Significance
Many proteins contain disordered linkers that join different functional units. Such linkers are crucial to many multidomain proteins, but are rarely studied in their own right. Here we study how intramolecular binding reactions are controlled by disordered protein linkers via effective concentrations. We show that the effective concentrations follow geometric models developed to describe polymers. The physical properties encoded into the linker sequence control the compaction of the linker and thus the ensuing effective concentrations. The system developed here provides a strategy to systematically investigate the relationship between sequence and compaction of intrinsically disordered proteins. We show here that such relationships directly affect biochemical binding equilibria by controlling the effective concentration.
Keywords: effective concentration, intrinsically disordered protein, flexible linker, polymer physics, fluorescent biosensor
Abstract
Many multidomain proteins contain disordered linkers that regulate interdomain contacts, and thus the effective concentrations that govern intramolecular reactions. Effective concentrations are rarely measured experimentally, and therefore little is known about how they relate to linker architecture. We have directly measured the effective concentrations enforced by disordered protein linkers using a fluorescent biosensor. We show that effective concentrations follow simple geometric models based on polymer physics, offering an indirect method to probe the structural properties of the linker. The compaction of the disordered linker depends not only on net charge, but also on the type of charged residues. In contrast to theoretical predictions, we found that polyampholyte linkers can contract to similar dimensions as globular proteins. Hydrophobicity has little effect in itself, but aromatic residues lead to strong compaction, likely through π-interactions. Finally, we find that the individual contributors to chain compaction are not additive. We thus demonstrate that direct measurement of effective concentrations can be used in systematic studies of the relationship between sequence and structure of intrinsically disordered proteins. A quantitative understanding of the relationship between effective concentration and linker sequence will be crucial for understanding disorder-based allosteric regulation in multidomain proteins.
Protein interactions are tightly regulated. The specificity is not only determined by the protein structure, but also by which molecules a protein encounters. Molecular encounters are often enhanced by a direct physical connection between the interaction partners, which changes the protein–protein interaction from a bi- to a unimolecular process. Many proteins thus primarily function by connecting other proteins, for example, an enzyme to its substrate or weakly interacting proteins to each other (1–3). Furthermore, multidomain proteins often contain long linkers that play a similar role for intramolecular interactions (4) and modulate phase separation (5). A physical connection often increases encounter rates by several orders of magnitude, which shifts equilibrium position of binding reactions and the rates of biochemical reactions by a similar amount (6, 7). The encounter frequency of such linked reactions is concentration-independent. Instead, it depends on the architecture of the connection, and therefore linker properties directly affect protein function.
Interdomain linkers often belong to the family of intrinsically disordered proteins (IDPs) (8). IDPs do not fold into stable structures, but form an ensemble of interconverting structures that may contain a range of transient interactions. Sufficiently long disordered linkers allow the tethered domains to contact in any orientation, and therefore they represent a generic mechanism for fusing domains, which cannot be accomplished by rigid proteins. For example, p300 coactivators consist of an enzymatic domain coupled to protein–protein interaction domains via disordered linkers (4). The connection between molecules is not static, but can vary in time and space to regulate the functions of the tethered domains. This variation may result in allosteric regulation via, e.g., phosphorylation or protein binding in the linkers. Allostery usually describes the thermodynamic interaction between 2 ligands that are not in direct contact. Usually, this allostery is transmitted through structural changes in folded domains, but may also occur via changes in the structural ensembles of IDPs (9, 10). Structural changes of linker regions can affect the function of the tethered domains, and linkers thus provide a generic mechanism whereby an event can affect distant parts of a protein. To understand such allosteric effects, it is crucial to study the role of linkers quantitatively.
The functions of linkers can be understood quantitatively in terms of effective concentrations. For an intramolecular reaction, the encounter rate between tethered domains equals the rate of the same untethered reaction at a given concentration (11). This concentration is known as the effective concentration. Formally, the effective concentration is defined as the ratio of the equilibrium constants for 2 equivalent binding reactions, where one occurs intra- and one intermolecularly (12). When the linker is sufficiently long to join the binding sites without strain, the effective concentration is independent of what is linked and solely a property of the linker (13, 14). Intriguingly, this suggests that effective concentrations can be measured in a convenient model system and extrapolated to other systems. Effective concentrations can be measured by competition experiments, where a free ligand displaces a tethered ligand (12). Such measurements have mostly been used in efforts to optimize multivalent drugs through avidity (15, 16). In molecular biology, effective concentrations are rarely measured experimentally, but usually estimated theoretically from the volume in which the tethered ligand is free to diffuse (17–21). While such simple geometric models are commonly used, they have not been tested in complex biological systems.
The simplest linker is a fully disordered chain. Fully disordered linkers are an attractive model for understanding effective concentrations, as they are well-described by theories borrowed from polymer physics. When the sizes of disordered proteins are measured as, e.g., end-to-end distances or the radius of hydration or gyration, they scale with the chain length following a power law such as:
where N is the number of residues and ν is a scaling exponent determined by chain compaction. Such scaling laws underpin theoretical calculations of effective concentrations, as the chain length defines the radius of the accessible volume. The scaling exponent for effective concentrations is thus usually assumed to be −3ν, although such models have not been verified experimentally. Prediction of effective concentrations thus depend on the scaling exponent ν. On average, IDPs have been found to have ν values from 0.51 to 0.58 (22–24), but the scaling exponents of disordered proteins varies from about 0.4 for disordered states of foldable proteins to about 0.72 for highly charged IDPs (25). For reference, globular proteins and rigid rods have scaling exponents of 0.33 and 1, respectively. The sequence–compaction relationship of IDPs has been studied by correlating chain size with variations in sequence. Net charge dominates chain compaction through intrachain repulsion (22, 25–27). Furthermore, compaction is weakly correlated to hydrophobicity and weakly anticorrelated to proline content (22). The literature depicts a complicated relationship between polyampholyte strength and compaction, as the overall effect of polyampholyte interactions can cause compaction or expansion (28, 29). The complexity arises due to the patterning of charged residues (29–31), which leads to attractive interactions between some parts of the chain and repulsive interactions with others.
Here we investigate how effective concentrations in multidomain proteins depend on linker architecture. We directly measure effective concentrations for many disordered linkers with systematic changes in the physical properties of the linker. Our fluorescent biosensor for measurement of the effective concentrations provides a way to probe sequence–compaction relationships in intrinsically disordered proteins and relating these to biochemical function.
Materials and Methods
Preparation of DNA Constructs.
DNA constructs were obtained from GenScript by insertion of synthetic genes between the NdeI and BamHI sites of a pET15b vector and subcloning of new linkers using unique NheI and KpnI sites flanking the linkers. Full protein sequences are given in SI Appendix, Supplementary Materials.
Protein Expression and Purification.
All fusion protein constructs were expressed in BL21(DE3) cells in 50 mL ZYM-5052 autoinduction medium (32) supplied with 100 μg/mL ampicillin and shaking at 120 rpm. The temperature was kept at 37 °C for 3 h and thereafter decreased to 18 °C. The cells were harvested by centrifugation (15 min, 6,000 × g) after 40 to 48 h, when the cultures had changed color, indicating mature fluorescent proteins. Bacterial pellets were lysed using B-PER Bacterial Protein Extraction Kit (Thermo Scientific) according to the manufacturer’s protocol, and the lysate was applied to gravity flow columns packed with nickel Sepharose. After washing with 20 mM NaH2PO4, pH 7.4, 0.5 M NaCl, 20 mM imidazole, fusion proteins were eluted by increasing the imidazole concentration to 0.5 M. Fusion proteins were subsequently purified using Strep-Tactin XT Superflow columns (IBA) according to the manufacturer’s instructions and dialyzed overnight into Tris-buffered saline (TBS). The MBD2 peptide was expressed overnight in BL21(DE3) cells at 37 °C in ZYM-5052 autoinduction medium with 100 μg/mL ampicillin with shaking at 120 rpm. The cells were resuspended in 20 mM NaH2PO4, pH 7.4, 0.5 M NaCl, 20 mM imidazole and lysed by heating to 80 °C for 20 min (33), and debris pelleted by centrifugation (15 min, 14,000 × g). The peptide was purified by nickel Sepharose as before except a stepwise elution up to 0.5 M imidazole was used before dialysis into TBS. It was critical to prepare and concentrate the MBD2 peptide freshly and store it on ice until use. Protein concentrations were measured using A280.
Measurement of Effective Concentrations.
A total of 0.1 μM of each fusion protein was titrated with the MBD2 peptide through 16 serial 2-fold dilutions in TBS. The starting concentration was in the range of 1.6 to 2 mM for WT MBD2 and 3.3 mM for the V227A MBD2. Samples were analyzed in triplicate in black 386-well plates with 1 g/L bovine serum albumin (BSA; Fisher Scientific) added to prevent sticking. The FRET measurements were performed in a SpectraMax I3 plate reader using donor excitation at 500 nm, and emission detected in 25-nm-wide bands centered at 535 and 600 nm. The titration data were analyzed by nonlinear fitting in MATLAB to the standard fitting equation for 1:1 binding reactions with Kd replaced by ce,app:
where E1 and E2 are the apparent FRET values in the open and closed states and P is the concentration of the fusion protein. For titration with the WT MBD2 peptide, this determines an “apparent effective concentration,” which was multiplied by the affinity ratio of the WT and V227A peptides to produce the true effective concentration. The correction factor was established to be 30 by titration of the fusion protein containing the GS120 linker with the V227A MBD2 peptide. Polymer scaling parameters were extracted by a linear fit to log(ce) vs. logN to allow an equal weighting of the errors in ce.
Diffusion Measurements by Fluorescence Correlation Spectroscopy (FCS).
FCS measurements were recorded on samples containing ∼10 nM of the 40-residue linker variant of each linker composition in TBS at 23 °C. The fluorescent proteins were excited using the 488-nm laser, and the donor dye emission was acquired for 5 min in 10-μs time bins using a home-built confocal single molecule FRET instrument described previously (34). Autocorrelation curves were generated using multipletau 0.3.3. Datasets with high molecular weight bursts were eliminated. The dimensions of the confocal volume were determined with Atto488 as reference (35). The autocorrelation curves were fitted globally using Igor Pro-6.37 to the equation describing free diffusion through a Gaussian volume with a triplet state contribution (36):
The parameters describing the confocal volume (r0, zz) were determined from the reference sample, and the parameters describing the triplet state dynamics (T, τtrip) were fitted as global parameters across all diffusion measurements.
Results
Reporter Design.
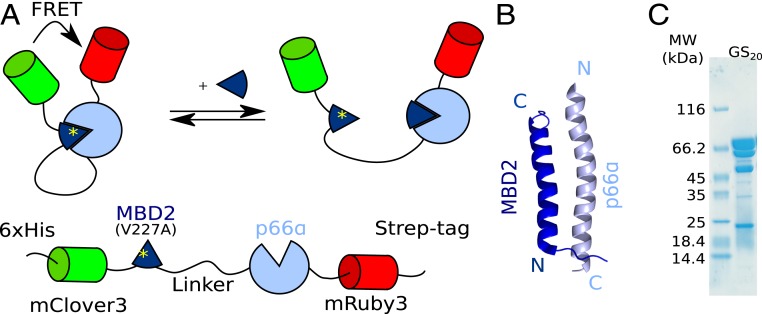
We designed a fusion protein inspired by FRET biosensors (37) to measure effective concentrations for different linker architectures. An exchangeable linker joins 2 protein domains that form an interaction pair. These domains are flanked by the fluorescent proteins mClover3 and mRuby3, which form a FRET pair (38) (Fig. 1A). When the intramolecular complex is formed, the fluorescent proteins are brought into close contact, resulting in efficient FRET. The effective concentration is measured by following the FRET efficiency in a titration (12), where a free ligand displaces the intramolecular interaction. The ideal interaction pair is small, has a known 3D structure, is easy to produce in Escherichia coli, and binds tightly to ensure full ring-closing. Furthermore, in the bound state, the N terminus of one protein should be close to the C terminus of the other and vice versa. This ensures a high FRET efficiency in the closed state and allows even a short linker to join the domains without strain. These constraints are ideally fulfilled by an antiparallel heterodimeric coiled coil such as the complex between MBD2 and p66α used here (Fig. 1B) (39).
Fig. 1.
A FRET biosensor for measuring effective concentrations. (A) Titration with free MBD2 peptide displaces the intramolecular interaction and results in a decrease in the FRET efficiency. (B) Structure of the interaction pair in the fusion protein: the antiparallel coiled coil formed between MBD2 and p66α (PDB ID code 2L2L) (39). (C) SDS/PAGE gel of the purified fusion protein reveals internal cleavage sites in mRuby3. The cleavage products cannot be removed, but do not affect measurements.
The fusion proteins have purification tags at both the N and C termini to allow parallel purification of many constructs by sequential affinity chromatography. After purification, SDS/PAGE revealed a major band corresponding to the expected molecular weight of the fusion protein (Fig. 1C). The 4 minor bands corresponded to proteolytic cleavage in the mRuby3 domain as revealed by mass spectrometry. These bands could not be removed by gel filtration. This suggests that the cleaved fluorescent proteins remain in a stable complex, although it is not clear whether it is fluorescent. A fraction of inactive fluorophores will decrease the FRET amplitude, but will not affect the midpoint of the titration and the measurements of effective concentrations.
Measurement of Effective Concentration.
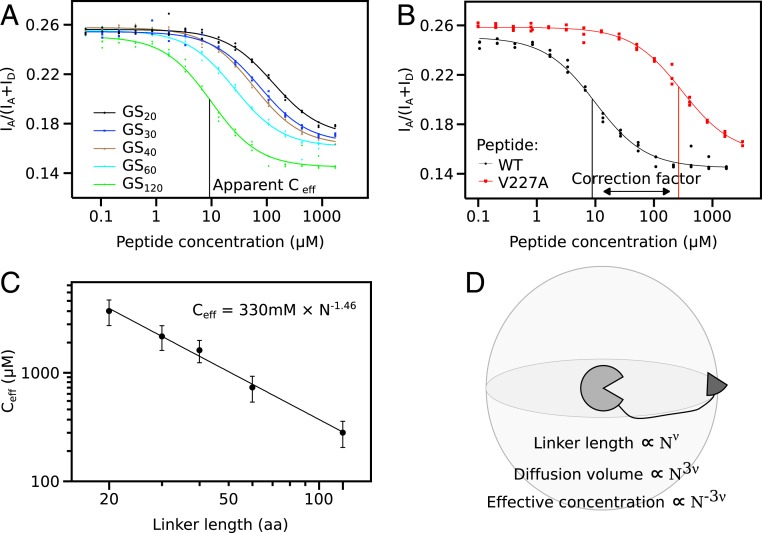
Fusion proteins were initially constructed with linkers consisting of (GS)n repeats ranging from 20 to 120 residues. A 240-residue GS-linker was also tested, but resulted in insoluble protein. Titration of all constructs with free WT MBD2 peptide resulted in a sigmoidal decrease of the proximity ratio (Fig. 2A), where the titration midpoint marks the apparent effective concentration. For increasing linker lengths, the titration midpoint was reached at lower peptide concentrations consistent with a decrease in effective concentration. Simultaneously, the proximity ratio of the posttitration baseline decreased with linker length consistent with a more expanded open form (Fig. 2A). Across the whole dataset, the pre- and posttransition proximity ratios varied unsystematically, which we believe was due to small differences in fluorophore maturation. This prevents determination of intramolecular distances from FRET values, but does not affect the midpoint of the titration and thus the measurement of the effective concentration.
Fig. 2.
Measurement of effective concentrations. (A) Titration curves of fusion proteins containing GS-linkers of variable length reveal that the effective concentration scales monotonously with linker length. (B) The correction factor corresponding to the difference in affinity between WT and V227A was determined by titration of fusion protein GS120 containing the V227A mutation with both competitor peptides. (C) Effective concentrations follow a power law as revealed by the straight line in a log-log plot. The scaling exponent that is the focus of the remainder of the manuscript is derived from nonlinear fitting as indicated. (D) The expected geometric relation between scaling exponents for protein size, ν, and effective concentration. The linker defines a volume in which tethered ligand can diffuse, and the effective concentration is inversely proportional to this diffusion volume. Error bars represent 95% confidence interval.
Fitting of titration data requires the concentration of the free ligand to exceed the midpoint by a factor of 10. This is impractical for effective concentrations that were expected to reach the millimolar range. Therefore, the fusion proteins contained a weakened mutant MBD2 (V227A) and were titrated with WT MBD2 peptide. This shifted the midpoint by the ratio between WT and V227A affinities, which subsequently was applied as a correction factor. To determine the correction factor, we titrated the fusion protein with the GS120 linker with both WT and V227A peptide (Fig. 2B). The titration midpoint was shifted by a factor of 30, which was applied to the titration midpoint of all variants to produce the effective concentration. As it was impractical to prepare competitor peptides for each linker composition, we used the titrant peptide with a flanking GS-segment for all other linker compositions. As the flanking linker residues may affect the stability of the complex, the correction factor may differ for other linker compositions. A mismatched correction factor results in a constant shift of the polymer scaling law, but should not affect the scaling exponent.
The effective concentration scales with linker length following a power law as shown by the straight line in Fig. 2C. Notably, this conclusion does not require any assumptions of the distribution of the linker conformations. Relative to one binding partner, geometric considerations suggest that the volume accessible to the other partner scales with the linker length with an exponent of 3ν (Fig. 2D). Accordingly, the effective concentrations should scale with an exponent of −3ν. Fitting of effective concentrations from the GS-linker series gave a scaling exponent of −1.46 corresponding to a ν of 0.49. The GS-linker is a polar tract in a recent systematic classifications (29), and is thus expected to form relatively compact globules due to backbone interactions (40). Accordingly, the GS-linker was slightly more compact than denatured proteins (41) and IDPs (22–24). GS-repeats are frequently used as linkers in protein design as they are assumed to behave as ideal Gaussian chains. The scaling exponent determined here agrees well with the predicted ν of 0.5 for a Gaussian chain, thus validating this assumption.
Variation of the Linker Sequence.
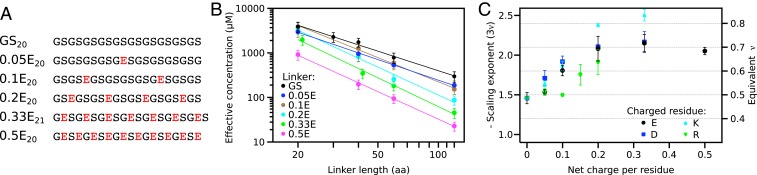
The excellent agreement with a power law suggested that effective concentrations could be used to probe the sequence–compaction relationship of IDPs. To probe how the effective concentration depends on linker sequence, we systematically varied the properties that were likely to affect linker compaction: charge, ampholyte strength, rigidity, hydrophobicity, and aromaticity. All sequences used have predicted helical propensities below 5%, suggesting that secondary structure formation does not contribute appreciably to the linker properties (SI Appendix, Table S1). We systematically increased the net charge of the linker by introduction of charged residues into a GS-linker. All linkers used here have a uniform pattern throughout the sequence (Fig. 3A), which allows linkers of different lengths to be described by a single scaling exponent. For each linker composition, we generated linkers with totals of 20, 40, 60, and 120 residues and measured the effective concentrations through titration experiments. In a few cases near the limit of solubility, the longest linker was shortened to 80 residues or the series only contained the 3 shorter linkers (SI Appendix, Table S1). Each linker composition followed a power law as illustrated for linkers containing glutamate residues (Fig. 3B and SI Appendix, Fig. S1). Both the prefactor and the scaling exponent from the fit varied with linker composition. Whereas scaling exponents vary in physically meaningful ways, we cannot rationalize the variation in the prefactors (SI Appendix, Table S2). The prefactor absorbs any variations in the strength of the protein interaction as a systematic error, e.g., due to the effect of the linker close to the interaction pair. Such errors are likely constant for different linker lengths and do thus not affect the scaling exponent. Therefore, in the following experiments we concentrated on the scaling exponent that reports on linker compaction.
Fig. 3.
Net charge per residue dominates the scaling of effective concentration with linker length. (A) Net charge was uniformly distributed into a background of GS-linkers. The same pattern is used for subsequent linker series. (B) Measurement of effective concentration allows determination of scaling exponents for each linker composition. Error bars represent 95% CI. (C) Scaling exponents demonstrate a linker expansion with increasing net charge. Compared to the negatively charged residues, arginine expands the chain less and lysine more. Error bars represent SE.
Disordered linkers can interact with adjacent folded domain (42) or with linkers from other molecules, and such interactions might be present in our biosensor. To test whether such interactions contribute to the changes in scaling coefficients, we evaluated the effects of a hypothetical interaction between the linker and a fluorescent domain using the ensemble optimization method (43, 44). We generated an ensemble consisting of rigid folded domains connected by disordered linkers and selected a subset with a contact between the linker and a fluorescent protein based on a distance <8 Å between any atom of the linker and the fluorescent domain. The average Rg of the contact subset decreased by 4% (SI Appendix, Fig. S2A), which represents a conservative estimate of the effect of an interaction. To evaluate whether any of the linkers perturbed the biosensor structure, we measured the diffusion coefficients by fluorescence correlation spectroscopy (SI Appendix, Fig. S2B and Table S2). As we wish to exclude effects on the biosensor structure, we sought to minimize the effect of the linker expansion itself by using variants with relatively short linkers (40 residues). The diffusion coefficients do not follow changes in scaling exponent for any linker series (SI Appendix, Fig. S2C) and are uncorrelated with scaling coefficients for the entire dataset (Fig. 2D). The errors of the FCS measurements are about the same size as the predicted effect of a linker interaction with a fluorescent domain, so these experiments cannot conclusively rule out interactions between the linkers and the folded domains. However, the lack of an overall correlation (SI Appendix, Fig. S2D) suggested that the trends in the scaling coefficients are unlikely to be caused by aggregation or severe linker-induced collapse of the biosensor.
Are All Charged Residues Equal?
To test if all charged residues affect linker compaction equally, we measured effective concentrations for linkers containing increasing amounts of the 4 charged residues. For each residue type, increased net charge per residue lead to chain expansion (Fig. 3C). Glutamate and aspartate caused an equal expansion: the scaling exponent changed gradually from −1.46 (ν = 0.49) in the uncharged linker to ∼−2.1 (ν = ∼0.7) at a net charge per residue of 0.2. The scaling exponent did not increase further with increasing net charge, suggesting an upper limit that the linker does not expand beyond. This limit corresponds to the ν previously observed for highly charged IDPs (25). Lysine-containing linkers initially followed the expansion of negatively charged linkers, but continued up to a scaling exponent of ∼−2.4 (ν = ∼0.8). This is higher than the ν-values reported for any other disordered protein. In contrast, arginine-containing linkers have the opposite effect: residue fractions up to 0.1 had the same scaling exponent as GS-linkers. At higher fractions, the scaling exponent increased, but did not reach the same expansion as the other residue types even at the highest fractions permitted by solubility. The difference between arginine and lysine mirror their different roles in intermolecular interactions between disordered proteins (45). In total, the titration series demonstrated that charged residues types have a different impact on IDP compaction.
Polyampholyte Linkers.
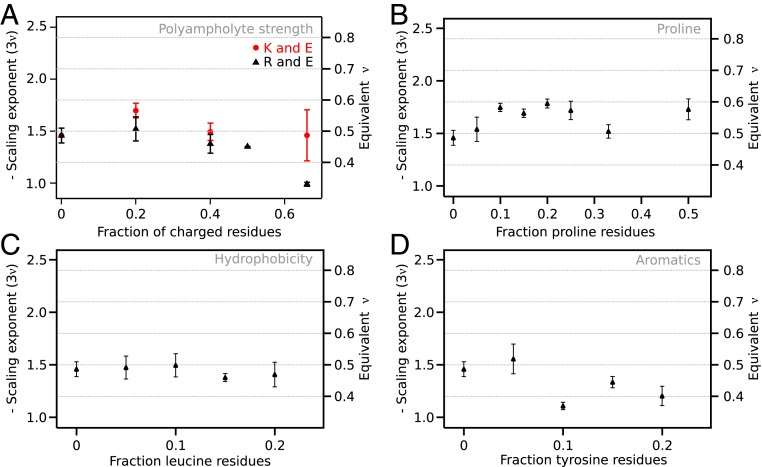
Most IDPs are polyampholytes, which entails that chain compaction is determined by the balance between attractive and repulsive interactions. Both simulation (30, 46) and experiments (46, 47) have shown that the patterning of charged residues have a large effect on polyampholyte compaction. To investigate the pure effect of ampholyte strength in the absence of charge patterning, we created linkers with a uniform distribution of equal numbers of positively and negatively charged residues. With the difference between lysine and arginine in mind, we also compared polyampholytes containing each of these 2 residues combined with glutamate. At low fractions of charged residues, the scaling exponent was roughly constant for both polyampholyte series. However, there appeared to be a threshold after which the chain contracts dramatically for arginine-containing polyampholytes (Fig. 4A). The last data point at a fraction of charged residues of 0.67 suggested a scaling exponent of −1 (ν = 0.33), which is the same compaction as a globular protein. This observation agrees qualitatively with the contraction caused by screening of charges in a polyampholyte (27), but contradicts the diagram-of-states representation of IDP classes (29, 30). In contrast, polyampholyte linkers containing lysine residues had approximately the same compaction for all fractions of charged residues.
Fig. 4.
The effect of polyampholyte strength and proline, leucine, and tyrosine residues on scaling exponents. (A) Scaling exponents from neutral polyampholyte linkers with an equal proportion of glutamate residues and either lysine or arginine. The scaling exponents show a strong compaction to a globular chain for polyampholytes containing arginine only. (B) Chain expansion shows a complex dependence on proline content. At low fractions of proline residues, proline residues lead to linker expansion, which may be reversed at high proline fractions. (C) Hydrophobicity was increased by introduction of leucine residues, but led to practically no change in chain compaction. (D) Tyrosine residues led to strong compaction, likely due to π-interactions. Error bars are SE estimated from the fit.
The Effect of Proline Residues.
Proline content is one of the main determinants of IDP compaction (22), in part because proline residues have less conformational freedom as they are restricted to certain ϕ/φ-angles. Therefore, we gradually increased the fraction of proline residues in the linker (Fig. 4B). The scaling exponent increased to a plateau at ∼−1.7 (ν = ∼0.57), which is reached at a proline fraction of 0.1, which is similar to the dimensions of chemically denatured proteins (41). Proline thus expanded the linker to a lesser extent than charged residues, in agreement with previous studies (22). Unlike the previous series, the expansion with increasing proline content appeared nonmonotonic. This could suggest that different effects dominate at different fractional content of proline, e.g., the propensity to form polyproline II helices, although it is difficult to draw a firm conclusion due to the experimental errors.
The Effect of Hydrophobic Interactions.
It is unclear how hydrophobicity affects compaction of disordered proteins. In IDPs, hydrophobicity is weakly anticorrelated with ν (22). However, disordered states of foldable protein, which are much more hydrophobic than IDPs, form both compact (25) and expanded states (48). To assess the effect of hydrophobicity systematically, leucine residues were introduced into the linker. We chose leucine as it is among the most hydrophobic of the nonaromatic residues (49), but is not β-branched and thus less likely to perturb backbone dihedral distributions. Due to solubility, the linker sequence can only be extended up to leucine fractions of 0.2. Introduction of leucine residues led to a small decrease in scaling exponent to −1.4 (ν = 0.47; Fig. 4C). This qualitatively agreed with a weak anticorrelation between size and hydrophobicity.
π-Interactions between Aromatic Residues.
Interactions between π-electrons in aromatic side chains have a strong potential to induce intrachain interaction, most recently demonstrated by their effect on liquid–liquid phase separation (45). We introduced tyrosine residues as it is the least hydrophobic aromatic amino acid and has the largest effect on phase separation (45, 50, 51). Tyrosine residues caused a noticeable reduction in the scaling exponents already at a fractional content of 0.1, where the scaling exponent was ∼−1.2 (ν = 0.4; Fig. 4D). The contraction was smaller than the contraction observed for polyampholytes, although the fractional content of aromatic residues cannot be increased as far due to insolubility. As for proline, the contraction appears to be nonmonotonous, although it is difficult to state decisively due to experimental errors. However, the effect of tyrosine residues was larger than that of leucine, which suggests that π-interactions are more important to chain compaction than hydrophobicity.
Are Contributions to Chain Compaction Additive?
Previous studies have identified individual factors that affect IDP compaction, but have not clarified whether they are additive. We therefore constructed linkers simultaneously increasing net charge and proline content. When probed alone, proline expanded linkers measurably already at a fraction of 0.05. However, when proline was introduced together with a charged residue, it did not lead to further expansion, but simply resulted in identical scaling exponents as glutamate-only linkers (Fig. 5A). This suggested that expansion caused by chain rigidity and net charge are not additive. Instead, the mixed sequences were simply dominated by the strongest individual effect.
Fig. 5.
Lack of additivity of individual contributions. (A) Proline or leucine residues were introduced in equal proportion to glutamate residues, but the scaling coefficient follows that of the glutamate-only series. This suggests that the expansion caused by net charge and chain stiffness are not additive. (B) Charge expansion caused by glutamate against a constant fraction of 0.1 of leucine residues shows that hydrophobicity has a negligible effect. (C) Increasing fractions of leucine only has a negligible effect on the dimensions of the linkers expanded by charge. Error bars represent SE.
Does Hydrophobicity Modify Charge Expansion?
Hydrophobicity had a surprisingly small effect when investigated alone. To further test this conclusion in the context of additivity, we created mixed linker series containing glutamate and leucine residues in equal proportion. Again, the scaling exponent followed that of charge series alone. This series changed 2 parameters at once, so we designed a linker series where either the glutamate content or the leucine content was varied and the other held constant at a residue fraction of 0.1. When the charge expansion was probed in the context of a fraction of 0.1 leucine residues, the curve again followed the glutamate-only series (Fig. 5B). When the fraction of leucine residues was increased within the limits permitted by solubility, the scaling exponent decreases slightly, although within error (Fig. 5C). In total, these data suggest that hydrophobicity in itself has a vanishing effect on the compaction of IDPs, at least within the limits of protein solubility. Furthermore, they suggest that the factors that affect IDP compaction are not necessarily additive.
Discussion
Linkers control many biochemical reactions via the effective concentration. Here we have shown that effective concentrations in multidomain proteins with disordered linkers follow polymer scaling laws. We have thus experimentally validated the geometric models commonly used to estimate effective concentrations, but also show that the effective concentration depends strongly on linker sequence. For a 100-residue linker, the difference between ν values of 0.4 and 0.7 corresponds to a 63-fold change in effective concentration assuming a constant prefactor. This can be the difference between an intramolecular interaction being saturated or hardly formed at all. Changes in the linkers following, e.g., ligand binding or posttranslational modification may thus be directly transmitted into allosteric regulation of the domains tethered at the end. Linkers may thus be one of the most direct examples of how the structural properties of intrinsically disordered proteins affect biochemical function, underscoring the need to understand the relationship between IDP sequence and compaction. In the short term, direct measurement of the effective concentrations using the system developed here may help us understand allostery in IDPs (10).
Sequence–Structure Relationships in IDPs.
Polymer models have been used successfully to describe the structural properties of IDPs and are the foundation for theoretical predictions of effective concentrations. Here we show that the relationship can be reversed: measurement of effective concentrations is an efficient way to parametrize polymer descriptions of IDPs and describe the relationship between sequence and compaction. The range of scaling exponents observed here agrees well with the range of scaling exponents of disordered proteins described previously. This validates the assumption of a scaling coefficient for effective concentrations equal to −3ν, and bridges the present work to the existing literature on IDP compaction. While we study interdomain linkers, it is likely that the conclusions can be generalized to other types of IDPs, although it is necessary to keep a number of caveats in mind. Fused domains may artificially affect the properties of a disordered segment, and it is thus important to exclude such effects. In our biosensors, the linker constitutes 3 to 17% of the protein, and its contributions are thus swamped in most measurements. Therefore, we cannot exclude interactions between the linker and the folded domains, but believe that the systematic changes with linker length and composition make contributions from spurious interactions unlikely.
Net Charge.
Net charge is the strongest predictor of the compaction of IDPs (22, 25, 26). Previous studies suggest that the maximal expansion is reached at a net charge per residue of ∼0.4. In contrast, we find a value of ∼0.2 (Fig. 3C). This difference may be due to sequence differences or a fundamental difference between a linker and an isolated chain. In previous studies, the charge expansion occurred against a complex background sequence with compensating attractive interactions. This can likely not be avoided, but is minimized in our linker series. Our value may thus represent a minimal estimate of how much charge density it takes to fully expand an otherwise inert IDP, whereas the value of around 0.4 may be more relevant for complex sequences with additional attractive interactions. Alternatively, linkers may be special because, e.g., the excluded volume of the attached domains favor expanded chains. Additionally, in the closed biosensor, the ends of the linkers are brought into close proximity, which may increase the linkers’ sensitivity to charge repulsion compared to an isolated chain. Another key difference is the role of arginine residues. Arginine form attractive interactions that partially compensate for the charge–charge repulsion. This recapitulates the role of arginine in self-association of IDPs during liquid–liquid phase separation, and may thus be due to its capacity to form π-interactions (45, 51). Conveniently, for most positively charged protein sequences, this effect may be offset by the higher than average repulsion of lysine residues. Therefore, chain properties may be well-described in terms of the net charge density as long as lysine and arginine are equally common.
Polyampholyte Sequences.
The compaction of polyampholytes is determined by the balance between attractive and repulsive interactions. As controlled experiments on polyampholytic IDPs have been scarce, organic polyampholytes have been used as models to understand the impact of ampholyte strength on IDP structure. Organic polyampholytes form a range of compact structures and, at high charge densities, they eventually become almost globular, albeit with a liquid-like internal structure (52). It is not clear how well such models describe proteins, especially because many proteins have a patchy distribution of charged residues. In one case, increase of the ionic strengths led to expansion of a polyampholytic protein (27), which suggested that polyampholyte interactions are overall attractive. This conclusion is also supported by the compact state adopted by the complex between 2 oppositely charged IDPs (53). On the contrary, a computational study of polyampholyte sequences suggested that the compact state only arises if charges are unevenly distributed, whereas a well-mixed polyampholyte was predicted to form expanded coils (30). This is summarized in the diagram-of-states description of IDPs, where increase of the strength of neutral polyampholyte leads to a globule-to-coil transition (29). Here we find that increase of the polyampholyte strength led to compaction (Fig. 4A), but only for polyampholytes containing arginine. This mirrors the difference we see between the positively charged residues probed in isolation. In the presence of the attractive π-interactions in arginine residues, even a perfectly mixed polyampholytes can contract, whereas the lysine-containing ampholytes remain relatively expanded as predicted previously (30). These polyampholytes resembles the mixed charge domains of nuclear speckle proteins, which contain repeats of positively and negatively charged residues. Increasing the fractions of arginine residues increases the propensity to condensate, which is consistent with the greater propensity for intramolecular self-association observed here (54). Intriguingly, this shows that the ratio between the 2 positively charged residues can tune the functional properties of IDPs in vivo.
Hydrophobic Side Chains in IDPs.
Hydrophobic side chains in IDPs are mostly solvent-exposed. Contraction may bring such side chains in proximity to interact and thus form a partial protection from the solvent. Therefore, it was expected that hydrophobicity would be anticorrelated with ν in disordered proteins (22, 25). Recently, this conclusion was questioned, as several unfolded, but foldable, proteins form relatively expanded chains (ν = 0.54) despite their hydrophobicity (48). By testing the effect of hydrophobicity in a variety of sequence contexts, we found that increasing the hydrophobicity did not lead to a noticeable contraction of the linker. One explanation for this is that the disorder of the chain prevents the proteins from forming even partially desolvated hydrophobic interactions. In complex sequences, hydrophobicity correlates with other factors that could cause chain compaction. Such confounders could explain the correlation of hydrophobicity with the compaction of unfolded states of foldable proteins (25). A key candidate for such a confounder is aromatic residues, which we found to induce strong compaction of the linkers. A tyrosine fraction of 0.1 was thus sufficient to contract the linker to most compact dimensions observed for unfolded, but foldable, proteins (25). Similarly, tyrosine drives phase separation of IDPs to a much higher extent than leucine (55), providing the equivalent finding in intermolecular IDP interactions. We note that the most compact unfolded, but foldable, proteins in previous studies also have a higher fraction of aromatic residues. Aromatic residues thus potentially explain the disagreement (25, 49) on the effect of hydrophobicity in disordered proteins.
The Additivity of IDP Compaction.
Several properties affect chain compaction, but how do they add up? Both charge and proline residues expand the linker, but the combination of the two did not expand the linker more than charge alone. This demonstrates that not all contributions to chain compaction are additive. A likely explanation is that rigidity added by proline residues can be accommodated inside the ensemble expanded by charge–charge repulsion. The reason net charge density is such a good predictor of the compaction of IDPs may thus be that the strongest effect dominates in the absence of additivity. The most likely explanation for the different threshold for charge expansion is thus the presence of compensating attractive interactions. This suggests an additivity between some factors, but likely not all, and demonstrates that much remains to be uncovered about sequence–compaction relationship of IDPs.
The Value of Synthetic IDPs.
Our present understanding of the relationship between sequence and structure in IDPs is mainly based on the study of natural proteins. However, it is inherently difficult to draw general conclusions from a small set of examples. In contrast, we have studied synthetic linkers never seen in nature. The synthetic linkers allow tight control over the physical properties of the linker, which is crucial for hypothesis testing. Experiments on synthetic IDPs are thus a natural step for critically evaluating our understanding of IDPs. Synthetic DNA has removed the need for ingenious cloning strategies used previously (56), leaving protein preparation as the major bottleneck. For artificial proteins spanning a range of physical properties, this can, however, be a major challenge. We have, for example, not been able to make our linkers in isolation yet. The fusion protein used here serves both as solubility tags and a reporter system operating at nanomolar concentrations. These are likely the key factors that have allowed us to study a broader range of IDPs than previous studies. Although the potential of spurious interactions are intrinsic to such fusion proteins, they may provide useful tools for future investigations of sequence–structure relationships in IDPs. An exciting future direction would thus be a direct comparison of the scaling exponents determined from size and effective concentrations on the same sequences. Such measurements could help delineate in which ways the sequence–structure relationship of linkers differ from isolated IDPs.
Data Availability.
All data discussed in the paper can be found in the SI Appendix and Figshare, doi:10.6084/m9.figshare.10029254.
Supplementary Material
Acknowledgments
This work was supported by grants to M.K. from the “Young Investigator Program” of the Villum Foundation; the AIAS COFUND program funded by the EU FP7 Cofund programme (Agreement no. 754513); and PROMEMO – Center for Proteins in Memory, a Center of Excellence funded by the Danish National Research Foundation (Grant Number DNRF133). We thank Birthe B. Kragelund, Mateusz Dyla, and Xavier Warnet for critical comments to this manuscript; and Anna Marie Nielsen and Tanja Klymchuk for technical assistance.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All data for this paper have been deposited in Figshare, https://doi.org/10.6084/m9.figshare.10029254, and in the SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904813116/-/DCSupplemental.
References
- 1.Langeberg L. K., Scott J. D., Signalling scaffolds and local organization of cellular behaviour. Nat. Rev. Mol. Cell Biol. 16, 232–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nussinov R., Ma B., Tsai C. J., A broad view of scaffolding suggests that scaffolding proteins can actively control regulation and signaling of multienzyme complexes through allostery. Biochim. Biophys. Acta 1834, 820–829 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Good M. C., Zalatan J. G., Lim W. A., Scaffold proteins: Hubs for controlling the flow of cellular information. Science 332, 680–686 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dyson H. J., Wright P. E., Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Harmon T. S., Holehouse A. S., Rosen M. K., Pappu R. V., Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife 6, 1–31 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitazawa T., et al. , A bispecific antibody to factors IXa and X restores factor VIII hemostatic activity in a hemophilia A model. Nat. Med. 18, 1570–1574 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Greenwald E. C. M., Redden J. M., Dodge-Kafka K. L., Saucerman J. J., Scaffold state switching amplifies, accelerates, and insulates protein kinase C signaling. J. Biol. Chem. 289, 2353–2360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortese M. S., Uversky V. N., Dunker A. K., Intrinsic disorder in scaffold proteins: Getting more from less. Prog. Biophys. Mol. Biol. 98, 85–106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papaleo E., et al. , The role of protein loops and linkers in conformational dynamics and allostery. Chem. Rev. 116, 6391–6423 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Tompa P., Multisteric regulation by structural disorder in modular signaling proteins: An extension of the concept of allostery. Chem. Rev. 114, 6715–6732 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Page M. I., Jencks W. P., Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc. Natl. Acad. Sci. U.S.A. 68, 1678–1683 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnamurthy V. M., Semetey V., Bracher P. J., Shen N., Whitesides G. M., Dependence of effective molarity on linker length for an intramolecular protein-ligand system. J. Am. Chem. Soc. 129, 1312–1320 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gargano J. M., Ngo T., Kim J. Y., Acheson D. W. K., Lees W. J., Multivalent inhibition of AB(5) toxins. J. Am. Chem. Soc. 123, 12909–12910 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Li M., Cao H., Lai L., Liu Z., Disordered linkers in multidomain allosteric proteins: Entropic effect to favor the open state or enhanced local concentration to favor the closed state? Protein Sci. 27, 1600–1610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mack E. T., et al. , Dependence of avidity on linker length for a bivalent ligand-bivalent receptor model system. J. Am. Chem. Soc. 134, 333–345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou H. X., Quantitative account of the enhanced affinity of two linked scFvs specific for different epitopes on the same antigen. J. Mol. Biol. 329, 1–8 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Timpe L. C., Peller L., A random flight chain model for the tether of the Shaker K+ channel inactivation domain. Biophys. J. 69, 2415–2418 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diestler D. J., Knapp E. W., Statistical mechanics of the stability of multivalent ligand-receptor complexes. J. Phys. Chem. C 114, 5287–5304 (2010). [Google Scholar]
- 19.Diestler D. J., Knapp E. W., Statistical thermodynamics of the stability of multivalent ligand-receptor complexes. Phys. Rev. Lett. 100, 178101 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Borcherds W., et al. , Optimal affinity enhancement by a conserved flexible linker controls p53 mimicry in MdmX. Biophys. J. 112, 2038–2042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherry K. P., Johnson S. E., Hatem C. L., Majumdar A., Barrick D., Effects of linker length and transient secondary structure elements in the intrinsically disordered notch RAM region on notch signaling. J. Mol. Biol. 427, 3587–3597 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh J. A., Forman-Kay J. D., Sequence determinants of compaction in intrinsically disordered proteins. Biophys. J. 98, 2383–2390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernadó P., Svergun D. I., Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol. Biosyst. 8, 151–167 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Fuertes G., et al. , Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proc. Natl. Acad. Sci. U.S.A. 114, E6342–E6351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofmann H., et al. , Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 16155–16160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao A. H., Crick S. L., Vitalis A., Chicoine C. L., Pappu R. V., Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 8183–8188 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller-Späth S., et al. , From the Cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U.S.A. 107, 14609–14614 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuler B., Soranno A., Hofmann H., Nettels D., Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins. Annu. Rev. Biophys. 45, 207–231 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Das R. K., Ruff K. M., Pappu R. V., Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 32, 102–112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das R. K., Pappu R. V., Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. U.S.A. 110, 13392–13397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin E. W., et al. , Sequence determinants of the conformational properties of an intrinsically disordered protein prior to and upon multisite phosphorylation. J. Am. Chem. Soc. 138, 15323–15335 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Kalthoff C., A novel strategy for the purification of recombinantly expressed unstructured protein domains. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 786, 247–254 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Kjaergaard M., et al. , Oligomer diversity during the aggregation of the repeat region of Tau. ACS Chem. Neurosci. 9, 3060–3071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapusta P., Absolute Diffusion Coefficients: Compilation of Reference Data for FCS Calibration. (Application note rev. 1, PicoQuant GmbH, Berlin, 2010). [Google Scholar]
- 36.Schwille P., Haustein E., Fluorescence correlation spectroscopy–An introduction to its concepts and applications. Spectroscopy (Springf.) 94, (2001). [Google Scholar]
- 37.Zhang J., Ma Y., Taylor S. S., Tsien R. Y., Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc. Natl. Acad. Sci. U.S.A. 98, 14997–15002 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bajar B. T., et al. , Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 6, 20889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gnanapragasam M. N., et al. , p66Alpha-MBD2 coiled-coil interaction and recruitment of Mi-2 are critical for globin gene silencing by the MBD2-NuRD complex. Proc. Natl. Acad. Sci. U. S. A. 108, 7487–7492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holehouse A. S., Garai K., Lyle N., Vitalis A., Pappu R. V., Quantitative assessments of the distinct contributions of polypeptide backbone amides versus side chain groups to chain expansion via chemical denaturation. J. Am. Chem. Soc. 137, 2984–2995 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohn J. E., et al. , Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12491–12496 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mittal A., Holehouse A. S., Cohan M. C., Pappu R. V., Sequence-to-Conformation relationships of disordered regions tethered to folded domains of proteins. J. Mol. Biol. 430, 2403–2421 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., Svergun D. I., Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Tria G., Mertens H. D. T., Kachala M., Svergun D. I., Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ 2, 207–217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., et al. , A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das R. K., Huang Y., Phillips A. H., Kriwacki R. W., Pappu R. V., Cryptic sequence features within the disordered protein p27Kip1 regulate cell cycle signaling. Proc. Natl. Acad. Sci. U.S.A. 113, 5616–5621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherry K. P., Das R. K., Pappu R. V., Barrick D., Control of transcriptional activity by design of charge patterning in the intrinsically disordered RAM region of the Notch receptor. Proc. Natl. Acad. Sci. U.S.A. 114, E9243–E9252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riback J. A., et al. , Innovative scattering analysis shows that hydrophobic disordered proteins are expanded in water. Science 358, 238–241 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wimley W. C., White S. H., Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3, 842–848 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Kato M., et al. , Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nott T. J., et al. , Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 57, 936–947 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobrynin A. V., Theory and simulations of charged polymers: From solution properties to polymeric nanomaterials. Curr. Opin. Colloid Interface Sci. 13, 376–388 (2008). [Google Scholar]
- 53.Borgia A., et al. , Extreme disorder in an ultrahigh-affinity protein complex. Nature 555, 61–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greig J. A., et al. , Arginine-enriched mixed-charge domains provide cohesion for nuclear speckle condensation. bioRxiv:10.1101/771592 (16 September 2019). [DOI] [PMC free article] [PubMed]
- 55.Lin Y., Currie S. L., Rosen M. K., Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 292, 19110–19120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evers T. H., van Dongen E. M., Faesen A. C., Meijer E. W., Merkx M., Quantitative understanding of the energy transfer between fluorescent proteins connected via flexible peptide linkers. Biochemistry 45, 13183–13192 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper can be found in the SI Appendix and Figshare, doi:10.6084/m9.figshare.10029254.