Abstract
Bromodomain and extra-terminal motif (BET) protein BRD4 binds to acetylated histones at enhancers and promoters through its bromodomains (BDs) to regulate transcriptional elongation. Here, we reveal in human colorectal cancer cells that BRD4 is recruited to enhancers that are co-occupied by mutant p53 and support the synthesis of enhancer-directed transcripts (eRNAs) in response to chronic immune signaling. We identify that BRD4 selectively associates with eRNAs that are produced from BRD4 bound enhancers. Through biochemical and biophysical characterizations, we show that BRD4 BDs function cooperatively as docking sites for eRNAs and that the BDs of BRD2, BRD3, BRDT, BRG1, and BRD7 directly interact with eRNAs. BRD4-eRNA interactions increase BRD4 binding to acetylated histones and promote enhanced BRD4 recruitment at specific enhancers which augments BRD4 transcriptional activities. This work highlights a mechanism by which eRNAs play a direct role in gene regulation by modulating enhancer interactions and transcriptional functions of BRD4.
Main
Enhancers shape gene expression programs by coalescing information from environmental stimuli and the coordinated activities of transcription factors and cofactors1–5. Our ability to annotate enhancers and their activity states stems from the identification of epigenomic signatures that include an accumulation of the histone mark, histone H3 lysine 4 monomethylation (H3K4me1) that together with histone H3 lysine 27 acetylation (H3K27ac) demarcates active enhancers3,6,7–9. A number of transcriptional coregulators bind to acetylated histones through bromodomain (BD) modules to affect chromatin accessibility and transcriptional activation at active enhancers10,11. The affinity and selectivity of BD interactions with acetylated lysines are typically weak, but can be increased by BD interactions with multiple acetylation sites within the histone tail12–14, suggesting that additional mechanisms regulating BD binding at enhancers are likely to have important implications for chromatin and gene regulation.
Bromodomain and extra-terminal motif (BET) proteins bind to histone acetylation marks through their tandem BDs (BD1 and BD2)10,15. BRD4, a well-studied member of the BET family binds to acetylated histones and non-histone proteins at enhancers and promoters10,16–18 to regulate gene expression programs that play pivotal roles in inflammation and cancer development19–21. BRD4 has been shown to associate with transcriptional regulators, that include TWIST, p53, C/EBPα, C/EBPβ, ERG, and NFκB, which has provided increasing support for its role in enhancer and gene regulation22–28. Selective inhibitors of the BET family including JQ129 have demonstrated that BRD4 binding at enhancers supports super-enhancer formation25,30,31. BRD4 also promotes RNAPII elongation through its recruitment of active Positive Elongation Factor-b (P-TEFb)32,33 and its histone chaperone activity34, which affects the production of enhancer-derived transcripts (eRNA)34,35. While there are clear links between BRD4 and gene regulation, the mechanisms underlying the enhancer and gene-specific targeting and functions of BRD4 remain elusive.
eRNAs have been implicated in the regulation of gene expression in multiple cell types and in response to different stimuli36–44. Recent studies have linked eRNAs to the regulation of enhancer-promoter interaction interfaces38,41, chromatin remodeling43, RNAPII pause release45, and the recruitment of general cofactors that include cohesin38,41, Mediator46, and CBP47. While eRNAs are well positioned to add a layer of complexity to enhancer and gene regulation, the molecular basis by which they function remains poorly understood.
In this study, we show that BDs function as RNA binding modules. Notably, we find that eRNAs exhibit cooperativity with acetylated histone lysines in vitro and at active enhancers to augment BRD4 binding and transcriptional activity. Collectively, our results provide evidence for a feedforward epigenetic mechanism in which eRNAs through BRD4 interactions support an active enhancer landscape that modulates proinflammatory gene expression.
Results
BRD4 binds and functions at p53R273H,P309S bound enhancers in response to chronic immune signaling
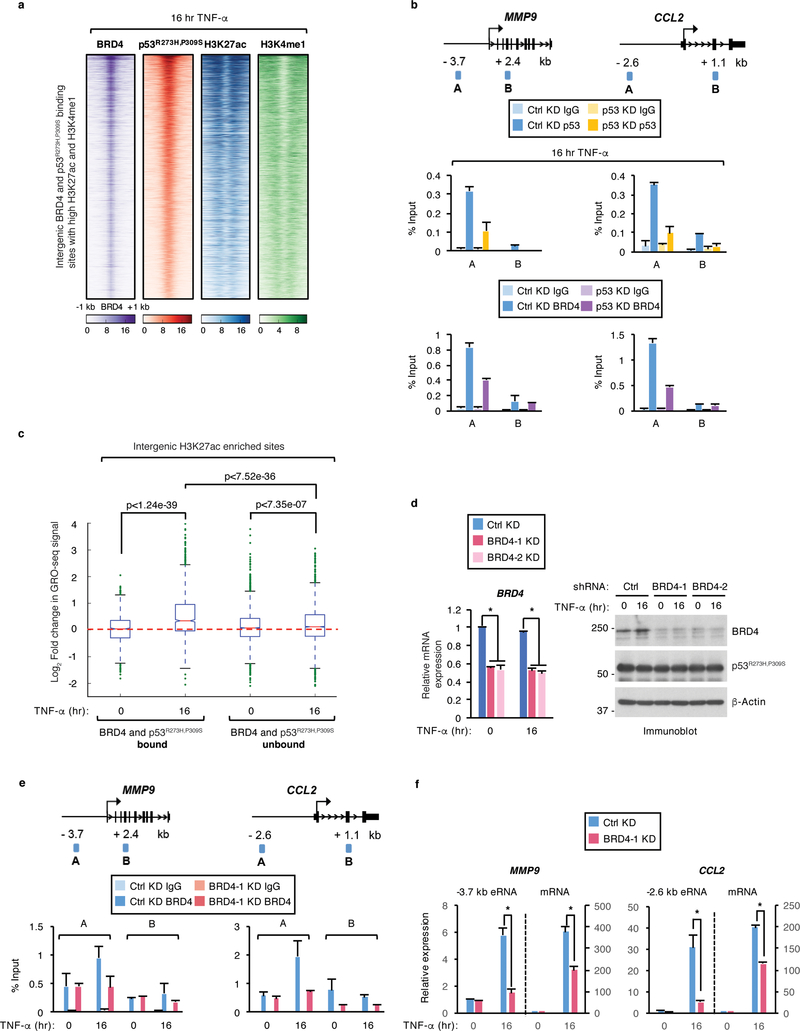
We sought to investigate whether BRD4 regulates recently identified enhancers that are activated by p53R273H,P309S in response to chronic immune signaling44. Using genome-wide chromatin immunoprecipitation followed by sequencing (ChIP-seq) in human SW480 colon cancer cells, expressing p53R273H,P309S (hereafter mutp53) and treated with tumor necrosis factor alpha (TNF-α) for 16 hr, we identified stringent BRD4 binding peaks (n=21,528, p-value < 10−5). Comparative analyses of the BRD4 peaks with previously published mutp53, H3K4me1, and H3K27ac ChIP-seq data in response to chronic TNF-α signaling44 showed colocalization of BRD4 binding peaks with the peak sites for mutp53 at intergenic sites showing enrichment of H3K4me1 and H3K27ac (Fig. 1a). We identified that approximately a third (28%, n=5,949, p-value < 10−5) of the total BRD4 binding sites occur at intergenic sites overlapping with active enhancers and that over 80% (n= 4,884, p-value < 10−5) of the BRD4 bound enhancers are also occupied by mutp53 (Supplementary Fig. 1a). Furthermore, de novo motif analysis identified that among the most highly enriched motifs to overlap with BRD4 and mutp53 peaks are those recognized by NFκB/p65 and EWS:ERG fusion (ETS) (Supplementary Fig. 1b), which is consistent with our previous findings that NFκB recruits mutp53 to active enhancers in response to chronic immune signaling44. We also performed sequential ChIP followed by quantitative PCR (qPCR) using primer sets against downstream control regions (amplicon B) and the enhancer (amplicon A) regions of MMP9 and CCL2, which are among the enhancers that we previously found to be activated by mutp53 and NFκB in response to chronic TNF-α signaling44. These assays revealed simultaneous binding of mutp53 and BRD4 at the MMP9 and CCL2 enhancers, but not the control regions in response to chronic TNF-α signaling (Supplementary Fig. 1c). We identified by Co-IP that BRD4 and mutp53 form physiological associations in SW480 cells before and following TNF-α treatment (Supplementary Fig. 1d). Using purified proteins, we also found that BRD4 forms direct interactions with p53 R273H and wild-type (WT) p53 (Supplementary Fig. 1e), which is similar to previous reports showing BRD4 interactions with WT p5324. We next assessed mutp53 contributions to BRD4 binding at enhancers by ChIP-qPCR analyses in SW480 cells expressing doxycycline-inducible short hairpin RNAs (shRNAs) against mutp53 following TNF-α treatment for 16 hr. Relative to a non-targeting shRNA against LacZ (Ctrl), p53 shRNA markedly reduced mutp53 protein levels after TNF-α treatment without affecting BRD4 protein levels (Supplementary Fig. 1f). As shown in Fig. 1b, mutp53 knockdown resulted in a comparable loss of mutp53 binding at the MMP9 (66%) and CCL2 (72%) enhancer regions that resulted in a significant reduction in BRD4 binding at the MMP9 (52%) and CCL2 (65%) enhancers. Together, these results establish functional mutp53-BRD4 interactions and underscore the requirement for mutp53 in regulating BRD4 binding at active enhancers in response to chronic TNF-α signaling.
Figure 1. BRD4 co-occupies and regulates mutp53 bound enhancers in response to chronic TNF-α signaling.
a, Heatmaps of BRD4 and p53R273H,P309S binding at intergenic regions enriched for H3K27ac and H3K4me1 in SW480 cells treated with TNF-α for 16 hr. Each row shows ±1 kb centered on BRD4 peaks. b, (Top) schematics of ChIP-qPCR amplicons and (bottom) ChIP-qPCR analyses of IgG, p53R273H,P309S, and BRD4 enrichment at the enhancer (amplicon A) and non-specific (amplicon B) regions of the MMP9 and CCL2 gene loci in SW480 cells expressing LacZ (Ctrl) or p53 shRNA and treated with TNF-α for 16 hr. c, Log2 fold change in GRO-seq signal in response to TNF-α signaling at intergenic regions enriched for H3K27ac that were co-bound or unbound by both BRD4 and p53R273H,P309S. Boxplots enclose values between first and third quartiles, midlines show medians, and whiskers extend to data points within 1.5x the interquartile range from the box. Statistical significance was determined by Mann-Whitney U test. d, qRT-PCR and immunoblot analysis of SW480 cells expressing control (Ctrl) or two BRD4 shRNAs (BRD4–1, BRD4–2) and treated with TNF-α for 0 or 16 hr. n = 3 experiments. Uncropped blots are shown in Supplementary Data Set 1. e, ChIP-qPCR analysis of BRD4 binding in SW480 cells treated as described in (d) at the MMP9 and CCL2 gene loci. In (b) and (e), data represent the mean and s.e.m. of n = 2 independent ChIP-qPCR experiments that are representative of at least three replicates. f, qRT-PCR analyses of MMP9 and CCL2 eRNAs and mRNAs in SW480 cells treated as described in (d). Expression levels shown after TNF-α are relative to the levels before treatment and data represent the mean and s.e.m. of n = 3 independent experiments. Statistical significance was determined by two-tailed Student’s t-test. * indicates p-value <0.05.
We next examined the global levels of nascent transcription (+/− TNF-α) centered upon the enhancer-specific BRD4 and mutp53 ChIP-seq peaks using our global run-on sequencing (GRO-seq) data44. As revealed in Fig. 1c, markedly induced eRNA levels following 16 hr TNF-α treatment were identified at H3K27ac-enriched intergenic sites that are co-occupied by BRD4 and mutp53. Notably, the TNF-induced eRNA levels observed at the mutp53 and BRD4 co-bound enhancers were significantly higher relative to the TNF-induced eRNA levels at H3K27ac-enriched intergenic sites that were not co-bound by BRD4 and mutp53 (Fig. 1c). To examine a direct role for BRD4 in the regulation of TNF-α-induced enhancer transcription, we established SW480 cells stably expressing nontargeting (Ctrl) or two different shRNAs specific to BRD4 (BRD4–1 and BRD4–2). Relative to the control knockdown, both BRD4 shRNAs decreased BRD4 mRNA and protein levels (Fig. 1d). ChIP-qPCR analyses in control knockdown cells (+/− TNF-α) revealed that BRD4 binding at the MMP9 and CCL2 enhancers is increased (53% and 70%, respectively) in response to chronic TNF-α signaling (Fig. 1e). As expected, BRD4 knockdown resulted in a significant loss of TNF-α-inducible BRD4 binding at the MMP9 and CCL2 enhancers (55% and 63%, respectively) (Fig. 1e) that resulted in a significant decrease in MMP9 and CCL2 eRNA levels (four-fold and six-fold, respectively) and a two-fold decrease in the mRNA levels of both genes (Fig. 1f). Under uninduced conditions, BRD4 knockdown had little to no effect on BRD4 binding and the eRNA/mRNA expression levels at the MMP9 and CCL2 enhancers (Figs. 1e,f). The decreased expression of MMP9 and CCL2 eRNAs/mRNAs following BRD4 knockdown was independent of a decrease in mutp53 binding at the MMP9 and CCL2 enhancers (Supplementary Fig. 1h), which taken together with our mutp53 knockdown analyses (Fig. 1b) reveals that while mutp53 affects BRD4 binding, BRD4 is not required for mutp53 binding at the MMP9 and CCL2 enhancers. Consistent with the BRD4 knockdown data (Fig. 1e,f), a second shRNA oligo against BRD4 (Supplementary Fig. 1g) and BRD4 inhibition with JQ1 treatment also revealed a significant decrease in the TNF-α-inducible MMP9 and CCL2 eRNA/mRNA levels (Supplementary Fig. 1i). This identified role for BRD4 in regulating the TNF-induced expression of eRNAs and tumor promoting genes is consistent with our previous observations revealing that mutp53 is also required to support TNF-α-inducible eRNA/mRNA expression levels in SW480 colon cancer cells44. Collectively, these data suggest that BRD4 acts coordinately with mutp53 at active enhancers to support potent levels of eRNA synthesis and gene expression in response to chronic TNF-α signaling.
BRD4 associates with RNAs synthesized from genomic regions occupied by BRD4
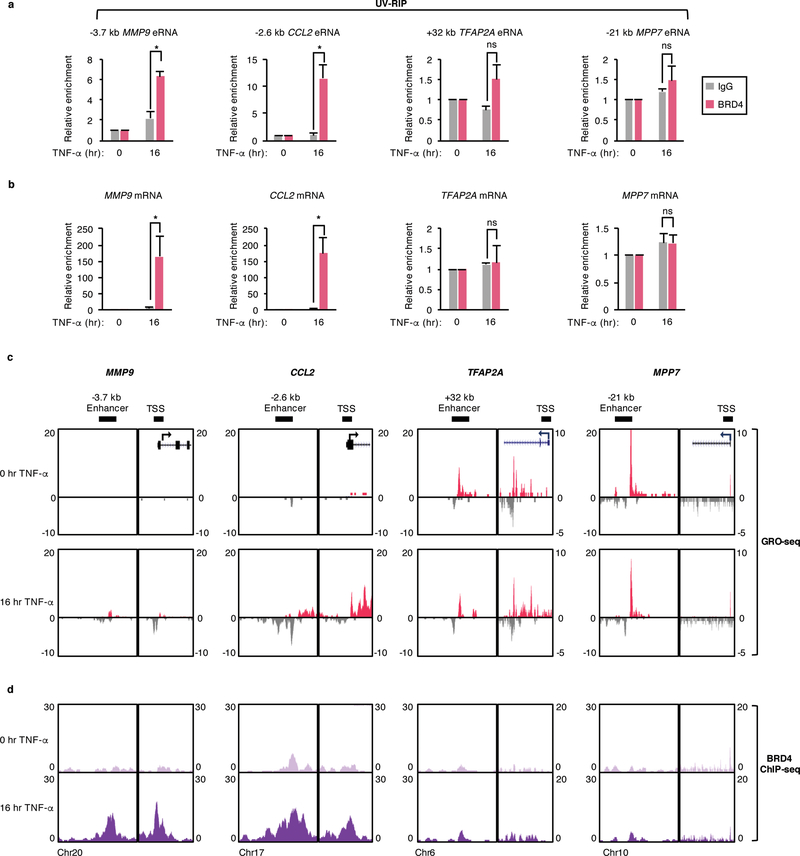
Given the overlap between BRD4 enrichment and eRNA synthesis in response to chronic TNF-α signaling and the previously described BRD4 association with RNAPII elongation complexes34, we examined whether BRD4 associates with RNAs by performing UV-crosslinked RNA immunoprecipitation (UV-RIP) in SW480 cells (+/− TNF-α). BRD4 mRNA and protein levels were unaffected by chronic TNF-α treatment in this cell line (Supplementary Fig. 2a). In TNF-α-treated cells, a BRD4 antibody specifically co-immunoprecipitated eRNAs produced from the MMP9, CCL2, CSF2, and TNFAIP3 but not the TFAP2A and MPP7 enhancers (Fig. 2a; Supplementary Fig. 2b) despite the TFAP2A and MPP7 eRNAs being expressed at comparable or higher levels relative to the MMP9, CCL2, CSF2, and TNFAIP3 eRNAs (Fig. 2c; Supplementary Fig. 2c). We also found that BRD4 associates with MMP9 and CCL2 but not the TFAP2A and MPP7 mRNAs in response to chronic TNF-α signaling (Fig. 2b). Under uninduced conditions, BRD4 was not found to associate with any of the analyzed RNAs (Figs. 2a,b; Supplementary Fig. 2b). The specific association of BRD4 with RNAs in response to chronic TNF-α signaling is consistent with the significantly higher TNF-α-induced versus uninduced RNA expression levels (Fig. 2c; Supplementary Fig. 2c). Notably, BRD4-RNA associations are also consistent with BRD4 binding levels, which were found to be significantly higher at the MMP9 and CCL2 relative to the TFAP2A and MPP7 enhancers and transcription start sites (TSSs) in response to chronic TNF-α signaling (Fig. 2d). Also consistent, the TNF-α-induced associations of BRD4 with the CSF2 and TNFAIP3 eRNAs (Supplementary Fig. 2b) parallels with the high levels of BRD4 binding at the corresponding enhancers (Supplementary Fig. 2c).
Figure 2. BRD4 associates with RNAs synthesized from genomic regions occupied by BRD4.
a, qRT-PCR analysis of MMP9, CCL2, TFAP2A, and MPP7 eRNAs and b, mRNAs following UV-RIP with IgG or BRD4 antibodies in SW480 cells treated with TNF-α for 0 or 16 hr. Enrichment levels for each TNF-α treated IP is relative to the levels before TNF-α treatment and data represent the mean and s.e.m. of n = 3 independent experiments. Statistical significance was determined by two-tailed Student’s t-test. * indicates p-value <0.05. ns: no significance. c, UCSC genome browser images of GRO-seq and d, BRD4 ChIP-seq signals in SW480 cells treated as described in (b) at active enhancer regions and TSSs adjacent to the MMP9, CCL2, TFAP2A, and MPP7 gene loci.
We next explored direct interactions between recombinant BRD4 full-length (FL) and several RNAs and revealed that BRD4 interacts with the in vitro transcribed MMP9 and CCL2 eRNAs, MEG3 long noncoding RNA (lncRNA), and p21 exonic RNA (Supplementary Fig. 2d). Together with our UV-RIP analyses, these findings suggest that BRD4 displays broad rather than sequence-specific RNA interactions and that BRD4-RNA associations occur in an enhancer/locus-specific manner with BRD4 forming associations with RNAs synthesized from genomic regions that are enriched for BRD4 binding.
BRD4 directly interacts with eRNAs through its tandem bromodomains
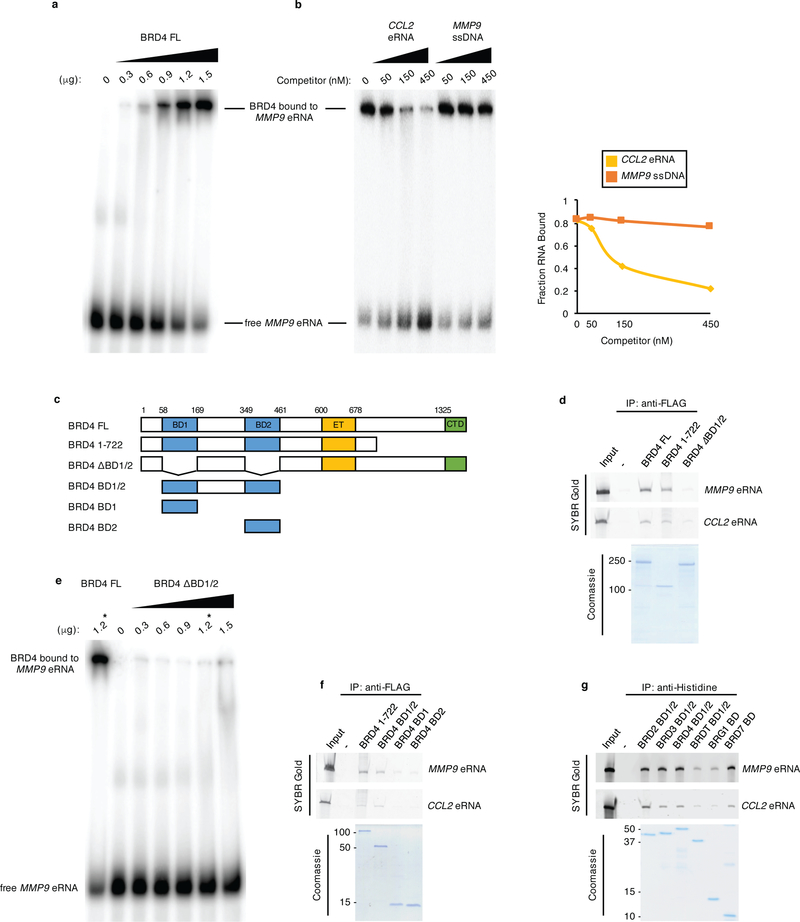
To further investigate the direct binding of BRD4 to eRNAs, electrophoretic mobility shift assays (EMSAs) were performed. As revealed in Fig. 3a, the incubation of purified BRD4 FL and a 32P-labeled MMP9 eRNA probe revealed a single prominent BRD4-MMP9 eRNA complex. Quantification of the RNA EMSA with increasing doses of BRD4 FL revealed that ~80% of the MMP9 eRNA was bound by BRD4 FL at the highest titration of BRD4 FL (Supplementary Fig. 2e). Competition EMSAs revealed that BRD4 binding to the labeled MMP9 eRNA probe was specifically competed by excess amounts of unlabeled CCL2 eRNA (Fig. 3b) but that comparable amounts of cold MMP9 single stranded DNA (ssDNA) was notably less efficient (Fig. 3b), which demonstrates that BRD4 binds preferentially to RNA, in vitro.
Figure 3. BRD4 directly interacts with eRNAs through its tandem bromodomains.
a, RNA EMSA performed with in vitro transcribed, 32P-labeled MMP9 eRNA and titrations of recombinant BRD4 FL. b, (Left) competition binding RNA EMSAs in which binding of 32P-labeled MMP9 eRNA to 0.9 μg of BRD4 FL was competed with 50, 150, and 450 nM unlabeled CCL2 eRNA and MMP9 ssDNA. (Right) quantification of the fraction of RNA bound to BRD4 FL in the presence of unlabeled competitors as denoted, corresponding to the autoradiogram shown in the left panel. c, Schematics of BRD4 proteins expressed, purified, and used in RNA EMSA and in vitro RNA binding assays. d, f, In vitro pull-down of MMP9 and CCL2 eRNAs with the indicated FLAG-tagged BRD4 proteins and g, the His-tagged tandem BDs of BRD2, BRD3, BRD4, and BRDT and the single BDs of BRG1 and BRD7 as revealed by SYBR Gold staining and SDS-PAGE/Coomassie staining analyses of the purified proteins. e, RNA EMSA performed with in vitro transcribed, 32P-labeled MMP9 eRNA and titrations of recombinant BRD4 ΔBD1/2. One reaction with 1.2 μg of BRD4 FL was also performed as control and denoted with *. n = 3 independent experiments for all RNA EMSAs and in vitro binding assays with representative images for each assay shown in the corresponding panels. Uncropped gel images are shown in Supplementary Data Set 1.
To identify the eRNA-interacting domains of BRD4, we performed RNA pulldowns using BRD4 deletion mutants (Fig. 3c) and in vitro transcribed MMP9 and CCL2 eRNAs. Both BRD4 FL and the naturally existing BRD4 isoform spanning amino acids 1–72248 bind comparably to the MMP9 and CCL2 eRNAs (Fig. 3d). We next tested the contributions of the BRD4 BDs since these domains localize within the eRNA interacting BRD4 (1–722) protein. Notably, BRD4 that is devoid of both BDs (Fig. 3c, BRD4 ΔBD1/2) showed a nearly complete loss of binding to the MMP9 and CCL2 eRNAs (Fig. 3d). Similarly, RNA EMSAs revealed that radiolabeled MMP9 eRNA did not display a prominent mobility shift with increasing titrations of BRD4 ΔBD1/2 protein (Fig. 3e). Specifically, less than 5%, relative to the nearly 80% of the labeled eRNA was bound by BRD4 ΔBD1/2 as compared to BRD4 FL, respectively (Fig. 3e; Supplementary Fig. 2e).
We next tested whether the individual BRD4 BDs (Fig. 3c, BD1 or BD2) are sufficient to support BRD4-eRNA interactions. We found that neither BD1 nor BD2 are able to bind to the MMP9 and CCL2 eRNAs as strongly as the BRD4 (1–722) protein (Fig. 3f). In comparison, a BRD4 protein (Fig. 3c; BRD4 BD1/2) consisting of both BDs and the linker sequence that joins them revealed a level of eRNA binding that is comparable to BRD4 (1–722) (Fig. 3f). As shown in Fig. 3g, the tandem BDs (BD1/2) of the BET family members BRD2, BRD3, BRD4, BRDT, and the single BDs of the non-BET proteins, BRG1 and BRD7 bind to MMP9 and CCL2 eRNAs with the eRNA interaction levels of BRG1 and BRDT BDs relatively lower as compared to the BDs of the other proteins. Together, these findings demonstrate that BDs directly interact with eRNAs and that the tandem BRD4 BDs function cooperatively to facilitate BRD4-eRNA interactions.
eRNAs enhance BRD4 binding to acetylated histone H3 and H4 peptides and histone octamers
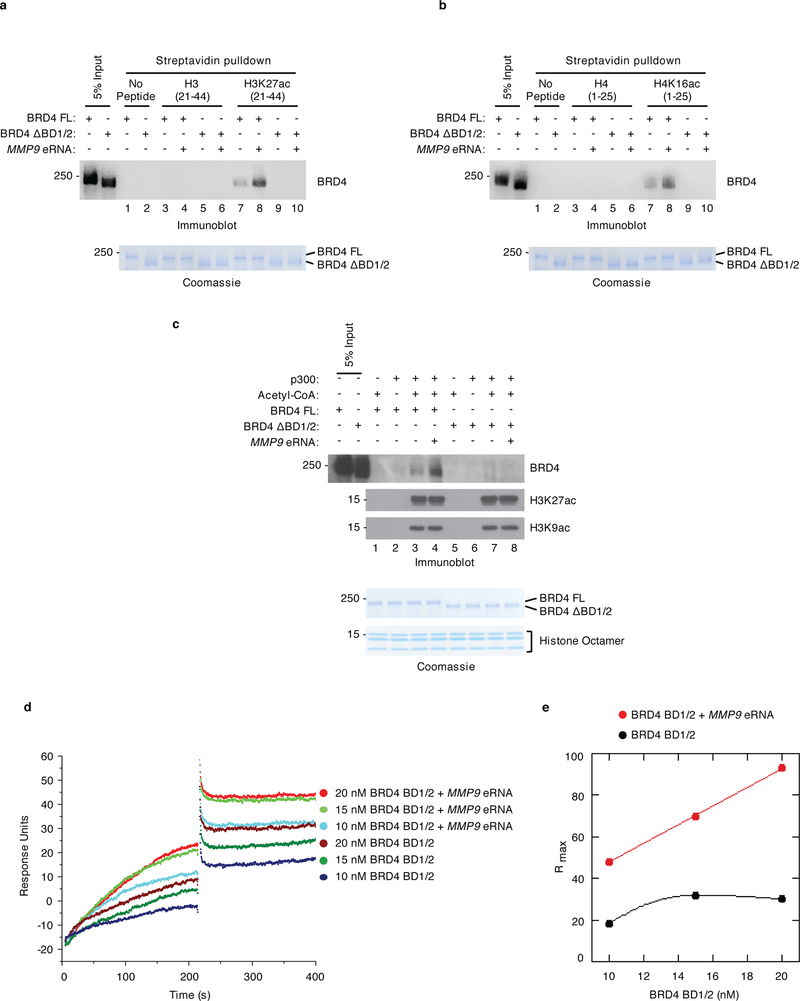
To investigate the significance of eRNA-BRD4 interactions in the regulation of BRD4 BD binding to acetylated histones, in vitro binding assays with unmodified or acetylated H3 and H4 histone peptides were performed. As expected, BRD4 FL (Figs. 4a,b lane 7 versus 3), but not BRD4 ΔBD1/2 (Figs. 4a,b lane 9 versus 5) bound more strongly to H3K27ac- and H4K16ac-modified versus unmodified peptides. Notably, the binding of BRD4 FL to acetylated H3K27 and H4K16 peptides was increased in the presence of MMP9 eRNA (Figs. 4a,b lane 8 versus 7). In addition, eRNAs were not found to promote BRD4 binding in the absence of BRD4 interactions with acetylated peptides, as demonstrated by the inability of eRNAs to support BRD4 FL binding to unmodified histone H3 and H4 peptides (Figs. 4a,b lane 4 versus 3) and the lack of an eRNA effect on BRD4 ΔBD1/2 binding to acetylated H3K27 and H4K16 peptides (Figs. 4a,b lane 10 versus 9).
Figure 4. eRNAs cooperate with acetylated histones to enhance BRD4 binding, in vitro.
Immobilized peptide pulldown assays using biotinylated a, H3 peptides (unmodified or K27acetylated-modified) and b, H4 peptides (unmodified or K16acetylated-modified) with either recombinant BRD4 FL or BRD4 ΔBD1/2 in the absence or presence of refolded MMP9 eRNA as indicated. Recombinant BRD4 FL and BRD4 ΔBD1/2 were detected by immunoblotting with an antibody specific to BRD4. c, Immunoblot analysis of in vitro binding assays with unacetylated or acetylated histone octamers, recombinant BRD4 FL or ΔBD1/2, and refolded MMP9 eRNA as indicated. Immunoblot analysis with H3K27ac and H3K9ac antibodies confirmed the p300/acetyl-CoA-mediated acetylation of the histone octamers. Lower panels in (a), (b), and (c) represent the loading of the indicated proteins by Coomassie staining. d, Analysis of BRD4 BD1/2 binding to immobilized, biotin-labeled H3K27ac peptides in the absence or presence of refolded MMP9 eRNA as measured in response unit (RU) by surface plasmon resonance. Representative sensorgrams were obtained from injections of 10, 15, and 20 nM of BRD4 BD1/2 with and without 0.2 nM of MMP9 eRNA in binding buffer for 300 s at 50 μl min−1. e, Rmax plot depicting the maximal binding capacity of BRD4 BD1/2 at various concentrations in the absence (black) and presence (red) of refolded MMP9 eRNA. n = 3 independent experiments for all in vitro binding assays with representative images for each assay shown in the corresponding panels. Uncropped blot and gel images are shown in Supplementary Data Set 1.
We also examined the effects of eRNAs on BRD4 binding to histone octamers that were either unacetylated or acetylated by the histone acetyltransferase p300 (Supplementary Fig. 3a). As predicted, p300 efficiently acetylated the histone octamers as demonstrated by the increased acetylation of H3K27 and H3K9 (Fig. 4c, lanes 3, 4, 7, and 8) and little to no acetylation was detected in the absence of p300 or acetyl-CoA (Fig. 4c, lanes 1, 2, 5, and 6). Acetylated histone octamers were coupled to magnetic beads using H3K27ac and H3K9ac antibodies and then incubated with BRD4 in the presence or absence of MMP9 eRNA. Similar to the histone peptide binding assay results, we found that MMP9 eRNA increased BRD4 FL (Fig. 4c, lane 4 versus 3), but not BRD4 ΔBD1/2 binding to acetylated histone octamers (Fig. 4c, lane 8 versus 7). The enhanced binding of BRD4 to acetylated histone peptides and octamers was confirmed in the presence of the CCL2 eRNA (Supplementary Figs. 3b,c). Moreover, eRNA titration experiments revealed that several concentrations of the MMP9 eRNA enhanced BRD4 binding to acetylated histone octamers (Supplementary Fig. 3d, lanes 3, 4, and 5 versus 2) while high molar ratios of MMP9 eRNA inhibited BRD4 binding (Supplementary Fig. 3d, lanes 6 and 7 versus 2).
To gain insight into the steady state binding of BRD4 to acetylated histone H3 in the presence or absence of eRNAs, surface plasmon resonance (SPR) analyses were performed. Sensorgrams resulting from various concentrations of BRD4 BD1/2 protein flowed over immobilized H3K27ac-modified peptides in the presence or absence of MMP9 eRNA revealed that the amount of bound BRD4 BD1/2 was enhanced in the presence of the eRNA to yield an absolute level of binding approximately threefold higher than observed in the absence of eRNA (Fig. 4d). An appreciable dissociation was not detected as BRD4 BD1/2 remained bound to the H3K27ac-modified peptide during the time course of the experiments (Fig. 4d). Notably, the sensorgrams also demonstrated that all concentrations of BRD4 BD1/2 reached a significantly higher maximum binding capacity (Rmax) in the presence versus absence of eRNA (Fig. 4e). These biophysical and biochemical findings indicate that eRNA interactions with BRD4 BDs play a pivotal role in the enhanced binding of BRD4 to acetylated histones, in vitro.
eRNAs modulate BRD4 enhancer occupancy to maintain enhancer and gene activation
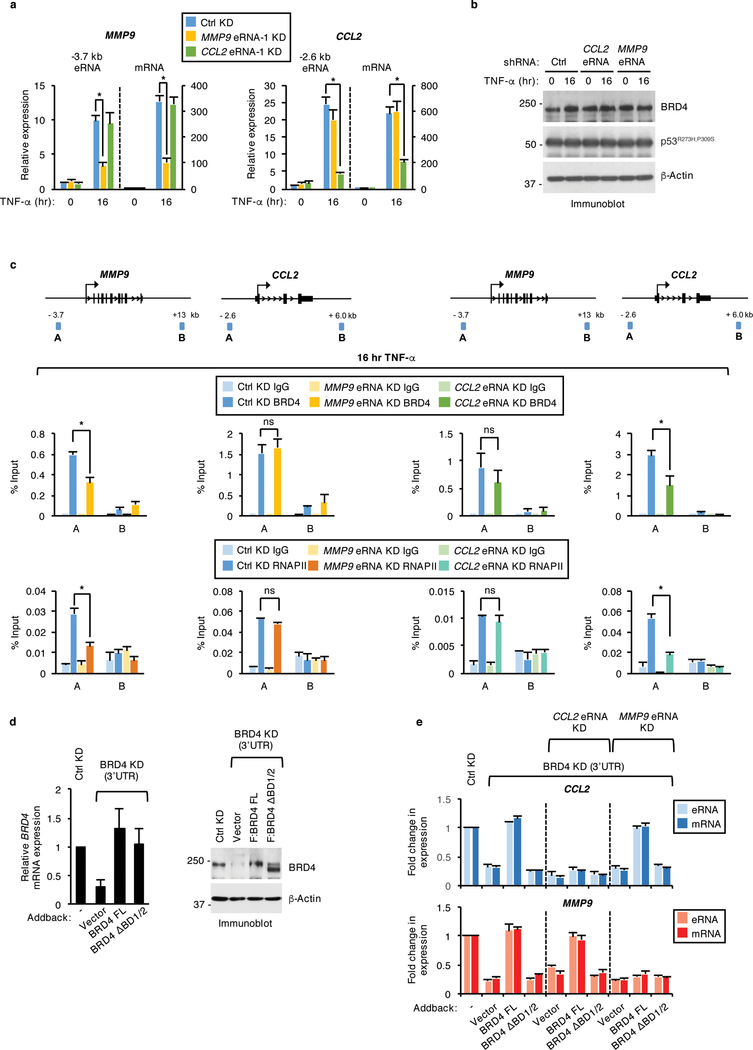
To investigate the biological significance of BRD4-eRNA interactions, we first employed shRNA-mediated knockdown of eRNAs in SW480 cells (+/− TNF-α). qRT-PCR analysis revealed that relative to a nontargeting control, shRNAs that target the MMP9 (MMP9 eRNA-1) and CCL2 (CCL2 eRNA-1) eRNAs specifically decrease the TNF-α-induced levels of MMP9 and CCL2 eRNAs by approximately three-fold and six-fold, respectively (Fig. 5a). Notably, this decrease in MMP9 and CCL2 eRNA levels resulted in a three-fold decrease in mRNA levels of MMP9 and CCL2, respectively, thereby revealing a direct role for these eRNAs in the induction of proinflammatory gene expression (Fig. 5a). This reduction in TNF-α-induced MMP9 and CCL2 mRNA levels was further confirmed with a second shRNA targeting the MMP9 (MMP9 eRNA-2) and CCL2 eRNAs (CCL2 eRNA-2) (Supplementary Fig. 4a). Consistent with the specificity of the eRNA knockdown effects, depletion of the MMP9 and CCL2 eRNAs did not affect the eRNA and mRNA expression levels of CCL2 and MMP9, respectively (Fig. 5a and Supplementary Fig. 4a) or that of CPA4 and CYP24A1 that are also significantly induced following TNF-α treatment (Supplementary Fig. 4b). Also, the gene-selective effects of the MMP9 and CCL2 eRNAs are not due to altered mutp53 or BRD4 protein levels, which were found to be unaffected in the eRNA knockdown cells (Fig. 5b).
Figure 5. BRD4 enhancer occupancy and regulation of select eRNAs and genes is modulated by BRD4 interactions with eRNAs.
a, qRT-PCR analysis of MMP9 and CCL2 eRNAs and mRNAs and b, immunoblot analysis of SW480 cells stably expressing control (Ctrl) or MMP9 and CCL2 eRNA shRNAs and treated with TNF-α for 0 or 16 hr. n = 3 independent experiments. c, IgG, BRD4, and RNAPII ChIP-qPCR analyses at the enhancers (A) and nonspecific (B) regions of MMP9 and CCL2 gene loci in SW480 cells described in (b) and treated with TNF-α for 16 hr. Data represent the mean and s.e.m. of n = 2 independent ChIP experiments that are representative of at least three replicates. d, qRT-PCR and immunoblot analysis of BRD4 mRNA and protein levels in SW480 cells that expressed control or BRD4 shRNAs together with empty vector control or shRNA-resistant plasmids expressing BRD4 FL or BRD4 ΔBD1/2 and treated with TNF-α for 16 hr. n = 3 independent experiments. e, qRT-PCR analysis of MMP9 and CCL2 eRNA and mRNA expression levels in single BRD4 or double BRD4 and MMP9 or CCL2 eRNA knockdown SW480 cells that were transiently transfected with shRNA-resistant constructs expressing vector control, BRD4 FL, or BRD4 ΔBD1/2 and treated with TNF-α for 16 hr. For all eRNA and mRNA expression analyses in (a), (d), and (e), the expression levels following TNF-α treatment are relative to the levels before TNF-α exposure and data represent the mean and s.e.m. of n = 3 independent experiments. Statistical significance was determined by two-tailed Student’s t-test. * indicates p-value <0.05. ns: no significance. Uncropped blot images are shown in Supplementary Data Set 1.
We next performed ChIP-qPCR in TNF-α-treated MMP9 and CCL2 eRNA knockdown cells to examine a direct role for these eRNAs in the regulation of BRD4 enhancer occupancy. Notably, BRD4 binding in the MMP9 and CCL2 eRNA-depleted cells was significantly (46% and 49%) reduced at the MMP9 and CCL2 enhancer regions, respectively (Fig. 5c). Consistent with the specificity of eRNA function, MMP9 eRNA knockdown did not affect BRD4 binding at the CCL2 enhancer region and vice versa (Fig. 5c). The decrease in BRD4 binding following eRNA knockdown parallels with a comparable (54% and 66%) reduction in RNAPII binding at the MMP9 and CCL2 enhancers, respectively (Fig. 5c), which is consistent with the previously described roles of BRD4 in supporting RNAPII binding32,33. These results were further confirmed by treating SW480 cells with Actinomycin D (Act D), which inhibits transcription without affecting BRD4 protein levels (Supplementary Fig. 4c). The reduced MMP9 and CCL2 eRNA levels following Act D treatment (Supplementary Fig. 4d) correlated with a notable decrease in TNF-α-induced BRD4 binding by 61% and 77%, at the MMP9 and CCL2 enhancers, respectively (Supplementary Fig. 4e).
We next investigated the effects of eRNAs on BRD4-dependent eRNA and mRNA expression levels. ShRNA-resistant, BRD4 FL or BRD4 ΔBD1/2 constructs were transiently expressed in SW480 cells co-transduced with shRNAs against MMP9 or CCL2 eRNA and BRD4. Relative to the vector control, both BRD4 FL and BRD4 ΔBD1/2 reconstituted BRD4 mRNA and protein levels in BRD4 knockdown cells (Fig. 5d). As expected, BRD4 FL, but not BRD4 ΔBD1/2 was able to rescue TNF-α-inducible MMP9 and CCL2 eRNA/mRNA expression levels in the single BRD4 knockdown cells (Fig. 5e). Notably however, the ability of BRD4 FL to rescue CCL2 eRNA/mRNA expression levels in the double CCL2 eRNA and BRD4 knockdown cell lines was significantly (two to three-fold) less efficient. Moreover, BRD4 FL was comparably less efficient in rescuing MMP9 eRNA/mRNA levels in the double BRD4 and MMP9 knockdown cells (Fig. 5e). BRD4 ΔBD1/2 was unable to rescue MMP9 and CCL2 eRNA/mRNA levels in the eRNA/BRD4 double knockdown cells (Fig. 5e). These rescue experiments, taken together with our in vitro analyses, demonstrate that eRNAs play a pivotal role in regulating BRD4 enhancer binding that supports BRD4-dependent enhancer transcription and gene activation in response to chronic TNF-α signaling.
Discussion
BRD4 regulates a class of mutp53 enhancers and target genes in response to chronic immune signaling
BRD4 localizes at enhancers to regulate the expression of oncogenes that include MYC, which has important implications in human disease19–21,49,50. Our studies advance our understanding of BRD4’s role in human cancer through the identification of this coactivator at enhancers that are co-occupied by the oncogene mutp53. Among the enhancers co-bound by mutp53 and BRD4 are several enhancers that are regulated by mutp53 and NFκB and are linked to the activation of a proinflammatory gene expression program that leads to increased colon cancer cell invasion potential44. In support of a functional relationship between mutp53 and BRD4, we found that BRD4 forms direct interactions and cellular associations with mutp53 and is simultaneously bound with mutp53 at active enhancers in response to chronic TNF-α signaling. We also demonstrated that mutp53 affects BRD4 recruitment at a subset of enhancers following chronic immune signaling. Moreover, our data supports a role for BRD4 in regulating the activation of mutp53 and BRD4 co-bound enhancers as demonstrated by the significantly higher eRNA levels found at BRD4 and mutp53 co-bound versus unbound enhancers and the downregulation of eRNA levels following BRD4 knockdown or inhibition by JQ1. Given the mutp53 and BRD4 cooperativity in colon cancer cells reported in this study, future studies testing BRD4 regulation of mutp53-dependent gene expression programs in other systems are likely to have clinical significance given the widespread roles of mutp53 and BRD4 in cancer and due to the potential for pharmacological inhibition of BRD4.
eRNAs interact with BRD4 to affect transcriptional activation
Given the prevalence of eRNAs in various cell types and in response to stimuli36–44, a number of functions for eRNAs in transcriptional regulation can be envisioned. Our results provide strong evidence that eRNAs stimulate transcription by enhancing BRD4 binding to acetylated histones. Specifically, we demonstrate that eRNAs form physiological associations and direct interactions with the tandem BDs of BRD4, which in turn promotes enhanced BRD4 binding to acetylated forms of histone H3 and H4 peptides, acetylated histone octamers, and active enhancers. Strikingly, eRNA depletion results in a notable decrease in the enhancer binding of BRD4, which coincides with reduced RNAPII occupancy and the downregulation of inflammation-induced gene expression in colon cancer cells. Our studies also underscore the significance of BRD4-eRNA interactions in regulating enhancer-dependent gene activation by revealing the recovery of eRNA and tumor promoting gene expression by BRD4 FL (and not BRD4 ΔBD1/2) in BRD4 single but not BRD4 and eRNA double knockdown cells. Taken together, our studies support a model (Fig. 6) for a positive feedback loop in which eRNAs are involved in the cis recruitment of BRD4 at mutp53 bound enhancers for augmented enhancer and gene activation. Furthermore, this model supports the functional convergence between histone modifications and eRNAs, which emphasizes that eRNAs impact BRD4’s binding potential after it has bound to acetylated histones. This observed cooperativity between epigenetic mechanisms is consistent with previous studies showing that the methyl-lysine recognition chromodomain of CBX7CD binds lncRNA ANRIL51 and further suggest that eRNAs, similar to lncRNAs are likely to function through chromatin effector domains to impact gene regulation. Importantly, our studies did not rule out whether eRNAs regulate other BRD4 functions including its atypical histone modifying activity52.
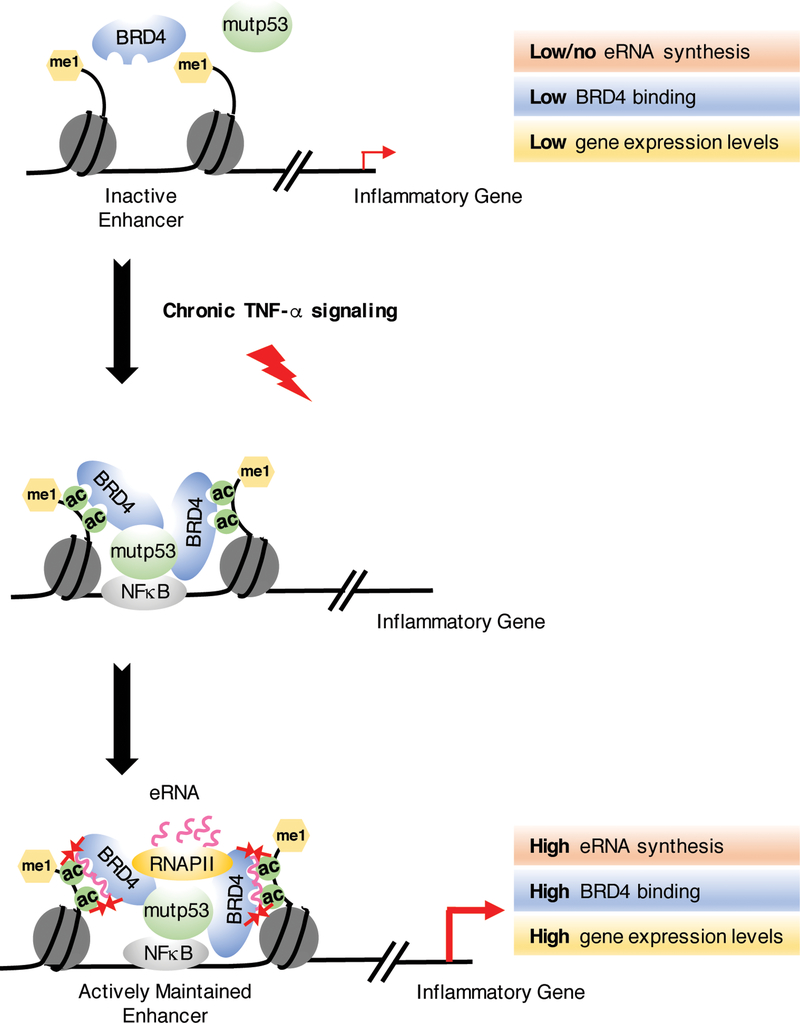
Figure 6. Proposed model for eRNA-mediated BRD4 tethering to acetylated histones at active enhancers.
Upon chronic TNF-α signaling, NFκB recruits mutp53 to a subset of enhancers as we have previously described44. In our current study, we show that BRD4 forms direct interactions with mutp53 and is recruited by mutp53 to these mutp53/NFκB-activated enhancers following chronic immune signaling. Subsequently, eRNAs synthesized from the activated enhancers bind to the bromodomains of BRD4 to enhance its binding to acetylated chromatin. This stabilization of BRD4 at active enhancers contributes to TNF-α-induced production of eRNAs and potent expression of the nearby tumor promoting genes.
We demonstrate that BRD4 binds a broad spectrum of RNAs, which suggests a promiscuous relationship with RNAs. Although RNA sequence does not appear to contribute to the specificity of BRD4-RNA interactions, and while we cannot rule out RNA structural contributions, we have found that BRD4 forms associations with RNAs in an enhancer/locus-specific manner. These results are supported by our BRD4 UV-RIP, BRD4 ChIP-seq, and GRO-seq analyses that together demonstrate BRD4’s ability to form associations with RNAs that are produced specifically from genomic regions that are highly enriched for BRD4. This observation is consistent with a recent study showing that CBP also associates with RNAs that are produced at sites where there exists high levels of CBP binding47. Our findings further demonstrate the ability of BRD4 FL to reconstitute MMP9 and CCL2 eRNA levels in the CCL2 and MMP9, but not the MMP9 and CCL2 eRNA and BRD4 double knockdown cells, respectively, which is further consistent with enhancer/locus-specific eRNA-BRD4 associations.
eRNAs interact with the highly conserved acetyl-lysine recognition bromodomain
Our identification of eRNA function in regulating the chromatin binding and transcriptional coactivator functions of BRD4 could be widespread given that BDs are evolutionarily conserved12 and were mapped in this study as the eRNA-interacting domains of BRD4. In support of this possibility, we identified that the tandem BDs of all BET family members and the single BD-containing, non-BET proteins, BRG1 and BRD7 form interactions with eRNAs. Our observation of variable level of RNA binding by various BDs is consistent with known differences in BD structure and conformation, which are linked to differential BD interactions with acetylated residues53. Moreover, given the development of inhibitors that are specific to the BDs of various members of the BET family54, eRNAs that bind to BDs could be potent therapeutic targets to repress pro-tumorigenic gene expression programs.
Our observation that eRNAs enhance BRD4 binding to acetylated histones and octamers suggest that eRNAs are likely to interact with the BD in a region that is independent of the acetyl lysine-recognition domain. While BDs are categorized into eight distinct classes based on their diverse structures, they share four conserved α-helices that are connected to one another by loops of variable lengths15,53. Conserved amino acids located within these loops facilitate the docking of acetylated lysine residues and a patch of basic amino acids surrounds the acetyl binding pocket that further prompts interactions with acetylated substrates10,53. It is possible that this positively charged patch could facilitate interactions with negatively charged nucleic acids, including eRNAs. This possibility is consistent with recent studies that have shown that the positively charged patch facilitates interactions of the BRDT BD155 and the BRG1/hBRM BDs56 with DNA. Furthermore, point mutations in the positively charged patch of hBRM BD were found to significantly reduce hBRM’s binding to DNA without disrupting its interactions with acetylated peptides56. This finding suggests that BD interactions with nucleic acids are likely to occur outside of the acetyl-lysine recognition domain and thereby could allow for RNA/DNA interactions that enhance BD associations with acetyl-lysines. In addition, differences in the amino acids within the BD loops could contribute to variations in the electrostatic charge that together with variable structural/conformational contributions may predict the variable levels of interaction that we observed between eRNAs and different BDs assessed in our study.
Our demonstration that each of the tandem BRD4 BDs are required to reconstitute interactions with eRNAs and the significance of bivalent interactions of BDs with acetylated lysines and eRNAs is consistent with previous reports revealing the importance of multivalency in chromatin regulation. Specifically, the overall affinity and specificity of chromatin binding can be enhanced by multiple chromatin interacting domains within a single chromatin reader or through multiple chromatin interacting proteins that are present in multiprotein complexes57. Consistent, is the identification that the BDs of the BET family exhibit modest affinity for monoacetylated lysine that is significantly increased when the BDs are anchored to multiple acetylation sites10,14,58. Future studies are needed to extend the implications of BD-RNA interactions and test how BDs engage these epigenetic modifiers to transduce downstream function.
Taken together, our findings are consistent with the emerging notion that eRNAs are functional molecules, rather than merely reflections of enhancer activation or simply transcriptional noise. Our studies provide mechanistic insights into the role of eRNAs that directly interact with BRD4 at active enhancers, which leads to increased RNAPII binding, eRNA synthesis, and the transcription of proinflammatory genes. These findings provide a framework for understanding eRNAs and their convergence with histone modifications in the regulation of transcriptional coactivators.
METHODS
Cell culture and treatments
Human colorectal adenocarcinoma SW480 cells and human embryonic kidney 293T (HEK293T) cells were purchased from American Type Culture Collection (ATCC) and grown in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco). SW480 cells that stably and inducibly express short hairpins against LacZ or p53 were kindly provided by Xinbin Chen (UC Davis) and were grown in standard DMEM medium containing penicillin and streptomycin (Gemini Bio-Products), 1.5 μg ml−1 puromycin (Sigma), and were induced with 1 μg ml−1 doxycycline (Sigma). Lentivirus infected SW480 cells were propagated in growth medium containing DMEM, 10% FBS, 100 units ml−1 penicillin, 100 μg ml−1 streptomycin, and 1.5 μg ml−1 puromycin. All cell lines were tested for mycoplasma contamination. For experiments with TNF-α treatment, the indicated cells were treated with 12.5 ng ml−1 recombinant TNF-α (Shenandoah Biotechnology) for the indicated time points before harvesting for gene expression or ChIP analyses. SW480 cells were treated with a final concentration of 500 nM JQ1 (Cayman Chemical), 2 μg ml−1 Actinomycin D (ApexBio), or vehicle (DMSO, Fisher Scientific) before harvesting for gene expression and immunoblot analyses.
Immunoblotting
Protein samples were incubated at 95 °C for 5 min, separated by SDS-PAGE, and transferred to PVDF membranes (EMD Millipore) that were probed with the indicated antibodies. Reactive bands were detected by ECL (Thermo Scientific Pierce), and exposed to Blue Devil Lite ECL films (Genesee Scientific) or visualized by LI-COR (LI-COR Biosciences).
Antibodies
Antibodies used for ChIP assays were obtained commercially as follows: anti-BRD4 (A301–985A100, 1 μg) from Bethyl laboratories; anti-p53 (DO1, sc126, 1 μg), anti-RNAPII (N20, sc899, 1 μg), anti-IgG (sc2027, 1 μg) from Santa Cruz Biotechnology. Antibodies used for immunoblotting were obtained as follows: anti-BRD4 (1:3000 dilution) from Bethyl laboratories, anti-H3K27ac (ab4729, 1:5000 dilution) from Abcam; anti-H3K9ac (13–0020, 1:5000 dilution) from EpiCypher; and anti-p53 (DO1, sc126, 1:2000 dilution) and anti-β-Actin (sc47778, 1:2000 dilution) from Santa Cruz Biotechnology.
Lentivirus production and transduction
pLKO.1 TRC control and target shRNA plasmids were generated with annealed primers to knockdown BRD4 and the MMP9 and CCL2 eRNAs. shRNA primers used in this study are listed in Supplementary Table 1. For lentivirus production and transduction, 50–60% confluent HEK293T cells were transfected with Lipofectamine 3000 (Invitrogen) with TRC control, target shRNAs, and packaging plasmids psPAX2 and pMD2.G. Virus-containing medium was collected 48 and 72 hr post transfection, filtered with a 0.45μm pore size filter, and used for viral infection. SW480 cells were transduced with the viral supernatants containing 8 μg ml−1 polybrene (Sigma-Aldrich). After 8 hr infection, virus-containing medium was removed and replaced with fresh medium. After 48 hr, puromycin was added at a final concentration of 1.5 μg ml−1. Cells were cultured for an additional 72 hr before harvesting the cells for qRT-PCR and immunoblot to confirm successful knockdown efficiency.
In vitro RNA synthesis
Primers were designed to amplify desired genomic regions that correspond to maximal eRNA (GRO-seq) peaks at MMP9 and CCL2 enhancers. The MEG3 and p21 exonic sequences were identical to those used in a previous study47. Primers sequences can be found in Supplementary Table 2. The T7 promoter sequence was included in the forward primer and genomic fragments were PCR amplified from HeLa genomic DNA (NEB), confirmed by sequencing, and subsequently used for RNA synthesis using T7 RiboMAX Express Large Scale RNA Production System (Promega). Synthesized RNAs were purified per manufacturer’s instructions and quantitated by Nanodrop (Invitrogen). RNA probes were refolded by incubation at 95 ºC for 5 min followed by snap-cooling on ice for 5 min. Cold RNA refolding buffer (10 mM Tris-HCl at pH 7, 100 mM KCl, 10 mM MgCl2) was then added and the samples were transferred to an ice-cold metal rack. RNAs were allowed to refold by warming the sample to room temperature for 20–30 min.
RNA purification and quantitative real-time PCR
Total RNA was extracted with TRIzol LS reagent (Invitrogen) from SW480 stably expressing control, BRD4, or CCL2 and MMP9 eRNA shRNAs and treated with 12.5 ng ml−1 TNF-α for the indicated time points. Total RNA (0.5–1 μg) was used for cDNA synthesis using ProtoScript II First Strand cDNA Synthesis Kit (NEB) with random hexamers. PCR reactions were performed on an Applied Biosystems Step One Plus real-time PCR systems using SYBR Green PCR Master Mix (Applied Biosystems) in duplicate using samples from at least three independent cell harvests and the specificity of amplification was examined by melting curve analysis. The relative levels of eRNA and mRNA expression were calculated according to the (ΔΔCt) method and individual expression data was normalized to GAPDH. The gene expression levels determined after TNF-α treatment are relative to the levels before TNF-α treatment. Primers for qRT-PCR are listed in Supplementary Table 3.
Ultraviolet-RNA Immunoprecipitation (UV-RIP)
SW480 cells treated with TNF-α for 0 or 16 hr were crosslinked by UV irradiation (150 mJ per cm2 at 254 nm) using a Stratalinker and lysed in RIP lysis buffer [25 mM HEPES-KOH at pH 7.5, 150 mM KCl, 0.5% NP40, 1.5 mM MgCl2, 10% glycerol, 1 mM EDTA, 0.4 U RNase inhibitor (Promega), protease inhibitor cocktails (PICs)] on ice for 30 min. Cleared cell lysates were used for IP with BRD4 and IgG-antibody bound Protein A Dynabeads (Invitrogen) overnight. Beads were subsequently washed three times with RIP lysis buffer and RNA samples were eluted using TRIzol LS reagent. cDNA samples were prepared as described above and analyzed by qRT-PCR primers listed in Supplementary Table 3.
Histone peptide binding assays
Unmodified H3 (21–44), K27ac-modified H3 (21–44), unmodified H4 (1–25), or K16ac-modified H4 (1–25) peptides (Innopep Inc.) were immobilized on Dynabeads MyOne Streptavidin C1 (Life technologies) and subsequently incubated with 200 ng of recombinant FLAG-BRD4 FL or FLAG-BRD4 ΔBD1/2 in the absence or presence of 0.2 nM refolded MMP9 or CCL2 eRNA in binding buffer (150 mM NaCl, 20 mM HEPES-KOH at pH 7.9, 0.1% NP40, 0.25 mg ml−1 bovine serum albumin (BSA), 1 mM Sodium butyrate, 1mM PMSF, PICs, RNase inhibitor) for 4 hr at 4°C. Beads were washed ten times with binding buffer containing 400 mM NaCl. Bound proteins were eluted in sample buffer and analyzed by SDS-PAGE and immunoblotting.
Histone acetyltransferase (HAT) assays
Recombinant histone octamers (0.5 μg) that were purified as previously described59 were incubated with p300 (30 ng) and acetyl-CoA (20 μM) in reaction buffer (50 mM HEPES pH 7.8, 30 mM KCl, 0.25 mM EDTA, 5.0 mM MgCl2, 5.0 mM sodium butyrate, 2.5 mM DTT). Reactions were incubated at 30˚C for 30 min, resolved by SDS-PAGE, and analyzed by immunoblotting.
Histone octamer binding assays
p300-acetylated histone octamers were isolated by incubating the HAT reactions with H3K9ac and H3K27ac antibody-coupled Protein A Dynabeads in binding buffer (150 mM NaCl, 50 mM Tris-HCl at pH 8.0, 0.1% NP40, 1 mM Sodium butyrate, 1 mM PMSF, PICs) for 3 hr at 4 ºC. Following the incubation, the acetylated histone octamer bound beads were washed three times with binding buffer and then incubated with 200 ng of recombinant FLAG-BRD4 FL or FLAG-BRD4 ΔBD1/2 in the absence or presence of 0.2 nM refolded eRNA or increasing doses in the range of 0.06 nM to 2 nM in binding buffer with 1% BSA for 12 hr at 4 ºC. The protein complexes were washed eight times with wash buffer (400 mM NaCl, 50 mM Tris-HCl at pH 8.0, 0.1% NP40, 0.5% BSA, 1 mM Sodium butyrate, 1 mM PMSF, PICs). Bound proteins were eluted in sample buffer and analyzed by SDS-PAGE and immunoblotting with the indicated antibodies.
Surface plasmon resonance (SPR) assays
Sensorgrams were recorded on a BIAcore 3000 instrument using streptavidin (SA) chips. Biotinylated H3K27ac histone peptide was immobilized on the SA chip in binding buffer (150 mM NaCl, 20 mM HEPES-KOH at pH 7.9, 0.1% NP40). Sensorgrams were run in the automatic subtraction mode using flow cell 1 (FC 1) as an unmodified reference. Data was collected for FC’s 2, 3, and 4, which contained varying amounts of the BD1/2 domain of BRD4 and in the presence or absence of 0.2 nM refolded MMP9 eRNA with 100 Response Units (RU) immobilized on FC2, 200 RU on FC3, and 400 RU on FC4. Injections were made using the kinject injection mode with a 3 min contact time and a 3 min dissociation phase, at a flow rate of 50 μl min−1. The running buffer used for the binding experiments was 150 mM NaCl, 20 mM HEPES-KOH at pH 7.9, 0.1% NP40. The data was analyzed using the BIA Evaluation 4.1 software.
Co-immunoprecipitation
Assays were performed using whole cell lysates prepared from SW480 cells that were treated with 12.5 ng ml−1 TNF-α for 0 or 16 hr. Cleared lysates were first incubated with indicated antibodies for 2 hr at 4 ºC followed by an additional 2 hr incubation with Protein A Sepharose (Rockland Inc.). Beads were washed with wash buffer (20 mM Tris-HCl at pH 7.9, 0.1% NP40, 150 mM KCl) five times and analyzed by immunoblotting.
Chromatin Immunoprecipitation (ChIP) and ChIP-Seq
BRD4 ChIP assays were performed using SW480 cells that were (i) untreated or treated with 12.5 ng ml−1 TNF-α for the indicated time points or (ii) transduced with indicated shRNAs and treated with 12.5 ng ml−1 TNF-α for 0 or 16 hr. Cells were sequentially cross-linked using 6 mM DSG (disuccinimidyl glutarate; ProteoChem) for 30 min and a final concentration of 1% formaldehyde for 10 min at room temperature and stopped with 125 mM glycine (Fisher Scientific). Cells were lysed in lysis buffer (20 mM Tris-HCl at pH 7.5, 300 mM NaCl, 2 mM EDTA, 0.5% NP40, 1% Triton X-100, 1 mM PMSF, PICs) and incubated on ice for 30 min. The resuspended cells were then dounced in an ice-cold homogenizer. Nuclear pellets were collected and resuspended in shearing buffer (0.1% SDS, 0.5% N-lauroylsarcosine, 1% Triton X-100, 10 mM Tris-HCl at pH 8.1, 100 mM NaCl, 1 mM EDTA, 1 mM PMSF, PICs). Isolated chromatin was fragmented to an average size of 200–600 bp with a bioruptor Pico (Diagenode). Precleared chromatin was immunoprecipitated overnight at 4 ºC and immunocomplexes were collected with protein A Dynabeads. The immunocomplexes were washed eight times in wash buffer (50 mM HEPES-KOH at pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP40, 0.7% sodium deoxycholate, 1 mM PMSF, PICs), followed by two 1X TE washes, and eluted in elution buffer (50 mM Tris-HCl at pH 8.0, 10 mM EDTA, 1% SDS), crosslinks were reversed at 65 ºC for 4 hr or overnight, and DNA was purified using DNA Clean & Concentrator Kit according to the manufacturer’s instructions. PCR reactions were performed on an Applied Biosystems Step One Plus real-time PCR systems using SYBR Green PCR Master Mix in duplicate using samples from at least three independent cell harvests and the specificity of amplification was examined by melting curve analysis. The relative amounts of ChIP DNA were quantified relative to inputs. Primers for ChIP-quantitative PCR are listed in Supplementary Table 4. Sequential ChIP experiments were performed exactly as described above with minor modifications. Specifically, 150 μg of sheared chromatin was used to perform the IP. Following the washes after the first IP, immunocomplexes were eluted in re-IP elution buffer (50 mM Tris-HCl at pH 8.0, 1% SDS, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, PICs) and diluted 10 fold in dilution buffer (16.7 mM Tris-HCl at pH 8.0, 167 mM NaCl, 0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 1 mM PMSF, PICs). The second IP was performed with indicated antibodies overnight at 4 ºC, followed by additional washes and the final elution as described above.
For ChIP-seq experiments, the IP’s were performed as described. The eluted ChIP DNA was quantified using a Qubit 2.0 fluorometer (Invitrogen), and 2–5 ng of ChIP DNA was used to prepare the sequencing libraries from two biological replicates using the TruSeq ChIP Sample Prep Kit according to the manufacturer’s instructions (Illumina). Briefly, ChIP DNA was end-repaired and adaptors were ligated to the ends of the DNA fragments. Adaptor-ligated ChIP DNA fragments with average size of 350 bp were used to construct libraries and single-end sequenced (50 bp) on Illumina HiSeq 4000. Sequencing reads were mapped to the hg38 human genome using Bowtie2 software 38 and default parameters. The mapped reads were then processed to make Tag Directory module using HOMER for filtering. Briefly, PCR duplications were removed and only uniquely mapped reads were kept for further analysis. The genome browser files for the resulting reads were generated by using makeUCSCfile module from HOMER. Enriched regions for BRD4 were called using find Peaks module from HOMER by using preset options, factor or histone styles respectively and compared to the corresponding inputs. Deeptools were used to generate heat maps. De novo motif analysis was performed using the BRD4/mutp53 co-bound enhancer regions using “findMotifsGenome.pl” of Homer with ±100 bp window relative to the peak center.
Purification of recombinant proteins
Mutant p53 R273H was generated by site-directed mutagenesis (Agilent Technologies). FLAG-BRD4 BD1/2, BD1, BD2 were cloned into pET11d bacteria expression vector using NdeI and BamHI sites after amplification from BRD4 cDNA. His-tagged tandem BDs (BD1/2) of BRD2, BRD3, BRD4, and BRDT as well as the single BDs of BRG1 and BRD7 were cloned into pET6His bacteria expression vector using BamHI and EcoRI sites. p53 and BD constructs were expressed in E. Coli and purified on FLAG M2 agarose beads (Sigma) or TALON metal affinity resin (Clontech). The FLAG p300, BRD4 FL, and ΔBD1/2 proteins were expressed in Sf9 cells and purified on FLAG M2 agarose. Primers used for cloning are listed in Supplementary Table 5.
In vitro pull-down RNA binding assays
FLAG- and His-tagged proteins were incubated with 500 ng of refolded RNA while rotating at 4 ºC for 1 hr in RNA Binding Buffer (20 mM Tris-HCl at pH 7.4, 100 mM KCl, 0.2 mM EDTA, 0.05% NP40, 0.4 U RNase inhibitor, PICs). Protein-RNA complexes were recovered using FLAG M2 agarose beads or TALON resin for 1 hr at 4 ºC. Beads were washed three times with RNA wash buffer (20 mM Tris-HCl at pH 7.4, 200 mM KCl, 0.2 mM EDTA, 0.05% NP40, 0.4 U RNase inhibitor, PICs) and RNA samples were eluted using Trizol LS reagent. Purified RNA samples were resolved on a denaturing 5% TBE urea gel and stained with SYBR gold for 30 min before imaging using a Typhoon phosphorimager or LI-COR (LI-COR Biosciences).
RNA electrophoretic mobility shift assay (EMSA)
EMSAs were performed following established protocols60. Binding reactions were performed in 1x RNA EMSA buffer (20 mM Tris-HCl at pH 7.4, 100 mM KCl, 1 mM EDTA, 1% glycerol, 0.05% NP40, 0.5 mM ZnCl2, 0.1 mg ml−1 BSA (Fisher), 0.1 mg ml−1 yeast tRNA (Sigma), 2 mM DTT, 0.4 U RNase inhibitor). The binding reactions were initiated by adding different doses (0.3–1.5 μg) of FLAG-BRD4 protein in the presence of 12,000 cpm refolded RNA and allowed to proceed for 30 min at 4 ºC. Reactions were loaded immediately on 6% Native polyacrylamide gels that were pre-run for 1 hr at 100V in 0.5X TBE at 4 ºC. The gel was run for 4 hr at 100V and exposed to autoradiography screen before imaging with Typhoon phosphorimager. For competition assays, (0, 50, 150, or 450 nM) of unlabeled RNA or ssDNA was added in addition to the 32P radiolabeled RNA.
Supplementary Material
Acknowledgements
We are grateful to Cheng-Ming Chiang (UT Southwestern) for providing the pF:hBRD4 (1-722)-11d, pcDNA3-F:hBRD4 FL, pcDNA3-F:hBRD4 ΔBD1/2 plasmids and BRD4 FL and BRD4 ΔBD1/2 expressing baculovirus. We are also thankful to Xinbin Chen (UC Davis) for providing the SW480 shLacZ and shp53 cell lines. H.R. was supported by the UCSD Cellular and Molecular Genetics Training Program through an institutional grant from the National Institute of General Medicine (T32 GM007240). This work was supported by the Research Scholar Award from the Sidney Kimmel Foundation for Cancer Research #857A6A (S.M.L.), American Cancer Society ACS-IRG #70-002 (S.M.L.), and the University of California Cancer Research Coordinating Committee, CRN-17-420616 (S.M.L.).
Footnotes
Data availability
All sequencing data that support the findings of this study has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) and are accessible through the GEO Series Accession numbers GSE102796 and GSE110473. All other relevant data are available from the corresponding author upon reasonable request.
Conflict sof interest
The authors declare no conflict of interest.
References
- 1.Calo E. & Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinz S, Romanoski CE, Benner C. & Glass CK The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16, 144–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heintzman ND et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thurman RE et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visel A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chepelev I, Wei G, Wangsa D, Tang Q. & Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res 22, 490–503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creyghton MP et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107, 21931–6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rada-Iglesias A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippakopoulos P. & Knapp S. The bromodomain interaction module. FEBS Lett 586, 2692–704 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Marushige K. Activation of chromatin by acetylation of histone side chains. Proc Natl Acad Sci U S A 73, 3937–41 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippakopoulos P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umehara T. et al. Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain. J Biol Chem 285, 7610–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moriniere J. et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461, 664–8 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Dhalluin C. et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399, 491–6 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Mujtaba S, Zeng L. & Zhou MM Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26, 5521–7 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Yang XJ Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26, 1076–87 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Zeng L. & Zhou MM Bromodomain: an acetyl-lysine binding domain. FEBS Lett 513, 124–8 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Dawson MA et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delmore JE et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuber J. et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478, 524–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang B, Yang XD, Zhou MM, Ozato K. & Chen LF Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol 29, 1375–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou Z. et al. Brd4 maintains constitutively active NF-kappaB in cancer cells by binding to acetylated RelA. Oncogene 33, 2395–404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu SY, Lee AY, Lai HT, Zhang H. & Chiang CM Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell 49, 843–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown JD et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell 56, 219–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roe JS, Mercan F, Rivera K, Pappin DJ & Vakoc CR BET Bromodomain Inhibition Suppresses the Function of Hematopoietic Transcription Factors in Acute Myeloid Leukemia. Mol Cell 58, 1028–39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J. et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell 25, 210–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart HJ, Horne GA, Bastow S. & Chevassut TJ BRD4 associates with p53 in DNMT3A-mutated leukemia cells and is implicated in apoptosis by the bromodomain inhibitor JQ1. Cancer Med 2, 826–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filippakopoulos P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J. et al. BET Inhibition Attenuates Helicobacter pylori-Induced Inflammatory Response by Suppressing Inflammatory Gene Transcription and Enhancer Activation. J Immunol 196, 4132–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hah N. et al. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc Natl Acad Sci U S A 112, E297–302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang MK et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19, 523–34 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Winter GE et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol Cell 67, 5–18 e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanno T. et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 21, 1047–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagarajan S. et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep 8, 460–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arner E. et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347, 1010–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh CL et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A 111, 7319–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai F, Gardini A, Zhang A. & Shiekhattar R. Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam MT et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498, 511–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melo CA et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49, 524–35 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Mousavi K. et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 51, 606–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahnamoun H. et al. Mutant p53 shapes the enhancer landscape of cancer cells in response to chronic immune signaling. Nat Commun 8, 754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaukowitch K. et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell 56, 29–42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai F. et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bose DA et al. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 168, 135–149 e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu SY & Chiang CM The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282, 13141–5 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Mertz JA et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A 108, 16669–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ott CJ et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood 120, 2843–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yap KL et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38, 662–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devaiah BN et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol 23, 540–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fujisawa T. & Filippakopoulos P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat Rev Mol Cell Biol 18, 246–262 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Sanchez R, Meslamani J. & Zhou MM The bromodomain: from epigenome reader to druggable target. Biochim Biophys Acta 1839, 676–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller TC et al. A bromodomain-DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat Commun 7, 13855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison EA et al. DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat Commun 8, 16080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruthenburg AJ, Li H, Patel DJ & Allis CD Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8, 983–94 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dey A, Chitsaz F, Abbasi A, Misteli T. & Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A 100, 8758–63 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lauberth SM et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lauberth SM, Bilyeu AC, Firulli BA, Kroll KL & Rauchman M. A phosphomimetic mutation in the Sall1 repression motif disrupts recruitment of the nucleosome remodeling and deacetylase complex and repression of Gbx2. J Biol Chem 282, 34858–68 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.