Significance
Although amyloids are the hallmarks for several neurodegenerative diseases, it is becoming evident that small oligomers are the toxic species leading to disease rather than the amyloids themselves. The vital first step of amyloid formation, primary nucleation, is difficult to monitor, making it relatively underexplored. Here we combine aggregation assays and cutting-edge native mass spectrometry to study the recently discovered and very highly conserved protein SERF in its unusual ability to accelerate primary nucleation of disease-related amyloid-prone proteins. We find that SERF exhibits a high degree of plasticity and forms fuzzy, highly extended complexes with amyloid-prone proteins. Our findings suggest that proteins are much more structurally diverse in the early stages of amyloid formation than previously assumed.
Keywords: protein folding, Alzheimer’s, chaperone
Abstract
The assembly of small disordered proteins into highly ordered amyloid fibrils in Alzheimer’s and Parkinson’s patients is closely associated with dementia and neurodegeneration. Understanding the process of amyloid formation is thus crucial in the development of effective treatments for these devastating neurodegenerative diseases. Recently, a tiny, highly conserved and disordered protein called SERF was discovered to modify amyloid formation in Caenorhabditis elegans and humans. Here, we use kinetics measurements and native ion mobility-mass spectrometry to show that SERF mainly affects the rate of primary nucleation in amyloid formation for the disease-related proteins Aβ40 and α-synuclein. SERF’s high degree of plasticity enables it to bind various conformations of monomeric Aβ40 and α-synuclein to form structurally diverse, fuzzy complexes. This structural diversity persists into early stages of amyloid formation. Our results suggest that amyloid nucleation is considerably more complex than age-related conversion of Aβ40 and α-synuclein into single amyloid-prone conformations.
As the human population ages, the number of people affected by age-related diseases including Alzheimer’s disease (AD) and Parkinson’s disease (PD) is poised to increase from the grim total of ∼40 million worldwide who currently suffer from AD to over 100 million by 2050 (1, 2). In the United States, 10% of people 65 y or older and 32% of those 85 y or older suffer from Alzheimer’s dementia (3). About 5% of people over 85 y currently suffer from PD. No effective treatments or cures for either of these devastating diseases are currently available. AD and PD show similar phenotypic hallmarks: In AD, amyloid plaques assembled from the amyloid-β protein (usually Aβ40 and Aβ42) are visible in the brains of AD sufferers upon autopsy; in PD, α-synuclein forms somewhat similar aggregates into Lewy bodies in the brains of PD patients. In both cases, the small intrinsically disordered proteins amyloid-β and α-synuclein self-assemble into highly ordered amyloid fibrils.
The mechanism by which the self-assembly of Aβ peptides or α-synuclein dysregulates protein homeostasis and leads to the neurotoxicity found in AD and PD patients is under intense investigation. Prior studies have shown that the amyloid formation pathway is composed of multiple steps (4, 5). The initial step is primary nucleation. In this crucial but mysterious step, the monomeric peptide undergoes a poorly characterized conformational change that results in the formation of amyloid nuclei. The rate of primary nucleation is slow due to the high energy barrier for nuclei formation, and this generates a lag in amyloid formation. In in vitro amyloid formation experiments, a long lag phase occurs before amyloids become detectable via thioflavin T (ThT) fluorescence. In patients, this slow primary nucleation step is associated with the late onset of these diseases (6).
It is becoming evident that the toxic Aβ or α-synuclein species that lead to symptoms in AD or PD patients are more likely to be smaller oligomers than the large fibrils that the ThT assay reports in refs. 7–9. The flat line during the lag phase in a ThT assay therefore embodies a number of active conformational changes, including the assembly of nuclei that then go on to form mature fibrils. It is thus misleading to consider this “lag” phase as a period waiting for the reaction to start; it should instead be viewed as an active phase of amyloid formation, crucial because it both generates neurotoxic species but unfortunately is understudied because it is invisible to amyloid-specific dyes like ThT (10).
After some fibrils have been generated, Aβ monomers can attach to the end of fibrils to elongate the existing fibrils in an elongation reaction, or they can attach to the side of these fibrils to form new nuclei in a process known as secondary nucleation. These new nuclei can dissociate and elongate into new fibrils. Secondary nucleation creates a positive feedback loop that drives the autocatalytic nature of amyloid formation and is thought to be the dominant means of generating new Aβ fibrils (4).
The formation of amyloid fibrils is thought to tax the protein quality-control systems in aging tissues and disrupt the delicate equilibrium of protein homeostasis that is required for cells to be healthy. Cells respond to changes in protein homeostasis, in part, through a network of molecular chaperones, which are responsible for keeping the proteome stable and preventing cytotoxic protein aggregation (11, 12). Chaperones have been shown to influence a number of different steps of amyloid formation in vitro. For example, the Bri2 BRICHOS domain inhibits the secondary nucleation step in Aβ42 amyloid formation, whereas the chaperone DNAJB6 inhibits the rate of primary nucleation in Aβ42 amyloid formation (13–16). Chaperones Hsp70 and Hsp 90 also inhibit Aβ amyloid formation (17).
Interestingly, some molecules have actually been reported to accelerate amyloid formation. For example, polyions such as polyphosphate or heparin have long been known to accelerate the formation of amyloid fibrils; heparin, for instance, is commonly added to initiate the amyloid formation process in vitro (18, 19).
A very small 68-amino acid intrinsically disordered protein called MOAG-4 (so named due to its ability to modify aggregation [MOAG]) was recently discovered as an in vivo enhancer of amyloid formation in Caenorhabditis elegans. The deletion of MOAG-4, the gene for a protein which is well-conserved from Saccharomyces cerevisiae to Homo sapiens (SI Appendix, Fig. S1), leads to fewer amyloid puncta in a C. elegans model of Huntington’s disease (20). MOAG-4 and its human homolog the small EDRK-rich factor 1 (SERF1a) have been shown to accelerate amyloid formation of a broad range of amyloidogenic proteins in vitro (21). The detailed mechanism whereby MOAG-4 and its homologs influence amyloid formation is not yet clear.
One intriguing possibility is that by accelerating amyloid formation, MOAG-4 homologs rush amyloid-prone proteins through the steps that involve neurotoxic intermediates, speeding the transformation of these proteins into amyloids, which have been postulated to represent a relatively nontoxic storage form. The small size and biophysical accessibility of S. cerevisiae SERF, Aβ40, and α-synuclein present a unique opportunity to investigate the influence of a host factor such as MOAG-4 on the critical early steps of amyloid formation.
We therefore investigated the MOAG-4/SERF homolog YDL085C-A from S. cerevisiae, hereafter termed ScSERF, in order to gain mechanistic insights into how this family of proteins accelerates amyloid formation of Aβ40 and α-synuclein. We found that ScSERF is efficient in accelerating amyloid formation at substoichiometric concentrations. Using native ion mobility-mass spectrometry (IM-MS), we show that ScSERF engages in fuzzy complexes with Aβ40 and α-synuclein. These complexes keep the features of intrinsically disordered proteins in that they are structurally heterogeneous and also exist in an ensemble of stoichiometric assemblies. That ScSERF engages in fuzzy complexes while accelerating amyloid formation implies that the vital process of amyloid nucleation is more complex than frequently drawn simple models that involve the transition of proteins from a soluble form to a single amyloid-prone conformation. We propose a model based on structural and kinetic data in which ScSERF provides a binding surface that increases the pool of conformations that Aβ40 and α-synuclein can explore, which leads to the observed acceleration of primary amyloid nucleation.
Results
ScSERF Accelerates Amyloid Formation of Aβ40 and α-Synuclein.
The NMR structure of the amyloid accelerator MOAG-4, as determined at 4 °C, shows a significant amount of helical content (22). To test how much of this helical content is maintained at higher and more physiologically relevant temperatures, we monitored the secondary structure of ScSERF as a function of temperature by circular dichroism (SI Appendix, Fig. S2A). In agreement with the published NMR structure, ScSERF shows considerable α-helical content at low temperatures. However, as the temperature increases, ScSERF’s α-helical content decreases from about 24% of the spectra at 4 °C to 4% at 37 °C (23, 24), the temperature where we performed all of our in vitro amyloid assays. ScSERF is mostly disordered under these conditions, suggesting that this is the relevant structural state of this protein as it acts to influence amyloid formation in vivo.
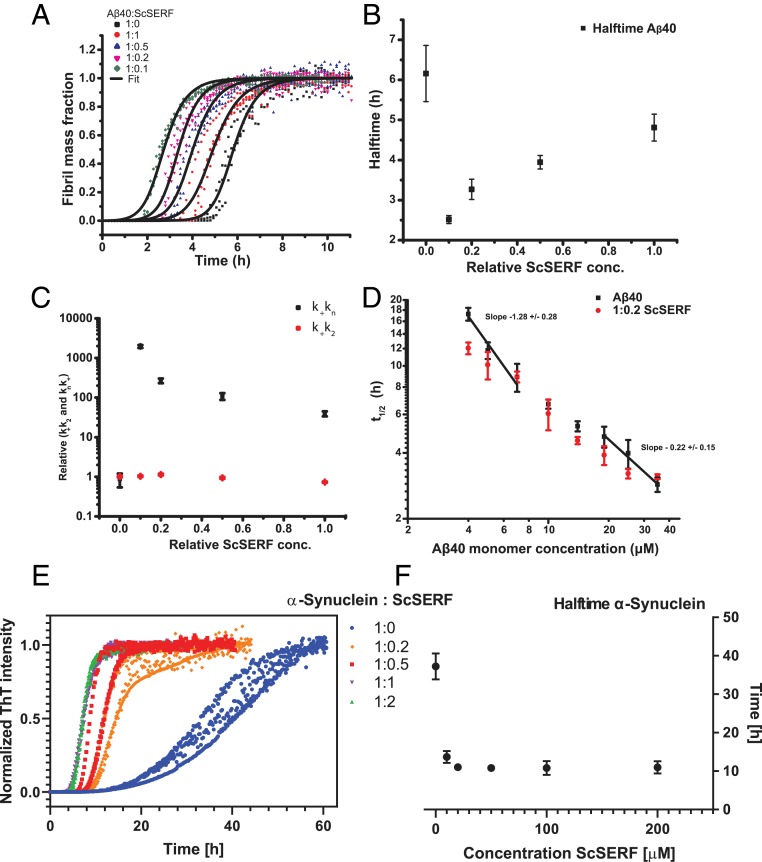
We next investigated how ScSERF affects the process of amyloid formation of Aβ40 and α-synuclein by using ThT fluorescence as a readout. ThT is an amyloid-specific dye that increases in fluorescence upon binding to amyloid fibrils, making it a reagent that is commonly used to monitor the kinetics of amyloid formation on a macroscopic scale (13). We found that the fibrillation process follows sigmoidal behavior at all ScSERF concentrations tested. Notably, ScSERF is very efficient in accelerating amyloid formation even at low substoichiometric concentrations. For instance, the half-time of amyloid formation for Aβ40 was reduced from 6 h to 2.6 h in the presence of a 1:0.1 ratio of Aβ40 to ScSERF (Fig. 1 A and B). We note that Aβ40 fibrillation performed in the presence of a large excess of ScSERF actually displayed slower fibrillation kinetics and a lower final ThT signal than that performed with substoichiometric ratios of ScSERF (SI Appendix, Fig. S2 B and C). Inspection of the fibrils at the end of the reaction by transmission electron microscopy (TEM) revealed the presence of amorphous aggregates in addition to the amyloid fibrils, which were not observable at substoichiometric ratios of ScSERF (SI Appendix, Fig. S2D). This suggests that some off-pathway amorphous aggregation of Aβ40 occurs when excess ScSERF concentrations are used, lowering the total Aβ40 concentration able to form ThT-detectable fibrils and resulting in the observed slower fibrillation kinetics. Whether or not these observations are of physiological significance is dependent on what concentration of SERF is present in vivo. For ease of comparison, protein concentrations in systematic databases are commonly reported in parts per million (ppm). In vivo ScSERF is present at 143 ppm in S. cerevisiae (25), human SERF1a is present at 60 ppm, human SERF2 is present at 318 ppm, and the C. elegans homolog MOAG-4 is expressed at 387 ppm (26). We see clear inhibition of Aβ40 fibrillar formation only at ScSERF concentrations of 125 μM (SI Appendix, Fig. S2B), which corresponds to 994 ppm in our assay. Thus, we consider the concentrations where we see slower fibrillation kinetics and amorphous aggregation to be unphysiologically high, particularly in yeast, and as a result we have excluded these data from further analysis.
Fig. 1.
ScSERF accelerates amyloid kinetics. (A) Fibril mass fraction of 25 μM Aβ40 incubated in the absence (black) or presence of different concentrations of ScSERF. Data were fit with a multistep secondary nucleation model, with k+kn combined primary nucleation rate constant and k+k2 combined secondary nucleation rate as free fitting parameters (SI Appendix, Table S1). (B) Half-times of Aβ40 plotted against the indicated ratios of ScSERF. Assays were conducted under quiescent conditions at 37 °C (20 mM NaPI, 200 μM EDTA, pH 7.4, and 25 μM ThT). (C) Dependencies of the relative combined rate constants on ScSERF ratio obtained from fitting the data. (D) Double logarithmic plot of average half-time of Aβ40 aggregation in the absence (black) or presence of 1:0.2 Aβ40:ScSERF (red). (E) Normalized ThT fluorescence of 100 μM α-synuclein incubated in the absence (blue) or presence of different concentrations of ScSERF. (F) Half-times of α-synuclein plotted against ScSERF concentrations. Assays were conducted with constant shaking at 37 °C (20 mM NaPI, 50 mM NaCl, 200 μM EDTA, pH 7.4, and 25 μM ThT). The data in A, B, and F present means and standard derivations of 3 or 4 replicates of the experiment, which have been repeated at least 3 times with qualitatively similar results. The errors in C present fitting errors.
Meisl et al. (5) described the aggregation mechanism for Aβ40 as a multistep secondary nucleation process. A recently developed approach allowed us to fit the Aβ40 amyloid kinetics and derive a set of combined microscopic rate constants that describe the individual steps, where kn is the rate of primary nucleation, k2 is the rate of secondary nucleation, and k+ is the rate constant for fibril elongation (5, 27).
A double logarithmic plot of half-time vs. monomer concentration gives useful insights into the underlying mechanism of the amyloid kinetic reaction (5, 28). Others have shown that for Aβ40 amyloid formation the plot has a positive curvature, which is indicative of the presence of a nucleation step that is saturated at high monomer concentrations (5). In our hands, Aβ40 aggregation kinetics in the absence of ScSERF exhibited the same previously observed positive curvature, which is a good sign that our aggregation kinetics fits the previously published mechanism (Fig. 1D and SI Appendix, Fig. S2E). In the presence of a substoichiometric concentration of ScSERF (1:0.2), the half-time of the Aβ40 reaction was reduced, consistent with previous reports of the ability of SERF homologs to accelerate amyloid formation (21). Notably both with and without added ScSERF, the half-time plot retained its positive curvature, indicating that the presence of ScSERF does not change the underlying mechanism for Aβ40 aggregation even though it accelerates a step in the aggregation process (Fig. 1D and SI Appendix, Fig. S2E).
Aβ40 aggregation in the presence of varying ScSERF concentrations was fitted to the multistep secondary nucleation process that Aβ40 is known to undergo, as was described by the Knowles group (5, 29). In the case of an unseeded aggregation reaction, the system depends only on the combination of the rate constants for primary nucleation, k+kn, and secondary nucleation, k+k2, not on the individual rate constants (5). The fit of our data for a range of ScSERF to Aβ40 ratios with k+k2 and k+kn as free fitting parameters revealed that the rate constant that describes the primary nucleation process, k+kn, is accelerated significantly in the presence of ScSERF, whereas the rate constant for secondary nucleation, k+k2, is only very slightly affected (Fig. 1 A and C).
ScSERF also accelerates the half-time of α-synuclein aggregation. The degree of acceleration, as reflected by the differences in half-times, was higher for α-synuclein than for Aβ40. The reaction half-time for α-synuclein was reduced from 37 h to 10 h in the presence of 1:0.5 α-synuclein:ScSERF (Fig. 1 E and F). We found ScSERF’s effectiveness in accelerating α-synuclein amyloid formation to be equivalent to that reported for its homologs SERF1a and MOAG-4 (21, 22). Unlike for Aβ40 where very high, evidently nonphysiological concentrations of ScSERF could cause some inhibition of aggregation, at no ratio tested could ScSERF inhibit α-synuclein aggregation (Fig. 1 E and F).
Notably, the α-synuclein samples in these experiments were subject to constant shaking at 300 rpm. This was done to ensure that aggregation occurred within a reasonable timescale. However, shaking also generates amyloid fragments, making it difficult to determine the rate constant for later steps in the α-synuclein aggregation process. To circumvent this issue and to distinguish between the process of primary nucleation, later nucleation, and elongation events, we decided to perform a self-seeded amyloid aggregation experiment.
ScSERF Has No Effect on Self-Seeded Amyloid Kinetics.
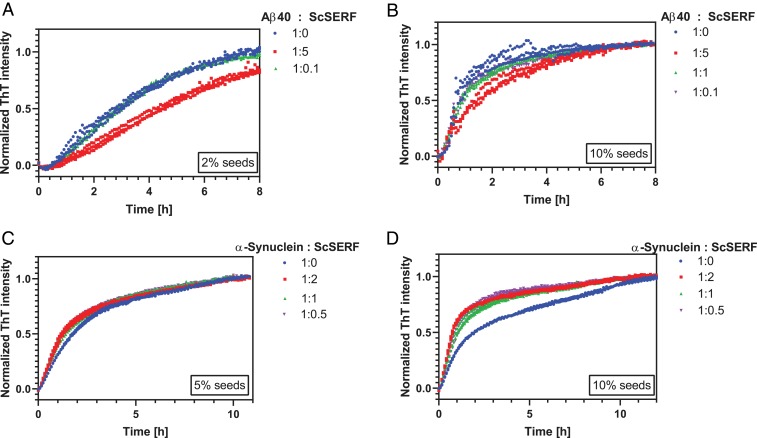
Lag phases caused by primary nucleation can be bypassed by seeding the fibrillation reaction with preformed oligomers of fibrils (30). In a seeded reaction, secondary nucleation and elongation can dominate the formation of new fibrils, depending on the concentration of seeds added. We can therefore probe whether ScSERF predominantly accelerates the pivotal primary nucleation step by simply adding seeds to the Aβ40 or α-synuclein amyloid reaction. If ScSERF only accelerates primary nucleation, we should be able to substitute for ScSERF by adding seeds; that is, we would expect to see a minimal effect upon adding ScSERF to self-seeded Aβ40 or α-synuclein experiments.
In our self-seeded experiments, the lag phase vanished. For both Aβ40 and α-synuclein self-seeded reactions, addition of ScSERF had no noticeable effect on the kinetics of amyloid formation (Fig. 2). This result strongly supports our hypothesis that ScSERF primarily affects the primary nucleation process for Aβ40 and α-synuclein amyloid formation.
Fig. 2.
ScSERF has minimal effect on self-seeded amyloid kinetics. (A) Normalized ThT fluorescence of 25 μM Aβ40 self-seeded with 0.5 μM preformed Aβ40 fibrils (2% seeds) incubated in the absence (blue) or presence of different concentrations of ScSERF. (B) Same as in A, but self-seeded with 2.5 μM preformed Aβ40 fibrils (10% seeds). (C) Self-seeding assay with 100 μM α-synuclein and 5% seeds. (D) Self-seeding assay with 100 μM α-synuclein and 10% seeds. In all experiments, assays were conducted under quiescent conditions at 37 °C (20 mM NaPI, 50 mM NaCl, 200 μM EDTA, pH 7.4, and 25 μM ThT).
To determine whether ScSERF affects fiber morphology, samples taken at the end of the reaction were examined by TEM. In the presence of equimolar or lower concentrations of ScSERF, the overall fiber morphology of Aβ40 and α-synuclein were, at least superficially, not greatly affected by the addition of ScSERF (SI Appendix, Fig. S3). In the self-seeded experiments, we observed some amorphous aggregation that was not evident in the absence of added seeds. We hypothesize that these amorphous aggregates could have resulted from the sonication of preformed fibrils or from secondary aggregation pathways active in the seeded reactions.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of the insoluble (P) and soluble (S) fractions at the end of the experiment showed that Aβ40 and α-synuclein fibrils were overwhelmingly in the insoluble fraction (P), whereas most of the ScSERF protein was in the soluble fraction (S) (SI Appendix, Fig. S4 C and E). These results are similar to those previously obtained using human SERF1a and α-synuclein (21). Notably, however, a small fraction of ScSERF was detected in the insoluble fraction, and this amount did not change with the ScSERF concentration, but instead appeared to be dependent on the total Aβ40 concentration. One possible explanation for this could be that a small number of ScSERF binding sites exist on amyloid fibrils, consistent with the observation that more ScSERF is found in the pellet at higher Aβ40 concentrations.
Given that a small amount of ScSERF was detected in the insoluble fraction containing the Aβ40 or α-synuclein amyloid fibrils, we next investigated whether ScSERF can bind to preformed amyloid fibrils. To accomplish this, we titrated AlexaFluor532-labeled ScSERF (AF532 ScSERF) with preformed fibrils and monitored changes in fluorescence anisotropy. An increase in fluorescence anisotropy would indicate an increase in the complex formed between the labeled protein and the added preformed fibrils (31). The addition of preformed Aβ40 fibrils resulted in an increase in anisotropy of AF532 ScSERF, consistent with other indications that ScSERF can bind to amyloid fibrils (SI Appendix, Fig. S4 A and B). Moreover, addition of unlabeled ScSERF to the AF532 ScSERF–Aβ40 or AF532 ScSERF–α-synuclein fibril complexes decreased their fluorescence anisotropy signal, indicating that ScSERF binding to fibrils is reversible (SI Appendix, Fig. S4 F and G). The interaction between AF532 ScSERF and Aβ40 fibrils could also be competed using unlabeled ScSERF even if AF532 ScSERF was present during the formation of the Aβ40 amyloid fibrils. This result indicates that ScSERF binds to Aβ40 fibrils but is not irreversibly integrated into the structure of the fibrils (SI Appendix, Fig. S4D).
Merle et al. (32) found that human SERF1a colocalizes with puncta of α-synuclein in human SH-SY5Y neuroblastoma cell experiments. Our observations that ScSERF can reversibly associate with preformed fibrils without integrating into their structures is consistent with these in vivo results.
ScSERF Forms Stoichiometrically Heterogeneous Complexes with Aβ40 and α-Synuclein.
Our kinetic studies indicate that ScSERF affects the lag time of Aβ40 and α-synuclein amyloid formation, as measured by ThT fluorescence. ThT primarily binds to β-sheet–rich conformations, like the cross β-sheet conformation present in proteins that compose amyloid fibrils. In some cases, like in the case of β-2-microglobulin, it can report on β-sheet–rich intermediate states (33); however, in the case of Aβ40/42 and α-synuclein no ThT-reactive stable intermediates have been observed during the lag time of the fibrillation process so we consider that, at least in the cases of Aβ40/42 and α-synuclein, ThT fluorescence is not an ideal way to monitor the critical nucleation step. We find it particularly unfortunate that ThT binding fails to monitor the critical conformational change(s) that initiate amyloid formation or the transient development of cytotoxic oligomers that occurs early in amyloidogenic processes (6, 34).
The very small size of the amyloid-modifying host factor ScSERF, along with its biophysically amenable properties, suggested that it might be ideally suited to provide detailed information about how host factors can affect the initial stages of amyloid formation. To investigate the influence of ScSERF on the early events in Aβ40 amyloid formation, we used native mass spectrometry (MS), an analytical technique capable of rapidly analyzing proteins in the gas phase while retaining native-like structure and preserving noncovalent protein–protein interactions that occur in solution. Ion Mobility (IM), a gas-phase technique that separates ions based on their orientationally averaged collision cross-section (CCS) and charge coupled to native MS has become a valuable structural MS method to study proteins and protein complexes (35). In IM separation, ions move through a neutral drift gas under the influence of a weak electric field. Larger ions collide with the drift gas more frequently than smaller ions, thus traversing the IM cell in a longer drift time. In native IM-MS experiments, structural information about protein analytes can be obtained from charge-state distributions (CSDs) and CCS distributions. Globular proteins usually yield narrow and low CSDs and CCS distributions. However, intrinsically disordered proteins have previously been shown to exhibit a broader and higher range of CSDs and CCS distributions (36, 37).
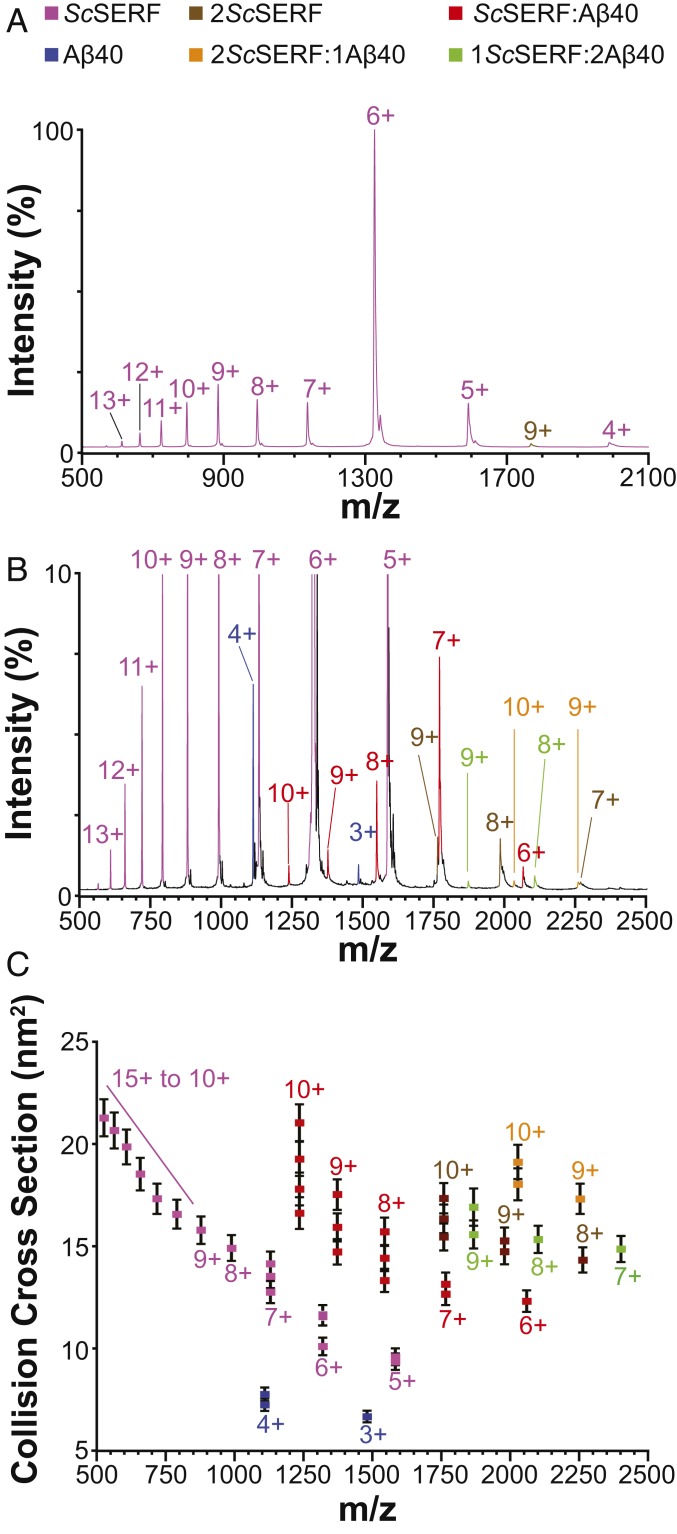
IM-MS experiments indicate that ScSERF is an intrinsically disordered protein with flexible structural properties. We observed a broad CSD (11 charge states) from 5+ to 15+, which is indicative of the various structures of ScSERF present in solution prior to the nanoelectrospray ionization (nESI) process (Fig. 3 A and B). The IM arrival time distributions (ATDs) of the 11 charge states portrayed an even more disordered ensemble of ScSERF structures. By fitting the ATDs to multiple Gaussian functions, we were able to define discrete structural conformers of ScSERF that range in CCS from 9 to 22 nm2 (Fig. 3C). Whereas the higher charge states (8 to 15+) consist of mainly one structural population, the lower-charge-state ions (5 to 7+) have multiple ATDs, indicating the presence of multiple structural populations. For example, 3 distinct structural conformational families are observed for the 7+ ScSERF ion (Fig. 3C). The structural heterogeneity of ScSERF seems to be preserved in interactions with other intrinsically disordered proteins, as evidenced by the various stoichiometries of complexes observed containing ScSERF with Aβ40 and α-synuclein (Figs. 3B and 4A).
Fig. 3.
Analysis of ScSERF interacting with Aβ40 using nESI IM-MS. (A) Native mass spectrum of ScSERF at 10 μM. The measured monoisotopic mass of 7,945 ± 1 Da is in close agreement with the average sequence mass of 7,949 Da (UniProtKB ID Q3E7B7). The 4 to 13+ charge states of ScSERF monomer (magenta) and the 7+ to 9+ charge states of ScSERF dimer (brown) are labeled. (B) The 25 μM ScSERF and Aβ40 were incubated together. Samples were diluted to 10 μM each and analyzed by native IM-MS. The mass spectrum shows charge states corresponding to 6 different complexes as indicated by the color key at the top of the figure. The spectrum is cropped to 10% relative intensity to enlarge the low-intensity signals correlating to ScSERF:Aβ40 complexes that are suppressed by the high intensity of ScSERF. (C) The arrival time profile of each ion was extracted and fit to Gaussian functions. The Gaussian centroids are converted to CCSs and plotted vs. m/z. The error bars are least squares errors, which account for calibration error as well as the SD for at least 3 samples.
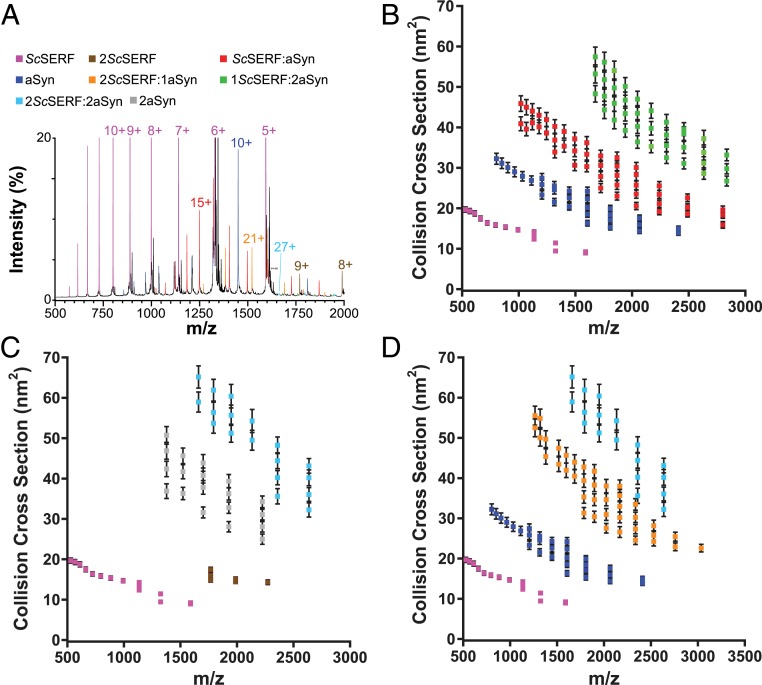
Fig. 4.
Analysis of ScSERF interacting with α-synuclein using nESI IM-MS. (A) The 25 μM ScSERF and α-synuclein (aSyn) were incubated together. Samples were diluted to 10 μM each and analyzed by native IM-MS. The mass spectrum shows highest intensity charge states corresponding to 8 different complexes as indicated in the color key at the top of the figure. The spectrum is cropped to 20% relative intensity to enlarge the low intensity signals correlating to ScSERF:α-synuclein complexes that are suppressed by the high intensity of ScSERF. The m/z range is cropped at 2,000 because signals corresponding to complexes beyond m/z = 2,000 are << 1% total intensity. These complexes can, however, be isolated and included for CCS analysis. The presence of these complexes is addressed in the CCS vs. m/z plots. (B–D) The arrival time profile of each ion was extracted and fit to Gaussian functions. The Gaussian centroids are converted to CCSs and plotted vs. m/z. The error bars are least squares error, which account for calibration error as well as the SD for at least 3 samples.
In reactions containing equimolar ScSERF with either Aβ40 or α-synuclein, we detected 1:1, 1:2, and 2:1 complexes of ScSERF:Aβ40 (Fig. 3C) and 1:1, 1:2, 2:1, and 2:2 complexes of ScSERF:α-synuclein (Fig. 4 B–D). The low micromolar dissociation constants (Kd) measured for the 1:1 ScSERF:Aβ40 complex by IM-MS (10 ± 2 μM) and by fluorescence anisotropy (32 ± 1.8 μM) allow us to estimate that under the reported experimental conditions Aβ40 should be 20 to 38% bound. The IM-MS data support this estimate, as we observe that ∼33% Aβ40 is bound in the 1:1 ScSERF:Aβ40 complex based on MS intensity quantitation. Similarly, the Kd measured for the 1:1 ScSERF:α-synuclein complex by fluorescence anisotropy was 9.5 ± 1.3 μM, in agreement with the MS data which indicate that the 1:1 complex of ScSERF:α-synuclein accounts for ∼40% of α-synuclein bound (estimated ∼39% based on Kd). Together these results indicate that ScSERF interacts weakly with at least 2 amyloidogenic proteins (SI Appendix, Fig. S5 A–C).
Typically, a power-law relationship exists between protein ion mass and CCS (38). However, some conformers of ScSERF are nearly 2- to 3-fold larger than expected for a 7,949-Da protein, demonstrating the abnormally extended nature of ScSERF structure (Fig. 3C). This tendency toward above-average CCS and broad CSDs is a common theme for most complexes that contain ScSERF. The 1:1 complexes of ScSERF:Aβ40 occupy extended conformations with CCSs ranging from 12 to 22 nm2 (Fig. 3C), and the 1:1 complexes of ScSERF:α-synuclein have CCSs in the range of 20 to 50 nm2 (Fig. 4 A and B). In both cases, the higher ranges of measured CCSs are nearly 2-fold above those expected for protein ions in the respective size ranges. The broad CSDs of the 1:1 complex of ScSERF and Aβ40 also indicate that the various conformers of ScSERF are capable of interacting with Aβ40, which typically only ionizes in 2 to 3 charge states (4+ to 2+) (39). The conformational heterogeneity seen in the 1:1 ScSERF:Aβ40 and 1:1 ScSERF:α-synuclein complexes is similar to that seen for ScSERF alone. This finding indicates that the structural heterogeneity of ScSERF likely dictates the observed structural heterogeneity of the 1:1 ScSERF:Aβ40 complex. In contrast, while the 1:1 complexes of ScSERF:α-synuclein are substantially more heterogeneous than ScSERF:Aβ40 complexes, they do not exhibit a gain of heterogeneity beyond that which was already observed for α-synuclein independently. This suggests that the preexisting structural heterogeneity of α-synuclein dictates the large CSDs and CCS distributions observed for ScSERF:α-synuclein complexes. The 1:2 and 2:1 complexes between ScSERF and Aβ40 or α-synuclein and 2:2 complexes of ScSERF:α-synuclein were also observed, and the trend of broad CSDs and CCS distributions remained consistent (Figs. 3C and 4 B and C).
Interestingly, the 1:2 and 2:1 ScSERF:Aβ40 complexes do not exhibit CSDs nearly as broad as those observed for the 1:1 complex, which suggests these complexes are less heterogeneous. However, the higher-order ScSERF:α-synuclein complexes (1:2, 2:1, and 2:2) continue to exhibit a high degree of structural heterogeneity as indicated by the broad CSDs and CCS distributions consistent with the 1:1 complex. Overall ScSERF:α-synuclein complexes are substantially more heterogeneous in stoichiometry, CSD, and CCS distribution in comparison to ScSERF:Aβ40 complexes, which we anticipate is due to the preexisting structural flexibility of α-synuclein.
The ScSERF:α-synuclein complexes are particularly interesting due to the presence of higher-stoichiometry complexes that were not observed between ScSERF and Aβ40. For example, ScSERF forms 2:2 complexes with α-synuclein, which range in CCS from 30 to 70 nm2 (Fig. 4D). Some of the larger 2:2 complexes have CCSs as large as those typically observed for antibodies, which are more than 3-fold larger in molecular weight (40, 41). This result demonstrates the degree to which ScSERF complexes adopt large extended conformations, often 2- to 3-fold greater than expected for ions of their size. Overall, the IM-MS experiments indicate that ScSERF is a highly disordered protein that interacts with other amyloidogenic proteins in various stoichiometries. These complexes occupy particularly extended structures, which might play a role in accelerating aggregation of the amyloidogenic proteins studied here.
Last, we also observed ScSERF dimers that have CCSs of 13 to 17 nm2. Whereas monomeric ScSERF exhibits large structural heterogeneity as demonstrated by the IM-MS data, dimeric ScSERF exists in a more homogenous structural population as indicated by the narrow CSDs and relatively compact CCS values (Fig. 3). These dimers are noticeably more compact than the ScSERF complexes containing Aβ40 or α-synuclein. These results suggest that some of the more extended conformers of ScSERF are more likely to interact with other intrinsically disordered proteins, resulting in the extended complexes observed, whereas some of the more compact forms of ScSERF might be prone to interacting with other ScSERF molecules to form dimers. The role of these ScSERF dimers is currently unknown. Some complexes of Aβ40 and α-synuclein were observed to be interacting with 2ScSERF; however, it is unclear whether these complexes were a result of Aβ40/α-synuclein interacting with a ScSERF dimer or 2 independent ScSERF proteins.
We further investigated if the distribution of any of the observed complexes changed during the early phase of Aβ40 amyloid formation. Therefore, we incubated an equimolar ratio of Aβ40:ScSERF under conditions identical to the kinetic measurements and analyzed samples every 30 min. The distribution of complexes remained constant up to the point where, due to formation of insoluble aggregates, all Aβ40 signal disappeared, and only ScSERF was detected by nESI (SI Appendix, Fig. S6A).
Additionally, we monitored the CCS populations of 1:1 ScSERF:Aβ40 complexes over time and observed that the general equilibrium between the complexes of varying CCSs remained mostly unchanged over the course of the experiment (SI Appendix, Fig. S6B).
The IM-MS data provide direct evidence of the interaction between ScSERF and 2 amyloidogenic proteins, Aβ40 and α-synuclein. The complexes are of varied stoichiometries and have extended structures with uncharacteristically large CCSs for proteins in this mass range. The complexity of the mixtures suggests that various conformers of ScSERF interact with various conformers of the amyloidogenic proteins. The broad charge state and CCS distributions of the various complexes indicate that these assemblies are flexible and dynamic in solution. Together, the IM-MS data provide insights into how ScSERF interacts with amyloidogenic proteins during the lag phase of aggregation.
Formation of Fuzzy Complexes Leads to Acceleration of Primary Nucleation.
Our kinetic observations together with our IM-MS data prompt us to propose a model in which early events that are driven by ScSERF binding lead to a change in the nucleation behavior of Aβ40 and α-synuclein, which substantially decreases the lag time of amyloid formation.
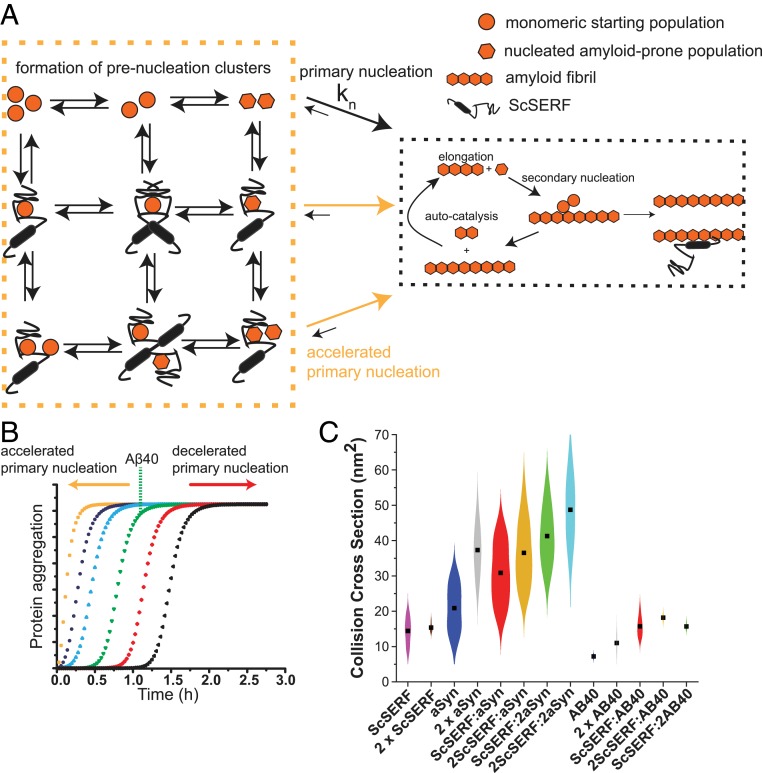
In a test of this model, we can simulate Aβ40 aggregation behavior based on the published kinetic mechanism for Aβ40 aggregation (SI Appendix, Fig. S7). Simulations are valuable because they allow one to visualize how the aggregation curves will respond to changes in the various rate constants for primary nucleation, elongation, or secondary nucleation. These simulations, and simple logic, show that a change in primary nucleation is effective in modulating the lag time of the aggregation curve (SI Appendix, Fig. S7). In contrast, changes in elongation or secondary nucleation mainly affect the slope of the sigmoidal aggregation curve, resulting in a change in the shape of the curve. By comparing the various simulations performed, it becomes evident that the profiles generated by changing the rate for primary nucleation best match our experimental ScSERF Aβ40 kinetic traces (Fig. 5B). This model fits well with our experimental observations. The high plasticity for ScSERF and the range of different stoichiometries in formed complexes with Aβ40 and α-synuclein are illustrated in the plots of the CCSs for all of the complexes we observe (Fig. 5C). A mixture of these diverse complexes is present in the early stages of amyloid formation. Taking all our data into account, we propose a model by which ScSERF acts as an accelerator for the rate-limiting step of primary nucleation, kn, whereby the monomeric disordered proteins Aβ40 and α-synuclein form aggregation-prone nuclei (Fig. 5A).
Fig. 5.
Schematic representation of ScSERF's effect on amyloid formation. (A) The Aβ40/α-synuclein amyloid formation pathway consists of primary nucleation, elongation, and secondary nucleation. During primary nucleation, Aβ40/α-synuclein forms its amyloid-prone conformation (hexagons) from the starting pool of disordered monomeric Aβ40 (circles). ScSERF engages in 3 different complexes with Aβ40 (1:1, 1:2, and 2:1) and in 4 different complexes with α-synuclein (1:1, 1:2, 2:1, and 2:2). In the presence of ScSERF, the primary nucleation step is accelerated by these binding events. (B) Simulation of 25 μM Aβ40 amyloid kinetics based on the published model; simulation with the published rate constants is shown in green (primary nucleation rate = 2 × 10−6, elongation rate = 3 × 105, and secondary nucleation rate = 3 × 103) (5). Increasing the primary nucleation rate (blue, purple, and orange plots) or decreasing it (red and black plots) changes the lag time, which matches our observed data in the presence of ScSERF. (C) Violin plots of the CCSs of the different ScSERF containing complexes observed in the native MS experiments. Black squares indicate the mean CCS of each complex. The kernel density distributions around the mean are a probability estimation of the number of complexes occupying that CCS bin based on the input set of experimental data.
Discussion
The spontaneous aggregation of Aβ40/42 or α-synuclein proteins into amyloid fibrils is a rather slow process in vivo, reflecting the age-dependent nature of the diseases that their amyloid formation is linked to—AD and PD. Understanding the nucleation process of amyloid formation is crucial to understanding these diseases and to eventually devising effective treatments. This is in part because primary nucleation is the rate-limiting step of the amyloid reaction and is required for the generation of small neurotoxic oligomers. Therefore, identification of factors that affect the rate of primary nucleation of amyloid-prone, disease-related proteins is of interest. The widely conserved protein MOAG-4/SERF is a host factor that is known to affect amyloid formation (20); we therefore decided to investigate how this factor exerts its influence in more detail.
Analysis of the kinetics of amyloid formation for Aβ40 or α-synuclein in the presence and absence of the yeast MOAG-4/SERF homolog ScSERF showed that addition of ScSERF led to a significant acceleration of amyloid formation, with ScSERF predominantly acting at the amyloid nucleation step. To arrive at this conclusion, we used the Knowles group’s framework to examine the macroscopic rate constants for Aβ40/42 kinetics assays (5, 27, 29). This analysis enabled us to describe in detail how ScSERF accelerates Aβ40 amyloid formation. The analysis revealed that ScSERF does not change the underlying mechanism of the multistep secondary nucleation process previously established for the aggregation of Aβ40. Using self-seeded aggregation assays, we independently confirmed that ScSERF does not affect elongation and secondary nucleation for both Aβ40 and α-synuclein. Our data revealed instead that ScSERF accelerates the rate of primary nucleation for Aβ40 by over 100-fold and also apparently acts on a similar step in α-synuclein aggregation, as indicated by a significant decrease in the α-synuclein lag phase.
There have been a few reports of compounds that can act generally to accelerate amyloid formation; these include the polyions heparin and polyphosphate. Heparin was found to integrate into α-synuclein fibrils and bind to them with a low micromolar Kd, and polyphosphate was found to accelerate amyloid formation of tau, Aβ42, and α-synuclein. Polyphosphate binds to fibrils and changes the fibril morphologies of α-synuclein fibrils (18, 19, 42). In contrast, ScSERF binds to the early-stage monomeric forms of Aβ40 and α-synuclein and acts on the primary nucleation step—a distinctively different mechanism from that hypothesized for polyions. Another difference from polyions is that ScSERF does not integrate into fibrils, although it does have the ability to reversibly bind to preformed Aβ40 and α-synuclein fibrils. In SH-SY5Y neuroblastoma cells, ScSERF was found to colocalize with aggregated puncta of α-synuclein, indicating that this binding could also take place in vivo (32).
ScSERF binds monomeric Aβ40 and α-synuclein with weak micromolar affinities in solution, raising the question as to how ScSERF can so effectively accelerate amyloid formation even when present in substoichiometric quantities. Notably, the fluorescence anisotropy approach that we used to measure binding affinities, like most equilibrium analysis techniques, samples an ensemble of populations and is therefore biased toward the most abundant conformer. Fortunately, the native MS method we employed allows us to detect low-abundance conformers that might be important for the dynamics and kinetic changes we observed. By applying native IM-MS, we found that ScSERF binds to Aβ40 and α-synuclein at the early steps of the process and in multiple stoichiometries and conformations. ScSERF:Aβ40 complexes appear in 3 stoichiometries (1:1, 1:2, and 2:1), and ScSERF:α-synuclein complexes exhibit 4 stoichiometries (1:1, 1:2, 2:1, and 2:2). These complexes were observed to have long multimodal ATDs, indicating that the complexes are present in a variety of conformations. ScSERF alone is an extensively disordered protein as evidenced by its broad ATDs and by previous NMR structural analysis (20); thus, structural heterogeneity in the complex is not entirely surprising. Clearly, our results suggest that the simple model of amyloid formation in which amyloid-prone proteins transition from a single soluble form to a single amyloid-prone form needs revision.
The long ATDs of the 1:1 ScSERF:Aβ40 complexes we observed by IM-MS suggest that ScSERF and Aβ40 both adopt extended structures when in complex. Interestingly, Aβ40 monomers in the same solution are compact in comparison, suggesting that when they form a complex with ScSERF, these amyloid-prone proteins may gain structural plasticity. Unlike Aβ40, ScSERF and α-synuclein monomers exhibit intrinsic structural heterogeneity prior to interaction. This conformational plasticity persists into the complexes formed between ScSERF and α-synuclein; this result supports the finding by Yoshimura et al. (22) that MOAG-4 disrupts intramolecular interactions in α-synuclein.
Note that NMR data for human SERF1a and α-synuclein also characterize the interaction of these proteins as a random fuzzy complex (32). Our data expand on this report, as we not only show complexes with multiple conformations but in multiple stoichiometries. The concept of fuzziness in the context of intrinsically disordered protein binding has been established recently, and there are now a few examples characterizing random complexes with extreme fuzziness in which binding does not induce any order (43). One example is the intrinsically disordered protein pair of prothymosin-α and histone 1.0 (44), which form a conformationally dynamic complex that is physiologically active. In contrast to our interactions, which show affinities in the micromolar range, the affinity between prothymosin-α and histone 1.0 is in the nanomolar range. In our case, the complexes are less tightly formed but very diverse both in stoichiometry and in conformation.
It appears that ScSERF shows a high degree of plasticity in having multiple conformations that interact with multiple amyloid proteins. Plasticity is an important feature for disordered regions in proteins because it allows them to adapt to different partners or recognition motifs (43). We hypothesize that ScSERF’s plasticity is one reason it is able to accelerate amyloid formation for both of the unrelated amyloid-prone proteins investigated in this study, Aβ40 and α-synuclein, and also for the additional SERF substrates that have been reported in the literature, namely Huntingtin exon1 and prion protein PrP (21).
Another class of proteins that show remarkably promiscuous binding behavior are molecular chaperones. Chaperones also often use intrinsically disordered regions to bind to and interact with their substrates (45). ScSERF is an example in which an at least superficially similar binding mode leads to the opposite result—instead of preventing aggregation, as chaperones do, SERF promotes amyloid formation.
Although Aβ40/42 aggregation in the brain is a gradual process, it is thought that toxic species that transiently develop during the process of amyloid formation are more problematic than the accumulation of amyloids themselves (46). The presence of SERF, by accelerating the overall process of amyloid formation, may reduce the concentration of these intermediate species and reduce the toxicity of the process, although it remains to be directly demonstrated that this is happening in neurons.
In conclusion, we have provided evidence that the conserved highly disordered protein ScSERF interacts with the monomeric and dimeric states of 2 unrelated amyloid-prone proteins, Aβ40 and α-synuclein, to form fuzzy complexes that have both multiple conformations and multiple stoichiometries. These interactions lead to the acceleration of the primary nucleation step for both of these amyloid-prone proteins. Our data demonstrate that weak interactions can change the kinetics of amyloid proteins and their tendency to form aggregates and fibrils. Future investigations into the effects of host factors like SERF are necessary to increase our understanding of amyloid formation and neurodegeneration in vitro and in vivo with the eventual hope of finding new drug targets.
Materials and Methods
Additional information can be found in SI Appendix, Material and Methods.
ThT Aggregation Kinetics.
To achieve reproducible Aβ40 kinetics, it is crucial to start with pure monomeric Aβ40. Therefore, the assay was conducted as previously described (29). Monomeric Aβ40 was dissolved in 7 M GdnHCl and 50 mM Tris (pH 8) buffer and loaded onto an analytical Superdex S75 10/300 column (17517401; GE Healthcare). The center fractions of the monomer peak (13.5-mL elution volume) were collected and iced. The protein concentration was determined by using an extinction coefficient ε 280 = 1,490 M−1⋅cm−1. The Aβ40 protein typically had a concentration of 80 to 160 μM at this step.
ScSERF (YDL085C-A) was buffer-exchanged into the assay buffer (20 mM NaPi 200 μM EDTA, pH 7.4) via a PD10 desalting column (GE Healthcare). Samples were prepared in low-binding Eppendorf tubes on ice, using careful pipetting to avoid air bubbles. ThT was added to a final concentration of 100 μM from a 10 mM ThT stock. Each sample was then pipetted into multiple wells in 3 or 4 technical replicates into a 96-well plate that featured low-binding polyethylene glycol coating (3881; Corning), 90 μL per well. The plate was sealed with an adhesive sealing sheet (125434; Thermo Scientific).
Aβ40 amyloid formation assays were initiated by placing the 96-well plate at the specified temperature of 37 °C in a Tecan Infinite M200 plate reader while reading the ThT fluorescence from the bottom using an excitation wavelength of 440 nm and an emission wavelength of 482 nm. Measurements were taken every 5 min under quiescent conditions, and shaking only occurred for 10 s before the fluorescence reading took place. The formation of fibrils was confirmed by TEM.
Immediately prior to performing α-synuclein amyloid formation assays, preexisting aggregates/amyloids and multimers were removed from the α-synuclein preparation by first resuspending the lyophilized α-synuclein in 7 M GdnHCl and 50 mM Tris (pH 8) buffer and loading the dissolved α-synuclein onto an analytical Superdex S75 10/300 GL column (17517401; GE Healthcare). α-Synuclein was eluted using assay buffer (20 mM NaPI, pH 7.4, 50 mM NaCl, and 200 μM EDTA); monomeric α-synuclein was collected, and its concentration was determined by using an extinction coefficient ε 280 = 5,600 M−1⋅cm−1. A 320-μL stock solution sufficient to allow for 3 technical replicates was prepared by diluting monomeric α-synuclein to the stated concentrations in assay buffer (20 mM NaPI, pH 7.4, 50 mM NaCl, and 200 μM EDTA) in low-binding Eppendorf tubes on ice; ThT was added to 25 μM final concentration. Then, 100-μL technical replicates were pipetted into multiple wells of a 96-well microtiter plate with a clear bottom (3631; Corning). Two glass beads were added per reaction well to accelerate the amyloid formation. The plate was sealed with an adhesive sealing sheet (Thermo Scientific). Assays were initiated by placing the 96-well plate at 37 °C in a Tecan Infinite M200 plate reader while reading the ThT fluorescence from the bottom using an excitation wavelength of 440 nm and an emission wavelength of 482 nm. Measurements were taken every 10 min under constant shaking at 300 rpm. The formation of fibrils was confirmed by TEM.
Kinetic Data Analysis.
The experimental data were normalized so that the relative mass concentration of aggregates was 0 at time 0 and 1 at the completion of the aggregation. Kinetic data were normalized and fitted using the platform on AmyloFit (https://www.amylofit.ch.cam.ac.uk/login) (29).
Aggregation traces of 25 μM Aβ40 with different concentrations of ScSERF in molar ratios of 0 to 100% were analyzed by a kinetic nucleation model that is defined by a set of microscopic rate constants, one for primary nucleation (kn), one for secondary nucleation (k2), and one for fibril elongation (k+) (4, 5) In the case of unseeded aggregation, the system depends only on the combination of the rate constants for primary nucleation k+kn and secondary nucleation k2k+, not on the individual rate constants.
To determine the nucleation model, the half-times of different initial monomer concentrations m0 in the absence or presence of 1:0.2 molar ratio ScSERF were determined. In both cases, a double logarithmic plot showed a slight positive curvature as previously described for Aβ40, indicating a multistep secondary nucleation model (5, 29). The kinetics were fitted by keeping the reaction orders for primary and secondary nucleation at the Aβ40 values nc = n2 = 2. The combined rate constants for primary nucleation and secondary nucleation were the only free fitting parameters. The web tool (AmyloFit) defines the multistep secondary nucleation mechanism as follows (5, 27):
where and Here, m0 is the initial monomer concentration, M0 is the initial fibril mass concentration, P0 is the initial fibril number concentration, kn is the primary nucleation rate constant, nc is the critical nucleus size for primary nucleation (2), n2 is the critical nucleus size for secondary nucleation (2), KM is the Michaelis–Menten constant, which determines the monomer concentration at which secondary nucleation begins to saturate, and k+ is the elongation rate constant. The integrated rate law for the normalized aggregate mass concentration is determined as (29)
where the definitions of the parameters are
Native IM-MS Measurements.
Native IM-MS experiments were carried out on a Synapt G2 traveling wave ion mobility-mass spectrometer (Waters). Aqueous samples were introduced into the gas phase by an nESI source operated in positive ion mode. Ions were generated using a capillary voltage of 1,100 to 1,200 V and sample cone voltage of 10 V. The instrument was operated with a backing pressure of 2.63 mbar, source pressure of 7.38 × 10−3 mbar, trap pressure of 3.34 × 10−2 mbar, IM pressure of 3.47 mbar, transfer pressure of 1.00 × 10−6 mbar, and time-of-flight pressure of 1.44 × 10−6 mbar. The IM wave height was 30 V and the IM wave velocity was 600 m/s.
ScSERF was coincubated with Aβ40 or α-synuclein to a final concentration of 25 μM each in 20 mM ammonium acetate, pH 7.4. A ThT assay was carried out in these buffer conditions to verify that the aggregation kinetics of Aβ40 was not changed due to the change in reaction buffer. Samples were incubated at 37 °C. Time-course experiments were conducted by setting up separate reactions that were removed from 37 °C at the indicated times for IM-MS analysis. All reactions were diluted 2.5-fold to final concentrations of 10 μM prior to IM-MS analysis to prevent nonspecific interactions due to nESI. Although various substoichiometric ratios of ScSERF to Aβ40/α-synuclein were screened by other methods, a 1:1 ratio was chosen for the IM-MS experiments to ensure that ScSERF bound complexes were sufficiently abundant to be observed. The 1:1 ratio was the highest ratio of ScSERF to Aβ40/α-synuclein where ScSERF was still observed to accelerate aggregation. Thus, we anticipate that the complexes observed at this ratio are representative of those present at substoichiometric ratios. The dissociation constant of ScSERF binding to Aβ40 monomer was determined as described in SI Appendix, Material and Methods and Native IM-MS Measurements.
ATDs were fit to a minimal number of Gaussian functions, and the peak centers of the Gaussian functions were converted to CCSs using a previously described protocol (47). CCS measurements were calibrated using native β-lactoglobulin, ubiquitin, cytochrome C, insulin, melittin, and denatured ubiquitin. Calibration was conducted using a database of known values in helium, and all CCS values reported are an average of at least 3 replicates. The associated least square error with each average CCS combines inherent calibrant error from drift tube measurements (3%), the calibration curve error (R2), and 2 times the replicate SD error as per a previously published protocol (48).
Raw data were analyzed using MassLynx (Waters). ATDs for all CCS calibrants as well as ScSERF complex samples were extracted from raw data using TWIMExtract (49), and IM ATDs were fit to Gaussian functions using CIUSuite2 (50).
Supplementary Material
Acknowledgments
We thank Sheena Radford for helpful advice, Dr. Michael Cianfrocco and Dr. Tony Ludlam for assistance in performing the electron microscopy experiments, and Ke Wan in the J.C.A.B. laboratory for protein purification. This work was funded by the Howard Hughes Medical Institute, at which J.C.A.B. is an Investigator. This work was also supported by the University of Michigan Protein Folding Diseases Initiative and the Agilent Thought Leader Award Program (B.T.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The datasets generated and/or analyzed during the current study (the assay data, raw mass spectrometry data, transmission electron microscopy pictures, and raw data used to construct the figures) have been deposited on figshare at https://doi.org/10.6084/m9.figshare.c.4684733.v1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913316116/-/DCSupplemental.
References
- 1.Alzheimer’s Association , 2016 Alzheimer’s disease facts and figures. Alzheimers Dement. 12, 459–509 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Prince M., et al. , World Alzheimer Report 2015: The global impact of dementia. An analysis of prevalence, incidence, cost and trends ( Alzheimer’s Disease International, 2015).
- 3.Hebert L. E., Weuve J., Scherr P. A., Evans D. A., Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology 80, 1778–1783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S. I. A., et al. , Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 9758–9763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meisl G., et al. , Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl. Acad. Sci. U.S.A. 111, 9384–9389 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haass C., Selkoe D. J., Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 8, 101–112 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Bucciantini M., et al. , Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Selkoe D. J., Folding proteins in fatal ways. Nature 426, 900–904 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Dobson C. M., Knowles T. P. J., Vendruscolo M., The amyloid phenomenon and its significance in biology and medicine. Cold Spring Harb. Perspect. Biol., 033878 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arosio P., Knowles T. P. J., Linse S., On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 17, 7606–7618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knowles T. P. J., Vendruscolo M., Dobson C. M., The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Hartl F. U., Protein misfolding diseases. Annu. Rev. Biochem. 86, 21–26 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Arosio P., et al. , Kinetic analysis reveals the diversity of microscopic mechanisms through which molecular chaperones suppress amyloid formation. Nat. Commun. 7, 10948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G., et al. , Bri2 BRICHOS client specificity and chaperone activity are governed by assembly state. Nat. Commun. 8, 2081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willander H., et al. , BRICHOS domains efficiently delay fibrillation of amyloid β-peptide. J. Biol. Chem. 287, 31608–31617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen S. I. A., et al. , A molecular chaperone breaks the catalytic cycle that generates toxic Aβ oligomers. Nat. Struct. Mol. Biol. 22, 207–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans C. G., Wisén S., Gestwicki J. E., Heat shock proteins 70 and 90 inhibit early stages of amyloid β-(1-42) aggregation in vitro. J. Biol. Chem. 281, 33182–33191 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Cremers C. M., et al. , Polyphosphate: A conserved modifier of amyloidogenic processes. Mol. Cell 63, 768–780 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart K. L., et al. , Atomic details of the interactions of glycosaminoglycans with amyloid-β fibrils. J. Am. Chem. Soc. 138, 8328–8331 (2016). [DOI] [PubMed] [Google Scholar]
- 20.van Ham T. J., et al. , Identification of MOAG-4/SERF as a regulator of age-related proteotoxicity. Cell 142, 601–612 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Falsone S. F., et al. , SERF protein is a direct modifier of amyloid fiber assembly. Cell Rep. 2, 358–371 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshimura Y., et al. , MOAG-4 promotes the aggregation of α-synuclein by competing with self-protective electrostatic interactions. J. Biol. Chem. 292, 8269–8278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Micsonai A., et al. , Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 112, E3095–E3103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Micsonai A., et al. , BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 46, W315–W322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghaemmaghami S., et al. , Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Wang M., Herrmann C. J., Simonovic M., Szklarczyk D., von Mering C., Version 4.0 of PaxDb: Protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics 15, 3163–3168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knowles T. P. J., et al. , An analytical solution to the kinetics of breakable filament assembly. Science 326, 1533–1537 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Meisl G., Yang X., Frohm B., Knowles T. P. J., Linse S., Quantitative analysis of intrinsic and extrinsic factors in the aggregation mechanism of Alzheimer-associated Aβ-peptide. Sci. Rep. 6, 18728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meisl G., et al. , Molecular mechanisms of protein aggregation from global fitting of kinetic models. Nat. Protoc. 11, 252–272 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Arosio P., Cukalevski R., Frohm B., Knowles T. P. J., Linse S., Quantification of the concentration of Aβ42 propagons during the lag phase by an amyloid chain reaction assay. J. Am. Chem. Soc. 136, 219–225 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Thompson M., Thermodynamic and kinetic analysis of bromodomain-histone interactions. Methods Enzymol. 466, 383–407 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Merle D. A., et al. , Increased aggregation tendency of alpha-synuclein in a fully disordered protein complex. J. Mol. Biol. 431, 2581–2598 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Antwi K., et al. , Cu(II) organizes β-2-microglobulin oligomers but is released upon amyloid formation. Protein Sci. 17, 748–759 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walsh D. M., et al. , Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416, 535–539 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Eschweiler J. D., Kerr R., Rabuck-Gibbons J., Ruotolo B. T., Sizing up protein–ligand complexes: The rise of structural mass spectrometry approaches in the pharmaceutical sciences. Annu. Rev. Anal. Chem. (Palo Alto, Calif.) 10, 25–44 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Beveridge R., et al. , A mass-spectrometry-based framework to define the extent of disorder in proteins. Anal. Chem. 86, 10979–10991 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Smith D. P., Radford S. E., Ashcroft A. E., Elongated oligomers in β2-microglobulin amyloid assembly revealed by ion mobility spectrometry-mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 107, 6794–6798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruotolo B. T., Benesch J. L. P., Sandercock A. M., Hyung S.-J., Robinson C. V., Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 3, 1139–1152 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Soper M. T., DeToma A. S., Hyung S.-J., Lim M. H., Ruotolo B. T., Amyloid-β-neuropeptide interactions assessed by ion mobility-mass spectrometry. Phys. Chem. Chem. Phys. 15, 8952–8961 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen K., et al. , A mass-spectrometry-based modelling workflow for accurate prediction of IgG antibody conformations in the gas phase. Angew. Chem. Int. Ed. Engl. 57, 17194–17199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez-Alba O., Wagner-Rousset E., Beck A., Cianférani S., Native mass spectrometry, ion mobility, and collision-induced unfolding for conformational characterization of IgG4 monoclonal antibodies. Anal. Chem. 90, 8865–8872 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Cohlberg J. A., Li J., Uversky V. N., Fink A. L., Heparin and other glycosaminoglycans stimulate the formation of amyloid fibrils from α-synuclein in vitro. Biochemistry 41, 1502–1511 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Tompa P., Fuxreiter M., Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 33, 2–8 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Borgia A., et al. , Extreme disorder in an ultrahigh-affinity protein complex. Nature 555, 61–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bardwell J. C. A., Jakob U., Conditional disorder in chaperone action. Trends Biochem. Sci. 37, 517–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selkoe D. J., Hardy J., The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richardson K., et al. , An improved calibration approach for travelling wave ion mobility spectrometry: Robust, high-precision collision cross sections. https://www.waters.com/webassets/cms/library/docs/2019asms_richardson_calibration.pdf. Accessed 6 June 2019.
- 48.Bush M. F., et al. , Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Haynes S. E., et al. , Variable-velocity traveling-wave ion mobility separation enhancing peak capacity for data-independent acquisition proteomics. Anal. Chem. 89, 5669–5672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polasky D. A., Dixit S. M., Fantin S. M., Ruotolo B. T., CIUSuite 2: Next-generation software for the analysis of gas-phase protein unfolding data. Anal. Chem. 91, 3147–3155 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.