Abstract
Deciphering the massive volume of complex electronic data that has been compiled by hospital systems over the past decades has the potential to revolutionize modern medicine, as well as present significant challenges. Deep learning is uniquely suited to address these challenges, and recent advances in techniques and hardware have poised the field of medical machine learning for transformational growth. The clinical neurosciences are particularly well positioned to benefit from these advances given the subtle presentation of symptoms typical of neurologic disease. Here we review the various domains in which deep learning algorithms have already provided impetus for change—areas such as medical image analysis for the improved diagnosis of Alzheimer’s disease and the early detection of acute neurologic events; medical image segmentation for quantitative evaluation of neuroanatomy and vasculature; connectome mapping for the diagnosis of Alzheimer’s, autism spectrum disorder, and attention deficit hyperactivity disorder; and mining of microscopic electroencephalogram signals and granular genetic signatures. We additionally note important challenges in the integration of deep learning tools in the clinical setting and discuss the barriers to tackling the challenges that currently exist.
Keywords: Artificial intelligence, Biomedical informatics, Computer vision, Connectome mapping, Deep learning, Genomics, Machine learning, Neurology, Neuroscience
Introduction
Twenty-first century healthcare is marked by an abundance of biomedical data and the development of high-performance computing tools capable of analyzing these data. The availability of data and increased speed and power of computer systems together present both opportunities and challenges to researchers and healthcare professionals. Most significantly, they provide the potential to discover new disease correlates and translate these insights into new data-driven medical tools that can improve the quality and delivery of care. However, such advancements require the navigation of high-dimensional, unstructured, sparse, and often incomplete data sources, with the outcomes being cumbersome to track. Identifying novel clinical patterns amidst this complexity is definitely not a trivial task [1–3].
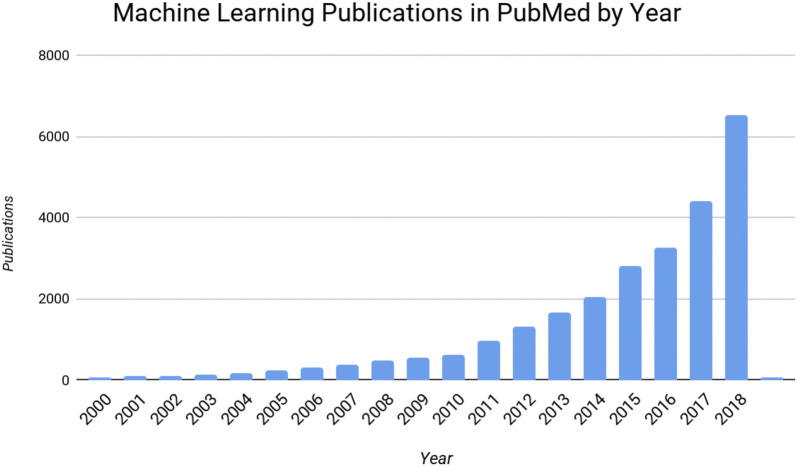
Modern representation learning methods enable the automatic discovery of representations needed to generate insights from raw data [4]. Deep learning algorithms are an example of such representation learning approaches that hierarchically compose nonlinear functions to transform raw input data into more sophisticated features that enable the identification of novel patterns [5]. Such approaches have proved to be essential in modern engineering breakthroughs—from face recognition and self-driving cars to chat-bots and language translation [6–12]. In medicine, the successful application of deep learning algorithms to routine tasks has enabled a flood of academic and commercial research, with publications on various applications growing from 125 published papers identified as machine learning publications in arXiv, the electronic scientific and engineering paper archive, in 2000, to more than 3600 by November of 2018 (see Fig. 1).
Fig. 1.
Machine learning publications in PubMed by year through 2018 showing the exponential growth of interest in the field, as reported by the US National Library of Medicine of the National Institutes of Health [13]
The multidiscipline of clinical neurosciences has similarly experienced the beginnings of an impact from deep learning, with movement towards the development of novel diagnostic and prognostic tools. Deep learning techniques are particularly promising in the neurosciences where clinical diagnoses often rely on subtle symptoms and complicated neuroimaging modalities with granular and high-dimensional signals. In this article, we discuss the applications of deep learning in neurology and the ongoing challenges, with an emphasis on aspects relevant to the diagnosis of common neurologic disorders. However, our aim is not to provide comprehensive technical details of deep learning or its broader applications. We begin with a brief overview of deep learning techniques followed by a review of applications in the clinical neuroscience field. We conclude the review with a short discussion on existing challenges and a look to the future. This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Fundamentals of Deep Learning
Machine learning is a subset of artificial intelligence that learns complex relationships among variables in data [14]. The power of machine learning comes from its ability to derive predictive models from large amounts of data with minimal or, in some cases, entirely without the need for prior knowledge of the data or any assumptions about the data. One of the most widely discussed modern machine learning algorithms, the artificial neural network (ANN), draws inspiration from biological neural networks that constitute mammalian brains. The functional unit of the ANN is the perceptron, which partitions input data into separable categories or classes [15]. When hierarchically composed into a network, the perceptron becomes an essential building block for modern deep neural networks (DNNs), such as multilayer perceptron classifiers. Similar examples of commonly used traditional machine learning algorithms include linear regression (LR), logistic regression, support vector machines (SVMs), and the Naïve Bayes classifier (Fig. 2).
Fig. 2.
Breakdown of algorithm types in the machine learning family that are commonly used in medical subdomain research and analyses
These traditional machine learning methods have been important in furthering advancements in medicine and genomics. As an example, LR has proven useful in the search for complex, multigene signatures that can be indicative of disease onset and prognosis, tasks which are otherwise too intricate and cumbersome even for researchers with professional training [16]. Although such tools have been very effective in parsing massive datasets and identifying relationships between variables of interest, traditional machine learning techniques often require manual feature engineering and suffer from overhead that limits their utility in scenarios that require near real-time decision-making.
Deep learning differs from traditional machine learning in how representations are automatically discovered from raw data. In contrast to ANNs, which are shallow feature learning techniques, deep learning algorithms employ multiple, deep layers of perceptrons that capture both low- and high-level representations of data, enabling them to learn richer abstractions of inputs [5]. This obviates the need for manual engineering of features and allows deep learning models to naturally uncover previously unknown patterns and generalize better to novel data. Variants of these algorithms have been employed across numerous domains in engineering and medicine.
Convolutional neural networks (CNNs) have garnered particular attention within computer vision and imaging-based medical research [17, 18]. CNNs gather representations across multiple layers, each of which learns specific features of the image, much like the human visual cortex is arranged into hierarchical layers, including the primary visual cortex (edge detection), secondary visual cortex (shape detection), and so forth [19]. CNNs consist of convolutional layers in which data features are learned: pooling layers, which reduce the number of features, and therefore computational demand, by aggregating similar or redundant features; dropout layers, which selectively turn off perceptrons to avoid over-reliance on a single component of the network; and a final output layer, which collates the learned features into a score or class decision, i.e., whether or not a given radiograph shows signs of ischemia. These algorithms have achieved rapid profound success in image classification tasks and, in some cases, have matched board-certified human performance [20–24].
Recurrent neural networks and variants, such as long short-term memory (LSTM) and gated recurrent units, have revolutionized the analysis of time-series data that can be found in videos, speech, and texts [25]. These algorithms sequentially analyze each element of input data and employ a gating mechanism to determine whether to maintain or discard information from prior elements when generating outputs. In this manner, they efficiently capture long-term dependencies and have revolutionized machine translation, speech processing, and text analysis.
Autoencoders (AEs) are a class of unsupervised learning algorithms that discover meaningful representations of data by learning a lower-dimensional mapping from inputs to outputs [26, 27]. They are composed of an encoder, which learns a latent representation of the input, and a decoder, which reconstructs the input from the latent representation. By constraining the latent representation to a lower dimensionality than the input, AEs are able to learn a compressed representation of data that contains only the features necessary to reconstruct the input. Such algorithms are often employed to learn features that can be subsequently utilized in conjunction with the deep learning techniques previously discussed.
Generative adversarial networks are a newer class of algorithms aimed at generating novel data that statistically mimic input data by approximating a latent distribution for the data [28]. Such algorithms are composed of two competing (“adversarial”) networks: a generator, which produces synthetic data from noise by sampling from an approximated distribution, and a discriminator, which aims to differentiate between real and synthetic instances of data. As the two networks engage in this adversarial process, the fidelity of the generated data gradually improves. In some contexts, the resulting data have been utilized to augment existing datasets [29].
These strides in deep learning are largely due to breakthroughs in computing capabilities and the open-source nature of research in the field. The application of graphics processing units to deep learning research has dramatically accelerated the size and complexity of algorithm architectures and simultaneously reduced the time to train such algorithms from months to the order of days. The consequence has been high-throughput research characterized by rapid experimentation, ultimately enabling more efficacious algorithms. In addition, the rise of open-source deep learning frameworks, such as TensorFlow, Keras, PyTorch, Caffe, and others, has increased accessibility to technical advances and facilitated the sharing of ideas and their rapid application across various domains [30, 31]. The truly collaborative nature of deep learning research has led to surprising innovations and changed the landscape of medical research and care.
Literature Review
In this article, we review and summarize published literature on the application of deep learning to the clinical neurosciences. We used search engines and repositories such as Google Scholar, PubMed, ScienceDirect, and arXiv to identify and review existing literature and performed keyword searches of these databases using the following terms: “deep learning,” “machine learning,” “neurology,” “brain,” and “MRI.” Following a comprehensive review of the literature initially retrieved, we identified 312 articles as containing one or more keywords associated with our queries. Of these articles, 134 were subsequently identified as being relevant to the subject of this review. Following collation of the relevant articles, we grouped articles first into broad modalities, namely image classification, image segmentation, functional connectivity and classification of brain disorders, and risk prognostication. Within these areas, we then grouped publications into disease applications. We focused our discussion on the clinical implications of the developments in the field.
Deep Learning in Neurology
The deep learning techniques described above are playing an increasingly crucial role in neurological research, tackling problems within several subdomains. First, radiological image classification and segmentation has been a traditional locus of deep learning development efforts. Image classification and segmentation tasks are uniquely suited to deep learning due to the high-dimensional nature of neuroimaging data which is unfavorable to manual analysis, combined with the naturally digital nature of most modern imaging. Secondly, deep learning has been applied to functional brain mapping and correlational studies using functional magnetic resonance imaging (fMRI) data for tasks such as prediction of postoperative seizure. Lastly, diagnostic prognostication with deep learning using multiple data types, including lab values, images, notes, among others, has been used to assign disease risk. In the following sections, we discuss the successes and challenges inherent in the deep learning approaches adopted towards these tasks, as well as the limitations and difficulties that such methods face within the field of neurology and within medicine as a whole.
Medical Image Classification
The first application of deep learning in medicine involved the analysis of imaging modalities, especially those for the detection of Alzheimer’s disease (AD) and other cognitive impairments. A variety of publicly available databases, such as the Alzheimer’s Disease Neuroimaging Initiative (ADNI) and Brain Tumor Segmentation Benchmark (BraTS), have become available to spur advancements in neuroimaging analysis [32, 33].
Early approaches used AEs in conjunction with a classifier to distinguish AD, mild cognitive impairments (MCI) and healthy controls. Among the first such applications, Suk and Shen utilized a stacked AE to learn multimodal brain representations from structural MRI and positron emission tomography (PET), and incorporated those features with cerebrospinal fluid biomarker data and clinical scores from the Mini-Mental State Examination (MMSE) and Alzheimer’s Disease Assessment Scale-Cognitive subscale (ADAS-Cog) to train an SVM classifier that improved diagnostic accuracy [34]. Other approaches pre-trained a stacked AE using natural images (everyday images) prior to training on brain MR images in order to learn more high-fidelity anatomical features, such as gray matter and structural deformities, for incorporation into a CNN [35]. Variations on these approaches have been used to incrementally improve diagnostic performance [36–42].
Whereas older approaches were limited to two-dimensional (2D) slices of medical images due to computational constraints, newer applications have been able to incorporate the full 3D volume of an imaging modality for AD detection. Among the first such examples was work by Payan and Montana in which they trained a sparse AE on 3D patches of MRI scans to learn a volumetric brain representation that was used to pre-train a 3D CNN for AD diagnosis [43]. More recently, Hosseini-Asl et al. used an adaptable training regime with a 3D CNN pre-trained by a convolutional AE to learn generalizable AD biomarkers [44, 45]. This approach was notable because it allowed the transfer of learned features from the source CADDementia dataset to the target ADNI dataset, resulting in state-of-the-art AD diagnosis accuracy on an external dataset. Analogous work with volumetric data has been conducted in the computed tomography (CT) domain to differentiate AD from brain lesions and the processes of normal aging [46].
The most recent work has built on existing work in AD diagnosis and focused on predicting the onset of AD in at-risk patients in order to stem progression of the disease. Ding et al. used fluorine-18-fluorodeoxyglucose PET scans of the brain derived from the ADNI database to train a CNN to diagnose AD [47]. Unlike many investigators before them, however, the authors evaluated the efficacy of their algorithm against data from the long-term follow-up of patients who did not have AD at the time. Interestingly, they found that the algorithm predicted onset of AD on average 75.8 months prior to the final diagnosis on an independent dataset, which surpassed the diagnostic performance of three expert radiologists.
Deep learning-based image classification has also been applied in the diagnosis of acute neurologic events, such as intracranial hemorrhage (ICH) and cranial fractures, with the aim of reducing time to diagnosis by optimizing neuroradiology workflows. Titano et al. trained a 3D CNN in a weakly supervised manner on 37,236 CT scans to identify ICH for the purposes of triaging patient cases [48]. They leveraged a natural language processing algorithm trained on 96,303 radiology reports to generate silver-standard labels for each imaging study and validated the efficacy of their CNN on a subset of studies with gold standard labels generated by manual chart review [49]. The investigators conducted a double-blind randomized control trial to compare whether the algorithm or expert radiologists could more effectively triage studies in a simulated clinical environment and found that the CNN was 150-fold faster in evaluating a study and significantly outperformed humans in prioritizing the most urgent cases. Subsequent studies have similarly demonstrated the potential for deep learning to optimize radiology workflows in the diagnosis of ICH and detect as many as nine critical findings on head CT scans with sensitivity comparable to that of expert radiologists [50–52].
Medical Image Segmentation
Segmentation of radiological brain images is critical for the measurement of brain regions, including shape, thickness, and volume, that are important for the quantification of structural changes within the brain that occur either naturally or due to various disease processes [53]. Accurate structural classification is particularly important in patients with gliomas, the most common brain tumor type, with less than a 2-year survival time [54, 55]. Manual segmentations by expert raters show considerable variation in images obscured by field artifacts or where intensity gradients are minimal, and rudimentary algorithms struggle to achieve consistency in an anatomy that can vary considerably from patient to patient [33]. In light of these factors, deep learning segmentation of neuroanatomy has become a prime target for efforts in deep learning research.
Measurement of the performance of neuroanatomical segmentation algorithms has been standardized by the BraTS, which was established at the 2012 and 2013 Medical Image Computing and Computer Assisted Interventions (MICCAI) conference [33]. Prior to the establishment of this challenge, segmentation algorithms were often evaluated on private imaging collections only, with variations in the imaging modalities incorporated and the metrics used to evaluate effectiveness. The establishment of BraTS has been critical in standardizing the evaluation of various models for the determination of which to pursue in clinical practice. At the time of BraTS establishment, the models being evaluated were largely simple machine learning models, including four random forest-based segmentation models [33]. Since then, there has been considerable advancement in performance, largely based on the adoption of CNNs for anatomical segmentation.
The traditional computational approach to segmentation is to employ an atlas-based segmentation, namely the FreeSurfer software, which assigns one of 37 labels to each voxel in a 3D MRI scan based on probabilistic estimates [56]. In a recent comparative study, Wachinger et al. designed and applied a deep CNN, called DeepNAT, for the purposes of segmenting neuroanatomy visualized in T1-weighted MRI scans into 25 different brain regions. The authors used the MICCAI Multi-Atlas Labeling challenge, consisting of 30 T1-weighted images, in addition to manually labeled segmentations [53, 57]. When the authors compared the current clinical standard, FreeSurfer, which uses its own anatomical atlas to assign anatomic labels, to DeepNAT, they found that DeepNAT showed statistically significant performance improvements. Performance in segmentation was measured using a Dice volume overlap score, with DeepNAT achieving a Dice score of 0.906, in comparison to FreeSurfer’s 0.817 [53].
In addition to tissue-based segmentation efforts, vascular segmentation has been an area of deep learning research aimed at quantifying brain vessel status. Traditional vessel segmentation relies on either manual identification or rule-based algorithms since there is no equivalent atlas-based method for brain vessels as there is for neuroanatomy. In their recent study on blood vessel segmentation, Livne et al. applied a U-net model to labeled data from 66 patients with cerebrovascular disease and then compared the method to the traditional vascular segmentation method of graph-cuts. The U-net model outperformed graph-cuts, achieving a Dice score of 0.891 compared to 0.760 for graph-cuts [58]. Of note, the model, which was trained on 3T MRI time-of-flight images, failed to generalize well to 7T images [58].
Quantification of changes in white matter as biomarkers for disease processes has been a third area of deep learning segmentation efforts in neurology. Perivascular spaces (PVSs) are small spaces surrounding blood vessels that can be caused by the stress-induced breakdown of the blood–brain barrier by various inflammatory processes [59, 60]. While PVSs have been implicated in a wide range of disease processes, the quantification of these spaces is difficult due to their tubular and low-contrast appearance even on those clinical MRI machines with the highest-approved resolution [61]. In one 2018 study, Lian et al. used a deep CNN to evaluate PVSs in 20 patients scanned on a 7T MRI machine, comparing these to gold-standard manual labels. Their deep CNN outperformed unsupervised algorithmic methods, such as a Frangi filter, as well as a U-net deep learning model, achieving a positive predictive value (PPV) of 0.83 ± 0.05, compared to a PPV of 0.62 ± 0.08 for the Frangi filter and 0.70 ± 0.10 for the U-net.
U-net models have also been leveraged in quantifying white matter hyperintensities as biomarkers for age-related neurologic disorders [62]. White matter changes have been shown to be involved in various forms of cortical dementia, such as AD, and manifest themselves as high-intensity regions in T2-fluid-attenuated inversion recovery (FLAIR) MRI scans [63]. In addition to quantifying PVSs, U-nets have been used in segmentation efforts to identify regions of abnormally intense white matter signals. In 2019, Jeong et al. proposed a sailiency U-net, a U-net combined with simple regional maps, with the aim to lower the computational demand of the architecture while maintaining performance in order to identify areas of signal intensity in T2-FLAIR MRI scans of patients with AD [62, 64]. Their model achieved a Dice coefficient score of 0.544 and a sensitivity of 0.459, indicating the utility of such a model to augment clinical image analysis [62]. The efforts described above in neuroanatomical segmentation and anomaly detection highlight the versatility of deep learning in analyzing an inherently complex organ system.
Functional Connectivity and Classification of Brain Disorders
Research in diagnostic support using multiple modalities has been a key area of focus in deep learning research, particularly in disease spaces such as AD, autism spectrum disorder (ASD), and attention deficit hyperactivity disorder (ADHD). For all of these diseases, the onset can be insidious, and diagnosis is reliant on non-specific symptoms, such as distractibility and hyperactivity in the case of ADHD, which results in poor sensitivity and specificity for clinical diagnostic testing; in fact, the sensitivity of the American Psychiatric Association’s Diagnostic and Statistical Manual testing for ADHD is between 70 and 90% [65]. Furthermore, delays in diagnosis inevitably delay treatment, resulting in the treatment being less effective or entirely ineffective [65]. Using fMRI and connectome mapping alongside clinical and demographic data points, multidisciplinary teams have sought to improve upon the accuracy of currently utilized neurological tests.
Within the realm of AD and disorders implicated in MCIs, deep learning has been increasingly adopted as a method to analyze neural connectivity information. Although much of the work in connectome mapping has relied on less complex classifiers, recent publications have explored the benefits of deep learning [66, 67]. When applied to fMRI data, deep learning has several advantages over simpler SVMs and Lasso models, and exhibits an exponential gain in accuracy over simpler models with increasing volumes of training data [5, 68]. Meszlenyi et al. utilized a variant of a convolutional neural network called a connectome convolutional neural network (CCNN) to classify MCI in a relatively small dataset of functional connectivity data from 49 patients [67]. Although accuracies were comparable between the deep learning and less complex classifiers (53.4% accuracy for the CCNN compared to 54.1% for the SVM), the authors postulate that the accuracy benefits of the CCNN architecture are well suited to fMRI tasks as dataset sizes expand [67].
Deep learning classifiers have been applied numerous times toward the accurate diagnosis of ASD using fMRI data. In one study published in 2015, Iidaka et al. selected 312 patients with ASD and 328 control patients from the Autism Brain Imaging Data Exchange (ABIDE), together with 90 regions of interest, and used a probabilistic neural network to classify individuals with ASD. Their method achieved a classification accuracy of 90% [69]. Additionally, Chen et al. published a classifier based on a constructed functional network and additional data from the ABIDE dataset in a clustering analysis aimed at grouping discriminative features and found that many discriminative features clustered into the Slow-4 band [70].
In the realm of ADHD, several efforts have been made to use publicly available imaging data and deep learning algorithms for diagnosis. In a study published in 2014, Kuang et al. attempted to classify ADHD using a deep belief network, comprised of stacked Boltzmann’s machines trained on the public ADHD-200 dataset [71]. Using time-series fMRI data, the deep belief network achieved an accuracy of 35.1%. While each of the above classifiers have achieved results that are either on-par or less accurate than clinical diagnoses using fMRI data, methods are expected to improve dramatically as the quantity of labeled data continues to grow [71].
Risk Prognostication
In addition to widespread research on deep learning applications for image classification and segmentation, researchers have applied deep learning to a variety of other neurology-specific and general medicine data for the purposes of risk prognostication. These efforts have been applied to electroencephalogram (EEG) signals and genetic biomarkers in the hope of predicting clinically meaningful events. Neurologists frequently rely on EEG data for the management and diagnosis of neurological dysfunction, in particular epilepsy and epileptic events. Several studies using deep learning methods have investigated its utility when applied to preictal scalp EEGs as a predictive tool for seizures [72–74]. The most successful of these efforts included a LSTM network, which is particularly useful for interpreting time-series data, allowing a model to allocate importance to previously seen data in a sequence when interpreting a given datapoint. These algorithms are uniquely suited to large sequences of data and have proved their efficacy in predicting epileptic events [73].
In their 2018 study, Tsiouris et al. used a two-layer LSTM-based algorithm to predict epileptic seizures using the publicly available CHB-MIT scalp EEG database. While previous efforts had been made using CNNs and scalp EEGs to predict epileptic events, the novel use of an LSTM set a new state-of-the-art over traditional machine learning algorithms and other deep learning algorithms. Following feature extraction, the LSTM was provided several meaningful features, including statistical moments, zero crossings, Wavelet Transform coefficients, power spectral density, cross-correlation, and graph theory, to use in the prediction of seizures. Notably, the authors compared the predictive ability of the raw EEG data to the extracted features and determined that feature extraction improved model performance [73]. This model configuration achieved a minimum of 99.28% sensitivity and 99.28% specificity across the 15-, 30-, 60-, and 120-min preictal periods, as well as a maximum false positive rate of 0.11/h. Similar experiments on the CHB-MIT scalp EEG database using CNNs, as opposed to LSTMs, achieved worse results, namely poorer sensitivity and a higher hourly rate of false positives [75, 76].
Genetic data has been another important area of research and development for precision medicine. Predictive tasks in large-scale genomic profiles face high-dimensional datasets that are often pared down by experts who hand-select a small number of features for training predictive models [77]. In ASD, deep learning has played a particularly important role in determining the impact of de-novo mutations, including copy number variants and point mutations, on ASD severity [78]. Using a deep CNN, Zhou et al. modeled the biochemical impact of observed point mutations in 1790 whole-genome sequenced families with ASD, on both the RNA and DNA levels [78]. This approach revealed that both transcriptional and post-transcriptional mechanisms play a major role in ASD, suggesting biological convergence of genetic dysregulation in ASD.
Genomic data, either alone or in conjunction with neuroimaging and histopathology, has provided cancer researchers a wealth of data on which to perform cancer-related predictive tasks [77, 79, 80]. Deep learning offers several advantages when working simultaneously with multiple data modalities, removing subjective interpretations of histological images, accurately predicting time-to-event outcomes, and even surpassing gold standard clinical paradigms for glioma patient survival [80]. Using high-powered histological slices and genetic data, namely IDH mutation status and 1p/19q codeletion, on 769 patients from The Cancer Genome Atlas (TCGA), Mobadersaney et al. used a survival CNN (SCNN) to predict time-to-event outcomes. The histological and genetic model performed on par with manual histologic grading or molecular subtyping [80]. In a second paper by this group, SCNNs were shown to outperform other machine learning algorithms, including random forest, in classification tasks using genetic data from multiple tumor types, including kidney, breast, and pan-glioma cancers [77]. The ability of deep learning algorithms to reduce subjectivity in histologic grading and disentangle complex relationships between noisy EEG or genetic data, has the potential to improve current standards for predicting clinical events.
Challenges
Despite the profound biomedical advances due to deep learning algorithms, there remain significant challenges that must be addressed before such applications gain widespread use. We discuss some of the most critical hurdles in the following sections.
Data Volume
Deep neural networks are computationally intensive, multilayered algorithms with parameters on the order of millions. Convergence of such algorithms requires data commensurate with the number of parameters. Although there are no strict rules governing the amount of data required to optimally train DNNs, empirical studies suggest that tenfold more training data relative to the number of parameters is required to produce an effective model. It is no surprise then that domains, such as computer vision and natural language processing, have seen the most rapid progress due to deep learning given the wide availability of images, videos, and free-form text on the Internet.
Biomedical data on the other hand is mostly decentralized—stored locally within hospital systems—and subject to privacy constraints that make such data less readily accessible for research. Furthermore, given the complexity of patient presentations and disease processes, reliable ground truth labels for biomedical applications are extremely expensive to obtain, often requiring the efforts of multiple highly specialized domain experts. This paucity of labeled data remains an important bottleneck in the development of deep learning applications in medicine.
Data Quality
Healthcare data are fundamentally ill-suited for deep learning applications. Electronic medical records are highly heterogeneous, being composed of clinical notes, a miscellany of various codes, and other patient details that may often be missing or incomplete. Clinical notes consist of nuanced language and acronyms that often vary by specialty and contain redundant information that provides an inaccurate temporal representation of disease onset or progression. Diagnosis codes suffer from a similar fate as they track billing for insurance purposes instead of health outcomes. This inherent complexity makes it impossible for deep learning algorithms to parse signal from noise.
Generalizability
Although existing deep learning applications have garnered success in silico, their widespread adoption in real-world clinical settings remains limited due to concerns over their efficacy across clinical contexts. Much of the concern is based on the tendency of deep learning algorithms to overfit to the statistical characteristics of the training data, rendering them hyper-specialized for a hospital or certain patient demographic and less effective on the population at-large [81, 82]. The siloed existence of healthcare data in hospitals and the heterogeneity of data across healthcare systems make the task of developing generalizable models even more difficult. And even when multi-institutional data are acquired, the data are often retrospective in nature, which prevents practical assessment of algorithm performance.
Interpretability
The power of deep learning algorithms to map complex, nonlinear functions can render them difficult to interpret. This becomes an important consideration in healthcare applications where the ability to identify drivers of outcomes becomes just as important as the ability to accurately predict the outcome itself. In the clinical setting, where clinical decision support systems are designed to augment the decision-making capacity of healthcare professionals, interpretability is critical to convince healthcare professionals to rely on the recommendations made by algorithms and enable their widespread adoption. As such, major efforts within the deep learning community to tackle problems of interpretability and explainability have the potential to be particularly beneficial for facilitating the use of deep learning methods in healthcare.
Legal
Medical malpractice rules govern standards of clinical practice in order to ensure the appropriate care of patients. However, to date, no standards have been established to assign culpability in contexts where algorithms provide poor predictions or substandard treatment recommendations. The establishment of such regulations is a necessary prerequisite for the widespread adoption of deep learning algorithms in clinical contexts.
Ethical
Incidental introduction of bias must be carefully evaluated in the application of deep learning in medicine. As discussed previously, deep learning algorithms are uniquely adept at fitting to the characteristics of the data on which they are trained. Such algorithms have the capability to perpetuate inequities against populations underrepresented in medicine and, by extension, in the very healthcare data used to train the algorithms. Furthermore, recent research evaluating algorithmic bias in a commercial healthcare algorithm provides a cautionary tale on the importance of critically evaluating the very outcomes algorithms are trained to predict [83].
Conclusion
Deep learning has the potential to fundamentally alter the practice of medicine. The clinical neurosciences in particular are uniquely situated to benefit given the subtle presentation of symptoms typical of neurologic disease. Here, we reviewed the various domains in which deep learning algorithms have already provided impetus for change—areas such as medical image analysis for improved diagnosis of AD and the early detection of acute neurologic events; medical image segmentation for quantitative evaluation of neuroanatomy and vasculature; connectome mapping for the diagnosis of AD, ASD, and ADHD; and mining of microscopic EEG signals and granular genetic signatures. Amidst these advances, however, important challenges remain a barrier to integration of deep learning tools in the clinical setting. While technical challenges surrounding the generalizability and interpretability of models are active areas of research and progress, more difficult challenges surrounding data privacy, accessibility, and ownership will necessitate conversations in the healthcare environment and society in general to arrive at solutions that benefit all relevant stakeholders. The challenge of data quality, in particular, may prove to be a uniquely suitable target for addressing using deep learning techniques that have already demonstrated efficacy in image analysis and natural language processing. Overcoming these hurdles will require the efforts of interdisciplinary teams of physicians, computer scientists, engineers, legal experts, and ethicists working in concert. It is only in this manner that we will truly realize the potential of deep learning in medicine to augment the capability of physicians and enhance the delivery of care to patients.
Acknowledgements
Funding
No funding or sponsorship was received for this study or publication of this article.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Aly Al-Amyn Valliani, Daniel Ranti and Eric Karl Oermann have nothing to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any studies with human participants or animals performed by any of the authors.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Footnotes
Enhanced digital features
To view enhanced digital features for this article go to 10.6084/m9.figshare.9272951.
Aly Al-Amyn Valliani and Daniel Ranti contributed equally to this article.
References
- 1.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 2.Luo J, Wu M, Gopukumar D, Zhao Y. Big data application in biomedical research and health care: a literature review. Biomed Inform Insights. 2016;19(8):1–10. doi: 10.4137/BII.S31559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohli MD, Summers RM, Geis JR. Medical image data and datasets in the era of machine learning-whitepaper from the 2016 C-MIMI meeting dataset session. J Digit Imaging. 2017;30(4):392–399. doi: 10.1007/s10278-017-9976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengio Y, Courville A, Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35(8):1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 5.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Lin Z, Shen X, Brandt J, Hua G. A convolutional neural network cascade for face detection. In: Proceedings of IEEE conference on computer vision and pattern recognition. Boston, MA. 2015. pp. 5325–34.
- 7.Gilani SZ, Mian A. Learning from millions of 3D scans for large-scale 3D face recognition. 2017. http://arxiv.org/abs/1711.05942.
- 8.Ramanishka V, Chen Y-T, Misu T, Saenko K. Toward driving scene understanding: a dataset for learning driver behavior and causal reasoning. In: Proceedings of IEEE conference on computer vision and pattern recognition. Salt Lake City, UT. 2018. pp. 7699–707.
- 9.Maqueda AI, Loquercio A, Gallego G, Garcia N, Scaramuzza D. Event-based vision meets deep learning on steering prediction for self-driving cars. 2018. http://arxiv.org/abs/1804.01310.
- 10.Mazaré P-E, Humeau S, Raison M, Bordes A. Training millions of personalized dialogue agents. 2018. http://arxiv.org/abs/1809.01984.
- 11.Zhang S, Dinan E, Urbanek J, Szlam A, Kiela D, Weston J. Personalizing dialogue agents: I have a dog, do you have pets too? 2018. http://arxiv.org/abs/1801.07243.
- 12.Wu Y, Schuster M, Chen Z, et al. Google’s neural machine translation system: bridging the gap between human and machine translation. 2016. http://arxiv.org/abs/1609.08144.
- 13.US National Library of Medicine National Institutes of Health. PubMed. 2019. https://www.ncbi.nlm.nih.gov/pubmed/?term=Machine+Learning.
- 14.Mitchell TM. The discipline of machine learning, vol. 9. Pittsburgh: School of Computer Science, Carnegie Mellon University; 2006.
- 15.Rosenblatt F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol Rev. 1958;65:386–408. 10.1037/h0042519. [DOI] [PubMed]
- 16.Ogutu JO, Schulz-Streeck T, Piepho H-P. Genomic selection using regularized linear regression models: ridge regression, lasso, elastic net and their extensions. BMC Proc. 2012;6(Suppl 2):S10. doi: 10.1186/1753-6561-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in neural information processing systems. New York: Curran Associates, Inc.; 2012. pp. 1097–1105. [Google Scholar]
- 18.Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. 2014. http://arxiv.org/abs/1409.4842.
- 19.Saba L, Biswas M, Kuppili V, Cuadrado Godia E, Suri HS, Edla DR, et al. The present and future of deep learning in radiology. Eur J Radiol. 2019;114:14–24. doi: 10.1016/j.ejrad.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 20.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 21.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haenssle HA, Fink C, Schneiderbauer R, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29(8):1836–1842. doi: 10.1093/annonc/mdy166. [DOI] [PubMed] [Google Scholar]
- 23.De Fauw J, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 24.Poplin R, Varadarajan AV, Blumer K, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng. 2018;2(3):158–164. doi: 10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- 25.Lipton ZC, Berkowitz J, Elkan C. A critical review of recurrent neural networks for sequence learning. 2015. http://arxiv.org/abs/1506.00019.
- 26.Rumelhart DE, McClelland JL. Learning internal representations by error propagation. In: Parallel distributed processing: explorations in the microstructure of cognition: foundations. Wachtendonk: MITP Verlags-GmbH & Co. KG; 1987. pp. 318–62.
- 27.Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 28.Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks. 2014. http://arxiv.org/abs/1406.2661.
- 29.Shin H-C, Tenenholtz NA, Rogers JK, et al. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In:Proc Third International Workshop, SASHIMI 2018, held in conjunction with MICCAI 2018, Granada, Spain, September 16, 2018. In: Gooya A, Goksel O, Oguz I, Burgos N, editors. Simulation and synthesis in medical imaging. Cham: Springer International Publishing; 2018:1–11.
- 30.Shi S, Wang Q, Xu P, Chu X. Benchmarking state-of-the-art deep learning software tools. 2016. http://arxiv.org/abs/1608.07249.
- 31.Liu J, Dutta J, Li N, Kurup U, Shah M. Usability study of distributed deep learning frameworks for convolutional neural networks. 2018. https://www.kdd.org/kdd2018/files/deep-learning-day/DLDay18_paper_29.pdf.
- 32.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menze BH, Jakab A, Bauer S, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) IEEE Trans Med Imaging. 2015;34(10):1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suk H-I, Shen D. Deep learning-based feature representation for AD/MCI classification. Med Image Comput Comput Assist Interv. 2013;16(Pt 2):583–590. doi: 10.1007/978-3-642-40763-5_72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta A, Ayhan M, Maida A. Natural image bases to represent neuroimaging data. In: Proceedings of 30th international conference on machine learning. vol. 28. Atlanta, GA. 2013. pp. 987–94.
- 36.Li F, Tran L, Thung K-H, Ji S, Shen D, Li J. Machine learning in medical imaging. New York: Springer International Publishing; 2014. Robust deep learning for improved classification of AD/MCI Patients; pp. 240–247. [Google Scholar]
- 37.Liu S, Liu S, Cai W, Pujol S, Kikinis R, Feng D. Early diagnosis of Alzheimer’s disease with deep learning. In: 2014 IEEE 11th international symposium on biomedical imaging (ISBI). Beijing, China. 2014. pp. 1015–8. http://ieeexplore.ieee.org.
- 38.Liu S, Liu S, Cai W, et al. Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease. IEEE Trans Biomed Eng. 2015;62(4):1132–1140. doi: 10.1109/TBME.2014.2372011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suk H-I, Lee S-W, Shen D. Alzheimer’s disease neuroimaging initiative. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct Funct. 2015;220(2):841–859. doi: 10.1007/s00429-013-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarraf S, Tofighi G. Classification of Alzheimer’s disease using fMRI data and deep learning convolutional neural networks. 2016. http://arxiv.org/abs/1603.08631.
- 41.Suk H-I, Lee S-W, Shen D. Alzheimer’s disease neuroimaging initiative. Deep sparse multi-task learning for feature selection in Alzheimer’s disease diagnosis. Brain Struct Funct. 2016;221(5):2569–2587. doi: 10.1007/s00429-015-1059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valliani A, Soni A. Deep residual nets for improved Alzheimer’s diagnosis. In: BCB. Boston, MA. 2017. p. 615.
- 43.Payan A, Montana G. Predicting Alzheimer’s disease: a neuroimaging study with 3D convolutional neural networks. 2015. http://arxiv.org/abs/1502.02506.
- 44.Hosseini-Asl E, Gimel’farb G, El-Baz A. Alzheimer’s disease diagnostics by a deeply supervised adaptable 3D convolutional network. 2016. http://arxiv.org/abs/1607.00556. [DOI] [PubMed]
- 45.Hosseini-Asl E, Ghazal M, Mahmoud A, et al. Alzheimer’s disease diagnostics by a 3D deeply supervised adaptable convolutional network. Front Biosci. 2018;1(23):584–596. doi: 10.2741/4606. [DOI] [PubMed] [Google Scholar]
- 46.Gao XW, Hui R. A deep learning based approach to classification of CT brain images. In: 2016 SAI computing conference (SAI). London, UK. 2016. pp. 28–31. http://ieeexplore.ieee.org.
- 47.Ding Y, Sohn JH, Kawczynski MG, et al. A deep learning model to predict a diagnosis of Alzheimer disease by using 18F-FDG PET of the brain. Radiology. 2019;290(2):456–464. doi: 10.1148/radiol.2018180958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Titano JJ, Badgeley M, Schefflein J, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med. 2018;24(9):1337–1341. doi: 10.1038/s41591-018-0147-y. [DOI] [PubMed] [Google Scholar]
- 49.Zech J, Pain M, Titano J, et al. Natural language-based machine learning models for the annotation of clinical radiology reports. Radiology. 2018;30:171093. doi: 10.1148/radiol.2018171093. [DOI] [PubMed] [Google Scholar]
- 50.Arbabshirani MR, Fornwalt BK, Mongelluzzo GJ, et al. Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. NPJ Digit Med. 2018;1(1):9. doi: 10.1038/s41746-017-0015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chilamkurthy S, Ghosh R, Tanamala S, et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet. 2018;392(10162):2388–2396. doi: 10.1016/S0140-6736(18)31645-3. [DOI] [PubMed] [Google Scholar]
- 52.Lee H, Yune S, Mansouri M, et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng. 2018;5:6. doi: 10.1038/s41551-018-0324-9. [DOI] [PubMed] [Google Scholar]
- 53.Wachinger C, Reuter M, Klein T. DeepNAT: deep convolutional neural network for segmenting neuroanatomy. Neuroimage. 2018;15(170):434–445. doi: 10.1016/j.neuroimage.2017.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64(6):479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 55.Holland EC. Progenitor cells and glioma formation. Curr Opin Neurol. 2001;14(6):683–688. doi: 10.1097/00019052-200112000-00002. [DOI] [PubMed] [Google Scholar]
- 56.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/S0896-6273(02)00569-X. [DOI] [PubMed] [Google Scholar]
- 57.Landman B, Warfield S. MICCAI 2012 workshop on multi-atlas labeling. In: Medical image computing and computer assisted intervention conference. Nice, France. October 1–5, 2012.
- 58.Livne M, Rieger J, Aydin OU, et al. A U-Net deep learning framework for high performance vessel segmentation in patients with cerebrovascular disease. Front Neurosci. 2019;28(13):97. doi: 10.3389/fnins.2019.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loftis JM, Huckans M, Morasco BJ. Neuroimmune mechanisms of cytokine-induced depression: current theories and novel treatment strategies. Neurobiol Dis. 2010;37(3):519–533. doi: 10.1016/j.nbd.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menard C, Pfau ML, Hodes GE, et al. Social stress induces neurovascular pathology promoting depression. Nat Neurosci. 2017;20(12):1752–1760. doi: 10.1038/s41593-017-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lian C, Zhang J, Liu M, et al. Multi-channel multi-scale fully convolutional network for 3D perivascular spaces segmentation in 7T MR images. Med Image Anal. 2018;46:106–117. doi: 10.1016/j.media.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeong Y, Rachmadi MF, Valdés-Hernández MDC, Komura T. Dilated saliency U-Net for white matter hyperintensities segmentation using irregularity age map. Front Aging Neurosci. 2019;27(11):150. doi: 10.3389/fnagi.2019.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gootjes L, Teipel SJ, Zebuhr Y, et al. Regional distribution of white matter hyperintensities in vascular dementia, Alzheimer’s disease and healthy aging. Dement Geriatr Cogn Disord. 2004;18(2):180–188. doi: 10.1159/000079199. [DOI] [PubMed] [Google Scholar]
- 64.Karargyros A, Syeda-Mahmood T. Saliency U-Net: A regional saliency map-driven hybrid deep learning network for anomaly segmentation. In: Medical imaging 2018: computer-aided diagnosis. International Society for Optics and Photonics. Houston, TX. 2018. 105751T.
- 65.Kuang D, He L. Classification on ADHD with deep learning. In: 2014 international conference on cloud computing and big data. Wuhan, China. 2014. pp. 27–32. http://ieeexplore.ieee.org.
- 66.Suk H-I, Wee C-Y, Lee S-W, Shen D. State-space model with deep learning for functional dynamics estimation in resting-state fMRI. Neuroimage. 2016;1(129):292–307. doi: 10.1016/j.neuroimage.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meszlényi RJ, Buza K, Vidnyánszky Z. Resting state fMRI functional connectivity-based classification using a convolutional neural network architecture. Front Neuroinform. 2017;17(11):61. doi: 10.3389/fninf.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montufar GF, Pascanu R, Cho K, Bengio Y. On the number of linear regions of deep neural networks. In: Ghahramani Z, Welling M, Cortes C, Lawrence ND, Weinberger KQ, editors. Advances in neural information processing systems. Red Hook: Curran Associates, Inc.; 2014. pp. 2924–2932. [Google Scholar]
- 69.Iidaka T. Resting state functional magnetic resonance imaging and neural network classified autism and control. Cortex. 2015;63:55–67. doi: 10.1016/j.cortex.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 70.Chen H, Duan X, Liu F, et al. Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity—a multi-center study. Prog Neuropsychopharmacol Biol Psychiatry. 2016;4(64):1–9. doi: 10.1016/j.pnpbp.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 71.Kuang D, Guo X, An X, Zhao Y, He L. Intelligent computing in bioinformatics. New York: Springer International Publishing; 2014. Discrimination of ADHD based on fMRI data with deep belief network; pp. 225–232. [Google Scholar]
- 72.Tjepkema-Cloostermans MC, de Carvalho RCV, van Putten MJAM. Deep learning for detection of focal epileptiform discharges from scalp EEG recordings. Clin Neurophysiol. 2018;129(10):2191–2196. doi: 10.1016/j.clinph.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 73.Tsiouris ΚΜ, Pezoulas VC, Zervakis M, Konitsiotis S, Koutsouris DD, Fotiadis DI. A long short-term memory deep learning network for the prediction of epileptic seizures using EEG signals. Comput Biol Med. 2018;1(99):24–37. doi: 10.1016/j.compbiomed.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 74.Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput Biol Med. 2018;1(100):270–278. doi: 10.1016/j.compbiomed.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 75.Truong ND, Nguyen AD, Kuhlmann L, Bonyadi MR, Yang J, Kavehei O. A generalised seizure prediction with convolutional neural networks for intracranial and scalp electroencephalogram data analysis. 2017. http://arxiv.org/abs/1707.01976. [DOI] [PubMed]
- 76.Khan H, Marcuse L, Fields M, Swann K, Yener B. Focal onset seizure prediction using convolutional networks. IEEE Trans Biomed Eng. 2018;65(9):2109–2118. doi: 10.1109/TBME.2017.2785401. [DOI] [PubMed] [Google Scholar]
- 77.Yousefi S, Amrollahi F, Amgad M, et al. Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Sci Rep. 2017;7(1):11707. doi: 10.1038/s41598-017-11817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou J, Park CY, Theesfeld CL, et al. Whole-genome deep-learning analysis identifies contribution of noncoding mutations to autism risk. Nat Genet. 2019;51(6):973–980. doi: 10.1038/s41588-019-0420-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buda M, Saha A, Mazurowski MA. Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm. Comput Biol Med. 2019;109:218–225. doi: 10.1016/j.compbiomed.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 80.Mobadersany P, Yousefi S, Amgad M, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci USA. 2018;115(13):E2970–E2979. doi: 10.1073/pnas.1717139115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15(11):e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Confounding variables can degrade generalization performance of radiological deep learning models. 2018. http://arxiv.org/abs/1807.00431. [DOI] [PMC free article] [PubMed]
- 83.Obermeyer Z, Mullainathan S. Proceedings of conference on fairness, accountability, and transparency. New York: ACM; 2019. Dissecting racial bias in an algorithm that guides health decisions for 70 million people; p. 89. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.