Abstract
Melanoma is a highly aggressive tumor with a poor prognosis, and half of all melanoma patients harbor BRAF mutations. A BRAF inhibitor, vemurafenib (PLX4032), has been approved by the US Food and Drug Administration (FDA) and European Medicines Agency (EMA) to treat advanced melanoma patients with BRAFV600E mutation. However, the efficacy of vemurafenib is impeded by adaptive resistance in almost all patients. In this study, using a cell-based SELEX (systematic evolution of ligands by exponential enrichment) strategy, we obtained a DNA aptamer (named LL4) with high affinity and specificity against vemurafenib-resistant melanoma cells. Optimized truncated form (LL4A) specifically binds to vemurafenib-resistant melanoma cells with dissociation constants in the nanomolar range and with excellent stability and low toxicity. Meanwhile, fluorescence imaging confirmed that LL4A significantly accumulated in tumors formed by vemurafenib-resistant melanoma cells, but not in control tumors formed by their corresponding parental cells in vivo. Further, a transmembrane protein CD63 was identified as the binding target of aptamer LL4A using a pull-down assay combined with the liquid chromatography-tandem mass spectrometry (LC-MS/MS) method. CD63 formed a supramolecular complex with TIMP1 and β1-integrin, activated the nuclear factor кB (NF-кB) and mitogen-activated protein kinase (MAPK) signaling pathways, and contributed to vemurafenib resistance. Potentially, the aptamer LL4A may be used diagnostically and therapeutically in humans to treat targeted vemurafenib-resistant melanoma.
Keywords: melanoma, vemurafenib-resistant, aptamer, LL4A, CD63
Introduction
Melanoma is a highly aggressive tumor with a poor prognosis in the metastatic stage. Approximately 50% of melanoma patients harbor a BRAF mutation, the most common being BRAF(V600E).1 BRAF inhibitors, such as vemurafenib (PLX4032) or dabrafenib, were approved by the US Food and Drug Administration (FDA) and European Medicines Agency (EMA) to treat advanced melanoma patients with BRAFV600E mutation.2 Unfortunately, nearly 100% of the patients exhibited drug resistance and disease progression within 6–7 months after treatment with BRAF inhibitors.3,4 Even with current development of immune therapy, BRAF-targeted therapy remains an important therapeutic method.5 Developing novel target treatment methods is still urgently needed.
Aptamers are single-stranded DNA (ssDNA) or RNA molecules that evolved from random oligonucleotide pools by a process called systematic evolution of ligands by exponential enrichment (SELEX).6,7 Compared with traditional antibody probes, aptamers have several advantages, including relatively simple synthesis and modification, low potential for immunogenicity, long-term stability, reversible denaturation, low toxicity, efficient tissue penetration, and low variability among different batches.8, 9, 10 Because of these advantages, aptamers show great application prospects in drug development, clinical diagnosis, and targeted therapy.11 Aptamers recognize and bind specifically to their targets with high affinity, acting as carriers to deliver therapeutic agents to their targets, which is the main way for the potential exploitation of these molecules for diagnosis and therapy.12, 13, 14 Aptamers not only can detect previously known tumor markers but also can be utilized for discovery of potential novel biomarkers in human cancers.15 Many aptamers have been used to detect a variety of cancers by targeting tumor markers, such as nucleolin,16 tenascin,17 PTK7,18 MUC1,19 c-Met,20 and matrix metalloprotease-9 (MMP-9).21 Therefore, generating a group of aptamers with high specificity is an effective and realistic method to discover novel tumor biomarkers and to construct a targeted delivery system.15
Given the importance of vemurafenib in cancer therapy and its well-recognized nature of drug resistance, we have developed vemurafenib-resistant melanoma cell lines15 and, for the first time, identified a group of DNA aptamers that can specifically recognize the melanoma PLX4032-resistant cell lines. One of the selected aptamers, LL4, and its truncated version aptamer LL4A showed high specificity and affinity for resistant cells Mel28-PLX and A375-PLX. Furthermore, in vivo and ex vivo fluorescence imaging demonstrated that aptamer LL4A could specifically target the tumors formed by resistant melanoma cells after intravenous injection into nude mice. Our subsequent studies indicated that LL4A targeted a cell surface protein CD63, which is a member of the tetraspanin superfamily.
Melanoma patients showed higher plasma levels of CD63 compared with healthy controls,22 suggesting that CD63 could serve as a potential biomarker for melanoma. CD63 was known to transmit protein kinase signals in melanoma cells.23 Recently, CD63 was reported to be involved in a supramolecular complex with TIMP1 and β1-integrin, conferring melanoma anoikis resistance.24 In breast cancer cells, CD63 prevented chemo-induced apoptosis, which contributes to chemoresistance.25 In this study, our results suggest that upregulation of CD63 in vemurafenib-resistant cells may contribute to cell survival and vemurafenib resistance by activating the nuclear factor кB (NF-κB)/TIMP1/CD63/β1-integrin/extracellular signal-regulated kinase (ERK) pathway. CD63 is a key LL4A binding protein whereby LL4A could specifically recognize and bind to melanoma PLX4032-resistant cell lines.
Results
Selection of the DNA Aptamer LL4 against Mel28-PLX Cells
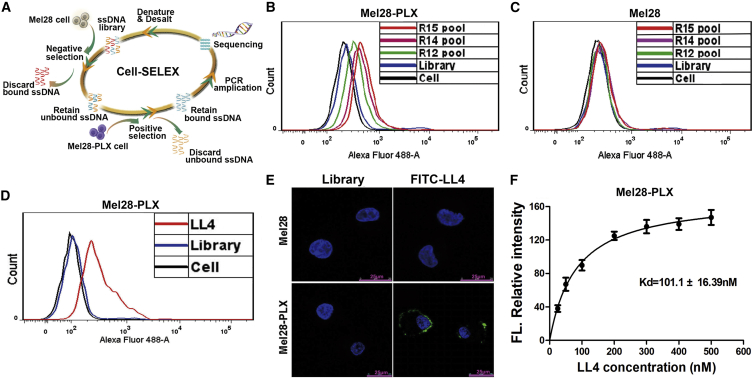
In a previous study, we generated two PLX4032-resistant melanoma cell lines, Mel28-PLX and A375-PLX, which were able to proliferate in the presence of 5 μM PLX4032 and exhibited an IC50 value approximately 20-fold higher than the parental cells Mel28 and A375, respectively. Meanwhile, RNA sequencing (RNA-seq) and qRT-PCR arrays data showed that the molecular profiles and signaling pathways have been changed dramatically between PLX4032-resistant cells versus control.26 Therefore, we aimed to obtain a DNA aptamer with high affinity and specificity against vemurafenib-resistant melanoma cells, further identify the binding target of the aptamer, and subsequently investigate the underlying mechanism for vemurafenib-induced drug resistance in melanoma cells. To generate aptamers against PLX4032-resistant melanoma cells, Mel28-PLX cells were used for positive selection, and the parental Mel28 cells were used for counterselection. The cell-SELEX process is schematically shown in Figure 1A. The enrichment of the ssDNA library was monitored by flow cytometry during selection. The fluorescence intensity reflected the binding capacity of the enriched pools. As shown in Figure 1B, the fluorescence intensity on target Mel28-PLX cells gradually increased after incubation with increasing rounds of fluorescein isothiocyanate (FITC)-labeled ssDNA pools. In contrast, almost no increase in fluorescence signal was observed for control cells Mel28 with increasing rounds of incubation (Figure 1C). The target cell-binding DNA sequences were gradually enriched during the selection process and finished after the 15th round of selection. The final ssDNA pool was cloned and subjected to high-throughput sequencing with Illumina MiSeq.
Figure 1.
Selection of the DNA Aptamer LL4 against Mel28-PLX Cells
(A) Schematic representation of the cell-SELEX process for PLX4032-resistant melanoma cell Mel28-PLX. (B and C) Flow cytometry assay to monitor the binding of the selected pool with Mel28-PLX cells (target cells) (B) and Mel28 cells (control cells) (C). The final concentration of the sequences was 250 nM. R, round of selection. (D) Flow cytometry assays for the binding capacity of LL4 with Mel28-PLX cells. The final concentration of the FITC-labeled sequence was 250 nM. (E) The binding site of FITC-labeled aptamer LL4 to Mel28 and Mel28-PLX cells was investigated by confocal microscopy imaging. Scale bars, 25 μm. (F) Determining the binding capacity of LL4 with Mel28-PLX cells (dissociation curve for estimating the dissociation constant [KD]).
After sequencing, the aptamer candidates were grouped based on their sequential repeatability, homogeneity, and the abundance of each sequence using the MEGA software (Table S1). The six most enriched sequences from the six highest confidence groups (six largest the Bootstrap value) were selected and chemically synthesized for further research (Table S2). Flow cytometry results demonstrated that one of these sequences, termed LL4, showed the highest affinity toward Mel28-PLX cells, in comparison with the control Mel28 cells (Figure 1D; Figures S1A and S1B). Confocal microscopy imaging was used to investigate the binding specificity of aptamer LL4 to Mel28-PLX cells. After incubation with LL4, the fluorescence signal was observed mainly on the surface of Mel28-PLX cells, but not on Mel28 cells (Figure 1E). These results indicated that aptamer LL4 was able to recognize resistant Mel28-PLX cells, but not parental Mel28 cells. To quantitatively evaluate the binding affinity of LL4 to Mel28-PLX cells, we measured the equilibrium dissociation constant (KD). As shown in Figure 1F, the KD of aptamer LL4 for Mel28-PLX was approximately 101.10 nM, indicating that the selected aptamer LL4 could specifically recognize target Mel28-PLX cells with high affinity (a KD of nanomolar level).
Truncation of Aptamer LL4
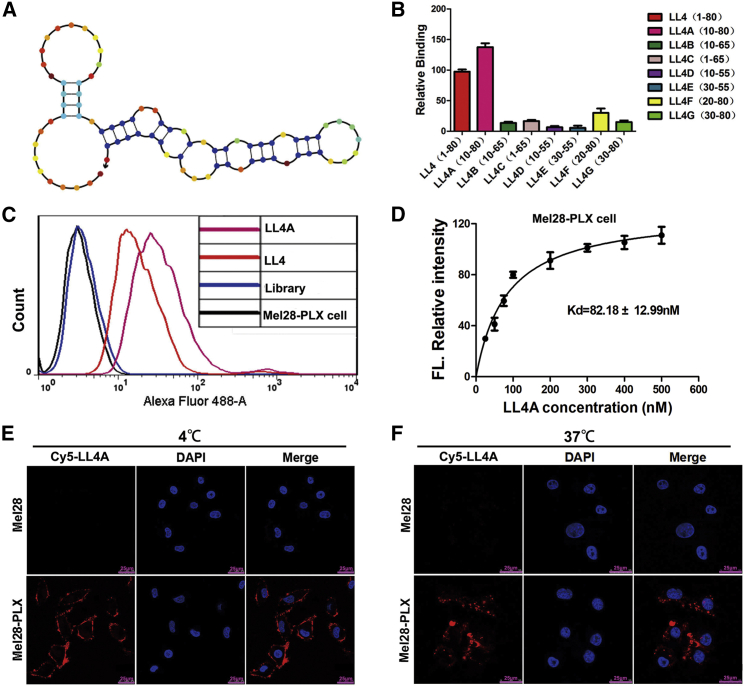
The full-length aptamer LL4 generated by Cell-SELEX contains 80 nt, including two flanked primer sequences on each end for PCR amplification. However, not all nucleotides of one aptamer are required for target binding. To reduce the cost of DNA synthesis and further enhance LL4 specificity and affinity, the full-length aptamer LL4 was truncated to a minimal functional sequence for further applications.26 Based on the secondary structure of LL4, predicted by NUPACK (nucleic acid package) (Figure 2A), seven kinds of truncated sequences from LL4 were synthesized by gradually removing the nucleotides at the 5′ and/or 3′ termini (Table S3). Flow cytometry analysis showed that the truncated LL4A sequence could bind to Mel28-PLX cells with a higher affinity than other variants or the full-length aptamer LL4. None of the modified variants bound to Mel28 cells (Figures 2B and 2C; Figures S2A and S2B). Quantitative binding assay showed that aptamer LL4A possessed a KD of 82.18 ± 12.99 nM (Figure 2D).
Figure 2.
Truncation of Aptamer LL4 and Characterization of LL4A
(A) Secondary structure of LL4 predicted by NUPACK. (B) Binding ability of different truncated versions of LL4 generated by removing nucleotides at the 5′ and 3′ termini (250 nM) on target Mel28-PLX cells. (C) Flow cytometry assays for the binding capacity of LL4 and LL4A (250 nM) to Mel28-PLX cells. (D) Dissociation constant (KD) curve of LL4A for Mel28-PLX cells. (E and F) The binding site (E) and the internalization (F) of Cy5-labeled aptamer LL4A to Mel28 and Mel28-PLX cells at 4°C and 37°C, respectively, were investigated by confocal microscopy imaging. Scale bars, 25 μm.
We also found that aptamer LL4 and its truncated variant LL4A strongly bind to another PLX4032-resistant melanoma cell line A375-PLX, but were negative for the parental A375 cells (Figures S3A and S3B). These results indicate that the LL4A aptamer targeted specifically to PLX4032-resistant melanoma cell lines, which suggests that the binding targets of LL4A are perhaps highly expressed in the resistant cells.
To investigate the LL4A binding site and internalization properties, PLX4032-resistant melanoma cells (Mel28-PLX and A375-PLX) and their parental cells (Mel28 and A375) were incubated with Cy5-labeled LL4A at 4°C and 37°C for 1 h. Confocal imaging showed that strong fluorescence was observed on the plasma membrane of Mel28-PLX and A375-PLX cells after incubation at 4°C (Figure 2E; Figure S4A). However, after incubation at 37°C, the fluorescence signal was mainly located in the cytoplasm of Mel28-PLX and A375-PLX cells (Figure 2F; Figure S4B). These results suggest that LL4A could bind to the plasma membrane and rapidly internalize into Mel28-PLX and A375-PLX cells.
Characterization of LL4A
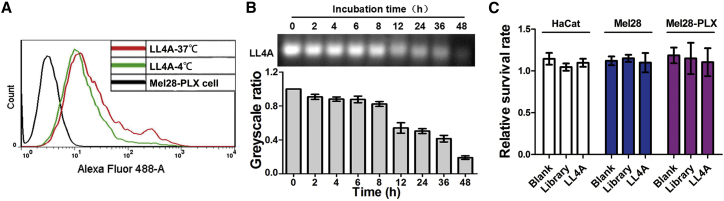
Cell-SELEX was performed at 4°C to avoid enrichment of nonspecific DNA sequences caused by endocytosis, which is more active at 37°C. However, selection at 4°C may result in poor binding capacity of aptamers at the physiological temperature.27 To investigate whether incubation temperature could affect the binding capacity of LL4A, Mel28-PLX cells were incubated with LL4A at different temperatures and then analyzed by flow cytometry. As illustrated in Figure 3A, Mel28-PLX cells showed similar fluorescence intensity and patterns after incubation with aptamer LL4A at 4°C or 37°C, indicating that the incubation temperature had little effect on the binding capacity of LL4A. Moreover, serum stability is usually a concern for the application of nucleic acid-based molecules, including DNA and RNA aptamers. Therefore, we examined the stability of LL4A when incubated with Minimum Essential Medium (MEM) supplemented with 10% human serum. During the incubation with human serum, LL4A was stable with up to 8 h of incubation at 37°C, showing no signs of degradation. LL4A started to degrade after 12 h, but a significant portion remained even after 36 h (Figure 3B). Moreover, LL4A also showed a good serum stability in mouse serum and fetal bovine serum (FBS) at 37°C (Figure S5). In addition, toxicity is also a key limiting factor in the clinical application of aptamers. As shown in Figure 3C, MTT results demonstrated that after incubation with 10 μM LL4A for 48 h, Mel28, Mel28-PLX, and human normal keratinocyte HaCaT cells showed no significant survival rate change, indicating that aptamer LL4A has no in vitro toxicity. Given the above results, the DNA aptamer LL4A is stable enough for subsequent in vivo studies and also will be useful for future clinical applications.
Figure 3.
Characterization of LL4A
(A) Flow cytometry assays for the binding capacity of LL4A to Mel28-PLX cells at 4°C or 37°C, respectively. (B) Aptamer LL4A was incubated in human serum, and its biostability was evaluated by gel electrophoresis of the residual products at the indicated time points. (C) The in vitro toxicity of LL4A in Mel28, Mel28-PLX, and HaCaT cells was investigated by MTT assay.
In Vivo and Ex Vivo Xenograft Fluorescence Imaging
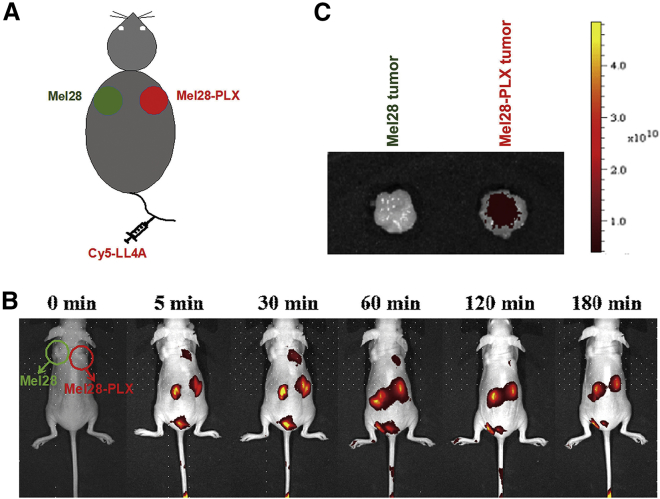
To test whether aptamer LL4A retained its recognition ability in vivo, a systematic comparative investigation28 was performed to validate the selectivity of Cy5-labeled LL4A in the same mouse model, in which Mel28 and Mel28-PLX cells were implanted on the left and right sides of the back, respectively (Figure 4A). Imaging studies showed that after injecting Cy5-labeled LL4A through the tail vein, aptamer LL4A specifically targeted and accumulated in tumors formed by Mel28-PLX cells, but not in tumors formed by control Mel28 cells in the same mouse. A fluorescent signal was also observed in kidney tissue, which suggested that LL4A was excreted and cleared by the kidneys. The specific imaging signal lasted up to 120 min post systemic administration (Figure 4B). Aptamer LL4A also presented more fluorescence signal accumulation in excised Mel28-PLX tumors than in Mel28 tumors (Figure 4C). These results demonstrated that aptamer LL4A was able to recognize and target to the PLX4032-resistant melanoma cells in vivo, which established a base for future use as a molecular probe for prognosis, especially use in monitoring development of PLX4032 resistance during treatment.
Figure 4.
In Vivo and Ex Vivo Xenograft Fluorescence Imaging
(A) The animal model bearing both a Mel28 tumor (left) and a Mel28-PLX tumor (right) will be systemically administered with Cy5-labeled aptamer LL4A through the tail veins. (B) The two sides of tumor-bearing mice were intravenously injected with Cy5-labeled aptamer LL4A and then imaged at the indicated time points. (C) The excised Mel28 tumor (left) and Mel28-PLX tumor (right) tissues were harvested, and the fluorescence signals of tumor tissues were collected by using the IVIS Lumina II in vivo imaging system.
CD63 Is the Binding Target of Aptamer LL4A
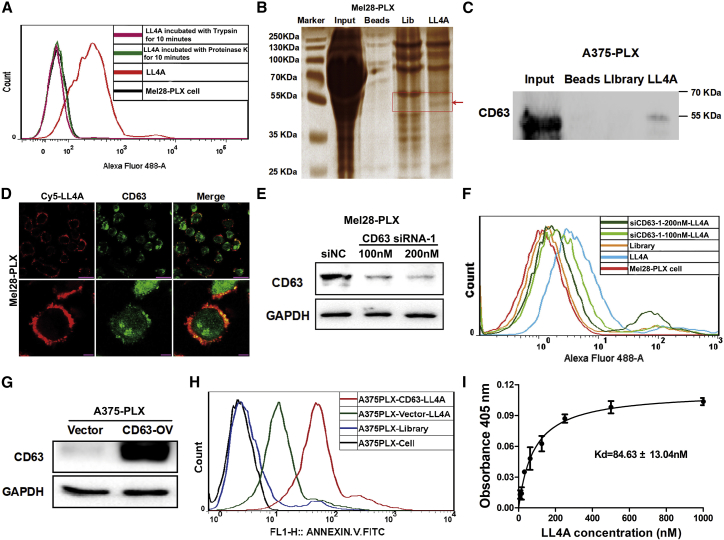
Next, in order to discover the mechanism of aptamer LL4A, we identified its potential targets that could be proteins, lipids, saccharides or other organic molecules.29 After Mel28-PLX cells were incubated with trypsin or Proteinase K, aptamer LL4A completely lost its binding capacity (Figure 5A), suggesting that the target of LL4A was a membrane protein. Based on this finding, we incubated biotin-tagged LL4A with solubilized membrane protein from Mel28-PLX cells, and streptavidin-coated magnetic beads were used to extract biotin-labeled LL4A binding complexes (the initial ssDNA library was used as control). The beads along with the capture proteins were then eluted and separated by SDS-PAGE (Figure 5B). Compared with control experiments, the characteristic protein bands (∼50 kDa) on the gel captured by biotin-labeled LL4A were digested and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) QSTAR analysis. A MASCOT database search was used to assign possible protein candidates to the MS results (Table S4). Interestingly, CD63 is highly enriched among the surface transmembrane protein candidates (Figure S6).
Figure 5.
CD63 Is the Binding Target of Aptamer LL4A
(A) Binding capacity of LL4A to Mel28-PLX cells pretreatment Proteinase K or trypsin was measured by flow cytometry. (B) Colloidal silver-stained SDS-PAGE used to analyze the LL4A-assisted target purification. (C) The direct interaction between LL4A and CD63 in Mel28-PLX cells was detected using an aptamer pull-down assay, and the CD63 protein was examined by western blotting. (D) The co-localization of Cy5-labeled LL4A and anti-CD63 in Mel28-PLX cells was investigated by confocal microscopy imaging. Top panels: scale bars, 25 μm; bottom panels: scale bars, 10 μm. (E) The knockdown efficiency of CD63 was determined by western blotting. (F) Binding capacity of LL4A in CD63 knockdown Mel28-PLX cells and control cells was measured by flow cytometry. (G) The overexpression efficiency of CD63 in A375-PLX cells was determined by western blotting. (H) Binding capacity of LL4A in CD63 overexpressed A375-PLX cells and control cells was measured by flow cytometry. (I) Dissociation constant of LL4A for purified His-CD63 recombinant protein was determined by ELONA.
To validate that LL4A binds to CD63, first an aptamer-mediated pull-down assay was performed in A375-PLX cells. As shown in Figure 5C, biotin-labeled LL4A, but not Library-captured proteins, could bind to CD63 antibody, indicating a direct interaction between LL4A and CD63. Subsequently, an immunofluorescence experiment was used to examine the co-localization between LL4A and CD63 on the Mel28-PLX cells (Figure 5D). CD63 showed both plasma membrane and cytoplasmic locations as found in other cells reported before.30 LL4A showed complete overlapping signal with CD63 on the plasma membrane of Mel28-PLX cells (Figure 5D). We next used the small interfering RNA (siRNA) to knock down CD63 in Mel28-PLX cells, with random siRNA as negative control (Figure 5E). These cells were analyzed with flow cytometry after incubation with LL4A; the results showed a dramatic reduction of LL4A fluorescence in the CD63 knocked down Mel28-PLX cells in a siRNA concentration-dependent manner as compared with the negative control cells (Figure 5F). Conversely, overexpression of CD63 in A375-PLX cells (where CD63 was at a lower level than in Mel28-PLX cells) dramatically increased the binding affinity of LL4A to A375-PLX cells (Figures 5G and 5H), indicating that CD63 is an important LL4A binding protein on the Mel28-PLX and A375-PLX cells. To investigate whether LL4A shares the same binding site with anti-CD63 antibody to multiple transmembrane protein CD63, we performed a competition assay by flow cytometry. Cy5-labeled LL4A and FITC-labeled anti-CD63 antibody were co-incubated with Mel28-PLX cells, and flow cytometry results showed few changes in the binding of LL4A to Mel28-PLX cells; also, LL4A had no obvious influence on anti-CD63 antibody binding (Figure S7), indicating that the different sites of the extracellular domain of CD63 were simultaneously bound by LL4A and antibody. Finally, the binding affinity by enzyme-linked oligonucleotide assay (ELONA) revealed that the KD of LL4A against purified His-CD63 was 84.63 ± 13.04 nM (Figure 5I). Taken together, we identified that CD63 was the binding target of aptamer LL4A.
CD63 Contributes to PLX4032 Drug Resistance in Melanoma Cells
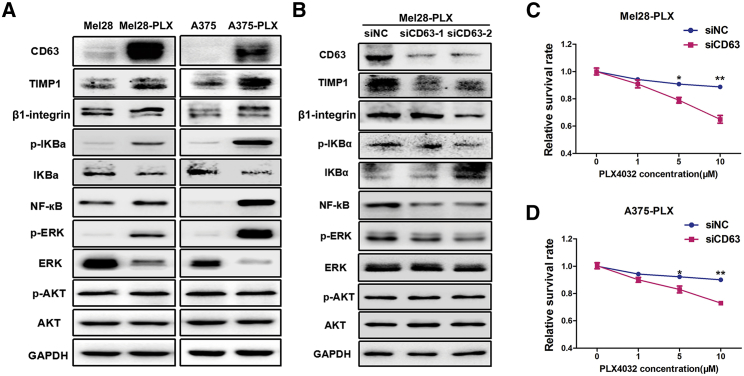
Previous studies indicated that TIMP1 bound to the tetraspanin membrane receptor CD63 and assembled a supramolecular complex with β1-integrin, further triggering the hyperactivation of NF-κB and phosphorylation of ERK1/2, and eventually leading to promotion of cell growth and invasion.31,32 In this study, we found that PLX4032 resistance cells formed a TIMP1/CD63/β1-integrin supramolecular complex through co-localization fluorescent assay and co-immunoprecipitation assay (Figure S8). Therefore, we next examined the role of CD63 in the NF-κB and mitogen-activated protein kinase (MAPK) pathway, and its impact in response to PLX4032. As shown in Figure 6A, we observed the elevated expression of the TIMP1/CD63/β1-integrin axis and activated NF-κB and ERK signaling pathways in both PLX4032-resistant melanoma cells (Mel28-PLX and A375-PLX), as compared with the parental cells (Mel28 and A375). Furthermore, the PLX4032-resistant cells also showed enhanced β1-integrin-mediated signaling, perhaps through TIMP1/CD63, but not PI3K/AKT, signaling pathways, because there was no difference in p-AKT levels between PLX4032-resistant melanoma cells and their parental cells. To further determine whether the elevated NF-κB/TIMP1/CD63/β1-integrin/ERK pathway was involved in PLX4032 resistance in melanoma cells, CD63 was knocked down in Mel28-PLX cells, and a significant reduction in TIMP1 expression and ERK1/2 phosphorylation was observed as compared with the control cells treated with negative control siRNA (siNC) (Figure 6B). Furthermore, when CD63 was knocked down, Mel28-PLX and A375-PLX cells were much more sensitive to PLX4032 treatment (Figures 6C and 6D). Taken together, our results suggest that the elevated expression of the TIMP1/CD63/β1-integrin axis mediates the activation of the NF-κB and ERK1/2 signaling pathways, contributing to PLX4032 resistance, but this regulation is independent of AKT phosphorylation.
Figure 6.
CD63 Contributes to PLX4032 Drug Resistance in Melanoma Cells
(A) Western blotting analysis of the expression of CD63, TIMP1, β1-integrin, NF-κB, ERK, and PI3K/AKT signaling pathways in Mel28/Mel28-PLX and A375/A375-PLX cells. GAPDH served as a loading control. (B) Mel28-PLX cells were knocked down with siCD63-1, siCD63-2, and siNC for 48 h, and the CD63, TIMP1, β1-integrin, NF-κB, ERK, and PI3K/AKT signaling pathways were analyzed by western blotting. (C and D) Mel28-PLX (C) and A375-PLX (D) cell viability as measured by the MTT assay after the cells were transfected with either siCD63 or siNC for 48 h.
Discussion
In this study, using the cell-based SELEX strategy, we discovered a DNA aptamer LL4 and its truncated form LL4A, which could bind to vemurafenib-resistant melanoma cells with high affinity, both in vitro and in vivo. Furthermore, we characterized LL4A to be highly stable under physiological conditions with super-low toxicity. We further identified CD63 as the cell surface target for LL4/LL4A and explored the role of CD63 in vemurafenib resistance. These findings paved the way to develop aptamers LL4/LL4A in versatile biomedical applications such as bioimaging and targeted therapy, and suggested a new mechanism for vemurafenib-induced drug resistance in melanoma cells.
A potential use of LL4/LL4A is in imaging for patient prognosis monitoring during vemurafenib treatment by optical bioimaging, positron emission tomography (PET), single-photon emission computed tomography (SPECT), or magnetic resonance imaging (MRI) using the modified aptamers with fluorescent dyes, radioisotopes, or magnetic nanomaterial reporters. This will help doctors to make early decisions of switching patients to other treatments. A second potential use of these aptamers is to be developed as therapeutic agents in the form of aptamer-drug conjugates (ApDCs). Because aptamer LL4A itself is not toxic to melanoma cells and human normal keratinocytes, it can be internalized into vemurafenib-resistant cells. This opens the therapeutic windows for LL4A as targeting ligands for drug delivery in the form of ApDCs. The ApDC-mediated chemotherapy could specifically deliver drugs to diseased tissues or cells, and thus reduce side effects and improve therapeutic efficacy.33 In addition, ApDC could be used in mediating gene therapy. Specifically, aptamers can be conjugated with nucleic acid gene therapeutics, ranging from siRNA, small hairpin RNA (shRNA), and microRNA (miRNA) by molecular engineering of nucleic acids, which reduces the potential “off-target” effect in gene delivery.33
A third potential application of LL4/LL4A aptamers is for discovery of biomarkers. The cell-SELEX process is simple, fast, and reproducible, and can be done without prior knowledge of target molecules. The selected aptamers can specifically recognize target cancer cells mixed with normal cells.34 In this study, we used aptamer-mediated pull-down assay combined with LC-MS/MS QSTAR analysis and identified CD63 as a potential biomarker for vemurafenib resistance. CD63 was an abundant surface antigen in early-stage human melanoma cells.35 TIMP1 is a CD63 ligand, and CD63 acts as a regulator of the PI3K, FAK, Src, and Akt signaling pathways. CD63-mediated signaling is implicated in the anti-apoptotic activity of TIMP1.36 The interaction of CD63 and TIMP1 is also involved in survival and increased chemoresistance in different human cancers, which may be mediated by hyperactivation of NF-κB and phosphorylation of ERK1/2.25,37,38 In melanoma, the BRAFV600E mutation is associated with the hyper-activation of NF-κB and upregulation of both TIMP1 and its receptor CD63.31 Here, we show evidence that elevated NF-κB/TIMP1/CD63/β1-integrin axis facilitates melanoma drug resistance through activation of NF-κB and ERK1/2 signaling pathways without AKT phosphorylation. Our results appear contradictory with a recent study by Kudo et al.,39 which reported that CD63 was able to sensitize melanoma cell lines to PLX4032. These seeming conflicts may be caused by several factors, including different drug selection method for resistant cells, leading to differences in the genetic background of resistant cell lines, cell heterogeneity during the course of drug screening, and measurement method of drug resistance. In fact, CD63 was found to be one of the best genes that could efficiently distinguish BRAF mutant versus BRAF-wild-type melanomas, and the correlation between V600E mutation and CD63 overexpression was extremely strong in the melanoma.40 In addition, our results are consistent with another previous study that described an increased level of CD63-Timp1-β1-integrin supramolecular complex along melanoma progression.24 More work is warranted in fully characterizing the role of CD63-related signaling in melanoma progression and drug resistance.
Furthermore, CD63 abundantly localizes at extracellular vesicle exosomes released from melanoma cells. Logozzi et al.22 found that plasma exosomes expressing CD63 were significantly increased in melanoma patients compared with healthy donors. Since then, CD63 is commonly used as an exosome marker for melanoma. Exosomes have been shown to promote tumor development, such as metastatic niche formation, tumor immune escape, and drug resistance.41 Peinado et al.42 reported that exosomes derived from melanoma cells enhanced metastasis through education of bone marrow-derived cells in order to prime the pre-metastatic niche and increase vascularization, indicating that exosomal CD63 may be a potential therapeutic target for melanoma. Indeed, Nishida-Aoki et al.43 demonstrated that treatment with human-specific anti-CD63 antibodies significantly suppressed tumor metastasis in a human breast cancer xenograft mouse model. In particular, anti-CD63 antibody-tagged cancer-derived exosomes were preferentially internalized and eventually eliminated by macrophages.43 Similar to antibodies, aptamers bind to their targets with high affinity and selectivity. However, aptamers are advantageous over antibodies in terms of their low potential for immunogenicity, efficient tissue penetration, and relatively simple synthesis. Based on the LL4A antigenic, biological, and molecular properties, LL4A could be developed as a promising molecular tool for targeting applications in melanoma, such as target identification of Plx4032-resistant cells, targeting killing resistant cells via loading with chemotherapy drugs, targeting labeling, and isolation of plasma exosomes expressing CD63. Furthermore, developing a bi-specific/bivalent aptamer that targets both CD63 and co-inhibitory molecules, like PD-L1, CTLA-4, and B7-H4, may enhance tumor-immune interactions.
Taken together, this study identified a novel aptamer, LL4A, which could serve as a potential imaging probe for prognosis or as a carrier for targeted therapeutics against PLX4032-resistant melanoma with high expression of CD63. Our results suggest future therapy methods can be developed based on blocking CD63 by antibodies or small-molecule inhibitors, or using LL4A as a CD63 siRNA delivery tool for developing novel combination therapy with vemurafenib.
Materials and Methods
Cell Culture and Reagents
Human malignant melanoma cells SK-Mel28 (Mel28), A375, and human normal keratinocyte HaCaT were purchased from the ATCC. Mel28-PLX and A375-PLX (PLX4032-resistant Mel28 and A375 parental) cells were obtained and cultured as described in a previous study.26 The cells were incubated in a humid atmosphere maintained at 37°C with 5% CO2. Mel28/Mel28-PLX and A375/A375-PLX cells were cultured in MEM (HyClone, USA) and DMEM (HyClone, USA), respectively, supplemented with 10% FBS (GIBCO, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies, USA). Mel28-PLX and A375-PLX were generated by prolonged cell culture in the presence of PLX4032. PLX4032 was purchased from Selleck Chemicals (Houston, TX, USA) and dissolved in DMSO at a concentration of 5 mM. For each cell passage cultivation, the final concentration of PLX4032 (5 μM) was freshly prepared in MEM. SELEX washing buffer for aptamers was prepared with Dulbecco’s PBS (DPBS; Thermo Fisher Scientific) supplemented with 4.5 mg/mL glucose (Sigma, Germany) and 5 mM MgCl2 (Sigma, Germany). SELEX binding buffer for aptamers was prepared by adding 0.1 mg/mL yeast tRNA (Sigma, Germany) and 1 mg/mL BSA (Sigma) into the washing buffer to diminish background binding.
DNA Library and Primers
The DNA library used in the cell-SELEX was constructed from oligonucleotide sequences with a length of 80 nt based on the following generic sequence: 5′-ACCGACCGTGCTGGACTCA(N)42
ACTATGAGCGAGCCTGGCG-3′. The specific oligonucleotide sequence contained a 19-nt fixed sequence on both sides and the middle 42-nt randomized region, where N represents a randomized nucleotide (A, T, G, or C). A FITC-labeled forward primer (5′-FITC-ACCGACCGTGCTGGACTCA-3′) and a biotin-labeled reverse primer (5′-biotin-CGCCAGGCTCGCTCATAGT-3′) were used in the PCR amplification for the synthesis of double-labeled, double-stranded DNA (dsDNA) sequences. The DNA library and primers used in the cell-SELEX were synthesized and high-pressure liquid chromatography (HPLC) purified by Sangon Biotech (Shanghai, China).
Cell-SELEX Procedures
The cell-SELEX process was performed using Mel28-PLX cells as the target cells and Mel28 cells as the negative control cells, as reported previously with a few modifications.44 For the first and second rounds of SELEX, the initial library pool (10 OD/optical density) was incubated with Mel28-PLX cells at 4°C for positive selection. After incubation, the supernatant solution was discarded, and the cells were washed to remove unbound sequences. Then Mel28-PLX cells were collected, and the cell-bound ssDNA was harvested by heating the cell-DNA complexes. The harvested cell binding ssDNA was used as a template for PCR amplification, and the dsDNA product was separated from the PCR solution by streptavidin-coated Sepharose beads (GE Healthcare, USA) and then treated with 0.2 M NaOH, followed by desalting with NAP-5 column (GE Healthcare, USA) to obtain the FITC-labeled ssDNA. Finally, after lyophilization, the FITC-labeled ssDNA pool was used for the next round of selection.
Starting from the third round of selection, the negative control Mel28 cells were first incubated with the selected ssDNA pool evolved from the last round for negative selection. The unbound ssDNAs were then removed and applied onto Mel28-PLX cells to enrich the specific binders to the target. As the number of selection rounds increased, the negative incubation time was increased from 30 min to 2 h, whereas the positive incubation time was gradually shortened from 2 h to 30 min to increase the stringency and accuracy of the selection. Additionally, the washing times gradually increased from two to three times, and the amount of the ssDNA library used for subsequent rounds of selection was decreased gradually. The enrichment of the selected FITC-labeled ssDNA pools was monitored by detecting the FITC fluorescence signal with flow cytometry (BD FACSCalibur flow cytometer, USA). Following 15 rounds of selection, the final ssDNA pool was subjected to high-throughput sequencing using Illumina MiSeq by Sangon Biotech (Shanghai, China).
Flow Cytometric Analysis
Target and negative cells were cultured in 100-mm-diameter culture dishes to 80%–90% confluence. Both cell lines were washed twice with 2 mL of DPBS, followed by digestion with 1 mL of 0.02% EDTA. Then 2 × 105 Mel28/Mel28-PLX cells and A375/A375-PLX cells were respectively incubated with 250 nM FITC-labeled selected ssDNA pools and the initial library in 400 μL of binding buffer at 4°C for 1 h in the dark place to monitor the enrichment of the evolved ssDNA pools. In the same way, each cell line (2 × 105) was incubated with aptamer or initial library to analyze the binding specificity of the aptamer. The FITC-labeled ssDNA library was used as a negative control to distinguish nonspecific binding from specific binding. After incubation, each cell sample was washed twice with 500 μL of washing buffer, re-suspended in 400 μL of binding buffer, and then analyzed with a BD FACSCalibur flow cytometer. All experiments for the binding specificity assay were repeated three times independently.
Aptamer Truncation
The nucleotide secondary structure of aptamer LL4 was predicted by using the bioinformatics software NUPACK. Seven truncated sequences were synthesized by gradually removing the nucleotides at the 5′ and 3′ termini based on the secondary structure analysis of LL4 and then performing a binding assay. All experiments were repeated three times independently.
Affinity Analysis
To quantitatively assess the affinity of aptamer to target Mel28-PLX cells, we measured the equilibrium KD of aptamers by GraphPad software. Target Mel28-PLX cells (2 × 105) were incubated with increasing concentrations of FITC-labeled aptamers and the initial library in 400 μL of binding buffer at 4°C for 1 h in the dark and then analyzed by flow cytometry. The FITC-labeled initial library was used as a negative control. The KD of the aptamers on Mel28-PLX cells was determined by measuring the dependence of the fluorescence intensity of aptamers by different aptamer concentrations with the equation Y = Bmax X/(KD+X). All experiments for the binding affinity assay were repeated three times independently, and data are expressed as means ± SEM.
Aptamer Internalization Analysis
A total of 1 × 105 Mel28/Mel28-PLX cells and A375/A375-PLX cells were respectively incubated with 250 nM Cy5-labeled aptamer at 37°C for 1 h in the dark. After incubation, the cells were washed twice with washing buffer, fixed with 4% paraformaldehyde for 10 min, and then stained with 400 μL of DAPI for 5 min. The images were captured by a Leica SP5 II scanning confocal microscope (Leica, Bannockburn, IL, USA). All experiments were performed in triplicate, and data shown in the figures are representative of all of the experimental replicates.
Trypsin and Proteinase K Dissociating Analysis
Target Mel28-PLX cells were washed twice with 2 mL of DPBS and then dissociated with 200 μL of 0.02% EDTA, 0.25% trypsin, or 0.2 mg/mL Proteinase K for 10 min at room temperature. After adding complete culture medium to inhibit proteinase activity, the cells were washed and then incubated with FITC-labeled aptamer (250 nM) in 400 μL of binding buffer at 4°C for 40 min in the dark. After washing, all cell samples were analyzed by flow cytometry as described in the previous section. All experiments were performed in triplicate, and data shown in the figures are representative of all the experimental replicates.
Effects of Temperature on Aptamer Binding
To investigate whether the temperature will affect the binding ability of aptamers, target Mel28-PLX cells were dissociated with 0.02% EDTA and incubated with FITC-labeled aptamer (250 nM) in 400 μL of binding buffer for 1 h in the dark placed at 4°C or 37°C, respectively. The binding affinity was measured by flow cytometry. All experiments were performed in triplicate, and data shown in the figures are representative of all of the experimental replicates.
Aptamer Biostability Assays
Next, 3 μM FITC-labeled aptamer was incubated in 400 μL of MEM supplemented with 10% human serum, mouse serum, or FBS at 37°C at the indicated time points. After incubation, 40 μL of sample was collected for each time point and denatured immediately at 95°C for 10 min, followed by storage at −80°C. After finishing the collection, the samples were thawed at 4°C and then subjected to electrophoretic separation on a 3% agarose gel. The amount of undigested aptamer was visualized and further analyzed by a molecular imager (Bio-Rad Laboratories, China). Each bar represents the mean ± SEM of three independent experiments.
In Vivo Fluorescence Imaging
All animal procedures were approved by the Animal Care and Use Committee of the Third Xiangya Hospital of Central South University (Changsha, Hunan, China) and performed strictly in accordance with institutional policies and approved guidelines of experiment operations. The in vivo fluorescence imaging assay was performed as reported previously28 with a few modifications. In detail, 4- to 6-week-old female athymic BALB/c (BALB/c-nude) mice were purchased from Hunan SJA Laboratory Animal. Then, 4 × 106 Mel28 cells and Mel28-PLX cells were co-implanted subcutaneously in the opposite flanks of BALB/c-nude mice. When the tumors reached 0.5–1.0 cm3, tumor-bearing BALB/c nude mice were anesthetized and then injected intravenously with 6.5 nM Cy5-labeled aptamer dissolved in DPBS via the tail vein. At the indicated time points, the fluorescence signal was monitored on an IVIS Lumina II in vivo imaging system (Caliper Life Sciences, USA). For the in vitro fluorescence experiment, tumor-bearing BALB/c nude mice were intravenously injected with Cy5-labeled aptamer via the tail vein. Then the mice were anesthetized and euthanized by cervical dislocation 1 h after injection. The excised Mel28-PLX and Mel28 tumor tissues were harvested, and the fluorescence signals of tumor tissues were collected by using the IVIS Lumina II in vivo imaging system. All experiments were performed in triplicate, and data shown in the figures are representative of all of the experimental replicates.
Aptamer-Mediated Pull-Down Assay
A total of 5 × 106 Mel28-PLX or A375-PLX cells were washed three times with precooling DPBS and lysed in 5 mL of hypotonic buffer (washing buffer containing 10× cocktail, 1 M Tris-HCl, and 100× PMSF) at 4°C for 30 min. After centrifugation, the debris was washed three times with 5 mL of hypotonic buffer and dissolved in 2 mL of lysis buffer (hypotonic buffer containing 1% Triton X-100) at 4°C for 30 min. After centrifugation, the supernatant was collected and then incubated with 150 pmol nonbinding biotin-labeled library sequences for 1 h at 4°C as a nonspecific competitor. The protein-DNA complex was captured by incubating it with 2 mg (200 μL) of streptavidin-coated magnetic beads for 1 h at 4°C and collecting the beads on a magnetic stand. After centrifugation, the resulting supernatant was incubated with 150 pmol biotin-labeled LL4A for 1 h at 4°C, and the protein-LL4A complex was captured by incubating it with 2 mg (200 μL) of streptavidin-coated magnetic beads at 4°C for 1 h and then collecting the beads on a magnetic stand. The collected magnetic beads were washed five times with 1 mL of SELEX washing buffer. The beads along with the captured proteins were eluted by heating in 30 μL of loading buffer and then separated by PAGE (12%, SDS-PAGE), followed by staining with colloidal silver or immunoblotting with specific CD63 antibody. The aptamer-purified protein bands were digested and subjected to LC-MS/MS QStar analysis. A MASCOT database search was used to assign possible protein candidates to the MS results. All experiments were performed in triplicate, and data shown in the figures are representative of all of the experimental replicates.
Confocal Microscopy Imaging
For confocal imaging of the aptamers bound with the cells, 1 × 105 Mel28/Mel28-PLX cells and A375/A375-PLX cells were respectively seeded onto glass coverslips in 24-well dishes and cultured for 24 h. After washing twice with cold washing buffer, the cells were incubated with FITC/Cy5-labeled aptamer (250 nM) in 400 μL of binding buffer at 4°C for 1 h in the dark. Subsequently, the cells were washed twice, fixed with 4% paraformaldehyde for 10 min, and then stained with 400 μL of DAPI for 5 min. The images were captured by a Leica SP5 II scanning confocal microscope (Leica, Bannockburn, IL, USA).
For confocal imaging of the co-localization of the LL4A aptamer and the anti-CD63 antibody, Mel28-PLX cells were directly labeled with Cy5-LL4A and then incubated with anti-CD63 monoclonal antibodies at a 1:100 dilution for 2 h at 37°C. The cells were then incubated with a 1:100 dilution of TRITC (Tetramethylrhodamine-5-(and-6)-isothiocyanate)-conjugated goat anti-mouse immunoglobulin G (IgG; Jackson ImmunoResearch Laboratories) for 1 h at 37°C in the dark. The images were captured using a Leica SP5 II scanning confocal microscope (Leica, Bannockburn, IL, USA).
ELONA
Experimental protocols were conducted according to a previous description.45 In brief, His-CD63 recombinant protein (11271-H08H; Sino Biological, China) was diluted to 1 μg/mL with binding buffer, and 200 μL of the solution was added to a 96-well microtiter plate (NUNC, Rochester, NY, USA) and then incubated overnight at 4°C. Then the wells were washed four times with washing buffer and blocked with 1% BSA in PBS for 2 h at 37°C and then washed three times with washing buffer. Afterward, biotin-labeled aptamer LL4A or Library (Sangon Biotech, China) was denatured for 10 min at 95°C and cooled for 10 min on ice, then diluted in binding buffer at concentrations indicated in the figures. Next, 200 μL of the solution was added to each well, and the plate was incubated at 37°C for 1 h and then washed four times with washing buffer to remove unbound ssDNA. Afterward, 200 μL of a 1:2,000 dilution of streptavidin labeled with horseradish peroxidase (D3308; Beyotime, China) was added to each well, and then the plate was incubated at room temperature for 2 h. At last, the plate was washed four times and developed by EL-ABTS Chromogenic Reagent kit (C510031; Sangon Biotech, China), and then 405-nm single-wavelength absorbance was detected by using a microplate reader.
Western Blotting
Experimental protocols were conducted according to our previous description.46 In brief, the cells were lysed with radioimmunoprecipitation assay (RIPA) buffer containing a protease or phosphatase inhibitor mixture; 25 μg of proteins was separated by SDS-PAGE, followed by immunoblotting with specific antibodies (Table S5).
Co-immunoprecipitation
For endogenous co-immunoprecipitation assays, cells were lysed in cold IP (immunoprecipitation) buffer (P0013; Beyotime, China) supplemented with protease inhibitor cocktail (Roche, France), 5%–10% of the cell extract was saved as the input, and the rest was incubated with primary antibody at 4°C overnight and then with protein A/G agarose beads (Santa Cruz, USA) for 2 h at 4°C. After washing three times with the IP buffer, bound proteins were eluted by boiling with 2× SDS loading buffer and then separated by PAGE (10%, SDS-PAGE), followed by immunoblotting with specific antibody (Table S5).
Small Interfering RNAi and Transfection
siRNAs targeting CD63 and an unrelated sequence as a negative control were synthesized by GenePharma (Shanghai, China), and the sequences of the siRNAs are listed in Table S6. CD63 overexpression plasmid and null vector were purchased from Sino Biological (Beijing, China). Mel28-PLX cells were transfected with CD63 siRNA or overexpressed plasmid using the riboFECTTM CP Transfection Kit (RiboBio, Guangzhou, China) according to the manufacturer’s protocol. The knockdown efficiency was measured by western blotting. The binding ability of LL4A to Mel28-PLX cells transfected with CD63 siRNAs was assessed by flow cytometric analysis.
Competition Assay of Aptamer and Antibody
A total of 250 nM Cy5-labeled LL4A and FITC-labeled anti-CD63 antibody was prepared and incubated alone or co-incubated with the Mel28-PLX cells in 200 μL of binding buffer at 4°C for 1 h. After incubation, non-bound aptamers or antibody was washed out, and then cells were resuspended in 500 μL of binding buffer. Flow cytometer was used to measure the fluorescence intensity of each sample.
MTT Assay
The cells were seeded into flat-bottom 96-well plates (3 × 103 cells/well) and grown for 24, 48, and 72 h. At each time point, 20 μL of MTT working solution was added to the medium and incubated for 4 h. Subsequently, the medium was removed, and 200 μL of DMSO was added to dissolve the formazan crystals. The absorbance at 570 nm (A570) was read on a microplate reader (Bio-Rad, Hercules, CA, USA). All experiments were performed in triplicate, and data are expressed as means ± SEM.
Statistical Analyses
All statistical analyses were performed using SPSS 16.0 statistical software. Student’s t test was used to determine the significance of the differences between the control and the experimental groups. A p value <0.05 was considered statistically significant.
Author Contributions
Conceptualization: J.L. (Juan Liu), Z.W. and H.L.; Formal Analysis: X.X., M.Y., Z.W., S.S., B.Z., and H.L.; Funding Acquisition: F.L.-S., Z.W. and J.L. (Jing Liu); Investigation: H.L., J.L. (Juan Liu), H.Z., Y.Z., W.Z., and Z.W.; Methodology: H.L., J.L. (Jing Liu), F.L.-S. and M.Y.; Project Administration: F.L.-S. and J.L. (Jing Liu); Resources: F.L.-S., J.L. (Jing Liu), and Z.W.; Supervision: J.L. (Jing Liu); Visualization: J.L. (Jing Liu); Writing – Original Draft Preparation: H.L.; Writing – Review & Editing: F.L.-S., J.L. (Jing Liu), and M.R.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81702722, 81872229, and 81772496); the Science and Technology Key Project of Hunan Province (2018SK21212 and 2018SK2128); National Postdoctoral Program for Innovative Talents (BX201700292); Natural Science Foundation of Hunan Province (2018JJ3703); and the Fundamental Research Funds for the Central Universities of Central South University (2017zzts078, 2017zzts352, and 2018zzts830).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.005.
Contributor Information
Zi Wang, Email: zhongnanwangzi@126.com.
Feng Liu-Smith, Email: liufe@uci.edu.
Jing Liu, Email: liujing2@sklmg.edu.cn.
Supplemental Information
References
- 1.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Stadler S., Weina K., Gebhardt C., Utikal J. New therapeutic options for advanced non-resectable malignant melanoma. Adv. Med. Sci. 2015;60:83–88. doi: 10.1016/j.advms.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O’Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sosman J.A., Kim K.B., Schuchter L., Gonzalez R., Pavlick A.C., Weber J.S., McArthur G.A., Hutson T.E., Moschos S.J., Flaherty K.T. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dummer R., Ascierto P.A., Gogas H.J., Arance A., Mandala M., Liszkay G., Garbe C., Schadendorf D., Krajsova I., Gutzmer R. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2018;19:1315–1327. doi: 10.1016/S1470-2045(18)30497-2. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 8.Osborne S.E., Matsumura I., Ellington A.D. Aptamers as therapeutic and diagnostic reagents: problems and prospects. Curr. Opin. Chem. Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 9.Navani N.K., Li Y. Nucleic acid aptamers and enzymes as sensors. Curr. Opin. Chem. Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Famulok M., Hartig J.S., Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 11.Esposito C.L., Catuogno S., de Franciscis V., Cerchia L. New insight into clinical development of nucleic acid aptamers. Discov. Med. 2011;11:487–496. [PubMed] [Google Scholar]
- 12.Xiang D., Shigdar S., Qiao G., Wang T., Kouzani A.Z., Zhou S.F., Kong L., Li Y., Pu C., Duan W. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: the next generation of cancer medicine. Theranostics. 2015;5:23–42. doi: 10.7150/thno.10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanwar J.R., Shankaranarayanan J.S., Gurudevan S., Kanwar R.K. Aptamer-based therapeutics of the past, present and future: from the perspective of eye-related diseases. Drug Discov. Today. 2014;19:1309–1321. doi: 10.1016/j.drudis.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H., Li J., Zhang X.B., Ye M., Tan W. Nucleic acid aptamer-mediated drug delivery for targeted cancer therapy. ChemMedChem. 2015;10:39–45. doi: 10.1002/cmdc.201402312. [DOI] [PubMed] [Google Scholar]
- 15.Hori S.I., Herrera A., Rossi J.J., Zhou J. Current Advances in Aptamers for Cancer Diagnosis and Therapy. Cancers (Basel) 2018;10:e9. doi: 10.3390/cancers10010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noaparast Z., Hosseinimehr S.J., Piramoon M., Abedi S.M. Tumor targeting with a (99m)Tc-labeled AS1411 aptamer in prostate tumor cells. J. Drug Target. 2015;23:497–505. doi: 10.3109/1061186X.2015.1009075. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson O., Yan X., Niu G., Weiss I.D., Ma Y., Szajek L.P., Shen B., Kiesewetter D.O., Chen X. PET imaging of tenascin-C with a radiolabeled single-stranded DNA aptamer. J. Nucl. Med. 2015;56:616–621. doi: 10.2967/jnumed.114.149484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shangguan D., Cao Z., Meng L., Mallikaratchy P., Sefah K., Wang H., Li Y., Tan W. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H., Zhao J., Zhang M., Yang H., Ma Y., Gu Y. MUC1 aptamer-based near-infrared fluorescence probes for tumor imaging. Mol. Imaging Biol. 2015;17:38–48. doi: 10.1007/s11307-014-0763-y. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Gao H., Zhou W., Sun S., Zeng Y., Zhang H., Liang L., Xiao X., Song J., Ye M. Targeting c-met receptor tyrosine kinase by the DNA aptamer SL1 as a potential novel therapeutic option for myeloma. J. Cell. Mol. Med. 2018;22:5978–5990. doi: 10.1111/jcmm.13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Da Rocha Gomes S., Miguel J., Azéma L., Eimer S., Ries C., Dausse E., Loiseau H., Allard M., Toulmé J.J. (99m)Tc-MAG3-aptamer for imaging human tumors associated with high level of matrix metalloprotease-9. Bioconjug. Chem. 2012;23:2192–2200. doi: 10.1021/bc300146c. [DOI] [PubMed] [Google Scholar]
- 22.Logozzi M., De Milito A., Lugini L., Borghi M., Calabrò L., Spada M., Perdicchio M., Marino M.L., Federici C., Iessi E. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iida J., Skubitz A.P., McCarthy J.B., Skubitz K.M. Protein kinase activity is associated with CD63 in melanoma cells. J. Transl. Med. 2005;3:42. doi: 10.1186/1479-5876-3-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toricelli M., Melo F.H., Peres G.B., Silva D.C., Jasiulionis M.G. Timp1 interacts with beta-1 integrin and CD63 along melanoma genesis and confers anoikis resistance by activating PI3-K signaling pathway independently of Akt phosphorylation. Mol. Cancer. 2013;12:22. doi: 10.1186/1476-4598-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung K.K., Liu X.W., Chirco R., Fridman R., Kim H.R. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 2006;25:3934–3942. doi: 10.1038/sj.emboj.7601281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zecena H., Tveit D., Wang Z., Farhat A., Panchal P., Liu J., Singh S.J., Sanghera A., Bainiwal A., Teo S.Y. Systems biology analysis of mitogen activated protein kinase inhibitor resistance in malignant melanoma. BMC Syst. Biol. 2018;12:33. doi: 10.1186/s12918-018-0554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daniels D.A., Chen H., Hicke B.J., Swiderek K.M., Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen J., Tao W., Hao S., Iyer S.P., Zu Y. A unique aptamer-drug conjugate for targeted therapy of multiple myeloma. Leukemia. 2016;30:987–991. doi: 10.1038/leu.2015.216. [DOI] [PubMed] [Google Scholar]
- 29.Rong Y., Chen H., Zhou X.F., Yin C.Q., Wang B.C., Peng C.W., Liu S.P., Wang F.B. Identification of an aptamer through whole cell-SELEX for targeting high metastatic liver cancers. Oncotarget. 2016;7:8282–8294. doi: 10.18632/oncotarget.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duffield A., Kamsteeg E.J., Brown A.N., Pagel P., Caplan M.J. The tetraspanin CD63 enhances the internalization of the H,K-ATPase beta-subunit. Proc. Natl. Acad. Sci. USA. 2003;100:15560–15565. doi: 10.1073/pnas.2536699100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bommarito A., Richiusa P., Carissimi E., Pizzolanti G., Rodolico V., Zito G., Criscimanna A., Di Blasi F., Pitrone M., Zerilli M. BRAFV600E mutation, TIMP-1 upregulation, and NF-κB activation: closing the loop on the papillary thyroid cancer trilogy. Endocr. Relat. Cancer. 2011;18:669–685. doi: 10.1530/ERC-11-0076. [DOI] [PubMed] [Google Scholar]
- 32.Dong J., Ma Q. TIMP1 promotes multi-walled carbon nanotube-induced lung fibrosis by stimulating fibroblast activation and proliferation. Nanotoxicology. 2017;11:41–51. doi: 10.1080/17435390.2016.1262919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y.F., Shangguan D., Liu H., Phillips J.A., Zhang X., Chen Y., Tan W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem. 2009;10:862–868. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shangguan D., Li Y., Tang Z., Cao Z.C., Chen H.W., Mallikaratchy P., Sefah K., Yang C.J., Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang H.I., Lee H. A decrease in the expression of CD63 tetraspanin protein elevates invasive potential of human melanoma cells. Exp. Mol. Med. 2003;35:317–323. doi: 10.1038/emm.2003.43. [DOI] [PubMed] [Google Scholar]
- 36.Berditchevski F., Odintsova E. Characterization of integrin-tetraspanin adhesion complexes: role of tetraspanins in integrin signaling. J. Cell Biol. 1999;146:477–492. doi: 10.1083/jcb.146.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi L., Forte D., Migliardi G., Salvestrini V., Buzzi M., Ricciardi M.R., Licchetta R., Tafuri A., Bicciato S., Cavo M. The tissue inhibitor of metalloproteinases 1 increases the clonogenic efficiency of human hematopoietic progenitor cells through CD63/PI3K/Akt signaling. Exp. Hematol. 2015;43:974–985.e1. doi: 10.1016/j.exphem.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Lee S.Y., Kim J.M., Cho S.Y., Kim H.S., Shin H.S., Jeon J.Y., Kausar R., Jeong S.Y., Lee Y.S., Lee M.A. TIMP-1 modulates chemotaxis of human neural stem cells through CD63 and integrin signalling. Biochem. J. 2014;459:565–576. doi: 10.1042/BJ20131119. [DOI] [PubMed] [Google Scholar]
- 39.Kudo K., Yoneda A., Sakiyama D., Kojima K., Miyaji T., Yamazaki M., Yaita S., Hyodo T., Satow R., Fukami K. Cell surface CD63 increased by up-regulated polylactosamine modification sensitizes human melanoma cells to the BRAF inhibitor PLX4032. FASEB J. 2019;33:3851–3869. doi: 10.1096/fj.201800664RR. [DOI] [PubMed] [Google Scholar]
- 40.Kannengiesser C., Spatz A., Michiels S., Eychène A., Dessen P., Lazar V., Winnepenninckx V., Lesueur F., Druillennec S., Robert C., EORTC Melanoma group Gene expression signature associated with BRAF mutations in human primary cutaneous melanomas. Mol. Oncol. 2008;1:425–430. doi: 10.1016/j.molonc.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whiteside T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., García-Santos G., Ghajar C. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishida-Aoki N., Tominaga N., Takeshita F., Sonoda H., Yoshioka Y., Ochiya T. Disruption of Circulating Extracellular Vesicles as a Novel Therapeutic Strategy against Cancer Metastasis. Mol. Ther. 2017;25:181–191. doi: 10.1016/j.ymthe.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X., Zhao Z., Bai H., Fu T., Yang C., Hu X., Liu Q., Champanhac C., Teng I.T., Ye M., Tan W. DNA Aptamer Selected against Pancreatic Ductal Adenocarcinoma for in vivo Imaging and Clinical Tissue Recognition. Theranostics. 2015;5:985–994. doi: 10.7150/thno.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.García-Recio E.M., Pinto-Díez C., Pérez-Morgado M.I., García-Hernández M., Fernández G., Martín M.E., González V.M. Characterization of MNK1b DNA Aptamers That Inhibit Proliferation in MDA-MB231 Breast Cancer Cells. Mol. Ther. Nucleic Acids. 2016;5:e275. doi: 10.1038/mtna.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H., Liu J., Cao W., Xiao X., Liang L., Liu-Smith F., Wang W., Liu H., Zhou P., Ouyang R. C-myc/miR-150/EPG5 axis mediated dysfunction of autophagy promotes development of non-small cell lung cancer. Theranostics. 2019;9:5134–5148. doi: 10.7150/thno.34887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.