Significance
Generation of heat by brown and beige fat cells is a potential avenue to increased energy expenditure, and thus management of obesity and metabolic syndrome. PM20D1 plays a role in thermogenesis based on mouse studies, but its expression had not been investigated in human adipocytes. Here we show that human PM20D1 expression is genetically variable at 2 levels. Genotype at certain distant variants correlates with overall PM20D1 expression levels across all human tissues (an “on/off switch”), while a different variant near the gene determines its regulation specifically in adipocytes by the PPARγ receptor and the antidiabetic drugs that target it (a “rheostat”). Human regulatory genetic variation in PM20D1 expression is associated with obesity and may ultimately inform individualized medicine approaches.
Keywords: genetics of gene regulation, adipocyte browning, PM20D1, PPARγ, obesity
Abstract
PM20D1 is a candidate thermogenic enzyme in mouse fat, with its expression cold-induced and enriched in brown versus white adipocytes. Thiazolidinedione (TZD) antidiabetic drugs, which activate the peroxisome proliferator-activated receptor-γ (PPARγ) nuclear receptor, are potent stimuli for adipocyte browning yet fail to induce Pm20d1 expression in mouse adipocytes. In contrast, PM20D1 is one of the most strongly TZD-induced transcripts in human adipocytes, although not in cells from all individuals. Two putative PPARγ binding sites exist near the gene’s transcription start site (TSS) in human but not mouse adipocytes. The −4 kb upstream site falls in a segmental duplication of a nearly identical intronic region +2.5 kb downstream of the TSS, and this duplication occurred in the primate lineage and not in other mammals, like mice. PPARγ binding and gene activation occur via this upstream duplicated site, thus explaining the species difference. Furthermore, this functional upstream PPARγ site exhibits genetic variation among people, with 1 SNP allele disrupting a PPAR response element and giving less activation by PPARγ and TZDs. In addition to this upstream variant that determines PPARγ regulation of PM20D1 in adipocytes, distinct variants downstream of the TSS have strong effects on PM20D1 expression in human fat as well as other tissues. A haplotype of 7 tightly linked downstream SNP alleles is associated with very low PMD201 expression and correspondingly high DNA methylation at the TSS. These PM20D1 low-expression variants may account for human genetic associations in this region with obesity as well as neurodegenerative diseases.
Given the worldwide epidemics of obesity and diabetes, stimulating energy expenditure by brown and beige adipose tissue has emerged as a promising avenue to weight loss and treatment of metabolic diseases (1, 2). Uncoupling protein 1 (UCP1) is key to brown adipocyte function, residing in the inner mitochondrial membrane and dissipating the proton gradient as heat rather than generating ATP. However, there are UCP1-independent mechanisms of thermogenesis (3, 4), several of which have been described recently, including futile cycling of calcium (5) and creatine (6). PM20D1 also mediates UCP1-independent thermogenesis (7). Mouse Pm20d1 was identified based on its expression enriched in brown versus white adipocytes. PM20D1 is a secreted enzyme that condenses fatty acids and amino acids to generate N-acyl amino acids (NAAs), and bidirectionally catalyzes the reverse hydrolysis reaction as well. These NAA metabolites may act in a paracrine or even endocrine manner as endogenous uncouplers, binding mitochondria of cells without UCP1 to stimulate energy expenditure. NAA analogs are thus in the early stages of pharmacological development (8).
PM20D1 has also emerged from human genetic studies, most notably in neurodegenerative diseases. PM20D1 is 1 of 5 candidate genes falling in the PARK16 locus on chromosome 1, one of the first identified and strongest Parkinson’s disease-associated loci in genome-wide association studies (GWAS) (9). Genetically determined differential expression and methylation of PM20D1 was found in human brain samples, with the high methylation and low-expression genotypes showing increased risk of Alzheimer’s disease (10). Furthermore, mouse studies with over- or underexpression of PM20D1 support its protective role in neuropathology (10).
We sought to define the expression and regulation of PM20D1 in human adipocytes, in particular by peroxisome proliferator-activated receptor-γ (PPARγ), a nuclear receptor transcription factor and master regulator in adipocytes (11). We show strong induction of PM20D1 by PPARγ agonist drugs, although this effect is species-specific, occurring in humans but not mice due to a segmental duplication event in the primate genome. Furthermore, the effect of PPARγ on PM20D1 in adipocytes differs among people due to natural genetic variation in a PPARγ binding site upstream of the gene. This variant thus functions as a “rheostat” to change adipose PM20D1 levels in response to PPARγ ligands. However, different variants downstream of PM20D1 also strongly correlate with its expression in fat and across most other human tissues, affecting methylation of the promoter and thus serving as an “on/off” switch for overall expression. Therefore, common natural genetic variation determines human PM20D1 expression at 2 levels, with downstream variants correlating with expression in all tissues, and an unlinked upstream variant determining induction by PPARγ in adipocytes. Differential PM20D1 expression due to genetic variation may drive disease risk, making it a potential target for individualized medicine approaches.
Results
PM20D1 Expression Is Activated by Thiazolidinedione in Human but Not Mouse Adipocytes.
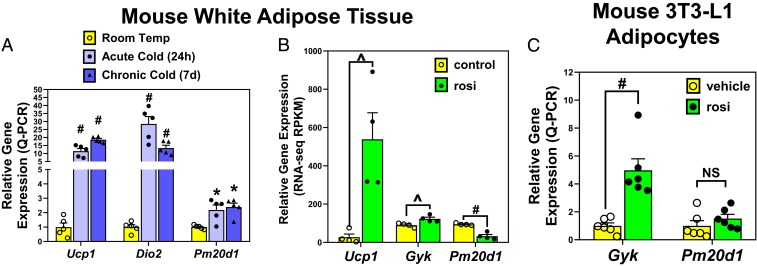
In mice, Pm20d1 was identified as a candidate thermogenic gene, with its mRNA levels cold-induced and enriched in brown and beige versus white adipocytes (7). We confirmed the ∼2-fold induction by acute or chronic cold in mouse inguinal white adipose tissue (WAT), although this induction was less than classic thermogenic genes Ucp1 and Dio2 (Fig. 1A). Thiazolidinedione (TZD) PPARγ agonist drugs are another strong stimulus of adipocyte browning, which may also be predicted to induce Pm20d1. Remarkably, however, the TZD rosiglitazone decreased Pm20d1 expression in mouse gonadal WAT, despite inducing other PPARγ target genes, including Ucp1 and glycerol kinase (Gyk) (Fig. 1B). TZD repression of Pm20d1 was also observed in mouse inguinal WAT (SI Appendix, Fig. S1A), as previously reported in immortalized white adipocytes from this adipose depot (12). In mouse 3T3-L1 adipocytes, treatment with rosiglitazone fails to increase Pm20d1 expression, while Gyk is significantly induced (Fig. 1C). Consistent with the lack of induction by rosiglitazone, the mouse Pm20d1 locus shows no PPARγ binding regions based on published chromatin immunoprecipitation sequencing (ChIP-seq) from mouse WAT or 3T3-L1 adipocytes, while Ucp1 shows the expected occupancy (SI Appendix, Fig. S1B).
Fig. 1.
Mouse adipose Pm20d1 is induced by cold but not PPARγ agonist drug. (A) Wild-type mice were housed at room temperature versus 4 °C for 24 h (acute cold) or 7 d (chronic cold, n = 5 per group), and gene expression measured by qPCR in inguinal WAT. (B) Wild-type mice were treated with rosiglitazone (rosi) versus control diet (n = 4 per group), and gene expression measured by RNA-seq in epididymal WAT. (C) Mouse 3T3-L1 adipocytes were treated for 24 h with 1 μM rosiglitazone versus vehicle (DMSO, n = 6 wells each). Mean and SEM, #P < 0.001; *P < 0.01; ^P < 0.05; NS, not significant by 2-tailed type 2 Student’s t-test.
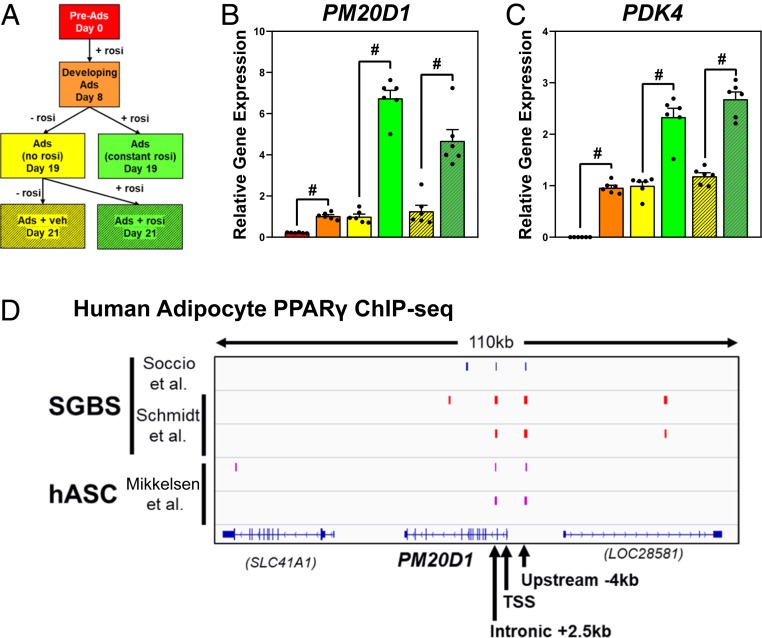
The regulation of PM20D1 differs in human adipocytes. In a microarray gene-expression analysis of human Simpson–Golabi–Behmal syndrome (SGBS) adipocytes at passage 35, PM20D1 was the second most-highly rosiglitazone-induced gene, with other known targets relevant to adipocyte browning, like UCP1 and glycerol kinase, also strongly induced (Table 1). In a more detailed analysis of SGBS adipocyte differentiation and rosiglitazone treatment (Fig. 2A), PM20D1 expression was induced during 8 d in differentiation media, which included rosiglitazone (Fig. 2B, red vs. orange). For the next 10 d, cells were cultured in maintenance media with or without rosiglitazone, and mature day 18 adipocytes had higher PM20D1 expression in the constant presence of rosiglitazone (Fig. 2B, yellow vs. green). When rosiglitazone was reintroduced to the mature adipocytes without it, PM20D1 expression was similarly induced (Fig. 2B, yellow vs. green striped). This pattern of TZD regulation is the same as the classic PPARγ target PDK4 (Fig. 2C) and distinct from other genes, like ACER3, which are induced in adipogenesis but TZD-independent (SI Appendix, Fig. S2A). Similarly, strong PM20D1 activation by TZD was also observed in primary human adipocytes differentiated using stem cells from omental or subcutaneous fat, and with either rosiglitazone or pioglitazone as the TZD drug (SI Appendix, Fig. S2B). Consistent with regulation by PPARγ and TZDs, the human PM20D1 locus shows apparent PPARγ binding regions near the transcription start site (TSS) based on independent ChIP-seq analyses of SGBS adipocytes (13, 14) as well as human primary adipocytes (15) (Fig. 2D).
Table 1.
Top 10 genes induced by rosiglitazone in human SGBS adipocytes
| Affymetrix ID | RefSeq ID | Gene symbol | Gene | DMSO | Rosi | Fold-induction | P value |
| 7939056 | NM_003986 | BBOX1 | γ-Butyrobetaine hydroxylase 1 | 49.9 (6) | 267.5 (30) | 5.37 | 0.0000031 |
| 7923837 | NM_152491 | PM20D1 | Peptidase M20 domain containing 1 | 82.5 (9.5) | 437.4 (88.6) | 5.24 | 0.0000113 |
| 8081564 | NM_198196 | CD96 | CD96 molecule | 98.5 (13.4) | 476.6 (60.2) | 4.85 | 0.0000072 |
| 7958884 | NM_016816 | OAS1 | 2′-5′-oligoadenylate synthetase 1 | 289.2 (18.5) | 1269.4 (89.1) | 4.39 | 0.0000020 |
| 7938951 | NM_213599 | ANO5 | Anoctamin 5 | 46.9 (4.4) | 162.8 (47.9) | 3.35 | 0.0000768 |
| 8025402 | NM_139314 | ANGPTL4 | Angiopoietin-like 4 | 590 (15.2) | 1977 (87.6) | 3.35 | 0.0000010 |
| 7963545 | NM_175834 | KRT79 | Keratin 79 | 242 (5.4) | 811.1 (81.6) | 3.34 | 0.0000061 |
| 8166632 | NM_001128127 | GK | Glycerol kinase | 149.5 (30) | 492.4 (42.1) | 3.34 | 0.0000256 |
| 8054722 | NM_000576 | IL1B | Interleukin 1β | 261.5 (15.5) | 842.7 (78.6) | 3.22 | 0.0000082 |
| 8102904 | NM_021833 | UCP1 | Uncoupling protein 1 | 68.4 (14) | 198.8 (50.2) | 2.89 | 0.0001106 |
SGBS cells were differentiated to adipocytes then cultured in maintenance media without a TZD for 10 d. Vehicle (DMSO) or 1 uM rosiglitazone (Rosi) was then added for 36 h prior to RNA preparation and Affymetrix microarray analysis; n = 4 wells per condition, with mean (SD) shown, along with fold-induction and P value from Robust Multi-Array Average and Significance Analysis of Microarrays analysis.
Fig. 2.
PM20D1 expression is highly induced by PPARγ agonist in human adipocytes. (A) SGBS cells were cultured in adipocyte differentiation media including rosiglitazone (rosi) for 8 d, then in maintenance media with or without rosi for 11 d. Rosiglitazone or vehicle was then reintroduced to mature adipocytes for 2 d. n = 6 wells per condition, and this schema is the color key for samples in B and C. (B and C) Expression of PM20D1 or PDK4 was assayed by qPCR and normalized to 36B4 expression, with levels in mature adipocytes set equal to 1. Mean and SEM, #P < 0.001 by Student’s t-test. (D) Browser track of published human adipocyte PPARγ ChIP-seq at the PM20D1 locus, from either SGBS or primary adipocytes from human adipose stem cells (hASC) (13–15).
Segmental Duplication in the Human PM20D1 Locus Encompassing PPARγ Binding Regions.
These 2 regions of PPARγ ChIP-seq binding are located ∼2.5 kb downstream of the PM20D1 TSS in intron 2 (henceforth called “intronic”) and ∼4 kb upstream of the TSS (“upstream”). Comparisons of the intronic and upstream regions surprisingly revealed 97% sequence identity (only 6 annotated mismatches in the central 200 bp), suggesting a duplication event. Indeed, a DNA sequence of ∼1.8 kb within the PM20D1 gene (spanning from intron 1 to intron 2 and thus encompassing exon 2) appears with ∼97% overall identity in the human genome assembly upstream of the TSS in the reverse orientation (Fig. 3A). This duplication includes exon 2 sequence in reverse orientation at 100% identity, and the putative PPARγ binding is just adjacent to this exon in both the original/intronic and the duplicated/upstream region. This upstream duplication found in humans is also present in other great apes (chimp, bonobo, gorilla, orangutan, gibbon) and Old World monkeys (rhesus, baboon), but not New World monkeys (marmoset, squirrel monkey) or other primates (bush baby, lemur, tarsier), and not in mice or any other mammals (pig, cow, dog, and so forth) (Fig. 3B). This indicates that the duplication event occurred in the common ancestor of apes and Old World monkeys, after divergence from other primate lineages, thus ∼25 to 50 million y ago (16). This duplicated sequence is not annotated as a transposon or repetitive element, and the current GRCh38/hg38 genome assembly characterizes it among segmental duplications (“Duplications of >1000 Bases of Non-RepeatMasked Sequence” in SI Appendix, Fig. S3 A and B).
Fig. 3.
A segmental duplication in the human PM20D1 locus. (A) The upstream and intronic regions of PPARγ binding (red) were highly similar to each other, and fall in a larger ∼1.8-kb duplicated region (black arrows) encompassing exon 2 of PM20D1 (blue). The 97% identical duplicated DNA is ∼5 kb away and in reverse orientation. (B) Available mammalian genomes were interrogated on the UCSC genome browser, and the duplicated upstream DNA was limited to Old World monkeys and great apes. (C) Schematic zoom-in on the central 250-bp PPARγ binding region. There are 7 variable nucleotide positions in intron 2, with 2 (stars) falling in PPREs. Based on sequencing of PCR products, 3 differences distinguish the upstream and intronic paralogs (yellow), while 4 differences are true polymorphisms (green).
Segmental duplications are challenging for genome sequence annotation, as it is difficult to distinguish a true allelic difference at 1 of the paralogous regions (i.e., A:G SNP in intronic region) versus a difference between the 2 paralogous regions (i.e., A intronic and G upstream). Indeed, many paralogous sequence variants “contaminate” SNP databases (17). We defined the central 250 bp of the PPARγ binding region as spanning from positions 101 to 350 of PM20D1 intron 2, and noted 2 potential PPARγ response elements (PPREs) in this region. This region also has 3 annotated differences from the paralogous upstream sequence, all of which also have SNP annotations in dbSNP150, and other SNPs are also annotated in this region (SI Appendix, Table S1). To distinguish paralogous differences from true SNPs, we designed a PCR to selectively amplify the putative intronic paralog with a primer discriminating at the last difference (intron 2, 336-C). Genomic DNA from 13 individuals uniformly contained the intronic variants at 229-T and 242-G (SI Appendix, Fig. S3C). In addition, 2 true SNPs were identified in the intronic paralogue (120-G/A and 215-T/C), where sequencing of different individuals revealed all 3 genotypes (i.e., G/G, G/A, A/A at position 120). We also selectively amplified the upstream paralogue with the equivalent primer (T at position paralogous to 326-C), and sequencing showed the upstream variants (C paralogous to 229-T, and A to 242-G), as well as 2 true SNPs that only appeared in this upstream-selective PCR product (180-A/C and 332-C/T). In summary, these results show 7 total variants in the PM20D1 PPARγ binding region, 3 differing between the paralogs, 2 true SNPs only in the intronic paralog, and 2 true SNPs only in the upstream paralog (Fig. 3C).
PPARγ Binds Selectively to the Duplicated Region Upstream of PM20D1.
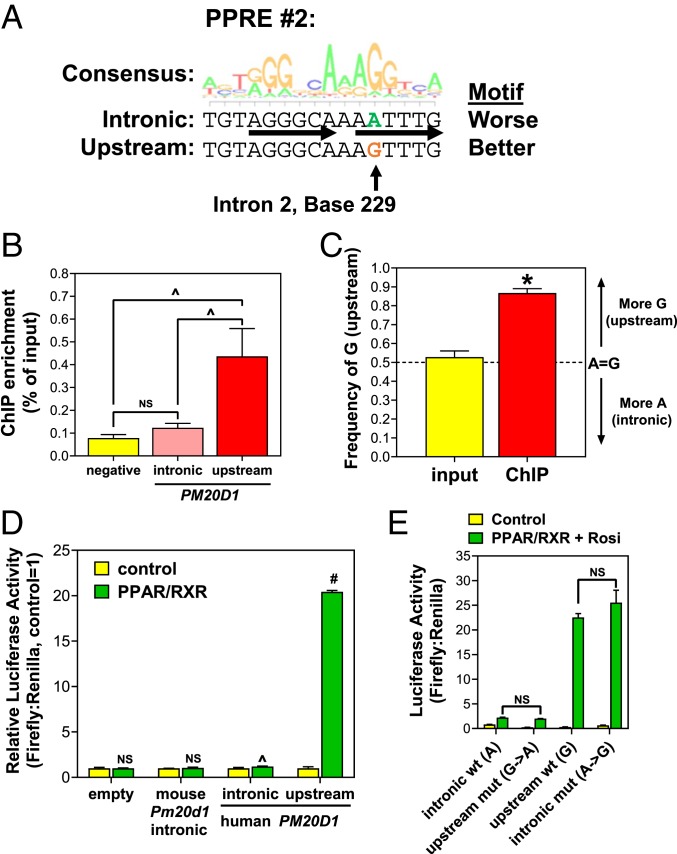
One of differences between the intronic and upstream paralogs (intron 229-T versus upstream-C) falls in the second PPRE (Fig. 3C). The consensus PPRE is defined as a nuclear receptor direct repeat 1 motif (AGGTCA separated by 1 nucleotide), and at this PPRE the base present in the upstream paralog (C) gives much better agreement with the consensus PPRE motif than the intronic base (T) (Fig. 4A; note the PPRE is on the reverse strand, and the second position of the AGGTCA half-site prefers G to A). The first PPRE motif does not differ between the intronic and upstream reference sequences (although in the upstream region this motif has a true SNP) (Fig. 3C and below), so based on this second PPRE difference, the upstream region would be predicted to bind PPARγ better. While the 2 peak heights are similar in ChIP-seq data, most reads that aligned to either the intronic or upstream paralog could have been assigned to the other given their high-sequence identity. A strict requirement for unique read alignment at a single genomic location might have been expected to eliminate such reads from bioinformatic analysis, yet the 3 independent PPARγ ChIP-seq analyses in human adipocytes using different computational approaches all identified both peaks (Fig. 2C).
Fig. 4.
The upstream duplicated region mediates PPARγ activation of PM20D1. (A) The consensus motif logo for a PPRE is shown, along with the sequences of the second PPRE in the intronic and upstream putative PPARγ binding regions near PM20D1. There is 1 A:G difference (arrow), which predicts better PPARγ binding to the upstream G. (B) PPARγ ChIP was performed in SGBS cells, followed by qPCR to amplify a negative site, versus selective primers that distinguish the intronic or upstream sites in PM20D1. (C) Pyrosequencing analysis of PPARγ ChIP DNA, with the PCR not distinguishing the intronic and upstream paralogues, yet the ChIP but not input DNA showing enrichment of the upstream G nucleotide. (D) Luciferase reporter assay showing that mouse Pm20d1 intronic sequence is not activated by PPARγ, while the human upstream sequence is much more strongly activated than the intronic paralog. (E) Mutagenesis of luciferase reporters shows that the A:G difference in PPRE#2 is responsible for this difference. Mean and SEM, #P < 0.001; *P < 0.01; ^P < 0.05; NS, not significant by 2-tailed type 2 Student’s t-test.
Based on the ChIP-seq reads alone it was unclear whether PPARγ binding occurs at both locations or selectively to 1 paralog, so we used 2 other methods to distinguish them. First, we designed selective ChIP qPCR primers based on intron position 242, to test in PPARγ ChIP DNA from SGBS adipocytes. The intron-selective primer pair failed to show PPARγ binding here, with the same occupancy as a negative control site in the ALB gene, while the upstream-selective primer pair showed significant PPARγ occupancy (Fig. 4B). Second, we used pyrosequencing to quantify the amount of upstream versus intron sequence based on the second PPRE difference (intron 229-A, upstream G). PPARγ ChIP in SGBS adipocytes was followed by a PCR to amplify the intronic and upstream paralogues nonselectively, then this mixed product was subjected to pyrosequencing. Input DNA (total chromatin not selected for PPARγ binding), showed equal ∼50% representation of the upstream (G) and intronic (A), as expected, while ChIP DNA (PPARγ binding) had significant enrichment of the upstream region (∼90% G) (Fig. 4C). These results demonstrate that, on chromatin in adipocyte nuclei, PPARγ binds near PM20D1 almost exclusively to the upstream paralogue relative to the intronic 1, consistent with a sequence difference in the second PPRE, and despite both regions having the same first PPRE.
We also tested upstream-selective PPARγ binding to PM20D1 by transient transfection luciferase reporter assays. Luciferase reporters were cloned driven by 344 bp of human PM20D1 intronic or upstream sequence, differing only by the 3 nucleotides distinguishing the paralogs (intron positions 229, 242, 336). A reporter driven by 383 bp of syntenic mouse Pm20d1 intron 2 sequence was also cloned (the upstream segmental duplication does not exist in rodents), although it does not appear to have either PPRE present in humans and would thus not be predicted to bind PPARγ (SI Appendix, Fig. S4). Reporters were transfected into 293T cells with or without expression plasmids for PPARγ and its heterodimeric partner RXRα. The empty vector and mouse intron reporter were not activated by PPARγ/RXRα, the human intron reporter was weakly activated by only ∼20%, while the human upstream reporter was strongly activated ∼20-fold (Fig. 4D). Thus, the PM20D1 intron 2 region from mice and humans does not confer strong PPARγ activation, while the primate-specific segmental duplication upstream of human PM20D1 does.
To confirm that this difference between the human paralogous reporters was due to the PPRE difference (position 229) rather than the other 2 paralogous differences (positions 242 and 336), we performed site-directed mutagenesis and determined the transcription of luciferase reporters in the presence of PPARγ (and its ligand rosiglitazone to maximize activity). Mutating the upstream PPRE G to A reduced maximal activity to resemble the wild-type intronic reporter with A, while mutating the intronic reporter PPRE from A to G increased activity to similar levels to the wild-type upstream with G (Fig. 4E). Therefore, a single nucleotide difference in 1 of the 2 PPREs drives PPARγ binding and activation of the upstream segmental duplicated region rather than the paralogous original intronic region.
Human Genetic Variation in PPARγ Activation of PM20D1 in Adipocytes.
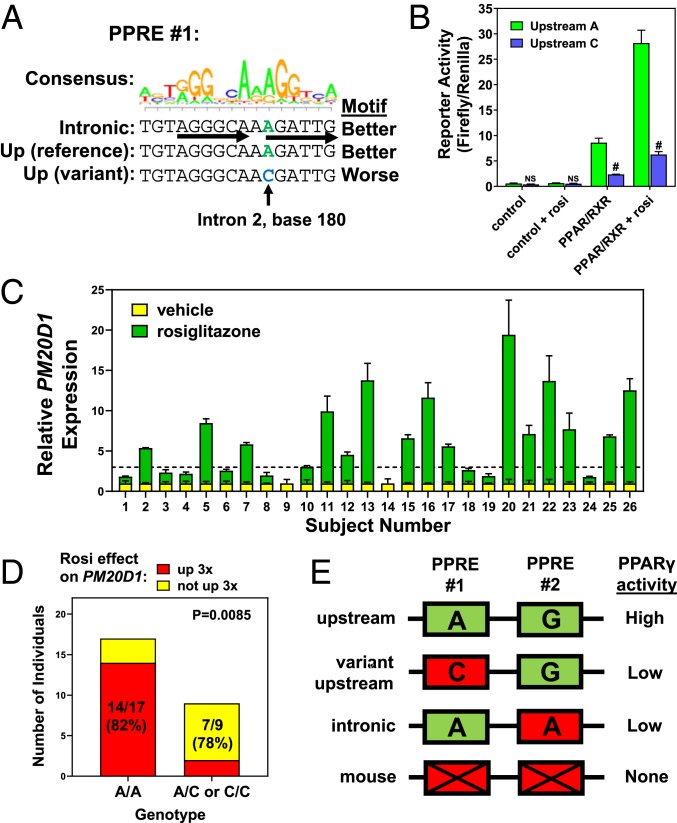
This functional PPARγ binding region upstream of PM20D1 has the second PPRE that distinguishes it from the intronic region, but also the first PPRE, which has a true SNP that differs among people (rs6667995) (Fig. 3C). The reference A allele agrees with consensus at the first position of an AGGTCA half site, while the alternate C allele does not (Fig. 5A). We hypothesized that if the first PPRE contributes to PPARγ binding, then the C allele would reduce reporter activity. We cloned an upstream luciferase reporter from an individual with the C allele, and its activation by PPARγ/RXRα was markedly reduced, both in the presence and absence of rosiglitazone (Fig. 5B). Therefore, some individuals have a single polymorphic nucleotide in the upstream PM20D1 region first PPRE that reduces PPARγ transcriptional activity.
Fig. 5.
An upstream PPARγ motif-altering polymorphism drives genetic variation in TZD activation of PM20D1. (A) The consensus PPRE motif logo is shown above the first PPRE sequences in the intronic and upstream regions near PM20D1. A C variant in the upstream site is predicted to reduce PPARγ binding activity. (B) Luciferase reporter assay of upstream reporters, showing that the C variant has reduced activation by PPARγ and rosiglitazone (rosi). Mean and SEM, #P < 0.001 versus A allele; NS, not significant; 2-tailed type 2 Student’s t-test. (C) Human adipose stem cell-derived adipocytes were cultured from 26 individuals and treated with rosiglitazone, and induction of PM20D1 as measured by qPCR was variable. Mean and SEM. (D) Individuals with low response of PM20D1 to rosi enriched for C alleles at this PPRE SNP. Contingency P value by 2-sided Fisher’s exact test. (E) Schematic model showing requirement for both PPREs for maximal PPARγ activity.
We hypothesized that individuals with the C variant at this PPRE would show reduced transcriptional response of PM20D1 to rosiglitazone. Primary adipocytes were differentiated using stem cells from subcutaneous fat biopsies of 26 individuals. Cultured adipocytes were treated with rosiglitazone versus vehicle control, and 16 individuals showed strong activation of PM20D1 as expected, yet 10 showed less than 3-fold activation (Fig. 5C). This defect was specific to PM20D1, as rosiglitazone activated the PPARγ target gene FABP4 similarly and >3-fold in adipocytes from all 26 individuals (SI Appendix, Fig. S5A). Only 2 individuals had the homozygous C/C genotype at the rs6667995 A/C SNP, and both had <2-fold activation of PM20D1 by rosiglitazone despite strong activation of FABP4 (SI Appendix, Fig. S5 B and C). Activation was more variable in A/A and A/C individuals, but there was a trend toward higher-fold activation in A/A versus the other 2 genotypes (SI Appendix, Fig. S5C). Defining an activation threshold of 3-fold, there was a highly significant genotype effect, with 82% (14 of 17) of A/A homozygous individuals showing >3-fold rosiglitazone activation of PM20D1, compared to only 22% (2 of 9) of individuals with the alternate allele (A/C or C/C) (Fig. 5D). Therefore, presence of a C allele at rs6667995 disrupts a PPRE and predicts poor transcriptional response of PM20D1 to PPARγ agonist drug. Overall, the data thus far are consistent with both PPREs being necessary for full PPARγ binding activity, such that it is reduced by disruption of either the first PPRE (via the SNP in the upstream region) or the second PPRE (due to the paralogous variant in the intronic region) (Fig. 5E).
Human Genetic Variation in PM20D1 Expression in Adipose and Other Tissues.
Since the upstream PPRE SNP rs6667995-C allele reduces the effect of PPARγ agonists on PM20D1 expression in cultured adipocytes, we tested its effect on PM20D1 expression in human adipose tissue samples. Abdominal subcutaneous fat biopsies from 50 individuals showed wide variation in PM20D1 expression as measured by qPCR. Some samples had threshold cycle (Ct) values of 28, indicating robust expression, while 6 samples had undetectable expression (Ct > 35). rs6667995 genotyping was performed on genomic DNA from these fat samples, yet genotypes at this SNP did not correlate with expression levels (SI Appendix, Fig. S6A).
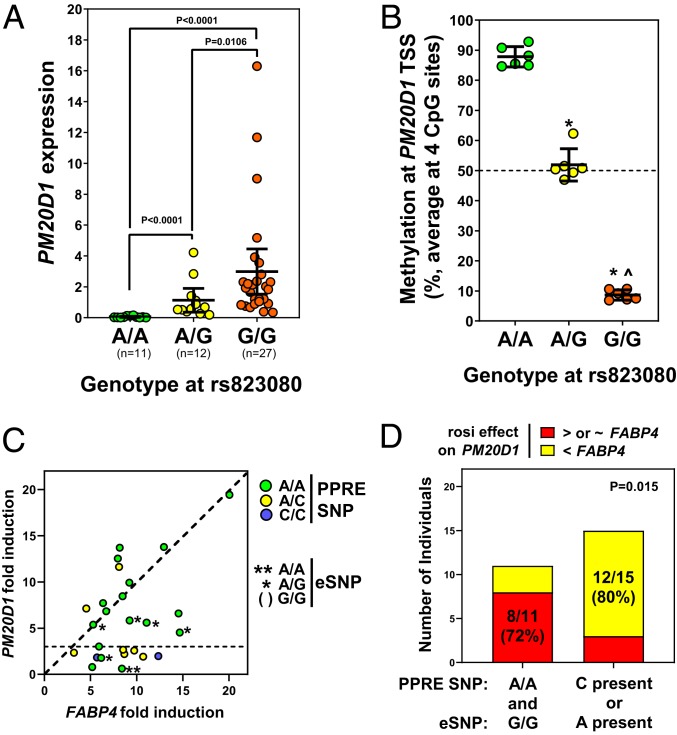
Given the wide variation in adipose PM20D1 expression, we looked at other genetic variants in published expression quantitative trait loci (eQTL) studies. An SNP rs708727 located ∼51 kb downstream of the PM20D1 TSS shows very strong association with PM20D1 expression in adipose tissue. This signal was 1 of the strongest in a survey of ∼1,000 adipose biopsies from obese subjects (18), with P values less than 10−70 ranking in the top 200 most significant eQTLs (of >23,000 total) in both subcutaneous and visceral adipose tissue. This strong eQTL signal was confirmed in other adipose datasets, including MuTHER (19), METSIM (20), and GTEx (21). rs708727 itself is a synonymous substitution in a coding exon of the downstream gene SLC41A1, thus unlikely to causally affect PM20D1 expression. Six other SNPs downstream of PM20D1 are also tightly linked to rs708727 (r2 > 0.95), including rs823080 in the intergenic region between PM20D1 and SLC41A1 (SI Appendix, Fig. S6B). We genotyped rs823080 in the 50 adipose tissue biopsies, and confirmed the strong correlation of genotype with PM20D1 expression. A/A individuals had very low to undetectable adipose PM20D1 expression, G/G individuals had much higher expression, while average expression in A/G heterozygous was intermediate, as expected for cis-acting codominant effects (Fig. 6A). Genotype at this SNP does not correlate with expression of an adipocyte marker gene, adiponectin (SI Appendix, Fig. S6C).
Fig. 6.
Genetically variable human PM20D1 expression in adipose and other tissues. (A) Expression of PM20D1 was measured by qPCR in subcutaneous WAT biopsies from 50 individuals, divided based on their genotype at SNP rs823080. Pairwise P values calculated by 2-tailed type 2 Student’s t-test. (B) DNA methylation near the PM20D1 start site was assayed by bisulfite pyrosequencing in a subset of these biopsies (n = 6 per genotype, *P < 0.01 versus A/A, ^P < 0.01 versus G/G by student’s t-test). (C) In the cultured adipocytes from 26 individuals, the rosiglitazone (rosi) response of PM20D1 (in this scatterplot relative to the control gene FABP4) is reduced by the A allele at this expression SNP (eSNP), as well as the C allele at the PPRE SNP. (D) Either minor allele predicts low response of PM20D1 to rosiglitazone. Contingency P value by 2-sided Fisher’s exact test.
It is unlikely that gene activation by PPARγ accounts for the variation in PM20D1 expression among people. First, no PPARγ binding is apparent by ChIP-seq in the downstream region where eQTL SNPs reside (Fig. 2D). Second, there is a recombination hotspot (red rectangle in SI Appendix, Fig. S6B) in the PM20D1 gene separating the downstream eQTL from the TSS and upstream, such that none of the eQTL SNPs are linked to the PPARγ binding region we identified above. Third, and most importantly, the eQTL signal occurs not only in fat, where PPARγ is abundant, but also in nearly every other human tissue surveyed in GTEx (SI Appendix, Fig. S6D), including tissues that do not express PPARγ. These tissues include some brain regions, where PM20D1 genotype-dependent expression and imbalance was recently reported (10). These authors also showed that genotype at upstream eQTL SNPs correlates with methylation status at a CpG island near the PM20D1 TSS in brain. We quantified TSS methylation in our adipose tissue DNA. Adipose tissue samples with the A/A genotype and low PM20D1 expression showed nearly complete >90% methylation, while G/G gave <10% methylation, and A/G showed intermediate ∼50% methylation consistent with 1 methylated and 1 unmethylated copy (Fig. 6B). We also assayed these methylation levels in SGBS adipocytes, which are A/G at rs823080 and thus have intermediate methylation. We found that methylation was unchanged during adipogenic differentiation (when expression of PPARγ and PM20D1 increases), and furthermore was not decreased when PM20D1 expression is induced by the PPARγ agonist rosiglitazone (SI Appendix, Fig. S6E). Therefore, basal PM20D1 expression in multiple tissues, including fat, correlates strongly with genotype at downstream eQTL SNPs and CpG methylation at the TSS, independent of PPARγ.
We also genotyped the downstream eQTL SNP rs823080 in the cultured human adipocytes treated with rosiglitazone. In contrast to genotype at the upstream PPRE SNP (Fig. 5D), genotype at this expression SNP alone did not predict rosiglitazone response (SI Appendix, Fig. S6F). Only 1 sample had the A/A low-expression genotype, and these adipocytes had low PM20D1 expression that was not induced by rosiglitazone (despite the favorable genotype at the upstream PPRE SNP) (Fig. 6C). Furthermore, while G/G adipocytes typically showed PM20D1 rosiglitazone induction equal to or higher than FABP4, most G/A heterozygous failed to induce PM20D1 to same extent. There was a genotype interaction such that adipocytes with the reference homozygous genotypes (AA at upstream PPRE and GG downstream) showed strong PM20D1 induction, while the presence of either alternate allele (upstream C disrupting the PPRE or downstream A correlating with TSS methylation) reduced this effect (Fig. 6D). Therefore, by affecting methylation and silencing global expression of PM20D1, the downstream SNPs also modulate the degree of induction of PM20D1 by TZD in adipocytes.
Potential Human Phenotypes Due to PM20D1 Genetic Variation.
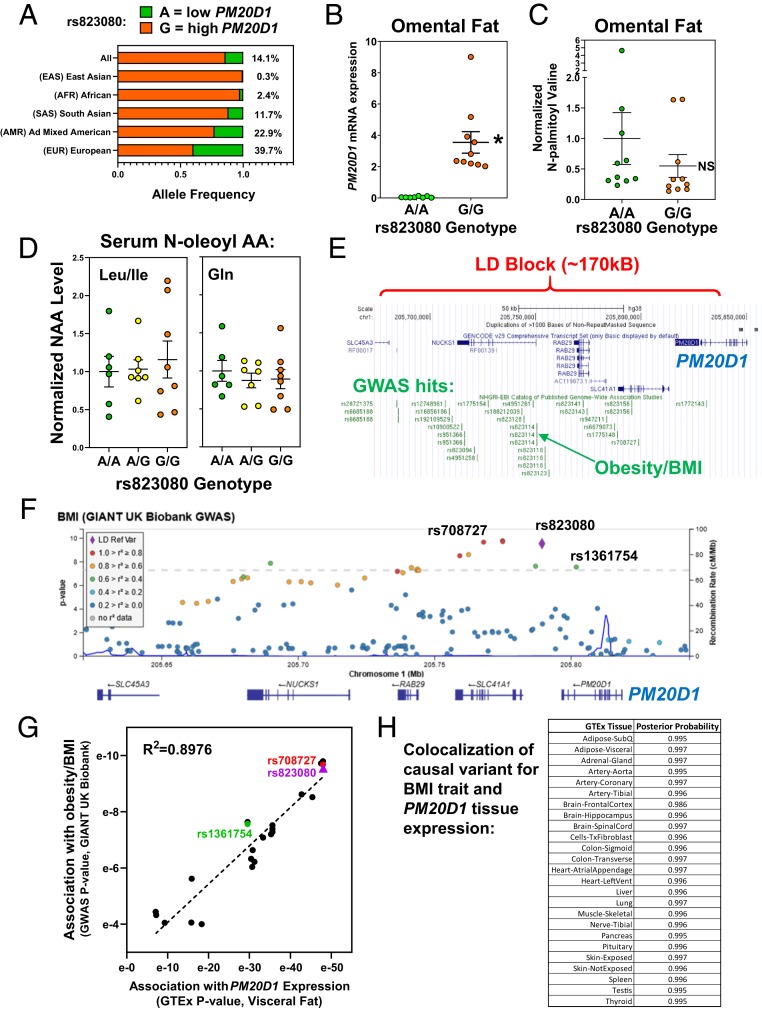
The rs823080 A allele, which correlates with PM20D1 high methylation and low expression, is very rare in African and East Asian populations (under 3%), but quite common in European populations (∼40%), with an overall frequency of ∼14% (Fig. 7A). Given the prevalence of genetically silenced PM20D1, we sought human phenotypes associated with PM20D1 expression SNPs.
Fig. 7.
Potential phenotypes of human PM20D1 genetic variation. (A) Population-specific allele frequencies were calculated for the PM20D1 eSNP rs823080 using LDhap. (B) PM20D1 mRNA expression was quantified by qPCR in 20 human omental fat samples, divided by rs823080 genotype with n = 10 A/A (green) and n = 10 G/G (orange). Mean and SEM, *P < 0.01 by student’s t-test. (C) The metabolite N-palmitoyl-valine was measured in these same 20 omental fat samples. Mean and SEM, *P < 0.01. (D) The indicated NAAs were measured in human serum samples from 21 individuals differing by rs823080 genotype, n = 6 A/A (green), n = 7 A/G (yellow), and n = 8 G/G (orange). Mean and SEM. (E) The linkage disequilibrium (LD) block containing the PM20D1 expression SNPs was visualized on the UCSC genome browser, along with a catalog of lead SNPs in published GWAS studies (green). (F) Locus zoom plot showing association of BMI with SNPs in the PM20D1 locus, colored by their linkage to the eSNP rs823080 (purple). (G) Scatterplot correlating the significance of each SNP’s association with BMI (via GWAS query, y axis) versus PM20D1 expression (via GTEx query, x axis). (H) Colocalization analysis was performed to calculate the likelihood that the same causal SNP drives both PM20D1 expression in various tissue and the BMI GWAS signal in this locus.
Since PM20D1 has an enzymatic activity to synthesize or hydrolyze NAAs (7), we quantified levels of these metabolites in human biospecimens using liquid chromatography-mass spectrometry (LC-MS). First, we assayed NAAs in omental adipose tissue samples from 20 individuals with high or low PM20D1 expression levels (G/G versus A/A genotype at downstream rs823080 SNP) (Fig. 7B), and failed to find significant genotype-dependent effects on levels of various NAAs (SI Appendix, Fig. S7A). For example, N-palmitoyl valine was detectable in human omental fat, and valine NAAs were previously shown to increase in mice overexpressing PM20D1 (7), yet levels did not differ in human fat with genotype-dependent PM20D1 expression (and the trend is in the opposite direction, with higher expression G/G genotype having the lower mean NAA level) (Fig. 7C). A similar lack of genotype effect was found in paired subcutaneous and omental fat from 10 other subjects (SI Appendix, Fig. S7B). While there were no significant genotype effects, differences were found that validate our NAA measurement by LC-MS. All 3 taurine NAAs showed significant ∼2-fold higher levels in omental versus subcutaneous fat (SI Appendix, Fig. S7B). This difference between fat depots is consistent with the prominent role of taurine in bile acid physiology (22), with likely more taurine in the portal circulation to the omental fat. For many other NAAs, there was a tight correlation between the levels measured in omental and subcutaneous fat in each individual (SI Appendix, Fig. S7C), showing that there are consistent NAA level differences across individuals that do not correlate with PM20D1 expression due to rs823080 genotype.
It is unknown which tissues are the main source of circulating NAAs, but the rs823080 SNP genotype correlates with PM20D1 expression across tissues and thus could affect serum levels, regardless of source. However, serum NAA levels also did not differ depending on genotype (SI Appendix, Fig. S7D). For example, N-oleoyl leucine/isoleucine or glutamine, which change significantly in mouse models of PM20D1 overexpression or knockout (7, 23), did not differ in serum samples from humans with genotype-dependent differences in PM20D1 expression (Fig. 7D).
We also surveyed the PM20D1 locus for signals in human genetic studies. The 7 SNPs highly associated with PM20D1 expression were also tightly linked to each other. Haplotype analysis of 5,008 chromosomes showed perfect linkage of all 7 major or minor alleles in 98.8%, with only 1.2% of haplotypes showing any recombination (SI Appendix, Fig. S8A). The linkage disequilibrium block containing these SNPs spans ∼170 kb encompassing 4 genes (PM20D1, SLC41A1, RAB7L1/RAB29, and NUCKS1). Remarkably, 24 SNPs in this region are cataloged as significant lead SNPs in GWAS (Fig. 7E). To see which of these GWAS phenotypes may be caused by differences in PM20D1 expression, we looked for linkage among these GWAS SNPs and the 7 expression SNPs. Several GWAS traits, including body anthropometrics (height, body mass index [BMI]), lipids (HDL cholesterol), neurological phenotypes (Parkinson’s disease, tremor, reaction time), and blood cell counts (monocytes), were linked at r2 > 0.5 to 1 or more PM20D1 expression SNPs (SI Appendix, Fig. S8B). Therefore, it is plausible that genetic differences in PM20D1 expression could causally drive these phenotypes.
Given the potential role of PM20D1 in adipose tissue biology, we also interrogated the expression SNPs in GWAS metaanalyses for cardiometabolic traits in European populations with the highest minor allele frequencies. Associations with BMI, a measure of obesity, were found for rs823080 and the other tightly linked expression SNPs for PM20D1 (SI Appendix, Fig. S8C, red), as well as all other linked SNPs (SI Appendix, Fig. S8C, orange and green), all showing a prominent regional association at P ∼ 10−4 (SI Appendix, Fig. S8C). Obesity is associated with other components of the metabolic syndrome (24), and similar associations were found in this locus for type 2 diabetes and HDL cholesterol levels. The rs823080-A allele was associated with silenced PM20D1 expression, lower BMI, higher HDL-C, and lower diabetes risk.
While the signals in these individual GWAS analyses did not reach the accepted threshold of genome-wide significance (P < 5 × 10−8), this did emerge in the largest current GWAS metaanalysis for BMI, the GIANT UK Biobank study (25). There are genome-wide significant associations for rs823080 (P = 2.8 × 10−10) and the other tightly linked expression SNPs for PM20D1 (Fig. 7F, red), as well as all other linked SNPs (Fig. 7F, orange and green). Notably, 1 of these moderately linked SNPs, rs1361754 (SI Appendix, Fig. S6B), is a missense coding variant in PM20D1 exon 11, resulting in an Ile380Thr amino acid substitution in the M20 dimerization domain (not in the catalytic domain). Pairwise linkage analysis shows that this missense SNP is linked to rs823080 at r2 = 0.6, with the Ile-coding T allele occurring only in haplotypes with the rs823080-G high PM20D1 expression allele (SI Appendix, Fig. S9A). Consistent with this, the missense SNP T/T genotype gives high PM20D1 expression that is indistinguishable from the rs823080-G/G genotype (SI Appendix, Fig. S9B). However, the missense SNP C allele encoding Thr occurs in haplotypes with either rs823080-A (low PM20D1) or -G (high PM20D1). The missense SNP C/C genotype thus gives higher and more variable expression than the rs823080-A/A, such that rs823080 genotype shows much more significant association with PM20D1 expression than rs1361754 (SI Appendix, Fig. S9B).
Compared to our lead SNP rs823080, the missense SNP rs1361754 is less significantly associated with both PM20D1 expression (by GTEx query) (SI Appendix, Fig. S9C) and with BMI (by GWAS query) (Fig. 7F), leading us to investigate this correlation for all SNPs in this locus. For the 24 SNPs with BMI P values calculated in the GIANT UK Biobank GWAS, we found a striking correlation with the P values for PM20D1 expression in adipose tissue (Fig. 7G). Thus, each SNP’s effect on PM20D1 expression correlates tightly with its effect on BMI. Furthermore, colocalization analysis was performed and showed that the signal corresponding to variation that changes the expression of PM20D1 was indistinguishable from variation associated with changes in BMI (posterior probability ≥0.995 across tissues) (Fig. 7H), supporting the hypothesis that PM20D1 expression and BMI traits map to a single, underlying causal variant. These results are inconsistent with a causal role in obesity for the PM20D1 I380T coding difference due to rs1361754. Instead, all results are highly consistent with the hypothesis that noncoding regulatory genetic variation leads to PM20D1 silencing and drives the association with body weight.
Discussion
The regulation of human PM20D1 expression is complex and differs genetically among people at 2 levels. Downstream on/off switch variants control overall expression across all tissues, while an upstream variant affects the “rheostat” for PPARγ-regulated expression in adipose tissue.
The adipose tissue rheostat is a PPARγ binding site ∼4 kb upstream of the PM20D1 TSS. Interestingly, this regulatory element occurs in an ∼1.8-kb segmental duplication (Fig. 3), also called a low-copy repeat, defined as long (>1 kb) regions of nearly identical sequence (90 to 100% identity) that exist in multiple locations. These are distinct from more abundant interspersed repeats that arise from transposable elements, and the mechanism of segmental duplication is unknown. However, segmental duplications occurred at high frequency in the human–great ape lineage (>5% of sequence), and they are implicated in the evolution of novel genes and regulatory elements by allowing “mutational tinkering” of copies (26). Here we show that PM20D1 is a clear example of this, with PPARγ regulatory function appearing in an ∼1.8-kb duplicated upstream region but not the original intronic region (Fig. 4). The absence of this duplication, and lack of PPARγ binding to the intronic region, explains why mouse Pm20d1 is not PPARγ-activated (Fig. 1).
The example of human PM20D1, with actual PPARγ binding to only 1 of 2 duplicated paralogs, shows that segmental duplications can complicate the interpretation of genomic data. Of 62,187 potential PPARγ sites previously identified in human subcutaneous fat (27), 2.6% (1,612) overlap with segmental duplications. (Conversely, with 51,599 annotated segmental duplications, 3.1% of them have apparent PPARγ binding.) Therefore, for genome-wide ChIP-seq analyses, this relatively small number of sites in segmental duplications is unlikely to affect overall conclusions. However, focused study of an individual binding site (like PM20D1 here) must account for any segmental duplications and potential differences between the paralogs.
One of our main findings is that PPARγ activation of PM20D1 in adipocytes differs among individuals, due to a single genetic difference at rs6667995 in which the C allele disrupts a PPARγ binding motif (Fig. 5). Two such PPRE motifs exist in the PPARγ binding region identified by ChIP-seq, and we show that both are necessary for full activation in reporter assays. Most importantly, in cultured primary adipocytes derived from 26 different individuals, we show that this variant predicts transcriptional response of PM20D1 to TZD drugs. We used the same approach in our recent report showing another variant driving TZD regulation of ABCA1 (28). Such examples confirm that natural noncoding genetic variation in nuclear receptor genomic binding sites can determine differences in transcriptional response to drugs (27, 29).
While this upstream PPARγ binding rheostat fine-tunes PM20D1 expression in adipocytes, other unlinked downstream variants correlate more strongly with overall PM20D1 expression levels in adipose tissue, and in nearly every other tissue. A haplotype of 7 tightly linked variants downstream of the gene shows striking correlations with PM20D1 promoter methylation and mRNA expression levels in many human tissues (Fig. 6). The causal variant driving PM20D1 expression among these 7 SNPs remains uncertain. Interrogation of RegulomeDB (30) reveals no clear markers of regulatory DNA at any of these SNPs, except rs9438393, which falls in the proximal promoter of the adjacent gene SLC41A1. Similarly, rs708727 falls in the coding region of SLC41A1, although it is a synonymous variant that does not change protein sequence. A chromatin loop has been reported between a CTCF site near rs708727 and the TSS of PM20D1 (10), although the interacting DNA does not include rs708727, and it is unclear whether the loop is genotype-dependent. Given the tight linkage among these 7 SNPs, determining causality will likely require isolation of variants, for example by study of individuals with rare haplotypes (SI Appendix, Fig. S8A) or by genome-editing approaches. Here we use the rs823080 A allele as a marker of the methylated and silenced PM20D1 haplotype, which occurs in 40% of Caucasians but rarely in African or East Asian populations (Fig. 7A).
Differential methylation of the PM20D1 TSS CpG island was striking in our adipose tissue samples (Fig. 6B), and has emerged in multiple studies assessing differentially methylated regions. In 2012, this methylation in peripheral blood was first shown to correlate with genetic variation (31), and a subsequent study confirmed the strong effect of rs823080 and suggested a link of lower methylation to higher birth weight (32). PM20D1 differential methylation in peripheral blood has been reported in infants exposed to maternal asthma (33), or in adults who suffered childhood abuse (34). Differential PM20D1 methylation in salivary DNA was recently associated with childhood wheezing (35), and in bone marrow mesenchymal stem cells associated with acute myeloid leukemia (36). It is uncertain how to interpret so many positive associations with PM20D1 methylation. One possibility is that because this human genetic difference in methylation is so strong and common (at least in Caucasians), differential methylation often occurs by chance in many small studies of the methylome.
Some caveats must also be noted regarding these PM20D1 expression/methylation SNPs. Based on GTEx queries, genotype at the PM20D1 expression SNPs (i.e., rs823080 or rs708727) correlates weakly with expression of 3 other downstream genes, although with variability among tissues and much lower magnitude and significance (SI Appendix, Fig. S9C). The rs823080-A allele that correlates with very low PM20D1 levels also correlates with lower NUCKS1, yet with higher RAB29 and SLC41A1. Therefore, while PM20D1 expression is most strongly associated with these genetic variants, variable expression of other nearby genes cannot be ruled out as causally related to any phenotype. Indeed, ascribing causal variants and genes to GWAS signals is 1 of the major challenges in human genetics today. Our colocalization analysis shows with extremely high likelihood that the same causal variant drives PM20D1 expression and variation in BMI (Fig. 7H). While 1 common variant (rs1361754, I380T) affects PM20D1 protein coding, the less significant effects of this missense variant are consistent with its linkage to regulatory variants rather than it being causal. Overall, PM20D1 expression is strongly associated with BMI, consistent with the hypothesis that noncoding expression SNPs for PM20D1 are causally related to obesity, although (like most other human genetic associations) this remains to be proven definitively.
PM20D1 encodes a secreted enzyme that reversibly condenses amino and fatty acids to generate bioactive lipids, called NAAs (7). Silenced PM20D1 expression, as found in humans with the rs823080 A/A genotype (Fig. 7B), could thus affect the levels of these metabolites. However, we found no apparent correlation of rs823080 genotype with NAA levels in human serum or WAT (Fig. 7 C and D). Our NAA measurements by LC-MS were able to detect significant 2-fold differences in N-acyl taurines in visceral versus subcutaneous white adipose depots. Furthermore, within each individual the levels of many NAAs correlated between these 2 depots, such that some people had consistently higher or lower levels than others, yet these differences in NAA levels did not correlate with rs823080 genotype and thus PM20D1 expression.
This result is consistent with the recent report of Pm20d1 whole-body knockout mice (23), which are analogous to rs823080 A/A genotype people. While the knockout mice had reduced NAA hydrolase activity measured across many tissues, the levels of most NAAs did not differ markedly in blood, liver, or adipose tissue, although some NAAs were significantly higher in certain tissues. Related to human metabolic GWAS associations, it is notable that the knockout mice did not have changes in body weight, adiposity, or blood sugar, although they did have mild glucose intolerance and insulin resistance. Given the potential role of NAAs in thermogenesis, Pm20d1 knockout mice also had slightly higher body temperature and improved cold tolerance. Furthermore, NAAs were originally identified in mammals as modulators of pain (37), and Pm20d1 knockout mice showed reductions in certain nociceptive behaviors. Now with our identification of humans with genetically silenced multitissue PM20D1 expression, future studies can investigate effects on body temperature and nociception in people. Furthermore, the Pm20d1 knockout animals can be studied in models of neurological diseases.
Given that NAA levels are largely unchanged in knockout mice or humans homozygous for the silenced haplotype, it is likely that other enzymes besides PM20D1 contribute to the generation of NAA metabolites and control their levels. Four other enzymes of the peptidase M20A family in mammalian genomes are most closely related to PM20D1: Aminoacylase-1 (ACY1) is best-studied and a hydrolase for N-acetyl (not acyl) amino acids (38), carnosine dipeptidase 1 (CNDP1) selectively hydrolyzes carnosine (β-alanyl-l-histidine), and closely related dipeptides, peptidase A (PEPA, also known as CNDP2) is both a carnosinase and nonspecific dipeptidase (39), while PM20D2 (also known as ACY1L2) is a dipeptidase but not a carnosinase (40). These enzymes are not known to act on long-chain NAAs, and based on their effects on dipeptides they are more likely to serve as hydrolases than synthases. Indeed, the biosynthesis of NAAs remains quite uncertain, although several pathways have been proposed (41).
The metabolism of NAAs is complex and incompletely understood, yet our study clearly suggests that PM20D1 expression can affect disease phenotypes without affecting NAA levels. While the PM20D1 protein has no annotated domains beside the M20 catalytic and dimerization domains, it is plausible that it has other enzymatic or even nonenzymatic functions. Indeed, the phenomenon of “moonlighting proteins” is increasingly recognized (42), as examples abound of enzymes with noncatalytic roles in unrelated biological processes. A classic case is the crystallin proteins in the lens of the eye, in which different species have recruited various enzymes of intermediary metabolism to serve as structural proteins, thus modifying the lens properties (43). Therefore, future studies may uncover noncatalytic roles for PM20D1 accounting for its human disease associations.
There is a robust connection of the PM20D1 locus to neurodegenerative disease. A large study combining GWAS and epigenome-wide association studies of brain tissue methylation revealed a significant association between PM20D1 and Alzheimer’s disease (10). PM20D1 falls in the PARK16 locus, 1 of the first and strongest GWAS loci associated with Parkinson’s disease and 1 with likely allelic heterogeneity, with multiple causal alleles (44). We show here that some of these top-hit alleles are highly linked to PM20D1 expression and methylation (SI Appendix, Fig. S8C).
The PM20D1 locus also shows a genome-wide significant GWAS association with BMI (Fig. 7F), and weaker associations with obesity-related conditions like type 2 diabetes and HDL cholesterol levels (SI Appendix, Fig. S8C). These are directionally consistent, as lower BMI correlates with lower diabetes risk and higher HDL cholesterol. However, it is notable that this protection from obesity and metabolic disease was conferred by the SNP allele haplotype with silenced PM20D1 expression (i.e., rs823020-A). Based on mouse studies, the opposite might have been predicted, with metabolic benefits of higher PM20D1: Knockout led to insulin resistance (23), while overexpression led to protection against diet-induced obesity (7). Benefits of higher PM20D1 expression were also shown in Alzheimer’s disease (10), such that the silenced PM20D1 haplotype may reduce risk of metabolic disease yet increase risk of neurodegenerative disease.
PM20D1 function is only beginning to be understood, including the role of its NAA synthase/hydrolase activity. Here we report in detail the human genetics of PM20D1 gene regulation, which is interesting in that different variants affect gene expression at 2 levels. The rs6667995 variant, which falls upstream of PM20D1 in a segmental duplication, determines the activation of PM20D1 selectively in adipose tissue by the PPARγ nuclear receptor and its agonist drugs. Another distinct group of linked variants downstream of PM20D1 define a common haplotype with high promoter methylation and very low expression. The rs823080 A/A genotype identifies individuals with silenced PM20D1 expression across tissues and different risks of developing metabolic and neurological disease, yet without major differences in NAA levels. Regardless of the precise biological role of PM20D1, then, these genotype-dependent effects on its expression could contribute to individualized medicine approaches for disease prediction and treatment.
Materials and Methods
Animals.
Male wild-type mice were purchased from Jackson Laboratories. Rosiglitazone-treated samples were from 129S1/SvImJ mice, while cold exposure was performed on F1 mice (B6129SF1/J, progeny of C57BL/6J and 129S1/SvImJ), as described previously (45). Housing was in cages of 5 in a temperature-controlled specific pathogen-free facility with 12-h light/dark and ad libitum access to water and food. Breeding and weaning of mice was on standard rodent chow. Rosiglitazone treatment and RNA sequencing of these samples was previously published (45). For cold exposure, mice were housed at room temperature (22 °C) for 1 wk, then exposed to cold at 4 °C for 1 d or 1 wk. All mice were killed at 4:00 to 5:00 PM by CO2 asphyxiation followed by cervical dislocation, and the epididymal and inguinal white fat pads were dissected and snap-frozen in liquid N2. All mouse care and use procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Human Adipose Tissue Samples.
Human fat samples were obtained from the Human Metabolic Tissue Bank of the University of Pennsylvania Institute for Diabetes, Obesity, and Metabolism, which obtains preoperative informed consent from surgical patients for biopsies to be taken, banked, and distributed to investigators with de-identified patient characteristics. All protocols were approved by the University of Pennsylvania’s Institutional Review Board.
ChIp.
ChIP using anti-PPARγ antibody (Santa Cruz, sc-7196, 5 μg per immunoprecipitation) from adipose tissue or cultured adipocytes was performed as previously described (14, 27). Published ChIP-seq datasets were visualized on the Integrated Genomics Viewer (46).
Gene-Expression Analysis.
RNA was isolated from culture adipocytes or adipose tissue using TRIzol reagent (Life Technologies) followed by RNeasy mini columns (Qiagen). For microarray analysis of SGBS adipocytes, total RNA (n = 4 rosiglitazone-treated and n = 4 control DMSO vehicle-treated) was amplified using the Affmetrix wild-type expression kit (Ambion) and hybridized to a Human Gene 1.0ST chip (Affymetrix). Microarray analysis was performed by University of Pennsylvania Molecular Profiling Facility, using standard tools Robust Multi-Array Average for normalization and Significance Analysis of Microarrays for differential expression analysis (47). For quantitative real-time PCR analysis, total RNA was DNase-treated and reverse transcribed using SuperScript IV VILO Master Mix with ezDNase Enzyme (ThermoFisher), then amplified using ABI QuantStudio 5 Real-Time PCR System and Power SYBR Green PCR Master Mix (Applied Biosystems). All primers for qPCR, as well as reporter cloning and pyrosequencing, are in SI Appendix, Table S2.
Cell Culture.
Mouse 3T3-L1 adipocytes and human SGBS adipocytes were differentiated as previously described (14). Primary human adipose stem cell-derived adipocytes were differentiated from the stromal vascular fraction of fat biopsies and treated with rosiglitazone, as previously described (28). Note that both SGBS cells and primary adipocytes were generated in the presence of rosiglitazone, as TZD is required for differentiation, but they were cultured in maintenance media lacking this drug for 11 or 7 d prior to the rosiglitazone treatment at 1 μM for 24 to 48 h. 293T cells (ATCC) were maintained at 37 °C in 5% CO2 in high glucose DMEM supplemented with 10% fetal bovine serium and l-glutamine (Gibco).
Luciferase Reporter Assays.
For luciferase reporter assays, 344 bp of human DNA fragments from the PM20D1 locus were PCR-amplified from genomic DNA, using reverse primers distinguishing between the intronic and upstream duplicated regions. Similarly, mouse syntenic sequence was also amplified, and PCR products were cloned into the XhoI site of the pGL4.24 luciferase reporter (Promega) using a Gibson Assembly Cloning Kit (New England Biolabs). Mutations were introduced into the second PPRE of the human intronic and upstream reporters using the Q5 site-directed mutagenesis kit (New England Biolabs). All reporters were sequence-verified. Transient transfections of 293T cells were performed in 24-well plates, n = 3 wells per condition, using Lipofectamine 3000 (Invitrogen) to add 300 ng of pGL4 luciferase reporter, 1 ng of Renilla luciferase for normalization, and 100 ng of pCMX expression plasmid (empty or 50 ng each of pCMX+PPARγ and pCMX+RXRα), as previously described (27). Cells were cultured for 24 h after transfection (sometimes in the presence of 1 μM rosiglitazone), then a Firefly and Renilla Luciferase Enhanced Assay (Goldbio) was used to measure luciferase activities on a Synergy HT plate reader (Biotek).
Genotyping, Allele, and Methylation Quantification.
SNP rs823080 was genotyped either using a SNaPshot assay, as previously described (27), or by TaqMan SNP genotyping (Life Technologies). SNP rs6667995 was genotyped and allele quantification performed by pyrosequencing, with primers were designed used PyroMark Assay Design software (Qiagen). PCR was performed using the PyroMark PCR kit, and pyrosequencing with PyroMark Gold reagents on a PyroMark Q96 MD instrument (Qiagen) per the manufacturer’s instructions. To assay CpG methylation at the PM20D1 TSS, bisulfite pyrosequencing was performed using EZ DNA Methylation-Gold Kit (Zymo Research, D5005) and primers previously reported (10). Average methylation at 4 CpGs was calculated in each sample.
Metabolite Measurement.
Stearoyl chloride, palmitoyl chloride, oleoyl chloride, and all amino acids were purchased from Sigma Aldrich. NAA standards were synthesized as previously reported (7). To extract NAAs from human adipose tissue samples, frozen tissues were ground at liquid nitrogen temperature with a Cryomill (Retsch). The resulting tissue powder was weighed (∼20 mg). Then 500 µL −20 °C extraction solvent (50:50 methanol:acetonitrile) was added (with 2-µM internal standard) to the powder and incubated at 4 °C for 10 min, followed by vortexing and centrifugation at 16,000 × g for 30 min at 4 °C. Then, 15 µL of the supernatant was loaded to LC-MS. To extract NAAs from human serum samples, 50 µL −20 °C extraction solvent was added to 50 µL of serum sample with inclusion of internal standard (U-13C-palmitate, 2 µM) and incubated on ice for 10 min, followed by vortexing and centrifugation at 16,000 × g for 30 min at 4 °C. Then, the supernatant was dried down under N2 flow and resuspended into 50 µL extract solvent, followed by vortexing and centrifugation at 16,000 × g for 30 min at 4 °C. Next, 15 µL of the final supernatant was loaded to LC-MS. LC-MS measurement was performed by reverse-phase chromatography coupled with negative-mode electrospray high-resolution MS on a quadrupole orbitrap mass spectrometry (Q Exactive Plus, Thermo Fisher Scientific). The MS scan range was m/z 180 to 650. LC separation was achieved on an Agilent EC-C18 column (2.1 × 150 mm, 2.7-µm particle size) using a gradient of solvent A (1 mM NH4OAc+0.2% acetic acid in 90:10 H2O:MeOH) and solvent B (1 mM NH4OAc+0.2% acetic acid in 90:10 MeOH:isopropanol). Flow rate was 150 µL min−1. The gradient was: 0 min, 25% B; 2 min, 25% B; 4 min, 65% B; 16 min, 100% B; 20 min, 100% B; 21 min, 25% B; 25 min, 25% B. Peak identification and integration used ElMaven software, with compounds identified on the basis of exact mass, retention time, and MS/MS spectra match to the synthesized standards. The relative levels of each NAA in each sample were normalized to the mean of the rs823080 A/A (low PM20D1 expression) genotype samples, which was set equal to 1.
Human Genetic Analyses.
The University of California, Santa Cruz (UCSC) genome browser (48) was used for the identification and cross-species comparison of the PM20D1 segmental duplication, as well as localization of published GWAS lead SNPs. Human genetics of tissue-specific gene expression was interrogated using the GTEx portal (21). Haplotype and linkage analysis were performed using LDLink (49). Regional visualization of GWAS data were performed using LocusZoom (50), with BMI, HDL, and type 2 diabetes associations from published metaanalyses (51–53). Colocalization of eQTL and GWAS data were performed using Coloc (54) reporting PP4, the posterior probability that a single, causal variant underlies both statistical association signals.
Statistics.
Prism (Graphpad) was used for graphing and statistical tests, described in figure legends.
Supplementary Material
Acknowledgments
We thank Mitch Lazar for sharing datasets obtained in the course of studies funded by NIH Grant R01-DK49780, the Metabolomics Core of the University of Pennsylvania Diabetes Research Center (P30-DK19525), and the laboratory of Marisa Bartolomei for use of a pyrosequencer. R.E.S. initiated these studies while supported by NIH Grant K08-DK094968. B.F.V. was supported by NIH Grant R01-DK101478 and a Linda Pechenik Montague Investigator award. W.H. was supported by American Diabetes Association training Grant 1-18-PDF-132. A.H.B. was supported by the National Institute of Diabetes and Digestive and Kidney Diseases Medical Student Research Program in Diabetes.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913199116/-/DCSupplemental.
References
- 1.Kajimura S., Spiegelman B. M., Seale P., Brown and beige fat: Physiological roles beyond heat generation. Cell Metab. 22, 546–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee P., Wasting energy to treat obesity. N. Engl. J. Med. 375, 2298–2300 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Chouchani E. T., Kazak L., Spiegelman B. M., New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab. 29, 27–37 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Ukropec J., Anunciado R. P., Ravussin Y., Hulver M. W., Kozak L. P., UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1-/- mice. J. Biol. Chem. 281, 31894–31908 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Ikeda K., et al. , UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat. Med. 23, 1454–1465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kazak L., et al. , A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163, 643–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long J. Z., et al. , The secreted enzyme PM20D1 regulates lipidated amino acid uncouplers of mitochondria. Cell 166, 424–435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin H., et al. , Discovery of hydrolysis-resistant isoindoline N-acyl amino acid analogues that stimulate mitochondrial respiration. J. Med. Chem. 61, 3224–3230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satake W., et al. , Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat. Genet. 41, 1303–1307 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Mut J. V., et al. , PM20D1 is a quantitative trait locus associated with Alzheimer’s disease. Nat. Med. 24, 598–603 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Soccio R. E., Chen E. R., Lazar M. A., Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab. 20, 573–591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keipert S., et al. , Long-term cold adaptation does not require FGF21 or UCP1. Cell Metab. 26, 437–446.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Schmidt S. F., et al. , Cross species comparison of C/EBPα and PPARγ profiles in mouse and human adipocytes reveals interdependent retention of binding sites. BMC Genomics 12, 152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soccio R. E., et al. , Species-specific strategies underlying conserved functions of metabolic transcription factors. Mol. Endocrinol. 25, 694–706 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikkelsen T. S., et al. , Comparative epigenomic analysis of murine and human adipogenesis. Cell 143, 156–169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steiper M. E., Young N. M., Primate molecular divergence dates. Mol. Phylogenet. Evol. 41, 384–394 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Bailey J. A., et al. , Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Greenawalt D. M., et al. , A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 21, 1008–1016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nica A. C., et al. ; MuTHER Consortium , The architecture of gene regulatory variation across multiple human tissues: The MuTHER study. PLoS Genet. 7, e1002003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Civelek M., et al. , Genetic regulation of adipose gene expression and cardio-metabolic traits. Am. J. Hum. Genet. 100, 428–443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.GTEx Consortium , Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Ciaula A., et al. , Bile acid physiology. Ann. Hepatol. 16 (suppl. 1), S4–S14 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Long J. Z., et al. , Ablation of PM20D1 reveals N-acyl amino acid control of metabolism and nociception. Proc. Natl. Acad. Sci. U.S.A. 115, E6937–E6945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grundy S. M., Brewer H. B. Jr, Cleeman J. I., Smith S. C. Jr, Lenfant C.; American Heart Association ; National Heart, Lung, and Blood Institute , Definition of metabolic syndrome: Report of the national heart, lung, and blood institute/american heart association conference on scientific issues related to definition. Circulation 109, 433–438 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Yengo L., et al. ; GIANT Consortium , Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis M. Y., Eichler E. E., Human adaptation and evolution by segmental duplication. Curr. Opin. Genet. Dev. 41, 44–52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soccio R. E., et al. , Genetic variation determines PPARγ function and anti-diabetic drug response in vivo. Cell 162, 33–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu W., et al. , Patient adipose stem cell-derived adipocytes reveal genetic variation that predicts antidiabetic drug response. Cell Stem Cell 24, 299–308.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo C., D’Ippolito A. M., Reddy T. E., From prescription to transcription: Genome sequence as drug target. Cell 162, 16–17 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Boyle A. P., et al. , Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser H. B., Lam L. L., Neumann S. M., Kobor M. S., Population-specificity of human DNA methylation. Genome Biol. 13, R8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haworth K. E., et al. , Methylation of the FGFR2 gene is associated with high birth weight centile in humans. Epigenomics 6, 477–491 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Gunawardhana L. P., et al. , Differential DNA methylation profiles of infants exposed to maternal asthma during pregnancy. Pediatr. Pulmonol. 49, 852–862 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Suderman M., et al. , Childhood abuse is associated with methylation of multiple loci in adult DNA. BMC Med. Genomics 7, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popovic M., et al. , Differentially methylated DNA regions in early childhood wheezing: An epigenome-wide study using saliva Pediatr. Allergy Immunol. 30, 305–314, (2019). [DOI] [PubMed] [Google Scholar]
- 36.Huang J., et al. , Use of methylation profiling to identify significant differentially methylated genes in bone marrow mesenchymal stromal cells from acute myeloid leukemia. Int. J. Mol. Med. 41, 679–686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S. M., et al. , Identification of a new class of molecules, the arachidonyl amino acids, and characterization of one member that inhibits pain. J. Biol. Chem. 276, 42639–42644 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Sommer A., et al. , The molecular basis of aminoacylase 1 deficiency. Biochim. Biophys. Acta 1812, 685–690 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Teufel M., et al. , Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J. Biol. Chem. 278, 6521–6531 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Veiga-da-Cunha M., Chevalier N., Stroobant V., Vertommen D., Van Schaftingen E., Metabolite proofreading in carnosine and homocarnosine synthesis: Molecular identification of PM20D2 as β-alanyl-lysine dipeptidase. J. Biol. Chem. 289, 19726–19736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burstein S. H., N-acyl amino acids (elmiric acids): Endogenous signaling molecules with therapeutic potential. Mol. Pharmacol. 93, 228–238 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Chen C., Zabad S., Liu H., Wang W., Jeffery C., MoonProt 2.0: An expansion and update of the moonlighting proteins database. Nucleic Acids Res. 46, D640–D644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wistow G., The human crystallin gene families. Hum. Genomics 6, 26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pihlstrøm L., et al. , Fine mapping and resequencing of the PARK16 locus in Parkinson’s disease. J. Hum. Genet. 60, 357–362 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Soccio R. E., et al. , Targeting PPARγ in the epigenome rescues genetic metabolic defects in mice. J. Clin. Invest. 127, 1451–1462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S., A comprehensive evaluation of SAM, the SAM R-package and a simple modification to improve its performance. BMC Bioinformatics 8, 230 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casper J., et al. , The UCSC genome browser database: 2018 update. Nucleic Acids Res. 46, D762–D769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Machiela M. J., Chanock S. J., LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31, 3555–3557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pruim R. J., et al. , LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Locke A. E., et al. ; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium , Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott R. A., et al. ; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium , An expanded genome-wide association study of type 2 diabetes in europeans. Diabetes 66, 2888–2902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willer C. J., et al. ; Global Lipids Genetics Consortium , Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giambartolomei C., et al. , Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.