Significance
Dysregulated metabolism is one of the hallmarks of pancreatic ductal adenocarcinoma, which is the major subtype of pancreatic cancer considered as the deadliest malignancy worldwide. This led us to search the potent therapeutic target for regulating cancer metabolism in treatment. By uncovering that phosphoglycerate mutase 1 (PGAM1), a critical metabolic enzyme involved in glycolysis and biosynthesis, was frequently up-regulated in patients with pancreatic ductal adenocarcinoma (PDAC), we developed a series of allosteric PGAM1 inhibitors which showed efficacious in multiple preclinical models of PDAC, especially with high PGAM1 expression. Of note, PGAM1 inhibition cosuppressed several metabolic and cancerous pathways, of which the suppression level was correlated with efficacy. This work strongly suggests that inhibition of cancer metabolism would be a strategy for treating pancreatic cancer.
Keywords: PGAM1 inhibitor, PDAC, glycolysis, cancer metabolism
Abstract
Glycolytic enzyme phosphoglycerate mutase 1 (PGAM1) plays a critical role in cancer metabolism by coordinating glycolysis and biosynthesis. A well-validated PGAM1 inhibitor, however, has not been reported for treating pancreatic ductal adenocarcinoma (PDAC), which is one of the deadliest malignancies worldwide. By uncovering the elevated PGAM1 expressions were statistically related to worse prognosis of PDAC in a cohort of 50 patients, we developed a series of allosteric PGAM1 inhibitors by structure-guided optimization. The compound KH3 significantly suppressed proliferation of various PDAC cells by down-regulating the levels of glycolysis and mitochondrial respiration in correlation with PGAM1 expression. Similar to PGAM1 depletion, KH3 dramatically hampered the canonic pathways highly involved in cancer metabolism and development. Additionally, we observed the shared expression profiles of several signature pathways at 12 h after treatment in multiple PDAC primary cells of which the matched patient-derived xenograft (PDX) models responded similarly to KH3 in the 2 wk treatment. The better responses to KH3 in PDXs were associated with higher expression of PGAM1 and longer/stronger suppressions of cancer metabolic pathways. Taken together, our findings demonstrate a strategy of targeting cancer metabolism by PGAM1 inhibition in PDAC. Also, this work provided “proof of concept” for the potential application of metabolic treatment in clinical practice.
In recent years, research interest in tumor-specific altered metabolism, discovered by Otto Warburg about a century ago, has been growing worldwide (1). Targeting cancer metabolism has emerged during the past decade as a promising strategy for the development of anticancer agents by aiming at a tumor’s sweet spot during the past decade (2). It has been proposed that accelerated aerobic glycolysis is capable of incorporating nutrients into the biomass (e.g., nucleotides, amino acids, and lipids) for rapidly growing tumor cells (3). Thus glycolysis and subsequent biosynthesis pathways have not only been considered as an integral part of cancer biology, but have also become a promising target of cancer therapy (1, 3–12). Among all metabolic enzymes involved in cancer metabolism, our previous study has shown that phosphoglycerate mutase 1 (PGAM1) plays an important role in coordinating glycolysis and biosynthesis including pentose phosphate pathway and serine synthesis pathway to promote tumor growth (4). Furthermore, it has been suggested that blocking of PGAM1 has potential for controlling cancer cell growth by targeting both energetic and anabolic processes with a single drug, as a way of “killing two birds with one stone” (13).
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal malignancies, with 5-y survival rates about 6% (14). Surgery is the major therapy for treating early patients with PDAC; the 5-y survival rate of resected patients is about 20% (15, 16). Unfortunately, most patients with PDAC have late-stage disease and are unable to have resection when they are diagnosed. Combination chemotherapies have been used as first-line regimens for treating patients with late-stage PDAC, including a combination therapy of gemcitabine with nab-paclitaxel (17) and FOLFIRINOX, which is a 4-drug combination consisting of leucovorin, fluorouracil, irinotecan, and oxaliplatin (18). The median overall survival of these regimens is only 8.5 and 11.1 mo, respectively (17, 18). Hence, there is a strong demand to identify new therapeutic targets since PDAC is resistant to most of the currently used therapies (14, 15). In PDAC, it has been shown that reprogramming of cell metabolism is driven by frequently mutated genes (16, 19), which make cancer cells more dependent on key metabolites (20) and more vulnerable to inhibition of metabolite-related enzymes. This is due to the strategy of collateral lethality which occurs when deletion of tumor suppressor genes results in codeletion of a large number of genes commonly known without direct role in cancer development, due to proximity of chromosome. Collateral lethality is commonly found in cancers and considered in favor of expanding therapeutic strategy to nononcogenic targets, partially because several oncogenic targets are not druggable (21–23).
To date, several drugs have demonstrated promising anticancer efficacy by targeting cancer metabolism (11); however, the therapeutic candidates specifically targeting PGAM1 still remain limited (2, 24, 25). Here, we developed a potent PGAM1 inhibitor and investigated its efficacy and inhibitory mechanism in multiple PDAC preclinical models to evaluate its potential application in PDAC therapy.
Results
PGAM1 Is a Potential Target for PDAC Therapy.
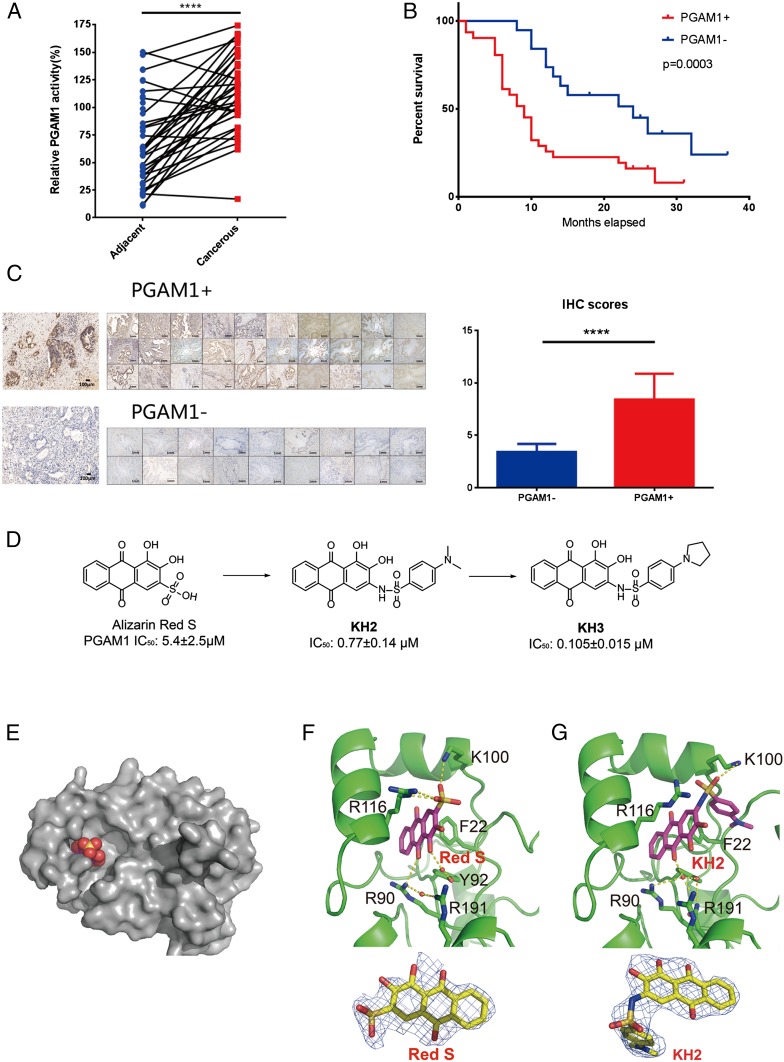
Based on our previous study showing that PGAM1 was elevated in lung cancer (4), we first checked the clinical relevance of PGAM1 in patients with PDAC to demonstrate the potential of targeting PGAM1 in their therapy. We assessed the activities of PGAM1 in cancerous and adjacent normal tissue collected from 50 patients. It was demonstrated that the activities of PGAM1 in cancerous tissue samples were significantly higher than that in the matched adjacent normal tissue samples (Fig. 1A), which is consistent with the previous finding in lung cancer (4). Moreover, we found that the patients showing elevated expression level of PGAM1 demonstrated remarkably poorer survival than the patients with low expression level of PGAM1 (Fig. 1 B and C and SI Appendix, Tables S4 and S5), which is similar to the phenomenon observed in hepatocellular carcinoma (26). We also assessed the expressions of PGAM1 in patients with PDAC of TCGA database, showing that higher expression level of PGAM1 was detected in the tumor tissue of the TCGA cohort. The patients with high PGAM1 expression level showed worse prognosis than those with low PGAM1 level (SI Appendix, Fig. S1 A and B). These findings suggest that PGAM1 inhibition may be able to improve the outcome of patients with PDAC who have high PGAM1 expression in their tumors.
Fig. 1.
PGAM1 is a potential target for PDAC therapy. (A) PGAM1 activity in cancerous tissue was higher than adjacent normal tissue in patients with PDAC (n = 50, P < 0.0001, P values were obtained from paired t test). (B) Patients with high PGAM1 expression level showed worse prognosis (overall survival) than those with low PGAM1 level (P = 0.0003). The P values were obtained from Kaplan-Meier survival analysis. (C) PGAM1 expression levels of corresponding tissue from patients with PDAC shown in B were analyzed. The IHC scores were displayed between the samples of high and low levels of PGAM1 expression. (Magnification: 200×.) (D) Lead optimization from Alizarin Red S to KH3. (E) Alizarin Red S (spacefill) binds to a novel allosteric pocket of PGAM1 (gray). (F) Cocrystal structures of PGAM1 and Alizarin Red S and Fo-Fc maps of Alizarin Red S (blue mesh, contoured at 2.0σ). Indicated residues and water molecules involved in small-molecule binding. (G) Cocrystal structures of PGAM1 and KH2 and Fo-Fc maps of KH2 (blue mesh, contoured at 2.0σ). Indicated residues and water molecules involved in small-molecule binding.
To testify the hypothesis that PGAM1 could be targeted in PDAC therapy, we next developed a series of potent PGAM1 inhibitors for further pharmocological study (Fig. 1D). We first cocrystallized PGAM1 with our previously reported lead compound Alizarin Red S. The X-ray crystal structures of full-length PGAM1 in complex with the compound revealed a binding mode for PGAM1 inhibitors (Fig. 1E). The 9,10-anthraquinone core scafford of lead compounds interacts with Arg90 and Arg191 through water bridges, along with electrostatic interaction between the sulfonic acid group and residues of PGAM1 (Lys100 and Arg116; Fig. 1F). To improve the cell permeability of lead compound, KH2 was then synthesized by replacing sulfonic acid group to sulfonamide group (Fig. 1D). The sulfonamide group of KH2 forms a cation–π interaction with Arg116 and a hydrogen bond with Lys100. Additionally, the phenyl ring of KH2 interacts with Phe22 through hydrophobic interaction (Fig. 1G), leading to better binding affinity with a 15-fold improvement of in vitro potency over Alizarin Red S. Compared with apo-form, residues 109 to 117 of PGAM1 in PGAM1-KH2 complex change its conformation, implying that KH2 is an allosteric inhibitor (SI Appendix, Fig. S2). With further modification on phenyl ring by N-substituted pyrrolidine derivative compound, KH3 showed a dramatic enhancement in both enzymatic (50-fold) and cell proliferation assay (>100-fold) over Alizarin Red S, probably due to its bigger N-substituted group for better binding and cell permeability. For cell-based assay, KH3 is the most potent PGAM1 inhibitor reported to date.
Structural and Biochemical Analysis of PGAM1 Inhibitors.
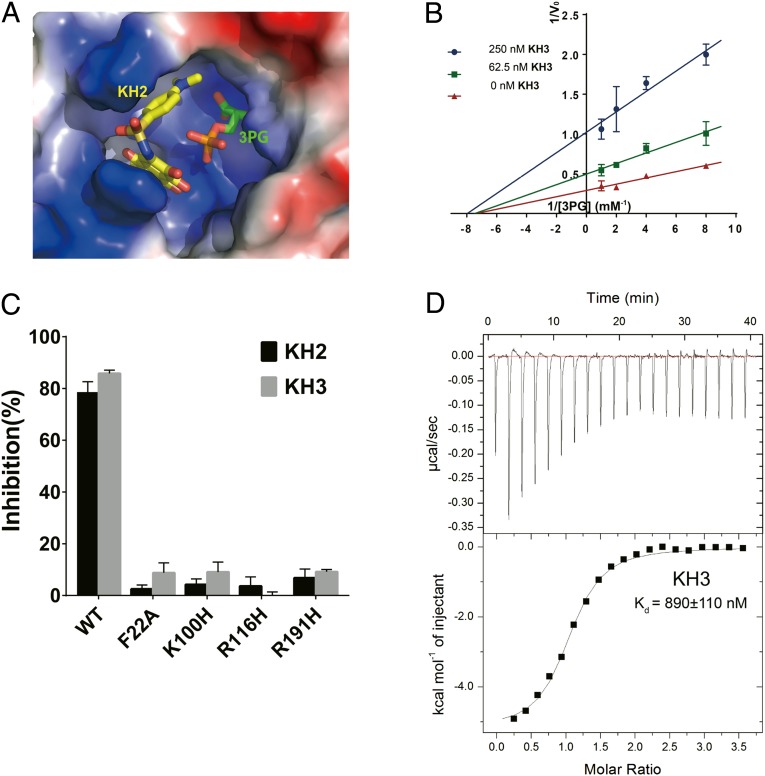
By developing a series of PGAM1 inhibitors, we next moved to analyze their stuctural and chemical charateristics. Comparison of cocrystal structures containing 3PG (PDB:2F90) and KH2 (PDB:5Y65) revealed that the compound KHs located at the gate of substrate entrance (Fig. 2A), acting as an allosteric inhibitor. The noncompetitive inhibition mechanism of KHs was further confirmed by enzymatic assay (Fig. 2B). Inhibition of KHs against PGAM1 were greatly impaired when surrounding residues of PGAM1 were mutated, indicating that these residues were important for small-molecule binding (Fig. 2C). Moreover, KH3 bound to PGAM1 with Kd of 890 nM which was measured by isothermal titration calorimetry, showing that it was a spontaneous process driven by both enthalpy (−5.22 kcal/mol) and entropy (10.2 cal/mol/deg; Fig. 2D).
Fig. 2.
Structural and biochemical analysis of PGAM1 inhibitors. (A) Structure overlay of PGAM1-KH2 complex (5Y65) and BPGM-3PG complex (2F90). Protein was shown in surface electrostatics. KH2 and 3PG were shown in yellow and green stick, respectively. (B) Inhibition mechanism analysis of KH3 determined by Lineweaver-Burk plots. (C) Inhibition of PGAM1 by KH2 and KH3 (5 μM) was reduced by the mutated residues around the inhibitor-binding pocket. (D) Binding affinity measurement of KH3 with PGAM1 by isothermal titration calorimetry (ITC).
In Vitro Analysis of KH3 Efficacy in PDAC Cells.
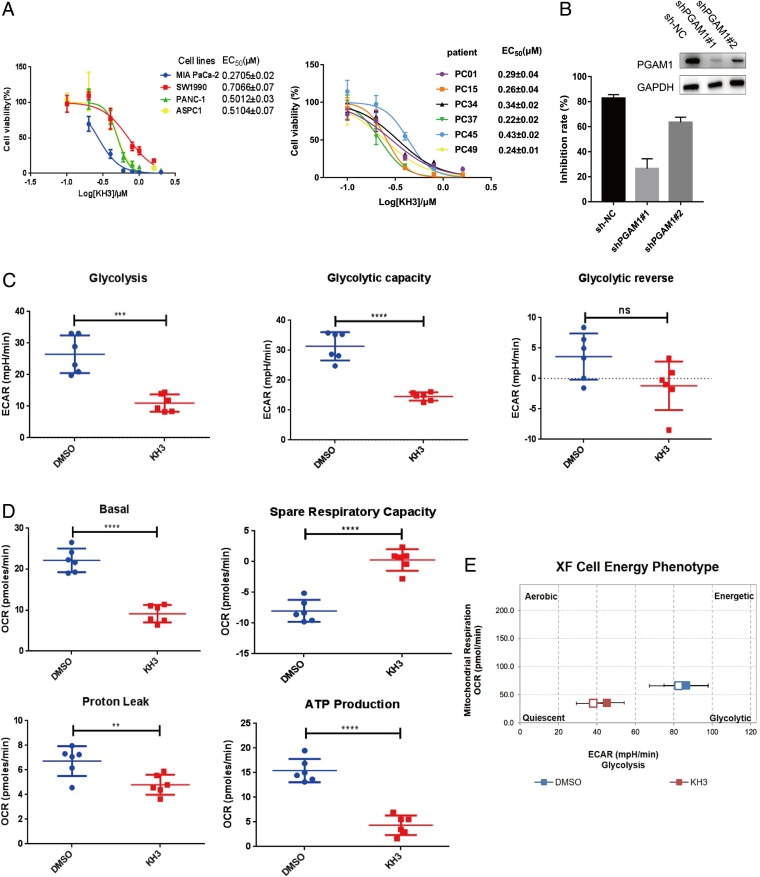
To determine the efficacy of PGAM1 inhibitors on cell proliferation, we first treated 4 commercial pancreatic cancer cell lines SW1990, PANC-1, AsPC-1, and MIA PaCa-2 with KH3. The result showed that KH3 effectively suppressed pancreatic cancer cell proliferation with EC50 ranging from 0.27 to 0.70 μM (Fig. 3A). Next, we tested the ability of KH3 for inhibiting primary pancreatic cancer cells isolated from 6 patients. Likewise, the primary pancreatic cancer cells were slightly more sensitive to KH3 with EC50 ranging from 0.22 to 0.43 μM (Fig. 3A) compared with the commercial cancer cells. We found that higher PGAM1 expression was associated with lower EC50 in both commercial and primary pancreatic cancer cells (SI Appendix, Fig. S3B), indicating that increased PGAM1 expression may confer better drug response.
Fig. 3.
KH3 inhibited PDAC cell growth by reducing glycolysis and mitochondrial respiration via targeting PGAM1. (A) KH3 suppressed the proliferation of PDAC cell lines and PDAC primary cells isolated from patients. (B) Knockdown of PGAM1 impaired the inhibition of KH3 (1 μM) on proliferation of PANC-1 cells when compared to control. Knockdown of PGAM1 in PANC-1 cells was verified by Western blot. (C) KH3 suppressed glycolysis of PANC-1 cells. (D) KH3 suppressed mitochondrial respiration of PANC-1 cells. (E) Cell energy phenotype of PANC-1 cells was altered after KH3 treatment. The data were presented as mean ± SD (n = 5), and P values were obtained from unpaired t test (n.s., not significant. *: 0.01 < P < 0.05, **: 0.001 < P < 0.01, ***: 0.0001 < P < 0.001, ****P < 0.0001). OCR, oxygen consumption rate; ECAR, extracellular acidification rate; XF, seahorse XF system.
In order to assess the efficiency of target engagement of KH3 in PDAC cells, we generated the stable PGAM1 knockdown (KD) cells for evaluating the off-target effect of KH3 by infecting with the lentiviral shRNAs (Fig. 3B). By monitoring the KH3 (1 μM) treatment on the proliferation of PGAM1 KD cells for 6 d (Fig. 3B), in contrast to control, the PGAM1 down-regulation markedly abolished the growth inhibition induced by KH3 (inhibition rate of shPGAM1#1 vs. shNC: 17.75 vs. 74.69%). In the meantime, we found PGAM1 overexpression partially impaired the growth inhibition induced by KH3 (SI Appendix, Fig. S3A). Taken together, our results illustrated that KH3 effectively suppresses pancreatic cancer cell proliferation mainly by blocking PGAM1.
Our previous work reported that blocking of PGAM1 down-regulated glycolysis by altering the levels of key metabolites such as 3PG (4); we here analyzed the levels of glycolysis and mitochondrial respiration of PDAC cells after KH3 treatment. Similar to the previous study, the current work confirmed that KH3 suppressed glycolysis and mitochondrial respirationof PDAC PANC-1cells (Fig. 3 C and D). The KH3-treated PANC-1 cells displayed low level of energy determined by the Seahorse System (Fig. 3E). We further assessed whether low cell energy was associated with increased cell death in KH3-treated PDAC cells. It was shown that KH3 inhibition induced apoptosis and cell-cycle arrest at G2 stage in PDAC cells (SI Appendix, Fig. S2 A and B). Altogether, KH3-induced inhibition of PGAM1 has significantly limited cell energy and growth by down-regulating glycolysis.
KH3 Targeted Similar Signaling Pathways in PANC-1 PDAC Cells Compared to PGAM1 Knockdown.
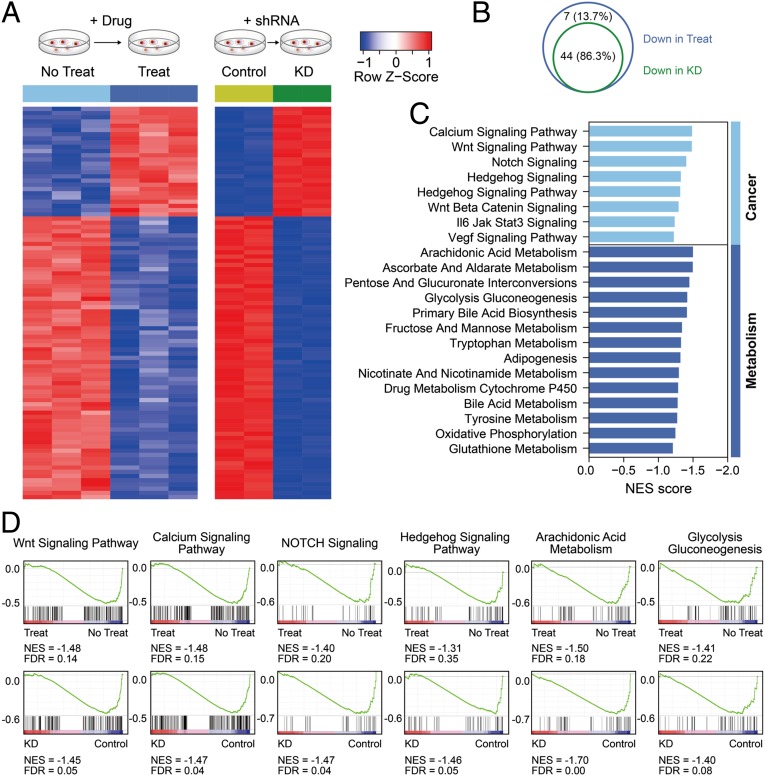
To further investigate which genes and signaling pathways were responsible for the inhibitory effects of KH3 on PDAC cell proliferation, transcriptome-wide RNA-sequencing analysis was performed in PGAM1 KD and KH3-treated PANC-1 cells (Fig. 4A). We used fragments per kilobase million (FPKM) to evaluate expression levels of individual genes of the different comparisons and demonstrated that the groups of PGAM1 KD and KH3 treatment harbored similar gene expression pattern (Fig. 4A). The global gene set enrichment analysis (GSEA) indicated that the PGAM1 KD and KH3-treated PANC-1 cells shared the vast majority of pathways (44 out of 51; 86.3%) among the decreased signaling pathways (Fig. 4B). Among these pathways, groups of PGAM1 KD and KH3 treatment both caused the significant suppression of glycolysis gluconeogenesis, arachidonic acid metabolism, Hedgehog signaling, NOTCH signaling, calcium signaling, and Wnt signaling, etc., which could be classified into metabolic and cancerous pathways, respectively (Fig. 4 C and D). These results indicate that PGAM1 KD and KH3 treatment down-regulate the pathways mainly involved in cancer metabolism and development, of which the inhibition is capable of limiting PDAC cell growth. Additionally, the similar pattern of global gene expressions shared by the PGAM1 KD and KH3 treatment confirms that the inhibition conferred by KH3 is mainly via targeting PGAM1.
Fig. 4.
Transcriptome-wide RNA-sequencing assays to identify on-target ability of KH3 in PANC-1 cells. (A) Transcriptome strategy of RNA sequencing conducted on PANC-1 cells exposed to 2 μm KH3 for 12 h is shown in schematic diagram. shPGAM1 and shNS groups contained 2 biological replicates; KH3 and DMSO groups contained 3 replicates. (B) Venn diagram of the shared pathways among the decreased signaling pathways in PGAM1 KD and KH3-treated PANC-1 cells. (C) The common core-enriched signaling pathways down-regulated in the groups of KH3 treatment and shPGAM1 knockdown. (D) GSEA was used to analyze the signaling pathway enrichment in the groups of KH3 treatment and shPGAM1 knockdown. Normalized enrichment score (NES) indicated the analysis results across gene sets. False discovery rate (FDR) presented if a set was significantly enriched. ES, enrichment score.
Signature Pathways Regulated by KH3 Treatment in Multiple PDAC Cells.
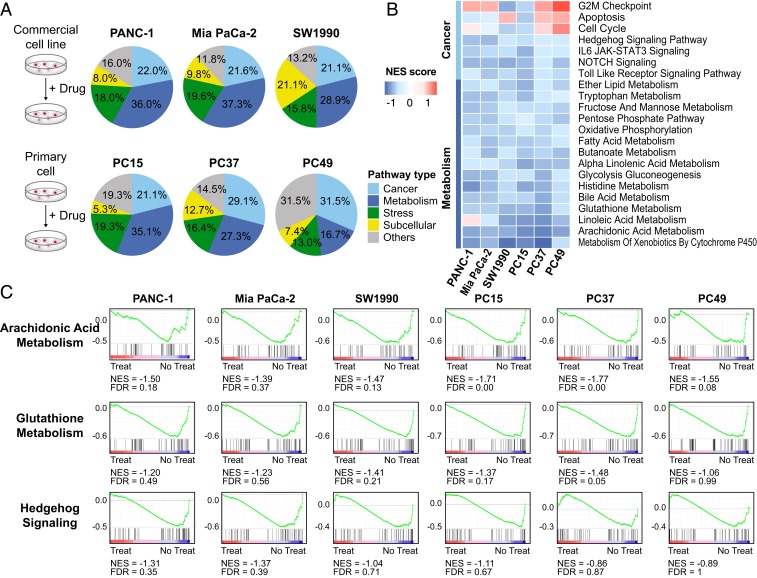
As previously described, KH3 treatment caused significant suppression of several cancerous and metabolic pathways in PANC-1 cells. To fully understand the molecular features of KH3 inhibition and its regulated vital pathways in PDAC, we implemented transcriptome analysis of 3 PDAC commercial cell lines (PANC-1, MIA PaCa-2, and SW1990) and 3 primary cells (PC15, PC37, and PC49) after KH3 treatment (Fig. 5A). The global GSEA indicated that transcriptome changes were mainly manifested in a series of signaling pathways decreased by KH3 treatment in 6 types of PDAC cells. All of the signaling pathways regulated by KH3 in individual cell type were classified into 5 subtypes (cancer, metabolism, stress, subcellular, and others). We found that the vast majority of pathways (∼50%) regulated by KH3 were classified into cancerous and metabolic pathways among all of the types of cells (Fig. 5A). According to the efficacy results, better drug responses (lower EC50) were associated with increased percentage of metabolic pathways regulated by KH3 in both commercial and primary cells (commercial cells: 36.65% average of PANC-1 and MIA PaCa-2 vs. 28.9% of SW1990; primary cells: 31.2% average of PC15 and PC37 vs. 16.7% of PC49). Additionally, the KH3 commonly regulated and core-enriched signaling pathways in cancer development and metabolism were analyzed among 6 types of cells. It was shown that, in particular, the suppression levels of metabolic pathways which were mainly glycolysis and lipid metabolisms were associated with PGAM1 expression level and KH3 efficacy (Fig. 5B and SI Appendix, Fig. S6). The representative signaling pathways such as arachidonic acid metabolism, glutathione metabolism, and Hedgehog signaling that were commonly down-regulated by KH3 among 6 types of PDAC cells were also shown by GSEA (Fig. 5C). Taken together, our results indicate that the inhibition of KH3 highly down-regulates the pathways playing crucial roles in cancer metabolism and development when treating PDAC cells. The common signature pathways found in multiple PDAC cells suggest that the efficacy of KH3 might be mainly conducted by regulating these gene expressions.
Fig. 5.
Transcriptome analysis to clarify the common and distinct pathways of multiple PDAC commercial cell lines and primary cells after KH3 treatment. (A) Three PDAC commercial cell lines (PANC-1, MIA PaCa-2, and SW1990) and 3 primary cells (PC15, PC37 and PC49) were exposed to 2 μM KH3 for 12 h as shown in the schematic diagram. All of the signaling pathways down-regulated by KH3 in individual cells were classified into 5 subtypes. (B) The core-enriched signaling pathways decreased by KH3 treatment in 6 PDAC cells were shown. (C) The representative signaling pathways such as Arachidonic Acid Metabolism, Glutathione Metabolism, and Hedgehog signaling that were commonly down-regulated by KH3 among 6 PDAC cells were shown by GSEA. Normalized enrichment score (NES) indicated the analysis results across gene sets. False discovery rate (FDR) presented if a set was significantly enriched. ES, enrichment score.
KH3 Significantly Attenuated PDAC Growth in Multiple Animal Models.
We next investigated the pharmacological profile of KH3 in vivo. By i.p. administration in Institute of Cancer Research mice at dose of 90 mg/kg KH3 loaded with poly (lactic-coglycolic acid) (PLGA), the blood was collected at different time points for analyzing the plasma level of KH3 (SI Appendix, Fig. S5). The pharmacological data showed that 5.2 h of the half-life (t1/2), as well as the 55 μg/mL of the maximum concentration (Cmax) and 214 μg*h/mL of the area under the curve (AUC0–24h) of KH3, suggesting that KH3 was well released in circulation (SI Appendix, Table S2). Subsequently, the PLGA-coated formulation of KH3 was employed to treat PDAC in the different types of mouse models shown below.
We evaluated the efficacy of KH3 in the orthotopic mouse model of PDAC by implating MIA PaCa-2 cells in the pancreas of nude mice. Six days after implantation, the mice were randomly grouped and treated with vehicle control (i.p.), gemcitabine (Gemzar; i.v.:intravenously), and KH3 (i.p.), respectively, at the time points indicated in SI Appendix, Fig. S7A. On day 20 post treatment, the orthotopic tumors were harvested, and it showed that the volumes and weights of tumors were significantly reduced in both Gemzar and KH3-treated mice, with a safe range of weight loss (SI Appendix, Fig. S7 B–D). It is suggested that KH3 exibits efficacious inhibition of cancer growth in the orthotopic model.
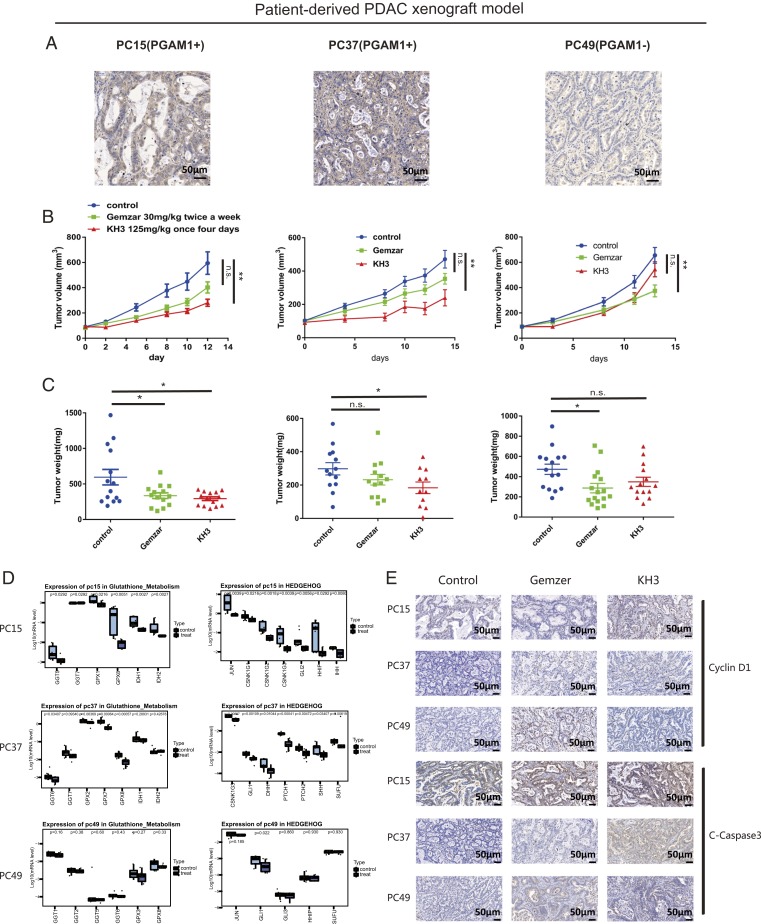
We next moved to develop the patient-derived xenograft (PDX) models of PDAC by implanting the tissue of resected patients, of which the primary cells have been used in cell proliferation assay (PC15, PC37, and PC49), in nod-scid mice s.c. Immunohistochemical staining of the PDX tissues revealed that PGAM1 expression levels were higher in PC15 and PC37 than in PC49 (Fig. 6A). Two weeks after engrafting the grown-out PDX tissue (passage# 2) in nude mice, the animals were treated with vehicle control (i.p.), Gemzar (i.v.), and KH3 (i.p.), respectively. In the PDX models of PC15 and PC37, the Gemzar-treated groups showed limited efficacy in both models when compared to control group (Fig. 6B). Whereas, the tumor volumes and weights were significantly reduced in the KH3-treated group (Fig. 6 B and C) with acceptable weight loss (SI Appendix, Fig. S8 C and D). The proliferation index also showed that KH3 was capable of inhibiting tumor growth effectively (SI Appendix, Fig. S8B). In the PDX PC49 expressing low level of PGAM1, the KH3-treated group showed very limited efficacy, whereas the Gemzar-treated group showed significant tumor growth inhibition (Fig. 6 B and C), which further comfirmed that the efficacy of KH3 was associated with PGAM1 expression level.
Fig. 6.
KH3 attenuated PDAC growth in patient-derived xenograft (PDX) models. (A) Immunochemical staining of PGAM1 in tumor tissue from patient PC15, PC37, and PC49. (B) Tumor volumes in PDAC PDXs (PC15, PC37, and PC49) were shown following 14-d treatment of Gemzar or KH3. (C) Tumor weights in PDXs (PC15, PC37, and PC49) were calculated at day 14 post Gemzar or KH3 treatment. (D) Suppressions of representative cancerous and metabolic pathways found in the PDX matched primary cells 12 h after KH3 dosing were validated at day 14 post in vivo treatment. (E) Immunochemical staining of C-Caspase3 and Cyclin D1 in tumor tissue of PDAC PDXs (PC15, PC37, and PC49) at day 14 post Gemzar or KH3 treatment. (Magnification: 200×.) The data were presented as mean ± SEM, and P values were obtained from unpaired t test (n.s., not significant. *: 0.01 < P < 0.05, **: 0.001 < P < 0.01, ***: 0.0001 < P < 0.001, ****P < 0.0001).
To determine whether cell growth inhibition was induced by KH3 in PDXs, immunohistochemistry (IHC) staining of C-Caspase3 and Cyclin D1 in tumor tissue of PDXs (PC15, PC37, and PC49) at day 14 post Gemzar or KH3 treatment was performed. In correspondence with the drug response data, we found the KH3-induced cell-cycle arrest and apoptosis were more extensive in PC15 and PC37 than in PC49, of which the cell growth inhibition level was in correlation with PGAM1 expression (Fig. 6E). Furthermore, we validated the common pathways discovered in KH3-treated PDAC cells and found that the cancer metabolism and development pathways such as glutathione metabolism and Hedgehog pathways were still highly suppressed in the PDXs of PC15 and PC37 at day 14 post KH3 treatment (Fig. 6D). In contrast, suppressions of these pathways were not detected in PC49.
Apart from evaluating the efficacy of KH3 in the animal models, the adverse effects of the compound were also examined in vivo. We evaluated the weight loss of all KH3-treated groups of mice after dosing, discovering that the weight change was mild (SI Appendix, Figs. S7D and S8 C–E). Also, by performing the peripheral blood analysis, the hematopoietic properties and liver function (such as ALT, AST, and WBC) of KH3-treated nude mice were detected in a safe range (SI Appendix, Fig. S9 A–C and Table S3). Moreover, no significant differences in histomorphology of kidneys and livers between control and KH3-treated groups were observed (SI Appendix, Fig. S9D). Altogether, the animal data indicate that, with tolerant toxicity, KH3 is capable of suppressing PDAC growth by inducing cell-cycle arrest and apoptosis. The inhibitory level is correlated with PGAM1 expression and associated with the down-regulated gene expressions in cancer metabolism and development.
Discussion
For treating PDAC, gemcitabine has been used as the first-line therapy for more than 15 y (17). Recently, additional treatments such as FOLFIRINOX (fluorouracil, leucovorin, irinotecan, and oxaliplatin), abraxane, and albumin-bound paclitaxel along with gemcitabine have been introduced for managing advanced PDAC (17, 18). Nevertheless, the prognosis of PDAC has not been improved significantly under the currently used therapeutics which showed limited efficacy in patients with either resectable or nonresectable PDAC (27). To date, the available targeted therapy in pancreatic cancer is extremely poor. The EGFR inhibitor erolotinib failed to extend the survival rate of patients with both resected or nonresected PDAC (28, 29). Also, the other targeted regiments such as MEK inhibitor and/or PI3K inhibitor are unable to improve the clinical outcome of PDAC in contrast to standard therapy (30, 31). KRAS-activating mutations are frequently detected in patients with PDAC; however, the development of an effective KRAS-targeted drug is still a struggle (27). Thus, a new direction of discovering a therapeutic target for PDAC is required.
Collateral lethality has recently been explored for discovering novel therapeutic targets which are not directly involved in cancer development. A recent study has shown that SMAD4 deletion causes eradication of a nearby metabolic enzyme gene ME2, resulting in up-regulation of the paralogue gene ME3 which inhibition suppresses pancreatic cancer progression via regulating branched-chain amino acid metabolism (22). This finding suggests that reprogrammed metabolism, considered as one of the hallmarks of pancreatic cancer, may be caused by collateral lethality. Thus, to discover the potent therapeutic targets, we looked into the metabolism of pancreatic cancer cells which growth depends heavily on glucose flux and some critical amino acid pathways such as glutamine (20) and alanine (32). Therefore, some key regulators in metabolism are considered to be promising targets in pancreatic cancer therapy. Among them, lactate dehydrogenase (LDH) caught the attention with its small-molecule inhibitor FX11 demonstrating potent efficacy in patient-derived xenograft models (23). Recently, PGAM1 was reported to promote homologous recombination repair by regulating dNTP pool (33) through its metabolic function. Additionally, a nonmetabolic function of PGAM1 in promoting cell migration was reported through interaction with ACTA2 (34) in breast cancer. These findings provided insights into the role of PGAM1 in cancer progression, which may also occur in pancreatic cancer.
Our study demonstrates that PGAM1 is a potential target for pancreatic cancer therapy in preclinical models. Based on clinical data that increased PGAM1 expression was associated with poor prognosis of patients with PDAC, the in vitro assay confirmed that KH3 was capable of inhibiting growth of multiple PDAC cells in correlation with PGAM1 expression level. These data suggest that PGAM1 is a druggable and promising target for PDAC therapy. Also, the on-target analysis of KH3 was confirmed by PGAM1 KD assay and gene expression profiling, indicating that inhibition of KH3 is mainly conferred by targeting PGAM1. Given the in vitro assay has validated the potential efficacy of PGAM1 inhibition, we moved to the in vivo study by testing the efficacy of KH3 in animal models of PDAC. Consistent with the in vitro assay, the efficacy of PGAM1 inhibition in terms of inducing tumor repression and cell-cycle arrest/apoptosis was associated with PGAM1 expression level of PDXs. Interestingly, by analyzing the gene expression profiling, the cancer metabolism and development pathways were mainly down-regulated by the PGAM1 inhibitor, and stronger suppression of metabolic pathways which were mainly glycolysis and lipid metabolism was associated with better response. Also, the suppression level of the signature pathways was correlated with PGAM1 expression. These findings indicate that inhibition of PGAM1 turns the cancer cell to low level of energy resulting in down-regulation of several major metabolites utilized by the PDAC cells. Moreover, the similar metabolic pathways reduced by PGAM1 inhibition in PDAC were also detected in HCC development (35), indicating PGAM1 inhibition might be a promising treatment for gastro-intestinal cancer. In the clinical scenario, it is likely that PGAM1 inhibitor could be a promising targeted regimen for treating the patients with PDAC showing high expression level of PGAM1, particularly for treating the patients who do not respond well or even demonstrate resistance to gemcitabine, which shows limited efficacy in most patients with PDAC (17, 18). In future study, we will further analyze the function of metabolic and nonmetabolic pathways discovered by RNA-seq during PGAM1 inhibition to clarify the molecular features of KH3's specificity. Moreover, a large number of PDXs will be used to further validate the efficacy, toxicity, and drug resistance of KH3 before launching on clinical study.
In this work, we have validated PGAM1 as a vital player in cancer development of patients with PDAC, and inhibitions of PGAM1 are regarded as a potent treatment strategy for PDAC. Our allosteric PGAM1 inhibitors have shown desirable drug-like property of satisfactory efficacy and limited toxicity. Both in vitro and in vivo studies showed that the compound KH3 was efficacious to inhibit PDAC growth in correlation with PGAM1 expression. Also, the PGAM1 inhibition suppressed several significant pathways in cancer metabolism and development for conferring its efficacy. Based on our data supported by the proof-of-principle study, we have provided solid evidence to consider initiation of developing PGAM1 inhibitor as a potential therapeutic for targeting cancer metabolism in PDAC.
Materials and Methods
Human Studies.
The clinical data and samples were obtained from patients who had been diagnosed as PDAC between 2012 and 2017 in Ruijin Hospital. The patient information was deidentified prior to use in our study. All of the studies with human subjects were approved by the Shanghai Jiaotong University School of Medicine Ethics Committee. The tumor samples were diagnosed by the Department of Pathology for the following sequencing and IHC staining. IHC staining was performed as previously described (36). More details are provided in SI Appendix, SI Materials and Methods.
In Vitro PGAM1 Inhibitors Screening Assay.
One microliter indicated inhibitor incubated with 49 μL 4.6 nM recombinant PGAM1 then 50 μL enzyme mixture containing 3 units/mL enolase (Sigma–Aldrich), 3 units/mL recombinant pyruvate kinase M2 (Sigma–Aldrich), 0.6 units/mL recombinant LDH (Sigma–Aldrich), 100 mM Tris⋅HCl, 100 mM KCl, 5 mM MgCl2, 1 mM ADP (adenosine diphosphate), 0.2 mM NADH (nicotinamide adenine dinucleotide), and 4 mM 3PG were added. The decrease in OD at 340 nm was measured as PGAM1 activity. We also performed a counter screening in which 50 μL 4 mM 2PG was added to 50 μL reaction mix containing the indicated inhibitor, 3 units/mL enolase (Sigma–Aldrich), 3 units/mL recombinant pyruvate kinase M2 (Sigma–Aldrich), 0.6 units/mL recombinant LDH (Sigma–Aldrich), 100 mM Tris⋅HCl, 100 mM KCl, 5 mM MgCl2, 1 mM ADP, and 0.2 mM NADH. The decrease in OD at 340 nm was measured as the activity of 3 enzymes mentioned above (4).
Cocrystallization of PGAM1 with Small Molecules and Data Collection and Refinement.
Crystals of PGAM1 were obtained as previously described (32). More details are provided in SI Appendix, SI Materials and Methods.
RNA-Seq Alignment and Preprocess.
STAR (v2.7.0d) (37) were used to align raw RNA-seq sequence to hg19 human reference genomes, which were downloaded from the UCSC (University of California, Santa Cruz) Genome Browser (https://hgdownload.soe.ucsc.edu/downloads.html) (38). Name-sorted and indexed BAM files were generated by Samtools (v1.8–47) (39). Transcript counts table files were generated by the HTSeq (40) htseq-count subprogram using the GENCODE annotation database and processed with the BAM files. Afterward, count matrix was obtained using DESeq2 (41).
Gene Expression Analysis.
We used FPKM to evaluate expression levels of individual genes (42). FPKM values were further produced using counts table files after normalizing the length of transcripts or genes. Differentially expressed genes were obtained using DESeq2 (v1.18.1) (41). R package gvmap (https://github.com/ytdai/gvmap) was used for visualization. Gene Ontology enrichment analysis of differentially expressed genes was performed using DAVID (https://david.ncifcrf.gov/) with default parameters. GSEA was performed using the GSEA (v3.0, http://software.broadinstitute.org/gsea) with MSigDB-hallmark gene sets (H) and MSigDB-curated gene sets (C2) (43) at 1,000-gene-set 2-sided permutation.
Data Availability.
All data can be viewed in NODE (http://www.biosino.org/node) by pasting the accession OEP000515 into the text search box or through the URL: http://www.biosino.org/node/project/detail/OEP000515.
Cell assay, plasmid construction, isothermal titration calorimetry, Seahorse, PLGA entrapment assay, pharmacokinetic study, HPLC for quantifying KH3, xenograft studies, and statistical analysis are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank the staff at beam line BL17U, BL18U1, and BL19U1 at the Shanghai Synchrotron Radiation Facility for support with data collection. We thank Dr. Qingyang Gu and Dr. Xuzhen Tang from Oncology and Immunology Unit, Research Service Division, WuXi AppTec, for assisting generation of primary cells and PDX models. We also thank Dr. Jing Xie from Department of Pathology, Ruijin Hospital for preparing the FFPE slides of resected tumor tissue and evaluating their pathological features. Work in the L.Z. laboratory is supported by the National Natural Science Foundation of China (No. 21472026, 21877014, 91853206). Work in the Y.F. and B.-Y.S. laboratories was supported by the Shanghai Municipal Committee of Science and Technology (grant#: 15JC1402800), the National Natural Science Foundation of China (No. 81502695, 81802358, 81871906, 81672325), the Shanghai Municipal Education Commission (grant#: 14ZZ103), and the Shanghai Eastern Youth Scholar Program (to Y.F.; grant#: DQ2015010).
Footnotes
The authors declare no competing interest.
Data deposition: All data can be viewed in NODE (http://www.biosino.org/node) by pasting the accession OEP000515 into the text search box or through the URL: http://www.biosino.org/node/project/detail/OEP000515.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914557116/-/DCSupplemental.
References
- 1.Pavlova N. N., Thompson C. B., The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones N. P., Schulze A., Targeting cancer metabolism–Aiming at a tumour’s sweet-spot. Drug Discov. Today 17, 232–241 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hitosugi T., et al. , Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell 22, 585–600 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kroemer G., Pouyssegur J., Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell 13, 472–482 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Tennant D. A., Durán R. V., Gottlieb E., Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10, 267–277 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Levine A. J., Puzio-Kuter A. M., The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330, 1340–1344 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Vander Heiden M. G., Targeting cancer metabolism: A therapeutic window opens. Nat. Rev. Drug Discov. 10, 671–684 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Schulze A., Harris A. L., How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 491, 364–373 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Ward P. S., Thompson C. B., Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay N., Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 16, 635–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luengo A., Gui D. Y., Vander Heiden M. G., Targeting metabolism for cancer therapy. Cell Chem. Biol. 24, 1161–1180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaneton B., Gottlieb E., PGAMgnam style: A glycolytic switch controls biosynthesis. Cancer Cell 22, 565–566 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Miller K. D., et al. , Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 66, 271–289 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Mayers J. R., et al. , Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Von Hoff D. D., et al. , Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 369, 1691–1703 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conroy T., et al. ; Groupe Tumeurs Digestives of Unicancer ; PRODIGE Intergroup , FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 364, 1817–1825 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Ying H., et al. , Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son J., et al. , Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller F. L., et al. , Passenger deletions generate therapeutic vulnerabilities in cancer. Nature 488, 337–342 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dey P., et al. , Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 542, 119–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajeshkumar N. V., et al. , Therapeutic targeting of the warburg effect in pancreatic cancer relies on an absence of p53 function. Cancer Res. 75, 3355–3364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans M. J., Saghatelian A., Sorensen E. J., Cravatt B. F., Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat. Biotechnol. 23, 1303–1307 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Li X., et al. , Identification of epigallocatechin-3- gallate as an inhibitor of phosphoglycerate mutase 1. Front. Pharmacol. 8, 325 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren F., et al. , Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol. Cancer 9, 81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neoptolemos J. P., et al. , Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 15, 333–348 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Sinn M., et al. , CONKO-005: Adjuvant chemotherapy with gemcitabine plus erlotinib versus gemcitabine alone in patients after R0 resection of pancreatic cancer: A multicenter randomized phase III trial. J. Clin. Oncol. 35, 3330–3337 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Moore M. J., et al. ; National Cancer Institute of Canada Clinical Trials Group , Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: A phase III trial of the National Cancer Institute of Canada clinical trials group. J. Clin. Oncol. 25, 1960–1966 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Lorusso P. M., et al. , Phase I and pharmacodynamic study of the oral MEK inhibitor CI-1040 in patients with advanced malignancies. J. Clin. Oncol. 23, 5281–5293 (2005). [DOI] [PubMed] [Google Scholar]
- 31.O’Neil B. H., et al. , A phase II/III randomized study to compare the efficacy and safety of rigosertib plus gemcitabine versus gemcitabine alone in patients with previously untreated metastatic pancreatic cancer. Ann. Oncol. 27, 1180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sousa C. M., et al. , Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu J., et al. , Phosphoglycerate mutase 1 regulates dNTP pool and promotes homologous recombination repair in cancer cells. J. Cell Biol. 216, 409–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D., et al. , Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene 36, 2900–2909 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Aizarani N., et al. , A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 572, 199–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J., et al. , Snail recruits Ring1B to mediate transcriptional repression and cell migration in pancreatic cancer cells. Cancer Res. 74, 4353–4363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyner C., et al. , The UCSC genome browser database: 2017 update. Nucleic Acids Res. 45, D626–D634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen B., et al. , Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U.S.A. 115, 373–378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data can be viewed in NODE (http://www.biosino.org/node) by pasting the accession OEP000515 into the text search box or through the URL: http://www.biosino.org/node/project/detail/OEP000515.
Cell assay, plasmid construction, isothermal titration calorimetry, Seahorse, PLGA entrapment assay, pharmacokinetic study, HPLC for quantifying KH3, xenograft studies, and statistical analysis are described in detail in SI Appendix.