Abstract
Purpose:
To identify genetic risk factors associated with risk of stroke among survivors of childhood cancer treated with cranial radiation therapy (CRT).
Experimental Design:
We analyzed whole–genome sequencing (36.8-fold) data of 686 childhood cancer survivors of European ancestry [median (range): 40.4 (12.4–64.7) years old; 54% male] from the St. Jude Lifetime Cohort study treated with CRT, of whom 116 (17%) had clinically–diagnosed stroke. Association analyses (single-variant and Burden/SKAT tests) were performed, adjusting for demographic characteristics and childhood cancer treatment exposures.
Results:
We identified a genome–wide significant association between 5p15.33 and stroke [rs112896372: hazard ratio (HR)=2.55; P=1.42×10–8], with a stronger association (HR=3.68) among survivors treated with CRT dose 25–50 Gray (Gy) and weaker associations among those treated with CRT doses <20 or 20–25 or >50 Gy (HRs=2.14, 2.40 and 2.28). The association was replicated in 90 CRT-exposed African survivors (HR=3.05; P=0.034). In CRT–exposed Europeans, rs112896372 significantly (P<0.001) improved predictive ability (AUC=0.717) for determining stroke risk than non-genetic factors alone (AUC=0.663) at 30 years since diagnosis, with significant improvement among African survivors (P=0.047). rs112896372 was further evaluated in three independent datasets including 1641 European (HR=1.54; P=0.055) and 316 African survivors (HR=1.88; P=0.283) not treated with CRT, and 166988 males in the UK Biobank (odds ratio=1.0012; P=0.042).
Conclusions:
A novel locus 5p15.33 is associated with stroke risk among childhood cancer survivors, with a possible CRT dose–specific effect. The locus is of potential clinical utility in characterizing individuals who may benefit from surveillance and intervention strategies.
INTRODUCTION
Long–term survivors of childhood cancer are at increased risk of stroke, a major cause of physical disability and cognitive impairment. Compared to sibling controls, childhood cancer survivors are approximately eight-times more likely to develop stroke(1) and this risk is strongly associated with cranial radiation therapy (CRT) used to treat the childhood cancers(1–4). In a report by the Childhood Cancer Survivor Study(2), CRT was significantly associated with an increased stroke risk in a dose–dependent manner, with a 5.9-fold risk for survivors exposed to 30–49 Gy and an 11.0-fold risk for survivors treated with ≥50 Gy, relative to those not treated with CRT.
The mechanisms by which CRT increases the risk of stroke in childhood cancer survivors are not well understood. Current literature has focused on radiation–induced vasculopathy where CRT can induce direct vascular injury to both large and small vessels, including accelerated intracranial atherosclerosis and vascular insufficiency(5–8). However, there is variation in vascular response to CRT with minimal vessel changes in some patients with stenosis and aneurysms occurring in others(6), suggesting role of genetic factors in risk of stroke in childhood cancer survivors treated with CRT.
The St. Jude Lifetime Cohort (SJLIFE) study provides a unique opportunity to study genetic contributions to clinically–defined late effects in pediatric cancer survivors. Leveraging deep–coverage (36.8-fold) whole–genome sequencing (WGS) data in the SJLIFE cohort(9), we comprehensively investigated the potential role of germline genetic factors to risk of stroke among childhood cancer survivors treated with CRT. Genetic associations that achieved statistical significance were examined in four independent samples including childhood cancer survivors from the SJLIFE and participants from the UK Biobank(10).
MATERIALS AND METHODS
Study Population
SJLIFE is a retrospective cohort study with prospective clinical follow–up and ongoing enrollment of survivors of childhood cancer treated since 1962 and followed up at St. Jude Children’s Research Hospital (SJCRH)(11). The SJLIFE study was initiated in late 2007 with eligibility for participation including diagnosis of pediatric malignancy treated at SJCRH, survived ≥10 years from diagnosis, and attained age of ≥18 years. Study participation at that time involved a three to four-day outpatient visit on the SJCRH campus during which biologic specimens were collected; metabolic, cognitive, and neuromuscular functional status was systematically evaluated; and risk-based screening of organ function was implemented, per the Children’s Oncology Group Guidelines recommendation. In 2015, recruitment eligibility was expanded to ≥5-year survivors and the study protocol modified to include systematic clinical assessments for all participants (i.e., screening is no longer risk-based except for selected cancer screening prior to ages recommended for the general population). Since activation of the expanded enrollment, participation rates have remained excellent. As of February 1, 2019, for survivors where attempts to contact have been initiated, 80.4% (5753/7152) have been successfully enrolled and 87.2% have enrolled or have confirmed their interest in participating. Of all eligible survivors who are alive, 68.8% (5753/8363) have thus far been enrolled and 74.6% have enrolled or confirmed their interest in participating. Currently, because of the volume of survivors relative to the clinical capacity to perform the extensive SJLIFE evaluation, the study is recruiting approximately 20–30 new survivors per month. Survivor participants who have completed their baseline clinical assessment are invited to return for subsequent systematic evaluations at a minimum of every 5 years. As of February 1, 2019, 47% (2257/4760) of survivors who have completed a baseline clinical assessment have returned for one or more subsequent follow-up assessments. Following the SJLIFE evaluations, clinical outcomes are validated, graded for severity according to a modified CTCAEv4.03 rubric(11) and codified for research analyses. To facilitate timely analysis and publication of the most current SJLIFE data, a mechanism has been established to clean, update and freeze analytic data files every year. This study was approved by the SJCRH institutional review board and all participants provided written informed consent, and the study was conducted in accordance with the Declaration of Helsinki.
Stroke definition
Survivors were clinically assessed for the presence of various chronic health conditions (CHCs) with physical and neurological examinations and detailed review of medical records from SJCRH and outside institutions. CHCs were graded using a modified version(11) of the National Cancer Institute’s Common Terminology Criteria for Adverse Events (CTCAE) version 4.03, and classified into six categories, including none (grade 0), mild (grade 1), moderate (grade 2), severe or disabling (grade 3), life–threatening (grade 4), or fatal (grade 5). Stroke was defined based on three CTCAE–graded neurological CHCs (first occurrence), which consisted of cerebrovascular accident (includes lacunes, hemorrhagic stroke and ischemic stroke), intracranial hemorrhage (includes subdural, subarachnoid and intraventricular hemorrhages) and cerebrovascular disease (includes stenotic or occlusive large vessel disease and moyamoya).
Survivors with grade 1 or higher for any of the three CHCs (clinically confirmed through review of medical records including reports of brain imaging) were classified as stroke cases and survivors with (grade 0) for all three of the CHCs were categorized as stroke–free.
Whole–genome sequencing data
WGS data with mean coverage at 36.8-fold were obtained from a larger effort to sequence whole genomes of 3006 SJLIFE survivors. Details of DNA sample extraction, WGS, quality control (QC), mapping, variant identification and annotation are described in Supplementary Methods. Additional QC on the genotype data resulted in exclusion of 20 samples with excess heterozygosity rate, leaving 2986 samples for final analysis (hereafter referred to as SJLIFE WGS dataset). Variants were annotated across the genome and functional domains using ANNOVAR(12) based on the RefSeq gene model and in–house pipelines, which include data reported by the FANTOM5 project(13,14) to map promoters and enhancers (Supplementary Methods).
Analytic discovery sample
Of the 2986 survivors in the SJLIFE WGS dataset, 243 had stroke and the remaining 2743 were stroke–free, as of June 30, 2017. A total of 46 survivors were excluded because of stroke prior to CRT (n=14) or congenital conditions (n=32), such as Down syndrome, Turner syndrome, polycystic kidney disease and neurofibromatosis type 1. Based on principal component analysis(15) (Supplementary Figure 1), 2327 and 406 survivors were of European and African ancestries, respectively. Among 2327 survivors of European descent, 686 were treated with CRT, including 116 survivors with and 570 survivors without stroke, which formed the discovery sample for this study (“SJLIFE European-descent discovery sample”). Of the 116 survivors with stroke, the majority (n=80 [69.0%]) had cerebrovascular accident and the remaining 22 (19.0%) and 14 (12.0%) had intracranial hemorrhage and cerebrovascular disease, respectively (Supplementary Table 1).
Discovery analysis
Association between QC–passed variants and stroke risk was assessed using an in–house analysis pipeline (Supplementary Figures 2 and 3 and Supplementary Methods). Briefly, the associations of common variants [minor allele frequency (MAF)>0.05 in SJLIFE European-descent discovery sample] were examined by a single–variant approach using Cox regression assuming an additive genetic model. Years since diagnosis was used as the underlying time scale, with individuals followed from the age at diagnosis of childhood cancer until the earliest of stroke diagnosis, death or last follow-up. Each survivor’s follow-up time was divided into segments so that we can adjust for time-dependent attained age, used in a counting-process format with start and end time points of segments. Covariates included sex, attained age, CRT dose and the top principal components (PCs). Burden test and Sequence Kernel Association Test (SKAT)(16) were used, initially with the large–sample–approximate test whose top results followed by randomization test with 100 million permutations, to assess associations of rare/low–frequency variants (MAF≤0.05 in SJLIFE European-descent discovery sample). Rare/low–frequency variants were aggregated into four analytical sets with respect to the RefSeq gene model [protein truncating variants (PTVs), non–synonymous–strict variants (NSstrict), non–synonymous–broad variants (NSbroad) and regulatory variants (REGMOTIF)], and 4–kb sliding window across the genome (see Supplementary Methods for more details). Common variants showing associations with P<5×10–8 were considered statistically significant at the genome–wide level. Rare/low–frequency variant associations were considered significant with P<6.5×10–7 accounting for the 38542 analytical sets with respect to the RefSeq gene model (see Supplementary Data and Supplementary Methods for more details) and the two tests (Burden and SKAT tests) for each set. For the sliding window approach, we considered two statistical tests for 668035 contiguous non–overlapping windows, leading to a statistical significance threshold at P<3.7×10–8 (equal to 0.05/668035/2).
Replication
Replication sample included 90 childhood cancer survivors (20 with and 70 without stroke) of African ancestry in the SJLIFE study (“SJLIFE African-descent replication sample”), independent of those in the SJLIFE European-descent discovery sample. Demographic characteristic of survivors in the replication sample are provided in Supplementary Table 1. For the genome-wide significant associations in the discovery analysis, replication analyses were performed utilizing the same statistical framework and covariates adjustment used in the discovery analysis. Results with P<0.05 were considered statistically significant.
SNP effects among survivors not exposed to CRT and in the general population
To assess specificity of the genetic associations identified in childhood cancer survivors of European ancestry exposed to CRT, we further evaluated the genome-wide significant results among survivors not exposed to CRT as well as in the general population. To do this, we employed three datasets consisting of individuals independent of those used in the discovery and replication analyses. Of these, the first independent dataset consisted of 1641 survivors of European ancestry not exposed to CRT in the SJLIFE study, including 60 survivors with stroke post-diagnosis of cancer and 1581 without stroke. The second dataset consisted of SJLIFE survivors of African ancestry not exposed to CRT, including 15 with stroke post-diagnosis of cancer and 301 survivors without stroke. Demographic characteristics of survivors in the first and second independent datasets are provided in Supplementary Table 1. The third dataset consisted of individuals from the general population in the UK Biobank(10). Specifically, we looked up pre-computed association summary results (http://www.nealelab.is/uk-biobank/) for stroke [Non-cancer illness code 20002, self-reported stroke (http://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=20002)], which consisted of 361141 individuals (166988 males) of European ancestry including 4836 (2933 males) with stroke.
Effect of SNPs with respect to CRT dose
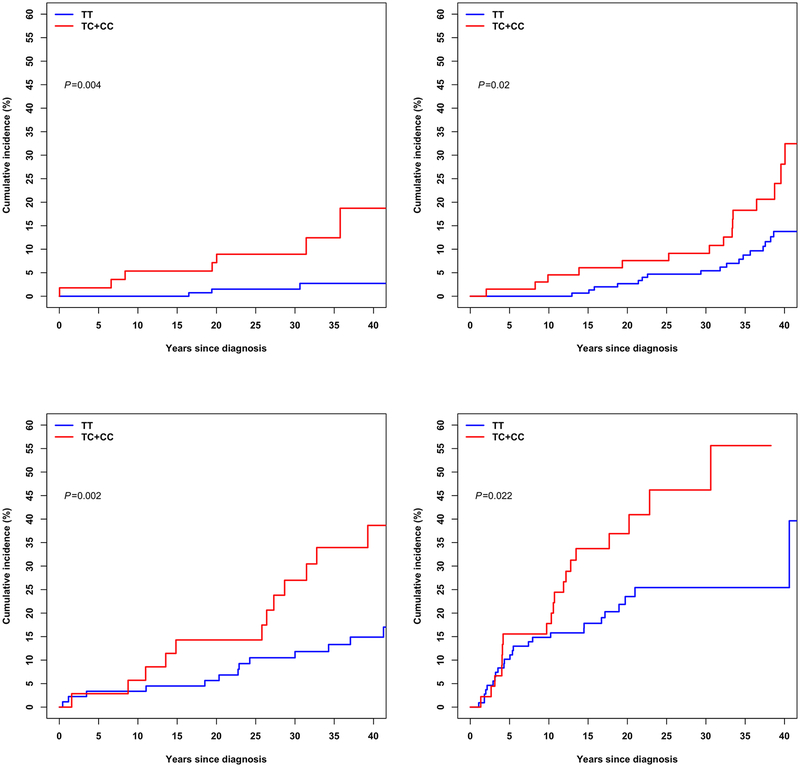
SNPs showing genome–wide significance in the SJLIFE European-descent discovery sample followed by successful replication were further examined with respect to the CRT dose, to assess CRT dose–specific genetic effect on the risk of stroke. Specifically, we examined the SNP’s effect across four subgroups of survivors in the SJLIFE European-descent discovery sample who were exposed to CRT doses of 0–20 Gy, 20–25 Gy, 25–50 Gy and 50 Gy or greater, using the Cox regression models. Cumulative incidence of stroke among survivors carrying at least one copy of the effect allele [C] of rs112896372 versus none is shown graphically stratified by CRT doses.
Association of established stroke risk loci from the general population in survivors
A recent multi–ancestry GWA study including 520000 subjects identified 32 loci associated with stroke and its subtypes(17). Of these, data were available in our SJLIFE European-descent discovery sample at 29 loci – the remaining three SNPs did not pass QC. We looked up association results of the lead (the most significant) SNP at the 29 loci in our SJLIFE European-descent discovery sample. We also examined the individual associations between the 29 stroke risk loci among survivors in the replication sample as well as among those in the first and second independent datasets. Additionally, we also constructed a genetic risk score (GRS) using the reported weights of the lead SNPs at the 29 risk loci from the published study per the following formula and evaluated the polygenic effect of the 29 stroke risk loci captured by the GRS (categorically: five quintiles with first quintile as a reference) in the SJLIFE European-descent discovery sample.
Where βi is the effect (log–odds) of the ith SNP for stroke reported in Malik et al.(17) and SNPi is the number of the effect allele (range: 0 to 2) of the ith SNP.
Clinical risk prediction modeling of stroke
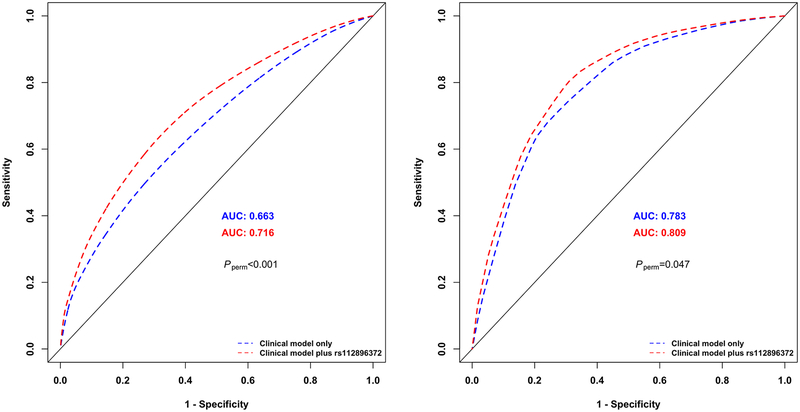
We performed receiver operating characteristics (ROC) analyses to evaluate predictive utility of the clinical model and the model with the addition of lead SNP(s) at genome-wide significant loci for risk of stroke among survivors treated with CRT. The predictive performance of each model was measured by the area under the ROC curve (AUC). Specifically, we obtained predicted probabilities of stroke for each survivor in SJLIFE European-descent discovery and SJLIFE African-descent replication samples separately, based on the Cox regression model with and without the rs112896372 genotype and calculated corresponding AUC values for the two models at 10, 20 and 30 years since childhood-cancer diagnosis, using the method of Heagerty and Zhang(18) with their risksetROC R package. We conducted 1000 random permutations of the genotypes of rs112896372 and calculated Pperm as the proportion of the 1000 AUC differences with and without the rs112896372 genotype calculated from the 1000 permuted datasets that were greater than the AUC difference with and without the rs112896372 genotype from the observed dataset.
RESULTS
Demographics of SJLIFE participants included in the discovery analysis are provided in Supplementary Table 1. The 686 survivors exposed to CRT in the SJLIFE European-descent discovery sample included slightly more males (56.9% and 53.9% in survivors with and without stroke, respectively). The proportion of survivors exposed to higher CRT dose (>25 Gy) was greater (62.9%) among those with stroke than survivors without stroke (35.8%).
The clinical prediction model of CRT–related stroke
Results from multivariable Cox regression examining associations between potential non-genetic risk factors and stroke among survivors exposed to CRT in the SJLIFE European-descent discovery sample are shown in Supplementary Table 2. Attained age and sex were not associated with stroke risk. Compared to survivors exposed to CRT doses >0–20 Gy, an increased risk of stroke was observed among survivors exposed to CRT doses >20–25 Gy [hazard ratio (HR)=2.16; 95% confidence interval (CI)=1.06–4.43; P=0.034], >25–50 Gy [HR=3.20; 95% CI=1.53–6.69; P=1.95×10–3], >50–70 Gy [HR=9.96; 95% CI=4.86–20.41; P=3.34×10–10] and >70 Gy [HR=14.62; 95% CI=5.18–41.26; P=4.05×10–7].
QC-passed WGS dataset for discovery analysis
A total of 29.9 million autosomal variants passed QC in the SJLIFE European-descent discovery sample, which included 6.5 million common and 23.4 million rare/low–frequency variants (MAF≤0.05). Of these, 63.6% of the variants had MAF≤0.005, 5.0% had 0.005<MAF≤0.01, 9.7% had 0.01<MAF≤0.05 and 21.7% had MAF>0.05 (Supplementary Figure 4). Among the 23.4 million rare/low–frequency variants, there were 38542 analytical sets with respect to the RefSeq gene with a distribution of 2 to 265 variants per set (Supplementary Figure 5 and Supplementary Methods), while among the 1.33 million 4–kb sliding windows, there were 2 to 392 variants per window (Supplementary Figure 6).
Common variants associated with CRT–related stroke
Single-variant analysis of the 6.5 million common variants in the SJLIFE European-descent discovery sample using Cox regression identified 104 SNPs showing associations with stroke risk at P<1×10–5 (Supplementary Table 3), including a locus on 5p15.33 achieving genome-wide statistical significance (Table 1 and Supplementary Figures 7 and 8). The quantile–quantile plot and genomic inflation factor of 1.076 (Supplementary Figure 9) indicate minimal influence of population stratification on the association results, but top results from the discovery analysis were further evaluated in independent cohorts to rule out potential spurious associations arising from population stratification. The 5p15.33 locus was marked by rs112896372 showing nearly 2.5–fold increased risk of CRT–related stroke [HR=2.55; 95% CI=1.85–3.51; P=1.42×10–8] (Table 1). This association was further supported by four SNPs in high linkage disequilibrium (LD; r2>0.9 in the 1000 Genomes Project European data) with rs112896372, including three producing genome-wide significance (P=2.24×10–8) and the fourth one with near genome–wide statistical significance (P=1.13×10–7). The MAF of rs112896372 among CRT–treated survivors of European ancestry with and without stroke was 0.27 and 0.14, respectively.
Table 1 |.
Association between chromosome 5p15.33 locus and stroke among survivors from the St. Jude Lifetime Cohort (SJLIFE) study exposed to cranial radiation therapy
| SNP | BP | EA | NEA | SJLIFE Europeans exposed to CRT (Discovery sample) | SJLIFE Africans exposed to CRT (Replication sample) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAFaff | EAFnon-aff | HR (95% CI) | P | EAFaff | EAFnon-aff | HR (95% CI) | P | |||||
| rs112896372 | 3756587 | C | T | 0.27 | 0.16 | 2.55 (1.85–3.51) | 1.42×10−8 | 0.23 | 0.13 | 3.05 (1.10–8.50) | 0.034 | |
| rs79103129 | 3760131 | A | G | 0.27 | 0.16 | 2.50 (1.82–3.44) | 2.24×10−8 | 0.05 | 0.04 | 1.43 (0.25–7.99) | 0.688 | |
| rs4391171 | 3760305 | C | G | 0.27 | 0.16 | 2.50 (1.82–3.44) | 2.24×10−8 | 0.05 | 0.04 | 1.43 (0.25–7.99) | 0.688 | |
| rs17652293 | 3760914 | G | C | 0.27 | 0.16 | 2.50 (1.82–3.44) | 2.24×10−8 | 0.05 | 0.04 | 1.43 (0.25–7.99) | 0.688 | |
| rs35710183 | 3753773 | C | T | 0.28 | 0.17 | 2.36 (1.72–3.23) | 1.13×10−7 | 0.05 | 0.04 | 1.43 (0.25–7.99) | 0.688 | |
Genomic positions are shown relative to GRCh38 (hg38); EA, effect allele; NEA, non-effect allele; EAF, effect allele frequency; EAFaff, EAF in survivors with stroke; EAFnon-aff, EAF in stroke-free survivors; HR, hazard ratio; CI, confidence interval; SJLIFE European-descent discovery sample (116 survivors with and 570 without stroke); SJLIFE African-descent replication sample (20 survivors with and 70 without stroke)
Role of rare/low–frequency variants in CRT–related stroke
Analyses of rare/low–frequency variants using the burden and SKAT tests identified two chromosomal regions tested using the 4–kb sliding windows showing significant association with risk of stroke (P<3.7×10–8). However, both rare/low–frequency variant associations did not remain statistically significant based on the 100 million permutations (P=3.0×10–7) (Supplementary Table 4).
Replication results
SNP rs112896372 at the 5p15.33 locus showed a statistically significant association with stroke survivors of African ancestry exposed to CRT (HR=3.05; 95% CI=1.10–8.50; P=0.034) (Table 1). Association of the remaining four SNPs with stroke was not statistically significant (P=0.688), with attenuated magnitude of their effects (HR=1.43) to that of rs112896372. This is likely due to very weak LD (r2=0.06 in 1000 Genomes Project African data) between the lead SNP rs112896372 and the remaining four SNPs at the 5p15.33 locus – thereby constituting two independent LD blocks.
Association of rs112896372 with stroke in survivors not exposed to CRT and in the UK Biobank
In first independent dataset including SJLIFE survivors of European ancestry not exposed to CRT, all five SNPs at 5p15.33 locus showed borderline nominally significant associations (P-values=0.055–0.061) but with attenuated effects on stroke risk (HRs=1.51–1.54) (Table 2). Among SJLIFE African survivors not exposed to CRT in the second independent dataset, all the five SNPs were not statistically significant (P>0.283); however, the effect size was slightly greater (HRs=1.79–1.88) relative to those among SJLIFE European survivors not exposed to CRT (HRs=1.51–1.54). In the third independent dataset consisting of the general population from the UK Biobank participants (Table 2), all the five SNPs at the 5p15.33 locus showed nominal significant associations, albeit with negligible effect, in males [odds ratio (OR)=1.0012; P<0.042]. In UK Biobank results for stroke, including both males and females, the 5p15.33 locus showed borderline associations (OR=1.0005; P<0.164) with no associations among females only (OR=0.9999; P>0.674) (Supplementary Table 5).
Table 2 |.
Association between chromosome 5p15.33 locus and stroke among independent survivors from the St. Jude Lifetime Cohort (SJLIFE) study not exposed to cranial radiation therapy (CRT) and in the UK Biobank participants
| SNP | BP | EA | NEA | SJLIFE European-descent survivors not exposed to CRT (Independent dataset 1) | SJLIFE African-descent survivors not exposed to CRT (Independent dataset 2) | UK Biobank European males (Independent dataset 3) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAFaff | EAFnon-aff | HR (95% CI) | PCOX | EAFaff | EAFnon-aff | HR (95% CI) | PCOX | EAFaff | EAFnon-aff | OR (95% CI) | PLOGISTIC | ||||||
| rs112896372 | 3756587 | C | T | 0.25 | 0.18 | 1.54 (0.99–2.39) | 0.055 | 0.20 | 0.15 | 1.88 (0.60–5.88) | 0.283 | 0.18 | 0.18 | 1.0012 (0.9999–1.0024) | 0.042 | ||
| rs79103129 | 3760131 | A | G | 0.25 | 0.18 | 1.53 (0.99–2.37) | 0.059 | 0.07 | 0.04 | 1.81 (0.28–11.87) | 0.536 | 0.18 | 0.18 | 1.0012 (0.9999–1.0024) | 0.039 | ||
| rs4391171 | 3760305 | C | G | 0.25 | 0.18 | 1.53 (0.99–2.38) | 0.058 | 0.07 | 0.03 | 1.82 (0.28–11.87) | 0.536 | 0.18 | 0.18 | 1.0012 (0.9999–1.0024) | 0.039 | ||
| rs17652293 | 3760914 | G | C | 0.25 | 0.18 | 1.53 (0.99–2.38) | 0.058 | 0.07 | 0.03 | 1.82 (0.28–11.87) | 0.536 | 0.18 | 0.18 | 1.0012 (0.9999–1.0024) | 0.040 | ||
| rs35710183 | 3753773 | C | T | 0.25 | 0.18 | 1.51 (0.98–2.32) | 0.061 | 0.07 | 0.04 | 1.79 (0.27–11.77) | 0.545 | 0.18 | 0.18 | 1.0014 (0.9998–1.0025) | 0.022 | ||
Genomic positions are shown relative to GRCh38 (hg38); EA, effect allele; NEA, non-effect allele; EAF, effect allele frequency; EAFaff, EAF in survivors with stroke; EAFnon-aff, EAF in stroke-free survivors; HR, hazard ratio; OR, odds ratio; CI, confidence interval; PCOX, p-value from Cox regression; PLOGISTIC, p-value from logistic regression; Independent dataset 1 (60 survivors with 1581 without stroke); Independent dataset 2 (15 survivors with and 301 without stroke); Independent dataset 3 (2933 male stroke cases and 164055 male controls from the UK Biobank)
Effect of rs112896372 with respect to CRT dose
Based on results of stratified analysis in the discovery analysis, the lead SNP rs112896372 at 5p15.33 locus showed the greatest association (HR=3.68; 95% CI=1.82–7.46; P=2.9×10–4) with risk of stroke among survivors treated with CRT dose 25–50 Gy. The risk association was around two-fold among survivors treated with lower CRT doses of ≤20 Gy (HR=2.14; 95% CI=1.01–4.53; P=0.047) and 20–25 Gy (HR=2.40; 95% CI=1.45–3.97; P=6.5×10−4), and among those exposed to the highest CRT dose of >50 Gy (HR=2.28; 95% CI=1.27–4.09; P=5.7×10–3). Cumulative incidence curves with respect to the rs112896372’s effect allele [C] among survivors SJLIFE European-descent discovery sample stratified by CRT doses are provided in Figure 1.
Figure 1 |.
Cumulative incidence curves among survivors in the St. Jude Lifetime Cohort European-descent discovery sample are shown, stratified by cranial radiation therapy doses including survivors exposed to >0–20 Gy (top left panel), 20–25 Gy (top right panel), 25–50 Gy (bottom left panel) and ≥50 Gy (bottom right panel). Cumulative incidence of stroke among survivors carrying at least one copy of the rs112896372’s effect allele (TC+CC) is shown as red and among survivors without the effect allele (TT) is shown as blue.
Established stroke risk loci from the general population in survivors
Of the 29 lead SNPs showing genome–wide significant association with stroke and/or its subtypes in the general population, rs7304841 in PDE3A showed nominal statistical significance (P=0.048) in the SJLIFE European-descent discovery sample, and further two SNPs near HDAC9–TWIST1 and FOXF2 also showed borderline significance (P<0.080) (Supplementary Table 6) with the same direction of effect. We did not observe statistically significant association (P>0.102 for all quintiles relative to the lowest quintile of GRS; Ptrend=0.305) between the GRS and stroke among survivors in the SJLIFE European-descent discovery sample (Supplementary Table 7). In the SJLIFE African-descent replication sample, rs4959130 near FOXF2, rs6825454 near FGA, rs17612742 in EDNRA and rs12445022 near ZCCHC14 showed nominal significance (P<0.05) with the same direction of effect. Among SJLIFE European-descent survivors not exposed to CRT, no SNPs showed nominal significance but a SNP near SLC22A7-ZNF318 showed borderline statistical significance (P=0.052) with the same direction of effect. Only one SNP near SMARCA4–LDLR showed borderline associations among SJLIFE African-descent survivors not exposed to CRT.
Clinical utility of genetic findings
In the SJLIFE European-descent discovery sample, AUC of the clinical model including sex, attained age and CRT dose at 10, 20 and 30 years since childhood-cancer diagnosis were 0.726, 0.696 and 0.663, respectively. Adding the rs112896372 genotype to the clinical model significantly improved the predictive ability of stroke incidence by 0.040, 0.047 and 0.054, respectively (Pperm<0.001 at all three time points). In the SJLIFE African-descent replication sample, AUC values based on the clinical model were 0.848, 0.845 and 0.783 at 10, 20 and 30 years since childhood-cancer diagnosis, respectively, and adding the top SNP to the clinical model improved the AUC values by 0.008 (Pperm=0.238), 0.019 (Pperm=0.065) and 0.026 (Pperm=0.047), respectively. ROC curves showing the probabilities of stroke at 30 years since childhood cancer diagnosis with at least one copy of rs112896372’s effect allele [C] versus none among childhood cancer survivors exposed to CRT in the SJLIFE European-descent discovery and SJLIFE African-descent replication samples are shown in Figure 2.
Figure 2 |.
Receiver operating characteristic (ROC) curves showing the probabilities of stroke among childhood cancer survivors at 30 years since childhood cancer diagnosis in the St. Jude Lifetime Cohort European-descent discovery sample (left) and St. Jude Lifetime Cohort African-descent replication sample (right) exposed to cranial radiation therapy. Probability of stroke based on the clinical model including non-genetic risk factors alone is shown in blue and the probability of stroke based on both non–genetic risk factors and the lead SNP rs112896372 at the 5p15.33 locus is shown in red. Random permutations (1000-times) of the rs112896372’s genotype within each sample were conducted to calculate p-value (Pperm) as the proportion of the 1000 permuted datasets that were greater than the area under the ROC curve difference with and without the rs112896372 genotype from the observed dataset.
DISCUSSION
We performed a comprehensive analysis of high quality WGS data with mean coverage of 36.8-fold among 686 childhood cancer survivors of European descent who had received CRT and identified a novel locus at 5p15.33 showing genome–wide significant association with risk of stroke, which was replicated in an independent sample. Our results showed significant improvement in prediction of CRT-related stroke using the genotype of the lead SNP rs112896372 at the 5p15.33 locus relative to using only non–genetic risk factors, thereby supporting potential clinical utility in identifying CRT-exposed survivors at high–risk of developing stroke.
Our data indicate that the association between rs112896372 and stroke was present among survivors of both European and African ancestries treated with CRT, with the magnitudes of the risk association approximately twice (HR estimates of 2.55 and 3.05, respectively) among survivors exposed to CRT relative to that among survivors not exposed to CRT (HR estimates of 1.54 and 1.88, respectively). Notably, the magnitude of the rs112896372’s stroke risk association among African survivors was greater than those among survivors of European descent, regardless of CRT exposure. Epidemiological data show greater risk of stroke among Africans relative to Caucasians in the general population(19,20) and in childhood cancer survivors(21). More importantly, the effect of the rs112896372 on stroke risk in the general population was the lowest and negligible (OR estimate of 1.00), suggesting that the association between rs112896372 and stroke is much more pronounced among childhood cancer survivors, particularly among those treated with CRT.
A possible CRT dose–specific association of rs112896372 was observed among European survivors exposed to CRT. The magnitude of risk association conferred by rs112896372 was similar to the overall analysis (HR estimate of 2.55) among those exposed to the lower CRT doses of <20 Gy (HR estimate of 2.14) and 20–25 Gy (HR estimate of 2.40), but the magnitude of association increased to an HR estimate of 3.68 among survivors exposed to an intermediate CRT dose of 25–50 Gy, which then attenuated to an HR estimate of 2.28 among those who received the highest CRT dose of >50 Gy. These data may imply that the influence of rs112896372 is higher among survivors exposed to low–to–intermediate doses of CRT, whereas in survivors exposed to the high doses, the overwhelming detrimental effect of the CRT may have played a stronger role in determining the risk of stroke, reducing the SNP’s risk association in this subgroup. Because of limited sample size for some subcategories in our study, however, our data are not conclusive regarding the dose-response pattern between the 5p15.33 locus and CRT dose. If confirmed by future larger studies, these findings may have important clinical implications in identifying survivors at higher risk of stroke based on rs112896372 genotype who, according to their low or intermediate (or no) CRT doses, would otherwise not be considered high–risk individuals compared to those who were exposed to higher CRT doses (≥50 Gy). In addition, we showed that rs112896372 is a marker with potential clinical utility as it significantly improved the predictive ability for determining risk of stroke among CRT–exposed European survivors, improving AUC values by up to 5%; the predictive performance among CRT-exposed African survivors had consistent results.
SNP rs112896372 maps to an intergenic region located approximately 155 kb downstream of IRX1 (iroquois homeobox 1) and alters DNA binding motif of Maf(22), which is a transcription factor with an oncogenic role and is also known to play active roles in many organs, tissues, cells for the development, differentiation and establishment of specific functions(23,24). IRX1 is a member of IRX family of transcription factors with roles in heart development and function and all the six IRX genes are expressed with distinct and overlapping patterns in the mammalian heart(25).
We observed minimal shared genetics contributing to stroke between the general population and childhood cancer survivors, presumably due to the unique exposure to a known strong risk factor of stroke, CRT, and other cancer-related exposures, that may alter the pathogenetic mechanisms underlying stroke in survivors of childhood cancer. Nonetheless, additional investigation utilizing larger studies are required to provide a more conclusive interpretation about generalizability of stroke risk loci from the general population in survivors of childhood cancers.
In summary, we identified a novel locus at 5p15.33 robustly associated with risk of stroke among childhood cancer survivors exposed to CRT, with a possible CRT-dose specific effect. The lead SNP rs112896372 showed significant improvement in the predictive ability for determining risk of stroke among CRT–exposed survivors. rs112896372 may be clinically useful in characterizing individuals who may benefit from surveillance and intervention strategies. Understanding of biological mechanisms underlying the rs112896372 association will provide important information about the causal genes and/or pathway, thereby informing potential therapeutic targets.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Survivors of childhood cancer are at increased risk of stroke, with a well–established dose–risk association with cranial radiation therapy (CRT). We analyzed whole-genome sequencing data among survivors of European ancestry treated with CRT from the St. Jude Lifetime Cohort study and identified a locus at 5p15.33 associated with increased risk of clinically-diagnosed stroke, followed by replication in CRT-treated survivors of African ancestry. Further analyses of the top SNP rs112896372 including additional three independent datasets (consisting of childhood cancer survivors and participants in the UK Biobank) indicated CRT dose-specific effect with the strongest association observed among survivors treated with CRT dose of 25–50 Gray. The 5p15.33 locus improved predictive ability for determining stroke risk over non-genetic factors alone, thereby supporting potential utility of the 5p15.33 locus in characterizing survivors at risk of stroke who may benefit from surveillance and intervention strategies including minimizing the risk of modifiable cardiovascular risk factors.
ACKNOWLEDGEMENTS
The St. Jude Lifetime Cohort (SJLIFE) study is supported by the National Cancer Institute (U01 CA195547: MMH and LLR, Principal Investigators; Cancer Center Support CORE grant CA21765: C. Roberts, Principal Investigator). This study is also supported by R01 CA216354 (YY and JZ, Principal Investigators) from the National Cancer Institute and the American Lebanese Syrian Associated Charities, Memphis, Tennessee.
Footnotes
DECLARATION OF INTERESTS
None declared.
References
- 1.Mueller S, Fullerton HJ, Stratton K, Leisenring W, Weathers RE, Stovall M, et al. Radiation, Atherosclerotic Risk Factors, and Stroke Risk in Survivors of Pediatric Cancer: A Report From the Childhood Cancer Survivor Study. Int J Radiat Oncol 2013;86(4):649–55 doi 10.1016/j.ijrobp.2013.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fullerton HJ, Stratton K, Mueller S, Leisenring WW, Armstrong GT, Weathers RE, et al. Recurrent stroke in childhood cancer survivors. Neurology 2015;85(12):1056–64 doi 10.1212/Wnl.0000000000001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haddy N, Mousannif A, Tukenova M, Guibout C, Grill J, Dhermain F, et al. Relationship between the brain radiation dose for the treatment of childhood cancer and the risk of long-term cerebrovascular mortality. Brain 2011;134:1362–72 doi 10.1093/brain/awr071. [DOI] [PubMed] [Google Scholar]
- 4.Mueller S, Sear K, Hills NK, Chettout N, Afghani S, Gastelum E, et al. Risk of First and Recurrent Stroke in Childhood Cancer Survivors Treated With Cranial and Cervical Radiation Therapy. Int J Radiat Oncol 2013;86(4):643–8 doi 10.1016/j.ijrobp.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorresteijn LDA, Kappelle AC, Scholz NMJ, Munneke M, Scholma JT, Balm AJM, et al. Increased carotid wall thickening after radiotherapy on the neck. Eur J Cancer 2005;41(7):1026–30 doi 10.1016/j.ejca.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 6.Keene DL, Johnston DL, Grimard L, Michaud J, Vassilyadi M, Ventureyra E. Vascular complications of cranial radiation. Child Nerv Syst 2006;22(6):547–55 doi 10.1007/s00381-006-0097-4. [DOI] [PubMed] [Google Scholar]
- 7.Omura M, Aida N, Sekido K, Kakehi M, Matsubara S. Large intracranial vessel occlusive vasculopathy after radiation therapy in children: Clinical features and usefulness of magnetic resonance imaging. Int J Radiat Oncol 1997;38(2):241–9 doi Doi 10.1016/S0360-3016(97)82497-2. [DOI] [PubMed] [Google Scholar]
- 8.Ullrich NJ, Robertson R, Kinnamon DD, Kieran MW, Turner CD, Chi SN, et al. Moyamoya following cranial irradiation for primary brain tumors in children. Neurology 2007;68(12):932–8 doi DOI 10.1212/01.wnl.0000257095.33125.48. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Wilson CL, Easton J, Thrasher A, Mulder H, Liu Q, et al. Genetic Risk for Subsequent Neoplasms Among Long-Term Survivors of Childhood Cancer. J Clin Oncol 2018;36(20):2078–87 doi 10.1200/JCO.2018.77.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. Plos Med 2015;12(3) doi ARTN e1001779 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudson MM, Ehrhardt MJ, Bhakta N, Baassiri M, Eissa H, Chemaitilly W, et al. Approach for Classification and Severity Grading of Long-term and Late-Onset Health Events among Childhood Cancer Survivors in the St. Jude Lifetime Cohort. Cancer Epidem Biomar 2017;26(5):666–74 doi 10.1158/1055-9965.Epi-16-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li MY, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38(16) doi ARTN e164 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lizio M, Harshbarger J, Abugessaisa I, Noguchi S, Kondo A, Severin J, et al. Update of the FANTOM web resource: high resolution transcriptome of diverse cell types in mammals. Nucleic Acids Res 2017;45(D1):D737–D43 doi 10.1093/nar/gkw995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 2015;16 doi ARTN 22 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38(8):904–9 doi 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 16.Wu MC, Lee S, Cai TX, Li Y, Boehnke M, Lin XH. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. Am J Hum Genet 2011;89(1):82–93 doi 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet 2018;50(4):524–37 doi 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heagerty PJ, Zheng YY. Survival model predictive accuracy and ROC curves. Biometrics 2005;61(1):92–105 doi DOI 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 19.Howard G, Moy CS, Howard VJ, McClure LA, Kleindorfer DO, Kissela BM, et al. Where to Focus Efforts to Reduce the Black-White Disparity in Stroke Mortality: Incidence Versus Case Fatality? Stroke 2016;47(7):1893–8 doi 10.1161/Strokeaha.115.012631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacco RL, Boden-Albala B, Gan R, Chen X, Kargman DE, Shea S, et al. Stroke incidence among white, black, and Hispanic residents of an urban community - The Northern Manhattan Stroke Study. Am J Epidemiol 1998;147(3):259–68. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Leisenring WM, Ness KK, Robison LL, Armstrong GT, Yasui Y, et al. Racial/Ethnic Differences in Adverse Outcomes Among Childhood Cancer Survivors: The Childhood Cancer Survivor Study. J Clin Oncol 2016;34(14):1634–43 doi 10.1200/Jco.2015.66.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40(D1):D930–D4 doi 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdellatif AM, Ogata K, Kudo T, Xiafukaiti G, Chang YH, Katoh MC, et al. Role of large MAF transcription factors in the mouse endocrine pancreas. Exp Anim Tokyo 2015;64(3):305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C, Guo ZM. Multiple functions of Maf in the regulation of cellular development and differentiation. Diabetes-Metab Res 2015;31(8):773–8 doi 10.1002/dmrr.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim KH, Rosen A, Bruneau BG, Hui CC, Backx PH. Iroquois Homeodomain Transcription Factors in Heart Development and Function. Circ Res 2012;110(11):1513–24 doi 10.1161/Circresaha.112.265041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.