Significance
Correct selection of nascent proteins into the appropriate biogenesis pathways is crucial for protein homeostasis. N-terminal methionine excision (NME) is a ubiquitous nascent protein modification that impacts >50% of the proteome. This enzymatic reaction occurs near the peptide exit tunnel of the ribosome, where multiple other ribosome-associated protein biogenesis factors also vie for access to the nascent protein. How timely and selective NME occurs in this crowded environment is unclear. Here, kinetic measurements with bacterial NME enzymes combined with mathematical modeling show that the crowded macromolecular environment at the ribosome exit site enhances both the efficiency and specificity of this fundamental enzymatic reaction via a combination of timing and allosteric mechanisms.
Keywords: nascent protein modification, ribosome, protein biogenesis, peptide deformylase, methionine aminopeptidase
Abstract
The nascent polypeptide exit site of the ribosome is a crowded environment where multiple ribosome-associated protein biogenesis factors (RPBs) compete for the nascent polypeptide to influence their localization, folding, or quality control. Here we address how N-terminal methionine excision (NME), a ubiquitous process crucial for the maturation of over 50% of the bacterial proteome, occurs in a timely and selective manner in this crowded environment. In bacteria, NME is mediated by 2 essential enzymes, peptide deformylase (PDF) and methionine aminopeptidase (MAP). We show that the reaction of MAP on ribosome-bound nascent chains approaches diffusion-limited rates, allowing immediate methionine excision of optimal substrates after deformylation. Specificity is achieved by kinetic competition of NME with translation elongation and by regulation from other RPBs, which selectively narrow the processing time window for suboptimal substrates. A mathematical model derived from the data accurately predicts cotranslational NME efficiency in the cytosol. Our results demonstrate how a fundamental enzymatic activity is reshaped by its associated macromolecular environment to optimize both efficiency and selectivity, and provides a platform to study other cotranslational protein biogenesis pathways.
Proper protein biogenesis is essential for the generation and maintenance of protein homeostasis in the cell. Emerging evidence shows that protein biogenesis begins early, while the nascent protein is still being synthesized by the ribosome. As a nascent polypeptide emerges from the ribosome tunnel exit, a variety of ribosome-associated protein biogenesis factors (RPBs) bind at overlapping docking sites near the tunnel exit and compete for the nascent protein to influence its folding, localization, maturation, and quality control (1–4). Efficient and accurate selection of nascent proteins into their proper biogenesis pathways is crucial, but how this is achieved in the crowded environment of the ribosome exit site is not well understood.
N-terminal methionine excision (NME) is an essential and ubiquitous process conserved across all kingdoms of life. NME generates essential functional groups for some enzymes (5–7) and introduces diverse N-termini for cellular proteins to enable their quality control via the N-degron pathways (8–10). In eukaryotic cells, NME also allows subsequent N-terminal modifications such as N-acetylation, which can serve as a degron to enable protein quality control (11, 12), and N-myristoylation, which enables localization of the protein to membrane compartments for participation in signal transduction pathways (13, 14). In bacteria and prokaryote-derived organelles, where protein synthesis initiates with formyl methionine, NME is carried out by 2 essential enzymes, peptide deformylase (PDF) and methionine aminopeptidase (MAP). Over 90% of the proteome in bacteria is deformylated, while ∼50% of the proteins are subject to methionine excision (14). Classic enzymological studies of PDF and MAP, based on short peptide substrates, provided rich information on the NME process (15–18). Deformylation by PDF is required for methionine excision by MAP (19). PDF displays relaxed sequence specificity except for the initiator methionine (15, 17), whereas MAP strongly favors substrates with small amino acid side chains at the second position (16, 18, 20, 21). These results were corroborated by studies in cell lysate and in vivo (20–22), providing useful tools for the qualitative prediction of NME (16, 23, 24). Nevertheless, the MAP reactions on peptide substrates are slow, with kcat/Km values of ∼103–105 M−1s−1 even for optimal substrates (16), raising questions as to how cotranslational NME could occur effectively for ∼50% of the proteome.
NME is one of the earliest events encountered by nascent proteins during their biogenesis and could occur cotranslationally as soon as the nascent chain reaches 44 to 48 residues (25). Earlier works have demonstrated the interaction between the ribosome and the NME enzymes in bacteria and eukaryotes (3, 4, 25–28). Recent measurements showed that Escherichia coli PDF and MAP bind the ribosome with apparent equilibrium dissociation constants (Kd) of 1.8 μM and 2.4 µM, respectively (25, 29). Ribosome binding of PDF is mediated by its C-terminal helix, which docks in a groove between uL22 and bL32 near the ribosome exit site (4, 29). The same site also mediates ribosome binding of MAP via a positively charged loop at the periphery of the active site (refs. 4 and 25 and SI Appendix, Fig. S1). Disruption of the ribosome interactions of PDF or MAP led to inefficient NME in cell extract and defective cell growth, suggesting the importance of the ribosome interaction for enzymatic activity (25, 29). However, whether and how the ribosome influences the NME reactions of the nascent protein was only recently explored for PDF (26) and remains elusive for MAP.
In addition to MAP and PDF, multiple other RPBs also engage newly synthesized proteins near the ribosome exit site. In bacteria, these include the Signal recognition particle (SRP) and the SecA ATPase that mediate the cotranslational targeting of membrane and secretory proteins (30–34) and trigger factor (TF) that cotranslationally chaperones numerous cytosolic, secretory, and outer membrane proteins (35–38). The binding sites of the RPBs overlap heavily with one another on the ribosome (refs. 1 and 2 and SI Appendix, Fig. S1). SRP binds free ribosomes at uL23 and uL29 near the ribosome exit site (39, 40) and, upon the emergence of a transmembrane domain (TMD) or signal sequence on the nascent polypeptide, further contacts bL32, uL22, and uL24, resulting in substantial overlaps with the PDF and MAP binding sites (4, 29, 40). The TF binding site on the ribosome spans uL22, uL23, and uL29, which partially overlaps with those of SRP, PDF, and MAP (4, 41). SecA also contacts the ribosome via uL23 (31, 32), where both SRP and TF can bind. The molecular crowding at the ribosome exit necessitates spatial and temporal coordination between the multiple RPBs, which must compete for not only ribosome binding but also access to the nascent polypeptide.
Recent works began to explore the interplay between the RPBs at the crowded ribosome exit site. Excess PDF or MAP reduces the ribosome occupancy of one another (25). On the contrary, a recent structural study showed that PDF and MAP can cobind at the ribosome, with MAP relocalizing to a secondary binding site near uL23 and uL29 in the presence of PDF (4). Pairwise cobinding has also been observed for TF and SRP with one another and with PDF or MAP on the ribosome and ribosome–nascent chain complex (RNC) (3, 4, 42). In addition, TF modestly reduced NME efficiency in vivo and in vitro (25, 38), and SRP slows the deformylation of RNCs bearing a TMD or a hydrophobic signal sequence on the nascent chain in vitro (26). These results demonstrate the ability of RPBs to influence the activity of one another. Finally, the PDF reaction became slower when the nascent chain elongated from 75 to 100 amino acids (26), and TF strongly inhibited SRP function when the nascent polypeptide exceeded a critical length (42). These observations suggest that translation elongation could act as a competing branch that limits the action of individual RPBs. Nevertheless, how the crowded environment at the ribosome exit site and competition with ongoing protein synthesis impact the timing, activity, and selectivity of nascent protein modification remains an outstanding question.
In this work, we combined pre–steady-state kinetic measurements and numerical simulations to address these questions. We showed that the MAP reaction on ribosome-bound nascent chains is 2 to 4 orders of magnitude faster than previously measured on peptide substrates, allowing the NME for optimal substrates to occur upon each encounter of MAP with the translating ribosome. TF and SRP are critical for ensuring the specificity of nascent protein modification by selectively narrowing the window for the reactions of MAP on suboptimal substrates during translation. Our experimental data allowed the construction of a computational model that accurately predicts the cotranslational NME efficiency of model substrates under near-in vivo conditions.
Results
MAP Reaction on Ribosome-Bound Nascent Chains Is Diffusion-Limited.
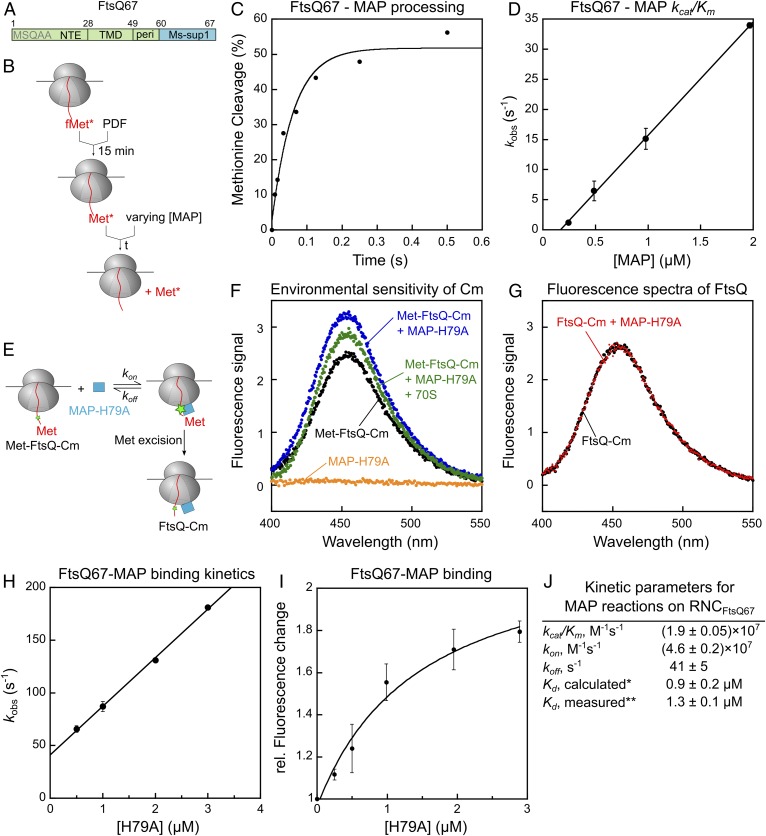
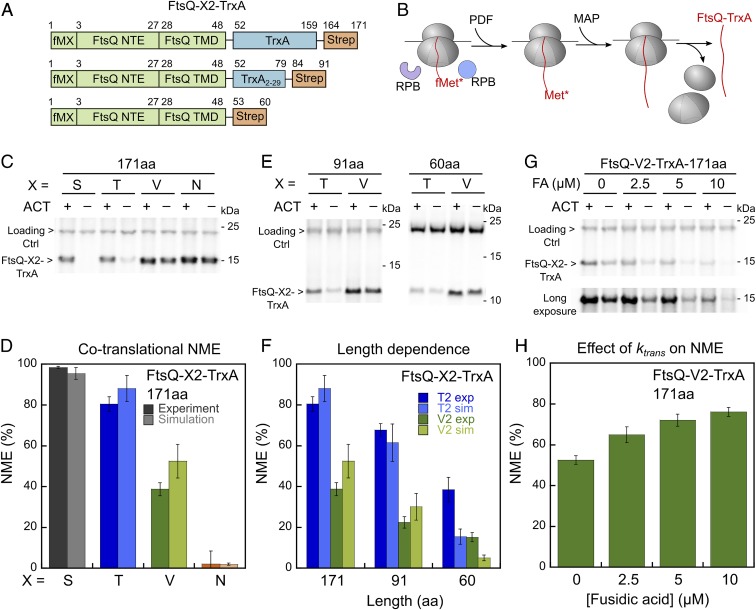
To characterize the NME reactions in the context of ribosomes, we generated and purified translation-arrested RNCs displaying FtsQ, an inner membrane protein with a 27-residue N-terminal extension (NTE) preceding its TMD. The unstructured NTE minimizes potential effects of nascent chain folding on enzymatic processing, and the N-terminal sequence of FtsQ (MSQAA) is predicted to be favored by both PDF and MAP (15, 16). A C-terminal 8-amino acid translation stall sequence, Ms-sup1 (43), allows generation of RNCs with defined nascent chain lengths (Fig. 1A and SI Appendix, Fig. S2A). RNCs labeled with 35S-Met were generated by in vitro translation in E. coli S30 extract in the presence of actinonin (ACT), a PDF inhibitor, to prevent NME by endogenous PDF and MAP, and purified by sucrose-gradient centrifugation.
Fig. 1.
MAP-mediated methionine excision of nascent chains on the ribosome is diffusion-limited. (A) Scheme of the model substrate FtsQ67 for generation of RNC, which contains the N-terminal extension (NTE), transmembrane domain (TMD), and part of the periplasmic region (peri) followed by the Ms-sup1 translation stall sequence. The N-terminal sequence is indicated. (B) Scheme of the methionine cleavage assay to measure the rate constant of the MAP reaction. Purified RNC with a single 35S-labeled N-terminal methionine was preincubated with PDF for 15 min, before initiation of reaction by addition of MAP. Reactions contained 10 nM RNC, 50 nM PDF, and varying concentrations of MAP. (C) A representative time trace for cleavage of 10 nM RNCMet-FtsQ67 by 1 μM MAP. Single exponential fit of the data (equation 1 in SI Appendix, Methods) gave a kobs value of 16.4 s−1. (D) The observed rate constants for methionine cleavage of RNCMet-FtsQ67 were plotted as a function of effective MAP concentration. The line is a linear fit of the data, and the obtained kcat/Km value is summarized in J. (E) Scheme of the fluorescence-based assay to measure the binding between RNCMet-FtsQ67Cm and MAP. The FtsQ67 nascent chain was labeled with coumarin (Cm) at the fifth residue. (F) Fluorescence emission spectra to demonstrate the enhancement of Cm fluorescence upon binding of RNCMet-FtsQ67Cm to MAP-H79A. Where indicated, reactions contained 8 nM deformylated RNCMet-FtsQ67Cm, 0.5 μM MAP-H79A, and 1.4 μM 70S ribosome. (G) MAP-H79A does not affect the fluorescence emission spectra of RNCFtsQ67Cm after methionine excision. RNCMet-FtsQ67Cm was pretreated with 20 nM wild-type MAP to yield RNCFtsQ67Cm, and fluorescence emission spectra were recorded before and after the addition of 0.5 µM MAP-H79A. (H) Observed rate constants (kobs) of RNC-MAP association, measured using the fluorescence assay in E, was plotted as a function of MAP-H79A concentration. The reactions contained 8 nM RNCMet-FtsQCm and indicated concentrations of MAP-H79A. The line is a linear fit of the data to equation 6 in SI Appendix, Supplementary Methods, and the obtained kon and koff values are summarized in J. (I) Equilibrium titration to measure the binding affinity of RNCMet-FtsQCm for MAP-H79A. The data were fit to equation 5 in SI Appendix, Supplementary Methods, and the obtained Kd value is summarized in J. (J) Summary of the kinetic parameters obtained from the measurements in D, H, and I. *Calculated from Kd = koff/kon. **Kd measured from the equilibrium titration in I. All values are reported as mean ± SD with n = 2.
We first measured methionine cleavage by MAP on RNCFtsQ with a nascent chain length of 67 residues (Fig. 1 B–D). As deformylation is a prerequisite for the MAP reaction, we preincubated RNCFtsQ67 with PDF to ensure complete removal of the formyl group on 35S-labeled N-terminal methionine. We then carried out the reaction under single turnover conditions with purified MAP in excess of RNCFtsQ67, and quantified the reaction extent at indicated times by scintillation counting of free [35S]methionine released from the nascent chain (44). The reaction rate rises linearly up to 2 µM MAP with no indication of saturation, giving estimated limits for Km of >>2 µM and kcat of >>34 s−1. The MAP concentration dependence of the observed rate constants gave a kcat/Km value of (1.9 ± 0.05) × 107 M−1s−1. Measurements on RNC bearing luciferase, a cytosolic protein with an unstructured N terminus, gave a comparable kcat/Km value of (1.9 ± 0.06) × 107 M−1s−1 (SI Appendix, Fig. S2B). These rate constants on RNC substrates are 102 to 104 fold faster than those on peptide substrates (kcat/Km values of 103 to 105 M−1s−1) measured here (SI Appendix, Fig. S2C) and previously (16, 18, 45). The MAP-K226E mutation, which weakens the binding of MAP to the ribosome (25), reduced the reaction rate of RNCFtsQ67 ∼20-fold but affected the reaction of a tetra-peptide substrate <2-fold (SI Appendix, Fig. S2 C and D), supporting an important contribution of the ribosome in enhancing the efficiency of the MAP reaction (25).
The kcat/Km value of the MAP reaction on RNC substrates approaches the rate constants of protein associations, suggesting that this reaction follows Briggs–Haldane kinetics whereby the reaction is rate-limited by enzyme–substrate association (46). To test this model, we developed a fluorescence-based assay to directly measure the binding of MAP and RNCFtsQ (Fig. 1E). To specifically probe the interaction between MAP and the substrate, we used mutant MAP-H79A, which reduced the rate of the chemical step >104-fold but has only minor effects on the binding of peptide substrates (47, 48) and the 70S ribosome (SI Appendix, Fig. S2 E–G). A fluorescent amino acid, L-(7-hydroxycoumarin-4-yl)ethylglycine (Cm), was incorporated at the fifth residue of the FtsQ nascent chain via amber suppression (49). Binding of MAP-H79A led to enhanced fluorescence intensity of RNCFtsQCm. The addition of excess ribosome reduced the fluorescence enhancement, in agreement with the competition of ribosomes with RNC for MAP binding (Fig. 1F). Importantly, removal of the N-terminal methionine on the nascent chain abolished the MAP-induced fluorescence enhancement of the Cm dye (Fig. 1G). This demonstrates that the fluorescence enhancement is specific to the interaction of MAP with the uncleaved nascent chain, and suggests that the nascent chain is released from the substrate-binding pocket of MAP after methionine excision.
Using this fluorescence assay, we measured the observed rate constants for MAP-RNCFtsQ67 assembly as a function of MAP-H79A concentration. Linear fit of the concentration dependence gave an association rate constant, kon, of (4.6 ± 0.2) × 107 M−1s−1 and an extrapolated dissociation rate constant, koff, of 41 ± 5 s−1 (Fig. 1H). The value of kon is comparable to the value of kcat/Km measured from the enzymatic reaction, supporting the model that the MAP reaction is rate-limited by the assembly of the MAP–RNC complex. Independent equilibrium titrations using the fluorescence assay gave an equilibrium dissociation constant (Kd) of 1.3 ± 0.1 μM for the MAP–RNCFtsQ67 complex, in good agreement with the Kd value of 0.9 ± 0.2 μM calculated from koff/kon (Fig. 1 I and J). Considering the sub- to low-millimolar Km values of MAP for peptide substrates (16, 45) and the apparent Kd value of 2.4 ± 0.4 μM for MAP–ribosome binding (25), these results indicate that MAP–RNC binding is dominated by interaction with the ribosome, with a modest contribution from the nascent chain.
Collectively, the results in this section showed that the MAP reaction for nascent proteins on the ribosome is substantially accelerated compared to peptide substrates and is rate-limited by the association between MAP and RNC for optimal substrates such as RNCFtsQ67. The diffusion-controlled kinetics imply that MAP can successfully process nascent chains on the RNC upon every encounter and thus ensure efficient NME of nascent proteins.
The PDF Reaction Is Rate-Limiting for NME of Optimal Substrates.
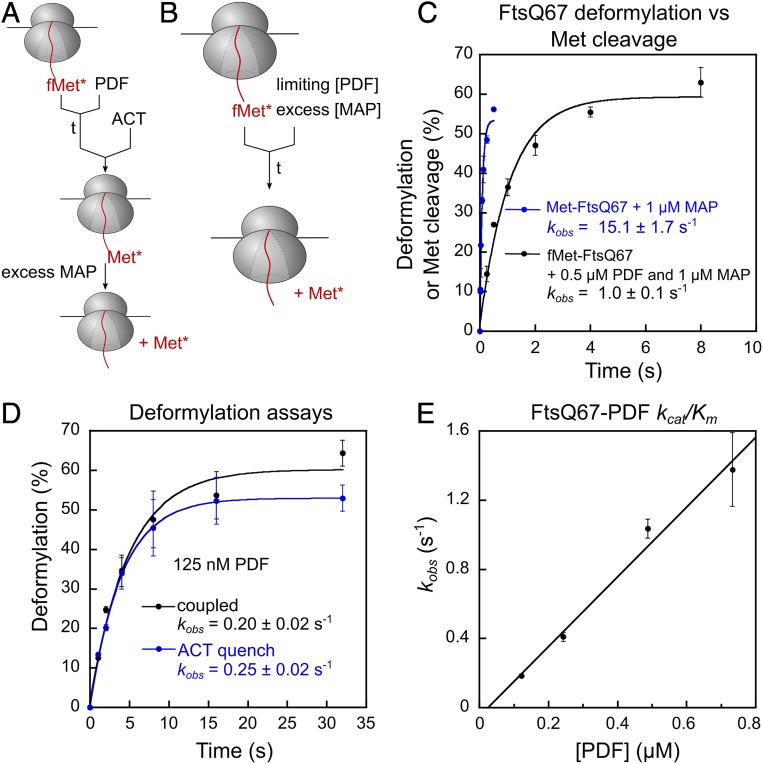
To measure PDF-mediated deformylation, the prerequisite for methionine excision, we designed 2 independent assays that use methionine excision as the readout. In the actinonin-quench assay, we quenched the PDF reaction at specified times using actinonin and released the deformylated methionine with excess MAP for quantification (Fig. 2A). In the coupled assay, limiting PDF and excess MAP were added simultaneously to initiate the reaction (Fig. 2B); this assay leverages the faster kinetics of the MAP versus the PDF reaction and was only used for RNCs bearing optimal MAP substrates. The results from the 2 assays agreed well (Fig. 2D), and, together, the 2 assays allowed us to measure the kinetics of the PDF reaction on a variety of substrates.
Fig. 2.
The PDF reaction is ∼10-fold slower than the MAP reaction. (A) Scheme of the actinonin quench-based deformylation assay. The reaction was initiated by adding PDF to RNCfMet-FtsQ67 and quenched with 40 μM actinonin at specified time points. MAP (100 nM for FtsQ-S2) was added thereafter to cleave the deformylated N-terminal methionine, allowing the reaction to be analyzed via scintillation counting of free [35S]methionine. (B) Scheme of the coupled deformylation assay. Indicated concentrations of PDF were added together with excess MAP (1 µM) to initiate NME of 10 nM RNCfMet-FtsQ67, and the generation of free [35S]methionine was monitored during the time course. (C) Comparison of the time courses for methionine excision of RNCfMet-FtsQ67, measured using the coupled deformylation assay in B, with that of deformylated RNCMet-FtsQ67 by MAP. (D) Comparison of the PDF reaction time courses measured by the coupled assay (black) and actinonin-quench assay (blue). The reactions used 10 nM RNCfMet-FtsQ67, 125 nM PDF, and 1 µM and 100 nM MAP in the coupled and actinonin-quench assay, respectively. The data were fit to equation 1 in SI Appendix, Methods, and the obtained rate constants are indicated. (E) Observed rate constants for deformylation of RNCfMet-FtsQ67 are plotted as a function of effective PDF concentration. Linear fit of the data gave a kcat/Km value of (2.0 ± 0.4) × 106 M−1s−1. All values are shown as mean ± SD with n = 2 to 3.
The observed methionine cleavage in the coupled PDF–MAP reaction with formylated RNCFtsQ was >10-fold slower than the MAP reaction with deformylated RNCFtsQ67 (Fig. 2C), suggesting that deformylation is rate-limiting in the coupled reaction. Varying the concentration of MAP from 0.5 to 2 μM did not affect the observed reaction rates (SI Appendix, Fig. S2H), providing an independent indication that the MAP reaction is not rate-limiting in the coupled assay. Using the coupled assay, the kcat/Km value for deformylation of RNCFtsQ67 was measured to be (2.0 ± 0.4) × 106 M−1s−1 (Fig. 2E), in good agreement with previous measurements by Ranjan et al. (26). These values are ∼10-fold lower than that of the MAP reaction, indicating that deformylation is the rate-limiting step for the overall NME of optimal MAP substrates.
Compromised Specificity of NME with Nascent Proteins on the Ribosome.
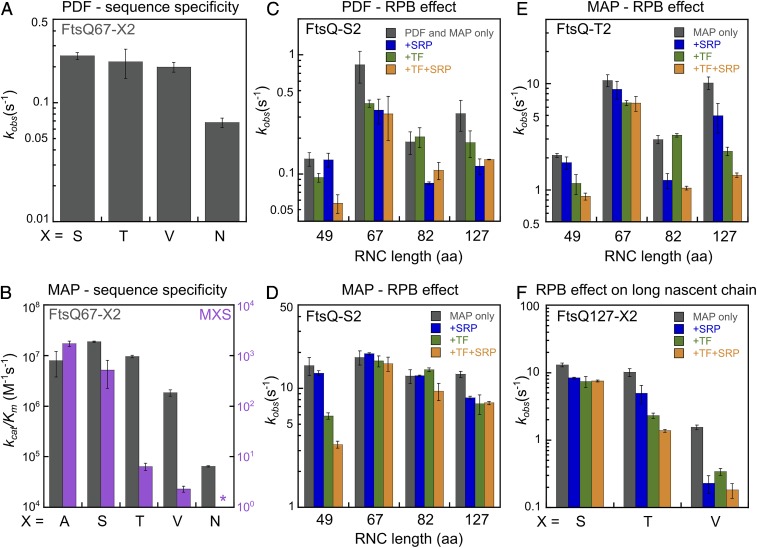
A prominent determinant of NME efficiency is the second residue on the nascent polypeptide (16, 18, 20–22). To test whether and how the ribosome modulates second residue specificity of NME, we prepared RNCFtsQ67-X2 in which Ser2 on the FtsQ nascent chain is mutated to different amino acids. Consistent with previous in vivo observations and in vitro studies using peptide substrates (17, 22), PDF remained insensitive to the second residue among all of the RNC substrates tested, except for a moderate 3-fold rate reduction with RNCFtsQ67-N2 (Fig. 3A).
Fig. 3.
Compromised second residue specificity of the NME reactions on RNCs is restored by length-dependent regulation from the RPBs. (A) Observed deformylation rate constants of RNCfMet-FtsQ67 with indicated amino acids at the second residue. Reactions were measured using the actinonin-quench assay (Fig. 2A) and contained 0.125 μM PDF. To release the deformylated methionine, 100 and 250 nM MAP were used for FtsQ-S2/T2 and FtsQ-V2/N2, respectively. (B) Comparison of MAP specificity toward the second residue for reactions with RNCFtsQ67-X2 (gray bars) versus tripeptide substrates (MXS, magenta bars; replotted from Frottin et al. [16]). The kcat/Km values for RNCFtsQ67-X2 were measured as in Fig. 1D. The asterisk denotes a kcat/Km value of <0.01 M−1s−1 for the MNS peptide. (C and D) Effects of TF and SRP on the deformylation (C) and methionine cleavage (D) of RNCs bearing FtsQ-S2 nascent chains of the indicated lengths. Observed PDF rate constants were measured with 10 nM RNCs and 0.5 μM PDF using the coupled assay. Observed MAP rate constants were measured with 10 nM RNCs and 1 μM MAP. (E and F) Effects of TF and SRP on methionine cleavage of RNCs bearing FtsQ-T2 nascent chains of the indicated lengths (E) and RNCs bearing FtsQ-127 nascent chains with the indicated second residue side chains (F). Observed rate constants were measured with 10 nM RNCs, 1 μM MAP, and 5 µM TF and 400 nM SRP where indicated. All values are shown as mean ± SD with n = 2 to 3.
In contrast to PDF, proteomics studies and previous kinetic measurements based on peptide substrates showed that the MAP reaction depends critically on the size of the second residue (16, 22). Peptides with medium-sized side chains at the second residue (T, V) define a “twilight zone” that display kcat/Km values 102 and 103 fold lower, respectively, than those of optimal substrates (G, A, S, P, C), and substrates with even larger second residues (N, K, R, etc.) react >105 fold more slowly (ref. 16 and Fig. 3B, magenta). In contrast to the observations with peptide substrates, the impact of second residues on the MAP reaction was substantially smaller for RNC substrates. Compared to optimal substrates such as RNCFtsQ67-A2 and RNCFtsQ67-S2, the kcat/Km value was comparable with RNCFtsQ67-T2 and reduced only 10-fold with RNCFtsQ67-V2 (Fig. 3B). RNCFtsQ67-N2, previously considered a very weak substrate, was still processed by MAP with a kcat/Km of ∼105 M−1s−1 (Fig. 3B).
The reduced specificity of the MAP reaction for RNCs can be attributed to the diffusion-limited nature of this reaction. As described in Fig. 1, the reaction of MAP with optimal substrates is rate-limited by its association with RNC, which would mask reductions in the rate of subsequent peptide bond hydrolysis. The slower rate of deformylation than methionine excision is expected to further reduce the specificity of overall NME. This is because modest rate reductions in the MAP reaction would be masked as long as the PDF reaction is rate-limiting, and the MAP reaction becomes rate-limiting only for substrates with second residues larger than valine (cf. kcat/Km values in Fig. 3B, dark bars, versus Fig. 2E). Both of these factors are expected to severely compromise the specificity of the NME reaction for nascent chains on the ribosome.
Nascent Chain Length and RPBs Define an Optimal Window for NME to Restore Specificity.
We reasoned that 2 factors could help restore the specificity of cotranslational NME. First, on actively translating ribosomes, NME must kinetically compete with elongation of the nascent polypeptide; this could impose a limited time window for NME if the reactions of these enzymes are sensitive to nascent chain length. Second, additional RPBs such as TF, SRP, and SecA are recruited to the ribosome exit site during translation and could modulate the reaction kinetics of the NME enzymes.
In vitro translation of stalled RNCs in cell extracts showed that the minimum nascent chain length required for NME is ∼45 residues (SI Appendix, Fig. S3 A and B), in agreement with previous reports (25) and with the predicted distances between the exit site and the actives sites of PDF and MAP (∼35 and ∼30 Å, respectively; refs. 25 and 29). To define the optimal window for NME, we performed single-turnover measurements of the PDF and MAP reactions on purified RNCs with different nascent chain lengths. The kcat/Km value for the PDF reaction of RNCFtsQ-S2 rose 20-fold when the nascent chain elongated from 45 to 67 amino acids and became 2–3 fold slower at longer nascent chain lengths (SI Appendix, Fig. S3C and Fig. 3C, gray bars). The MAP reaction varied <2-fold beyond a nascent chain length of 49 residues for an optimal substrate, RNCFtsQ-S2 (SI Appendix, Fig. S3D and Fig. 3D, gray bars), but became more sensitive to nascent chain length for substrates in the twilight zone, RNCFtsQ-T2 and RNCFtsQ-V2 (Fig. 3E, gray bars, and SI Appendix, Fig. S3E). Thus, both the PDF and MAP reactions showed modest dependences on nascent chain length, and, for MAP, this length dependence is larger for suboptimal substrates.
To test the effect of RPBs, we measured the length-dependent reactions of PDF and MAP in the presence of saturating concentrations of TF (SI Appendix, Fig. S4A) and SRP. The combination of TF and SRP largely reproduced the effect of the cytosol on the MAP reaction, and the additional presence of SecA had minimal effects on both reactions (SI Appendix, Fig. S4 B and C). The PDF reaction was slowed ∼2-fold across all of the tested nascent chain lengths by the combination of both factors (Fig. 3C, orange bars). A previous work reported a much larger inhibitory effect of SRP on the PDF reaction with membrane proteins (26) because the TMD or signal sequence was much closer to the N-termini of the substrates used in that study compared to FtsQ, in which the TMD is preceded by a 27-amino acid NTE. In contrast to PDF, the effects of RPBs on the MAP reactions are larger and more substrate-dependent. For an optimal substrate (RNCFtsQ-S2), SRP and TF reduced the rate of the MAP reaction 5-fold when the nascent chain is short (49 amino acids), but affected the reaction <2-fold at longer nascent chain lengths (Fig. 3D). For a suboptimal substrate, RNCFtsQ-T2, SRP and TF together slowed the MAP reaction ∼3-fold at both short (49 residues) and intermediate (82 residues) nascent chain lengths and ∼10-fold when the nascent chain reached 127 amino acids (Fig. 3 E and F). The inhibitory effects of SRP and TF on methionine excision of both short and long nascent chains were also observed with an SRP-independent cytosolic protein, luciferase (SI Appendix, Fig. S5). Finally, the inhibitory effect of TF or SRP on long-chain RNCs was larger with an even slower MAP substrate, FtsQ-V2 (Fig. 3F). Thus, RPBs selectively inhibit the MAP reaction on long nascent chains for RNCs bearing suboptimal MAP substrates.
Together, the results in this section show that the reactions of both PDF and MAP on the RNC are sensitive to the length of the nascent polypeptide, generating an optimal window for NME when the nascent chain is 49 to 82 amino acids long. For the MAP reaction, this window is largely imposed by the action of RPBs, which selectively inhibits methionine excision on suboptimal MAP substrates (Fig. 3 D–F) and therefore contributes to the restoration of NME specificity.
A Kinetic Model for Cotranslational NME Emphasizes the Contribution of RPBs to NME Specificity.
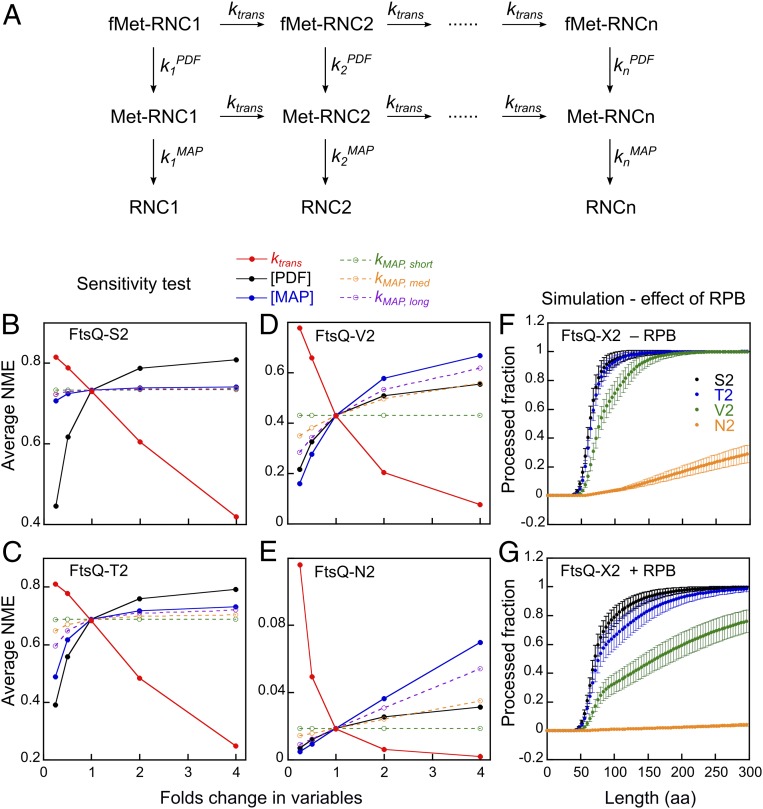
To quantitatively understand how the multiple factors that modulate the activity of NME enzymes impact the cotranslational N-termini modification of nascent proteins, we constructed an analytical kinetic model (Fig. 4A). Modeling of cotranslational NME in cell lysate is enabled by several features of this assay that simplify its mathematical description. First, ribosomes translating the reporter protein are at much lower concentrations compared to the NME enzymes in the lysate, rendering the reaction single-turnover and directly comparable to the single-turnover rate constants measured using purified RNCs. In addition, the concentration of NME enzymes are ∼10-fold lower than their Kd values for the ribosome and therefore subsaturating. This allows the NME reaction at a given nascent chain length to be described by pseudo–first-order rate constants for deformylation (kPDF) and methionine excision (kMAP; SI Appendix, Table S1) based on the experimentally determined or extrapolated kcat/Km values and enzyme concentrations determined by quantitative Western blot analyses in the E. coli lysate (SI Appendix, Fig. S6). The effect of ongoing protein synthesis is described by an estimated rate constant of translation elongation (ktrans). Based on differential equations constructed from this model, we used numerical integration to simulate the cumulative fraction of nascent proteins of n residues that successfully undergo deformylation and methionine excision (SI Appendix, Fig. S7 A–D). The robustness of the kinetic model was tested by applying an error of 20% in enzyme concentrations and reaction rate constants, which were randomly selected from normal distributions. The resulting deviations in simulation results were ∼10% (error bars in SI Appendix, Fig. S7 A–D), confirming the reliability of the simulation.
Fig. 4.
A kinetic model simulates cotranslational NME during protein synthesis. (A) A kinetic model for the cotranslational NME of a nascent protein with n amino acids. ktrans is the translation elongation rate. The PDF and MAP rate constants at nascent chain length i (kiPDF and kiMAP) are the products of effective enzyme concentration and kcat/Km values and, unless otherwise indicated, are also modulated by RPBs. The parameters used in the simulation are summarized in SI Appendix, Table S1. (B–E) Sensitivity test of the simulation results for FtsQ-X2. Each indicated parameter was varied independently by 2−2 to 22 fold of the value in SI Appendix, Table S1. “Average NME” denotes the mean value for the fraction of methionine-cleaved nascent chains throughout the reaction profile before 300 residues are synthesized. (F and G) Simulated cotranslational NME profiles for RNCFtsQ with different second residue side chains in the absence (F) and presence (G) of the regulatory effects of RPBs. Results from 100 runs are shown as mean ± SD.
We first asked how fast NME occurs during translation. Simulation based on the kinetic model showed that NME is 90% complete for FtsQ-S2 before ∼150 amino acids are synthesized (SI Appendix, Fig. S7A). As expected from the ∼10-fold faster reaction with MAP than with PDF for this substrate, methionine excision closely followed deformylation (SI Appendix, Fig. S7A). When the second residue is threonine and valine, methionine excision increasingly lagged behind deformylation (SI Appendix, Fig. S7 B and C). When the second residue is asparagine, deformylation was 90% complete when the nascent chain reached 250 amino acids, but no significant methionine excision occurred (SI Appendix, Fig. S7D). Notably, the MAP reaction profiles of both FtsQ-T2 and FtsQ-V2 exhibited biphasic behavior, with a steep rise before the nascent chain reached ∼80 residues followed by a slow phase (SI Appendix, Fig. S7 B and C), owing to the inhibition of MAP reactions at longer nascent chain lengths. Thus, the MAP reaction becomes increasingly rate-limiting for overall NME with increasing size of the second residue.
To dissect the contributions of different factors to the efficiency of cotranslational NME, we further tested whether and how the simulation outcome is sensitive to variations in the individual parameters in the kinetic model (Fig. 4 B–E and SI Appendix, Fig. S8). The tested parameters include the translation rate (solid red lines) and the concentrations of MAP or PDF (solid black and blue lines), which affect the enzymatic reaction rates at all nascent chain lengths (Fig. 4 B–E). The sensitivity test revealed a strong influence of translation elongation rate on NME efficiency (Fig. 4 B–E, red lines, and SI Appendix, Fig. S8 A–D) because faster translation shortens the time window available for the actions of PDF and MAP. This effect is substantial for all of the substrates tested and emphasizes the kinetic rivalry between NME and nascent chain synthesis.
On the contrary, the sensitivity of cotranslational NME to changes in enzyme concentration is substrate-dependent (Fig. 4 B–E, black and blue lines, and SI Appendix, Fig. S8 E–L). The NME efficiency of the optimal substrate, FtsQ-S2, is significantly affected only by reductions in PDF concentration (Fig. 4B and SI Appendix, Fig. S8 E and I). For suboptimal substrates such as FtsQ-T2, FtsQ-V2, and FtsQ-N2, NME efficiency can be modulated by changes in both PDF and MAP levels in both directions, and the MAP concentration becomes more dominant in dictating NME efficiency with increasing size of the second residue on the nascent chain (Fig. 4 C–E and SI Appendix, Figs. S8 F–H and J–L). These results reinforce the notion that the rate-limiting step of cotranslational NME shifted from the PDF to the MAP reaction, as predicted from SI Appendix, Fig. S7 A–D. As the MAP reaction is dependent on nascent chain length, we individually varied the MAP reaction rates for RNCs with short (45 to 56 residues), medium (56 to 82 residues), and long (>82 residues) nascent chains (Fig. 4 B–E, dashed lines). The simulation results showed that tuning kMAP,long had the largest effect on NME efficiency, and this effect is specific to FtsQ-V2 and FtsQ-N2.
Finally, we tested whether and how the RPBs, which reduce the MAP reaction rates in a substrate-dependent manner when the nascent chain exceeds 82 amino acids, impact the cotranslational NME profile (Fig. 4 F and G). The simulation results showed that the presence of RPBs moderately delayed the cotranslational NME of FtsQ-S2, which can be attributed to the 2-fold global effect of RPBs on the PDF reaction rate (Fig. 3C). On the contrary, the slow phase during the cotranslational processing of FtsQ-T2 and FtsQ-V2 becomes more prominent in the presence of RPBs, resulting in significantly delayed processing of FtsQ-T2 and incomplete NME of FtsQ-V2 for nascent chains up to 300 residues in length. These predictions are consistent with a previous study, which observed an inhibitory effect of TF on the cotranslational NME of barnase in cell lysate (25). By integrating the NME reaction kinetics with translation elongation, the kinetic model demonstrated how the presence of other RPBs improves the specificity of NME.
Cotranslational NME in Cell Extracts Agrees with Predictions from Kinetic Simulation.
To experimentally test the predictions from the kinetic model, we designed a cotranslational NME assay to measure the extent of NME during active translation. We fused the N-terminal extension and TMD of FtsQ to thioredoxin (TrxA), an unrelated E. coli cytosolic protein (Fig. 5A). In vitro translation of this 171-amino acid reporter protein in the E. coli S30 extract allowed us to assess its cotranslational NME by endogenous PDF and MAP in the presence of RPBs (Fig. 5B). The extent of NME was monitored by SDS/PAGE and autoradiography analyses of substrates labeled with a single [35S]methionine at the N terminus. Substrates with Ser and Thr at the second residue showed >80% NME (Fig. 5C). In contrast, over half of FtsQ-V2-TrxA retained the N-terminal [35S]methionine, and FtsQ-N2-TrxA was resistant to NME (Fig. 5C). These data agreed well with the simulation results for the individual substrates (Fig. 5D).
Fig. 5.
Measurement of cotranslational NME in cell lysate reproduces predictions from kinetic simulations. (A) Scheme of the model substrates used in the cotranslational NME assay. The 171-amino acid reporter protein contains full-length thioredoxin (TrxA), and the 60-amino acid reporter contains no thioredoxin. To generate the reporter with 91 residues, 80 residues were removed from the C terminus of thioredoxin. (B) Scheme of the cotranslational NME assay. FtsQ-X2-TrxA was translated in the S30 extract and processed by endogenous PDF and MAP during translation. (C) Representative SDS/PAGE and autoradiography analysis of the cotranslational NME of FtsQ-X2-TrxA (171 aa) in the S30 extract, measured as depicted in B. The reactions with actinonin (ACT, 5 μM) provide controls for the intensity of the FtsQ-X2-TrxA band without NME. (D) NME efficiency for FtsQ-X2-TrxA (171 aa) bearing different second-site residues. The values are from quantification of the data in C and their replicates (dark colors). The simulated data are from Fig. 4G at a nascent chain length of 171 aa (light colors). (E) Representative SDS/PAGE and autoradiograph of the cotranslational NME of shorter reporter proteins, measured as depicted in B. The second site residues are indicated. (F) Summary of the cotranslational NME efficiency for FtsQ-T2-TrxA (dark blue) and FtsQ-V2-TrxA (dark green) with different lengths. The values are from quantification of the data in C and E and their replicates. The simulated data (light blue and light green bars) are from Fig. 4G at the corresponding nascent chain lengths. (G) Representative SDS/PAGE and autoradiograph of cotranslational NME of FtsQ-V2-TrxA (171 aa) with indicated amount of fusidic acid (FA), which inhibits translation elongation. (H) Quantification of the data in G and their replicates. All values are reported as mean ± SD with n = 3.
Another prediction from the kinetic model is that the extent of NME is sensitive to the length of the protein, especially at 50 to 100 amino acids where NME products accumulate quickly (Fig. 4). To test this prediction, we varied the length of the reporter protein by truncating or removing thioredoxin (Fig. 5A). In agreement with predictions from the simulation, the extent of NME for substrates with Thr and Val at the second residue reduced with shorter protein length (Fig. 5 E and F). Finally, we tested the prediction that NME is sensitive to translation elongation rate by using fusidic acid (FA), which binds to EF-G and attenuates translation elongation (50, 51). Increasing amount of fusidic acid inhibited the translation of FtsQ-V2-TrxA while giving rise to more efficient NME (Fig. 5 G and H). The increase of overall NME from ∼50% to ∼80% was in good agreement with the simulation results for this substrate (cf. Figs. 5H and 4D), and supports the notion that slower elongation provides a longer time window for the reactions of PDF and MAP.
In conclusion, the results from the cotranslational NME assay agreed well with the predictions from the computational model, suggesting that kinetic modeling based on rate constants measured with translation-arrested RNCs provides an accurate description of cotranslational NME in the complex cytosolic environment.
Discussion
The majority of newly synthesized proteins undergo multiple maturation steps to ensure their proper function, localization, and turnover. As one of the earliest modification events during protein biogenesis, NME takes place in the crowded environment of the ribosome exit site, where multiple other RPBs compete for access to the nascent polypeptide. In this work, kinetic analyses demonstrate that NME is actively regulated by the ribosome, nascent chain length, and other RPBs at the ribosome tunnel exit. These factors, together with kinetic competition with translation elongation, dictate the efficiency, timing, and specificity of NME. Our results shed light on how the RPBs coordinate at the ribosome exit and demonstrate how a fundamental enzymatic reaction can be profoundly reshaped by its macromolecular environment. Moreover, our work provides a useful framework to conceptualize other cotranslational protein biogenesis events.
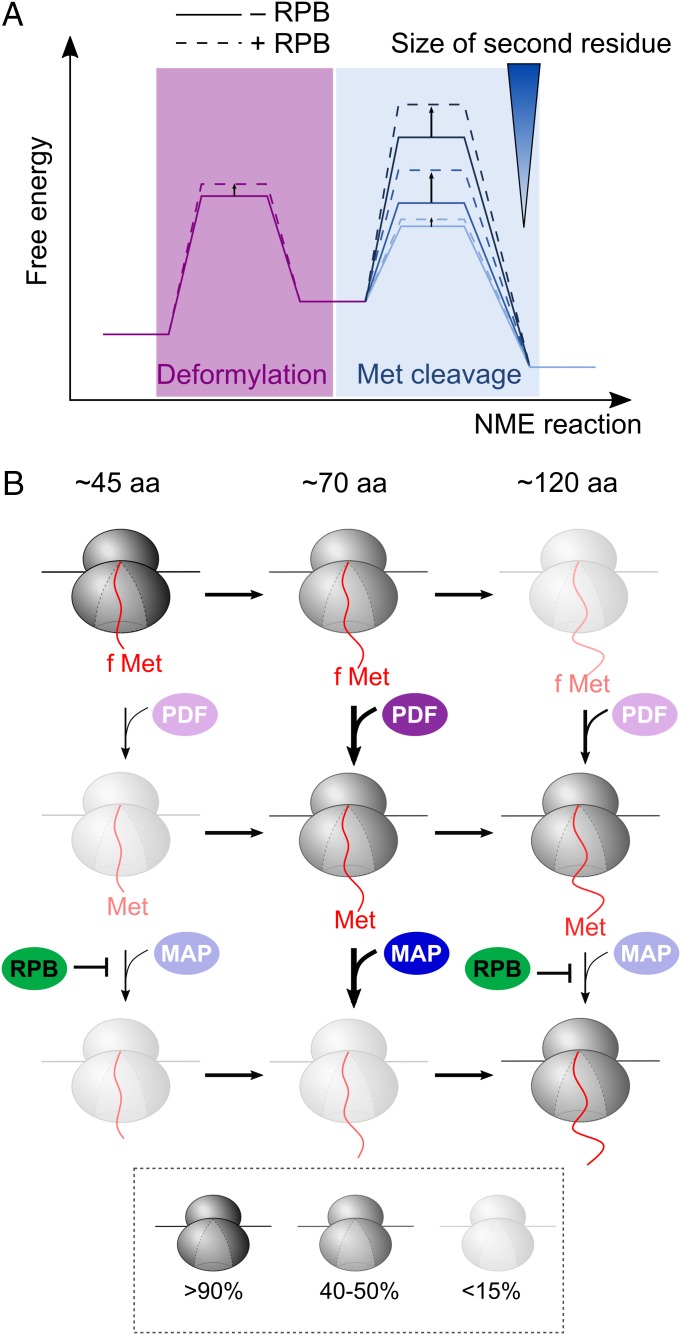
Our results show that the MAP reactions on RNCs are accelerated 102 to 104 fold compared to reactions with peptide substrates. This rate enhancement is likely due to the micromolar-affinity MAP binding site provided by the ribosome, which is ∼102- to 103-fold tighter than the affinity of MAP for peptides (16, 45). This interaction increases the local concentration of MAP and possibly preorients MAP to facilitate its access to the nascent chain, allowing methionine excision to proceed in a diffusion-limited manner for optimal substrates. Together with the rapid MAP–RNC association kinetics, over 90% of the nascent protein can be processed before ∼150 amino acids are synthesized under physiologically relevant conditions. However, the enhanced efficiency comes with a tradeoff in specificity, as any reductions in intrinsic reactivity would be masked by other steps that are rate-limiting, such as MAP-ribosome binding and deformylation (Fig. 6A). We found here that specificity is restored by other RPBs at the ribosome exit site, which selectively attenuates MAP reactions on suboptimal substrates when the nascent chain exceeds 82 amino acids. This imposes a limited time window on the NME of suboptimal substrates during translation, thus effecting their rejection (Fig. 6 A and B, dashed lines). These results exemplify the tradeoff between efficiency and specificity in biological systems, and reveal an unexpected role of the crowded environment at the ribosome exit in optimizing the balance between these 2 key parameters.
Fig. 6.
Model for cotranslational NME mediated by PDF and MAP. (A) Free energy profiles to depict the change in rate-limiting step during cotranslational NME due to both the sequence specificity of MAP and regulation by RPBs. As the second residue on the nascent protein becomes larger, the rate-limiting step shifts from deformylation to methionine cleavage. The RPBs moderately slows the PDF reaction, but significantly and selectively reduces the rates of the MAP reaction for substrates with larger second residue side chains. (B) RPBs define an optimal window for NME during translation. The relative abundance of individual ribosomal species at each nascent chain length was simulated during the cotranslational NME of FtsQ-V2 using the mathematical model and depicted using the indicated coloring scheme. PDF and MAP can act on the nascent proteins as soon as 45 residues are translated. PDF has an intrinsic preference for substrates of ∼70 residues in length (second row). The optimal window for the reaction of MAP is similar, but is imposed by the effects of RPBs (third row).
The results here also illustrate multiple mechanisms by which the RPBs coordinate in time and space at the ribosome exit site to ensure the proper functions of one another. First, given previous observations that TF and SRP can cobind with MAP or PDF on the ribosome (3, 25), the regulation observed here likely occurs allosterically rather than through exclusion of NME enzymes from the ribosome. This regulation could involve repositioning of these enzymes on the ribosome by TF or SRP that changes the access or orientation of the nascent chain relative to the enzyme active site. This model is consistent with the observation that the regulatory effects of TF and SRP on the MAP reaction are substrate-specific, whereas MAP–RNC binding is dominated by interaction with the ribosome. Second, TF regulates both NME enzymes when nascent proteins are as short as 49 amino acids. Given the enrichment of TF on nascent proteins longer than 100 amino acids in ribosome profiling data (38), our observation suggests that TF can begin regulating protein biogenesis before it stably engages with nascent polypeptides. Third, TF and SRP regulate the MAP reaction when the nascent polypeptide is short (49 residues) and long (127 residues), but not at intermediate chain lengths (67 residues). This suggests a remarkable degree of dynamics in the interplay between RPBs, which can adjust their relative positioning and/or accessibility to the nascent chain during translation. Most importantly, the length- and substrate-dependent regulation of RPBs defines an optimal time window for NME during translation and helps enhance its specificity, as discussed earlier. Analogous “timing” mechanisms that enhance fidelity was also observed between SRP with TF (3, 42), and could provide a general and effective mechanism to coordinate the different cotranslational biogenesis pathways.
Analogous to previous computational models that describe cotranslational protein folding (52–54), the kinetic model described here demonstrates how multiple parameters together contribute to NME, including enzyme concentration, nascent chain length, rivalry with translation elongation, and regulation by RPBs. Our simulation successfully predicted the NME efficiency of model substrates during active translation in cell extracts, as well as regulation of NME by the rates and duration of translation, demonstrating that the computational model derived from measurements on translation-arrested RNCs faithfully recapitulates cotranslational nascent protein modification by the NME enzymes. This model further predicts additional layers of control over NME that await to be tested, such as changes in the cellular level or activity of PDF during environmental changes (55, 56), SRP-induced inhibition of deformylation on nascent chains with TMDs or signal sequences close to the N terminus (26), and diverse factors that regulate translation elongation, including codon usage, translation stall sequences, and small molecule ligands that regulate this stalling (57–59). Importantly, quantitative kinetic measurements coupled to computational modeling under physiologically relevant conditions provide a powerful approach to elucidate how cotranslational protein biogenesis occurs in the crowded and complex cellular environment, and may be useful for understanding other biochemical pathways.
Materials and Methods
Material.
Recombinant PDF, MAP, and MAP mutants were expressed and purified from E. coli following published procedures (16, 60). RNCs were generated by in vitro translation in E. coli S30 extracts as described previously (49). Detailed purification procedures for proteins and RNCs are described in SI Appendix, Supplementary Methods.
Enzymatic Assays.
Pre–steady-state kinetic measurements were carried out under single-turnover condition using an RQF-3 Quench-flow instrument (KinTek) in assay buffer [50 mM Hepes-KOH, 150 mM KOAc, 10 mM Mg(OAc)2, and 1 mM TCEP, pH 7.5] with 0.1 mM CoCl2 at room temperature. Cotranslational NME assays were performed by coupled in vitro translation of reporter proteins in S30 extracts containing [35S]methionine at 30 °C for 30 min, and were analyzed by SDS/PAGE and autoradiography. Details of the procedure and data analysis are described in SI Appendix, Supplementary Methods.
Fluorescence Measurements.
All fluorescent measurements were carried out at room temperature in assay buffer containing 0.1 mM CoCl2. Emission spectra and equilibrium titrations were measured on a Fluorolog-3 spectrofluorometer (HORIBA). The association rate constant (kon) of the RNC–MAP complex was measured on a SF-2004 stopped-flow apparatus (KinTek). Details of the procedure and data analysis are described in SI Appendix, Supplementary Methods.
Simulation of Cotranslational NME.
A kinetic framework was constructed to describe the cotranslational NME of an n-residue nascent protein under single-turnover condition (Fig. 4A). Differential equations were constructed based on the framework, and a MATLAB script was used to simulate the time-dependent concentration of each species using the numerical integration method. Details of the model are described in SI Appendix, Supplementary Methods.
Data Availability.
Materials are available on request.
Supplementary Material
Acknowledgments
We thank T. F. Miller, M. H. Zimmer, and Y. L. Ni for critical advice on the simulation and T. F. Miller, A. Varshavsky, and members of the S.-o.S. laboratory for discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM078024 and a grant from the Weston Havens Foundation (to S.-o.S.) and a Think Global Education Trust Fellowship from Taiwan (to C.-I.Y.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All data discussed in the paper will be made available to readers.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912264116/-/DCSupplemental.
References
- 1.Kramer G., Boehringer D., Ban N., Bukau B., The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Kramer G., Shiber A., Bukau B., Mechanisms of cotranslational maturation of newly synthesized proteins. Annu. Rev. Biochem. 88, 337–364 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Bornemann T., Holtkamp W., Wintermeyer W., Interplay between trigger factor and other protein biogenesis factors on the ribosome. Nat. Commun. 5, 4180 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Bhakta S., Akbar S., Sengupta J., Cryo-EM structures reveal relocalization of MetAP in the presence of other protein biogenesis factors at the ribosomal tunnel exit. J. Mol. Biol. 431, 1426–1439 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Brannigan J. A., et al. , A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378, 416–419 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Kim J. H., Krahn J. M., Tomchick D. R., Smith J. L., Zalkin H., Structure and function of the glutamine phosphoribosylpyrophosphate amidotransferase glutamine site and communication with the phosphoribosylpyrophosphate site. J. Biol. Chem. 271, 15549–15557 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Bearne S. L., Active site-directed inactivation of Escherichia coli glucosamine-6-phosphate synthase. Determination of the fructose 6-phosphate binding constant using a carbohydrate-based inactivator. J. Biol. Chem. 271, 3052–3057 (1996). [PubMed] [Google Scholar]
- 8.Varshavsky A., The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varshavsky A., N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U.S.A. 116, 358–366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen K. T., Kim J.-M., Park S.-E., Hwang C.-S., N-terminal methionine excision of proteins creates tertiary destabilizing N-degrons of the Arg/N-end rule pathway. J. Biol. Chem. 294, 4464–4476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shemorry A., Hwang C.-S., Varshavsky A., Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell 50, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang C. S., Shemorry A., Varshavsky A., N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon J. I., Duronio R. J., Rudnick D. A., Adams S. P., Gokel G. W., Protein N-myristoylation. J. Biol. Chem. 266, 8647–8650 (1991). [PubMed] [Google Scholar]
- 14.Giglione C., Fieulaine S., Meinnel T., N-terminal protein modifications: Bringing back into play the ribosome. Biochimie 114, 134–146 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Hu Y. J., Wei Y., Zhou Y., Rajagopalan P. T., Pei D., Determination of substrate specificity for peptide deformylase through the screening of a combinatorial peptide library. Biochemistry 38, 643–650 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Frottin F., et al. , The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics 5, 2336–2349 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Ragusa S., Mouchet P., Lazennec C., Dive V., Meinnel T., Substrate recognition and selectivity of peptide deformylase. Similarities and differences with metzincins and thermolysin. J. Mol. Biol. 289, 1445–1457 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Xiao Q., Zhang F., Nacev B. A., Liu J. O., Pei D., Protein N-terminal processing: Substrate specificity of Escherichia coli and human methionine aminopeptidases. Biochemistry 49, 5588–5599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solbiati J., Chapman-Smith A., Miller J. L., Miller C. G., Cronan J. E. Jr, Processing of the N termini of nascent polypeptide chains requires deformylation prior to methionine removal. J. Mol. Biol. 290, 607–614 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Hirel P. H., Schmitter M. J., Dessen P., Fayat G., Blanquet S., Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U.S.A. 86, 8247–8251 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boissel J. P., Kasper T. J., Bunn H. F., Cotranslational amino-terminal processing of cytosolic proteins. Cell-free expression of site-directed mutants of human hemoglobin. J. Biol. Chem. 263, 8443–8449 (1988). [PubMed] [Google Scholar]
- 22.Bienvenut W. V., Giglione C., Meinnel T., Proteome-wide analysis of the amino terminal status of Escherichia coli proteins at the steady-state and upon deformylation inhibition. Proteomics 15, 2503–2518 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Meinnel T., Giglione C., Tools for analyzing and predicting N-terminal protein modifications. Proteomics 8, 626–649 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Martinez A., et al. , Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8, 2809–2831 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Sandikci A., et al. , Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding. Nat. Struct. Mol. Biol. 20, 843–850 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Ranjan A., Mercier E., Bhatt A., Wintermeyer W., Signal recognition particle prevents N-terminal processing of bacterial membrane proteins. Nat. Commun. 8, 15562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raue U., Oellerer S., Rospert S.; Association of Protein Biogenesis Factors at the Yeast Ribosomal Tunnel Exit Is Affected by the Translational Status and Nascent Polypeptide Sequence , Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 282, 7809–7816 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Fujii K., Susanto T. T., Saurabh S., Barna M., Decoding the function of expansion segments in ribosomes. Mol. Cell 72, 1013–1020.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bingel-Erlenmeyer R., et al. , A peptide deformylase-ribosome complex reveals mechanism of nascent chain processing. Nature 452, 108–111 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Akopian D., Shen K., Zhang X., Shan S. O., Signal recognition particle: An essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber D., et al. , SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol. Cell 41, 343–353 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Singh R., et al. , Cryo-electron microscopic structure of SecA protein bound to the 70S ribosome. J. Biol. Chem. 289, 7190–7199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawat S., Zhu L., Lindner E., Dalbey R. E., White S. H., SecA drives transmembrane insertion of RodZ, an unusual single-span membrane protein. J. Mol. Biol. 427, 1023–1037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S., Yang C.-I., Shan S. O., SecA mediates cotranslational targeting and translocation of an inner membrane protein. J. Cell Biol. 216, 3639–3653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deuerling E., Schulze-Specking A., Tomoyasu T., Mogk A., Bukau B., Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400, 693–696 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Fedyukina D. V., Cavagnero S., Protein folding at the exit tunnel. Annu. Rev. Biophys. 40, 337–359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beck K., Wu L. F., Brunner J., Müller M., Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19, 134–143 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh E., et al. , Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147, 1295–1308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu S.-Q., Peske F., Wieden H.-J., Rodnina M. V., Wintermeyer W., The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA 9, 566–573 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaffitzel C., et al. , Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature 444, 503–506 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Ferbitz L., et al. , Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431, 590–596 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Ariosa A., Lee J. H., Wang S., Saraogi I., Shan S. O., Regulation by a chaperone improves substrate selectivity during cotranslational protein targeting. Proc. Natl. Acad. Sci. U.S.A. 112, E3169–E3178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yap M.-N., Bernstein H. D., The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell 34, 201–211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gottesman S., Roche E., Zhou Y., Sauer R. T., The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12, 1338–1347 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D’souza V. M., Holz R. C., The methionyl aminopeptidase from Escherichia coli can function as an iron(II) enzyme. Biochemistry 38, 11079–11085 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Fersht A., “Measurement and magnitude of individual rate constants” in Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding, Julet M. R., Hadler G. L., Eds. (W. H. Freeman, ed. 6, 1998), pp. 158–167. [Google Scholar]
- 47.Lowther W. T., Zhang Y., Sampson P. B., Honek J. F., Matthews B. W., Insights into the mechanism of Escherichia coli methionine aminopeptidase from the structural analysis of reaction products and phosphorus-based transition-state analogues. Biochemistry 38, 14810–14819 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Watterson S. J., Mitra S., Swierczek S. I., Bennett B., Holz R. C., Kinetic and spectroscopic analysis of the catalytic role of H79 in the methionine aminopeptidase from Escherichia coli. Biochemistry 47, 11885–11893 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saraogi I., Zhang D., Chandrasekaran S., Shan S. O., Site-specific fluorescent labeling of nascent proteins on the translating ribosome. J. Am. Chem. Soc. 133, 14936–14939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okura A., Kinoshita T., Tanaka N., Formation of fusidic acid-G factor-GDP-ribosome complex and the relationship to the inhibition of GTP hydrolysis. J. Antibiot. (Tokyo) 24, 655–661 (1971). [DOI] [PubMed] [Google Scholar]
- 51.Dai X., et al. , Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2, 16231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Brien E. P., Vendruscolo M., Dobson C. M., Prediction of variable translation rate effects on cotranslational protein folding. Nat. Commun. 3, 868 (2012). [DOI] [PubMed] [Google Scholar]
- 53.O’Brien E. P., Vendruscolo M., Dobson C. M., Kinetic modelling indicates that fast-translating codons can coordinate cotranslational protein folding by avoiding misfolded intermediates. Nat. Commun. 5, 2988 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Ciryam P., Morimoto R. I., Vendruscolo M., Dobson C. M., O’Brien E. P., In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc. Natl. Acad. Sci. U.S.A. 110, E132–E140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Somerville G. A., et al. , Synthesis and deformylation of Staphylococcus aureus δ-toxin are linked to tricarboxylic acid cycle activity. J. Bacteriol. 185, 6686–6694 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkins J. C., Homer K. A., Beighton D., Altered protein expression of Streptococcus oralis cultured at low pH revealed by two-dimensional gel electrophoresis. Appl. Environ. Microbiol. 67, 3396–3405 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodnina M. V., The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci. 25, 1390–1406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buskirk A. R., Green R., Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seip B., Innis C. A., How widespread is metabolite sensing by ribosome-arresting nascent peptides? J. Mol. Biol. 428, 2217–2227 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Ragusa S., Blanquet S., Meinnel T., Control of peptide deformylase activity by metal cations. J. Mol. Biol. 280, 515–523 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials are available on request.