Significance
The posttranscriptional modification of messenger RNAs (mRNAs) is an emerging frontier in gene regulation. Understanding the biological implications of one of the most common mRNA modifications, pseudouridine, in cells is complicated by the substoichiometric occurrence of mRNA modifications and the difficulty of decoupling the effects on translation from mRNA stability. Here we used in vitro biochemical and structural studies together with cell-based assays to demonstrate that pseudouridine impedes translation elongation and increases the occurrence of amino acid substitutions. Our work supports the idea that mRNA modifications can modulate mRNA translatability and provides evidence that pseudouridine can alter tRNA selection by the ribosome. This study presents a biochemical foundation for better understanding of the consequences of modifications in mRNA coding regions.
Keywords: translation, pseudouridine, mRNA modification, ribosome
Abstract
Chemical modifications of RNAs have long been established as key modulators of nonprotein-coding RNA structure and function in cells. There is a growing appreciation that messenger RNA (mRNA) sequences responsible for directing protein synthesis can also be posttranscriptionally modified. The enzymatic incorporation of mRNA modifications has many potential outcomes, including changing mRNA stability, protein recruitment, and translation. We tested how one of the most common modifications present in mRNA coding regions, pseudouridine (Ψ), impacts protein synthesis using a fully reconstituted bacterial translation system and human cells. Our work reveals that replacing a single uridine nucleotide with Ψ in an mRNA codon impedes amino acid addition and EF-Tu GTPase activation. A crystal structure of the Thermus thermophilus 70S ribosome with a tRNAPhe bound to a ΨUU codon in the A site supports these findings. We also find that the presence of Ψ can promote the low-level synthesis of multiple peptide products from a single mRNA sequence in the reconstituted translation system as well as human cells, and increases the rate of near-cognate Val-tRNAVal reacting on a ΨUU codon. The vast majority of Ψ moieties in mRNAs are found in coding regions, and our study suggests that one consequence of the ribosome encountering Ψ can be to modestly alter both translation speed and mRNA decoding.
Nucleosides in messenger RNAs (mRNAs) can be enzymatically modified posttranscriptionally (1, 2) to expand the chemical and topological properties of these essential biomolecules. Transcriptome-wide mapping of individual modifications revealed the presence of modifications in both the untranslated and protein-coding regions of mRNAs (2, 3). The localization of modifications throughout mRNAs suggests that modifications could potentially alter protein production by multiple mechanisms, including affecting interactions of the translating ribosomal complex with the mRNA, mRNA structure, and mRNA stability. Among the mRNA modifications identified to date, N6-methyladenonsine (m6A) and pseudouridine (Ψ) are the most prevalent (2, 4). m6A modifications are estimated to occur in half of the human mRNAs and cells contain a complement of proteins reported to write, read, and erase the modification (5, 6).
Ψ has been mapped to hundreds of mRNA sequences (7–9), and mass spectrometry studies report the Ψ/U ratio in human cell lines to be comparable to that of m6A/A (∼0.3% for Ψ/U vs. ∼0.5% m6A/A) (10, 11). While the frequency of Ψ at most mapped sites has not been established, estimates of Ψ-frequency based on Ψ-seq experiments, and the direct measurement of Ψ occupancy at a discrete site (in EEF1A1) indicate that Ψ can be incorporated at frequencies (>50%) comparable to well-occupied m6A sites (8, 10). The preponderance of Ψ moieties in mRNA are in coding regions (>60%), and while a host of pseudouridinylating enzymes have been identified that incorporate Ψ into both mRNAs and noncoding RNAs in a reproducible, specific, and inducible manner (7–10, 12–14), no proteins that read or erase Ψ have been discovered. Consequently, the ribosome surely encounters Ψ in cells and it has been hypothesized that it could serve as a key cellular component to read Ψ in mRNAs (2). How, or even if, mRNA pseudouridinylation contributes to gene expression is not yet apparent. Reporter-based studies in human cells and bacterial lysates come to conflicting conclusions regarding the role of Ψ, with some studies suggesting that the presence of Ψ in mRNA codons increases protein production (15) and others reporting a reduction in protein synthesis (16, 17). The clearest evidence of a biological role for Ψ in mRNAs comes from studies in the parasite Toxoplasma gondii where Ψ increases mRNA stability and facilitates parasite differentiation (12, 13). Regardless of whether or not further studies reveal a significant role for Ψ in gene regulation, the ribosome surely translates Ψ-containing codons in cells and it is important to establish the possible outcomes of these events.
Since Ψ can alter the fundamental properties of RNAs, including their secondary structures and base-pairing abilities (18–20), it has been proposed that one consequence of Ψ could be to promote the incorporation of multiple amino acids on a single codon (2, 8). Indeed, Ψ-containing stop codons have been observed to direct the nonsense suppression of translation termination (14, 21), though the effect of Ψ in stop codons remains an unresolved question (22). Thus far, differential decoding of Ψ-containing sense codons has not yet been reported (16, 17). Establishing if Ψ can alter tRNA selection on the ribosome is a timely question given that a wide range of modified nucleosides (Ψ, N1-methyl-Ψ, 2-thiouridine, 5-methyl-cytosine) are being routinely inserted into synthetic mRNAs at high stoichiometric ratios for therapeutic applications (15).
Identifying the consequences of Ψ mRNA modification is complicated in cells because the enzymes that incorporate Ψ into mRNA also catalyze Ψ addition to noncoding RNA species. Furthermore, the impact of mRNA and protein stability on protein output can be difficult to deconvolute from effects on translation in cells. Here, we directly investigate the mechanistic effects of mRNA pseudouridinylation on translation using in vitro enzymology as well as X-ray crystallography, and support our in vitro conclusions with cell-based approaches. Our results demonstrate that the insertion of a single Ψ perturbs ribosome function and promotes the low-level synthesis of multiple peptide products from a single mRNA sequence in a context dependent manner. These studies provide a foundation for understanding the effects of Ψ modification on mRNA translation in cells.
Results
Ψ Reduces Rate Constants for Translation Elongation and EF-Tu GTPase Activation.
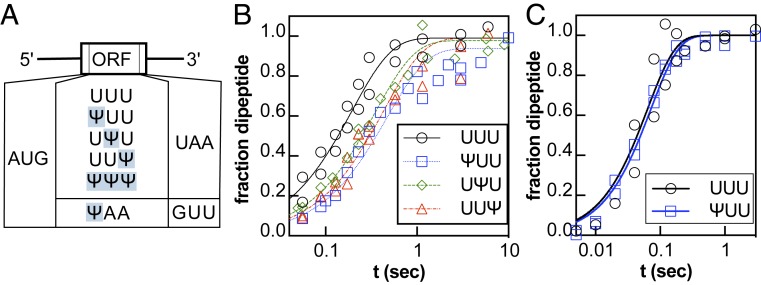
We assessed if Ψ impacts translation by performing kinetic assays with a well-established reconstituted Escherichia coli translation system (23, 24). In our assays, 70 nM of E. coli 70S ribosome complexes containing 35S-labeled formylmethionine-tRNAfMet in the P site and a UUU codon in the A site were reacted with 0.5–5 μM Phe-tRNAPhe•EF-Tu•GTP (ternary complex) at 37 °C and the products were visualized by electrophoretic TLC (SI Appendix, Fig. S1). We measured the rate of phenylalanine (Phe) addition on UUU, ΨUU, UΨU, and UUΨ codons because the rate constant for dipeptide formation on UUU codon is well established (23) and Ψ is found regularly in UUU codons in cells (7, 10) (Fig. 1; details of oligonucleotide quality assessment by UHPLC-MS/MS in SI Appendix). Phe was incorporated robustly on unmodified mRNAs with reaction end point and rate constants similar to those previously reported (23, 25) (Fig. 1 and SI Appendix, Table S1). fMet-Phe dipeptide formation catalyzed by ribosomes on Ψ-containing mRNAs also went to completion (SI Appendix, Table S1) and the kmax for Phe incorporation under 5 μM concentrations of Phe-tRNAPhe (15.7 ± 0.9 s−1) was unaffected (Fig. 1, SI Appendix, Table S1). However, the rate constant for fMet-Phe dipeptide formation was modestly reduced by 2-fold under reaction conditions with subsaturating concentrations of Phe-tRNAPhe (Fig. 1C and SI Appendix, Table S1). We approximated the K1/2 for Phe incorporation on UUU and ΨUU and found that the value is increased by 2-fold on ΨUU. Consistent with this, we find that the production of a full-length luciferase peptide in the reconstituted in vitro translation system (NEB PURExpress) is 3-fold slower on luciferase reporter mRNA with every U substituted for Ψ (SI Appendix, Fig. S2).
Fig. 1.
Ψ changes amino acid incorporation by the ribosome. (A) Coding sequences for the Ψ-containing mRNA constructs. (B and C) Time courses displaying the formation of fMet-Phe peptide on an unmodified and modified UUU codon [UUU (black circles), ΨUU (blue squares), UΨU (green diamonds), UUΨ (red triangles)]. Time courses were collected under single-turnover conditions (70–100 nM 70S ribosome initiation complexes, with either [B] near-saturating [1 μM] or [C] high [5 μM] levels of Phe-tRNAPhe).
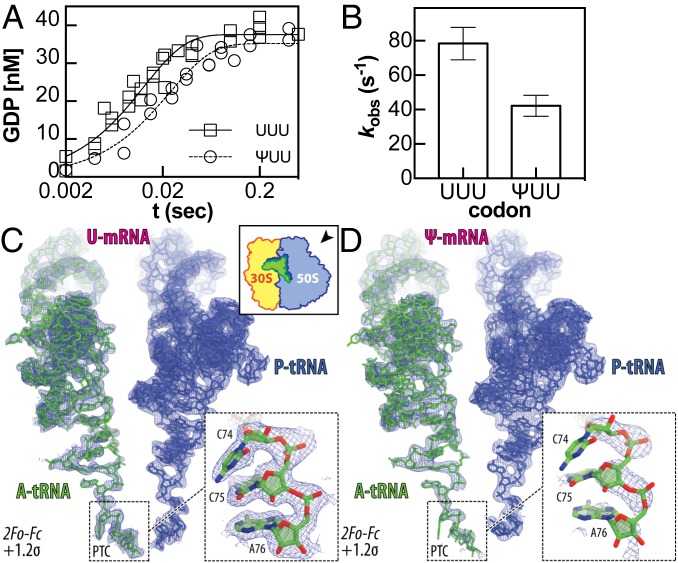
The decreased observed rate constants for amino acid incorporation on pseudouridinylated codons under sub- and near-saturating concentrations of Phe-tRNAPhe could reflect changes in the rate constants for one or more of the multiple upstream kinetic steps (23) (SI Appendix, Fig. S3). To gain further insight into which steps are affected by Ψ, we measured the rate constants for GTP hydrolysis by EF-Tu after binding of the aa-tRNA•EF-Tu•GTP ternary complex to the A site. In these assays, 1.8 μM 3H-fMet-labeled complexes were mixed with 100 nM of α-32P-GTP labeled ternary complex. The observed rate constant for GTP hydrolysis on the unmodified UUU codon (kGTP = 78 ± 10 s−1) was consistent with previously reported values (23), while the rate constant was slower on the ΨUU codon (kGTP = 42 ± 6 s−1) (Fig. 2 A and B, SI Appendix, Table S1).
Fig. 2.
Ψ alters GTP hydrolysis during ternary complex binding to the ribosome. (A) Time courses displaying the formation of GDP when 1.6 μM 3H-fMet-labeled complexes were mixed with 100 nM of γ-32P-GTP labeled ternary complex formed with Phe-tRNAPhe and nucleotide-free EF-Tu. Single-exponential curves were fitted to data collected in 3 independent experiments. (B) Observed rate constants for data fit in A. Error bars are the SE of the fitted value of kobs. (C and D) 2Fo-Fc electron difference Fourier maps (blue mesh) for the ribosome-bound A site (green) and the P site (dark blue) tRNAs interacting with unmodified (C) or Ψ-containing mRNA (D). In C, both the map and the model are from PDB entry 4Y4P. The direction of the view for both panels is indicated on the Upper Right Inset in C. The refined models of mRNA (magenta) and tRNA (green) are displayed in their respective electron densities contoured at 1.2σ. Close-up views of the CCA-ends of the A-site tRNAs are shown by Lower Right Insets in each of the panels. The electron density corresponding to the CCA-end of the tRNA interacting with the Ψ-containing mRNA is much weaker compared to the CCA-end of the tRNA interacting with the unmodified mRNA, while the electron density corresponding to the bodies of the A-site tRNAs are comparable between the 2 complexes.
tRNAPhe 3′CCA Is Not Ordered in the Crystal Structure of 70S Bacterial Ribosome Complex with ΨUU.
To investigate whether the presence of Ψ in the mRNA codon alters tRNA interactions with the ribosome during translation elongation, we solved a crystal structure of T. thermophilus 70S ribosome in complex with ΨUU-containing mRNA, P-site tRNAiMet, and A-site tRNAPhe (on a ΨUU A-site codon) at 2.95 Å resolution (Fig. 2 C and D, SI Appendix, Table S2, PDB 6OU1). We compared this structure to our previously published structure of the same 70S ribosome complex containing tRNAPhe from the same preparation in the ribosomal A site recognizing unmodified Phe codon. In our ΨUU-containing structure, we observed a strong electron density corresponding to the body and the anticodon stem-loop of the A-site tRNAPhe interacting with the mRNA codon (SI Appendix, Fig. S4), similar to the previous structures containing unmodified mRNAs (26). The RMSD value of 0.612 calculated for the entire body of the A-site tRNA (residues 1–73) indicates that it remains in its normal position.
The observed electron density corresponding to the CCA-end of tRNAs in the ribosomal A site is strong and well defined in most of the previously published structures (26). As expected, this is the case for the fully accommodated CCA-end of the tRNAPhe interacting with the unmodified mRNA (Fig. 2C). However, when the Ψ-containing mRNA is present we observed no electron density for the bases of the CCA-end of the same A-site tRNAPhe even though the rest of the tRNA body was visible (Fig. 2D). Even after the refinement of our X-ray data against a 70S ribosome model containing full-length tRNAPhe in the A site, no density for the bases of the CCA-end could be observed in the (2Fo-Fc) electron density map (Fig. 2 D, Inset). These data point to the flexibility of the CCA-end of the A-site tRNA interacting with the ΨUU codon. As a consequence, the CCA-end of this tRNA is unable to form canonical interactions in the A site of the peptidyl transferase center (PTC) on the large ribosomal subunit, which normally comprises the formation of the Watson–Crick base pair between the C75 nucleotide of the A-site tRNA and the G2553 of the 23S rRNA. Since the primary difference of the 70S complex crystallized in this study is the substitution of the uridine with Ψ in a canonical Phe codon, the absence of the CCA-end in the electron density is likely attributed to changes in codon decoding, which occur at the opposite end of the tRNA molecule in the decoding center and apparently propagates all of the way to the PTC (Fig. 2 C and D). The displacement of the A-site tRNA CCA-end has been observed in multiple structures of the ribosome bound to antibiotics [e.g., Madumycin II (27) or Hygromycin A (28)]. In these antibiotic-bound ribosome structures, the observed conformational changes in the CCA-end resulted from steric interference between the CCA-end and the drug, which prevented proper positioning of the tRNA acceptor stem in the 70S ribosome PTC.
Ψ Promotes Amino Acid Substitution in a Reconstituted E. coli Translation System.
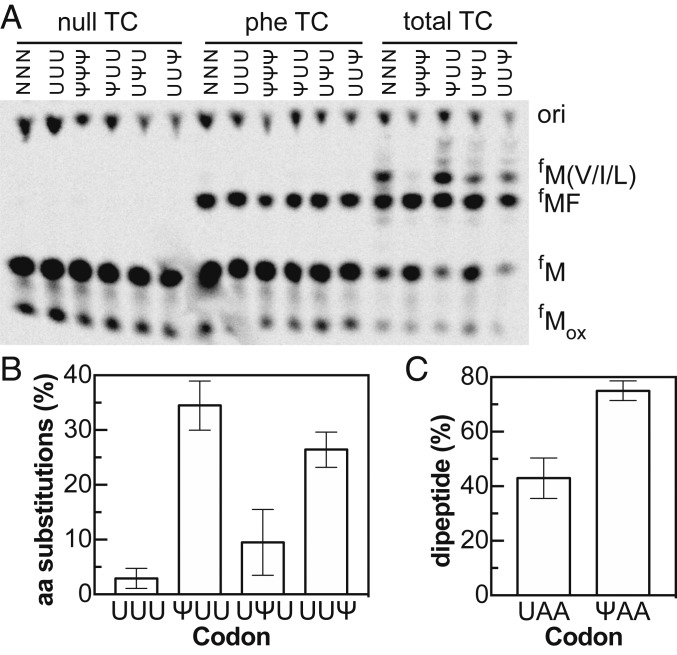
Ψ has the potential to change base-pairing interactions between tRNA anticodons and mRNA. This has raised the possibility that Ψ might, at some frequency, cause the ribosome to accept an aminoacyl-tRNA (aa-tRNA) that would not be cognate on a U-containing codon. To test this possibility, we prepared pools of total aa-tRNA by charging total E. coli tRNA using an S100 extract. We then presented 70S ribosome complexes with a dilute mixture of aa-tRNAs bound to EF-Tu instead of pure Phe-tRNAPhe. If aa-tRNA selection is not altered then we should see fMet-Phe dipeptide formation almost exclusively. As expected, 97% of the dipeptides formed on UUU codons were the fMet-Phe product (Fig. 3 and SI Appendix, Fig. S5). In contrast, mRNAs containing ΨUU or UUΨ directed the synthesis of multiple products (Fig. 3) with reasonable efficiency; nearly half of total peptides produced on ΨUU mRNAs were alternative nonfMet-Phe products (Figs. 3B and SI Appendix, S5A). The extent to which Ψ promotes amino acid substitution appears to be context dependent—we found different levels of amino acid substitution on ribosome complexes programmed with modified stop codon (ΨAA) in the A site (Fig. 3C). Significantly, these experiments were performed under conditions that mimic starvation and result in reduced translational fidelity. We do not expect near-cognate tRNAs to compete as effectively against appreciable concentrations of cognate aa-tRNAs.
Fig. 3.
Ψ promotes incorporation of alternative amino acids by the ribosome at limiting concentrations of aa-tRNA. (A) Electrophoretic TLC displaying the translation products a mixture of mRNAs containing a single randomized codon (NNN), and unmodified and Ψ-containing UUU messages in the presence of no tRNA (null), Phe-tRNAPhe tRNA (phe TC), and total aa-tRNA (total TC). Translation of the NNN pool of mRNAs with random codons in the A site demonstrates the presence of multiple aa-tRNAaa species in the total tRNA preparation. (B) Percent of amino acid substituted dipeptides, relative to the correct fMet-Phe product, on unmodified and modified UUU codons (e.g., % not forming expected MF peptide). (C) Percent of ribosomes that react with 2 μM Lys-tRNALys ternary complex on UAA and ΨAA stop codons to form a MK peptide after 10 min. The near-cognate Lys-tRNALys reacts to produce twice as much peptide on ΨAA than on UAA. All of the data displayed in plots reflect the averages and SEs of at least 3 experiments.
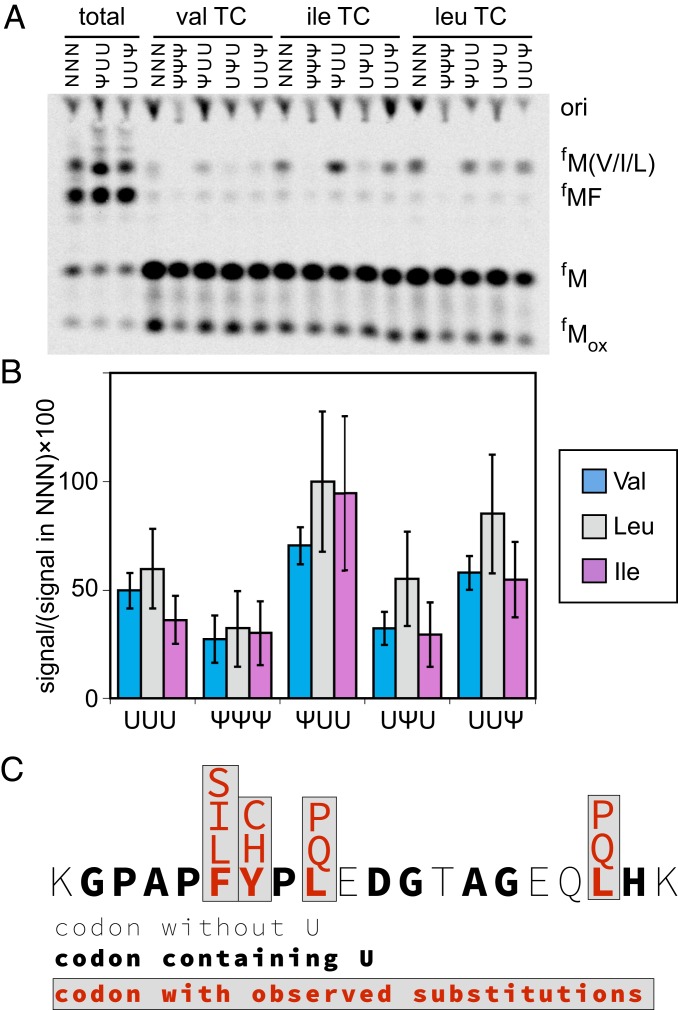
To determine which amino acids are incorporated on the Ψ-containing UUU codons, we performed translation reactions with total tRNA charged one at a time with either valine (Val), isoleucine (Ile), leucine (Leu), or serine (Ser) (Fig. 4 and SI Appendix, S5). These amino acids were selected for investigation based on the migration of amino acid substituted peptides in electrophoretic TLC experiments (Fig. 3A) and their tRNA anticodons. Val and Leu were incorporated on ΨUU and UUΨ codons, Ile was incorporated only on ΨUU, and Ser was not incorporated above background (Fig. 4B and SI Appendix, S5B). Our data are consistent with the known possible base-pairing interactions of Ψ (29–31), and additionally suggest that a Ψ:U base pair can satisfy the requirements for decoding at the first position (SI Appendix, Table S3). This is a surprising degree of flexibility for the decoding site, which as a general rule strictly monitors the codon-anticodon interaction.
Fig. 4.
Amino acids from near-cognate and noncognate tRNAs are incorporated on Ψ-containing codons. (A) Electrophoretic TLC displaying the translation products of NNN and Ψ-containing messages in the presence of total aa-tRNA (total), total tRNA aminoacylated with valine (val TC), total tRNA aminoacylated with isoleucine (ile TC), and total aa-tRNA aminoacylated with leucine (leu TC). (B) Percent of MV/ML/MI products generated on UUU and Ψ-containing codons relative to NNN. The values plotted are the mean of 4 experiments and the error bars reflect the SE. (C) Summary of amino acid substitutions observed by mass spectrometry in a luciferase peptide incorporated on Ψ-containing mRNAs translated in 293H cells.
To quantitatively determine if Ψ reduces the ability of E. coli ribosomes to discriminate between cognate and near-/noncognate aa-tRNAs, we performed kinetic assays with the near-cognate Val-tRNAVal. We reacted 10 μM Val-tRNAVal ternary complexes with 100 nM 70S ribosomes containing 35S-fMet-tRNAMet in the P site and UUU or ΨUU in the A site in the presence of EF-Ts and an energy regeneration mix (32). We observed a burst followed by a long linear phase (SI Appendix, Fig. S7). Both the burst and the rate constant were 2-fold greater on ΨUU (Figs. 5A and SI Appendix, S7). These differences could reflect changes in any step before and including peptidyl-transfer, but—given our experimental conditions ([Mg(II)]free = 10 mM)—likely report on the relative rates of accommodation and rejection of Val-tRNAVal on UUU and ΨUU, similar to what was previously observed for the incorporation of a Leu-tRNALeu on a UUU codon (33). These findings are consistent with the 2-fold increase we observe in Val incorporation on ΨUU reacted with Val-tRNAVal charged from a mixture of total tRNA (Fig. 4B).
Fig. 5.
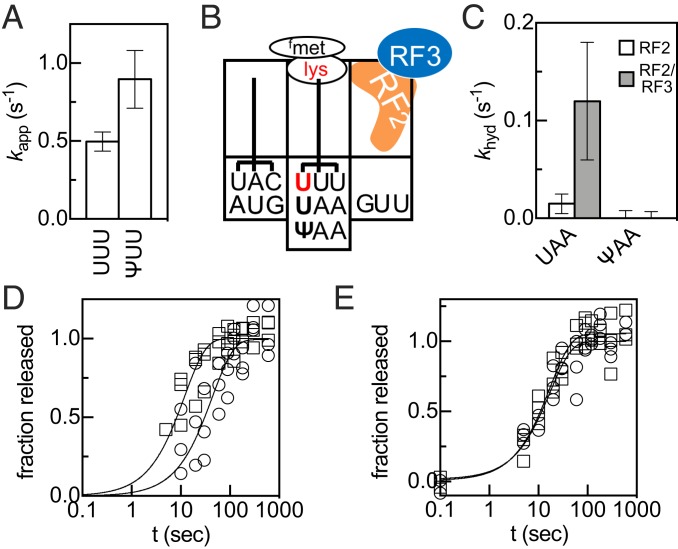
Ψ changes how codons are read. (A) kapp values for fMet-Val and formation on UUU and ΨUU codons in the presence of 10 nM EF-Tu and 10 µM Val-tRNAVal. (B) Position of the codons and peptidyl-tRNA in the purified ribosome elongation complexes prior to addition of RF2 and RF2/RF3 P-site mismatch surveillance assay. (C) Rate constants for premature hydrolysis of fMet-Lys from fMet-Lys-tRNAlys bound to UAA or ΨAA in the P site catalyzed by RF2 (white) and RF2/RF3 (gray). (D and E) fMet release on UAA (squares), ΨAA (circles) stop codons catalyzed by 500 nM RF1 (D) or RF2 (E).
Ψ Increases the Levels of Amino Acid Substitution in Human Embryonic Kidney Cells.
We next investigated the effect of Ψ on amino acid substitution during translation in eukaryotic cells. Luciferase mRNA was transcribed in vitro with either uridine or Ψ and transfected into 293H cells (SI Appendix, Fig. S8). Full-length luciferase protein was purified (SI Appendix, Fig. S9) and analyzed by mass spectrometry with a focus on a specific luciferase peptide with favorable ionization characteristics. Amino acid substitutions in this peptide, which totaled ∼1%, were only observed in peptides generated from Ψ-containing mRNAs (Figs. 4C and SI Appendix, Fig. S10, and Tables S4 and S5). We also extended our analyses to the entire luciferase dataset. Luciferase protein translated from Ψ-containing mRNAs possessed a significantly higher rate of amino acid substitution (totaling ∼1.5%, integrated over all Ψ-containing codons) relative to protein synthesized from uridine-containing mRNAs (substitutions totaling <0.05% were observed only on two Val codons) (SI Appendix, Table S6). The miscoding events that we observed in our unmodified uridine-containing samples (Val substitutions) are likely relatively common substitutions as they have also been seen by more sophisticated mass-spectrometry approaches investigating unmodified EF-Tu sequences (34). These observations are consistent with the expected levels of amino acid substitution that we would estimate from our in vitro kinetic studies with cognate Phe-tRNAPhe and near-cognate Val-tRNAVal, which suggest that under conditions where all tRNAs are equally well charged and available, the expected total level of amino acid substitution on ΨUU codons should be ∼1% (SI Appendix, Fig. S7).
mRNA:tRNA Mismatches on ΨAA in the P Site Not Surveilled by E. coli Ribosome.
Our data indicate that the ribosome can interact differently with near-/noncognate aa-tRNAs when Ψ is present within the A-site codons. To assess if Ψ also alters how the ribosome perceives mRNA:tRNA interactions in the P site, we investigated if the ribosome detects mRNA:tRNA mismatches on Ψ-containing P-site codons. On unmodified codons E. coli ribosomes sense P-site mismatches, and release factors 2 and 3 (RF2/3) catalyze the hydrolysis of truncated peptides containing substituted amino acids from sense (nonstop) codon to ensure translational fidelity (Fig. 5B) (35). We tested if mismatches involving a pseudouridinylated codon are similarly surveilled by reacting ribosome initiation complexes containing UAA or ΨAA in the A site with ternary complexes containing Lys-tRNALys in the presence of elongation factor G (EF-G). This generated a mixture of mismatched ribosome complexes containing either fMet-Lys-tRNALys or fMet-Lys-Lys-tRNALys in the P site (Figs. 5 and SI Appendix, Fig. S11 A and B). We then added RF2 or RF2/RF3 and measured the rate constants for fMet-Lys (MK) and fMet-Lys-Lys (MKK) peptide release from these mRNAs. If a mismatch is detected, we expect that RF2/RF3 will catalyze premature peptide release much faster than RF2 alone (35). On the unmodified mRNA we observed that MK and MKK peptides are released by RF/RF3 at rates comparable to those previously reported for other mismatched complexes (Figs. 5 and SI Appendix, Fig. S11 C and D) (35). In contrast, when ΨAA is in the P site, the MK peptide was not released (Figs. 5C and SI Appendix, Fig. S11E). However, when ΨAA is translocated into the E site, MKK peptide release was catalyzed by RF2/RF3; this means that the mismatch between the tRNALys and the GUU codon in the P site is surveilled on the Ψ-containing transcript (Fig. S11F). Our data demonstrate that mRNA:tRNA mismatches on ΨAA P-site codons are not sensed, suggesting that Ψ can alter how the ribosome interacts with near-/noncognate aa-tRNAs in the P site.
Class I Release Factor 1 (RF1) Is Modestly Impeded by the Presence of Ψ.
The presence of Ψ in stop codons has been reported to promote nonsense suppression, incorporating Ser or Thr instead of terminating translation on UAA codons, in both bacteria and yeast cells (14, 21). Computational studies have predicted that this is due to alterations in release factor activity on pseudouridinylated stop codons (36). To assess if Ψ alters the ability of class I release factors to catalyze the hydrolysis of the peptide from peptidyl-tRNA, we measured the rate constants for peptide release on mRNAs encoding methionine followed by the universal stop codon (UAA/ΨAA) (Fig. 5 D and E). At saturating concentrations of RF1 or RF2, peptide release on the UAA codon occurred with rate constants (kmax,release) between 0.06–0.24 s−1 at 22 °C, consistent with previously published values (35, 37) (Fig. 5 D and E and SI Appendix, Table S7). Peptide release on the ΨAA codon was only modestly perturbed: kmax,release for RF1 was decreased ∼3-fold, but kmax,release for RF2, and the K1/2 for both RF1 and RF2 were unchanged (Fig. 5 D and E and SI Appendix, Fig. S12 and Table S7). Our results are mostly consistent with a previous study utilizing RF1 and the A246T variant of RF2 (38), which found no difference in rate constants for peptide release on wild-type and Ψ-containing stop codons (22). The modest impact on RF1 activity appears to be specific to Ψ, as we find that m6A impedes RF2‒, but not RF1‒, mediated peptide release (SI Appendix, Figs. S13 and S14). Together, the low magnitude decrease in the rate constant for RF1 catalyzed release, our failure to incorporate Ser on ΨAA in the absence of release factors (SI Appendix, Fig. S15), and our inability to detect extended products from a fully Ψ substituted luciferase reporter in the NEB PURExpress in vitro translation system, suggest that Ψ is unlikely to significantly suppress translation termination under normal cellular conditions.
Discussion
The inability to knock out the enzymes that incorporate Ψ into mRNAs without also impacting noncoding RNA modification, coupled with the lack of known Ψ reader or eraser proteins, has made it difficult to investigate the biological consequences of Ψ mRNA modifications. We approached this challenge by using a fully reconstituted in vitro translation assay and asking how the function of one possible Ψ reader, the ribosome, is impacted by the pseudouridinylation of mRNAs. Our studies show that the presence of Ψ in codons subtly changes how the ribosome interacts with both cognate and non/near-cognate aa-tRNAs. We observed that Ψ-containing codons perturb the translation of cognate codons and promote the synthesis of a variety of peptide products from a single mRNA more often than from unmodified mRNAs both in a reconstituted bacterial translation system and human cells.
Consideration of our kinetic and structural data with respect to the established mechanistic paradigm for aa-tRNA binding to the A site (SI Appendix, Fig. S3) (23) provides some insight into the mechanistic effects of Ψ. We observed reductions in the rate constants for amino acid addition (kobs) and EF-Tu catalyzed GTP-hydrolysis (kGTP) on Ψ-containing codons, consistent with our finding that Ψ-substituted codons decrease the overall rate of full-length protein production. The changes to kobs, kGTP, and Mg(II) dependence of our observations (SI Appendix, Fig. S16) are reminiscent of those seen for near-cognate tRNAs (33), although more subtle, suggesting that pseudouridinylated codon recognition exists somewhere on a spectrum between cognate and near-cognate complexes. Our structural data provide further evidence that the ribosome interacts differently with tRNAs bound to Ψ-containing codons. We find that the 3′ CCA of the A-site tRNA becomes disordered, suggesting that the unconventional decoding of the ΨUU A-site codon at the decoding center in the small ribosomal subunit leads to long-range changes in the acceptor stem of the A-site tRNA located in the PTC on the large ribosomal subunit. Taken as a whole, our kinetic and structural results indicate that Ψ modestly impacts multiple steps in the translation kinetic pathway to exert an overall observed effect on amino acid addition.
Our kinetic and mass-spectrometry data demonstrate that while amino acid substitution is increased in the presence of Ψ, these events are relatively rare in unstressed cells (<1.5%). Furthermore, many amino acid substitutions would likely be neutral, so the incorporation of Ψ in mRNAs would not be expected to generate significant quantities of nonfunctional protein under normal cellular conditions. Nonetheless, there could be some conditions or sequence contexts in which Ψ-mediated amino acid substitution happens more robustly. Our observation that Ψ alters decoding by the ribosome in cells differs from previous reporter-based studies that did not observe amino acid substitutions at a single, defined position in a full-length ErmCL reporter peptide or in a fully Ψ-substituted GFP reporter (16, 17). There are several potential explanations for this discrepancy. First, our mass-spectrometry assays were able to detect multiple in vivo amino acid substitution events that occurred at frequencies (0.1–0.4%; SI Appendix, Tables S5 and S6) lower than the reported limit of detection for the GFP study (∼1%). Second, the effect of Ψ on decoding could depend strongly on local mRNA sequence and structure (39). This would be unsurprising, given that our studies indicate the degree of alternative amino acid incorporation depends both on codon identity and nucleotide position within a codon (Fig. 3). Such context dependence has been previously observed for inosine; both how inosine is decoded and the frequency of amino acid substitution (0.5–25%) range widely depending on sequence context (40). Third, cellular stresses change the available pool of aa-tRNAs and distribution of mRNA modifications (7, 41). The extent of amino acid substitution should be highly sensitive to the relative levels of aa-tRNAs, so different cellular conditions will likely modulate the degree of amino acid substitution. Lastly, it is possible that the observability of alternative decoding in different mRNAs could be quite distinct based on the identity of the mRNA and the posttranslational fate of the amino acid substituted peptide.
We observed that Ψ-containing codons modestly affect the ability of the ribosome to incorporate Phe, in line with studies by ourselves (SI Appendix, Fig. S2) and others demonstrating that overall protein production is reduced by on mRNA reporters containing either a single- (<2-fold) (16, 17) or fully Ψ substituted codons (3-fold) in a fully reconstituted E. coli translation system and human cells (17). We anticipate that the effect of Ψ on the rate of codon translation may depend on the sequence context of the codon; we have seen that incorporating Ψ into different mRNA sequences coding for identical luciferase peptides can have very different protein expression outcomes in 293H cells (SI Appendix, Fig. S17). Overall, our findings are similar to what has been reported for the decoding of m6A and 2′ O-methyl containing codons (32, 42) suggesting that mRNA modifications might generally alter aa-tRNA binding and accommodation. Given the propensity of the ribosome to react with near- and noncognate aa-tRNAs during the translation of Ψ-containing mRNAs (Figs. 3 and 4), these small rate defects could become important for cognate aa-tRNA selection under conditions of cellular stress or starvation. We speculate that it could be advantageous for cells to maintain a small reservoir of protein diversity for evolution and adaptation to environmental stresses (43, 44). Indeed, increased levels of amino acid substitution have been shown to increase cellular fitness under oxidative and temperature stress, and during transition from stationary to cell growth conditions (45–47). The idea that amino acid substitution levels might vary in response to cellular conditions is supported by a recent study demonstrating that the frequency of amino acid substitutions in the E. coli EF-Tu varies by as much as 2 orders of magnitude depending on protein expression level (34). Ultimately, the full suite of modern scientific tools—including ensemble and single-molecule biochemistry, deep sequencing, and cell biology—will be required to understand how a single modification is coupled to mRNA stability and protein synthesis in the cell.
Materials and Methods
Tight-couple 70S ribosomes were purified from E. coli MRE600. Unmodified mRNAs were prepared by run-off T7 transcription of DNA oligonucleotides. mRNAs containing modified nucleotides were synthesized and HPLC purified by Dharmacon and their quality was assessed by UHPLC-MS/MS (SI Appendix, Fig. S18). E. coli transfer RNAs were purchased from either MP Biomedical, Sigma, or tRNA Probes. All translation experiments were performed in 1× 219-Tris buffer (50 mM Tris pH 7.5, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, 5 mM β-ME) as previously described (25, 35). Initiation complexes were purified by ultracentrifugation. Detailed procedures and reaction conditions for all experiments are provided in the SI Appendix.
Data Availability.
Coordinates and structure factors were deposited in the RCSB Protein Data Bank.
Supplementary Material
Acknowledgments
This work was funded by the University of Michigan start-up funds (to K.S.K.), Rackham Merit Fellowship (to M.K.F.), National Institutes of Health awards R35 GM128836 (to K.S.K.), T32 GM008597 (to M.K.F.), University of Illinois startup funds (to Y.S.P.), and New England Biolabs (NEB) Inc. (M.Z.W. and B.R. are New England Biolabs, Inc. employees). The structural work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health [P30-GM124165 to NE-CAT]. The Pilatus 6M detector on 24ID-C beamline is funded by an NIH-ORIP HEI [S10-RR029205 to NE-CAT]. The Eiger 16M detector on 24ID-E beamline is funded by an NIH-ORIP HEI grant [S10-OD021527 to NE-CAT]. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We thank the staff at NE-CAT beamlines 24ID-C and 24ID-E for help with data collection and freezing of the crystals. We also thank Dr. Rachel Green for providing the translation factor plasmids, Dr. Robert Kennedy for the use of his HPLC and mass spectrometer, and Scott Shaffer and Markos Koutmos for their comments on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors were deposited in the RCSB Protein Data Bank with accession code 6OU1 for the Thermus thermophilus 70S ribosome in complex with ΨUU-mRNA, A-, P-, and E-site tRNAs.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821754116/-/DCSupplemental.
References
- 1.Zhao B. S., Roundtree I. A., He C., Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 18, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert W. V., Bell T. A., Schaening C., Messenger RNA modifications: Form, distribution, and function. Science 352, 1408–1412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arango D., et al. , Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175, 1872–1886.e24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao B. S., et al. , m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer K. D., Jaffrey S. R., Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y., Dominissini D., Rechavi G., He C., Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Carlile T. M., et al. , Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz S., et al. , Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovejoy A. F., Riordan D. P., Brown P. O., Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9, e110799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., et al. , Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Zheng G., et al. , ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamoto M. A., Lovejoy A. F., Cygan A. M., Boothroyd J. C., mRNA pseudouridylation affects RNA metabolism in the parasite Toxoplasma gondii. RNA 23, 1834–1849 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson M. Z., Brewer J., Singh U., Boothroyd J. C., A pseudouridine synthase homologue is critical to cellular differentiation in Toxoplasma gondii. Eukaryot. Cell 8, 398–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karijolich J., Yu Y. T., Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karikó K., et al. , Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16, 1833–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoernes T. P., et al. , Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 44, 852–862 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoernes T. P., et al. , Eukaryotic translation elongation is modulated by single natural nucleotide derivatives in the coding sequences of mRNAs. Genes (Basel) 10, E84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis D. R., Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 23, 5020–5026 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newby M. I., Greenbaum N. L., Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. U.S.A. 99, 12697–12702 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kierzek E., et al. , The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 42, 3492–3501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández I. S., et al. , Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 500, 107–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svidritskiy E., Madireddy R., Korostelev A. A., Structural basis for translation termination on a pseudouridylated stop codon. J. Mol. Biol. 428 (10 Pt B), 2228–2236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pape T., Wintermeyer W., Rodnina M. V., Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17, 7490–7497 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavlov M. Y., Ehrenberg M., Rate of translation of natural mRNAs in an optimized in vitro system. Arch. Biochem. Biophys. 328, 9–16 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Koutmou K. S., et al. , Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife 4, e05534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polikanov Y. S., Melnikov S. V., Söll D., Steitz T. A., Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 22, 342–344 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osterman I. A., et al. , Madumycin II inhibits peptide bond formation by forcing the peptidyl transferase center into an inactive state. Nucleic Acids Res. 45, 7507–7514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polikanov Y. S., et al. , Distinct tRNA accommodation intermediates observed on the ribosome with the antibiotics hygromycin A and A201A. Mol. Cell 58, 832–844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charette M., Gray M. W., Pseudouridine in RNA: What, where, how, and why. IUBMB Life 49, 341–351 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Ofengand J., Bakin A., Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266, 246–268 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Seelam P. P., Sharma P., Mitra A., Structural landscape of base pairs containing post-transcriptional modifications in RNA. RNA 23, 847–859 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi J., et al. , N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 23, 110–115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pape T., Wintermeyer W., Rodnina M., Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18, 3800–3807 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garofalo R., et al. , Broad range of missense error frequencies in cellular proteins. Nucleic Acids Res. 47, 2932–2945 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaher H. S., Green R., Quality control by the ribosome following peptide bond formation. Nature 457, 161–166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parisien M., Yi C., Pan T., Rationalization and prediction of selective decoding of pseudouridine-modified nonsense and sense codons. RNA 18, 355–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hetrick B., Lee K., Joseph S., Kinetics of stop codon recognition by release factor 1. Biochemistry 48, 11178–11184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinçbas-Renqvist V., et al. , A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19, 6900–6907 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauger D. M., et al. , mRNA structure regulates protein expression through changes in functional half-life. bioRxiv, 10.1101/549022 (13 February 2019). [DOI] [PMC free article] [PubMed]
- 40.Licht K., et al. , Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res. 47, 3–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Netzer N., et al. , Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi J., et al. , 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol. 25, 208–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Brien P. J., Herschlag D., Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6, R91–R105 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Drummond D. A., Wilke C. O., The evolutionary consequences of erroneous protein synthesis. Nat. Rev. Genet. 10, 715–724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan Y., et al. , Protein mistranslation protects bacteria against oxidative stress. Nucleic Acids Res. 43, 1740–1748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz M. H., Pan T., Temperature dependent mistranslation in a hyperthermophile adapts proteins to lower temperatures. Nucleic Acids Res. 44, 294–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan Y., et al. , Heterogeneity of stop codon readthrough in single bacterial cells and implications for population fitness. Mol. Cell 67, 826–836.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors were deposited in the RCSB Protein Data Bank.