Abstract
Background
Various evidences showed that abnormally expressed long non-coding RNAs (lncRNAs) play important roles in the tumorigenesis and progression of malignancies. However, the exact role and regulatory mechanism of lncRNA TP73-AS1 in the pathogenesis and progression of lung adenocarcinoma remain to be further elucidated.
Purpose
The aim of this study was to investigate the functional role and underlying mechanism of lncRNA TP73-AS1 in lung adenocarcinoma progression.
Methods
RT-PCR assay was employed to detect TP73-AS1 expression in lung adenocarcinoma tissues and cells. The function of TP73-AS1 in lung adenocarcinoma progression was estimated by MTT assay, EdU assay, flow cytometry, Western blot, wound-healing assay and transwell assay.
Results
LncRNA TP73-AS1 expression was significantly increased in lung adenocarcinoma tissues and cell lines. Moreover, functional assays revealed that silencing of lncRNA TP73-AS1 could attenuate cell proliferation, migration, invasion and epithelial–mesenchymal transition of lung adenocarcinoma, while enhanced expression of lncRNA TP73-AS1 led to the opposite results. Additionally, lncRNA TP73-AS1 knockdown could facilitate cell apoptosis and overexpression of lncRNA TP73-AS1 inhibited cell apoptosis. In addition, we further determined that lncRNA TP73-AS1 regulated cell metastasis through inducing the activation of Wnt/β-catenin signaling pathway in lung adenocarcinoma.
Conclusion
Our results indicated that lncRNA TP73-AS1 may play an oncogenic role in lung adenocarcinoma progression, which provided a promising therapy strategy for the treatment of lung adenocarcinoma.
Keywords: lncRNA TP73-AS1, proliferation, apoptosis, EMT, Wnt/β-catenin pathway, lung adenocarcinoma
Introduction
Lung cancer is the most common cause of cancer-related deaths worldwide.1 Small-cell lung cancer and non-small cell lung cancer (NSCLC) are two main histological categories of lung cancer. Lung adenocarcinoma is most common histological type of NSCLC.2 Despite advancements in treatments for lung adenocarcinoma, the 5-year survival rate remains as low as 16%. Lung adenocarcinoma can develop a high metastatic potential, which is the major cause of treatment failure. Tumor metastases in the progression of lung adenocarcinoma are responsible for approximately 90% of patients succumbed to lung cancer.3 Therefore, further investigation of the regulators and molecular mechanisms underlying lung adenocarcinoma pathogenesis is crucial for improving the therapy and prognosis in lung adenocarcinoma.
Cancer cell metastasis is a complex process by which involving many factors. The epithelial–mesenchymal transition (EMT) is regarded as one of the most critical molecular mechanisms during tumor metastasis.4–6 Cell migration and invasion abilities are critical to understand the cancer metastasis.7 In addition, the EMT is characterized by important molecules change including the expression of E-cadherin, N-cadherin and vimentin.8
Long non-coding RNA (lncRNA) is one kind of non-encoding RNA transcripts which is over 200 nucleotides (nt).9–11 Increasing evidences suggested that lncRNAs participate in a variety of cellular biological processes, including cell proliferation, differentiation and apoptosis.12–14 LncRNA P73 antisense 1T (TP73-AS1) is located on human chromosomal band 1p36.32.15 Previous studies reported that TP73-AS1 was closely associated with tumor progression and poor prognosis in esophageal squamous cell carcinoma, breast cancer, brain glioma, gastric cancer and hepatocellular carcinoma.16–23 These reports demonstrated that lncRNA TP73-AS1 acted as one oncogene in the tumorigenesis of cancer. Unfortunately, the biological function and underlying mechanism of lncRNA TP73-AS1 in lung adenocarcinoma are rarely explored and more researches are needed to be clarified. In the present study, we demonstrated that lncRNA TP73-AS1 exerted enhanced effects on tumor progression of lung adenocarcinoma. Function al assays showed that lncRNA TP73-AS1 could lead to significant promotion of cell proliferation and EMT and suppression of apoptosis by Wnt/β-catenin signaling pathway in lung adenocarcinoma. Our findings provide new insights into the biological function and underlying mechanism of lncRNA TP73-AS1 in lung adenocarcinoma and will give a novel therapeutic target for lung adenocarcinoma.
Materials And Methods
Patient Tissues Samples
A total of 30 tissues and paired adjacent noncancerous tissues were collected from Liyang People’s Hospital between January 2017 and July 2018 and this study was approved by the Liyang People’s Hospital Ethics Committee (Nanjing, People’s Republic of China). All the procedures of this work were in compliance with the criterions of the Declaration of Helsinki. All patients signed the written informed consent and did not receive any radiotherapy or chemotherapy treatment before surgery. These tissue samples were rapidly frozen in liquid nitrogen and stored in the –80°C refrigerator.
Cell Culture
All the cell lines used in our study consisted of three human lung adenocarcinoma cell lines (H1299, H1793 and A549) and human lung epithelial cell line BEAS-2B, which were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Then, A549 and H1299 cells were both cultured with RPMI-1640 medium (Gibco, USA) supplemented with 10% FBS (Gibco, USA). H1793 cells were grown in DMEM/F12 medium (Gibco, USA) containing 5% FBS, 0.005 mg/mL insulin, 0.01 mg/mL transferrin, 2 mM l-glutamine, 10 nM hydrocortisone and 30 nM sodium selenite. BEAS-2B cells were cultivated in DMEM (Gibco, USA) with 10% FBS and 1% penicillin/streptomycin (Sigma, USA). All cells were incubated in a humidified incubator at 37°C with 5% CO2.
Cell Proliferation Assay
MTT assay was used to detect cell proliferation ability according to the manufacturer’s instructions (Dojindo Laboratories, Japan). Briefly, 1×105 cells were plated into 96-well plates and cultured for 0, 24, 48, 72 and 96 hrs after transfected with si-TP73-AS1, si-NC, Lv-TP73-AS1 or Lv-NC. MTT solution (Sigma, USA) was added into each well and incubated for 4 hrs. The absorbance was measured by the EnSpire Multimode Plate Reader (PerkinElmer, CA, USA) at 570 nm. Data represent at least three separate experiments independently. For cell colony formation assay, cells were plated into 6-well plate (500 cells/well) and incubated for 7–14 days. At the endpoint of incubation, the cells were fixed with paraformaldehyde and stained with crystal violet. Colonies were counted under a light microscope. Each experiment was performed three times independently.
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
1×104 A549 and H1299 cells were seeded in a 96-well plate and cultured for 24 hrs at 37°C with 5% CO2. A quantity of 100 μL of EdU (50 μM) was added and cultured for another 2 hrs following fixation with 4% paraformaldehyde for 20 mins. The cells were stained by Cell-Light™ EdU Apollo®488 In Vitro Imaging Kit (Life Technologies, New York, USA) and DAPI staining was performed according to the instructions. EdU positive cells were detected under a fluorescence microscope.
RT-PCR Analysis
Total RNAs were extracted from cells or tumor tissues with Trizol reagent (Invitrogen) and were reversed transcribed into cDNA using the Reverse Transcription Kit (Applied Biosystems, USA). For RT-PCR, the cDNA was amplified using SYBR Premix Ex Taq (Takara, Dalian, People’s Republic of China) and run on LightCycler 480 II (Roche, Germany). Relative mRNA expression was counted using the 2−ΔΔCT method. All the reactions were performed in triplicates.
Western Blot Analysis
Proteins were extracted from cells using RIPA lysis buffer (Beyotine, Shanghai) and supernatant was collected by centrifuging at 12,000× g for 15 mins at 4°C. Protein concentrations were determined by BCA protein assay (Thermo, USA). Briefly, equal amounts of samples were transferred to PVDF membranes, which were blocked with 5% nonfat milk and incubated with primary antibodies at 4°C overnight. Then, the membranes were incubated with horseradish peroxidase-conjugated species-specific secondary antibodies. Bands were visualized using enhanced chemiluminescence (ECL, Beijing, People’s Republic of China) and all data were normalized to the mean value of GAPDH.
Cell Cycle And Apoptosis Analysis
H1299 and A549 cells were collected after si-TP73-AS1, si-NC, Lv-TP73-AS1 or Lv-NC transfection for flow cytometer analysis and then stained with 50 µg/mL propidium iodide (PI) for 15 mins at room temperature in the dark. Cell apoptosis was performed with Annexin V Apoptosis Detection kit Ⅰ (BD Biosciences, USA). Cells were processed as described early and incubated with FITC annexin V and PI for 15 mins in the dark. A total number of 1×104 cells were subjected to the cell cycle and apoptosis analysis by the flow cytometer (Becton Dickinson, NJ, USA).
Wound-Healing Assay
The migratory ability of H1299 and A549 cells was assessed by scratch wound assay. Cells (1×105/mL) were seeded in a 6-well plate and cultured overnight. Then, the cells were carefully wounded using a pipette tip and washed with DMEM. The crosses of each well were photographed with microscope at 0, 24 and 48 hrs after scratching. Each experiment was performed three times independently.
Transwell Assays
Transwell assay was used to evaluate the migratory and invasive capacities of H1299 and A549 cells using 24-well migration chamber with or without matrigel. Briefly, 1×105 cells were added to the upper chamber and 600 μL 10% FBS medium was added into the lower chamber. After incubation for 24 hrs, those cells not passing through the pore were wiped out with a cotton swab and the migrated or invaded cells were stained with crystal violet, following the cells were counted and photographed under a light microscope. Each experiment was performed three times independently.
Statistical Analysis
Data analyses were performed using the SPSS 18.0 statistical software. All data were presented as mean±SD. Student’s t-test and two-way ANOVA were performed to determine statistical significance. All of the experiments were repeated for at least three times, P<0.05 was considered to demonstrate statistically significant.
Results
LncRNA TP73-AS1 Was Upregulated In Lung Adenocarcinoma Tissues And Cell Lines
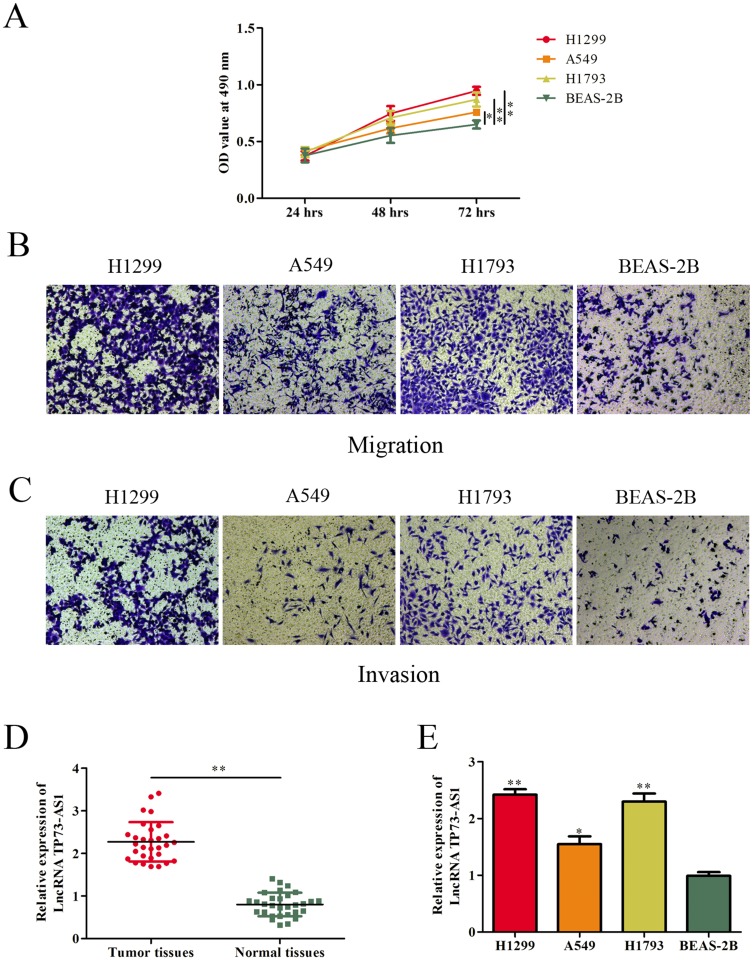
First, the proliferation, migration and invasion abilities of these cells were detected. As shown in Figure 1A, lung adenocarcinoma cells (H1299, H1793 and A549) presented stronger proliferative capacity compared with normal epithelial cells BEAS-2B. Likewise, transwell assays showed that lung adenocarcinoma cell lines expressed more positive abilities of migration and invasion than BEAS-2B cell line (Figure 1B and C). Subsequently, we examined the expression of lncRNA TP73-AS1 in lung adenocarcinoma tissues and normal adjacent tissues using RT-PCR. The data demonstrated that lncRNA TP73-AS1 was significantly upregulated in lung adenocarcinoma tissues compared with those in the matched adjacent normal tissues (Figure 1D). Furthermore, the expression of lncRNA TP73-AS1 in human lung adenocarcinoma cell lines and normal epithelial cell line was also evaluated. Similarly, the expression level of lncRNA TP73-AS1 in lung adenocarcinoma cell lines was higher than that in normal cell line (Figure 1E).
Figure 1.
LncRNA TP73-AS1 was upregulated in lung adenocarcinoma tissues and cell lines. (A–C) The proliferative, migratory and invasive abilities of cell lines were evaluated by MTT and transwell assays. (D) Expression levels of lncRNA TP73-AS1 in human lung adenocarcinoma tissues and corresponding adjacent tissues were determined by RT-PCR. (E) LncRNA TP73-AS1 expression in lung adenocarcinoma cell lines and human epithelial cell was examined by RT-PCR. *P<0.05, **P<0.01.
Silencing lncRNA TP73-AS1 Inhibited Lung Adenocarcinoma Cell Proliferation
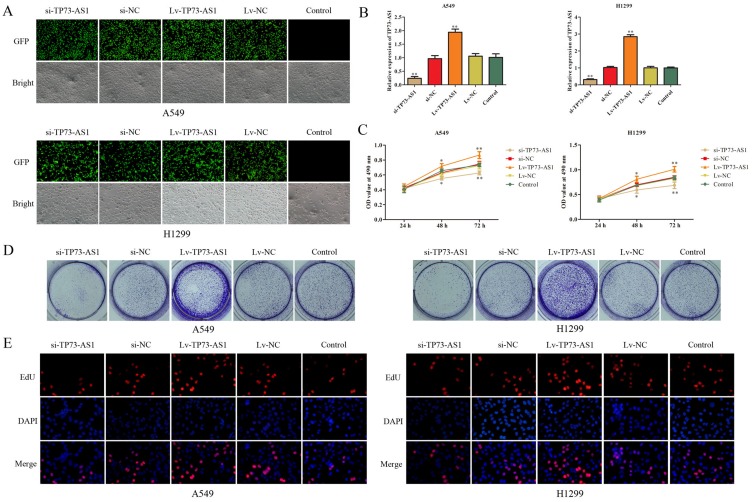
H1299 and A549 were chosen for the loss- or gain-of-function experiments to determine the role of lncRNA TP73-AS1 in lung adenocarcinoma cells. LncRNA TP73-AS1 knockdown or upregulation was achieved by si-TP73-AS1 or Lv-TP73-AS1 transfection. The transfection efficiency was validated by GFP and RT-PCR (Figure 2A and B). The effects of lncRNA TP73-AS1 on the proliferation of H1299 and A549 cell lines were examined by MTT, colony formation and EdU assays. Data of MTT assay exhibited that the knockdown of TP73-AS1 remarkably attenuated the proliferation rate of H1299 and A549 cells compared with matched control group, while ectopic expression of TP73-AS1 prompted cell proliferation (Figure 2C). Additionally, colony formation assay demonstrated that silencing of TP73-AS1 contributed to the suppression of colony formation, whereas cells transfected with Lv-TP73-AS1 exhibited stronger colony formative capacity than corresponding control group (Figure 2D). EdU assay further confirmed the promoting proliferative role of TP73-AS1 in lung adenocarcinoma cells (Figure 2E). These results revealed that lncRNA TP73-AS1 fortified the proliferation of H1299 and A549 cells and suggested that lncRNA TP73-AS1 functioned as an oncogenic molecule in lung adenocarcinoma.
Figure 2.
LncRNA TP73-AS1 promoted cell growth of lung adenocarcinoma cells. (A, B) The transfection efficiency was validated by GFP and RT-PCR. (C–E) Cell proliferation in H1299 and A549 cells was assessed by the MTT, colony formation and EdU assays. All data were shown as the mean fold change±SD from three experiments. *P<0.05, **P<0.01.
Effects Of lncRNA TP73-AS1 On Cell Cycle And Apoptosis
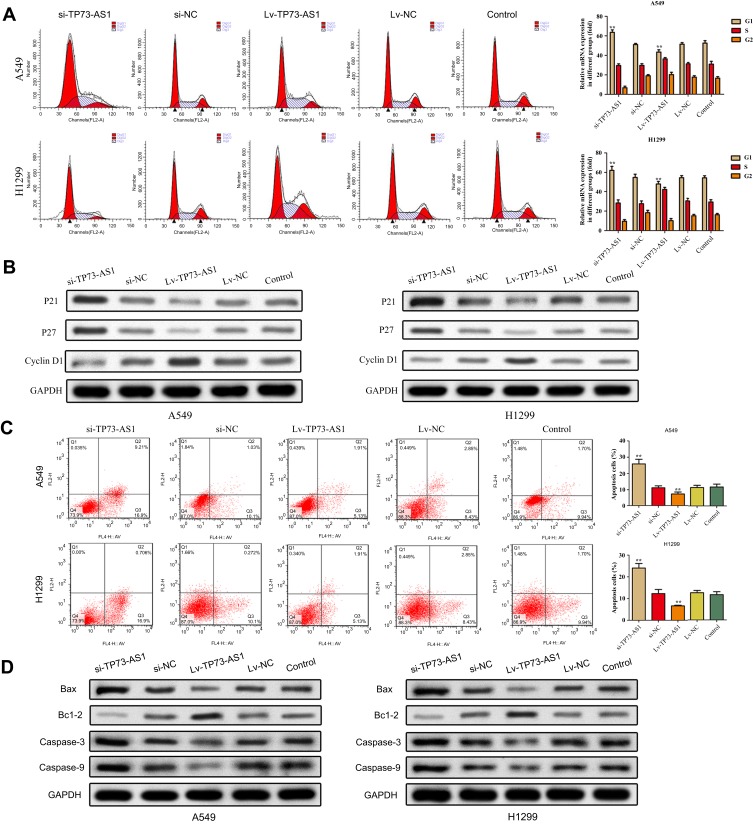
Effects of lncRNA TP73-AS1 on cell cycle distribution and apoptosis were analyzed using flow cytometry. The results illustrated that silencing expression of lncRNA TP73-AS1 significantly induced the cycle arrest at G1 phase compared with control group, while no significant alterations were viewed in S phase and G2/M phase. Inversely, upregulation of lncRNA TP73-AS1 caused the opposite results (Figure 3A). Meanwhile, the expression of cell cycle associated proteins p21, p27 and cyclinD1 was measured by Western blot. We observed that the levels of p21 and p27 were downregulated while cyclin D1 expression was increased in H1299 and A549 cells after transfection with Lv-TP73-AS1 compared with the matched control group. Adversely, the expression of p21 and p27 was remarkably promoted while cyclin D1 level was downregulated due to knocking down of lncRNA TP73-AS1 (Figure 3B).
Figure 3.
Effects of lncRNA TP73-AS1 on cell cycle and cell apoptosis. (A) The cell cycle progression was detected by flow cytometry. (B) The expression of cell cycle-related proteins was analyzed by Western blot. (C) The cell apoptosis was detected by flow cytometry. (D) The expression of cell apoptosis-related proteins was analyzed by Western blot. **P<0.01.
To confirm the quantity of cell apoptosis, Annexin V-FITC/PI dual staining was performed by flow cytometry. As shown in Figure 3C, the apoptosis rate of H1299 and A549 cells transfected with si-TP73-AS1 was higher than the control, whereas we observed that overexpression of TP73-AS1 lessened cell apoptosis. To confirm the impacts of TP73-AS1 on cell apoptosis, the expression levels of apoptosis-related proteins Bax, Bcl-2, Caspase-3 and Caspase-9 were estimated. It was indicated that TP73-AS1 knockdown remarkably declined Bcl-2 expression and increased the levels of Bax, Caspase-3 and Caspase-9. However, enhanced expression of TP73-AS1 produced the opposite results (Figure 3D). In summary, these data indicated that lncRNA TP73-AS1 had promotive effects on lung adenocarcinoma cells progression.
LncRNA TP73-AS1 Boosted Cell Migration And Invasion In Lung Adenocarcinoma
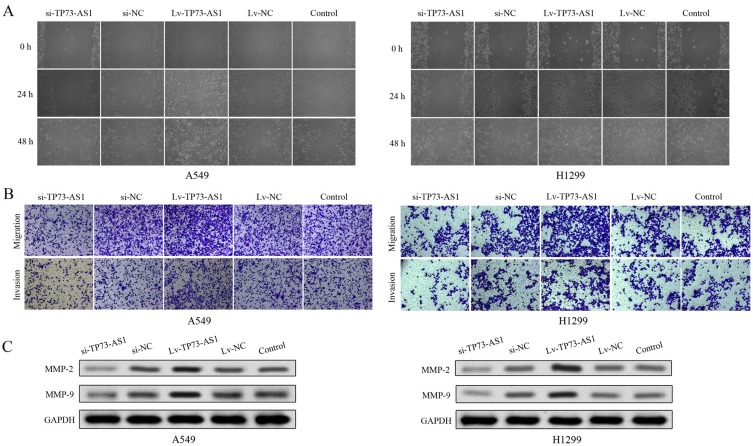
We next explored the effects of lncRNA TP73-AS1 on cell migratory and invasive abilities using a scratch wound assay and transwell assay in lung adenocarcinoma. The wound-healing and transwell assays demonstrated that silencing lncRNA TP73-AS1 markedly reduced the number of migrated and invaded cells compared with the control group. Contrarily, transfection with Lv-TP73-AS1 could enhance the cell migration and invasion abilities (Figure 4A and B). In addition, the expression of proteins (MMP-2 and MMP-9) which were related to migration and invasion was investigated using Western blot. Compared with control group, MMP-2 and MMP-9 levels were markedly heightened in cells transfected with Lv-TP73-AS1. In contrast, the protein expression of MMP-2 and MMP-9 was remarkably lowered in si-TP73-AS1 group (Figure 4C). These results revealed that lncRNA TP73-AS1 had an important role in promoting cell migration and invasion of lung adenocarcinoma.
Figure 4.
LncRNA TP73-AS1 regulated the migratory and invasive abilities of cells. (A) The migratory ability of H1299 and A549 was evaluated by wound-healing assay. (B) The cell migratory and invasive abilities were evaluated by transwell assay of H1299 and A549 cells. (C) The protein expression levels of MMP-2 and MMP-9 were analyzed by Western blot. All experiments were performed in triplicate and results were presented as the mean±SD.
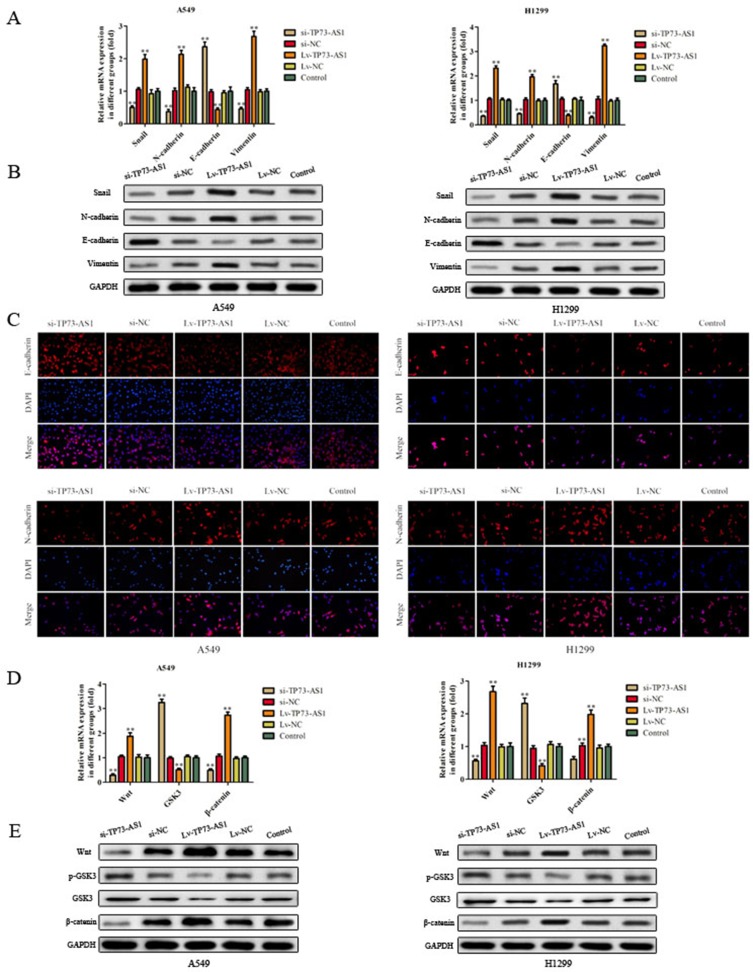
Effects Of lncRNA TP73-AS1 On Cell EMT In Lung Adenocarcinoma
EMT plays a critical role in cancer dissemination and metastatic spread, we investigated the expression of the EMT markers using RT-PCR, Western blot and immunofluorescence analysis. As shown in Figure 5A–C, the epithelial marker E-cadherin mRNA and protein levels were apparently upregulated in cells which transfected with si-TP73-AS1, while decreased in Lv-TP73-AS1 group compared with the corresponding control group. By contrast, the mesenchymal markers Snail, N-cadherin and vimentin were downregulated in cells which were transfected with si-TP73-AS1, while increased in Lv-TP73-AS1 group compared with matched control group. Taken together, these findings indicated that silencing of lncRNA TP73-AS1 inhibited the EMT progression in lung adenocarcinoma cells.
Figure 5.
Effects of lncRNA TP73-AS1 on EMT and Wnt/β-catenin signaling pathway in lung adenocarcinoma. (A–C) The expression of the EMT markers (E-cadherin, Snail, N-cadherin and vimentin) was detected by using RT-PCR, Western blot and immunofluorescence analysis. (D, E) The expression of some related mRNA and proteins was explored via RT-PCR and Western blot. The results were expressed as mean±SD of three independent experiments. **P<0.01.
LncRNA TP73-AS1 Activated Wnt/β-Catenin Signaling Pathway
The Wnt/β-catenin signaling pathway plays an important role in maintaining an epithelial cell phenotype, so we examined the expression of key proteins (Wnt, GSK3 and β-catenin) in Wnt/β-catenin signaling pathway in H1299 and A549 cells. As expected, RT-PCR experiment showed that knockdown of lncRNA TP73-AS1 significantly decreased the expression of Wnt and β-catenin while augmented the level of GSK3 in H1299 and A549 cells (Figure 5D). As shown in Figure 5E, the protein expression of Wnt and β-catenin was increased in lncRNA TP73-AS1 overexpressed cells, while the protein levels of p-GSK3 and GSK3 were diminished in Lv-TP73-AS1 group in contrast with matched control group. All these findings indicated that lncRNA TP73-AS1 regulated EMT and Wnt/β-catenin signaling pathway.
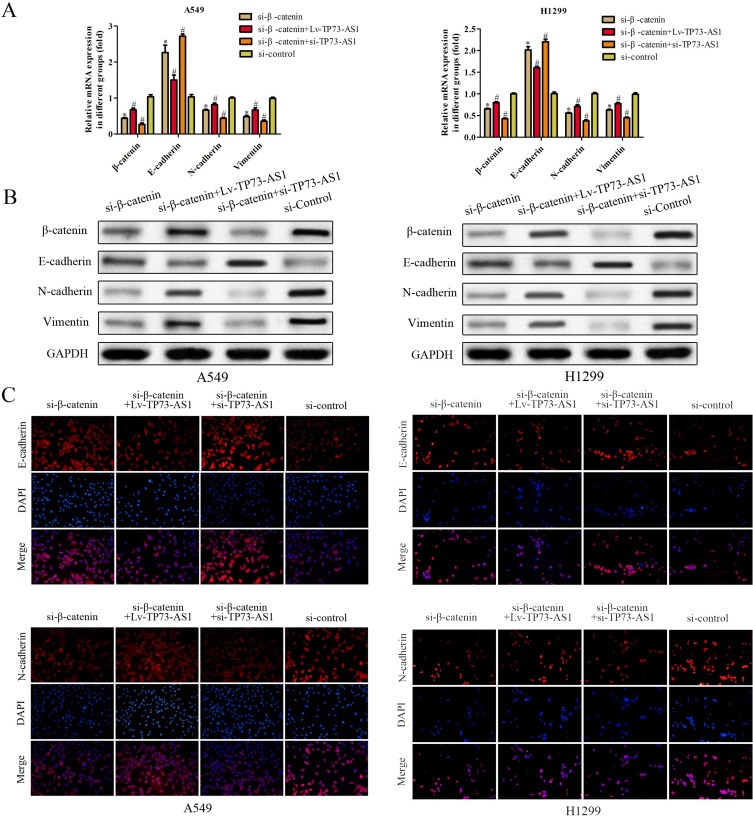
LncRNA TP73-AS1 Regulated Wnt/β-Catenin Signaling Pathway To Prompt EMT Of Lung Adenocarcinoma Cells
In order to validate whether TP73-AS1 executed its role via Wnt/β-catenin pathway, we further investigated the effects of si-β-catenin in the presence of Lv-TP73-AS1 and si-TP73-AS1 in H1299 and A549 cells using RT-PCR, Western blot and immunofluorescence assays. As shown in Figure 6, the expression of β-catenin was detected after transfected si-β-catenin and we observed that N-cadherin and vimentin expression was dramatically decreased. si-β-catenin in the presence of Lv-TP73-AS1 reversed the effects of si-β-catenin on H1299 and A549 cells. In contrast, the expression of E-cadherin was increased in cells transfected with si-β-catenin compared with control group. These observations were further verified the efficiency of lncRNA TP73-AS1 on EMT by regulating Wnt/β-catenin signaling pathway.
Figure 6.
LncRNA TP73-AS1 regulated Wnt/β-catenin signaling pathway to prompt EMT of lung adenocarcinoma cells. (A–C) The expression of EMT-related mRNA and proteins was explored via RT-PCR, Western blot and immunofluorescence assays. *P<0.05 versus control group. #P<0.05 versus si-β-catenin group.
Discussion
Lung cancer is the most common cause of cancer-related deaths worldwide, which is generally diagnosed at an advanced stage. So finding new molecular targets for its diagnosis and treatment has the potential to improve the clinical strategies and outcomes of lung cancer.24
LncRNAs are a class of non-coding RNA transcripts longer than 200 nucleotides. In recent years, many studies have shown that lncRNAs are aberrantly expressed in human cancer25 and participate in the regulation of cellular growth and metastasis.26–29 In addition, Wang et al found that overexpression of lncRNA HOTAIR promoted tumor growth and metastasis in human osteosarcoma30 and Zhang et al found that upregulation of lncRNA MALAT1 was correlated with tumor progression and poor prognosis in clear cell renal cell carcinoma.31 Therefore, the investigation of cancer-associated lncRNAs may provide novel prognostic biomarkers or therapeutic targets for lung adenocarcinoma. LncRNA antisense RNA to TP73 gene (TP73-AS1) is located on human chromosomal band 1p36.3232 and identified as an oncogenic RNA in breast cancer, esophagus cancer, glioma, hepatocellular carcinoma and lung cancer.16,17,19,33,34 Nevertheless, more detailed functional effects and associated molecular regulatory mechanism of lncRNA TP73-AS1 in lung adenocarcinoma remain largely to be explored.
In the present study, we found that lncRNA TP73-AS1 expression in lung adenocarcinoma tissues was significantly higher than that in matched adjacent non-tumor tissues. In addition, lncRNA TP73-AS1 knockdown significantly inhibited cellular proliferation through arresting cell cycle, migration, invasion as well as stimulating cell apoptosis of lung adenocarcinoma, implying that lncRNA TP73-AS1 may play an oncogenic role in lung adenocarcinoma.
Cancer metastasis is a complex process of cancer-related death.35 EMT is a critical process providing epithelial-derived tumor cells with the abilities of increased migration and invasion, contributing to tumor metastasis36 and characterized by loss of epithelial features and obtainment of mesenchymal characteristics. The dysregulation of the Wnt/β-catenin pathway has been observed in various cancers and regulated cancer metastasis.37,38 In this study, we observed that overexpression of lncRNA TP73-AS1 dramatically enhanced cell migratory and invasive abilities. Furthermore, the expression of EMT-associated proteins was explored. Our results showed that lncRNA TP73-AS1 knockdown increased E-cadherin expression and decreased the expression of Snail, N-cadherin, vimentin. What is more, our data indicated that overexpression of lncRNA TP73-AS1 could activate Wnt/β-catenin pathway to promote EMT of lung adenocarcinoma cells.
In summary, this study indicated that lncRNA TP73-AS1 may serve as an oncogene and may play an important role in lung adenocarcinoma development and progression. The results showed that lncRNA TP73-AS1 dramatically promoted cell proliferation, migration, invasion, EMT and inhibited cell apoptosis by Wnt/β-catenin pathway in lung adenocarcinoma. Therefore, our findings might provide a promising new therapeutic target and strategy for the treatment of lung adenocarcinoma.
Acknowledgments
This research was supported by Guiding Science and Technology Project of Changzhou Health Committee (wz201916) as well as Science and Technology Project of Liyang People's Hospital (2019YJKT008).
Disclosure
The authors have no conflicts of interest in this work.
References
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;67(1):7. doi: 10.3322/caac.21387 [DOI] [Google Scholar]
- 2.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088 [DOI] [PubMed] [Google Scholar]
- 3.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP, Acloque H, Huang RYJ, et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 5.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2015;119(6):1420–1428. doi: 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 7.Floor S, van Staveren WC, Larsimont D, et al. Cancer cells in epithelial-to-mesenchymal transition and tumor-propagating-cancer stem cells: distinct, overlapping or same populations. Oncogene. 2011;30(46):4609. doi: 10.1038/onc.2011.184 [DOI] [PubMed] [Google Scholar]
- 8.Perl AK, Wilgenbus P, Dahl U, et al. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392(6672):190–193. doi: 10.1038/32433 [DOI] [PubMed] [Google Scholar]
- 9.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126(8):2775. doi: 10.1172/JCI84421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 12.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 13.Li J, Meng H, Bai Y, et al. Regulation of lncRNA and its role in cancer metastasis. Oncol Res. 2016;23(5):205. doi: 10.3727/096504016X14549667334007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013 [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Xu Q, Liu F, et al. Identification and validation of long noncoding RNA biomarkers in human non–small-cell lung carcinomas. J Thorac Oncol. 2015;10(4):645–654. doi: 10.1097/JTO.0000000000000470 [DOI] [PubMed] [Google Scholar]
- 16.Zang W, Wang T, Wang Y, et al. Knockdown of long non-coding RNA TP73-AS1 inhibits cell proliferation and induces apoptosis in esophageal squamous cell carcinoma. Oncotarget. 2016;7(15):19960–19974. doi: 10.18632/oncotarget.6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou Q, Zhou E, Xu F, et al. A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J Cell Biochem. 2017;119(2):2189–2199. [DOI] [PubMed] [Google Scholar]
- 18.Zhang R, Jin H, Lou F. The long non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain glioma growth through HMGB1/RAGE pathway. J Cell Biochem. 2017;119(4):3007–3016. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Huang Y, Huang Y, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36(1):51. doi: 10.1186/s13046-017-0519-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao J, Xu F, Zhang D, et al. TP73-AS1 promotes breast cancer cell proliferation through miR-200a-mediated TFAM inhibition. J Cell Biochem. 2018;119(1). doi: 10.1002/jcb.27261. [DOI] [PubMed] [Google Scholar]
- 21.Ding Z, Lan H, Xu R, et al. LncRNA TP73-AS1 accelerates tumor progression in gastric cancer through regulating miR-194-5p/SDAD1 axis. Pathol Res Pract. 2018;214:1993–1999. doi: 10.1016/j.prp.2018.09.006 [DOI] [PubMed] [Google Scholar]
- 22.Tuo Z, Zhang J, Xue W. LncRNA TP73-AS1 predicts the prognosis of bladder cancer patients and functions as a suppressor for bladder cancer by EMT pathway. Biochem Biophys Res Commun. 2018;499:875–881. doi: 10.1016/j.bbrc.2018.04.010 [DOI] [PubMed] [Google Scholar]
- 23.Yufeng W, Shuai X, Bingyi W, et al. Knockdown of lncRNA TP73-AS1 inhibits gastric cancer cell proliferation and invasion via the WNT/β?catenin signaling pathway. Oncol Lett. 2018;16(3):3248–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowman E. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025 [DOI] [PubMed] [Google Scholar]
- 25.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 26.Lu K, Li W, Liu X, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13(1):461. doi: 10.1186/1471-2407-13-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatima R, Akhade VS, Pal D, et al. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015;3(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zang W, Wang T, Huang J, et al. Long noncoding RNA PEG10 regulates proliferation and invasion of esophageal cancer cells. Cancer Gene Ther. 2015;22(3):138–144. doi: 10.1038/cgt.2014.77 [DOI] [PubMed] [Google Scholar]
- 29.Xue Y, Ma G, Zhang Z, et al. A novel antisense long noncoding RNA regulates the expression of MDC1 in bladder cancer. Oncotarget. 2015;6(1):484–493. doi: 10.18632/oncotarget.2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Su Y, Yang Q, et al. Overexpression of long non-coding RNA HOTAIR promotes tumor growth and metastasis in human osteosarcoma. Mol Cell. 2015;38(5):432–440. doi: 10.14348/molcells.2015.2327 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zhang HM, Yang FQ, Chen SJ, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumor Biol. 2015;36(4):2947–2955. doi: 10.1007/s13277-014-2925-6 [DOI] [PubMed] [Google Scholar]
- 32.Lin ZY, Chuang WL. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother. 2012;66(6):454–458. doi: 10.1016/j.biopha.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 33.Jia Y, Feng X, Zhang D, et al. TP73-AS1 promotes breast cancer cell proliferation through miR-200a-mediated TFAM inhibition. J Cell Biochem. 2018;119(1):9444–9461. doi: 10.1002/jcb.27261 [DOI] [PubMed] [Google Scholar]
- 34.Zhu D, Zhou J, Liu Y, et al. LncRNA TP73-AS1 is upregulated in non-small cell lung cancer and predicts poor survival. Gene. 2019;710:98–102. doi: 10.1016/j.gene.2019.05.044 [DOI] [PubMed] [Google Scholar]
- 35.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. doi: 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- 36.Hugo H, Ackland ML, Blick T, et al. Epithelial–mesenchymal and mesenchymal–epithelial transitions in carcinoma progression. J Cell Physiol. 2010;213(2):374–383. doi: 10.1002/jcp.21223 [DOI] [PubMed] [Google Scholar]
- 37.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419 [DOI] [PubMed] [Google Scholar]
- 38.Choi YS, Shim YM, Kim SH, et al. Prognostic significance of E-cadherin and beta-catenin in resected stage I non-small cell lung cancer. Eur J Cardiothorac Surg. 2003;24(3):441–449. doi: 10.1016/s1010-7940(03)00308-7 [DOI] [PubMed] [Google Scholar]