Abstract
The activation of T cells by the T cell antigen receptor (TCR) results in the formation of signaling protein complexes (signalosomes), the composition of which has not been analyzed at systems-level. Here, we isolated primary CD4+ T cells from 15 gene-targeted mice each expressing one tagged-form of a canonical protein of the TCR signaling pathway. Using affinity purification coupled with mass spectrometry, we analyzed the signalosomes assembling around each of the tagged protein over 600 seconds of TCR engagement. We showed that the TCR signal-transduction network comprises at least 277 unique proteins involved in 366 high-confidence interactions, and that TCR signals diversify extensively at the level of the plasma membrane. Integrating the cellular abundance of the interacting proteins and their interaction stoichiometry provided a quantitative and contextual view of each documented interaction permitting to anticipate whether ablation of a single interacting protein can impinge on the whole TCR signal-transduction network.
Introduction
T cells express T cell antigen receptors (TCRs) on their surface through which they detect antigens. The initiation of TCR signals relies on the LCK and ZAP70 protein tyrosine kinases (PTKs) and generates protein assemblages of considerable complexity1, 2, 3. Most previous approaches aiming at disentangling such complexity addressed one protein at a time with limited quantitative insight. As a result, it remains difficult to understand how the TCR signal-transduction network processes signals and to predict the effects resulting from a mutation or a drug.
Affinity purification of a protein of interest (the ‘bait’) with its interacting partners (the ‘preys’), coupled with mass spectrometry (AP-MS), permits to define the composition of the corresponding protein complex as a set of binary bait-prey interactions termed an ‘interactome’. We provided proof-of-concept for interactomics in primary CD4+ T cells by determining the composition of the multiprotein complexes that formed around ZAP70 and the adaptors LAT and SLP-764. However, such pilot study was limited to 3 baits and relied on pervanadate-based T cell activation, a stimulation condition less physiological than that resulting from the engagement of the TCR in combination with the CD4 or CD8 coreceptors. Here, we extended our interactomics approach to signaling complexes (‘signalosomes’) that assemble around fifteen canonical proteins used by the proximal TCR signal-transduction network. We avoided pitfalls associated with transformed T cells5 by using primary CD4+ T cells, and we captured signaling dynamics by analyzing each of the 15 signalosomes prior to and at four different time points following anti-TCR plus anti-CD4 stimulation. 277 unique proteins involved in 366 high-confidence protein-protein (PPI) interactions were identified within the proximal TCR signal-transduction network, a complexity that led us to revisit the mode of action of several signalosomes used by the TCR.
TCR signals are classically described as proceeding from the TCR to the inside of T cells via the LAT transmembrane adaptor which is thought to serve as the earliest and often sole point of signal diversification downstream of the TCR2. In our original interactomics study, we showed that the transmembrane receptor CD6 was also able to nucleate its own signalosome in response to TCR signaling and independently of LAT4. However, the lack of information on the numbers of complexes nucleating around LAT and CD6 precluded assessing their respective quantitative contribution to early TCR signal propagation and diversification. Here, by capitalizing on the recent possibilities to measure both the numbers of copies per cell (cellular protein abundance) of each interacting proteins, and the quantitative relationship existing between a bait and a prey in a given complex (interaction stoichiometry)6, we succeeded identifying and quantifying the TCR-inducible signalosomes that form at the inner face of the plasma membrane. Unexpectedly, the CD5 and CD6 transmembrane receptors assembled signalosomes with kinetics and in numbers comparable to those nucleated by the LAT adaptor, demonstrating that the breadth of early TCR signal diversification is larger than expected. Finally, to decipher the function of the poorly characterized interacting proteins identified within the TCR signaling network, we developed a Crispr/Cas9-based pipeline that requires no mouse breeding and permits to analyze in 4 months and at organismal levels the immune phenotype of mice deprived of select interacting proteins.
Results
Mapping the TCR signal-transduction network of primary T cells
To make the TCR signal-transduction network of primary T cells amenable to quantitative AP-MS analysis, we developed 15 lines of gene-targeted mice each containing a canonical protein of the TCR signaling network tagged at its N- or C-terminus with an affinity Twin-Strep-tag (OST) (Fig. 1a). Mice expressing OST-tagged version of the cytosolic adaptors SLP-76 (LCP2) and GRB2, of the guanine nucleotide exchange factor VAV1, and of the evolutionary related E3 ubiquitin-protein ligases CBL and CBLB have been described4, 7, 8, 9. We introduced here 10 additional OST-tagged mouse lines corresponding to the phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 SHIP1 (INPP5D), the PTKs LCK and ITK, the phospholipase PLC-γ1, the cytosolic adaptors NCK1, THEMIS and FYB (ADAP), the PTPases SHP1 (PTPN6) and PTPN22, and the NFATc2 transcription factor (Supplementary Fig. 1a and Methods). Analysis of mice homozygous for each of the OST-tagged alleles showed that their T cells developped properly (Supplementary Fig. 1b), yielding normal numbers of mature CD4+ and CD8+ T cells (Supplementary Fig. 1c) that had no defect in proliferation (Supplementary Fig. 1d), cytokine production (Supplementary Fig. 1e), and tyrosine phosphorylation (Supplementary Fig. 2) in response to TCR stimulation. T cells expressed the bait proteins at physiological levels (Supplementary Fig. 3a), and, after lysing them with the nonionic detergent n-dodecyl-β-D-maltoside, bait proteins were efficiently purified using Sepharose beads coupled to Strep-Tactin (Supplementary Fig. 3b,c), obviating potential variations resulting from the use of antibody specific for each bait.
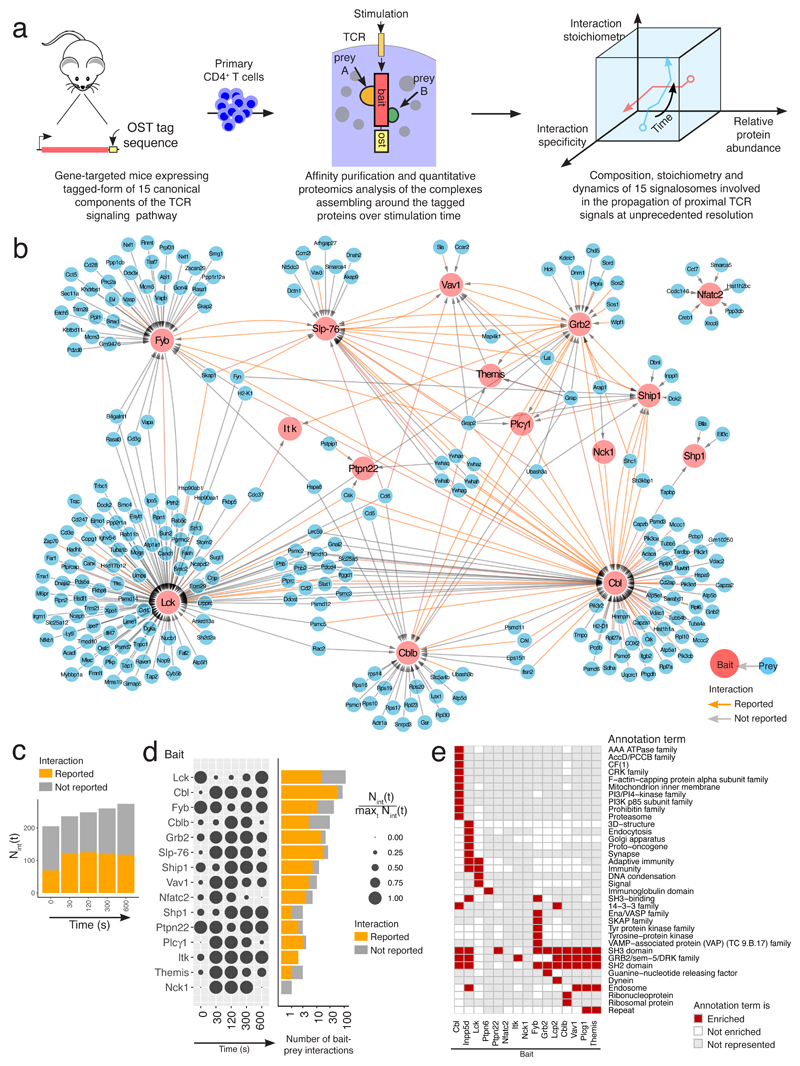
Fig. 1. Composition of the protein-protein network assembling around 15 canonical proteins of the TCR signaling pathway over 600 s of stimulation.
a, Workflow schematic for mapping the composition, stoichiometry and dynamics of the signaling complexes assembling around 15 canonical proteins of the TCR signaling network of primary CD4+ T cells prior to and following TCR activation for 30, 120, 300 and 600 s. b, High-confidence interactome forming around the 15 baits. High-confidence bait-prey interactions were defined as having FDR value ≤ 3% for two consecutive time points of stimulation (see ‘Statistics and data reproducibility’ in Methods). OST-tagged baits and preys are represented in red and blue, respectively. Interactions already reported in databases are highlighted using orange arrows. In most interactomes, several preys associate to a given bait and might contact it directly or indirectly through intermediary proteins. c, Evolution over the course of TCR stimulation of the number of high-confidence bait-prey interactions, a value termed Nint(t) with 't' corresponding to the time of stimulation. The proportion of them already reported in public databases is shown in orange. d, In the dot plot (left), the Nint(t) value corresponding to each bait has been row normalized to the maximum value it reached over the course of stimulation, a value termed maxt Nint(t) (see Key). Also shown (right) using a log base 10 scale is the number of high-confidence bait-prey interactions corresponding to each bait, and the proportion of which has been already reported. e, Annotations terms from Uniprot-Keywords and Uniprot-Protein families enriched in at least one of the 15 interactomes.
Purified primary CD4+ T cells were briefly expanded in vitro to reach the substantial cell numbers required for AP-MS (see Methods). Our results are thus reflecting the composition of the TCR-signaling network of antigen-experienced conventional CD4+ T cells. To capture signaling dynamics, we defined by AP-MS the preys assembling around each bait prior to or after stimulation through crosslinkage of the TCR and CD4 for 30, 120, 300 and 600 s (Fig. 1a). For each time point, three independent biological replicates were performed and each biological replicate was analysed in duplicate or triplicate by MS (technical replicates). To distinguish truly interacting proteins from nonspecific contaminants, we compared our data with control AP-MS experiments involving wild-type CD4+ T cells (Supplementary Fig. 4a,b). Data corresponding to 449 affinity purifications and 933 MS runs were analyzed using the MaxQuant software, and high-confidence interactors identified using a data-driven, false discovery rate (FDR)-controlled approach (Ref. 6 and Methods). FDR values were used to evaluate the statistical significance of the enrichment observed at a given time-point for a considered bait-protein interaction between CD4+ T cells isolated from OST-tagged and wild-type (control) mice (Supplementary Fig. 4c-e). High-confidence bait-prey interactions were defined has having FDR value ≤ 3% for two consecutive time points of stimulation. We also measured the stoichiometry of bait-prey interactions using intensity-based absolute quantification (iBAQ)10, and determined their changes after TCR engagement. The composition, stoichiometry and dynamics of the 15 interactomes are summarized in Supplementary Data Set 1.
Global analysis of the TCR signal-transduction network
Analysis of the interactomes assembling around each of the 15 baits revealed a bait-prey network composed of 277 unique proteins connected via 366 high-confidence interactions (Fig. 1b). Among them, 162 interactions have been already reported in human and mouse PPI databases compiling several cell types (Fig. 1c). As expected for a ligand-inducible signal-transduction PPI network, most of the 15 interactomes reached their highest complexity after TCR triggering (Fig. 1d). Such complexity peaked between 30 and 120 s in the case of the CBLB, GRB2, SLP-76, VAV1, and PLC-γ1 interactomes and persisted up to 600 s of stimulation for the CBL and SHIP1 interactomes. Conversely, several of the preys found in the LCK and FYB interactomes were quickly released upon TCR triggering.
The numbers of high-confidence bait-prey interactions varied widely across the analyzed baits, ranging from 1 (NCK1 bait) to 117 (LCK bait) (Fig. 1d). Only 20 % of the identified preys were found in more than one interactome, suggesting that each of the selected bait exerts specific functions within the TCR network. Along that line, annotation enrichment analysis showed that several interactomes were enriched for unique biological processes, molecular functions, interacting protein domains and families (Fig. 1e). The annotations 'SH2-domain' and ‘SH3-domain’ were, however, shared by 9 and 10 of the 15 interactomes, respectively, reflecting the key role both domains play in shaping the TCR PPI network. Therefore, by identifying 366 high-confidence PPI within the proximal TCR signal-transduction network our approach revealed a complexity higher than expected.
Bait-prey interaction stoichiometries over the course of TCR stimulation
The median value of the distribution of the stoichiometries observed for the bait-prey interactions increased following TCR activation (Fig. 2a), reflecting formation of protein assemblages of increasing complexity. The maximal interaction stoichiometries reached by the 366 documented bait-prey interactions over the course of TCR stimulation covered five orders of magnitude, ranging from 10–5 to 1.6 with a median value close to 10–3. Among the PPI described in the present study, those not reported in public databases showed a bias toward lower stoichiometry (Fig. 2a), highlighting the increased analytical depth afforded by AP-MS sensitivity as compared to conventional biochemical approaches. Therefore, low-stoichiometry PPI play a central role in the organization of the TCR-signaling network, making them prone to disruption in experiments relying on bait overexpression.
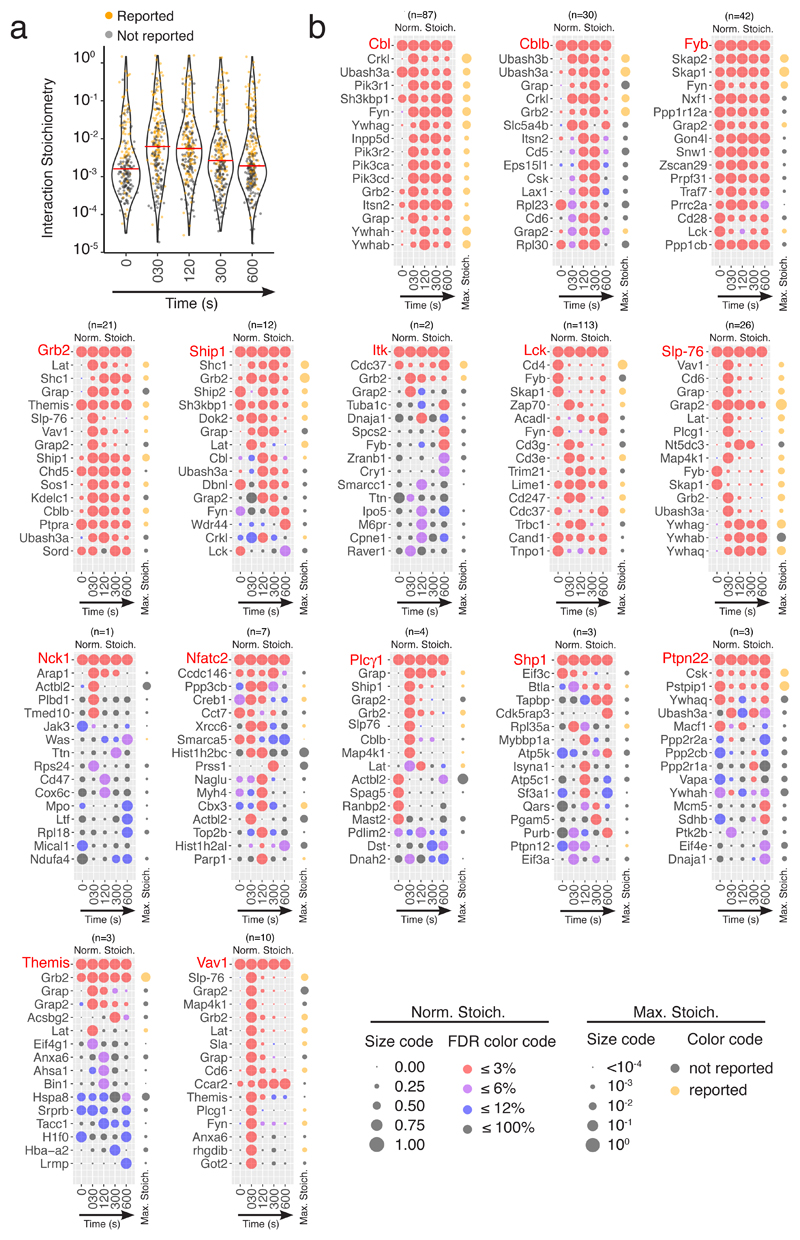
Fig. 2. Evolution over 600 s of TCR stimulation of bait-prey interaction stoichiometry among the 15 interactomes.
a, Distribution of the interaction stoichiometries of the 366 high-confidence bait-prey interactions observed for the 15 baits at each of the analyzed time points. Horizontal red lines correspond to median values and previously reported interactions are highlighted in orange. Note that 100=1. b, Each dot plot corresponds to a given bait (denoted in red) and shows its interaction stoichiometry over the course of TCR stimulation with its 16 highest-confidence preys (denoted in black and ranked according to their FDR value and maximum enrichment; see Key for FDR color code). The total number of high-confidence interactions (n) established by a given bait is shown at the top of each dot plot (for instance n = 87 in the case of the CBL bait and the full list of the 87 interactions can be found in tab ‘CBL’ of Supplementary Data Set 1). For a given bait-prey interaction, the interaction stoichiometry has been row normalized to its maximum value observed over the course of TCR stimulation (Norm. Stoich.). For instance, the FYB and LAT preys show a maximal binding to SLP-76 prior to and after 30 s of activation, respectively. Also shown on the right-side of each dot plot is the maximal interaction stoichiometry (Max. Stoich.) reached by each of the documented bait-prey interaction over the course of TCR stimulation (dot size is commensurate to the value of the maximal interaction stoichiometry; see Key). Orange and grey dots correspond to previously reported and undocumented interactors, respectively. In the case of a ‘rich’ interactome, as exemplified by the CBL interactome, all the 16 represented preys qualify as high-confidence preys in that they all show FDR ≤ 3% at two consecutive time points of stimulation. In contrast, in the case of a ‘sparse’ interactome, as exemplified by the NCK1 interactome, a single prey (ARAP1) fulfills our stringent high-confidence criteria. The ACTBL2, PLBD1 and TMED10 preys interacted with NCK1 with an FDR ≤ 3% at a single condition of stimulation (t30s), whereas preys such WAS and TTN have FDR ≤ 6%. Such bait-prey interactions of lower confidence are listed in tab ‘bait-prey interactions’ of Supplementary Data Set 1. Note that an FDR of 100% means that the prey does not interact with the bait at the considered time point.
Dot plots were used to depict over the course of TCR stimulation the normalized interaction stoichiometry of each bait with its 16-most enriched preys (Fig. 2b). The maximum interaction stoichiometry value reached over the course of stimulation was also shown. Note that interaction stoichiometry values can be stoichiometric (the bait and the prey interact with a 1 to 1 ratio), superstoichiometric (the bait and the prey interact with a greater than stoichiometric ratio), or substoichiometric (only a fraction of the bait interacts with a given prey). For instance, a bait such as VAV1 established transient and substoichiometric interactions with most of its preys. In contrast, SLP-76 interacted with GRAP2 (GADS) in a superstoichiometric and constitutive manner, whereas SLP-76 interactions with LAT, VAV1 and PLC-γ1 were transient and of relatively low stoichiometries (Fig. 2b).
Integrating cellular protein abundances and interaction stoichiometries
CD4+ T cells from the 15 OST-tagged mice and from wild-type mice were briefly expanded as for AP-MS analysis and the copy numbers per T cell (cellular protein abundance) of the distinct proteins they expressed determined using whole-cell proteome analysis11. We were able to quantify 6343 proteins that corresponded to 92% of the high-confidence preys identified here (Supplementary Data Set 2). The cellular abundances of the proteins used as baits were spread over two orders of magnitude, ranging from 1.3 × 103 for ITK to 2.6 × 105 for SHP1 (Fig. 3a). Consistent with immunoblot analysis (Supplementary Fig. 3a), the OST-tagged proteins showed cellular abundance comparable to that of their wild-type counterparts (Supplementary Fig. 5).
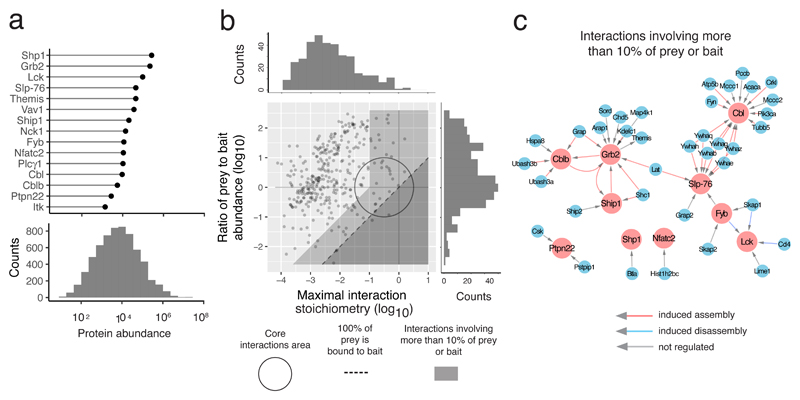
Fig. 3. Organizing the 15 interactomes using cellular protein abundance and interaction stoichiometry.
a, Cellular abundance distribution of the protein baits used for AP-MS (upper panel) and of the 6343 proteins that were quantified in CD4+ T cells used for AP-MS (lower panel). Cellular abundances are shown as mean protein copy number per CD4+ T cell. b, Stoichiometry plot organizes each interactome. For each documented bait-prey interaction the ratio of bait to prey cellular abundance (‘abundance stoichiometry’ in log base 10 scale) was plotted as a function of the maximal interaction stoichiometry the considered bait-prey interaction reached over the course of TCR stimulation (‘interaction stoichiometry’ in log base 10 scale)6. The stoichiometry plot represented in this panel corresponds to an overlay of the stoichiometry plots generated for each of the 15 baits (see Fig. 4). Each dot corresponds to one high-confidence bait-prey interaction. The circle (center: -0.5, radius: 1 in log base 10 scale) delimits the ‘core interactions area’ (see Results). The limit imposed on interaction stochiometries by the relative bait-prey cellular abundance is shown by a dashed diagonal that delimits a ‘forbidden’ area (dark grey). Dots lying on the dashed diagonal correspond to preys 100 % of which are bound to the corresponding bait in at least one of the stimulation conditions. The area including PPI involving more that 10% of the prey or of the bait is also indicated (light grey). The distribution of the maximal bait-prey interaction stoichiometry and of the ratios of the prey to bait abundance are shown in the top and right margins, respectively. c, Bait-prey interactions involving more than 10% of the prey or of the bait. Baits and preys are denoted in red and blue, respectively. Arrows are color coded to specify whether the interaction is positively or negatively regulated by TCR stimulation or not regulated.
By combining cellular protein abundances and interaction stoichiometries, each of the 15 signalosomes was organized into a ‘stoichiometry plot’6. Accordingly, for each documented bait-prey interaction the ratio of the bait to prey cellular abundance was plotted as a function of the maximal interaction stoichiometry reached by the considered bait-prey interaction over the course of TCR stimulation (Fig. 3b). It showed that a few (7%) of the identified high-confidence bait-prey interactions felt within a confined zone that is denoted as the ‘core interactions area’ and encompasses most of the permanent mammalian protein complexes collated in the CORUM database12 (Figure 3b). Among them standed the FYB-SKAP113 and the SLP-76-GRAP214 permanent complexes, as well as TCR-inducible complexes that remained stable for several minutes after TCR stimulation as illustrated by phosphoserine-based complexes involving SLP-76 and three members of the 14-3-3 protein family (YWHAG, YWHAB and YWHAQ), and a complex involving CBLB and the PTPases UBASH3A and UBASH3B (Fig. 3c).
A region corresponding to bait-prey interactions involving at least 10% of the bait or prey expressed in a given T cell is also highlighted (Fig. 3b,c; light grey). The functional relevance of such interactions can be illustrated by that observed between SHIP1 and its close relative SHIP2 (INPPL1) (Fig. 4), a PPI previously reported in platelets15. By catalyzing the hydrolysis of the phosphatidylinositol 3-kinase (PI3K) product PtdIns(3,4,5)P3 into PtdIns(3,4)P2, SHIP1 and SHIP2 regulate effectors containing PtdIns(3,4,5)P3- or PtdIns(3,4)P2-selective pleckstrin homology (PH) domains. Up to 1.3% of SHIP1 molecules were found associated with SHIP2 (Fig. 4), and such low interaction stoichiometry can be viewed as functionaly inconsequential. However, considering that SHIP2 is 31-fold less abundant than SHIP1 in CD4+ T cells, 41% of the SHIP2 molecules available in a CD4+ T cell are thus associated with SHIP1 (Fig. 4). Accordingly, a SHIP1 deficiency will not only ablate a key enzymatic activity but also freed from their interaction with SHIP1 approximately half of the SHIP2 molecules present in CD4+ T cells, allowing them to engage into rival interactions that might be beneficial or detrimental. Therefore, the possibility to integrate interaction stoichiometries and cellular abundances permits to anticipate whether genetic or pharmacological perturbation of a single PPI can result in global network rewiring.
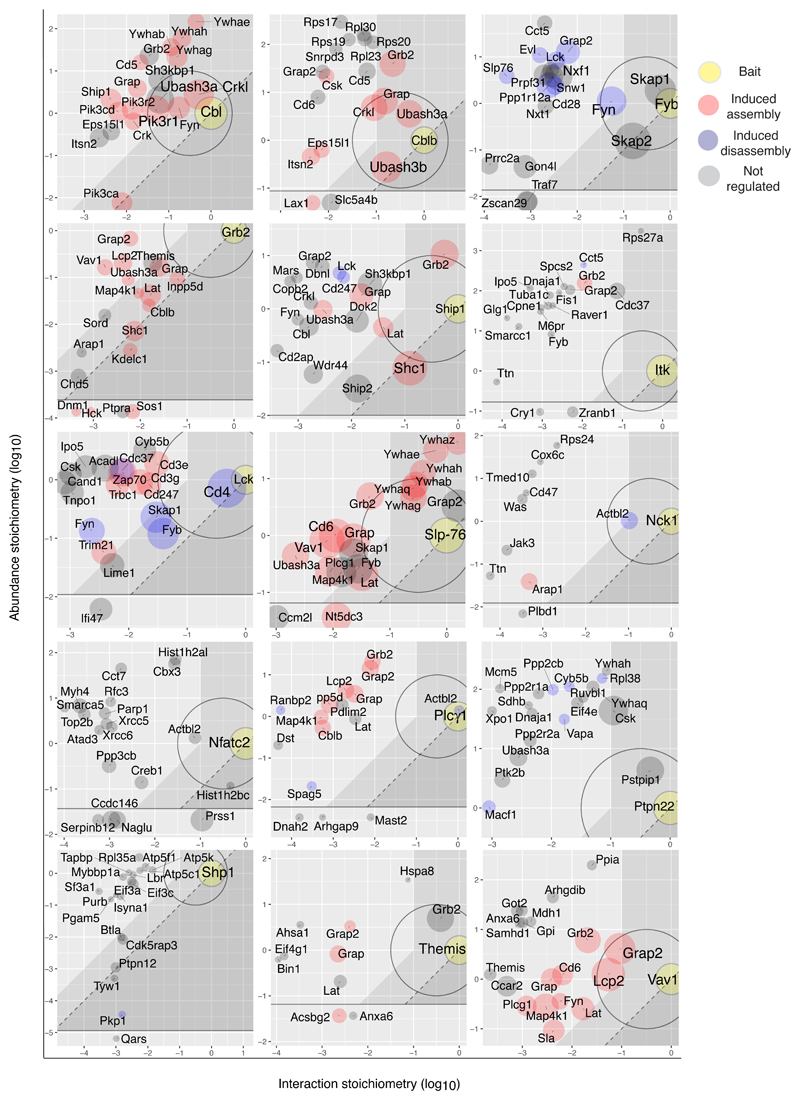
Fig. 4. Stoichiometry plots of the 15 baits.
For each bait, the 20 bait-prey interactions with the largest enrichment and an FDR value ≤ 6% for at least one condition of stimulation are shown and each stoichiometry plot is ‘zoomed’ on the area that includes them. Baits are shown as yellow dots. Red and blue dots correspond to preys that show an increased or decreased binding to the bait following TCR engagement. Grey dots correspond to preys whose association with the bait were not regulated by TCR stimulation. The size of the dots is commensurate to the maximal protein enrichment observed in OST-tagged CD4+ T cell samples as compared to wild-type control CD4+ T cell samples. For instance, the SLP-76 stoichiometry plot shows that GRAP2 is more abundant than SLP-76 and that it binds to SLP-76 in a constitutive and superstoichiometric manner. The six documented 14-3-3 family members (YWHAB, YWHAE, YWHAG, YWHAH, YWHAQ and YWHAZ) are more abundant than SLP-76 and interact with it in a TCR-induced manner and a stoichiometry in the 1.5-0.2 range. Cytosolic effectors such as VAV1 and PLC-γ1 are slightly less abundant than SLP-76 and interact with it in a TCR-induced manner and a stoichiometry in the 0.008 range. Preys for which it was not possible to determine the cellular abundance are shown at the bottom of each stoichiometry plot. See Figure 3b for a definition of the specified stoichiometry plot areas.
Accounting for the distinct function of the CBL and CBLB E3 ubiquitin-protein ligases
Our quantitative approach also permitted a high-resolution comparison of the signalosomes that form around CBL and CBLB (Fig. 4). Both comprised a receptor expressed at the plasma membrane (CD5), molecules involved in endocytosis (ITSN2, EPS15L1), adaptors (GRB2, GRAP, CRKL), and proteins that concur to CBL and CBLB negative regulatory role (UBASH3A and the PTK CSK). Consistent with the distinct phenotypes of T cells deprived of CBL or CBLB, only CBL associated with FYN, six 14-3-3 protein family members, SH3KBP1, SHIP1, and several catalytic and regulatory subunits of PI3K. The corecruitment of PI3K and SHIP1 by CBL likely sets in action proteins containing PH domain selective for PtdIns(3,4)P2 16, 17. Conversely, CBLB was the sole capable of sustained interaction with the UBASH3B PTPase. Importantly, 120 seconds after TCR engagement, CBLB associated with 58% of the available UBASH3B molecules, suggesting that the CD5-CBLB-UBASH3B axis provides a major quantitative contribution to TCR signal termination through ZAP70 dephosphorylation18.
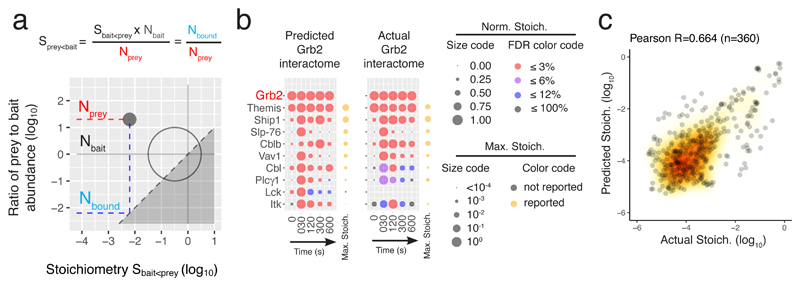
Accurate ab initio prediction of the GRB2 signalosome
The stoichiometry of a few bait-prey interactions identified here have been previously reported (FYB-SKAP113, SLP-76-GRAP214, and THEMIS-GRB219). Although our AP-MS-based stoichiometric calculations agreed with those reported values (Fig. 4), we further challenged their accuracy by testing their predictive power. Considering that the GRB2 adaptor constitutes a high-confidence prey in 9 of the 15 analyzed interactomes, we attempted to predict ab initio the stoichiometry of a simulated GRB2 interactome in which the 9 baits binding to GRB2 play the role of preys. By combining the experimentally determined interaction stoichiometry of the 9 baits with the GRB2 prey with cellular abundances, we calculated the stoichiometry of ‘reciprocal’ interactions in which GRB2 constituted the bait (Fig. 5a and Methods). The resulting GRB2 interactome showed interaction stoichiometries that were in good agreement over all time points with those of the actual GRB2 interactome (Fig. 5b). Similar analysis of all the high-confidence interactions involving protein pairs corresponding to the 15 baits further supported the accuracy of our measurements (Fig. 5c). Such accuracy allowed us to quantify the numbers of TCR-inducible complexes involved in TCR signal propagation and diversification.
Fig. 5. Ab initio prediction of the composition and stoichiometry of novel interactomes.
a, By combining experimentally determined interaction stoichiometry between a bait A and a prey B (SbaitA<preyB) and their cellular abundance it is possible to calculate a ‘reciprocal’ stoichiometry in which B and A behave as the bait and the prey, respectively. b, Dot plots representing the interaction stoichiometries of the predicted GRB2 interactome (left, FDR ≤ 3%, ranked by maximum predicted stoichiometries) and of the experimentally determined GRB2 interactome (right, no FDR restriction). Note that the repertoire of preys of the predicted GRB2 interactome is confined to the repertoire of OST-tag baits capable of binding to GRB2. See Key in Fig. 2b legend. c, Global comparison of experimentally determined and of predicted interaction stoichiometries corresponding to the 15 baits showed a correlation coefficient of 0.66 and a median ratio of measured against predicted interactions stoichiometries of 3.7.
Extensive TCR signal diversification occurs at the level of LAT
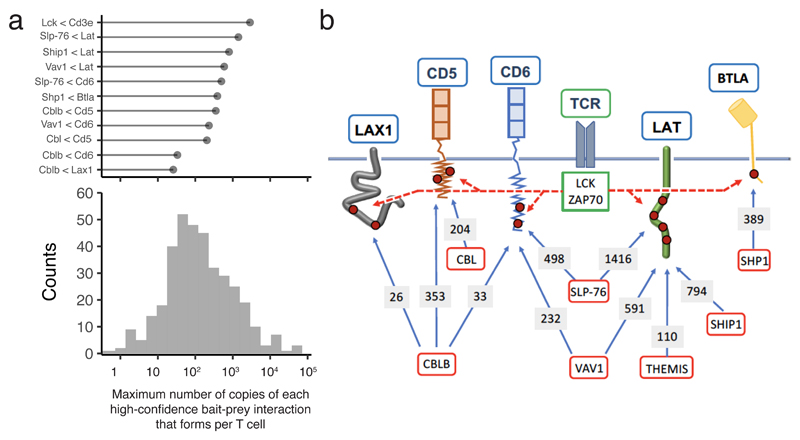
By combining interaction stoichiometries and cellular protein abundances, we enumerated the maximum number of copies of each high-confidence bait-prey interactions that forms per T cell over the course of TCR stimulation. Since TCR signals are initiated at the plasma membrane, we specifically focused on those high-confidence interactions that form at the inner face of the plasma membrane and constitute seeds for signal propagation (Fig. 6a). Consistent with the view that LAT serves as a early point of signal diversification downstream of the TCR2, 20, constitutive binary complexes made of GRAP2-SLP-76, GRB2-SHIP1 and GRB2-THEMIS docked onto LAT following TCR engagement and nucleated the assembly of SLP-76-, SHIP1- and THEMIS-based signalosomes (Fig. 6b). Such seeds reached maximal numbers of copies per T cell 30 s after TCR engagement, ranging from about a hundred (LAT-GRB2-THEMIS) to several hundreds (LAT-GRAP2-SLP-76 and LAT-GRB2-SHIP1) (Fig. 6b).
Fig. 6. TCR signals branch at the level of the plasma membrane leading to the assembly of multiple signalosomes.
a, For each of the 366 high-confidence bait-prey interactions, we calculated the maximun number of copies that form per CD4+ T cell over 600 s of TCR stimulation and showed their distribution in the lower panel. It ranged from 0.4 to 7.0 x 104 copies per T cell with a median close to 1.1 x 102 copies per T cell. The high-confidence bait-prey interactions that involve a prey corresponding to a receptor (CD3ε, CD5, CD6, BTLA) or an adaptor (LAT, LAX1) associated with the T cell plasma membrane are shown in the upper panel. b, Following TCR engagement, signalosomes involving CBL, CBLB and VAV1 nucleate around the CD5 and CD6 transmembrane receptors in numbers comparable to those that form around the LAT adaptor via SLP-76, SHIP1 and THEMIS intermediates. Also shown are the BTLA coinhibitory receptor and the LAX1 adaptor that assemble signalosomes involving SHP1 and CBLB, respectively. Tyrosine residues (red dots) present in the intracytoplasmic segments of the depicted receptors and adaptors are phosphorylated by the LCK or ZAP70 PTK that associate with active TCR (dashed red arrows). Baits and preys are denoted using red and blue boxes, respectively. The maximum number (copies per T cell) of bait-prey complexes reached over the course of 600 s of TCR stimulation is specified over the arrows connecting the baits and the preys. For instance, the maximm number of CD5-CBLB complexes reached per T cell over 600 s of TCR stimulation is approximately 353.
The signalosome assembling around the LAT-GRAP2-SLP-76 seed comprised PLC-γ1, the serine-threonine protein kinase MAP4K1 (HPK1), the FYB-SKAP1 binary complex, UBASH3A, GRB2, GRAP, 6 members of the 14-3-3 protein family, NCK1 and VAV1 (Fig. 7). The SLP-76-NCK1 interaction was of low stoichiometry and led to the recruitment of the ARAP1 RHO-GAP with high-confidence, of the WASp actin nucleation-promoting factor (FDR value of 4.7% at t600s), and of the beta-actin-like protein 2 (ACTBL2; FDR ≤ 3% at t30s). VAV1 recruited the SRC-like-adaptor protein 1 (SLAP-1), two RHO GDP-dissociation inhibitors (ARHGDIA and ARHGDIB; both with FDR ≤ 3% at t30s), and the small GTPase RHOA (FDR ≤ 3% at t30s) which activates WASp (Supplementary Data Set 1). Therefore, when probed by AP-MS under physiological conditions, the signalosome nucleating around the LAT-GRAP2-SLP-76 seed couples the TCR to the production of inositol trisphosphate and diacylglycerol and to F-actin dynamics, a finding consistent with recent imaging studies21, 22.
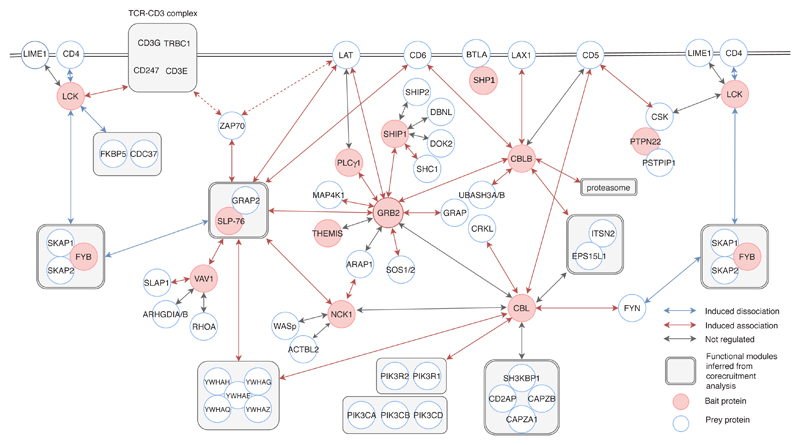
Fig. 7. A model summarizing the protein-protein interactions occurring during the first 10 minutes of TCR activation.
This diagram integrates data found in Fig. 2b and 4 and in Supplementary Data Sets 1 and 2. It focuses on those high-confidence bait-prey interactions (1) that are regulated upon TCR activation, (2) that engage a large fraction of the bait or prey present in a given T cell, or (3) that are part of a functional module identified using co-recruitment analysis (Supplementary Fig. 6). For a deeper exploratory analysis, we included interactions with an FDR ≤ 3% for one consecutive condition of stimulation. The interactions specified by red dotted arrows have been established in a former study4.
The signalosome that forms around the LAT-GRB2-THEMIS seed is thought to play a key role during T cell development in part due to its association with SHP123, 24, 25. SHP1 was, however, lacking in the THEMIS interactome of mature CD4+ T cells (Fig. 7 and Supplementary Data Set 1). To confirm such unanticipated finding, we analyzed whether THEMIS was present in the SHP1 interactome of mature CD4+ T cells. As expected, the B-and T-lymphocyte attenuator (BTLA26, 27) was found among the SHP1 preys, but THEMIS was still absent (Fig. 7). Along the same line, the GRB2 interactome of mature CD4+ T cells contained THEMIS but lacked SHP1 (Fig. 7), suggesting that THEMIS-GRB2-SHP1 ternary complexes primarily form in developing thymic T cells.
Consistent with the compound enzymatic and scaffolding role of SHIP1, the signalosome assembling around the LAT-GRB2-SHIP1 seed comprised several adaptors (GRB2, GRAP, SH3KBP1 (CIN85), SHC1, and DOK2), and associated with a lower interaction stoichiometry to molecules such as UBASH3A, DBNL (an adaptor protein that binds F-actin), FYN, and SHIP2 (Fig. 7 and Supplementary Data Set 1). Therefore, our quantitative and qualitative analysis of the SLP-76-, SHIP1- and THEMIS-based signalosomes emphasizes the unique contribution of LAT to TCR signal diversification, the breadth of which should be even greater when taking in account that the guanine nucleotide exchange factors SOS1 and SOS2 are also recruited via GRB2 to phosphorylated LAT molecules4.
CD5, CD6 and LAX1 contribute to TCR signal diversification at the plasma membrane
Akin to LAT, several receptors and adaptors embedded in the T-cell plasma membrane contain tyrosine residues that are capable of recruiting intracytoplasmic proteins following TCR-induced phosphorylation. Congruent with that view, CD6 was found in the SLP-76 and VAV interactomes, whereas CD5 was present in the CBL and CBLB interactomes (Fig. 7). The LAX1 transmembrane adaptor and CD6 were also found in the CBLB interactome, however with lower stoichiometries than CD5. The role of CD6 and LAX1 in T cell activation remains debated whereas CD5 negatively regulates TCR signaling. The numbers of copies per T cell of CD5-CBL (198), CD5-CBLB (329), CD6-SLP-76 (484), and CD6-VAV1 (214) complexes induced upon TCR activation were commensurable to those of LAT-SLP-76, LAT-SHIP1 and LAT-THEMIS complexes, unveiling their important quantitative contribution to TCR signal diversification. Moreover, the hundred of copies of CBL- and CBLB-based signalosomes assembling around CD5 after 120 s of stimulation strengthened the view that CD5 constitutes a major mediator of TCR-induced ubiquitylation9.
Assessment of the function of novel interacting proteins at organismal levels
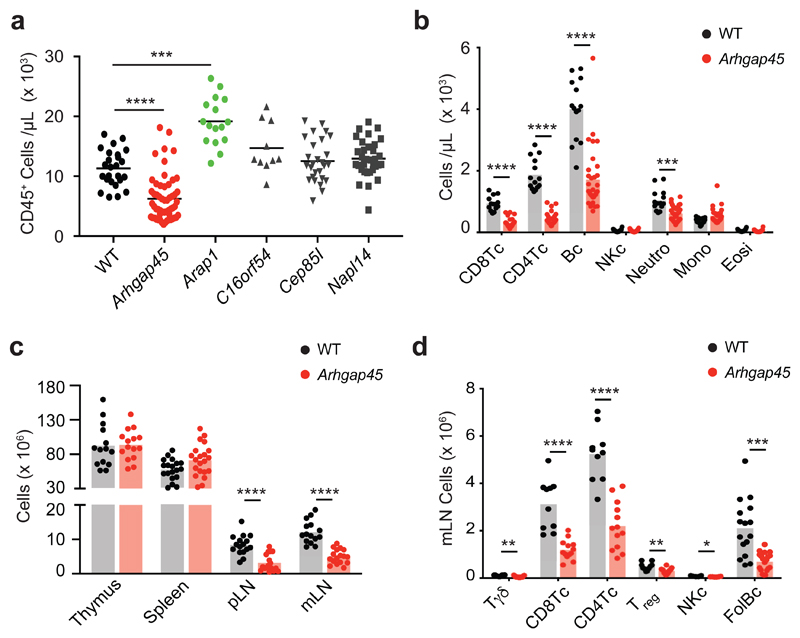
Genomic editing by CRISPR/Cas9-based approaches permits to study gene function in ex vivo primary T cells28. However, the role of genes involved in organismal function - such as lymphoid cell migration - cannot be assessed using ex vivo assays and requires developing mouse lines with homozygous loss-of-function mutations. To bypass such fastidious approach, we used CRISPR/Cas9 to establish F0 mice with biallelic deletions of a critical exon of genes coding for 5 poorly characterized preys identified in our interactomics dataset (see Methods). For each of the 5 tested genes (AI467606, Arap1, Arhgap45, Cep85l and Nap1l4), 3 single-guide RNAs were co-injected into zygotes and tail DNA from the 5 resulting F0 mice cohorts genotyped by PCR. 74% of the resulting F0 mice showed the expected biallelic DNA deletions and their blood was subjected to quantitative multiparametric analysis of myeloid and lymphoid cells (Fig. 8a). For instance, biallelic inactivation of the Arghap45 gene, which codes for a functionally uncharacterized RHO GTPase activating protein29 found in the LCK interactome (Supplementary Data Set 1), produced the strongest phenotype among the 5 analyzed genes. Arghap45 inactivation resulted in a severe reduction in blood CD45+ cell numbers (Fig. 8a). Consistent with the pattern of Arghap45 expression (http://www.immgen.org/databrowser/index.html), lymphocytes found in the blood were primarily affected by Arghap45 inactivation (Fig. 8b). Analysis of the progeny of F0 mice with biallelic Arghap45 gene inactivation showed that their thymus and spleen were normal in terms of composition and absolute numbers, suggesting that ARGHAP45 has no detectable role during T and B cell development (Fig. 8c). In contrast, their lymph nodes showed a reduction in T and B cell numbers similar to that of F0 mice (Fig. 8c,d). Considering that ARGHAP45 regulates actin cytoskeleton and cell spreading in transfected HeLa cells29, our data suggest that in absence of ARGHAP45, T and B cells have a reduced ability to enter into lymph nodes via diapedesis. Therefore, our reverse genetics approach constitutes a decision support tool permitting to identify, in 4 months and without mouse breeding, preys the ablation of which results in T cell phenotypes of interest when analyzed at organismal levels.
Fig. 8. ARGHAP45 is crucial for proper T and B cell migration.
a, Quantification of CD45+ cells in the blood of wild-type mice and of mice with biallelic deletions of the Arap1, AI467606, Arhgap45, Cep85l, and Nap1l4 genes. Each symbol corresponds to a mouse and the mean (horizontal bar) is indicated. Among the analyzed Arghap45 F0 mice, six contain one Arghap45 allele with a small deletion preserving its open reading frame and were used as internal control giving rise to normal blood phenotype. Wild-type: n = 25; Arhgap45: n = 56; Arap1, n=16; C16orf54: n = 27; Cep85l: n= 27; Nap1l4: n = 33. For each mutation, comparison was done relative to wild-type (WT) mice using unpaired Mann–Whitney’s t-test. ns, nonsignificant. *** P = 0.0002; **** P = 0.000004. In a to d, only significant values (P< 0.05) were specified. b, Quantification of CD8+ T cells, CD4+ T cells, B cells, NK cells, neutrophils, monocytes and eosinophils in the blood of wild-type mice (n = 14) and of F0 mice with biallelic deletions in the Arghap45 gene (n = 32). *** P = 0,00021; ****, P < 0.00001 (unpaired Holm-Sidak’s multiple t test corrected for multiple comparisons). c, Cellularity of the thymus, spleen, peripheral (p) and mesenteric (m) lymph nodes of the progeny of wild-type mice (n = 20) and of F0 mice with biallelic deletions in the Arghap45 gene (n = 20). ****, P< 0.00001 (unpaired Holm-Sidak’s multiple t test corrected for multiple comparisons). d, Quantification of γδ T cells, CD8+ T cells, conventional CD4+ T cells, regulatory CD4+ T cells, NK cells, and follicular B cells in the mesenteric lymph nodes of wild-type mice (n = 16) and of F0 mice with biallelic deletions in the Arghap45 gene (n = 16). *, P = 0.017; **, P = 0.004; ***, P = 0.00003; *****, P < 0.00001 (unpaired Holm-Sidak’s multiple t test corrected for multiple comparisons).
Discussion
We assessed the dynamics of the signalosomes assembling around 15 canonical proteins used by the TCR signal-transduction network in primary CD4+ T cells. The unique possibility to integrate interaction stoichiometries and cellular abundances over the course of TCR stimulation allowed us to reach key parameters for systems-level understanding of TCR signals propagation and diversification. For instance, we enumerated the number of signaling complexes that are used per T cell to convey TCR signals, and determined whether a fraction of the documented interacting proteins is left free to engage in alternative signalosomes over the course of stimulation. We also demonstrated that TCR signals divide into multiple branches at the level of the plasma membrane, resulting in the formation of CD5-, CD6-, BTLA- and LAX1-based signalosomes that assemble with kinetics comparable to the canonical LAT signalosome, and contribute to TCR signal diversification due to their unique composition in adaptor and effector molecules.
Our time-resolved analysis also illustrated the speed at which signalosomes assemble and disassemble following TCR triggering. For instance, full-blown LAT-SLP-76 signalosomes were already present 30 s after TCR triggering. A finding consistent with the view that TCR signals are transmitted in a rapid manner, resulting in phosphorylation of TCR proximal components in 4 s, and production of intracellular second messengers such as Ca2+ in 6-7 s30. The binding of MAP4K1 to SLP-76 peaked 30 s after TCR triggering. This led to serine-threonine phosphorylation of SLP-76 and GRAP231, and as documented here to the interaction of six 14-3-3 family members with SLP-76. These last interactions were of high stoichiometry and correlated with the dismantling of the LAT signalosome 120 s after TCR triggering, a finding consistent with the view that SLP-76 dissipates from TCR microclusters during their transport toward the center of immunological synapse32. We showed that the CD5-CBL and CD5-CBLB signalosomes that formed following TCR engagement in numbers comparable to the LAT signalosome are primarily endowed with negative regulatory role, likely contributing to the short temporal persistence of the LAT signalosome33.
Models accounting for the function of ITK, LCK, FYB and PTPN22 need to be revisited on the basis of the present quantitative interactomics study. Upon PtdIns(3,4,5)P3 production, ITK is recruited to the plasma membrane through its PH domain and after associating to the LAT-GRAP2-SLP-76 signalosome it phosphorylates and activates PLC-γ134. We found that LCK was 70-fold more abundant than ITK in CD4+ T cells, and that the LCK interactome was 60-fold larger than that of ITK. A reason for this last difference stems from the association of LCK with multiple transmembrane receptors and adaptors among which we identified CD4, the TCR-CD3 complex, ZAP70, LAT, CD2, CD6, CD28, CD45 (PTPRC), the interleukin 2 recpetor α and β subunits, and LIME1. In contrast to ITK which modulates the magnitude of calcium flux34, LCK is responsible for initiating the entire TCR signaling cascade and has T cell cancer promoting activities. Accordingly, several of the molecules unique to the LCK interactome are likely devoted to control its high activity state and degradation. They corresponded to chaperones (CDC37, DNAJA1 and 2, FKBP5 and 8, HSP90AA1, HSP90AB1, ST13), several 26S proteasome subunits and CAND1, a member of the SCF E3 ubiquitin ligase complex. Moreover, the presence of RAB11B and of several GTP-binding proteins (IFI47, IRGM1, IFGGD) in the LCK interactome likely fixes the activation threshold of naive T cells by controlling LCK subcellular localization35. The LCK interactome further differed from that of ITK by the presence of several proteins involved in nuclear export and import, suggesting unidentified roles of LCK.
FYB is rather unique among the analyzed baits in that it interacted with a large number of preys prior to TCR activation. Some of them, including FYB-SKAP1 and FYB-SKAP2, persisted after TCR activation, whereas other involving FYN, LCK and SLP-76 decreased upon TCR activation. Analysis of the LCK and SLP-76 interactomes confirmed that the FYB-LCK and FYB-SLP-76 interactions diminished after TCR engagement, a finding contrasting with results suggesting that FYB binds to SLP-76 in a TCR-inducible manner36. Unexpectedly, FYB constitutively interacted with several proteins involved in pre-mRNA splicing (corresponding to the SNW1-PPIL1 complex, PRPF3 and PRRC2A), nuclear RNA export (corresponding to DDX3X3 and the NXF1 and NXT1 complexes), and mRNA stability (KHDRBS1/SAM68), a finding consistent with the presence of two nuclear localization sequences in FYB and its cytoplasmic and nuclear distribution37. Therefore, the composition and dynamics of the FYB interactome point to as-yet-unidentified roles of FYB in T cells.
Genetic variants of PTPN22 are among the strongest genetic risk factors for several human autoimmunity diseases, the reasons of which remain unknown. We confirmed here the constitutive interaction between PTPN22 and the adaptor proline-serine-threonine phosphatase interacting protein 1 (PSTPIP1)38. Moreover, 7% of the PTPN22 molecules were associated to CSK prior to TCR stimulation. UBASH3A was found among the low-stoichiometry PTPN22 interactors, an observation to be related with genome wide association studies in which UBASH3A variants were linked to autoimmunity39. Moreover, after slightly relaxing cut-offs (FDR ≤ 3% for one condition of stimulation), the structural A (PPP2R1A), regulatory B (PP2R2A), and catalytic C (PP2CB) subunits of the serine-threonine-protein phosphatase 2A (PP2A) were found constitutively associated to PTPN22. PP2A has been recently implicated in several autoimmune disorders40, suggesting that PP2A contributes to the negative regulatory function of PTPN22 and its involvement in autoimmunity.
In conclusion, our study provides the most comprehensive analysis yet on the composition, stoichiometry and dynamics of the proximal TCR signal-transduction network in primary T cells. It illustrates the power of systems-level approaches to retrieve quantitative information on how TCR signals propagate and diversify at the level of the T cell plasma membrane for the successful initiation and termination of T cell activation. Moreover, the possibility to integrate the cellular abundance of the interacting proteins and their interaction stoichiometry provides a quantitative and contextual picture of each documented PPI that supersedes the view resulting from conventional co-immunoprecipitation approaches. Importantly, it constitutes a framework that should help rationalizing the phenotypic effect of genetic variations or drugs treatment intended to block T cell activation, and charting the redundant routes of signal propagation a T cell might use to bypass a drug-targeted component, which both constitute central issues in immune systems biology.
Methods
Mice
Mice were on C57BL/6 background, sex matched, and 9-week-old. They were maintained in specific pathogen-free conditions and used in accordance with institutional committee and European (Marseille) and Chinese (Xinxiang) guidelines for animal care. CBLOST mice (B6-Cbl tm1Mal), CBLBOST mice (B6-Cblb tm1Ciphe), GRB2OST mice (B6-Grb2 tm1Mal), SLP-76OST (B6-Lcp2 tm2Mal), and VAV1OST mice (B6-Vav1tm1Mal) have been described4, 7, 8, 9. Generation of FYBOST (B6-Fyb tm1Ciphe), SHIP1OST (B6-Inpp5d tm1Mal), ITKOST (B6-Itk tm1Ciphe), LCKOST (B6-Lck tm4Mal), NCK1OST (B6-Nck1 tm1Mal), NFATC2OST (B6-Nfatc2 tm1Ciphe), PLC-γ1OST (B6-Plcg1 tm1Ciphe), SHP1OST (B6-Ptpn6 tm1Mal), PTPN22OST (B6-Ptpn22 tm2Ciphe), and THEMISOST (B6-Themis tm1Ciphe) mice is described in Supplementary Note 1. Identification of OST-targeted alleles was performed by PCR using the pair of primers specified in Supplementary Table 1. Generation of F0 mice with biallelic null mutations in the Nap1l4, Arhgap45, AI467606, Cep85l, and Arap1 genes is described in Supplementary Note 2, and their analysis by flow cytometry described in Supplementary Note 3.
Biochemical validation of OST-tagged mice.
CD4+ T cells from wild-type mice and from each of the OST-tagged mice reported here were lysed in buffer (50 mM Tris, pH 7.5, 135 mM NaCl, 0.5 mM EDTA, 10% glycerol, 0.2% n-dodecyl-β-D-maltoside) supplemented with protease and phosphatase inhibitors. After 10 min of incubation on ice, cell lysates were centrifuged at 21,000 g for 5 min at 4°C. Post-nuclear lysates were used either as whole-cell lysates or immunopurified on Strep-Tactin (see below ‘Affinity purification of OST-tagged protein complexes’). Samples eluted from Strep-Tactin and whole-cell lysates were loaded on 8% SDS–PAGE gel and subsequently analyzed by immunoblot with specific antibodies: anti-ITK (sc-23902), anti-PTPN6 (sc-287), anti-SHIP1 (sc-8425), and anti-NFATc2 (sc-7296) were from Santa Cruz Biotechnology, anti-PLC-γ1 (2822), anti-VAV1 (2502), anti-SLP-76 (4958), anti-ZAP70 (2705) and anti-LCK (2752) from Cell Signaling Technology, anti-THEMIS (PA5-19288) was from Thermo Scientific, anti-NCK1 (610100) was from BD Biosciences, anti-FYB (07-546) was from Millipore and anti-PTPN22 was a gift from Genentech. Addition of the OST sequence had no effect on the expression of most tagged protein polypeptides (Supplementary Fig. 3a), with the exception of SHP1-OST polypeptides that were two-fold reduced as compared to their wild-type counterparts (see Supplementary Note 4).
Flow cytometry analysis of T cells from OST-tagged mice
Stained cells were analyzed using an LSRII system (BD Biosciences) and a Diva software (BD Biosciences). Cell viability was evaluated using SYTOX Blue (LifeTechnologies). The following antibodies were used: anti-CD5 (53-7.3), anti-CD4 (RM45) and anti-CD44 (Im7) were from BD Biosciences, anti-CD8 (53-6.7), anti-CD25 (PC61), anti-TCRβ (H57-597), anti-TCRγδ (GL3), and anti-CD19 (6D5) from BioLegend.
Functional analysis of CD4+ T cells from OST-tagged mice
For proliferation and IL-2 secretion assay, purified CD4+ T cells were stimulated with plate-bound anti-CD3 (145-2C11; Exbio) and soluble anti-CD28 (37-51; Exbio). After 48 h of culture, T-cell proliferation was assessed with CellTiter-Glo Luminescent (Promega). The resulting luminescence, which is proportional to the ATP content of the culture, was measured with a Victor 2 luminometer (Wallac, Perkin Elmer Life Science). IL-2 production was measured with a DuoSet ELISA test (R&D Systems).
CD4+ T cell isolation and short-term expansion for AP-MS analysis
CD4+ T cells were purified (> 95%) from pooled lymph nodes and spleens with Dynabeads Untouched Mouse CD4+ T Cell Kit (Life Technologies). Purified CD4+ T cells were expanded for 48 h with plate-bound anti-CD3 (145-2C11; 5 μg/ml) and soluble anti-CD28 (37-51; 1 μg/ml) both from Exbio. After 48 h of culture, CD4+ T cells were harvested and grown in the presence of IL-2 (10 U/ml) for 48 h prior to AP-MS analysis.
Stimulation and lysis of CD4+ T cells prior to AP-MS analysis
Short-term expanded CD4+ T cells (100 × 106) from wild-type mice and OST-tagged mice were incubated with anti-CD3 (0.2 μg per 106 cells; 145-2C11, Exbio) and anti-CD4 (0.2 μg per 106 cells; GK1.5, Exbio) on ice for 15 min, followed by one round of washing at 4°C. Cells were then incubated at 37°C for 5 min and then stimulated at 37°C with a purified rabbit anti-rat Ig (0.4 μg per 106 cells; Jackson Immuno-Research) for 30, 120, 300, and 600 s or left unstimulated. Stimulation was stopped by the addition of a twice-concentrated lysis buffer (100 mM Tris, pH 7.5, 270 mM NaCl, 1 mM EDTA, 20% glycerol, 0.4% n-dodecyl-β-D-maltoside) supplemented with protease and phosphatase inhibitors. After 10 min of incubation on ice, cell lysates were centrifuged at 21,000 g for 5 min at 4°C. Post-nuclear lysates were then used for affinity purification.
Affinity purification of OST-tagged protein complexes
Equal amounts of post-nuclear lysates were incubated with Strep-Tactin Sepharose beads (IBA GmbH) for 1.5 h at 4°C on a rotary wheel. Beads were then washed five times with 1 ml of lysis buffer in absence of detergent and of protease and phosphatase inhibitors. Proteins were eluted from the Strep-Tactin Sepharose beads with 2.5 mM D-biotin.
Tandem MS analysis
Following affinity purification, protein samples were partially air dried in a Speed-Vac concentrator, reconstituted in Laemmli buffer containing DTT (25 mM), and heated at 95°C for 5 min. Cysteines were alkylated for 30 min at room temperature by the addition of iodoacetamide (90 mM). Protein samples were loaded on an SDS-PAGE gel (0.15 × 3 × 8 cm) and subjected to electrophoresis. Migration was stopped as soon as the protein sample entered the gel. The gel was briefly stained with Coomassie blue, and a single slice containing the whole protein sample was excised. The gel slice was washed twice with 100 mM ammonium bicarbonate and once with 100 mM ammonium bicarbonate–acetonitrile (1:1). Proteins were in-gel-digested overnight at 37°C using 0.6 μg modified sequencing-grade trypsin (Promega) in 50 mM ammonium bicarbonate. The resulting peptides were extracted from the gel by one round of incubation (15 min, 37°C) in 50 mM ammonium bicarbonate and two rounds of incubation (15 min each, 37°C) in 10% formic acid–acetonitrile (1:1). The three extracted fractions were pooled and air- dried. Peptides were further purified on a C18 ZipTip (Millipore) and dried again. Tryptic peptides were resuspended in 20 μl of 2% acetonitrile and 0.05% trifluoroacetic acid and analyzed by MS. A mix of standard synthetic peptides (iRT Kit; Biognosys) was spiked in all samples to monitor the stability of the nano-LC-MS system during the analytical sequence. Peptides were analyzed by nano-liquid chromatography (LC) coupled to tandem MS, using an UltiMate 3000 system (NCS-3500RS Nano/Cap System; Dionex) coupled to an Orbitrap Velos Pro mass spectrometer (Thermo Fisher Scientific). 5 μl of each sample were loaded on a C18 precolumn (300 μm inner diameter × 5 mm; Dionex) in a solvent made of 2% acetonitrile and 0.05% trifluoroacetic acid, at a flow rate of 20 μl/min. After 5 min of desalting, the precolumn was switched online with the analytical C18 column (75 μm inner diameter × 50 cm, in-house packed with Reprosil C18) equilibrated in 95% solvent A (5% acetonitrile, 0.2% formic acid) and 5% solvent B (80% acetonitrile, 0.2% formic acid). Peptides were eluted using a 5-50% gradient of solvent B over 105 min and a flow rate of 300 nl/min. The LTQ Orbitrap Velos was operated in data-dependent acquisition mode with Xcalibur software. Survey scan MS was acquired in the Orbitrap on the 350-2,000 m/z range, with the resolution set to a value of 60,000. The 20 most intense ions survey scans were selected for fragmentation by collision-induced dissociation, and the resulting fragments were analyzed in the linear trap. Dynamic exclusion was used within 60 s to prevent repetitive selection of the same peptide. Technical LC-MS measurement replicates were performed for each sample (duplicate or triplicate runs depending on the series).
Protein identification and quantification for interaction proteomics
Raw MS files were processed with MaxQuant software (version 1.5.2.8) for database search with the Andromeda search engine and quantitative analysis. Data were searched against Mus musculus entries of the UniProt KB protein database (release UniProtKB/Swiss-Prot+TrEMBL 2017_01, 89297 entries including isoforms), the Biognosys iRT peptides, the One-Strep-tag peptide sequence, and the set of common contaminants provided by MaxQuant. Carbamidomethylation of cysteines was set as a fixed modification, whereas oxidation of methionine, protein N-terminal acetylation, and phosphorylation of serine, threonine, and tyrosine were set as variable modifications. Specificity of trypsin digestion was set for cleavage after K or R residues, and two missed trypsin cleavage sites were allowed. The precursor mass tolerance was set to 20 ppm for the first search and 4.5 ppm for the main Andromeda database search. The mass tolerance in tandem MS mode was set to 0.5 Da. Minimum peptide length was set to 7 amino acids, and minimum number of unique or razor peptides was set to 1 for validation. The I=L option of MaxQuant was enabled to avoid erroneous assignation of undistinguishable peptides belonging to very homologous proteins. Andromeda results were validated by the target decoy approach using a reverse database, with a FDR value set at 1% at both PSM (peptide sequence match) and protein level. For label-free relative quantification of the samples, the match between runs option of MaxQuant was enabled with a match time window of 1 min, to allow cross-assignment of MS features detected in the different runs, after alignment of the runs with a time window of 20 min. Protein quantification was based on unique and razor peptides. The minimum ratio count was set to 1 for LFQ calculation, and computation of the iBAQ metric was also enabled.
Statistics and data reproducibility
From the “proteinGroups.txt” files generated by MaxQuant with the options described above, proteins groups with negative identification scores were filtered as well as proteins identified as contaminants. Because protein groups were mapped using a redundant database (combining TrEMBL and Swiss-Prot), some protein groups corresponded to the same gene name. In such situations, protein intensities in a given sample were summed over the redundant protein groups. Protein intensities were log transformed before being normalized across all conditions (condition of stimulation, biological and technical replicates) by the median intensity. Normalized intensities corresponding to different technical replicates were averaged and missing values were replaced after estimating background binding from wild-type intensities. For each bait and each condition of stimulation, we used a two-tailed Welch t-test to compare normalized protein intensities detected in OST-tagged samples across all biological replicates with wild-type intensities pooled from all conditions of stimulation. Logarithmized fold-change and corresponding p-values were used to generate a global volcano plot representing interactions from all baits and all conditions of stimulation (Supplementary Fig. 4c). Asymmetry of this global volcano plot was used to define regions corresponding to different FDR values6. Briefly, the volcano plot was divided into different regions by a line of equation f(x)=c/(|x|-x0. For a given set of parameters (c, x0), the FDR is computed using the number of proteins falling in the upper left and right regions, denoted as nleft and nright respectively, according to the formula:
The FDR value of a given bait-prey interaction at a given stimulation time is then taken as the maximum FDR value calculated across all (c, x0) parameters. Bait-prey interactions with an FDR value ≤ 3% for two consecutive stimulation times were identified and referred to as high confidence interactions.
Calculation of interaction stoichiometries
For a given condition of stimulation (represented by the time of stimulation t, with t = 0 corresponding to the unstimulated condition), the stoichiometry of the interaction between a given bait and a prey x (denoted bait<x) was calculated using:
Npep corresponds to the number of tryptic peptides theoretically observables and brackets represent averages across all biological replicates. We also computed stoichiometries independently for each biological replicate, denoted Sbait<x(t; rep), and those values were used to quantify correlation in recruitment between different preys (see ‘Co-recruitment analysis’).
High-resolution MS characterization of the CD4+ T cell proteome
For proteome analysis, CD4+ T cells from wild-type and from OST-tagged mice were briefly expanded in vitro as described for AP-MS experiments. Cell pellets (5 × 106 cells) were incubated with a 150 μl of lysis buffer containing Tris 50 mM, pH 7.5, EDTA 0.5 mM, NaCl 135 mM, SDS 1% for 10 min on ice and subjected to sonication with a Bioruptor ultrasonicator. Protein concentration was determined using a detergent-compatible assay (DC assay, Bio-Rad) and total protein amounts were adjusted across samples. Each protein sample was migrated briefly on SDS-PAGE gel and processed for in-gel digestion as described for AP-MS. Resulting peptide mixtures were analyzed with a fast-sequencing QExactivePlus Orbitrap mass spectrometer (ThermoScientific, Bremen, Germany), using long gradient runs (5-50% gradient of solvent B over 240 min). Survey MS scans were acquired in the Orbitrap on the 350-2000 m/z range with a resolution of 70,000, the 10 most intense ions per survey scan were selected for HCD fragmentation and resulting fragments were analyzed at a resolution of 17,500 in the Orbitrap. Raw MS files were processes with MaxQuant as described above, with a search against Mus musculus entries of the UniProt KB/Swiss-Prot protein database, using a tolerance of 20 ppm for MS/MS in the Orbitrap.
Calculation of cellular protein abundance
Analysis of the proteome of CD4+ T cells from 14 mice lines (corresponding to 1 wild-type and 13 distinct OST-tagged genetic backgrounds) identified 4643 protein groups. Proteins entries from the MaxQuant “proteinGroups.txt” output were first filtered to eliminate entries from reverse and contaminant databases. Cellular protein abundances were determined from raw intensities using the protein ruler methodology11, using the following relationship: protein copies per cell = (protein MS signal × NA × DNA mass) / (M × Histone MS signal), where NA is Avogadro’s constant, M is the molar mass of the protein, and the DNA mass of a diploid mouse cell estimated to be 5.5209 pg. Cellular protein abundance were log transformed and averaged sequentially over technical replicates, and biological replicates for all genetic backgrounds. Overall, cellular protein abundance could be estimated for 4148 protein groups and averaged across 14 different backgounds.
Peptide fractionationwas further used to increase the depth of the CD4+ T cell proteome prior to and after stimulation through the TCR and CD4. This led to the identification of 6388 protein groups. Cellular rotein abundances for 6104 protein groups were determined as described above and averaged across all conditions of stimulation. To merge our two proteomes, the median averaged cellular protein abundance of the second proteome was adjusted to that of the first proteome. 6343 unique majority protein ID (which contained at least half of a protein groups razor + unique peptides) were then identified across all protein groups (Supplementary Data Set 2). In the case where a majority protein ID was identified in both proteomes, the cellular protein abundance value was taken from the proteome where the highest number of razor + unique peptides was associated to this protein ID.
Calculation of reciprocal stoichiometries
For all bait A-prey B interactions A<B, the stoichiometry of the reciprocal interaction B<A was calculated using the following relationship:
where NA and NB are cellular protein abundances of A and B respectively. Note that reciprocal stoichiometries could only be computed when the cellular abundances of both proteins were available. Comparison between experimentally determined stoichiometries and stoichiometries predicted from the reciprocal interaction was performed in all cases where both A and B were protein baits and both A<B and B<A interaction stoichiometries were quantified.
Co-recruitment analysis
We previously demonstrated that the occurrence of temporal correlations in the recruitment of two preys to a given bait can be used to infer functional or physical relationships between these preys and construct a co-recruitment network9. The rationale behind this reasoning is that physically interacting preys or preys cooperating toward a peculiar function should have similar profiles of recruitment to a given bait as a function of time of TCR stimulation. Here, we extended this approach to the analysis of recruitment correlations within the 15 interactomes. Within each interactome we quantified correlations in interaction stoichiometries for all pairs of high-confidence preys and selected strongly correlated pairs (Pearson R ≥ 0.8 with an associated p-value p ≤ 0.05). To limit the maximum degree of the resulting correlation network, we only kept the two preys that were the most strongly correlated with a given prey. Correlation networks from individual interactomes were then merged to form a single global co-recruitment network, the finer structure of which was divided into protein communities using a modularity optimization algorithm41.
Comparison with PPI reported in databases
Bait-prey interactions identified in this study were compared to PPI previously reported between human or mouse proteins in the publicly available BioGrid, IntAct, MINT and HPRD databases.
Annotation enrichment analysis
We annotated the different identified proteins using Uniprot-Protein families and Uniprot-Keywords. We used a hypergeometric test to determine if an annotation term was statistically enriched in a given interactome as compared to a background composed of all proteins identified in this interactome (without any filtering based on the FDR). Annotations that were represented by at least two high confidence preys (FDR ≤ 3% for two consecutive stimulation times) and had an enrichment greater than 2-fold with a p-value lower than 0.05 were selected as enriched terms. We used the same procedure to analyze annotation enrichment within functional communities identified in the co-recruitment network, the backgound corresponding to the set of high confidence preys.
Supplementary Material
Acknowledgements
We thank D. Mori, C. Wülfing (University of Bristol), and A. Zarubica for discussions, E. Bergot, S. Li, T. Chao, S. Durand and the late F. Danjan for technical help. This work was supported by CNRS, INSERM, the European Research Council (ERC) under FP7 program (grant agreement n° 322465 (INTEGRATE) to B.M.) and the European Union’s Horizon 2020 research and innovation program (grant agreement n° 787300 (BASILIC) to B.M.), Agence Nationale de la Recherche (Basilic project to M.M.), the MSDAVENIR Fund (to B.M.), the Investissement d’Avenir program of the French Ministry of Research ProFI (Proteomics French Infrastructure, ANR-10-INBS-08 to O.B.-S.), and PHENOMIN (French National Infrastructure for mouse Phenogenomics; ANR10-INBS-07 to B.M.), the National Natural Science Foundation of China (grants n° 81471595 and 31400759 to Y.L.) and the Education Department of Henan Province, China (16HASTIT030 to Y.L.) and by fellowships from the Integrate (M.G.M, G.V., K.K., E.B, S.D. and K.C.), MSDAVENIR (Y.O.), and PHENOMIN (L.G.) projects.
Footnotes
Author contributions
B.M., R.R. and G.V. conceived the project; B.M. and F.F supervised the construction of OST-tagged mice; R.R. and M.M. performed the experiments shown in Supplementary Fig. 1 to 3 with the help of L.G., Y.O. and M.G.M.; K.K. characterized the SHIP1OST, PLC-γ1OST and PTPN6OST mice with the help of J.C.; A.G. de P., K.C. and O.B.S. performed the MS experiments; G.V. designed the computational and bioinformatics analysis; Y.L., M.M., H.L. and B.M designed the experiments shown in Figure 8 and L.L., L.Z. and H.W. performed them. B.M., G.V, and R.R. wrote the manuscript.
Competing Interests Statement
The authors declare no competing interests.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) with the dataset identifiers PXD007660 and PXD003972.
Online content
Supplementary information includes 6 Figures, 2 Tables, 4 Notes and 2 Data Sets and can be found with this article on line.
Data Sets
Supplementary Data Set 1. List of the bait-prey interactions identified in the present study. In tab ‘bait-prey interactions’, each line shows an interaction between a bait and a prey and includes the corresponding FDRs, enrichments and stoichiometries prior to stimulation and at each stimulation time point. Cellular abundances (number of copies per cell) of the listed proteins are also specified. Where applicable, protein-protein interaction identified in public databases are indicated with the corresponding references and detection methods. The lists of the high-confidence bait-prey interactions identified for each bait are also shown in tabs CBL to VAV1.
Supplementary Data Set 2. Proteome of antigen-experienced conventional CD4+ T cells. The proteins identified in CD4+ T cells from wild-type and OST-tagged mice and their cellular abundance (number of copies per cell) are shown (see Methods).
References
- 1.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. 2014;15:798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. 2013;13:257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 3.Shah NH, et al. An electrostatic selection mechanism controls sequential kinase signaling downstream of the T cell receptor. Elife. 2016;5 doi: 10.7554/eLife.20105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roncagalli R, et al. Quantitative proteomics analysis of signalosome dynamics in primary T cells identifies the surface receptor CD6 as a Lat adaptor-independent TCR signaling hub. Nat Immunol. 2014;15:384–392. doi: 10.1038/ni.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Astoul E, Edmunds C, Cantrell DA, Ward SG. PI 3-K and T-cell activation: limitations of T-leukemic cell lines as signaling models. Trends Immunol. 2001;22:490–496. doi: 10.1016/s1471-4906(01)01973-1. [DOI] [PubMed] [Google Scholar]
- 6.Hein MY, et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 7.Caron E, et al. Precise Temporal Profiling of Signaling Complexes in Primary Cells Using SWATH Mass Spectrometry. Cell Rep. 2017;18:3219–3226. doi: 10.1016/j.celrep.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaud G, et al. The costimulatory molecule CD226 signals through VAV1 to amplify TCR signals and promote IL-17 production by CD4(+) T cells. Sci Signal. 2018;11 doi: 10.1126/scisignal.aar3083. [DOI] [PubMed] [Google Scholar]
- 9.Voisinne G, et al. Co-recruitment analysis of the CBL and CBLB signalosomes in primary T cells identifies CD5 as a key regulator of TCR-induced ubiquitylation. Mol Syst Biol. 2016;12:876. doi: 10.15252/msb.20166837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 11.Wisniewski JR, Hein MY, Cox J, Mann M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol Cell Proteomics. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruepp A, et al. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2010;38:D497–501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, et al. Deficiency of ADAP/Fyb/SLAP-130 destabilizes SKAP55 in Jurkat T cells. J Biol Chem. 2005;280:23576–23583. doi: 10.1074/jbc.M413201200. [DOI] [PubMed] [Google Scholar]
- 14.Seet BT, et al. Efficient T-cell receptor signaling requires a high-affinity interaction between the Gads C-SH3 domain and the SLP-76 RxxK motif. Embo J. 2007;26:678–689. doi: 10.1038/sj.emboj.7601535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giuriato S, et al. SH2-containing inositol 5-phosphatases 1 and 2 in blood platelets: their interactions and roles in the control of phosphatidylinositol 3,4,5-trisphosphate levels. Biochem J. 2003;376:199–207. doi: 10.1042/BJ20030581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu SL, et al. Quantitative Lipid Imaging Reveals a New Signaling Function of Phosphatidylinositol-3,4-Bisphophate: Isoform- and Site-Specific Activation of Akt. Mol Cell. 2018;71:1092–1104 e1095. doi: 10.1016/j.molcel.2018.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goulden BD, et al. A high-avidity biosensor reveals plasma membrane PI(3,4)P2 is predominantly a class I PI3K signaling product. J Cell Biol. 2018;218:1066–1079. doi: 10.1083/jcb.201809026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M, et al. K33-linked polyubiquitination of Zap70 by Nrdp1 controls CD8 T cell activation. Nat Immunol. 2015;16:1253–1262. doi: 10.1038/ni.3258. [DOI] [PubMed] [Google Scholar]
- 19.Paster W, et al. GRB2-mediated recruitment of THEMIS to LAT is essential for thymocyte development. J Immunol. 2013;190:3749–3756. doi: 10.4049/jimmunol.1203389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balagopalan L, Kortum RL, Coussens NP, Barr VA, Samelson LE. The linker for activation of T cells (LAT) signaling hub: from signaling complexes to microclusters. J Biol Chem. 2015;290:26422–26429. doi: 10.1074/jbc.R115.665869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumari S, et al. Actin foci facilitate activation of the phospholipase C-gamma in primary T lymphocytes via the WASP pathway. Elife. 2015;4 doi: 10.7554/eLife.04953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ditlev JA, et al. A composition-dependent molecular clutch between T cell signaling condensates and actin. Elife. 2019;8 doi: 10.7554/eLife.42695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi S, et al. THEMIS enhances TCR signaling and enables positive selection by selective inhibition of the phosphatase SHP-1. Nat Immunol. 2017;18:433–441. doi: 10.1038/ni.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehta M, et al. Themis-associated phosphatase activity controls signaling in T cell development. Proc Natl Acad Sci U S A. 2018;115:E11331–E11340. doi: 10.1073/pnas.1720209115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zvezdova E, et al. Themis1 enhances T cell receptor signaling during thymocyte development by promoting Vav1 activity and Grb2 stability. Sci Signal. 2016;9:ra51. doi: 10.1126/scisignal.aad1576. [DOI] [PubMed] [Google Scholar]
- 26.Murphy TL, Murphy KM. Slow down and survive: Enigmatic immunoregulation by BTLA and HVEM. Annu Rev Immunol. 2010;28:389–411. doi: 10.1146/annurev-immunol-030409-101202. [DOI] [PubMed] [Google Scholar]
- 27.Celis-Gutierrez J, et al. Quantitative Interactomics in Primary T Cells Provides a Rationale for Concomitant PD-1 and BTLA Coinhibitor Blockade in Cancer Immunotherapy. Cell Rep. 2019;27:3315–3330 e3317. doi: 10.1016/j.celrep.2019.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seki A, Rutz S. Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J Exp Med. 2018;215:985–997. doi: 10.1084/jem.20171626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Kreuk BJ, et al. The human minor histocompatibility antigen 1 is a RhoGAP. PLoS One. 2013;8:e73962. doi: 10.1371/journal.pone.0073962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huse M, et al. Spatial and temporal dynamics of T cell receptor signaling with a photoactivatable agonist. Immunity. 2007;27:76–88. doi: 10.1016/j.immuni.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 31.Lasserre R, et al. Release of serine/threonine-phosphorylated adaptors from signaling microclusters down-regulates T cell activation. J Cell Biol. 2011;195:839–853. doi: 10.1083/jcb.201103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashimoto-Tane A, Saito T. Dynamic Regulation of TCR-Microclusters and the Microsynapse for T Cell Activation. Front Immunol. 2016;7:255. doi: 10.3389/fimmu.2016.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi J, Balagopalan L, Nguyen T, McIntire KM, Samelson LE. TCR microclusters form spatially segregated domains and sequentially assemble in calcium-dependent kinetic steps. Nat Commun. 2019;10:277. doi: 10.1038/s41467-018-08064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreotti AH, Joseph RE, Conley JM, Iwasa J, Berg LJ. Multidomain Control Over TEC Kinase Activation State Tunes the T Cell Response. Annu Rev Immunol. 2018;36:549–578. doi: 10.1146/annurev-immunol-042617-053344. [DOI] [PubMed] [Google Scholar]
- 35.Bouchet J, et al. Rab11-FIP3 Regulation of Lck Endosomal Traffic Controls TCR Signal Transduction. J Immunol. 2017;198:2967–2978. doi: 10.4049/jimmunol.1600671. [DOI] [PubMed] [Google Scholar]
- 36.Geng L, Raab M, Rudd CE. Cutting edge: SLP-76 cooperativity with FYB/FYN-T in the Up-regulation of TCR-driven IL-2 transcription requires SLP-76 binding to FYB at Tyr595 and Tyr651. J Immunol. 1999;163:5753–5757. [PubMed] [Google Scholar]
- 37.Veale M, et al. Novel isoform of lymphoid adaptor FYN-T-binding protein (FYB-130) interacts with SLP-76 and up-regulates interleukin 2 production. J Biol Chem. 1999;274:28427–28435. doi: 10.1074/jbc.274.40.28427. [DOI] [PubMed] [Google Scholar]
- 38.Marcos T, et al. Proline-serine-threonine phosphatase interacting protein 1 inhibition of T-cell receptor signaling depends on its SH3 domain. Febs J. 2014;281:3844–3854. doi: 10.1111/febs.12912. [DOI] [PubMed] [Google Scholar]
- 39.Stanford SM, Mustelin TM, Bottini N. Lymphoid tyrosine phosphatase and autoimmunity: human genetics rediscovers tyrosine phosphatases. Semin Immunopathol. 2010;32:127–136. doi: 10.1007/s00281-010-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Q, et al. Phosphatase PP2A is essential for TH17 differentiation. Proc Natl Acad Sci U S A. 2019;116:982–987. doi: 10.1073/pnas.1807484116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clauset A, Newman ME, Moore C. Finding community structure in very large networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70 doi: 10.1103/PhysRevE.70.066111. 066111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) with the dataset identifiers PXD007660 and PXD003972.
Online content
Supplementary information includes 6 Figures, 2 Tables, 4 Notes and 2 Data Sets and can be found with this article on line.
Data Sets
Supplementary Data Set 1. List of the bait-prey interactions identified in the present study. In tab ‘bait-prey interactions’, each line shows an interaction between a bait and a prey and includes the corresponding FDRs, enrichments and stoichiometries prior to stimulation and at each stimulation time point. Cellular abundances (number of copies per cell) of the listed proteins are also specified. Where applicable, protein-protein interaction identified in public databases are indicated with the corresponding references and detection methods. The lists of the high-confidence bait-prey interactions identified for each bait are also shown in tabs CBL to VAV1.
Supplementary Data Set 2. Proteome of antigen-experienced conventional CD4+ T cells. The proteins identified in CD4+ T cells from wild-type and OST-tagged mice and their cellular abundance (number of copies per cell) are shown (see Methods).