Significance
Tc toxin complexes are virulence factors of many bacteria, including insect and human pathogens. Tc toxins are promising candidates for potential biopesticides as alternatives to Bacillus thuringiensis toxins. We report 2 high-resolution cryo-EM structures of a Tc holotoxin embedded in the membrane. These structures, along with biochemical studies, contribute to our understanding of key steps of the prepore-to-pore transition, membrane insertion, and toxin translocation mechanisms of this biotechnologically relevant type of bacterial toxin. Our study provides important insights into the mechanism of action of Tc toxins and can serve as a strong foundation for the development of biopesticides.
Keywords: toxin, cryo-EM, translocation, Tc, Photorhabdus
Abstract
Tc toxins are modular toxin systems of insect and human pathogenic bacteria. They are composed of a 1.4-MDa pentameric membrane translocator (TcA) and a 250-kDa cocoon (TcB and TcC) encapsulating the 30-kDa toxic enzyme (C terminus of TcC). Binding of Tc toxins to target cells and a pH shift trigger the conformational transition from the soluble prepore state to the membrane-embedded pore. Subsequently, the toxic enzyme is translocated and released into the cytoplasm. A high-resolution structure of a holotoxin embedded in membranes is missing, leaving open the question of whether TcB-TcC has an influence on the conformational transition of TcA. Here we show in atomic detail a fully assembled 1.7-MDa Tc holotoxin complex from Photorhabdus luminescens in the membrane. We find that the 5 TcA protomers conformationally adapt to fit around the cocoon during the prepore-to-pore transition. The architecture of the Tc toxin complex allows TcB-TcC to bind to an already membrane-embedded TcA pore to form a holotoxin. Importantly, assembly of the holotoxin at the membrane results in spontaneous translocation of the toxic enzyme, indicating that this process is not driven by a proton gradient or other energy source. Mammalian lipids with zwitterionic head groups are preferred over other lipids for the integration of Tc toxins. In a nontoxic Tc toxin variant, we can visualize part of the translocating toxic enzyme, which transiently interacts with alternating negative charges and hydrophobic stretches of the translocation channel, providing insights into the mechanism of action of Tc toxins.
Photorhabdus toxin complexes (Tc toxins) are large, heterotrimeric protein complexes produced by numerous insect and human pathogenic bacteria (1, 2). Tcs were originally discovered in Photorhabdus luminescens (3). Later, gene loci of Tc toxins were also found in Xenorhabdus nematophila (4, 5) and human pathogens, such as Photorhabdus asymbiotica (6) and different species of Yersinia (7, 8). While the entomopathogenic toxins are potential biopesticides and therefore the focus of crop protection research, understanding the mechanism of action of Tc toxins of human pathogens is medically relevant.
A Tc toxin is typically made up of 3 components: the ∼1.4-MDa TcA homopentamer, the ∼170-kDa TcB, and the ∼100-kDa TcC subunit. P. luminescens has different TcA, TcB, and TcC components (9, 10), enabling it to form various holotoxins that result in the translocation of different toxic payloads (11). The modularity of Tc toxins in insect pathogens may provide an alternative to Bt toxins for green biotechnology, overcoming the problem of rising resistance in insects (12).
TcA, a 18-nm wide and 24-nm long bell-shaped molecule, acts as a membrane permeation and protein translocation device. It is composed of a large outer shell, a central channel, and a linker that connects these 2 parts. The α-helical translocation channel at the center of TcA is hydrophobic at its tip and shielded by the shell (13). Receptor-binding domains at the periphery of the shell likely mediate the initial receptor-toxin interaction on target cells (14, 15). A shift to higher or lower pH results in the opening of the bottom of the shell, triggering its structural rearrangement and the release of the shielded translocation channel. Membrane permeation of the channel is driven by the proline-rich linker. The linker condenses from a stretched and unfolded conformation in the prepore to a partially helically folded, compacted conformation in the pore, resulting in a shifting-out movement of the channel from the shell (14). Once inside the membrane, the channel opens and releases the toxic enzyme into the cytosol (14).
TcB and TcC together form an ∼250-kDa cocoon composed mainly of rearrangement hotspot (RHS) repeats. The C-terminal end of the cocoon protrudes inward and forms an aspartyl autoprotease domain that cleaves off the C-terminal part of TcC (15). This generates a polypeptide of ∼30 kDa that resides inside the cocoon. In contrast to the highly conserved N-terminal region of TcC, the sequence of the C terminus of TcC is not conserved and thus is termed the hypervariable region (HVR) (11). The various HVRs are toxic enzymes that have different functions or targets. For example, while the toxic enzymes of TccC3 and TccC5 from P. luminescens both act as ADP-ribosyltransferases, they modify different proteins; TccC3 ADP-ribosylates actin and TccC5 ADP-ribosylates Rho GTPases (11). The hydrophobic inside of the cocoon creates an environment that stabilizes unfolded proteins, most likely resulting in an unfolded conformation of the toxic enzyme (15, 16).
The TcB-TcC cocoon binds through a distorted β-propeller that closes it at the bottom to the funnel-shaped end of TcA, resulting in the ABC holotoxin complex (15). During this process, parts of the β-propeller completely unfold and refold into an alternative conformation, resulting in the opening of the cocoon and release of the toxic enzyme into the channel with the C terminus first (17).
Several structures of TcA in its prepore and pore states, as well as structures of the ABC holotoxin complex, enabled us to understand the major steps in the mechanism of Tc toxin action (14–17). However, a structure of ABC in its pore state is missing, hampering our understanding of how the complete complex enters the membrane and translocates its toxic payload.
Here we present the high-resolution cryo-EM structure of an ABC holotoxin from P. luminescens embedded in lipid nanodiscs. The structure reveals that the shell domain of TcA forms a basin in which the TcB-TcC complex resides. The cocoon interacts directly with the α-helical shell domains of 2 out of 5 TcA subunits, which have to slightly adapt their conformation to fit around the TcB-TcC cocoon. All upper TcA domains are better resolved than in the previously obtained TcA complex alone, indicating that they are stabilized by the cocoon (14). Interestingly, the stable TcA-TcB interface is not affected by the prepore-to-pore transition of TcA, and TcB-TcC can also bind to the already membrane-embedded TcA pore to form a holotoxin. During this process, the toxic enzyme is released from the holotoxin, indicating that the translocation of the toxic enzyme is not driven by a proton gradient, another energy source, or chaperones. Furthermore, we show that the assembled ABC complex can enter the membrane in the absence of receptors. Lipids with zwitterionic head groups and mammalian lipids are preferred over lipids with negatively charged head groups and bacterial lipids. By using a nontoxic variant, we can visualize a part of the translocating toxic enzyme in the upper part of translocation channel, revealing that the toxic enzyme transiently interacts with alternating hydrophobic and negatively charged regions of the translocation channel.
Results and Discussion
Structure of the ABC Holotoxin Complex in Nanodiscs.
To obtain an ABC complex in which the toxic enzyme is still residing inside the translocation channel, we assembled the ABC holotoxin from the individually purified toxin components with proteolytically inactive TcC [TcdA1, TcdB2-TccC3(D651A) from P. luminescens, referred to collectively as ABC(D651A)] (15, 17), which is, in contrast to the wild-type ABC holotoxin [ABC(WT)], nontoxic to HEK 293T cells (SI Appendix, Fig. S1A). We induced pore formation and simultaneous reconstitution in lipid nanodiscs by dialyzing a mixture of ABC(D651A) and nanodiscs against a buffer at pH 11 (SI Appendix, Fig. S1 B and C). We then solved the structure of the complex by cryo-EM and single-particle analysis using SPHIRE (18) at an average resolution of 3.4 Å (Fig. 1 A and B, SI Appendix, Fig. S2 and Table S1 and Movie S1) (19). To improve the local resolution of the TcB-TcC cocoon, we shifted the center toward these components during image processing (Materials and Methods and SI Appendix, Fig. S3).
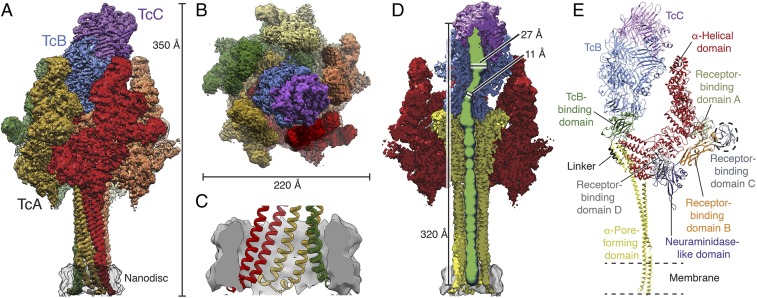
Fig. 1.
Structure of the Tc holotoxin in a lipid nanodisc. (A and B) Cryo-EM map of ABC(D651A) in side view (A) and top view (B). The individual protomers of TcA, TcB, and TcC are shown in different colors. The lipid nanodisc is shown in transparent gray. (C) Longitudinal section through the nanodisc region. The density of the nanodisc is shown in gray, and the molecular model of the TcA transmembrane domains is colored by protomer. (D) Longitudinal section through the cryo-EM structure displaying the central channel. The densities corresponding to the TcA outer shell, the TcA channel, TcB, TcC, and the nanodisc are shown in red, yellow, blue, violet and gray, respectively. The lumen of the translocation channel was determined in ChExVis (49) and is shown in green. Diameters inside the cocoon and at the constriction site are indicated. (E) Molecular models of a TcA protomer, TcB, and TcC in the context of the holotoxin. TcA is colored by domains. Receptor-binding domain C (dashed circle) is not resolved in the cryo-EM structure. TcB is in blue, and TcC is in violet.
The high quality of the map allowed us to build a model of 91% of the pore state of ABC(D651A). The final model contains residues 89 to 2,516 of TcA, residues 1 to 1,471 of TcB, and residues 1 to 683 of TcC with some loops missing. The only parts of the complex that could not be resolved are the receptor-binding domain (RBD) C of TcA (residues 1,382 to 1,491) and the ADP-ribosyltransferase of TcC (residues 684 to 960), indicating that these regions are highly flexible (Fig. 1E).
The structure of TcA in the 1.7-MDa ABC pore complex closely resembles the structure of the isolated TcA subunit embedded in lipid nanodiscs (Protein Data Bank [PDB] ID code 5LKI) (14) (SI Appendix, Fig. S4 A and B). The translocation channel is open and extends all of the way from the TcB-TcC cocoon to the transmembrane region in the nanodisc (Fig. 1 C and D).
The interface between the β-propeller of TcB and the TcB-binding domain of TcA is practically identical to that observed in the ABC prepore state (PDB ID code 6H6F) (SI Appendix, Fig. S4 E and F) (17). This and the rigidity of the rest of the cocoon result in the same relative position of the TcB-TcC cocoon to TcA, which sits at an angle of 32° on top of TcA (SI Appendix, Fig. S4E). As a consequence, 1 of the 5 α-helical domains of the shell that form a tight basin around TcB-TcC is shifted outward, which results in a break of the C5 symmetry of TcA at this position (Fig. 2 A and B, SI Appendix, Fig. S4 C and D, and Movie S2). The shift of TcA-A relative to the other TcA subunits is most pronounced at the uppermost part of the α-helical shell (residues 645 to 797) (Fig. 2B and SI Appendix, Fig. S4C). The other domains of TcA-A, such as the RBDs, the neuraminidase-like domain, and the α-helical pore-forming domain, remain unchanged (Fig. 2B), indicating that the conformational changes at the tip of the shell are not transmitted to the rest of the protein.
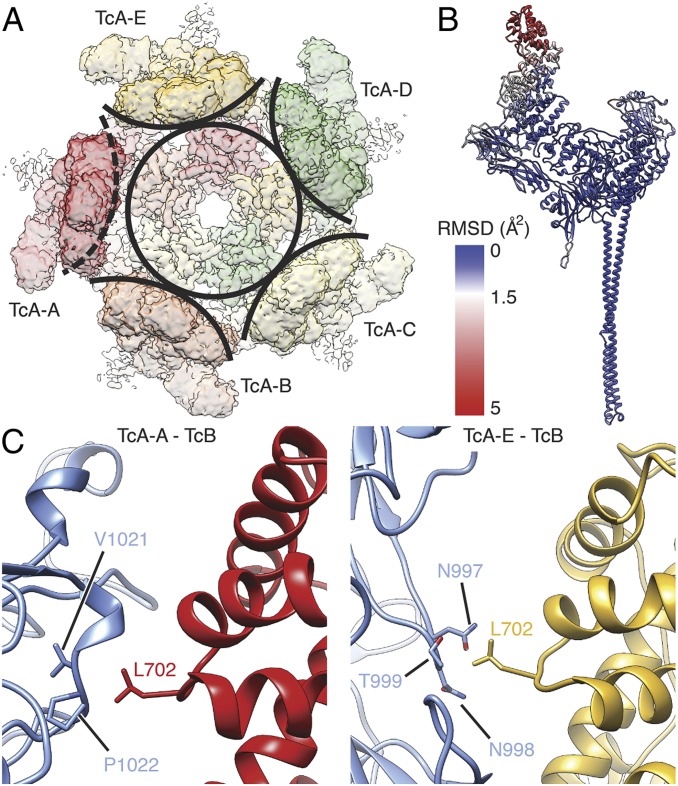
Fig. 2.
Interaction of the TcB-TcC cocoon with the TcA shell. (A) Top view of the density map corresponding to TcA in the context of ABC(D651A). A circle around the center of the TcA channel and arcs around the outer shell of individual TcA protomers indicate the disruption of C5 symmetry by TcA-A (dashed arc). (B) RMSD of Cα atoms of the individual TcA protomers, plotted on TcA-A. RMSD values (in Å2) increase from blue to red. (C) Interaction of the cocoon (TcB, blue) with 2 of 5 TcA protomers. (Left) Interaction of L702 of TcA-A (red) with V1021 and P1022 of TcB. (Right) Interaction of L702 of TcA-E (ochre) with N997, N998, and T999 of TcB.
The TcB-TcC cocoon directly interacts with the upper α-helical shell domain of 2 TcA chains. L702 of the displaced TcA protomer (TcA-A) and the same residue of the adjacent TcA-E interact with 2 hydrophobic patches at the surface of the cocoon (Fig. 2C). Although there are no close contacts between the cocoon and the other 3 TcA protomers, the entire upper α-helical shell of the pentamer is stabilized in the ABC pore, likely due to a decreased level of freedom of these domains in the holotoxin. Thus, in contrast to the structure of the TcA pore in the absence of the cocoon (14), the complete upper part of the outer shell is resolved (SI Appendix, Figs. S2E and S4 A and B).
The density corresponding to the ADP-ribosyltransferase does not reach the lower end of the translocation channel (Fig. 3B and Movie S3). Although the toxic enzyme is covalently bound to the cocoon at the N terminus, one would expect that the C-terminal part of the protein that enters the translocation channel first (17) would proceed through the entire channel and also leave it at the bottom. Instead, it resides in the cocoon and only in the very upper part of the TcA translocation channel (Fig. 3 B–E), comparable to the ADP-ribosyltransferase in the prepore form of the cleavage-deficient ABC(D651A) (17). This suggests that either the covalently linked toxic enzyme is locked in the cocoon and does not have the necessary flexibility to be further translocated or a missing driving force, such as a proton gradient between the cocoon and exit of the channel, is responsible for the halted translocation.
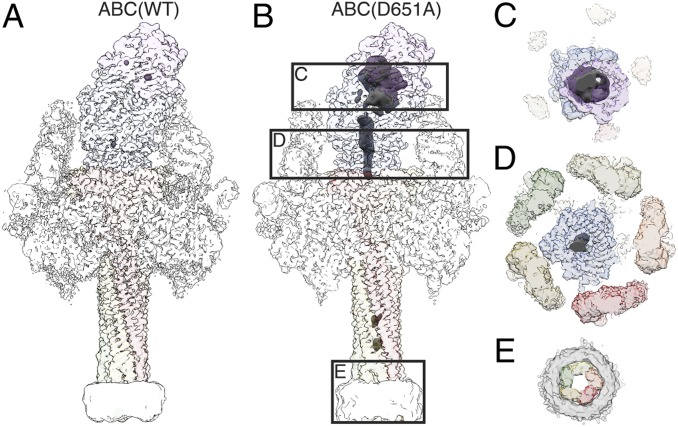
Fig. 3.
ADP-ribosyltransferase in the ABC(D651A) pore. (A and B) Reconstruction of ABC(WT) (A) and ABC(D651A) (B) with transparent surface. Density in which no atomic model was fitted, and the nanodisc were filtered to 10 Å and are shown at a lower binarization threshold. The small density blobs inside the cocoon in A and inside the TcA channel in B correspond to noise. Increasing the binarization threshold of the disordered density results in disappearance of the blobs before the density inside the cocoon disappears. See also Movie S3. (C–E) Cross-sections through ABC(D651A) at the cocoon, the TcB translocation channel, and the transmembrane region, as indicated in B. Continuous density of the ADP-ribosyltransferase is apparent in the cocoon (C) and in the narrow passage of TcB below the constriction site (D), but not in the transmembrane region embedded in the nanodisc (E).
Spontaneous Translocation of the ADP-Ribosyltransferase.
To find out whether the latter is true, we solved the structure of the ABC(WT) embedded in lipid nanodiscs at an average resolution of 3.4 Å (SI Appendix, Fig. S5) (20). If the translocation of the toxic enzyme is indeed dependent on an additional driving force, then the protein should also reside inside the cocoon and the TcA channel, comparable to the ABC(D651A). However, while the rest of the cryo-EM map did not differ from that of ABC(D651A) in its pore state, we did not find any density corresponding to the ADP-ribosyltransferase (Fig. 3A and Movie S3). This demonstrates that the ADP-ribosyltransferase is translocated and released from the holotoxin. Since we reconstituted the protein in vitro in the absence of other proteins, we conclude that no additional factors, such as molecular chaperones, or driving forces, such as a proton gradient, are necessary to ultimately translocate the ADP-ribosyltransferase. In addition, translocation of the ADP-ribosyltransferase requires that the protein is cleaved and flexible enough to move inside the TcB-TcC cocoon. Otherwise, its translocation can only be initiated, but not continued throughout the entire translocation channel.
ADP-Ribosyltransferase Inside the Cocoon.
The entire cocoon is resolved better than 4 Å in the ABC(D651A) pore complex, including the aspartyl protease cleavage site of TcC. As in the prepore complex, the density of the ADP-ribosyltransferase inside the cocoon and in the very upper part of the TcA translocation channel appears only at lower thresholds, and no secondary structure elements are recognizable (Fig. 3B). We estimated the 3D variance of the reconstruction in SPHIRE (18). Expectedly, the highest variance is found inside the cocoon and also at the upper domains of TcA, indicating high flexibility and disorder in these protein regions (SI Appendix, Fig. S6 A–C). 3D sorting focused on the inside of TcB-TcC did not improve the quality of the density.
Nevertheless, due to the overall improved local resolution in the cocoon after shifting the center of reconstruction (SI Appendix, Fig. S3E), we could identify the positions of the first 5 residues of TcC after the aspartyl protease cleavage site (M679 to A683). The residues form a loop that keeps the cleavage site between the catalytic aspartates (Fig. 4A, SI Appendix, Fig. S6 D and E, and Movie S4). G677 and P680 are the only conserved residues around the autoproteolytic cleavage site (Fig. 4 A and B). To identify whether these residues are part of the recognition sequence defining the specificity of the protease, we mutated both residues to alanines. Surprisingly, P680A does not affect autoproteolysis, and the mutation of G677 to alanine results in only ∼40% of the protein being uncleaved (Fig. 4C). Thus, although these residues are conserved in RHS repeat proteins that function as toxins [not in teneurins (21, 22)], neither of them is absolutely necessary for the activity of the aspartyl protease, indicating that there is no specific recognition sequence required. Similarly, aspartyl proteases of the digestive system, such as pepsin, that cleave in a rather unspecific manner adjacent to large hydrophobic and aromatic residues act independently of the residues 2 positions upstream and downstream of the cleavage site (23).
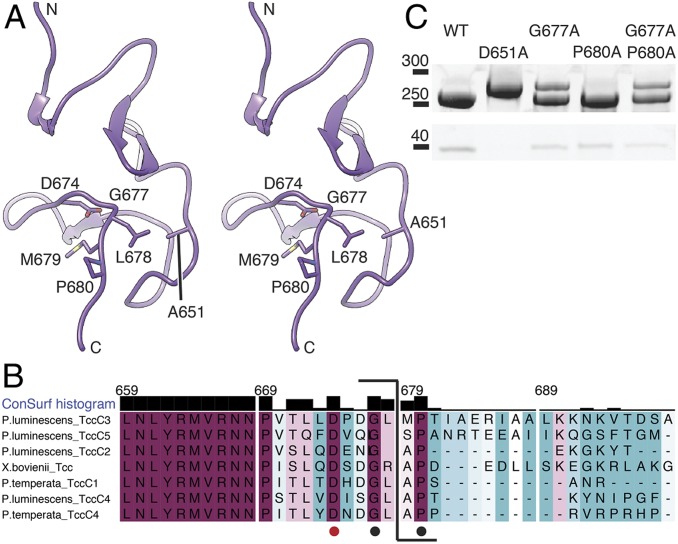
Fig. 4.
Structure and conservation of the aspartyl autoprotease domain of TcC. (A) Stereoview of the aspartyl protease domain (residues 618 to 678) with the N terminus of the ADP-ribosyltransferase in the TcB-TcC cocoon. The toxin is cleaved between L678 and M679. The residues forming the aspartyl protease cleavage site (D674 and A651, which is D651 in WT) and the conserved G677 and P680 are indicated. Representations that include the cryo-EM map are shown in SI Appendix, Fig. S6 D and E. (B) Sequence alignment of the C-terminal part of the aspartyl autoprotease site and the N-terminal region of the toxic enzyme, colored from minimum (cyan) to maximum (magenta) conservation. The cleavage site is indicated (bracket). D674 of the aspartyl protease domain is marked with a red dot, and the conserved G677 and P680 flanking the cleavage site are marked with black dots. (C) Cleavage of the toxin by different mutations in the TcC cleavage site, analyzed by SDS/PAGE. TcC variant D651A does not cleave the toxin, and variant G677A shows impaired cleavage, as does the double mutant G677A-P680A. Mutation P680A does not impair toxin cleavage significantly.
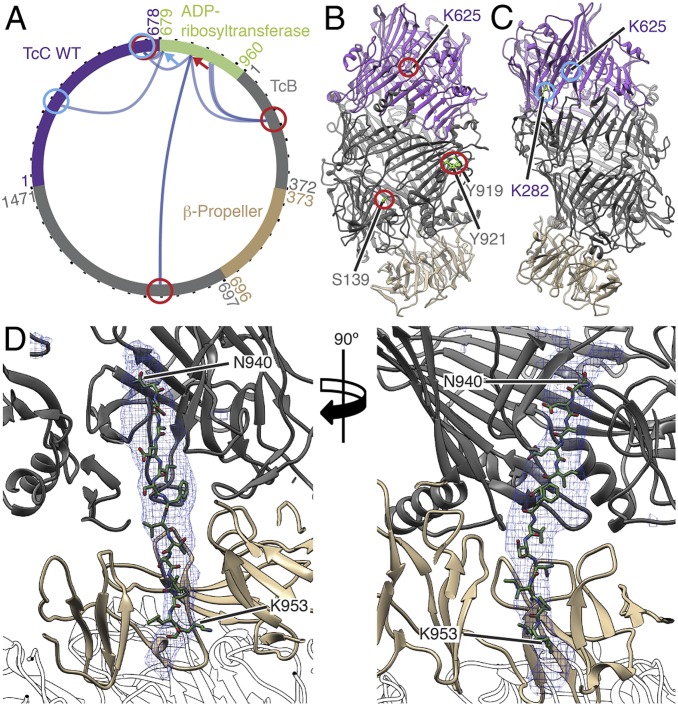
Since we could not determine the structure of the ADP-ribosyltransferase inside the TcB-TcC cocoon, we performed cross-linking mass spectrometry (XL-MS) using the 12-Å-long cross-linker disuccinimidyl dibutyric urea to obtain information on the orientation of the protein inside the TcB-TcC(WT) cocoon (SI Appendix, Table S2). We selectively screened for cross-links between the TcB-TcC cocoon and the ADP-ribosyltransferase and identified several of them at different positions of the molecules (Fig. 5A). For example, K785 of TcC (residue 107 of the ADP-ribosyltransferase) was cross-linked to S139, Y919, and Y921 of TcB and K625 of TcC (Fig. 5 A and B). The same cross-links were found for TcB-TcC(D651A) with noncleaved ADP-ribosyltransferase (SI Appendix, Fig. S6 F and G). These 3 positions are distributed over the entire inner surface of the TcB-TcC cocoon. In TcB-TcC(D651A), we found another prominent set of cross-links between 3 lysines in the central part of the ADP-ribosyltransferase (K853, K856, and K863 of TcC) and the residues S139 and Y921 of TcB and K11 and K282 of TcC. Again, they are distributed over the entire lumen of the cocoon, indicating that the toxic enzyme does not have a fixed position inside the cocoon (SI Appendix, Fig. S6 F and H).
Fig. 5.
XL-MS of the toxic enzyme inside the TcB-TcC WT cocoon and atomic model of the ADP-ribosyltransferase in the narrow passage of TcB. (A) Visualization of cross-links (dark-blue curves) between the TcB-TcC cocoon (gray and violet; β-propeller in sand) and the toxic enzyme (green). Two individual positions (K785 and K690 of TcC) and their cross-links to the TcB-TcC cocoon are indicated with red and blue arrows and circles, respectively. All cross-links with a score of ≥50 are shown. The plot was created using xVis (48). A complete list of all cross-links is shown in SI Appendix, Table S2. (B and C) The TcB-TcC cocoon (PDB ID code 4O9X) with all amino acids that undergo cross-links with K785 indicated and highlighted with red circles (B) and cross-links with K690 indicated and highlighted with blue circles (C). The cocoon is colored gray and violet, and the β-propeller (residues 376 to 691 of TcB) is in sand. (D) Atomic model of the ADP-ribosyltransferase (N940 to K953 of TcC) in the narrow passage of TcB. The density map corresponding to the ADP-ribosyltransferase is shown as a blue mesh. TcA is shown transparent.
However, since K690 of TcC, which is positioned close to the N terminus of the ADP-ribosyltransferase, cross-links to 2 prominent lysines close to the aspartyl protease site of the cocoon (K282 and K625 of TcC), the N terminus of the ADP-ribosyltransferase in TcB-TcC(WT) seems to not change its position after cleavage (Fig. 5 A and C).
Taken together, the XL-MS of TcB-TcC(WT) and TcB-TcC(D651A) indicate that the N terminus of the ADP-ribosyltransferase resides close to its cleavage site, whereas the rest of the protein takes random orientations inside the cocoon, suggesting that the enzyme is in an unfolded or semiunfolded state.
The C Terminus of the ADP-Ribosyltransferase Inside the Translocation Channel.
Before entering the TcA translocation channel, the toxic enzyme must pass a narrow passage at the lower part of the cocoon (17) (Fig. 1D and SI Appendix, Fig. S8 A–C). Although the density corresponding to the ADP-ribosyltransferase in this region was better resolved in the pore structure of ABC(D651A) than in our previous prepore structure (17), it was still difficult to build an atomic model of the protein. Since it is obvious that the protein enters the translocation channel with the C terminus first, we tested the optimal fit of 3 different C-terminal fragments of TcC (I932 to S945, N940 to K953, and L947 to R960) and found that the peptide N940-K953 fitted best into the density (Fig. 5D, SI Appendix, Fig. S7 A and B, and Movie S5). Although secondary structure predictions suggest the presence of a short β-strand at the center of the peptide (SI Appendix, Fig. S7A), it fits in the density only in its stretched, unfolded conformation, indicating that the protein passes the constriction site in its unfolded state, similar to translocated substrates in HSP104 (24) or AAA+ proteases (25).
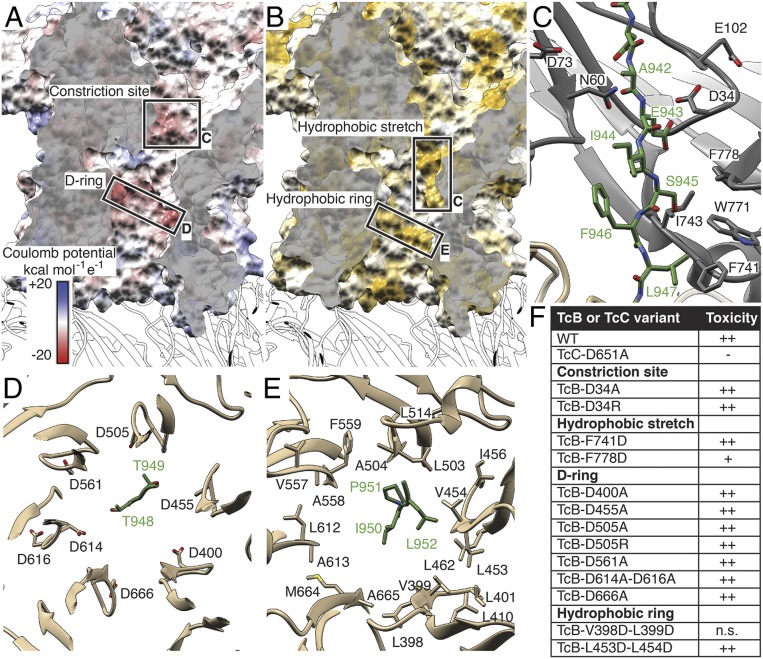
The structure of the C terminus of the ADP-ribosyltransferase inside the narrow passage of TcB reveals that along its translocation pathway, the enzyme passes first through a conserved negatively charged constriction site formed by D34, N60, D73, and E102 (17) and then through a conserved hydrophobic stretch (F741, I743, W771, and F778). Afterward, it passes through a negatively charged ring and finally a hydrophobic ring inside the β-propeller (Fig. 6 A–C and SI Appendix, Fig. S8 D and G). Interestingly, the protein does not locate at the axis of the channel but adheres to the channel wall (Fig. 6 D and E). As expected, the interactions are weak, but polar and hydrophobic residues of the ADP-ribosyltransferase are stabilized by interacting with hydrophilic and hydrophobic stretches of the channel, respectively (Fig. 6 C–E).
Fig. 6.
Alternating hydrophobic and negatively charged stretches in the narrow passage of TcB in ABC(D651A). (A) Surface representation of the narrow passage of TcB, colored according to the Coulomb potential (kcal mol−1 e−1) at pH 7.0. The negatively charged constriction site at the channel entrance and a negatively charged ring (D-ring) in the center of the channel are highlighted in dashed boxes. Translocation direction is from top to bottom. (B) The same view as in A, colored according to hydrophobicity as described by Hessa et al. (50). Hydrophobic regions are colored ochre, and nonhydrophobic regions are in white. A hydrophobic stretch at the upper part of the channel and a hydrophobic ring close to the TcB-TcA interface are highlighted. (C) Section of the narrow passage of TcB showing the hydrophobic stretch made up of F741, I743, W771, and F778. D34, N60, D73, and E102 form the negatively charged constriction site at the entrance of the TcB channel. The translocating ADP-ribosyltransferase (A942 to L947 of TcC) is shown in green. (D) The D-ring of TcB is formed by D400, D455, D505, D561, D614, D616, and D666. T948 and T949 of TcC interact with the D-ring. (E) The hydrophobic ring is formed by L398, V399, L401, L410, L453, V454, I456, L462, L503, A504, L514, V557, A558, F559, L612, A613, M664, and A665. I950 to L952 of TcC are at the height of the hydrophobic ring. (F) Toxicity of TcB-TcC variants against HEK293-T cells in the context of the holotoxin. n.s., nonsoluble expression of the respective variant.
The negatively charged ring inside the β-propeller is formed by 7 conserved aspartate residues: D400, D455, D505, D561, D614, D616, and D666 (Fig. 6 A and D and SI Appendix, Fig. S8 E and H). Therefore, we call it the D-ring. This ring interacts with the polar residues T948 and T949 of the ADP-ribosyltransferase (Fig. 6D).
The hydrophobic ring, which forms the central section of the β-propeller, is composed of many small and large hydrophobic residues facing the channel lumen (Fig. 6 B and E). They are either strictly conserved or replaced by nearly equal-sized hydrophobic side chains (SI Appendix, Fig. S8 F and H). The ring interacts transiently with 3 hydrophobic residues of the ADP-ribosyltransferase: I950, P951, and L952.
To demonstrate the role of these striking features in TcB-TcC, we mutated several key residues to change their property from hydrophobic to hydrophilic or negatively charged to either neutral or positively charged and probed the toxicity of the variants on HEK 293T cells (SI Appendix, Fig. S9). Surprisingly, the insertion of charged residues into the hydrophobic stretch has no or only minor impairing effects (Fig. 6F and SI Appendix, Fig. S9). Similarly, mutations of L453 and L454 to aspartate in the hydrophobic ring have no effect on the toxicity and mutations of L398 and L399 to alanine cause insoluble expression of TcB-TcC (Fig. 6F). The same is true for the constriction site and the D-ring, where mutations of the highly conserved D34 and D505 residues to alanine or arginine do not cause a decrease in toxicity (Fig. 6F and SI Appendix, Fig. S9). These results indicate that single point mutations of conserved residues in the narrow passage of TcB-TcC are not sufficient to change the property of the respective region, such that the translocation of the ADP-ribosyltransferase is negatively influenced. A combination of mutations is probably necessary to achieve this. Unfortunately, insertion of several mutations at the same time seem to have a strong effect on the overall fold of the protein, since it results in insoluble expression of TcB-TcC in almost all cases.
In summary, the structure of the C-terminal region of the ADP-ribosyltransferase in the narrow passage of TcB provides a snapshot of the translocation process, in which a temporary stabilization of the protein is achieved by transient interactions with residues of the channel wall.
Pore Formation of TcA in Different Lipid Environments.
The previously obtained cryo-EM structure and MD simulations of TcA in lipid nanodiscs showed that lipid headgroups intercalate between the TcA protomers (14). The natural flexibility of this region resulted in a lower resolution not only in the structure of the TcA pore (14), but also in the pore structures of ABC(D651A) and ABC(WT) (SI Appendix, Figs. S2E and S5E).
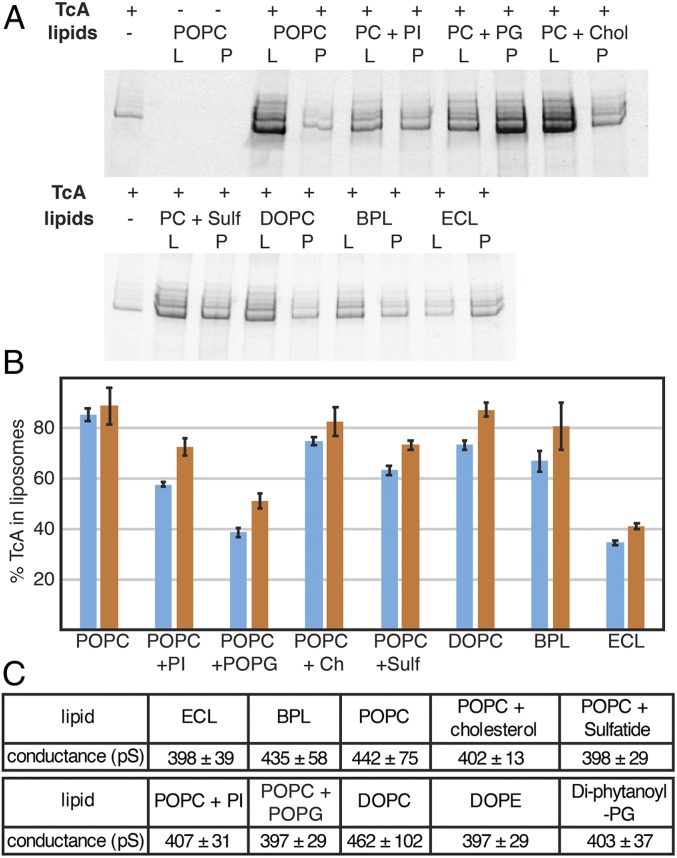
To find out whether membrane insertion of TcA is specific to certain lipids, we induced pore formation and checked how well the protein reconstituted into liposomes using different lipids. TcA integrated readily into 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC), di-oleoyl-phosphatidylcholine (DOPC), POPC with 20% cholesterol, and brain polar lipids (BPLs). In all cases, more than 70% of TcA is reconstituted in liposomes. The reconstitution efficiency in liposomes composed of POPC with 20% phosphatidylinositol (PI) or POPC with 20% sulfatide was slightly worse, and when POPC with 20% 1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG) or an Escherichia coli polar lipid extract (ECL) was used, considerably less proteoliposomes were formed (only 35% to 50% of the protein; Fig. 7 A and B). This demonstrates that although TcA has no pronounced affinity for certain lipids, it preferably integrates into membranes that do not contain negatively charged head groups. Furthermore, mammalian lipid mixtures seem to be preferred over bacterial lipids, which is expected since the natural targets of various bacterial Tcs are insects or mammals, not bacteria (2). Despite having different ratios of PE and PC and a different cholesterol content, the overall membrane fluidity is very similar in insect and mammalian cells (26). This and the lack of specificity for certain lipids makes it likely that Tc toxins integrate well into all eukaryotic cell membranes.
Fig. 7.
Lipid interaction and pore formation of TcA. (A) Integration of TcA in liposomes of various compositions. SDS/PAGE of the proteoliposome flotation of TcA incubated with various lipids (POPC, POPC + 20% liver PI, POPC + 20% POPG, POPC + 20% cholesterol, POPC + 20% sulfatide, DOPC, BPL extract, and E. coli polar lipid extract) at pH 11 for 48 h. L, liposome fraction; P, pellet fraction. (B) Percentage of TcA in the liposome fraction in 2 independent liposome flotation experiments determined by densitometry of SDS-gel bands (blue and orange bars, respectively). The error bars represent SDs of 3 samples. (C) Averages and SDs of single channel conductance values of TcA in different lipids in black lipid bilayer experiments (SI Appendix, Fig. S10).
To understand whether TcA indeed forms a pore in the different types of lipids used, we reconstituted TcA into black lipid bilayers composed of different lipids and measured the conductance of the channel. TcA readily integrated in all lipid bilayers; however, in contrast to the previously measured conductance of 500 to 600 pS for the open pore in diphytanoyl-phosphatidylcholine (13), the mean conductance values were between 397 and 462 pS in all conditions tested, even with bilayers formed exclusively by lipids with negatively charged headgroups (diphytanoyl-phosphatidylglycerol) (Fig. 7C and SI Appendix, Fig. S10). In addition, in bilayers consisting of phosphatidylcholine only or BPL, there is a considerable fraction of larger pores with a conductance of up to 600 pS (SI Appendix, Fig. S10 B, C, and H). While this indicates that TcA can indeed form pores in different lipid environments, it also demonstrates that the lipids have an influence on the TcA pore diameter. This is in line with our previous MD simulations that showed a strong interaction of lipids with the transmembrane region of TcA. Lipid head groups were even intercalating between TcA protomers, stabilizing the open conformation of the channel (14). Hence, different lipids likely have a direct influence on this part of the proteins and thus on the pore diameter.
An Alternative Sequence of Holotoxin Assembly.
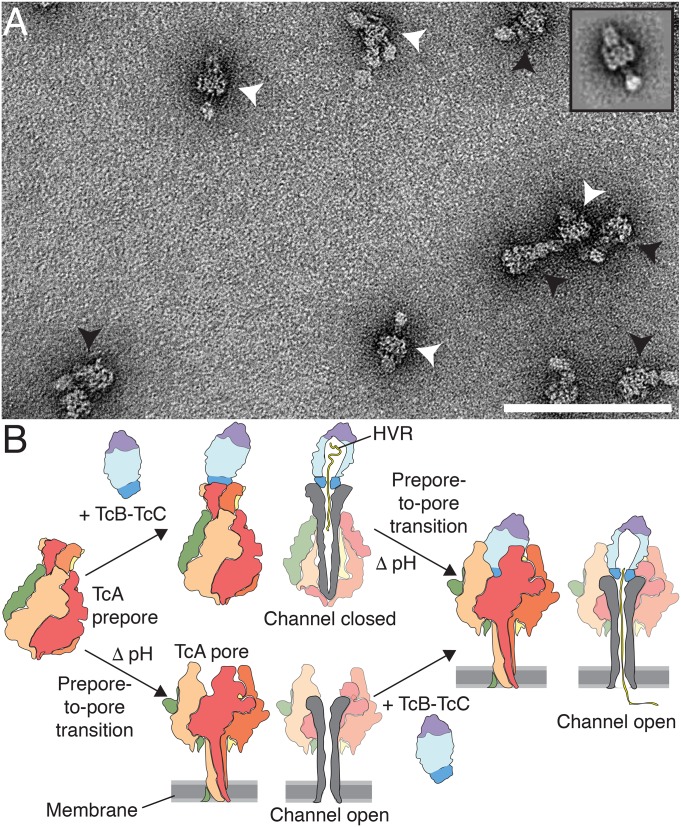
Since we have learned from the holotoxin pore structure that the TcB-TcC cocoon can easily be accommodated inside the basin formed by the shell domains of TcA, we asked ourselves whether TcB-TcC can also bind to TcA after it is already inserted into the membrane. To explore this possibility, we first formed TcA pores in nanodiscs and then added TcB-TcC. We indeed obtained holotoxin pores, clearly demonstrating that the complex can also be assembled in this order (Fig. 8A).
Fig. 8.
Model of 2 pathways of formation of the holotoxin pore. (A) Negative-stain electron micrograph of Tc holotoxin in nanodiscs formed by mixing TcA after the prepore-to-pore transition at pH 11 with TcB-TcC (80 and 500 nM, respectively), followed by size exclusion chromatography. Holotoxin prepores and pores are indicated with black and white arrowheads, respectively. (Inset) 2D class average of an ABC pore. (Scale bar: 100 nm.) (B) Cartoon representation of Tc holotoxin assembly, target cell interaction, and pore formation. The first pathway shows holotoxin formation with subnanomolar affinity from pentameric TcA and the TcB-TcC fusion protein (17), followed by membrane association and pH-induced prepore-to-pore transition. The second pathway shows membrane association and prepore-to-pore transition of TcA, followed by holotoxin formation of the TcA pore and TcB-TcC. Subsequently, the toxic enzyme is translocated to the target cells with its C terminus first.
Where and when Tc holotoxins assemble remains a matter of debate. The subnanomolar affinity of TcA and TcB-TcC (17) suggests that the components are likely secreted as a fully assembled holotoxin. This is also supported by the finding that TcC is needed for the proper secretion of TcA and TcB from P. luminescens (27). However, arguing against this secretion as a fully assembled complex is the relatively large size of the holotoxin (1.7 MDa). Our results demonstrate that alternative routes of holotoxin complex formation are possible and thus holotoxins could still be formed after TcA has penetrated the target cell membrane (Fig. 8B).
Conclusion
The pore structure of the Tc holotoxin complex from P. luminescens at 3.4-Å resolution presented in this study allowed us to clarify mechanistic details of the prepore-to-pore transition of the assembled complex and toxin translocation. During pore formation of ABC, the TcB-TcC cocoon is pulled into a basin formed by the outer shell of the TcA protomers, which adapt slightly to fit the cocoon. This adjustment causes a break in the symmetry of TcA, but no structural change at the interface between TcB and TcA (Movie S6). The structural flexibility of the upper part of the outer shell of TcA allows assembly of the ABC holotoxin after TcA has already entered the membrane (Fig. 8).
In cleavage-deficient ABC(D651A), the ADP-ribosyltransferase is still present inside the holotoxin, indicating that its full release from the cocoon requires autoproteolytic cleavage. Its major part remains unstructured in the TcB-TcC cocoon, as in our previous structures of the holotoxin in its prepore state (15, 17). However, the improved resolution of the present structure allowed us to model 2 regions of the ADP-ribosyltransferase for the first time. The first region is the N terminus, which follows the aspartyl autoprotease site. It forms a loop that keeps the cleavage site between the catalytic aspartates. The second part is a 13-residue-long stretch close to the C terminus that resides in the narrow passage of TcB below the constriction site. The C-terminal stretch is in an unfolded conformation and interacts with alternating charged and hydrophobic regions of the channel. A similar pattern has been described for chaperones (28) and the translocation pore of anthrax protective antigen (PA) (29) (SI Appendix, Fig. S11). There, the so-called ϕ-clamp forms a narrow (∼6 Å) hydrophobic constriction site (29). The details of how the substrates interact with the ϕ-clamp during translocation are not known. It has been proposed that the ϕ-clamp seals the translocation pore around the translocated polypeptide and helps to protect hydrophobic patches in the translocated protein (29, 30). The same is probably true for the Tc complex; however, a seal is not needed in this case, since in contrast to PA, the Tc translocation channel is closed by the TcB-TcC cocoon on one side.
Our results also show that translocation occurs spontaneously in vitro. It does not necessarily require driving forces such as pH gradients or chaperones, although folding of the toxin inside the host cell cytoplasm in vivo is likely supported by the chaperones of the host cell (31, 32), and translocation might be accelerated by a pH gradient. In anthrax toxin, the pH gradient over the endosomal membrane drives and accelerates the translocation of proteins with net negative charges (33); however, the translocated toxic enzymes of Tc toxins are positively charged, making a comparable enhancement of translocation speed unlikely. Based on our previous results (14), we have speculated that the direct entanglement with lipid head groups at the conserved tip of the TcA channel inside the membrane is partly responsible for the host specificity of Tc toxins; however, the results presented here demonstrate that this is likely not the case. While the type of lipid has an influence on the diameter of the TcA pore, it does not interfere with pore formation in general.
Materials and Methods
More details are provided in SI Appendix, Materials and Methods.
Protein Production and Nanodisc Integration.
P. luminescens TcdA1 (TcA), TcdB2-TccC3 (TcB-TcC), and the nanodisc scaffold protein MSP1D1 were expressed and purified as described previously (13, 14, 34). After holotoxin formation of TcdA1 and TcdB2-TccC3(WT) or TcdB2-TccC3(D651A), the complexes were integrated into MSP5ΔH5-POPC nanodiscs [ABD(D651A)] or into MSP1D1-POPC nanodiscs [ABC(WT)] in 20 mM CAPS-NaOH pH 11.2 and 250 mM NaCl for 2 d at 4 °C. Subsequently, ABC in nanodiscs was purified via SEC on a Superose 6 5/150 column equilibrated in 20 mM Tris⋅HCl pH 8.0 and 250 mM NaCl (SI Appendix, Fig. S1 B and C) and directly used for cryo-EM.
Single-Channel Conductance Measurements.
Single-channel conductance of TcA was measured with different lipid compositions as described previously (13). Solutions of different lipids and lipid mixes (SI Appendix, Fig. S10) at a total lipid concentration of 10 mg/mL were prepared in 80% n-decane/20% butanol. All measurements were performed with 20 pM TcA and a membrane potential of 20 mV in 20 mM CAPS-NaOH pH 11 and 1 M KCl.
Sample Vitrification and Cryo-EM Data Acquisition.
ABC(D651A) and ABC(WT) were applied to glow-discharged holey carbon grids (Quantifoil; QF 2/1, 300 mesh) covered with a 2-nm carbon layer at 0.12 mg/mL and 0.10 mg/mL, respectively, and vitrified in liquid ethane using a CP3 [ABC(D651A)] or a Vitrobot [ABC(WT)]. Datasets were collected with a Cs-corrected Titan Krios EM using a Falcon III direct electron detector operated in linear mode at a pixel size of 1.11 Å/pixel [ABC(D651A)] or a K2 direct electron detector at a pixel size of 1.05 Å/pixel [ABC (WT)].
Image Processing.
Images were inspected and drift-corrected using MotionCor2 (35). CTF parameters were estimated using CTER (36), implemented in SPHIRE (18). Particles were autopicked using Gautomatch for ABC(D651A) (37) or crYOLO (38) for ABC(WT). 2D classification was performed with ISAC (39) in SPHIRE, and particles showing a holotoxin pore were kept for 3D refinement. 3D refinement without symmetry was performed with MERIDIEN in SPHIRE (18), using the density of the TcdA1 pore (Electron Microscopy Data Bank ID code EMD-4068) docked with TcB-TcC (PDB ID code 4O9X) as the initial model. Subsequently, 3D classification using SORT3D in SPHIRE resulted in classes with different orientations of the asymmetric TcB-TcC cocoon. One 3D class with well-defined density of TcB-TcC was filtered to 5 Å and used as the initial template in a new 3D refinement with 1.875° initial angular sampling, resulting in a clearly defined density and a distinct orientation of TcB-TcC. The final density maps were postprocessed using a soft Gaussian mask and filtered according to its local resolutions. To achieve a better local resolution in the TcB-TcC part of the map of ABC(D651A), we shifted the center of the map to the TcB-TcC cocoon and re-extracted the particles (SI Appendix, Fig. S3 A–C), and we ran 3D refinement in MERIDIEN with the reboxed particles. 3D variability estimation was performed with sx3dvariability in SPHIRE with 25-Å low-pass–filtered images.
Atomic Model Building and Refinement.
Models of the TcdA1 pore (PDB ID codes 5LKH and 5LKI) and TcB-TcC (PDB ID code 6H6F) were fitted into the density map of ABC(651A) using Imodfit (40). Subsequently, we modeled the atomic structure using a combination of manual model building in Coot (41), Rosetta relaxation, and Phenix real-space refinement (42). Residues 1,382 to 1,491 of all TcA chains were removed, as the map showed insufficient density and resolution in that region. The model of TcB-TcC was further improved using the shifted map (SI Appendix, Fig. S6 D and E). We manually built a Poly-Ala peptide in C-N translocation direction into the density in the TcB channel, added side chains at regions with sufficient density of possible peptide stretches close to the C terminus of the ADP-ribosyltransferase, and chose the best-fitting model (SI Appendix, Fig. S7B). The final models were validated by Rosetta relaxation against 2 independent half-maps (43), EMringer (44), and MolProbity (45) (SI Appendix, Table S1).
XL-MS.
XL-MS was performed with 3 μM TcB-TcC (WT or D651A) in 20 mM Hepes-NaOH pH 7.5 and 150 mM NaCl using 3 mM disuccinimidyl dibutyric urea (Thermo Fisher Scientific) for 30 min at 25 °C. The cross-linking reaction was stopped by adding 100 mM Tris. Digestion of the samples, peptide separation, and LC-MS were performed as described previously (46). Cross-links were evaluated using MeroX (47) and visualized in xVis (48).
Supplementary Material
Acknowledgments
We thank A. Elsner and K. Vogel-Bachmayr for technical support, D. Pan and F. Müller for help preparing the cross-linked TcB-TcC complexes, and A. Brockmeyer and M. Metz for performing mass spectrometry and annotating the cross-linked peptides. This work was supported by funds from the Max Planck Society (to S.R.) and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013; Grant 615984, to S.R.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM densities of ABC(D651A) and ABC(WT) pore states have been deposited in the Electron Microscopy Data Bank (accession nos. 10312 and 10313, respectively). Coordinates of ABC(D651A) and ABC(WT) pore states have been deposited in the Protein Data Bank (ID codes 6SUE and 6SUF, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909821116/-/DCSupplemental.
References
- 1.Waterfield N. R., Bowen D. J., Fetherston J. D., Perry R. D., ffrench-Constant R. H., The tc genes of Photorhabdus: A growing family. Trends Microbiol. 9, 185–191 (2001). [DOI] [PubMed] [Google Scholar]
- 2.ffrench-Constant R., Waterfield N., An ABC Guide to the Bacterial Toxin Complexes (Elsevier, 2005). [DOI] [PubMed] [Google Scholar]
- 3.Bowen D., et al. , Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280, 2129–2132 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Sergeant M., Jarrett P., Ousley M., Morgan J. A. W., Interactions of insecticidal toxin gene products from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 69, 3344–3349 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ffrench-Constant R. H., Bowen D. J., Novel insecticidal toxins from nematode-symbiotic bacteria. Cell. Mol. Life Sci. 57, 828–833 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerrard J., Waterfield N., Vohra R., ffrench-Constant R., Human infection with Photorhabdus asymbiotica: An emerging bacterial pathogen. Microbes Infect. 6, 229–237 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Tennant S. M., Skinner N. A., Joe A., Robins-Browne R. M., Homologues of insecticidal toxin complex genes in Yersinia enterocolitica biotype 1A and their contribution to virulence. Infect. Immun. 73, 6860–6867 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterfield N., Hares M., Hinchliffe S., Wren B., ffrench-Constant R., The insect toxin complex of Yersinia. Adv. Exp. Med. Biol. 603, 247–257 (2007). [DOI] [PubMed] [Google Scholar]
- 9.ffrench-Constant R. H., et al. , A genomic sample sequence of the entomopathogenic bacterium Photorhabdus luminescens W14: Potential implications for virulence. Appl. Environ. Microbiol. 66, 3310–3329 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duchaud E., et al. , The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21, 1307–1313 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Lang A. E., et al. , Photorhabdus luminescens toxins ADP-ribosylate actin and RhoA to force actin clustering. Science 327, 1139–1142 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Tabashnik B. E., Brévault T., Carrière Y., Insect resistance to Bt crops: Lessons from the first billion acres. Nat. Biotechnol. 31, 510–521 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Gatsogiannis C., et al. , A syringe-like injection mechanism in Photorhabdus luminescens toxins. Nature 495, 520–523 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Gatsogiannis C., et al. , Membrane insertion of a Tc toxin in near-atomic detail. Nat. Struct. Mol. Biol. 23, 884–890 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Meusch D., et al. , Mechanism of Tc toxin action revealed in molecular detail. Nature 508, 61–65 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Busby J. N., Panjikar S., Landsberg M. J., Hurst M. R. H., Lott J. S., The BC component of ABC toxins is an RHS repeat-containing protein encapsulation device. Nature 501, 547–550 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Gatsogiannis C., et al. , Tc toxin activation requires unfolding and refolding of a β-propeller. Nature 563, 209–213 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Moriya T., et al. , High-resolution single particle analysis from electron cryo-microscopy images using SPHIRE. J. Vis. Exp. 2017, e55448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roderer D., Raunser S., Structure of Photorhabdus luminescens Tc holotoxin pore, Mutation TccC3-D651A. Protein Data Bank. https://www.rcsb.org/structure/6SUE. Deposited 13 September 2019. [Google Scholar]
- 20.Roderer D., Raunser S., Structure of Photorhabdus luminescens Tc holotoxin pore. Protein Data Base. https://www.rcsb.org/structure/6SUF. Deposited 13 September 2019. [Google Scholar]
- 21.Li J., et al. , Structural basis for teneurin function in circuit-wiring: A toxin motif at the synapse. Cell 173, 735–748.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson V. A., et al. , Structures of teneurin adhesion receptors reveal an ancient fold for cell-cell interaction. Nat. Commun. 9, 1079 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Athaudaa S. B. P., Takahashia K., Cleavage specificities of aspartic proteinases toward oxidized insulin B chain at different pH values. Protein Pept. Lett. 9, 289–294 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Gates S. N., et al. , Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 357, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puchades C., et al. , Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358, eaao0464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawaliby R., et al. , Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 291, 3658–3667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang G., Waterfield N. R., The role of TcdB and TccC subunits in secretion of the Photorhabdus Tcd toxin complex. PLoS Pathog. 9, e1003644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morán Luengo T., Kityk R., Mayer M. P., Rüdiger S. G. D., Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell 70, 545–552.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Jiang J., Pentelute B. L., Collier R. J., Zhou Z. H., Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature 521, 545–549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krantz B. A., et al. , A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science 309, 777–781 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang A. E., et al. , The chaperone Hsp90 and PPIases of the cyclophilin and FKBP families facilitate membrane translocation of Photorhabdus luminescens ADP-ribosyltransferases. Cell. Microbiol. 16, 490–503 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Ernst K., et al. , Hsp70 facilitates trans-membrane transport of bacterial ADP-ribosylating toxins into the cytosol of mammalian cells. Sci. Rep. 7, 2724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown M. J., Thoren K. L., Krantz B. A., Charge requirements for proton gradient-driven translocation of anthrax toxin. J. Biol. Chem. 286, 23189–23199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denisov I. G., Grinkova Y. V., Lazarides A. A., Sligar S. G., Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penczek P. A., et al. , CTER-rapid estimation of CTF parameters with error assessment. Ultramicroscopy 140, 9–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K., Li M, Sun F., Gautomatch: An efficient and convenient gpu-based automatic particle selection program (2011). https://www.mrc-lmb.cam.ac.uk/kzhang/. Accessed 21 June 2017.
- 38.Wagner T., et al. , SPHIRE-crYOLO: A fast and well-centering automated particle picker for cryo-EM. bioRxiv:10.1101/356584 (26 June 2018).
- 39.Yang Z., Fang J., Chittuluru J., Asturias F. J., Penczek P. A., Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopéz-Blanco J. R., Chacón P., iMODFIT: Efficient and robust flexible fitting based on vibrational analysis in internal coordinates. J. Struct. Biol. 184, 261–270 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonine P. V., et al. , Real-space refinement in Phenix for cryo-EM and crystallography. bioRxiv:10.1101/249607 (17 January 2018). [DOI] [PMC free article] [PubMed]
- 43.DiMaio F., et al. , Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods 12, 361–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barad B. A., et al. , EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Efremov R. G., Leitner A., Aebersold R., Raunser S., Architecture and conformational switch mechanism of the ryanodine receptor. Nature 517, 39–43 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Götze M., et al. , Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J. Am. Soc. Mass Spectrom. 26, 83–97 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Grimm M., Zimniak T., Kahraman A., Herzog F., xVis: A web server for the schematic visualization and interpretation of crosslink-derived spatial restraints. Nucleic Acids Res. 43, W362–W369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masood T. B., Sandhya S., Chandra N., Natarajan V., CHEXVIS: A tool for molecular channel extraction and visualization. BMC Bioinformatics 16, 119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hessa T., et al. , Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.