween and blocked in 5% BSA in 0.2% TBS-Tween. Primary antibodies are listed in Supplementary Table 2. Fluorescence signal from secondary antibodies (Li-Cor) was quantified using Image Studio software and statistical analysis was performed using Prism (GraphPad).
O- and S-GlcNAc PEGylation labelling using Gal-TY289L
The Click-iT™ O-GlcNAc enzymatic labelling system was used according to manufacturer’s instructions (Thermo Fisher Scientific) with some modifications. 50-100 μg of whole cell lysate proteins or 20 μg of in vitro GlcNAcylated TAB1 were subjected to Gal-TY289L labelling. The Gal-TY289L reaction components were added in the following order to 20 μl of chloroform/methanol precipitated proteins (in 20 mM HEPES, pH 7.9): 24.5 μl MiliQ water, 40 μl labelling buffer (component C), 5.5 μl MnCl2 (100 mM, component D) and vortexed briefly. 10 μl 0.5 mM UDP-GalNAz was added, mixed by pipetting and half of the reaction (50 μl) was removed as an unlabeled negative control. Finally, 4 μl Gal-TY289L was added to the reaction and mixed by pipetting. Both the reaction and the negative control were incubated at 4 °C overnight.
PEGylation of GalNAz-labelled proteins was performed using the Click-iT® Protein Reaction Buffer Kit according to manufacturer’s instructions (Thermo Fisher Scientific) with some modifications. MeO-PEG-alkyne (Mw = 5000 Da, IrisBiotech, PEG2830.0500) was prepared beforehand as a 10 mM solution in water and stored at -20 °C. Proteins (20 μl in 1% SDS, 50 mM Tris HCl, pH 8.0) prepared by GalNAz labelling were mixed with 40 μl Click-iT ® reaction buffer containing 3.3 mM PEG-alkyne (i.e. 100 μl of PEG-alkyne stock solution was mixed with 200 μl of Click-iT® reaction buffer, component A). 4 μl of CuSO4 was added, followed by 8 μl of Click-iT® reaction buffer additive 2 solution. The reaction was then incubated at room temperature with 1400 rpm shaking for 1-1.5 h. The samples were prepared in LDS buffer (containing 5% β-mercaptoethanol) and analyzed by Western blotting.
Cycloheximide treatment
mES cells were treated with cycloheximide (Cayman Chemical, dissolved in DMSO) at a final concentration of 20 μg/ml for different time points before lysis. DMSO treatment was used as a negative control.
OGA thermal stability assay in cell lysates
mES cells were lysed as described above and subjected to a range of temperatures in a thermomixer. After 5 min of incubation at each temperature, the lysates were centrifuged and soluble fraction was removed for subsequent Western blot analysis.
Extended Data
Extended Data Figure 1.
Extended Data Figure 2.
Extended Data Figure 3.
Extended Data Figure 4.
Extended Data Figure 5.
Extended Data Figure 6.
Extended Data Figure 7.
Supplementary Material
Acknowledgements
This work was funded by a Wellcome Trust Investigator Award (110061) to DvA and a Wellcome Trust 4-year PhD studentship (105310/Z/14/Z) to AG. We thank Robert Gourlay and David Campbell (MRC PPU) for assistance with mass spectrometry. We thank Olawale G. Raimi for purifying OGT-CK2 linear fusion proteins. We would like to thank ESRF (beamline ID29) for the synchrotron time.
Footnotes
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
Atomic coordinates and structure factors for the CpOGA–(S-GlcNAc peptide) complex structure are deposited in the Protein Data Bank under the accession code PDB 6RHE. Source data for Fig. 1c, 2c, 3a, 3b, 3c and Extended Data Fig. 1b, 6, 7 are available with the paper online. All other data are available upon reasonable request to the corresponding author.
Author contributions
AG and DvA conceived the study; AG, SGB, VSB, JV, ATF performed experiments and analyzed data, AG and DvA wrote the manuscript with input from all authors.
Competing interests
The University of Dundee holds a patent for the GlcNAcstatin inhibitor. The authors declare no other competing interests.
References
- 1.Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18:452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haltiwanger RS, Holt GD, Hart GW. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins: Identification of a uridine diphospho-N-acetylglucosamine: peptide beta-N-acetylglucosaminyltransferase. J Biol Chem. 1990 [PubMed] [Google Scholar]
- 3.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001 doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- 4.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingham PW. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell. 1984;37:815–823. doi: 10.1016/0092-8674(84)90416-1. [DOI] [PubMed] [Google Scholar]
- 6.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013;10:365–380. doi: 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuzwa SA, Vocadlo DJ. O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer’s disease and beyond. Chem Soc Rev. 2014;43:6839–6858. doi: 10.1039/c4cs00038b. [DOI] [PubMed] [Google Scholar]
- 9.Zachara NE. Critical observations that shaped our understanding of the function(s) of intracellular glycosylation (O-GlcNAc) FEBS Letters. 2018 doi: 10.1002/1873-3468.13286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwagerus S, Reimann O, Despres C, Smet-Nocca C, Hackenberger CP. Semi-synthesis of a tag-free O-GlcNAcylated tau protein by sequential chemoselective ligation. J Pept Sci. 2016;22:327–333. doi: 10.1002/psc.2870. [DOI] [PubMed] [Google Scholar]
- 11.Marotta NP, et al. O-GlcNAc modification blocks the aggregation and toxicity of the protein alpha-synuclein associated with Parkinson’s disease. Nat Chem. 2015;7:913–920. doi: 10.1038/nchem.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright TH, et al. Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science (80-. ) 2016 doi: 10.1126/science.aag1465. [DOI] [PubMed] [Google Scholar]
- 13.Khidekel N, et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 14.Whisenhunt TR, et al. Disrupting the enzyme complex regulating O-GlcNAcylation blocks signaling and development. Glycobiology. 2006 doi: 10.1093/glycob/cwj096. [DOI] [PubMed] [Google Scholar]
- 15.Teo CF, Wells L. Monitoring protein O-linked β-N-acetylglucosamine status via metabolic labeling and copper-free click chemistry. Anal Biochem. 2014 doi: 10.1016/j.ab.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rexach JE, et al. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol. 2010 doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao FV, et al. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25:1569–1578. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tarrant MK, et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat Chem Biol. 2012;8:262–269. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raj R, Lercher L, Mohammed S, Davis BG. Synthetic Nucleosomes Reveal that GlcNAcylation Modulates Direct Interaction with the FACT Complex. Angew Chem Int Ed Engl. 2016;55:8918–8922. doi: 10.1002/anie.201603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Leon CA, Levine PM, Craven TW, Pratt MR. The Sulfur-Linked Analogue of O-GlcNAc (S-GlcNAc) Is an Enzymatically Stable and Reasonable Structural Surrogate for O-GlcNAc at the Peptide and Protein Levels. Biochemistry. 2017 doi: 10.1021/acs.biochem.7b00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tegl G, et al. Facile Formation of β-thioGlcNAc Linkages to Thiol-Containing Sugars, Peptides, and Proteins using a Mutant GH20 Hexosaminidase. Angew Chemie - Int Ed. 2019;58:1632–1637. doi: 10.1002/anie.201809928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toleman CA, et al. Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins. Proc Natl Acad Sci U S A. 2018 doi: 10.1073/pnas.1722437115. 201722437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borodkin VS, et al. O-GlcNAcase Fragment Discovery with Fluorescence Polarimetry. ACS Chem Biol. 2018 doi: 10.1021/acschembio.8b00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selvan N, et al. A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat Chem Biol. 2017;13:882–887. doi: 10.1038/nchembio.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schimpl M, Borodkin VS, Gray LJ, van Aalten DM. Synergy of peptide and sugar in O-GlcNAcase substrate recognition. Chem Biol. 2012;19:173–178. doi: 10.1016/j.chembiol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maynard JC, Burlingame AL, Medzihradszky KF. Cysteine S-linked N-acetylglucosamine (S-GlcNAcylation), A New Post-translational Modification in Mammals. Mol Cell Proteomics. 2016;15:3405–3411. doi: 10.1074/mcp.M116.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathak S, et al. O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. EMBO J. 2012;31:1394–1404. doi: 10.1038/emboj.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafie K, et al. Recognition of a glycosylation substrate by the O-GlcNAc transferase TPR repeats. Open Biol. 2017;7 doi: 10.1098/rsob.170078. 170078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawa M, et al. GTDC2 modifies O-mannosylated α-dystroglycan in the endoplasmic reticulum to generate N-acetyl glucosamine epitopes reactive with CTD110.6 antibody. Biochem Biophys Res Commun. 2013;440:88–93. doi: 10.1016/j.bbrc.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Isono T. O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS One. 2011 doi: 10.1371/journal.pone.0018959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tashima Y, Stanley P. Antibodies that detect O-linked beta-D-N-acetylglucosamine on the extracellular domain of cell surface glycoproteins. J Biol Chem. 2014;289:11132–11142. doi: 10.1074/jbc.M113.492512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves RA, Lee A, Henry R, Zachara NE. Characterization of the specificity of O-GlcNAc reactive antibodies under conditions of starvation and stress. Anal Biochem. 2014;457:8–18. doi: 10.1016/j.ab.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorfmueller HC, et al. Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases. Chem Biol. 2010;17:1250–1255. doi: 10.1016/j.chembiol.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gloster TM, et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011 doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramakrishnan B, Qasba PK. Structure-based Design of β1,4-Galactosyltransferase I (β4Gal-T1) with Equally Efficient N -Acetylgalactosaminyltransferase Activity. J Biol Chem. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- 36.Boeggeman E, et al. Direct identification of nonreducing GlcNAc residues on N-glycans of glycoproteins using a novel chemoenzymatic method. Bioconjug Chem. 2007;18:806–814. doi: 10.1021/bc060341n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gambetta MC, Müller J. O-GlcNAcylation Prevents Aggregation of the Polycomb Group Repressor Polyhomeotic. Dev Cell. 2014;31:629–639. doi: 10.1016/j.devcel.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 38.Yuzwa SA, et al. Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat Chem Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 39.Chu C-S, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci. 2014;111:1355–1360. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rafie K, Gorelik A, Trapannone R, Borodkin VS, Aalten DMF. Thio-linked UDP-peptide conjugates as O-GlcNAc transferase inhibitors. Bioconjugate Chemistry. 2018 doi: 10.1021/acs.bioconjchem.8b00194>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu X, Pachamuthu K, Schmidt RR. Synthesis of S-linked glycopeptides in aqueous solution. J Org Chem. 2003 doi: 10.1021/jo034148n. [DOI] [PubMed] [Google Scholar]
- 42.Kabsch W. XDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vagin A, Teplyakov A, IUCr Molecular replacement with MOLREP. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D-Biological Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Delano WL, Bromberg S. PyMOL User’s. Guide DeLano Scientific LLC; 2004. [Google Scholar]
- 47.Van Den Ent F, Löwe J. RF cloning: A restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods. 2006 doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.