Summary
Quantitative mass spectrometry reveals how CD4+ and CD8+ T cells re-structure proteomes in response to antigen and mammalian target of rapamycin complex 1 (mTORC1). Analysis of copy numbers per cell of >9000 proteins provides new understanding of T cell phenotypes, exposing the metabolic and protein synthesis machinery and environmental sensors that shape T cell fate. We reveal that lymphocyte environment sensing was controlled by immune activation and that CD4+ and CD8+ T cells differed in their intrinsic nutrient transport and biosynthetic capacity. The data also revealed shared and divergent outcomes of mTORC1 inhibition in naïve versus effector T cells: mTORC1 inhibition impaired cell cycle progression in activated naïve cells, but not effector cells, whereas metabolism was consistently impacted in both populations. This study provides a comprehensive map of naïve and effector T cell proteomes and a resource for exploring and understanding T cell phenotypes and cell context effects of mTORC1.
Introduction
T lymphocytes respond to antigens, co-stimulators and cytokines by transcriptionally remodeling, proliferating, and differentiating to effector populations. T cell activation is also associated with dynamic changes in mRNA translation, amino acid transport and protein synthesis that shape how transcriptional programs are implemented1–3. The full effect of immune activation on T cells can thus only be understood by deep analysis of T cell proteomes. The use of high-resolution mass spectrometry for quantitative mapping of cellular protein signatures is thus a necessary tool for understanding lymphocyte phenotypes4–10.
One key signaling molecule that controls protein turnover in mammalian cells is the nutrient sensing protein kinase mTORC111. In this context, mTORC1 is a key regulator of T cell differentiation but molecular understanding of how mTORC1 controls T cell biology is incomplete and it is still unclear whether mTORC1 controls the same biological processes in different T cell populations12–15. For example, a comparison of how mTORC1 inhibition remodeled proteomes of polyclonally activated naïve CD4+ T cells as they exit quiescence and effector CD8+ cytotoxic T cells suggested shared and unique effects of losing mTORC1 activity5, 7. Moreover, mTORC1 inhibition restrains the first cell cycle entry of immune-activated naïve T cells, but has limited effect on the proliferation of rapidly cycling cells5, 12, 16, 17. The reasons for these differences is unresolved but could reflect intrinsic differences in mTORC1 function in different T cell populations. In the present study, high-resolution mass spectrometry (MS) was used to analyze proteomes of naïve and antigen activated murine CD4+ and CD8+ T cells and CD4+ TH1 and CD8+ cytotoxic effector T cells. We also compared how mTORC1 inhibition impacts CD4+ and CD8+ T cell exit from quiescence versus how mTORC1 reshapes differentiated effector CD4+ and CD8+ T cell proteomes. We quantify >9400 proteins providing a valuable resource that reveals how immune activation and mTORC1 reshape the proteomic landscape of naïve and effector CD4+ and CD8+ T cells. This open access data resource can be readily interrogated online via the Encyclopedia of Proteome Dynamics (EPD) (www.peptracker.com/epd). The data show how immune activation shapes CD4+ and CD8+ T cell metabolic processes and their ability to sense environmental stimuli. The data also reveal no major differences in mTORC1 function in CD4+ and CD8+ T cells but different consequences of mTORC1 inhibition at different stages of T cell differentiation. The data highlight the power of quantitative analysis of protein copy numbers and the stoichiometry of protein complexes for understanding how immune regulators control T cell function.
Results
Proteome re-modelling during T cell differentiation
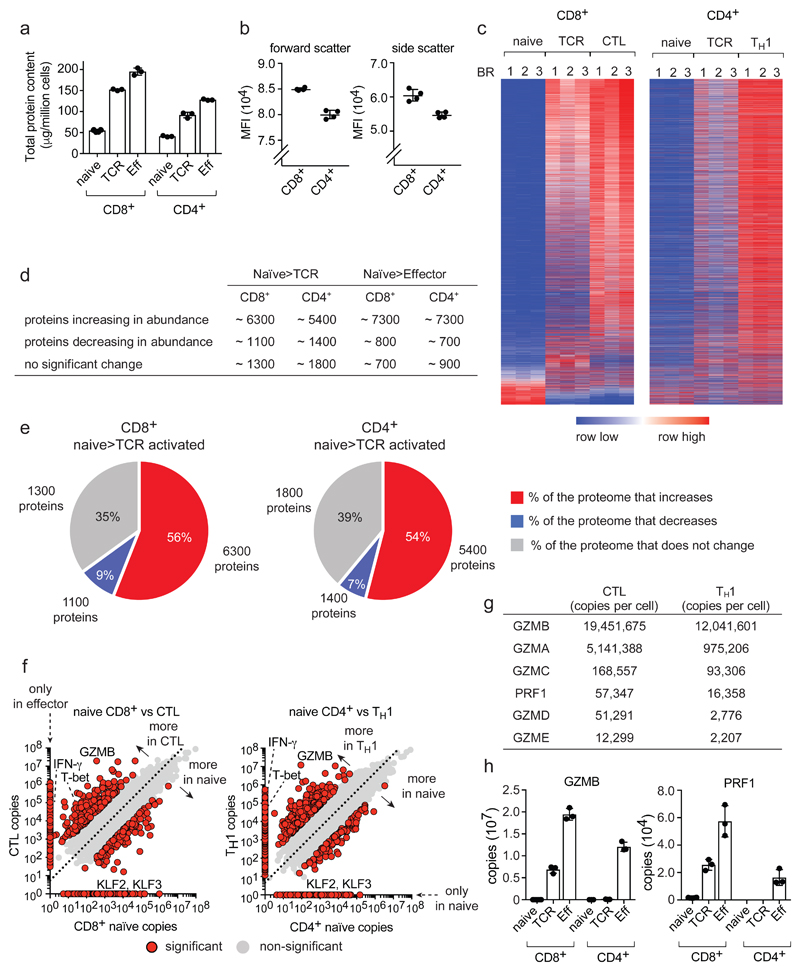
Quantitative high-resolution mass spectrometry resolved proteomes of naïve CD4+ and CD8+ T cells before and after 24 h antigen activation and proteomes of CD8+ cytotoxic T cell (CTLs) and CD4+ T helper (TH1) populations. Antigen activation models were P14 CD8+ T cells expressing TCRs specific for lymphocytic choriomeningitis virus glycoprotein peptide gp33-41 and OT-II CD4+ T cells expressing ovalbumin reactive TCRs. We also explored how mTORC1 regulates the proteomes of antigen activated naïve CD4+ and CD8+ cells compared to effects of mTORC1 inhibition in differentiated TH1 and CTLs. We identified 9400 T cell proteins and estimated absolute protein copies per cell using the ‘proteomic ruler’ method which uses the mass spectrometry signal of histones as an internal standard18. This method avoids error prone steps of cell counting and protein concentration evaluation and can be used to estimate protein abundance per cell18. These analyses revealed that CD8+ T cells triple their protein content within 24 h of antigen activation and CTLs have a 4-fold higher total protein content than naïve CD8+ cells (Fig. 1a). Immune activated CD4+ T cells also increase protein content but consistently had a lower (20%-30%) protein content than the corresponding CD8+ population (Fig. 1a). Note there was a slightly lower protein content of naïve CD4+ versus CD8+ T cells (Fig. 1a) which is consistent with forward and side light scattering analysis which indicates that naive CD4+ T cells are slightly smaller than naïve CD8+ T cells (Fig. 1b).
Figure 1. Proteome re-modelling during T cell differentiation.
(a) Total protein content of naïve, 24 h TCR-activated (TCR) and effector (Eff) populations. (b) Mean fluorescence intensity (MFI) of forward/side scatter for naïve CD4+ and CD8+ T cells. (c) Heat map of CD4+ and CD8+ proteomes during differentiation. The full list of proteins within the heat map is provided in Supplementary Data File 1. (d) Number of proteins changing in abundance during T cell differentiation: naïve to TCR activated and naïve to effector (P value <0.05, fold change >1.5 (two-tailed t-test with unequal variance) or presence/absence expression). (e) The proportion of the cell mass that corresponds to proteins increasing, decreasing or not changing in naïve cells in response to TCR triggering. The number of proteins in each category is also provided. Proteins were categorized as changing as described above. (f) Protein copy number comparisons between naïve and effector populations for CD4+ and CD8+ T cells. Proteins highlighted in red are significantly different between naïve and effector or show a presence/absence expression profile (P value <0.05, fold change >2 standard deviations from the mean fold change, two-tailed t-test with unequal variance). Proteins that were not detected in one population are positioned on the axis and are highlighted in red. The dashed line = mean fold change between naïve and effector. Interferon-γ (IFN-γ), granzyme B (GZMB), T-bet (TBX21) and Kruppel-like factor 2 and 3 (KLF2 and KLF3). (g) Abundance of effector molecules in CTLs and TH1 cells. (h) Abundance of GZMB and perforin (PRF1). For a, d, e, f, g and h, n = 6 biologically independent samples for CD8+ naïve cells and 3 biologically independent samples for each of the other T cell populations. For b, n = 4 biologically independent samples. For c, n = 3 biologically independent samples for each T cell population. Histogram bars represent the mean +/- SD.
To establish whether cell mass increases in activated T cells reflect scaled increases in expression of existing proteins or increased expression of new proteins, we used nearest neighbor analysis and Pearson correlation to align expression profiles of >8000 proteins in T cell populations. This analysis shows immune activation does not scale up all proteins but dynamically reshapes proteome landscapes (Fig. 1c and Supplementary Data File 1). There were increases in abundance of >6000 proteins in activated CD8+ cells and almost 5500 proteins in activated CD4+ cells compared to naïve cells (Fig. 1d). However, 1300-1800 proteins did not change abundance and a substantial proportion of the naïve cell proteome (7-9%) was downregulated as T cells respond to antigen (Fig. 1c,d,e). Proteins whose expression decreased upon immune activation had diverse functions (Supplementary Data File 2) and included translational repressors, cell cycle inhibitors and transcription factors.
Protein copy number comparisons illustrates proteins exclusively found at a particular stage of T cell differentiation and proteins expressed in all populations at either equal, enriched or depleted levels (Fig. 1f, Supplementary Data File 2 and 3). We detected >800 proteins in effector CD4+ and CD8+ cells that were not found in naive T cells (Fig. 1f) including effector molecules such as interferon-γ (IFN-γ), granzymes and perforin, (Fig. 1f, Supplementary Data File 2). Interestingly, effector TH1 cells expressed comparably high levels of granzymes and perforin as CTLs (Fig. 1g,h). The pattern of expression of transcription factors and chromatin regulators is also remodeled as T cells differentiate (Fig. 2a,b). For example, immune activation causes T cells to downregulate expression of Kruppel-like family (KLF) transcription factors which maintain pluripotency and cell quiescence (Fig. 2a). The complexity and extent of the transcription factor and proteome remodeling driven by immune activation was remarkable. However, it was striking that there were more similarities than differences between CD4+ and CD8+ T cells reflecting that the bulk of cellular proteins comprise core machinery essential to every cell (Supplementary Fig. 1).
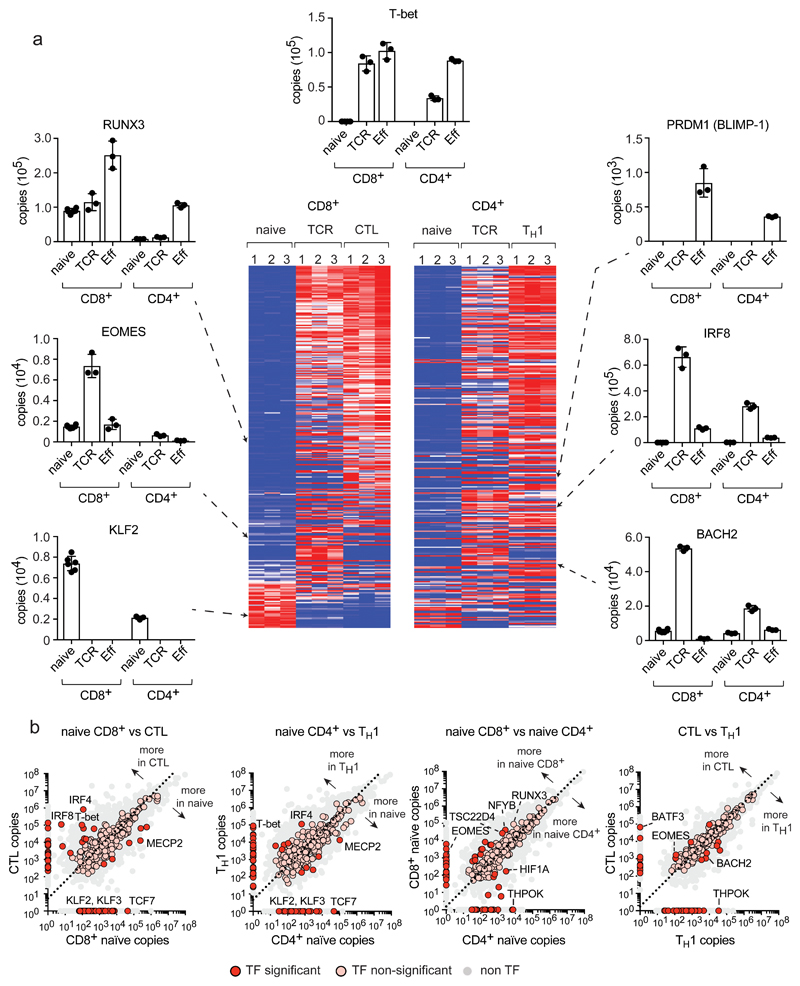
Figure 2. Expression profile of transcription factors during T cell differentiation.
Over 300 proteins annotated as DNA binding/transcription factor activity (GO:0003700) were identified in CD4+ and CD8+ T cell populations. Additional transcription factors without a GO annotation were added manually. (a) Transcription factor expression profiles during T cell differentiation. The full list of proteins included in heat maps is provided in Supplementary Data File 1. Histograms showing protein copy numbers per cell are provided for a selection of core transcription factors essential for T cell differentiation and activity: Runt Related Transcription Factor 3 (RUNX3); Eomesodermin (EOMES); Interferon Regulatory Factor 8 (IRF8); BTB Domain And CNC Homolog 2 (BACH2). (b) Copy number comparisons for transcription factors in different T cell populations. Scatter plots show the average copy number for transcription factors and allow a two-way comparison between T cell populations. Proteins that were not detected in one population are positioned on the axis. Transcription factors significantly different between 2 populations (P value <0.05 and a fold change >2 standard deviations from the mean fold change, two-tailed t-test with unequal variance) or showing a presence/absence expression profile, are represented with a red circle, while non-significant transcription factors are represented with a pink circle. The dashed line = the mean fold change between 2 populations. For heat maps in a, n = 3 biologically independent samples for each T cell population. For histograms in a and plots in b, n = 6 biologically independent samples for CD8+ naïve cells and 3 biologically independent samples for each of the other T cell populations. Histogram bars represent the mean +/- SD.
Scaling versus enrichment of core proteins and processes in differentiating T cells
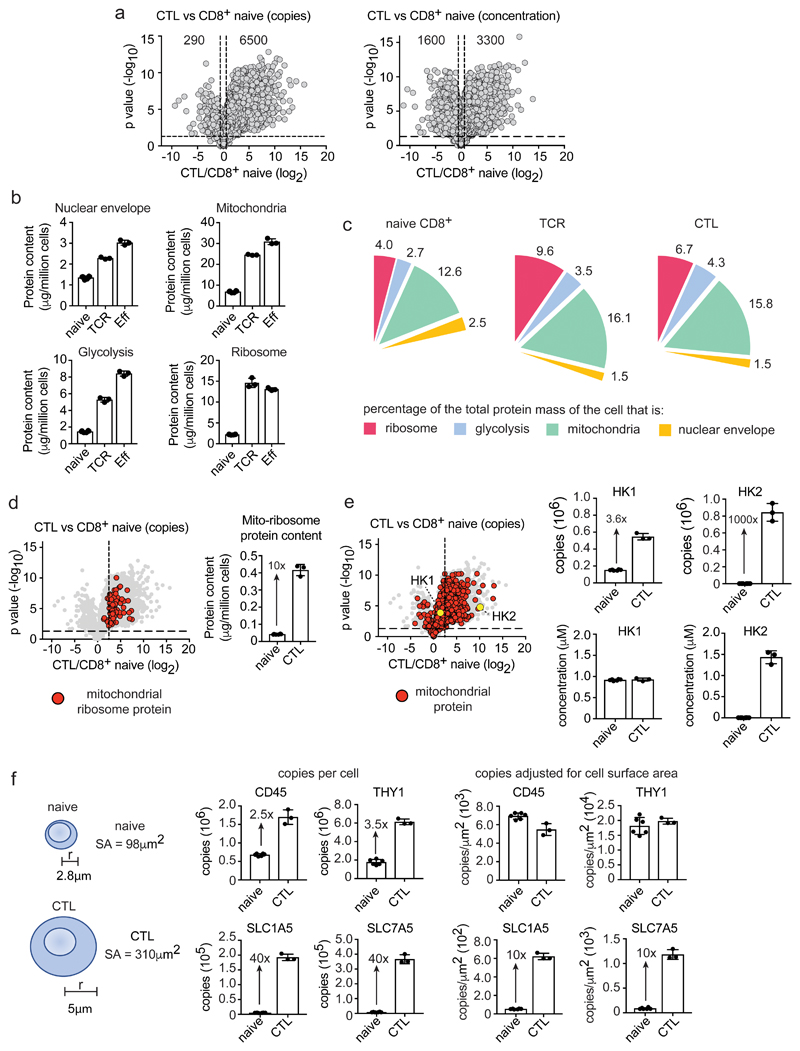
Many proteins change copy numbers in immune activated versus naïve T cells but as immune activation increases total T cell mass, only proteins whose abundance drops below or exceeds total cell mass will change concentration. Moreover, proteins with unchanged copy numbers in naïve versus activated cells decrease concentration. Hence, >6500 proteins increase copy number in CTLs versus CD8+ naïve cells, but only 3300 proteins increase concentration (Fig. 3a). One way to assess scaling versus enrichment or depletion of core subcellular compartments is to consider what percentage a protein group contributes to total protein mass in different populations. For example, nuclear envelope protein copy numbers increase in activated versus naive T cells but the percentage of cell mass that comprises nuclear envelope proteins decreases (Fig. 3b,c). Immune activated T cells thus do not equally scale up their nuclear envelope proteins compared to other proteins which is consistent with images showing increases in cytoplasm volumes of effector versus naïve cells. What about metabolic compartments? There is enrichment of total glycolytic pathway and mitochondrial proteins in activated versus naïve T cells (Fig. 3b,c Supplementary Fig. 2) although it should be highlighted that these protein groups are already abundant in naïve T cells. Glycolytic enzymes represent 2-3% of naïve T cell proteomes versus 4-5% in effectors; mitochondrial proteins are 12-13% of naïve T cells versus 15-16 % of effectors (Fig. 3c). An examination of mitochondrial proteins reveals that some increase in scale with the overall increase in cell mass that occurs during T cell differentiation while others increase beyond scaling. For example, mitochondrial ribosomal proteins are 10-fold more abundant in effector cells versus naïve cells (Fig. 3d). Another interesting example of scaling versus enrichment is that of hexokinase 1 and 2 (HK1 and HK2). HK2, one of the hexokinases that phosphorylate glucose to direct glucose metabolic pathways, shows a >1000-fold increase in effector versus naïve T cells (Fig. 3e). Interestingly, despite this huge increase in HK2 expression it proved non-essential for T cell activation19, 20. The current data explain why HK2 is redundant as they show that HK1 concentrations in naïve and activated T cells are very high and sufficient to compensate for HK2 loss (Fig. 3e and Supplementary Fig. 2).
Figure 3. Scaling versus selective enrichment of proteins and processes during T cell differentiation.
(a) Comparing protein copy number and concentration. Volcano plots show the ratio for proteins in CTLs versus naïve CD8+ T cells (CTL/naïve), using copies/cell or cellular protein concentration (μM). Horizontal dashed line indicates a P value = 0.05, vertical dashed lines indicate a fold change = 1.5 (b) Protein content of ribosomes (KEGG 03010), mitochondria (GO:0005739), nuclear envelope (GO:0005635) and the glycolytic pathway. (c) Protein content of cellular compartments relative to the total cellular protein mass (%). (d) Expression profile and protein content of mitochondrial ribosomal proteins in naïve CD8+ T cells and CTLs. The vertical dashed line on the volcano plot is the mean fold change (copy number CTL/naïve) of all proteins. (e) The expression profile of mitochondrial proteins in CTLs versus naïve CD8+ T cells (copy number CTL/naïve). Mitochondrial proteins are highlighted with red circles. Hexokinase 1 (HK1) and hexokinase 2 (HK2) are highlighted with yellow circles. Vertical dashed line is the mean fold change for all proteins. Copy number and concentration of HK1 and HK2 is also provided. (f) Protein copy numbers relative to cell surface area. The surface area of naïve and effector CD8+ T cells was estimated using the formula 4πr2, assuming the radius of a naïve cell to be 2.8 μm and a CTL to be 5 μm42, 43. Data shows protein copy numbers per cell and protein copy numbers adjusted for cell surface area (copies/μm2). For a-f, n = 6 biologically independent samples for CD8+ naïve cells and 3 biologically independent samples for each of the other T cell populations. Histogram bars represent the mean +/- SD. For a, d and e, P values calculated using a two-tailed t-test with unequal variance.
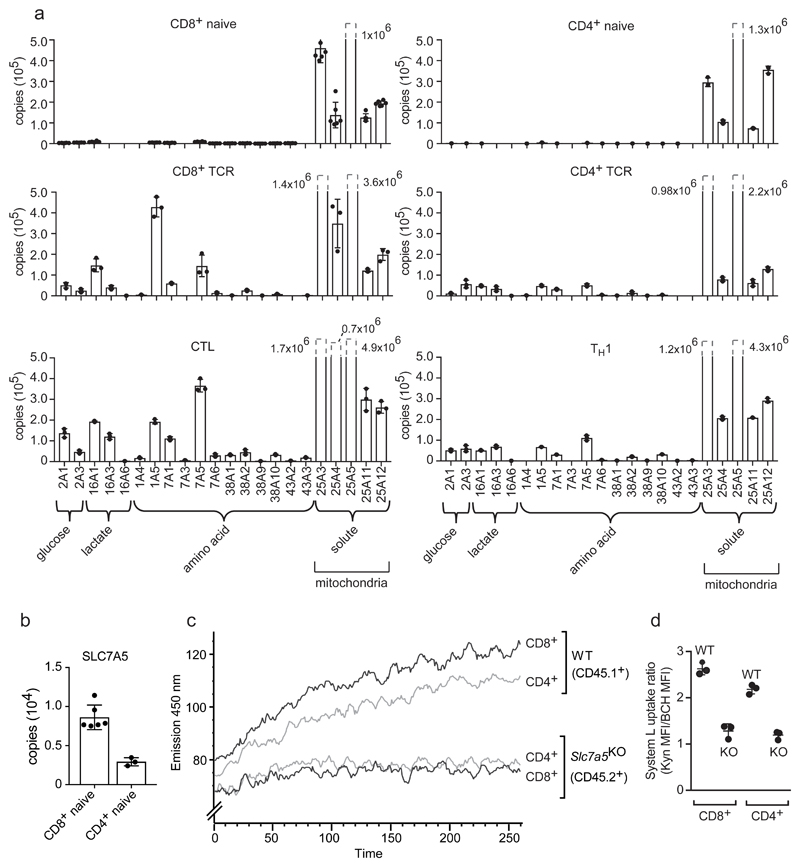
As activated T cells increase volume they increase cell surface area and may change membrane densities of plasma membrane proteins. Simple modeling, that does not model alterations in membrane ruffling, predicts a 3-fold increase in membrane surface area in CTLs versus naïve T cells (Fig. 3f). Molecules such as THY1 and CD45, have higher copy numbers in activated versus naïve CD8+ T cells, but do not increase membrane density after immune activation (Fig. 3f). In contrast, the amino acid transporters SLC1A5 and SLC7A5 increase 40-fold in copy number in CTLs versus naïve CD8+ cells; a 10-fold increase in membrane density (Fig. 3f). Further analysis shows naïve T cells have low expression of all amino acid and glucose transporters versus high levels of these transporters in activated T lymphocytes (Fig. 4a). In comparison, mitochondrial transporters were highly abundant in naïve T cells. Previous studies have shown increased amino acid and glucose transport in activated T cells compared to naïve T cells and noted lower nutrient transport in effector CD4+ versus CD8+ populations2, 21. The present data explain this result: activated CD4+ T cells have a similar nutrient transporter repertoire but consistently express lower copies of key amino acid and glucose transporters than activated CD8+ T cells (Fig. 4a). In this respect, although nutrient transporter expression in naive T cells is very low compared to activated T cells, transporters are not completely absent. For example, the System L amino acid transporter SLC7A5 was found at approximately 3000 copies in naïve CD4+ T cells and 8000 copies in naive CD8+ T cells (Fig. 4b). These data predict some System L transport activity in naïve T cells and that CD8+ T cells would have higher basal levels than CD4+ cells. To challenge this prediction we used a sensitive flow cytometry assay to assess System L amino acid transport activity in naïve T cells22. The data show naïve CD4+ and CD8+ T cells have a detectable basal System L transport activity mediated by SLC7A5. Crucially, the data show that naïve CD8+ cells have a higher System L transport capacity than naïve CD4+ cells (Fig. 4c,d). These results highlight the predictive value of the proteomic ruler methodology.
Figure 4. Nutrient and amino acid transport in T cells.
(a) Copy numbers for the major glucose, lactate and amino acid transporters and selected mitochondrial transporters in naïve, TCR activated and effector populations. (b) Copy numbers for the system L amino acid transporter SLC7A5 in naïve CD8+ and CD4+ T cells. For a and b, n = 6 biologically independent samples for CD8+ naïve cells and 3 biologically independent samples for each of the other T cell populations. Histogram bars represent the mean +/- SD. (c) Flow cytometric monitoring of System L dependent uptake in wild type and CD4Cre Slc7a5fl/fl naïve CD4+ and CD8+ cells. Lymph node cells from CD45.1+ (WT) and CD45.2+ (CD4Cre Slc7a5fl/fl, where SLC7A5 is deleted in all T cells) were mixed together and surface antibody stained for flow cytometric detection prior to the addition of the System L substrate kynurenine (KYN). Fluorescence emission was monitored over time using 405 nm excitation (violet laser) and band pass filter 450 ± 50. The representative trace shows the fluorescence data acquired post addition of 200 μM KYN plotted against time (seconds). (d) Kynurenine uptake in naïve CD4+ and CD8+ T cells +/- 10 mM 2-amino-2-norbornanecarboxylic acid (BCH); a System L transport inhibitor. The System L uptake ratio is calculated as the MFI after 5 min KYN uptake compared with the MFI after 5 min of KYN uptake in the presence of 10 mM BCH (KYN MFI / KYN+BCH MFI). For c and d, n = 3 biologically independent samples for WT and 3 biologically independent samples for CD4Cre Slc7a5fl/fl. Data presented in c is representative of one experiment and the experiment was carried out 3 times producing similar results. Bars shown in d represent the mean +/- SD.
Immune activation controls RNA translational machinery
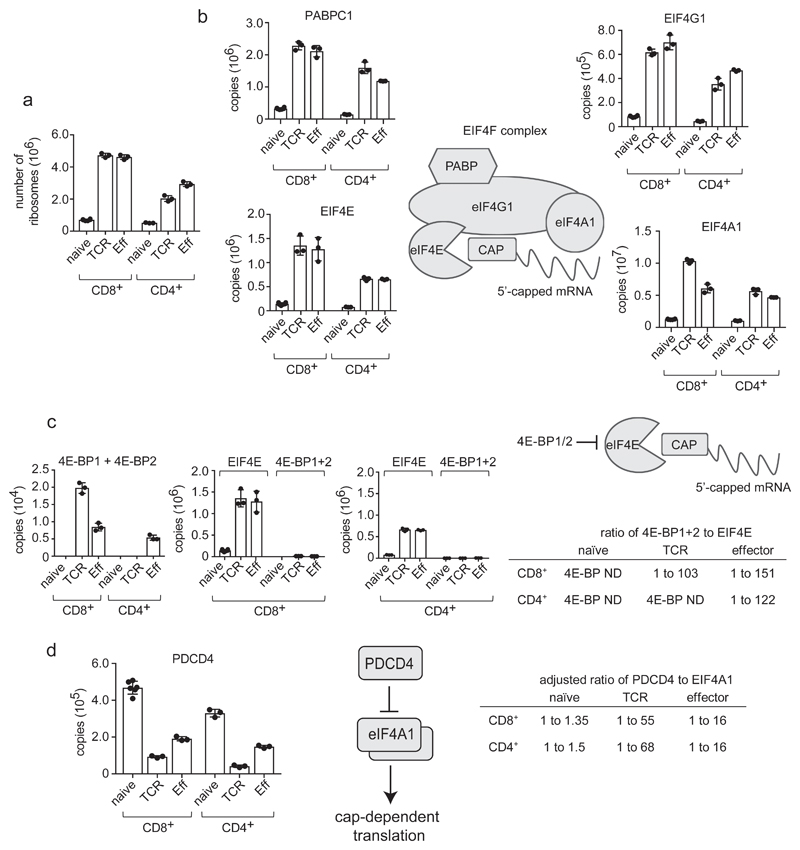
T cell exit from quiescence is associated with essential increases in mRNA translation1, 3. The current data show that immune activation results in a strong upregulation of ribosomes and mRNA translational machinery. Antigen activated T cells increase ribosome numbers almost 10 fold (Fig. 5a) and strongly increase levels of eukaryotic initiation factor 4 (eIF4F) complexes that translate methyl capped mRNAs (Fig. 5b) and EIF2 complexes which control tRNA transfer to ribosomes and tRNA synthetases (Supplementary Fig. 2). Hence, more of the cell mass of activated T cells is dedicated to protein synthesis than in naïve T cells. It was also notable that CD8+ cells consistently had higher ribosome and translational complex numbers than CD4+ T cells (Fig. 5a,b). However, how cells control mRNA translation also requires understanding translational repressor quantities in different T cell populations. T cells express the eIF4F inhibitors 4E-BP1 and 4E-BP2 (eukaryotic initiation factor 4E-binding proteins 1 and 2) and the translational repressor PDCD4 (programmed cell death 4)(Fig. 5c,d)23,24. 4EBPs bind to eIF4E and displace eIF4G to prevent active eIF4F translation initiation complex assembly11. PDCD4 represses translation via eIF4A1 binding25. Interestingly, 4E-BPs were only detected in activated but not naïve T cells (Fig. 5c). Increased expression of translational repressors from naïve to activated cells seems inconsistent with the increased translation capacity in activated T cells but critically 4E-BPs are only expressed at maximum of 2×104 copies per cell whereas eIF4E, their target has >106 copies in immune activated CD8+ cells and over 5×105 copies in activated CD4+ cells. eIF4E is thus always in large excess of 4EBPs (Fig. 5c). Hence any modeling of 4E-BP translation repression needs to consider repressor and effector stoichiometry: any effects will be restricted to a subpopulation of eIF4Es and not relevant in naïve T cells where these repressors cannot be detected. We next considered PDCD4:eIF4A1 ratios as to repress translation by eIF4F complexes, one PDCD4 molecule binds two eIF4A126. Naïve T cells have 3-5×105 PDCD4 copies per cell, sufficient to saturate the majority of eIF4A1 (Fig. 5d). In activated T cells PDCD4 is downregulated (Fig. 5d) whereas eIF4A1 copies increase to over 4×106 copies per effector cell (Fig. 5b) leaving very low PDCD4:eIF4A1 ratios and free eIF4A1 to promote cap-dependent translation (Fig. 5d)
Figure 5. Regulation of mRNA translation in T cells.
(a) Number of ribosomes in naive, TCR activated and effector CD4+ and CD8+ T cell populations. Number of ribosomes was estimated by calculating the mean number of ribosomal subunits within each cell using the KEGG annotation: 03010. (b) Expression profile of key components of the Eukaryotic Initiation Factor 4F (EIF4F) mRNA translation initiation complex during differentiation. EIF4F consists of Eukaryotic Translation Initiation Factor 4 Gamma 1 (EIF4G1), Eukaryotic Translation Initiation Factor 4A1 (EIF4A1), Eukaryotic Translation Initiation Factor 4E (EIF4E) and Poly(A) Binding Protein Cytoplasmic 1 (PABPC1). Data is presented as protein copies per cell in naïve, TCR activated and effector CD8+ and CD4+ T cell populations. (c) Stoichiometry of eukaryotic initiation factor 4E-binding proteins 1 and 2 (4E-BP1+2) to EIF4E in T cell populations. Copy numbers for 4E-BP1 and 2 were combined. 4E-BP1+2 are also plotted adjacent to copy numbers for EIF4E to assess whether inhibitor levels are adequate to block translation initiation, and the ratio of 4E-BP1+2 to EIF4E is presented (ND = not detected). (d) Stoichiometry of Programmed Cell Death 4 (PDCD4) and EIF4A1 during T cell differentiation. The ratio of PDCD4 to EIF4A1 in naïve, TCR triggered and effector CD4+ and CD8+ T cells is shown adjusted to account for 1 molecule of PDCD4 binding 2 molecules of EIF4A1 to inhibit CAP-dependent translation26. For a-d, n = 6 biologically independent samples for CD8+ naïve cells and 3 biologically independent samples for each of the other T cell populations. Histogram bars represent the mean +/- SD.
Immune activation shapes environment sensing pathways in T cells
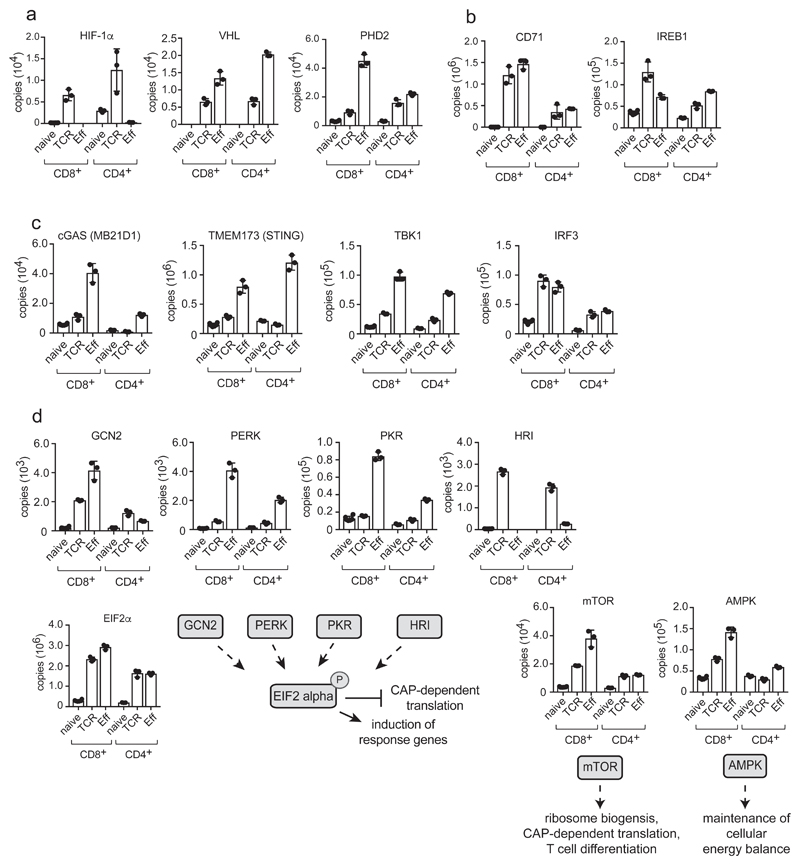
T lymphocyte function is regulated by environmental factors such as glucose, amino acids, oxygen and iron availability2, 27–29. Moreover, activated T cells must actively take up nutrients to fuel metabolic processes and macromolecule biosynthesis and to control nutrient sensing kinase activity. The present data show immune activation increases expression of a limited repertoire of 12 amino acid transporters, two glucose transporters and 3 lactate transporters to fuel and regulate activated T cells (Fig. 4a). Nutrient transporters are essential components of T cell environment sensing machinery as they act as ‘gatekeepers’ to control the activity of nutrient sensing kinases. However, one other key insight herein is that immune activation quite comprehensively regulates expression of other environment sensing molecules (Fig. 6). The expression of oxygen-sensing proline hydroxylase (PHD) proteins, hypoxia inducible factor 1 alpha (HIF-1α), along with iron transport and iron sensing proteins, is thus predominantly restricted to activated and effector T cells (Fig. 6a,b). Antigen activation also modulates expression of DNA sensing pathways. Cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) DNA sensing pathway is present in naïve T cells but highly enriched in effector TH1 and CTLs (Fig. 6c). One other noteworthy observation is that immune activation increases expression of nutrient-regulated protein kinases AMPKa1 and mTORC1 and amino acid-sensing kinases GCN2 and PERK1 of the integrated stress response pathway30 (Fig. 6d). There is also immune control of components of the nutrient sensing regulatory pathways that control mTORC1 activity including the lysosomal arginine sensor SLC38A9, the cytosolic arginine and leucine sensors CASTOR1 and SESTRIN2, the GTPase RHEB and GATOR complexes11, 31–35 (Supplementary Fig. 3). Collectively these data show immune activation comprehensively shapes T cell responsiveness to environmental stimuli.
Figure 6. Environmental sensing in T cells.
(a) Protein copy numbers for components of oxygen sensing pathways including the transcription factor hypoxia-inducible factor-1 alpha (HIF-1α), the oxygen sensing molecule prolyl hydroxylase domain-containing protein 2 (PHD2) and the E3 ligase that ubiquinates HIF-1α, von Hippel-lindau tumor suppressor (VHL). (b) The abundance of CD71 (Transferrin receptor) and IREB1 (Iron-Responsive Element-Binding Protein 1/Aconitase 1). (c) The impact of immune activation on the cGAS-STING DNA sensing pathway: Cyclic GMP-AMP Synthase (cGAS); Stimulator Of Interferon Genes Protein (STING or TMEM173); TANK Binding Kinase 1 (TBK1); Interferon Regulatory Factor 3 (IRF3). (d) Abundance of the amino acid sensing kinases Eukaryotic Translation Initiation Factor 2 Alpha Kinases 1, 2, 3 and 4 (HRI, PKR, PERK and GCN2 respectively) and mTOR (glucose and amino acid sensing) and AMPK (glucose sensing). For a-d, n = 6 biologically independent samples for CD8+ naïve cells and 3 biologically independent samples for each of the other T cell populations. Histogram bars represent the mean +/- SD.
mTORC1 control of CD4+ and CD8+ T cell proteomes
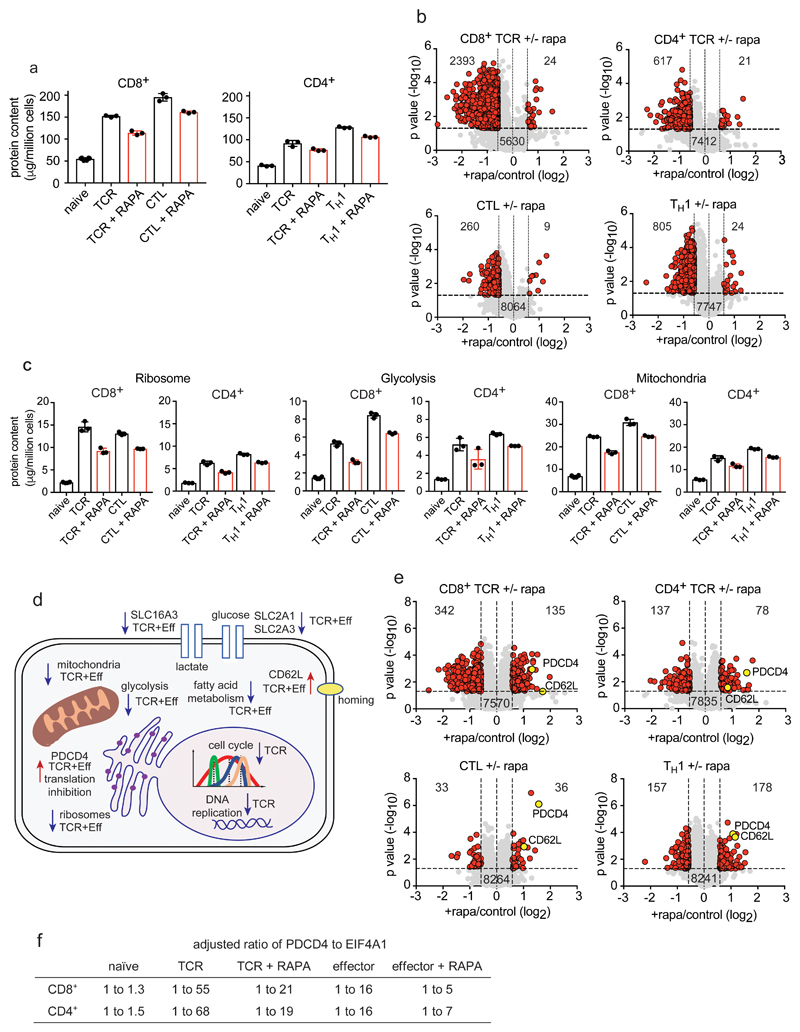
The nutrient sensing kinase mTORC1 is a critical regulator of both CD4+ and CD8+ T cell differentiation, and one current objective was to explore consequences of mTORC1 inhibition in different T cell populations12–15, 36, 37.The data reveal that mTORC1 controls cell growth in both CD4+ and CD8+ T cells: in TCR-activated CD8+ T cells mTORC1 inhibition with rapamycin caused a 26% loss of mass compared to a 16% loss of cell mass in the TCR activated CD4+ T cells (Fig. 7a). Using a >1.5-fold cut off, mTORC1 controlled expression of more than 2300 proteins in the TCR activated CD8+ T cells compared to 600 in the CD4+ T cells (Fig 7b). However, it is evident that immune activation still induces a substantial increase in cell mass in the absence of mTORC1 activity (Fig. 7a). This limited effect of mTORC1 inhibition on T cell growth was also seen in CTLs and TH1 cells where the impact of 24 h mTORC1 inhibition was an approximate 20% loss of cell mass but cells did not return to naïve cell size (Fig. 7a). In TH1 cells 800 proteins decreased expression whereas in CTLs the effects were restricted to around 260 proteins (Fig. 7b). One key point is that there were quantitative differences in the impact of mTORC1 inhibition on CD4+ and CD8+ proteomes, but no obvious qualitative differences. mTORC1 thus controls production of effector molecules IFN-γ, perforin and granzymes in both CD4+ and CD8+ cells (Supplementary Fig. 4) but does not radically prevent the dynamic remodeling of transcription factors that drives T cell differentiation (Supplementary Fig. 4). T cells activated in the presence of mTORC1 inhibition thus show effectively normal patterns of expression of T-bet, IRF4, BATF, MYC, and BLIMP-1 (Supplementary Fig. 4). mTORC1 control of HIF-1α has been described and the present data show this is a consistent effect in CD4+ and CD8+ T cells12 (Supplementary Fig. 4).
Figure 7. The impact of mTORC1 inhibition on CD4+ and CD8+ T cell proteomes.
(a) Protein content of T cells in response to mTORC1 inhibition. Naïve CD4+ and CD8+ T cells were TCR triggered for 24 h +/- rapamycin while effector CD4+ and CD8+ cells were incubated for 24 h +/- rapamycin. (b) Volcano plots show the protein ratios for rapamycin treated cells versus control (+rapamycin/control copy numbers). Proteins highlighted in red have a P value <0.05 and a fold change >1.5 while proteins highlighted in grey did not change significantly. (c) The impact of rapamycin on the glycolytic pathway, ribosomes and mitochondria in CD4+ and CD8+ cells. (d) Summary of cellular processes impacted by mTORC1 inhibition. Arrows pointing downwards indicate decreased abundance while arrows pointing upwards indicate increased abundance. Proteins/processes changing in TCR stimulated cells only are labelled “TCR” while those changing in TCR and effector populations are labelled “TCR+Eff”. (e) The impact of inhibiting mTORC1 on protein concentration. Volcano plots were generated as described above. (f) The ratio of PDCD4 to EIF4A1. 1 molecule of PDCD4 binds 2 molecules of EIF4A1 and the ratios have been adjusted to account for this binding stoichiometry. For a-f, n = 6 biologically independent samples for CD8+ naïve cells and 3 biologically independent samples for each of the other T cell populations. Histogram bars represent the mean +/- SD. For b and e, P values were calculated using a two-tailed t-test with unequal variance.
The main contribution to the loss of cell mass caused by mTORC1 inhibition in all populations was reduced expression of metabolic proteins notably ribosomes, glycolytic enzymes, mitochondrial proteins, glucose and lactate transporters and fatty acid and sterol metabolism proteins (Fig. 7c,d and Supplementary Fig. 4). It has been reported that loss of mTORC1 activity decreased expression of mitochondrial ribosomal proteins and oxidative phosphorylation enzymes in CD3 and CD28 activated CD4+ T cells7. The current data found a statistically significant decreased expression of mitochondrial proteins in both CD4+ and CD8+ populations but these were very low in magnitude such that mitochondrial proteins were highly abundant in mTORC1 inhibited T cells and unlikely to be rate limiting (Fig. 7c and Supplementary Fig. 5). Moreover, in no population did mTORC1 inhibition revert the expression levels of mitochondria or metabolic proteins back to naïve cell levels, indicating that the expression of these key components in T cells is also controlled by mTORC1 independent pathways. For example, rapamycin treatment caused expression of the glucose transporter SLC2A1 to drop from around 50,000 to 17,000 copies per cell in TCR activated CD8+ cells and from 137,000 to 77,000 copies per cell in CTLs (Supplementary Fig. 4). Glucose transport is rate limiting for glucose metabolism in T cells so reducing glucose transporter abundance would allow mTORC1 to control glucose metabolic pathways in T cells5. Nevertheless, a key insight is that mTORC1 is not an on or off switch for glucose metabolism nor is it an absolute on or off switch for cell growth.
Hence a salient point is that immune activated T cells still increase cell mass when mTORC1 activity is suppressed and loss of mTORC1 activity in effector cells does not revert their mass to that of a naïve T cell. In this context, the selectivity of mTORC1 control of T cell proteomes is emphasized by the large number of proteins that were unchanged in copy numbers in all rapamycin treated populations as well as the examples of proteins that increased in abundance and concentration in mTORC1 inhibited cells (Fig. 7e and Supplementary Data File 4). Proteins increasing in concentration include the translational repressor PDCD4, which increased in both TCR activated and effector cells treated with rapamycin (Fig. 7e). However, EIF4A1 did not decrease pro-rata and remained in excess of PDCD4 in these populations allowing continued protein synthesis (Fig. 7f and Supplementary Fig 5). Another protein consistently showing increased copy number in mTORC1 inhibited cells was the adhesion molecule CD62L (Fig. 7e). The increased expression of CD62L in mTORC1 inhibited CTLs restores the ability of these cells to traffic into secondary lymphoid tissue38. mTORC1 repression of CD62L expression in CD4+ T cells argues that mTORC1 could control CD4+ T cell positioning within lymphoid tissues.
Cell context effect of mTORC1 inhibition
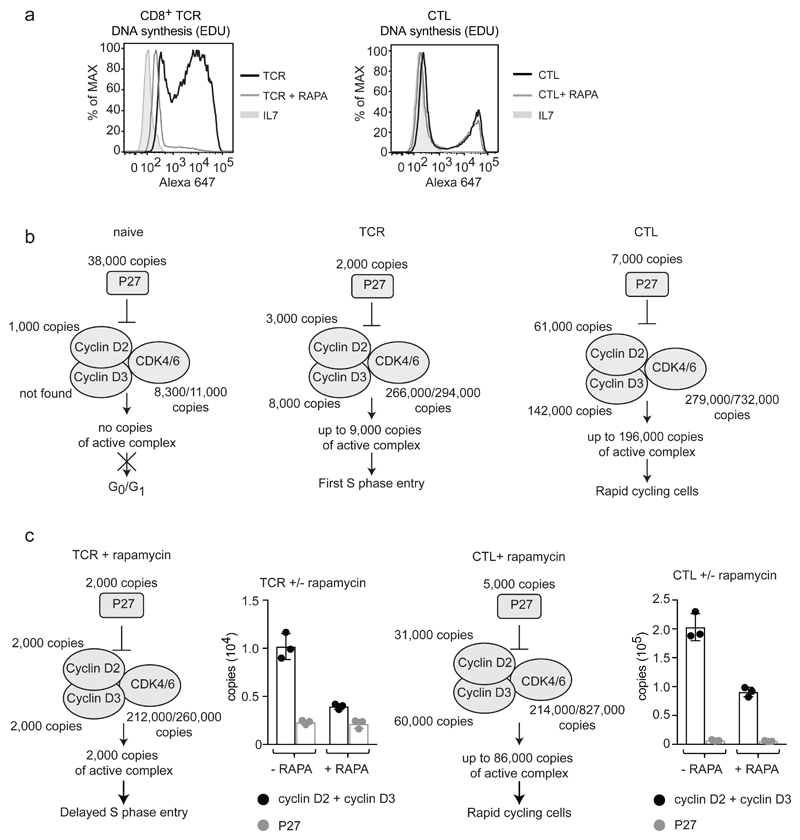
One objective of the present study was to identify cell context effects of mTORC1 signaling. In this respect, GO term enrichment analysis indicated dominant mTORC1 control of lipid metabolism in effector TH1 and CTLs (Table 1). mTORC1 control of lipid metabolism was also seen in antigen activated CD4+ and CD8+ T cells but GO term enrichment analysis indicated that the dominant effect of mTORC1 inhibition in these populations was on cell cycle and DNA replication pathways (Table 1). This data is consistent with early studies showing rapamycin delayed the first cell cycle entry of PHA activated human T cells but did not block proliferation once cells had committed to the cell cycle16, 17. To orthogonally challenge these predictions we monitored DNA synthesis in TCR activated and effector CD8+ populations and found that rapamycin inhibited DNA synthesis of antigen activated naïve CD8+ T cells but not CTLs (Fig. 8a). This is consistent with a decrease in expression of proteins implicated in DNA replication in rapamycin treated TCR activated cells but not in CTLs (Supplementary Fig. 6). In this respect, a critical cell cycle check point is mediated by D type cyclins and their associated kinases CDK4 and CDK6. Naive T cells have low levels of cyclin D’s and CDK4/6 but high levels of the cyclin dependent kinase inhibitor protein 1B (CDKN1B or p27) (Fig. 8b and Supplementary Fig. 6). Antigen activation increased cyclin D2 and D3 and CDK4/6 expression, resulting in excess D type cyclins, relative to the inhibitor CDKN1B. Interestingly, rapamycin treatment caused decreased cyclin D expression in both TCR activated CD8+ T cells and effectors (Fig. 8c) but why then were there no cell cycle progression defects in effectors? Here insights come from considering cyclin D copy numbers in different T cell populations relative to numbers of CDK4/6 and CDKN1B. For example, naïve CD8+ T cells express high levels of CDKN1B, undetectable cyclin D3 and low cyclin D2 and CDK4/6 (Fig. 8b). 24 h activated CD8+ cells upregulate cyclin D expression, increase CDK4/6 levels and downregulate CDKN1B: simple modeling predicts they have free cyclin D and CDK4/6 complexes. In this experiment, TCR activated CD8+ cells have approximately 3000 and 8000 copies per cell of cyclin D2 and 3 respectively and only 2000 copies of CDKN1B. As 60% of these cells are replicating DNA (Fig. 8a) this level of ‘free’ cyclin D and CDK complex (estimated as approximately 9000) must be sufficient to drive S phase entry. mTORC1 inhibition caused cyclin D2 and D3 levels to drop to 2000 copies per cell for each protein, while CDKN1B remained at 2000 copies, leaving little free cyclin complexes for cell cycle progression (Fig. 8c). In contrast, CTLs have high cyclin D2 and D3 levels: 60,000 and 140,000 copies per cell compared to 7000 CDKN1B copies. Rapamycin treatment decreased cyclin D levels 2 fold but cyclin D:CDKN1B stoichiometry remained > 10:1, ie more than 85,000 ‘inhibitor’ free cyclin D and CDK complexes which would be sufficient to drive cell cycle progression (Fig. 8c). These data illustrate that to fully understand the consequences of a change in expression for a particular protein it is necessary to understand protein quantity and relative levels of expression of its regulatory partners.
Table 1.
GO term enrichment analysis for proteins that significantly drop in abundance in rapamycin treated cells (significance = P value <0.05 (two-tailed t-test with unequal variance) and fold change >2 standard deviations from the mean fold change). The top 5 enriched GO terms for biological processes, are presented.
| enriched GO term | fold enrichment | P value |
|---|---|---|
| CD8+ TCR | ||
| GO:0007049~cell cycle | 6 | 1.7 x 10-35 |
| GO:0051301~cell division | 7 | 5.5 x 10-35 |
| GO:0007067~mitotic nuclear division | 8 | 6.4 x 10-31 |
| GO:0007059~chromosome segregation | 10 | 6 x 10-14 |
| GO:0007018~microtubule-based movement | 12 | 1.2 x 10-9 |
| CD4+ TCR | ||
| GO:0006260~DNA replication | 10 | 1.2 x 10-12 |
| GO:0007049~cell cycle | 3 | 6.5 x 10-8 |
| GO:0006270~DNA replication initiation | 14 | 2.9 x 10-4 |
| GO:0006974~response to DNA damage | 2 | 8.5 x 10-4 |
| GO:0051726~regulation of cell cycle | 5 | 1.2 x 10-3 |
| CTL | ||
| GO:0006636~unsaturated fatty acid biosynth | 35 | 1.3 x 10-4 |
| GO:0006633~fatty acid biosynthetic process | 10 | 2.3 x 10-4 |
| GO:0006629~lipid metabolic process | 3 | 1.0 x 10-3 |
| GO:0030154~cell differentiation | 3 | 1.3 x 10-3 |
| GO:0016239~regulation of macroautophagy | 14 | 2.3 x 10-3 |
| TH1 | ||
| GO:0016126~sterol biosynth process | 12 | 9.3 x 10-5 |
| GO:0008202~steroid metabolic process | 6 | 6.2 x 10-4 |
| GO:0006695~cholesterol biosynth process | 11 | 7.9 x 10-4 |
| GO:0006694~steroid biosynthetic process | 7 | 8.2 x 10-4 |
| GO:0035455~response to interferon-alpha | 18 | 1.1 x 10-3 |
Figure 8. The impact of mTORC1 inhibition on cell cycle proteins.
(a) Flow cytometric analysis of DNA synthesis in CD8+ T cells in response to rapamycin. Naïve CD8+ cells TCR triggered for 24 h and effector CTLs were treated +/- rapamycin. DMSO was used as a vehicle control. DNA synthesis was assessed by incorporation of EdU (a thymidine analogue) into newly synthesized DNA which is fluorescently labelled using a copper-catalyzed click reaction. Naïve cells (IL7 treated) were included as a control. n = 3 biologically independent samples for IL7 and TCR activated cells, n = 3 biologically independent samples for CTLs. The data shown is from 1 biologically independent sample and is representative of all replicates. (b) Stochiometric model for cell cycle entry and progression. Protein copy numbers are presented for cyclin D2 (CCND2), cyclin D3 (CCND3), cyclin dependent kinase 4 (CDK4), cyclin dependent kinase 6 (CDK6) and the cyclin dependent kinase inhibitor CDKN1B (P27). (c) The impact of rapamycin on the cyclin D/P27 model in CD8+ cells TCR triggered for 24 h and effector CD8+ cells incubated +/- rapamycin. Protein copy numbers are also presented in graphs for CCND2 + CCND3 adjacent to P27. For b and c, n = 6 biologically independent samples for CD8+ naïve cells and 3 biologically independent samples for each of the other T cell populations. Histogram bars represent the mean +/- SD. Copy numbers adjacent to cell cycle proteins are the average of replicates and were rounded to the nearest thousand.
Discussion
This study characterized murine naïve CD4+ and CD8+ T cell proteomes and mapped the impact of immune activation and mTORC1 inhibition on the expression of more than 9000 proteins. The data show how environment signaling pathways are integrated with antigen and cytokine selective pathways to ensure the immune specificity of T cell activation. New, biologically relevant insights include that antigen exposure increases expression of key oxygen and nutrient sensors and nutrient transporters, revealing that T cell sensing of environmental cues is not intrinsic but shaped by immune activation and restricted to activated T cells. The considerable upregulation of essential amino acid and glucose transporters in response to immune activation highlights a fundamental mechanism that ensures the targeted supply of nutrients to T cells participating in an immune response. Only T cells triggered by cognate antigen will be able to fuel the key metabolic pathways that drive T cell proliferation and differentiation. In this context one new insight was the consistent quantitative differences between CD4+ T cells and equivalently activated CD8+ T cells in terms of the expression of ribosomes, nutrient transporters and translational complexes. Immune activated CD4+ and CD8+ T cells have the same nutrient transporter repertoire but activated CD4+ cells expressed nutrient transporters at lower copies per cell and also have lower numbers of ribosomes and translational complexes than immune activated CD8+ cells. Collectively these data reveal a lower biosynthetic capacity in comparable activated CD4+ versus CD8+ cells which would explain why activated CD4+ cells have lower cell mass than activated CD8+ cells and could explain the competitive proliferative advantage of activated CD8+ versus CD4+ T cells. Other disparities and core parities between naïve and immune activated CD4+ and CD8+ T cell populations can be interrogated interactively via an online, searchable database the Encyclopaedia of Proteome Dynamics39 (www.peptracker.com/epd).
One experimental aim was to compare the impact of mTORC1 inhibition on CD4+ and CD8+ T cells since there are often general comments made about what mTORC1 does to control lymphocyte function based on experiments performed in a single cell population and assuming that observations can be extrapolated to all cells. In this context, a general comment frequently made about mTORC1 is that it regulates cell growth11, 40. The present data show this to be true but with the caveat that mTORC1 growth effects are relatively small and mTORC1 independent pathways also control T cell growth. Indeed, the selectivity of mTORC1 shaping of CD4+ and CD8+ T cell proteomes was striking. The present data give insights as to why mTORC1 effects on T cell protein mass are limited. For example, mTORC1 is proposed to control cell growth by phosphorylating and inactivating the translational repressors 4E-BP1 and 4E-BP211, 41. However, we show that 4E-BP copy numbers are very low relative to their target eIF4E, making it unlikely that releasing any 4E-BP repression would have a major impact on protein translation and would rather be highly selective to a small subset of proteins. Similarly, we have shown that mTORC1 inhibition increases expression of the translational repressor PDCD4 but that the PDCD4 target eIF4A1 remained in large excess to PDCD4 in rapamycin treated T cells. These considerations highlight the value of quantitative data that models protein complex stoichiometry for understanding how immunomodulatory stimuli effect T cell phenotypes.
What about cell context effects of mTORC1? A comparison of previous studies of how raptor deficient CD4+ T cells responded to CD3 and CD28 antibodies versus how CD8+ effector CTLs responded to rapamycin had indicated there might be differences between how mTORC1 controls CD4+ and CD8+ T cell proteomes5, 7. The present study enabled direct comparisons between populations and revealed no major qualitative differences in how mTORC1 controls comparable CD4+ and CD8+ T cell populations. mTORC1 inhibition had an impact on glucose transporters, glycolysis, mitochondria, fatty acid metabolism, ribosomes and translational machinery and cell adhesion molecules in both antigen-activated and effector CD4+ and CD8+ T cells. There were differences in how mTORC1 inhibition impacted antigen activated lymphocytes as they exit quiescence versus mTORC1 control of effector T cells. In particular, mTORC1 inhibition had a dominant effect on cell cycle progression in antigen activated naïve CD4+ and CD8+ cells but not in TH1 or CTL effector populations. The basis for this difference was that key cell cycle regulators are so highly abundant in effectors compared to T cells progressing into their first cell cycle that decreases in expression caused by loss of mTORC1 function are not rate limiting. These data indicate that for mTORC1 inhibitors to prevent T cell cycle progression it will be necessary to deliver the inhibitor before the T cell has accumulated high levels of D-type cyclins. The data reveal cell context dependent functions of mTORC1 and show that these can be understood when there is knowledge and understanding of the stoichiometry of critical protein complexes and their positive and negative regulators plus some understanding of functional thresholds for different pathways. The current data provide comprehensive quantitative information about protein copy number and concentration in T cell populations that are used extensively as models to probe T cell biology. We provide an easily interrogated resource for exploring the protein landscape of key immune cells and for predicting T cell responses to environment and other immune modulators.
Methods
Mice
For proteomics experiments P1444 and OT-II45 transgenic mice along with C57BL/6 (wild-type B6) mice were used. For the single-cell amino acid transport assay CD4Cre Slc7a5fl/fl (CD45.2+) and C57BL/6J Ly5.1 (CD45.1+) mice were used. Male and female mice aged between 50 and 120 days were used. Further details can be found in the Life Sciences Reporting Summary. All mice were maintained in the Biological Resource Unit at the University of Dundee using procedures that were approved by the University Ethical Review Committee and under the authorization of the UK Home Office Animals (Scientific Procedures) Act 1986.
Flow cytometry
Forward/side scatter analysis of naïve CD4+ and CD8+ T cells
Lymph node derived naïve CD4+ and CD8+ cells were stained with CD4-PE and CD8-PeCy7. Forward and side scatter profiles were acquired on a LSR Fortessa II with DIVA software and data was analyzed using FlowJo software v9 (TreeStar). 4 biologically independent samples were analyzed.
DNA Synthesis Assay
To measure DNA synthesis cells were fed 10 μM Click-iT EdU (Thermo Fisher) for 30 min. Cells were then harvested, stained with CD8-FITC (for TCR-activated cells), fixed with 1 % paraformaldehyde and permeabilized with 0.5 % Triton X 100 before undergoing a copper catalyzed click chemistry reaction with Alexa 647-azide (Thermo Fisher). Cells were analyzed by flow cytometry to determine the degree of incorporation of EdU. All flow cytometry data was acquired on either a LSR Fortessa II with DIVA software or a FACSVerse flow cytometer with FACSuite software (BD Biosciences). Data were analyzed using FlowJo software v9 (TreeStar). 3 biologically independent samples were analyzed for each experiment. The gating strategy for flow cytometry analysis is provided in Supplementary Fig. 7.
Monitoring System L amino acid transport
A single cell assay to monitor System L amino acid transport, with kynurenine as a fluorescent system L transport substrate, was performed as described previously22. In brief, wild-type (C57BL/6J Ly5.1, CD45.1+) and CD4Cre Slc7a5fl/fl (CD45.2+) ex vivo lymph node cells were mixed. Surface antibody staining for CD45.1 (PerCP Cy 5.5), CD45.2 (FITC), CD4 (PE) and CD8 (PeCy7) was performed (15 min, at 37 °C). After surface antibody staining, cells were washed in pre-warmed HBSS, and resuspended in 200 μl warmed HBSS (approx. 2.5 × 106 cells in FACS tubes). Cells were kept in a water bath at 37 °C. 100 μl of HBSS or the System L transport inhibitor BCH (2-amino-2-norbornanecarboxylic acid, Sigma) was added to appropriate samples. Finally, 100 μl pre-warmed kynurenine (stock 800 μM freshly made in HBSS; final concentration 200 μM; final sample volume 400 μl) was added. To monitor kynurenine uptake in live cells, data was acquired on flow cytometer immediately following addition of kynurenine and fluorescence plotted against time. The 405 nm laser and 450/50 BP filter were used for kynurenine fluorescence detection. To calculate the ratio of System L uptake, kynurenine uptake was fixed after 5 min by adding 125 μl 4% PFA for 30 min at room temperature, in the dark. Flow cytometry data was acquired on either a LSR Fortessa II with DIVA software or a FACSVerse flow cytometer with FACSuite software (BD Biosciences) and analysed using FlowJo software v9 (TreeStar) and following the gating strategy as shown in Supplementary Fig. 7. 3 biologically independent samples were collected and analyzed from wild-type and CD4Cre Slc7a5fl/fl mice.
Cells
All cells were activated and cultured in RPMI 1640 containing glutamine (Invitrogen) and supplemented with 10 % FBS (Gibco), 50 μM β-mercaptoethanol (Sigma) and penicillin/streptomycin (Gibco). Cells were cultured at 37 °C with 5 % CO2.
For all proteomics experiments 3 biological replicates were generated, except naïve CD8+ cells, for which 6 biological replicates were produced. For proteomics experiments pure populations of naïve CD8+ and CD4+ cells were generated by fluorescent-activated cell sorting (FACS). The gating strategy for cell sorting is provided in Supplementary Figs. 8–10. Cells were sorted to a purity >95%. For naïve CD8+ cells, lymph nodes were extracted from gender and age matched P14 mice and tissue mashed in RPMI media and filtered through a 70 μm cell strainer. FC receptors were blocked using 1 μg FC block (BD Pharmingen) per million cells. Cells were stained with the following fluorophore-conjugated antibodies; TCRβ-PerCP Cy 5.5, CD8-PE, CD44-APC, CD62L-FITC and DAPI and sorted on an Influx cell sorter (Becton Dickinson). Naïve CD8+ cells were collected (CD8+CD44loCD62Lhi) and cells were washed twice in HBSS before being snap frozen in liquid nitrogen. For naïve CD4+ cells, lymph nodes were isolated from age- and gender-matched C57BL/6 mice and processed as above but stained with the following antibodies; TCRβ-PerCP Cy 5.5, CD4-PeCy7, CD8-PE, CD44-APC, CD62L-FITC and DAPI. Naïve CD4+ cells were collected (CD4+CD44loCD62Lhi) and processed as above.
To activate primary T cells for generating a 24 h-activated CD8+ T cell population, lymph nodes were removed from P14 mice and mashed before suspending in media with 100 ng/ml antigenic peptide (glycoprotein amino acids 33-41 – GP33), 20 ng/ml IL-2 (Proleukin, Novartis) and 2 ng/ml IL-12 (PeproTech) and where needed were treated with 20 nM rapamycin (Merck). After 24 h cells were harvested by centrifugation and prepared for cell sorting to isolate a pure population of CD8+ cells. FC receptors were blocked using 1 μg FC block (BD Pharmingen) per million cells. Cells were stained with CD8-PE and DAPI and sorted cells were harvested as described above. TCR-activated CD4+ cells were generated from lymph nodes isolated from OT-II transgenic mice. To achieve effective TCR activation a preparation of antigen-presenting cells was first generated. Spleens were taken from wild-type B6 mice and T lymphocytes removed using anti-TCR-biotin antibody and an immunomagnetic isolation kit (Stemcell Technologies) to generate an antigen-presenting cell population. Antigen-presenting cells were incubated with lipopolysaccharide (4 μg/ml) and OVA peptide (323-339, 1 μg/ml, Sigma) overnight before being combined with cells from OT-II lymph nodes. Antigen-presenting cells were combined with OT-II lymph node cells at a ratio of 1:2 in RPMI media. Cells were incubated with OVA peptide (1 μg/ml) plus 2 ng/ml IL-12 (PeproTech) and activated for 24 h +/- 20 nM rapamycin. Once activated, cells were subject to cell sorting to generate a pure population of activated CD4+ cells. FC receptors were blocked using 1 μg FC block (BD Pharmingen) per million cells. Cells were stained with CD4-PerCP Cy 5.5, Vα2-PE and DAPI before cell sorting. CD4+ and Vα2+ cells were collected. Purified cells were washed twice in HBSS before being snap frozen in liquid nitrogen.
To generate CTLs, spleens were extracted from P14 mice and mashed in red blood cell lysis buffer before being suspended in RPMI media supplemented with GP33 peptide and IL-2 and IL-12 (as described for TCR activation above). Cells were activated for 48 h, washed out of activation buffer and then cultured for 3 days in media supplemented with 20 ng/ml IL-2 and 2 ng/ml IL-12. After 3 days cells were treated +/- 20 nM rapamycin for 24 h before being harvested. Dead cells were removed using a dead cell removal kit (Miltenyi Biotec), washed twice with HBSS and snap frozen. TH1 cells were cultured from wild-type C57BL/6 mice. Spleen and lymph nodes were extracted from mice, mashed, filtered through a 70-μm cell strainer and CD8+ cells depleted using immunomagnetic selection (Stemcell Technologies). Cells were activated with CD28 antibody (3 μg/ml), CD3 antibody (2 μg/ml), IL-2 (20 ng/ml) and IL-12 (10 ng/ml). Cells were cultured for a total of 6 days with the last 24 h +/- rapamycin at 20 nM. Dead cells were removed as described above and cells were washed twice with HBSS before being snap frozen in liquid nitrogen.
Proteomics sample preparation and TMT labelling
Cell pellets were lysed in 400 μL lysis buffer (4% SDS, 50 mM TEAB pH 8.5, 10 mM TCEP). Lysates were boiled and sonicated with a BioRuptor (30 cycles: 30 sec on, 30 sec off) before alkylation with 20 mM iodoacetamide for 1 h at 22 °C in the dark. The lysates were subjected to the SP3 procedure for protein clean-up46 before elution into digest buffer (0.1% SDS, 50 mM TEAB pH 8.5, 1mM CaCl2) and digested with LysC and Trypsin, each at a 1:50 (enzyme:protein) ratio. TMT labelling and peptide clean-up were performed according to the SP3 protocol. T cell populations were TMT labelled in the following six batches: naïve CD4+ cells (mass tag 126, 127C and 128N), TCR activated CD4+ cells +/- rapamycin (mass tag 126, 127N, 128C for control and 129N, 130C and 131 for rapamycin), TH1 +/- rapamycin (mass tag 126, 127N, 127C for control and 128N, 128C and 129N for rapamycin), naïve CD8+ cells (mass tag 128N, 128C, 129N, 129C, 130C and 131) , TCR-activated CD8+ cells +/- rapamycin (mass tag 126, 127N, 128C for control and 128N, 128C, 129N for rapamycin) and CTLs +/- rapamycin (mass tag 126, 127N, 128C for control and 129N, 130C and 131 for rapamycin). After labelling samples were eluted into 2% DMSO in water, combined and dried in vacuo.
Peptide fractionation
The TMT samples were fractionated using off-line high pH reverse phase chromatography: samples were loaded onto a 4.6 × 250 mm XbridgeTM BEH130 C18 column with 3.5 μm particles (Waters). Using a Dionex BioRS system, the samples were separated using a 25-minute multistep gradient of solvents A (10 mM formate at pH 9 in 2% acetonitrile) and B (10 mM ammonium formate pH 9 in 80% acetonitrile), at a flow rate of 1 mL/min. Peptides were separated into 48 fractions which were consolidated into 24 fractions. The fractions were subsequently dried and the peptides dissolved in 5% formic acid and analyzed by LC-MS.
Liquid chromatography electrospray tandem mass spectrometry analysis (LC-ES-MS/MS)
1 μg per fraction was analysed using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) equipped with a Dionex ultra high-pressure liquid chromatography system (nano RSLC). RP-LC was performed using a Dionex RSLC nano HPLC (Thermo Scientific). Peptides were injected onto a 75 μm × 2 cm PepMap-C18 pre-column and resolved on a 75 μm × 50 cm RP- C18 EASY-Spray temperature controlled integrated column-emitter (Thermo) using a 4 h multistep gradient from 5% B to 35% B with a constant flow of 200 nL min-1. The mobile phases were: 2% ACN incorporating 0.1% FA (Solvent A) and 80% ACN incorporating 0.1% FA (Solvent B). The spray was initiated by applying 2.5 kV to the EASY-Spray emitter and the data were acquired under the control of Xcalibur software in a data dependent mode using top speed and 4 s duration per cycle, the survey scan is acquired in the Orbitrap covering the m/z range from 400 to 1400 Thomson units (Th) with a mass resolution of 120,000 and an automatic gain control (AGC) target of 2.0 e5 ions. The most intense ions were selected for fragmentation using CID in the ion trap with 30 % CID collision energy and an isolation window of 1.6 Th. The AGC target was set to 1.0 e4 with a maximum injection time of 70 ms and a dynamic exclusion of 80 s. During the MS3 analysis for more accurate TMT quantifications, 10 fragment ions were co-isolated using synchronous precursor selection using a window of 2 Th and further fragmented using HCD collision energy of 55%. The fragments were then analyzed in the Orbitrap with a resolution of 60,000. The AGC target was set to 1.0 e5 and the maximum injection time was set to 300 ms.
Processing and analysis of proteomics data
The data were processed, searched and quantified with the MaxQuant software package, version 1.5.8.3, Proteins and peptides were identified using the UniProt mouse database (SwissProt and Trembl) and the contaminants database integrated in MaxQuant using the Andromeda search engine47, 48 with the following search parameters: carbamidomethylation of cysteine and TMT modification on peptide N-termini and lysine side chains were fixed modifications, while methionine oxidation, acetylation of N-termini of proteins were selected as variable modifications. The false discovery rate was set to 1% for positive identification at the protein and PSM level. The data set was filtered to remove proteins categorized as “contaminants”, “reverse” and “only identified by site”. Copy numbers were calculated as described18 after allocating the summed MS1 intensities to the different experimental conditions according to their fractional MS3 reporter intensities. The accuracy of quantification was established using the following guidelines: proteins categorized as high accuracy had more than 8 unique and razor peptides and a ratio for unique/unique + razor greater than or equal to 0.75, proteins categorized as medium accuracy had at least 3 unique and razor peptides and a ratio for unique/unique + razor greater than or equal to 0.5, any proteins below these thresholds were classified as low accuracy.
Statistics and calculations
P-values were calculated via a two-tailed, unequal variance t-test on log normalized data. Elements with P values lower or equal to 0.05 were considered significant. Fold-change thresholds were established using 2 methods. Method 1 established a fold-change cut-off ≥ 1.5 or ≤ 0.67. Method 2 used the standard deviation of the log2 fold-change. A cut-off was then set as two standard deviations from the mean log2 fold change, allowing one to focus on those proteins showing the greatest change in expression. The mass of individual proteins was estimated using the following formula: CN*MW/NA = protein mass (g/cell) where CN is protein copy number, MW is the protein molecular weight in Daltons and NA is Avogadro’s Constant. Heat maps were generated using the Morpheus tool from the Broad Institute (https://software.broadinstitute.org/morpheus). Proteins included in heat maps had a copy number of at least 1000 copies per cell in at least one population, and were found in at least one CD4+ and CD8+ T cell population. Heat maps were arranged with T-bet positioned at the top and with all proteins ranked according to similarity in expression using nearest neighbor analysis and Pearson correlation. CD4+ and CD8+ T cell heat maps were aligned in the same order to enable side-by-side comparison. Proteomics data were uploaded to the Encyclopedia of Proteome Dynamics (EPD – www.peptracker.com/epd) allowing public interrogation of the full data set.
Data Availability
All proteomics data is available for interrogation using the Encyclopedia of Proteome Dynamics (EPD – https://peptracker.com/epd). Analysed proteomics data used to generate figures is available in Supplementary Files 1-5. Raw mass spec data files and MaxQuant analysis files are available on the ProteomeXchange data repository (http://proteomecentral.proteomexchange.org/cgi/GetDataset) and can be accessed with identifier PXD012058. Flow cytometry data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Cantrell lab for comments on the manuscript, T. Ly for discussions on the data, A. Whigham and R. Clarke from the Flow Cytometry facility for cell sorting and advice on flow cytometry, and the biological sciences research unit at the University of Dundee. This research was supported by a Wellcome Trust Principal Research Fellowship to D.A.C. (205023/Z/16/Z), a Wellcome Trust Strategic Award to D.A.C. and A.I.L. (105024/Z/14/Z) and a Wellcome Trust Equipment Award to D.A.C. (202950/Z/16/Z). This work is dedicated to Olivia Mason, a beautiful person, always loved, always remembered.
Footnotes
Author Contributions
A.J.M.H., L.S., J.L.H. and L.V.S. performed experiments. J.L.H. performed LCMS analysis. A.J.M.H., J.L.H. and A.B. analyzed proteomics data. A.B. designed and implemented the Encyclopedia of Proteome Dynamics. A.I.L. and D.A.C. conceived the project and discussed the data. A.J.M.H and D.A.C. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
References
- 1.Araki K, et al. Translation is actively regulated during the differentiation of CD8(+) effector T cells. Nat Immunol. 2017;18:1046–1057. doi: 10.1038/ni.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinclair LV, et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricciardi S, et al. The Translational Machinery of Human CD4(+) T Cells Is Poised for Activation and Controls the Switch from Quiescence to Metabolic Remodeling. Cell Metab. 2018;28:895–906. doi: 10.1016/j.cmet.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geiger R, et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell. 2016;167:829–842. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hukelmann JL, et al. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat Immunol. 2016;17:104–112. doi: 10.1038/ni.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieckmann JC, et al. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat Immunol. 2017;18:583–593. doi: 10.1038/ni.3693. [DOI] [PubMed] [Google Scholar]
- 7.Tan HY, et al. Integrative Proteomics and Phosphoproteomics Profiling Reveals Dynamic Signaling Networks and Bioenergetics Pathways Underlying T Cell Activation. Immunity. 2017;46:488–503. doi: 10.1016/j.immuni.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Procaccini C, et al. The Proteomic Landscape of Human Ex Vivo Regulatory and Conventional T Cells Reveals Specific Metabolic Requirements. Immunity. 2016;44:712–712. doi: 10.1016/j.immuni.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duguet F, et al. Proteomic Analysis of Regulatory T Cells Reveals the Importance of Themis1 in the Control of Their Suppressive Function. Mol Cell Proteomics. 2017;16:1416–1432. doi: 10.1074/mcp.M116.062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuadrado E, et al. Proteomic Analyses of Human Regulatory T Cells Reveal Adaptations in Signaling Pathways that Protect Cellular Identity. Immunity. 2018;48:1046–1059. doi: 10.1016/j.immuni.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Valvezan AJ, Manning BD. Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat Metab. 2019;1:321–333. doi: 10.1038/s42255-019-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8(+) T cells. J Exp Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng H, et al. mTORC1 and mTORC2 Kinase Signaling and Glucose Metabolism Drive Follicular Helper T Cell Differentiation. Immunity. 2016;45:540–554. doi: 10.1016/j.immuni.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollizzi KN, Powell JD. Regulation of T cells by mTOR: the known knowns and the known unknowns. Trends Immunol. 2015;36:13–20. doi: 10.1016/j.it.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araki K, Youngblood B, Ahmed R. The role of mTOR in memory CD8+T-cell differentiation. Immunol Rev. 2010;235:234–243. doi: 10.1111/j.0105-2896.2010.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terada N, et al. Rapamycin blocks cell-cycle progression of activated t-cells prior to events characteristic of the middle to late g1 phase of the cycle. J Cell Physiol. 1993;154:7–15. doi: 10.1002/jcp.1041540103. [DOI] [PubMed] [Google Scholar]
- 17.Terada N, Franklin RA, Lucas JJ, Blenis J, Gelfand EW. Failure of rapamycin to block proliferation once resting cells have entered the cell-cycle despite inactivation of p70 s6 kinase. J Biol Chem. 1993;268:12062–12068. [PubMed] [Google Scholar]
- 18.Wisniewski JR, Hein MY, Cox J, Mann MA. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol Cell Proteomics. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta MM, et al. Hexokinase 2 is dispensable for T cell-dependent immunity. Cancer Metab. 2018;6:18. doi: 10.1186/s40170-018-0184-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varanasi SK, Jaggi U, Hay N, Rouse BT. Hexokinase II may be dispensable for CD4 T cell responses against a virus infection. Plos One. 2018;13:e0191533. doi: 10.1371/journal.pone.0191533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swamy M, et al. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat Immunol. 2016;17:712–720. doi: 10.1038/ni.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinclair LV, Neyens D, Ramsay G, Taylor PM, Cantrell DA. Single cell analysis of kynurenine and System L amino acid transport in T cells. Nat Commun. 2018;9:1981. doi: 10.1038/s41467-018-04366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.So L, et al. The 4E-BP-eIF4E axis promotes rapamycin-sensitive growth and proliferation in lymphocytes. Sci Signal. 2016;9:ra57. doi: 10.1126/scisignal.aad8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lingel H, et al. CTLA-4-mediated posttranslational modifications direct cytotoxic T-lymphocyte differentiation. Cell Death Differ. 2017;24:1739–1749. doi: 10.1038/cdd.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh PG, et al. Structural basis for translational inhibition by the tumour suppressor Pdcd4. Embo J. 2009;28:274–285. doi: 10.1038/emboj.2008.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki C, et al. PDCD4 inhibits translation initiation by binding to elF4A using both its MA3 domains. Proc Natl Acad Sci U S A. 2008;105:3274–3279. doi: 10.1073/pnas.0712235105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cham CM, Driessens G, O'Keefe JP, Gajewski TF. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8(+) T cells. Eur J Immunol. 2008;38:2438–2450. doi: 10.1002/eji.200838289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14:1173–U1199. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jabara HH, et al. A missense mutation in TFRC, encoding transferrin receptor 1, causes combined immunodeficiency. Nat Genet. 2016;48:74–78. doi: 10.1038/ng.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pakos-Zebrucka K, et al. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyant GA, et al. mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell. 2017;171:642–654. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saucedo LJ, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biology. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 33.Bar-Peled L, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfson RL, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chantranupong L, et al. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell. 2016;165:153–164. doi: 10.1016/j.cell.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang JL, et al. Critical roles of mTOR Complex 1 and 2 for T follicular helper cell differentiation and germinal center responses. eLife. 2016;5:e17936. doi: 10.7554/eLife.17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee K, et al. Mammalian Target of Rapamycin Protein Complex 2 Regulates Differentiation of Th1 and Th2 Cell Subsets via Distinct Signaling Pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinclair LV, et al. Phosphatidylinositol-3-OH kinase and nutrient-sensing mTOR pathways control T lymphocyte trafficking. Nat Immunol. 2008;9:513–521. doi: 10.1038/ni.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brenes A, Afzal V, Kent R, Lamond AI. The Encyclopedia of Proteome Dynamics: a big data ecosystem for (prote)omics. Nucleic Acids Res. 2018;46:D1202–D1209. doi: 10.1093/nar/gkx807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pause A, et al. Insulin-dependent stimulation of protein-synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 42.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3:1925–1938. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein-Hessling S, et al. NFATc1 controls the cytotoxicity of CD8(+) T cells. Nat Commun. 2017;8:511. doi: 10.1038/s41467-017-00612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific t-cell receptor transgenic mice varies with antigen. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 45.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 46.Hughes CS, et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol Syst Biol. 2014;10 doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotech. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 48.Cox J, et al. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All proteomics data is available for interrogation using the Encyclopedia of Proteome Dynamics (EPD – https://peptracker.com/epd). Analysed proteomics data used to generate figures is available in Supplementary Files 1-5. Raw mass spec data files and MaxQuant analysis files are available on the ProteomeXchange data repository (http://proteomecentral.proteomexchange.org/cgi/GetDataset) and can be accessed with identifier PXD012058. Flow cytometry data that support the findings of this study are available from the corresponding author upon request.