Significance
In nature, plants and microbes process substantial amounts of aromatic carbon for lignin biosynthesis and breakdown, respectively. These natural processes have important implications in the pursuit of lignin valorization, which is crucial for a vibrant, global bioeconomy. In both plant and microbial systems, an open question remains regarding how lignin-related aromatic compounds are transported across compartmental membranes, either by active membrane transporters or via passive membrane crossing. In this study, we predict that passive transport processes in plants and bacteria for uncharged aromatic compounds are likely sufficient for lignin biosynthesis and catabolism, thus implying that membrane translocation rates are controlled by compound delivery and utilization rates and membrane concentration gradients.
Keywords: molecular dynamics, lignin permeability, lignin biosynthesis, biological funneling, free energy calculation
Abstract
Lignin is an abundant aromatic polymer found in plant secondary cell walls. In recent years, lignin has attracted renewed interest as a feedstock for bio-based chemicals via catalytic and biological approaches and has emerged as a target for genetic engineering to improve lignocellulose digestibility by altering its composition. In lignin biosynthesis and microbial conversion, small phenolic lignin precursors or degradation products cross membrane bilayers through an unidentified translocation mechanism prior to incorporation into lignin polymers (synthesis) or catabolism (bioconversion), with both passive and transporter-assisted mechanisms postulated. To test the passive permeation potential of these phenolics, we performed molecular dynamics simulations for 69 monomeric and dimeric lignin-related phenolics with 3 model membranes to determine the membrane partitioning and permeability coefficients for each compound. The results support an accessible passive permeation mechanism for most compounds, including monolignols, dimeric phenolics, and the flavonoid, tricin. Computed lignin partition coefficients are consistent with concentration enrichment near lipid carbonyl groups, and permeability coefficients are sufficient to keep pace with cellular metabolism. Interactions between methoxy and hydroxy groups are found to reduce membrane partitioning and improve permeability. Only carboxylate-modified or glycosylated lignin phenolics are predicted to require transporters for membrane translocation. Overall, the results suggest that most lignin-related compounds can passively traverse plant and microbial membranes on timescales commensurate with required biological activities, with any potential transport regulation mechanism in lignin synthesis, catabolism, or bioconversion requiring compound functionalization.
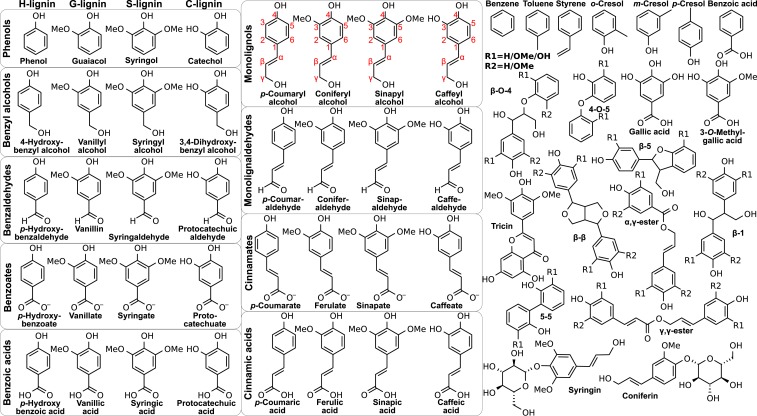
Lignin is an abundant aromatic polymer found in terrestrial plant secondary cell walls, where it is responsible, in part, for bundling together cell wall components and strengthening plant tissues. Lignin is known to enhance biomass recalcitrance, which can be mitigated via alterations in lignin biosynthesis or pretreatments (1). In plants, lignin biosynthesis begins as a branch of the shikimate pathway to create monolignols (Fig. 1) (1) and ends with their radical polymerization within the plant cell wall (2). Shikimate/monolignol pathway enzymes are localized within plant cells (3), whereas the peroxidases and laccases that form the radicals needed to polymerize lignin are found in the apoplastic space (4). Thus, monolignols must cross the cell membrane before incorporation into the growing lignin polymer.
Fig. 1.
LRCs examined, grouped by their chemistry. On the left are monomeric (single aromatic ring) compounds including monolignols and derivatives encountered via lignin deconstruction processes (H, G, S, and C substitutions from left to right). Other compounds enriched in fragmented lignin streams are also considered, including dimeric LRCs with monomer-dependent R1 and R2 groups, a representative flavonoid (tricin), glucosides (coniferin and syringin), and other aromatics (e.g., cresols). LRCs that can be derived from high-severity processes, such as benzene, are also considered. The red monolignol labeling defines lignin carbon nomenclature.
Lignin also represents an attractive feedstock for industrial processes to create renewable chemicals (5, 6). Extracellular lignin depolymerization processes result in heterogeneous mixtures, which create a downstream processing challenge to manufacture pure chemicals. Harnessing natural intracellular aromatic–catabolic pathways that convert lignin-related compounds (LRCs) to single intermediates (7) has emerged as a potential strategy to overcome this issue (8). LRCs thus also traverse cell membranes in microbial catabolic processes.
Understanding the movement of LRCs across cell membranes is therefore essential for lignin engineering in planta and for optimizing microbial lignin utilization. LRC transport across lipid membranes has been hypothesized to occur via membrane transporters or passive diffusion (9, 10). Membrane transporters have been identified for several charged LRCs (11–19), similar to other ATP-dependent transporters that compartmentalize lignin precursors within plant cells (20–22). However, membrane partitioning studies suggest that small uncharged LRCs may simply passively diffuse across biological membranes (23, 24). Specific LRC chemistry may dictate the predominant membrane transport mechanism for individual LRCs.
In this study, we specifically evaluate the passive diffusion hypothesis within a comprehensive set of LRC chemistries (Fig. 1) to determine passive permeability coefficients across 3 membrane bilayers using molecular dynamics simulation for comparison with potential transporter fluxes. The modeled lipid compositions are a homogenous eukaryotic membrane (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine [POPC]) for comparison with previous work (25), plant membranes (roots from Zea mays (26)) to model lignin biosynthesis, and bacterial membranes (from Pseudomonas putida (27)) to examine LRC uptake at the bacterial inner membrane. The lignin chemistries studied include the p-coumaryl, coniferyl, and sinapyl alcohol lignin monomers, that give rise to polymeric p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units (2), as well as the caffeyl alcohol precursors to C lignin units found in selected plant tissues (28). The substantial LRC chemical diversity naturally present in plant cells is also considered (Fig. 1, Left). Other LRCs in Fig. 1 include central intermediates in microbial lignin catabolism (catechol, protocatechuic acid, and gallic acid) (7), dimeric LRCs representative of lignin linkage chemistry (including carbohydrate conjugates, e.g., coniferin), a representative lignin-associated flavonoid (tricin (29)), and lignin-derived pyrolysis products considered for biological upgrading (30).
The replica-exchange umbrella sampling (REUS) simulations determined the membrane partitioning coefficient (P, measuring the compound distribution between membrane and solution) and membrane permeability coefficient (Pm, related to how quickly a compound crosses the membrane passively) for all 69 LRCs represented in Fig. 1 across the 3 different membrane compositions. LRCs are typically predicted to be enriched by - to -fold at the membrane interface independent of composition. These LRCs, including dimers but excluding charged or glycosylated species, are predicted to be readily permeable across biological membranes, obviating the requirement for membrane transporters in lignin biosynthesis or microbial conversion. Only lignin monomer glycosylation or carboxylation significantly reduces LRC permeability, slowing passive diffusion and permitting membrane permeation of these compounds to be regulated by transporters.
Results
Free Energy Profiles.
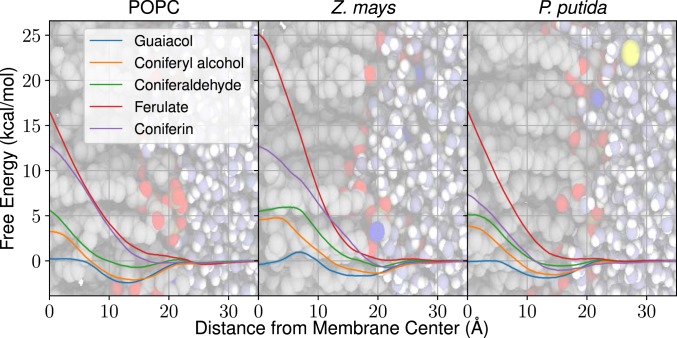
The inhomogeneous solubility diffusion model (31, 32) computes permeabilities through free energy and diffusivity profiles over the chosen reaction coordinate (SI Appendix, Eq. S1), in this case the LRC center of mass relative to the membrane midplane. The profiles in Fig. 2 highlight the range of energetic barriers possible with different LRCs, with all profiles provided in SI Appendix, Figs. S1–S17. An important consideration when comparing profiles across membranes is that the modeled Z. mays membrane is Å thicker than either the POPC or P. putida membranes (SI Appendix, Fig. S18).
Fig. 2.
Free energy profiles for diverse G-type LRCs in all 3 membrane compositions (POPC [Left], Z. mays [Middle], and P. putida [Right]), using the molecular center of mass distance from the membrane center as the reaction coordinate. Solid lines indicate the position-dependent free energy. Free energy uncertainties (shaded regions, typically on the order of 0.1 kcal mol−1) are determined from a population of 100 Gibbs samples drawn from the complete dataset. The profiles are superimposed on a simulation snapshot showing membrane lipid heavy atoms (gray, carbons; red, oxygens; blue, nitrogens; and brown, phosphorus atoms), water (white, hydrogens, and light blue, oxygens), and ions (yellow). SI Appendix, Figs. S1–S17 include profiles for all studied LRCs, including diffusivity profiles.
The bilayer-crossing free energy profiles are similar to other profiles for small hydrophobic molecules (25, 32). For most LRCs and membrane compositions, there is a small membrane entry barrier from solution. After crossing this barrier, the compounds arrive at their preferred penetration depth, typically near the lipid carbonyls or slightly toward the membrane midplane, depending on the functional groups present. There is usually a peak for the free energy profile at the center of the bilayer, representing the energetic cost of burying LRC hydrophilic moieties within the bilayer center, similar to prior simulations (25) (SI Appendix, Fig. S19). Notably, the tested glucosides and carboxylates have larger membrane crossing peaks than the other compounds, a significant barrier to exchange between membrane leaflets (Fig. 2).
Lignin Interaction and Orientation.
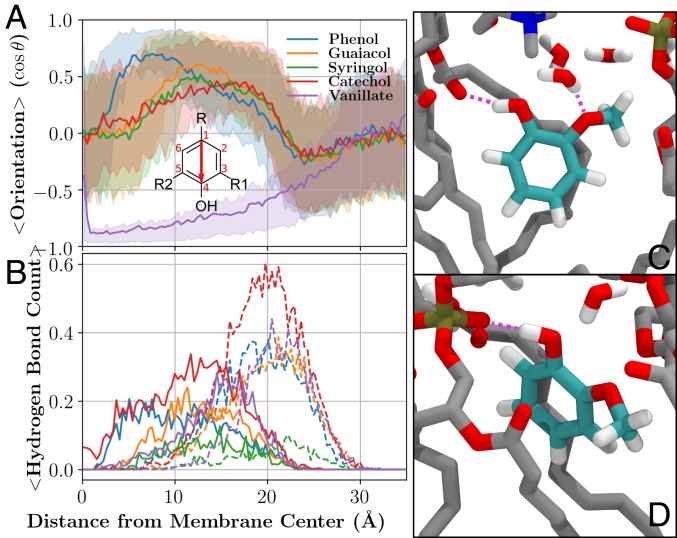
The free energy minimum at the lipid carbonyls is dictated by hydrogen bonding, as lipid carbonyls are exclusively hydrogen bond acceptors. Hydroxyl groups on in lignin monomers are observed to orient the aromatic ring within the membrane (7 to 20 Å) such that the hydroxyl can donate a proton to the lipid carbonyl until the molecule is effectively located at the membrane center ( Å; Fig. 3A and SI Appendix, Figs. S20–S36). The orientation preference is diminished in the headgroup region (20 to 25 Å); lipid headgroup oxygens provide competing hydrogen bonding sites (Fig. 3 B–D). Near the lipid carbonyls, LRC hydroxyls interact with lipid carbonyls, phosphates, and associated water molecules (Fig. 3 C and D), creating an optimal environment for LRCs. As isolated aromatic rings embed into the hydrophobic membrane environment without an orientational preference (SI Appendix, Fig. S29), the combination of aromatic embedding and hydroxyl interaction with the membrane interface proscribe the free energy profile shape and the preferred membrane insertion depth.
Fig. 3.
(A) Molecular orientation and (B) compound–lipid hydrogen bonding for phenols and vanillate from the REUS calculations in POPC, along with representative snapshots of (C) guaiacol interacting with lipid carbonyls and (D) lipid headgroup phosphates. (A) The mean molecular orientation at a specific distance from the membrane center is computed based on the vector from carbon 1 to carbon 4 (red arrow) and the cosine of the angle between that vector and the membrane normal. The R, R1, and R2 groups vary as shown in Fig. 1. The middle 50% of the distribution is shown using semitransparent shading. (B) Hydrogen bonds between LRCs and lipids depending on penetration depth. Solid lines indicate hydrogen bonds to lipid carbonyl groups, whereas dashed lines quantify hydrogen bonds to lipid headgroups. A hydrogen bond was defined by heavy atoms separated by less than 3.2 Å and a colinear hydrogen within . Comparable figures for other compound classes are presented in SI Appendix, Figs. S20–S36. (C) Small LRCs such as guaiacol (cyan colored carbons) can act as donors in hydrogen bonding interactions (purple dashed lines) with lipid carbonyl oxygens. Methoxy groups can similarly act as acceptors in interactions with water. (D) Depending on the position relative to the membrane center, the hydroxyl group can also act as a donor to phosphate groups in the lipid head. Both C and D use the same color scheme, with nonguaiacol carbons shown in gray, oxygens in red, hydrogens in white, nitrogens in blue, and phosphorus in brown.
Methoxy groups present in LRCs modulate these interactions through occluding and orienting adjacent hydroxyls. If the methoxyl is present, the hydroxyl may hydrogen bond to the adjacent methoxy oxygen, systematically reducing hydroxyl hydrogen bonding with the surrounding lipid carbonyls (Fig. 3B and SI Appendix, Figs. S20–S36), and has a weaker orientational preference. Therefore, G- and S-lignin LRCs systematically have shallower free energy wells, reducing the partition coefficient. Conversely, the additional hydroxyl in C-lignin LRCs improves partitioning through the formation of additional hydrogen bonds at the membrane interface.
Partitioning and Permeability.
P and Pm coefficients for all of the compounds and membrane compositions considered are reported in SI Appendix, Tables S1–S3, with coefficients for key molecules reported in Table 1. For selected rapidly permeable compounds, these permeabilities are compared with explicit transitions seen in equilibrium simulations used for cross-validation (SI Appendix, Table S4 and Fig. S37), and compare favorably with what REUS and the inhomogeneous solubility diffusion equation (31) compute. Most compounds have a positive logP, indicating that they are enriched within the membrane relative to solution. Generally, uncharged monomeric LRCs are 10- to 100-fold enriched, with methoxy groups reducing partition coefficients and hydroxyls increasing them (S G H C). Dimeric LRCs are typically further enriched, with membrane concentrations up to 25,000-fold higher than those in solution, similar to the representative flavonoid, tricin (Table 1 and SI Appendix, Fig. S13). Among carboxylates and glycosylated compounds, partitioning is significantly weaker, with limited concentration enhancement between membrane and solution. Benzene derivatives and cresols lack hydroxyls to aid solvation and are therefore seen to partition more strongly to the membrane than other LRCs. The computed partition coefficients generally follow the trends for octanol-water partitioning of the same compounds (SI Appendix, Fig. S38).
Table 1.
Selected membrane partition (P) and permeability (Pm) coefficients for LRCs of particular importance to lignin biosynthesis or utilization
| Membrane | Compound | ||
| Z. mays | p-Coumaryl alcohol | 1.4 | -1.4 |
| Coniferyl alcohol | 1.0 | -1.5 | |
| Sinapyl alcohol | 0.7 | -2.1 | |
| Caffeyl alcohol | 1.6 | -1.6 | |
| Tricin | 4.4 | -0.7 | |
| Coniferin | 0.5 | -6.8 | |
| p-Coumarate | 0.1 | -17.5 | |
| Ferulate | -0.1 | -15.3 | |
| P. putida | Phenol | 2.2 | 1.1 |
| Guaiacol | 1.3 | 1.2 | |
| Syringol | 1.3 | 1.0 | |
| Vanillyl alcohol | 0.4 | -1.3 | |
| Vanillin | 0.4 | -0.4 | |
| Vanillate | -0.3 | -10.2 | |
| p-Coumarate | 0.1 | -9.8 | |
| Ferulate | -0.1 | -9.0 | |
| G--O-4-G | 1.4 | -0.9 | |
| G--1-G | 0.7 | -4.0 | |
| G---G | 2.1 | 0.8 |
P and Pm coefficients for all membranes and compounds tested are reported in SI Appendix, Tables S1–S3.
The computed permeabilities varied by nearly 20 orders of magnitude, spread between the most permeable compounds (nonpolar aromatics) and the least (carboxylates in Z. mays bilayers). Most compounds tested were on the higher end of this permeability spectrum, with carboxylates and glycosylated LRCs being among the least permeable compounds. Breaking down the permeabilities into a interleaflet transition/membrane crossing step () and an extraction step () reveals that crossing rather than extraction is typically the rate-limiting step to membrane permeation (SI Appendix, Tables S1–S3), in contrast to earlier findings for fatty acyl products or terpenoids (33, 34). Only for highly processed aromatic compounds, such as benzenes or cresols, or specific LRC dimers with few hydroxy groups to retard membrane crossing (5-5 and 4-O-5 linkages), is . The exhibits the reverse pattern as logP, with C H G S due to carbonyl hydrogen bonds breaking during membrane transit.
Comparing P and Pm values across the membrane compositions tested highlights the impact sterols have on partitioning and permeability. Both the POPC and P. putida membranes were modeled without sterols, leading to greater membrane disorder and increased permeability. Z. mays bilayers, in contrast, exhibit both reduced partitioning and permeability previously associated with high sterol concentration (35) because sterols contribute hydrogen bonds to lipid carbonyls and compete for access to the same interaction sites.
Discussion
The computed permeability coefficients determine which specific compounds require transporters or channels to cross the membrane and which compounds instead may rely on passive diffusion through the membrane. Prior studies have characterized transporters for benzoates (11–13, 15), hydroxycinnamates (14–18), gallates (15, 19), and glycosides (20–22) considered in this work. Evidence for aldehyde and alcohol LRC transporters, however, has remained elusive (21). There may be a mechanistic bifurcation, with passive diffusion used for highly permeable molecules and transporters for the remaining impermeable compounds (SI Appendix, Fig. S39). The remaining discussion focuses on where the boundary between these mechanisms lies and its implications across biology.
Comparing Passive Permeability and Facilitated Transport.
Evaluating the transmembrane flux expressions provides a comparison between the effectiveness of passive diffusion and active membrane transport.
| [1] |
The flux across the membrane through passive diffusion () for a molecule is related to compound permeability (Pm), the surface area of the membrane (A), and the compound concentration difference between membrane compartments (). Similarly, transporter-mediated flux () is related to the rate of turnover or permeation events () and the number of transporters (N). Depending on Pm ( for interleaflet transport), , and some assumptions about transporter effectiveness, a relationship emerges to determine if active or passive fluxes from Eq. 1 are larger (SI Appendix, Fig. S39).
As an example relevant to bacterial lignin conversion, we consider a bacterium provided an LRC in a 1 to 2 mM concentration, as is done routinely in experimental cultures (36, 37), but below target product titers of 100+ mM (38). Assuming a 6 membrane surface area for the bacterial cell, molecules (Eq. 1) if the gradient can be maintained via enzymatic activity. For deprotonated carboxylates (; Table 1), only a single molecule per second will enter the cell via a passive route. Analogous ATP-driven pumps to those that import carboxylates may turn over as often as 10 to 20 (39, 40), implying that a single transporter outpaces passive diffusion and that transporters would accelerate membrane transport for compounds with similar permeabilities.
For other species such as guaiacol or coniferyl alcohol with substantially higher permeability (Table 1), diffusive flux alone can provide somewhere between and molecules (1 to 100 femtomoles) per second, likely saturating available metabolic enzymes with turnover rates of 1 to 10 (41), or nM-affinity export proteins at typical copy numbers (14). The mass of the ATP-driven pumps needed to match that permeation rate ( pg) exceeds total bacterial cell mass ( pg (42)). To avoid overburdening the cell membrane with only a single transporter, we estimate that active transport only contributes significantly to permeation once cm , with detailed discussion surrounding this cutoff choice provided in SI Appendix. Faster transporters, such as the barrel protein TodX found in the outer membrane of P. putida (43) or the hypothesized vanillin symporter (44), could potentially change this cutoff. Regardless, for uncharged LRCs, passive diffusion is likely sufficient to support rapid enzyme turnover in downstream conversion processes.
Toxicity Tolerance Implications.
Organisms maintain an array of polyspecific efflux transporters to export toxic compounds. If a compound is highly permeable, transporters are required to maintain a concentration gradient across the membrane, as can be seen by rearranging Eq. 1 to determine the concentration gradient that can be maintained against passive diffusion:
| [2] |
Based on Eq. 2, a cell will have significant difficulty maintaining a gradient for permeable species. Given the same assumptions as above (log Pm , 6 membrane area, and 10 transporter exchange), a single copy of a transporter could only maintain a pM-scale gradient between bilayer leaflets. As membrane partitioning is expected for these lipophilic LRCs, amplifying the LRC concentration within the membrane leaflets above the levels typically measured in solution, transporters acting alone likely would be insufficient to cope with high concentrations of toxic compounds.
Given bacterial studies showing tolerance to highly permeable LRCs such as phenol (45) and vanillin (46) at mM concentrations, alternative tolerance factors would be required. In these bacterial systems, lipopolysaccharide layers in the outer membrane may be one tolerance factor, with the hydrophilic polysaccharides not present in the inner membrane models considered here impeding the aromatic permeation without the appropriate porin (47). Due to the considerable thickness of the outer membrane and the associated computational cost, probing the impact of the glycosides on outer membrane permeability will be the subject of future studies. Other hypotheses for how bacteria tolerate permeable LRCs include detoxifying these compounds via fast metabolic enzymes or stress responses that change membrane composition or structure to modulate permeability. There may also be systematic errors in our computed permeabilities, regardless of their agreement with previous estimates (SI Appendix, Fig. S19) or alternative calculation methods (SI Appendix, Table S4). The tolerance for highly permeable LRCs notwithstanding, minimally permeable compounds would be easily exported, maximizing product titers without triggering cell toxicity and simplifies product collection.
Lignin Bioavailability.
The computed partition coefficients (SI Appendix, Tables S1–S3) suggest significant membrane enrichment of LRCs. Enzymes that metabolize these compounds could therefore potentially be more efficient if they were membrane associated to exploit the existing enrichment of these species within the lipid bilayer. Compared with strictly soluble enzymes without membrane interaction, integral membrane proteins embedded in the membrane or peripheral membrane proteins interacting with the membrane surface would experience higher substrate effective concentrations, thereby improving enzyme turnover. As a consequence, membrane binding of lignin-active enzymes may be common. The binding may be through direct membrane association as has been proposed for cytochrome P450s (48–50) or through complexation with other membrane-associated proteins, as observed for caffeoyl shikimate esterase (51) and cinnamoyl-CoA reductase (52).
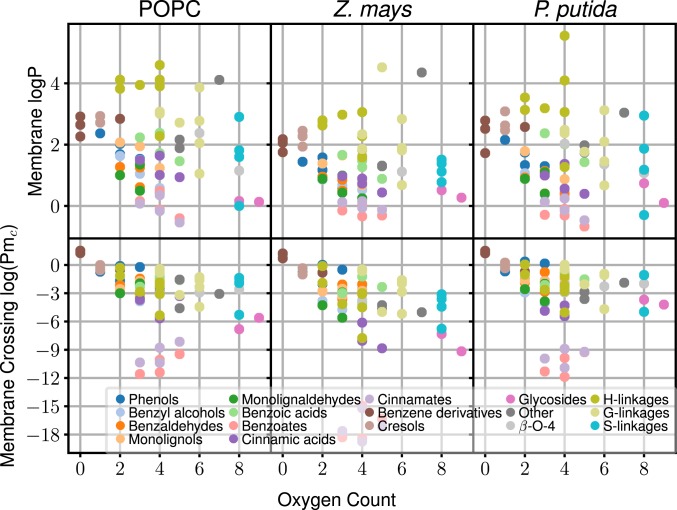
Interestingly, the partitioning behavior is nonadditive, with LRC dimers and monomers demonstrating approximately the same range of partition coefficients (Fig. 4, Top). The number of hydroxy or other oxidation sites is, however, a reasonable predictor of spontaneous permeation rates (Fig. 4, Bottom). Although monomeric and dimeric LRCs cross spontaneously, the hydrophillicity of larger LRCs demands a transporter for bilayer transit, due to the permeability reduction of additional hydroxyls in these compounds. Assuming that the linear trend observed in Fig. 4 for noncarboxylated LRCs continues, we estimate that beyond 10 to 12 oxygens present on the LRC, , and passive diffusion will routinely become slower than facilitated diffusion or active transport. This size roughly corresponds to lignin tetramers. Therefore, our results predict that for a cell to degrade larger LRCs, organisms must either break down the lignin externally to trimers or smaller or import the fragments through an active transport mechanism.
Fig. 4.
Comparison of partitioning (Top) and membrane permeability (Bottom; from SI Appendix, Tables S1–S3) depending on the number of oxygen atoms present in each molecule.
Lignin Accumulation and Synthesis Considerations.
There are significant uncertainties surrounding membrane transport of lignin monomers during synthesis processes. Glycosylated lignin precursors such as coniferin are actively transported and stored in intracellullar vacuoles in some plants as part of lignin biosynthesis (2, 10, 20, 53). Unglycosylated LRCs are not observed to accumulate anywhere within plant cells, suggesting that they are quickly incorporated into extracellular lignin polymers. Depending on the exact concentration gradients across plant membranes, either passive or active transport routes could carry more of the flux (SI Appendix, Fig. S39). Thus, there are 3 potential pathways for individual lignin building blocks for egress from the cell, store and release, passive transport, and active transport.
We begin with the impact of lignin glycosylation where, across the membranes studied, Pm for glycosylated LRCs coniferin or syringin are cm , implying significant potential for transporter-mediated accumulation. Prior kinetics measurements estimate that the coniferin transporter turnover rate is 500 at saturating ATP concentrations (20), which implies that a single transporter per would eventually establish a concentration gradient of 1 mM. The established gradient would scale linearly with transporter density, with modest transporter copy numbers resulting in measurable gradients. The concentrations built up in these vacuoles may permit regulation of lignin synthesis by controlling release rates from the vacuole through an unknown mechanism (10), as knocking out lignin glucosidases yields a normal lignin phenotype (54). We speculate that releasing the stored glycosylated lignins may require reversing the transporter or having a kinase-controlled intravacuolar hydrolase generate a large monolignol gradient that could passively diffuse.
However, as monolignol permeability is so high, monolignol diffusion across the membrane prior to glycosylation may compete with glycosylation and sequestration. After diffusion across the membrane, an extracellular laccase or peroxidase would form a lignin radical that polymerizes, consuming lignin monomers and creating a gradient across the membrane (Eq. 1), implying that the concentration gradient is similar in magnitude to the intracellular monolignol concentration. The 10 M substrate affinity for glycosyltransferases (55) provides an upper bound estimate for the intracellular monolignol concentration and is comparable to the concentrations used in exogenous fluorescent monolignol addition studies (56, 57). Coupled with cm for these molecules and a plant cell with surface area, the steady state monolignol flux at this concentration would be 0.1 fmol , the equivalent of ATP-saturated coniferin transporters, or 1,000 transporters per . This transporter density is possible; thus, we expect both passive diffusion and vacuolar lignin accumulation to occur, with perhaps more monolignol flux proceeding via a passive permeation route due to its lower metabolic cost.
Assuming the availability of a fast passive transport mechanism, an active transport mechanism for nonglycosylated monomers likely contributes little to the flux. This is demonstrated clearly by cinnamyl alcohol dehydrogenase (CAD) knockout studies, in which monomeric lignin aldehydes (monolignaldehydes) build the lignin polymer (58, 59) rather than the native monomeric lignin alcohols formed by CAD, whose fast turnover (60, 61) minimizes aldehyde toxicity (62) and limits aldehyde incorporation into lignin polymers during normal lignification. Putative plant transporters specific to monolignols would likely not transport aldehydes due to different hydrogen bonding patterns reducing substrate affinity, suggesting that the monolignaldehydes passively traversed the plant cell membrane prior to incorporation into the polymer. Given that monolignaldehydes and monolignols have broadly similar membrane permeabilities (SI Appendix, Tables S1–S3), monolignols would also be expected to transit passively. The natural consequence of this passive transport mechanism is that the intracellular monolignol concentration could be the same in all compartments within the plant cell, with increased concentration within membranes due to favorable membrane partitioning for most LRCs.
The caveat to these findings is that the actual concentration gradient across plant cell membranes created by lignin biosynthesis is currently unknown. If the concentration gradient were sufficiently small, passive transport would be reduced and the cell may rely on active transport mechanisms to increase flux (SI Appendix, Fig. S39). For context, a 1 concentration within a typical plant cell volume of around 160 (63) is approximately 100,000 molecules; 100,000 molecules could be generated on the timescale of hours by a single enzyme ( (60, 61)), suggesting that concentrations at this scale are at least plausible and could be reached in a relatively short time. This reduces evolutionary pressure to evolve specific transport proteins as plant growth responses take place on the timescale of hours or days. If lower concentrations are measured during lignin biosynthesis, it may be that a steady state between intracellular synthesis, extracellular polymerization, and passive membrane transport is reached below the theoretical limits due to nutrient limitations reducing synthetic pathway fluxes, but should not be taken as definitive evidence for the existence of specific membrane transport proteins.
Conclusion
The mechanism for LRC permeation has significant implications for lignin biosynthesis and valorization. Through molecular simulation, we estimate membrane permeability and partitioning to assess potential passive permeation across lipid bilayers. For most LRCs, the high permeability and associated passive flux suggest that passive diffusion mechanisms may outpace lignin synthesis and utilization rates, and conventional active transporters for these compounds may not exist, as evolutionary pressures to develop these transporters are eased by rapid passive transport across the membrane. Glycosylated or carboxylated LRCs are the exception, with low calculated membrane permeabilities and the potential for significant gradients to be generated across the membrane through membrane transporters (11–19) as might be required to regulate lignin biosynthesis or mitigate product toxicity in microbial lignin valorization.
Membrane partitioning of the different compounds showed a consistent trend, in which hydroxyl interactions with the membrane dictated LRC orientation within the membrane, and most compounds were enriched at the level of the lipid carbonyl groups. The enrichment of LRCs at the membrane interface suggests that enzymes involved in lignin metabolism may associate with biological membranes (48–50) and leverage the high substrate concentration within the membrane to improve efficiency. Together, these findings inform future engineering approaches to biological lignin valorization and address a longstanding question in lignin biosynthesis.
Materials and Methods
We determine lignin compound permeability coefficients through molecular simulation using the inhomogeneous-solubility diffusion equation (31, 32), using the free energy profile and diffusivity of the molecule as it crosses the membrane. We determine these quantities from REUS simulations carried out for 69 lignin-derived or associated compounds in 3 different membrane environments representative of a homogenous POPC membrane, the plasma membrane found in corn roots, and the inner membrane of P. putida. The simulations were performed in NAMD 2.12 (64) using the CHARMM36 force field for lipids (65), lignin (66), and the general force field (67), depending on the specific molecules. Extensive details regarding system preparation, simulation, and analysis can be found in SI Appendix, including equilibrium GROMACS simulations (68) prepared with TopoGromacs (69) used to validate the REUS results.
Supplementary Material
Acknowledgments
We thank the reviewers for encouraging an expanded discussion to connect with experimental observables and alternative validation metrics. This work was authored in part by Alliance for Sustainable Energy, LLC, the manager and operator of the National Renewable Energy Laboratory (NREL) for the US Department of Energy (DOE) under Contract DE-AC36-08GO28308. J.V.V. was supported by the NREL Director’s Fellowship funded by the Laboratory Directed Research and Development program. J.V.V., M.F.C., and G.T.B. acknowledge funding from the DOE Office of Energy Efficiency and Renewable Energy (EERE) Bioenergy Technologies Office. J.R. and S.D.M. acknowledge support from the DOE Great Lakes Bioenergy Research Center (DOE Office of Science DE-SC0018409). W.B. acknowledges the Research Foundation–Flanders for funding the projects G020618N and G0C1914N. R.A.D., F.C., and G.T.B. acknowledge funding from the Center for Bioenergy Innovation, which is a US DOE Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. A portion of the research was performed using computational resources sponsored by the DOE EERE and located at NREL. This work used the Extreme Science and Engineering Discovery Environment (XSEDE) (70) through grant MCB090159 (to G.T.B.) using the Bridges system, which is supported by NSF award ACI-1445606, at the Pittsburgh Supercomputing Center and the Stampede2 system at the University of Texas at Austin in the Texas Advanced Computing Center. XSEDE is supported by NSF grant ACI-1548562.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904643116/-/DCSupplemental.
References
- 1.Mottiar Y., Vanholme R., Boerjan W., Ralph J., Mansfield S. D., Designer lignins: Harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37, 190–200 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W., Lignin biosynthesis and structure. Plant Physiol. 153, 895–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzin V., Galili G., New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 3, 956–972 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Yi Chou E., et al. , Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J. Exp. Bot. 69, 1849–1859 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ragauskas A. J., et al. , Lignin valorization: Improving lignin processing in the biorefinery. Science 344, 1246843 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Beckham G. T., Johnson C. W., Karp E. M., Salvachúa D., Vardon D. R., Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 42, 40–53 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Fuchs G., Boll M., Heider J., Microbial degradation of aromatic compounds—From one strategy to four. Nat. Rev. Microbiol. 9, 803–816 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Linger J. G., et al. , Lignin valorization through integrated biological funneling and chemical catalysis. Proc. Natl. Acad. Sci. U.S.A. 111, 12013–12018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins M., Smith R. A., Samuels L., The transport of monomers during lignification in plants: Anything goes but how? Curr. Opin. Biotechnol. 56, 69–74 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Liu C. J., Miao Y. C., Zhang K. W., Sequestration and transport of lignin monomeric precursors. Molecules 16, 710–727 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols N. N., Harwood C. S., PcaK, a high-affinity permease for the aromatic compounds 4-hydroxybenzoate and protocatechuate from Pseudomonas putida. J. Bacteriol. 179, 5056–5061 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Argenio D. A., Segura A., Coco W. M., Bünz P. V., Ornston L. N., The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK. J. Bacteriol. 181, 3505–3515 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhry M. T., et al. , Genome-wide investigation of aromatic acid transporters in Corynebacterium glutamicum. Microbiology 153, 857–865 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Salmon R. C., Cliff M. J., Rafferty J. B., Kelly D. J., The CouPSTU and TarPQM transporters in Rhodopseudomonas palustris: Redundant, promiscuous uptake systems for lignin-derived aromatic substrates. PLoS One 8, e59844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michalska K., et al. , Characterization of transport proteins for aromatic compounds derived from lignin: Benzoate derivative binding proteins. J. Mol. Biol. 423, 555–575 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan K., et al. , Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids. Proteins. 81, 1709–1726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otani H., Lee Y. E., Casabon I., Eltis L. D., Characterization of p-hydroxycinnamate catabolism in a soil Actinobacterium. J. Bacteriol. 196, 4293–4303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori K., Kamimura N., Masai E., Identification of the protocatechuate transporter gene in Sphingobium sp. strain SYK-6 and effects of overexpression on production of a value-added metabolite. Appl. Microbiol. Biotechnol. 102, 4807–4816 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Reverón I., et al. , Differential gene expression by Lactobacillus plantarum WCFS1 in response to phenolic compounds reveals new genes involved in tannin degradation. Appl. Environ. Microbiol. 83, e03387-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao Y. C., Liu C. J., ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. U.S.A. 107, 22728–22733 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alejandro S., et al. , AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 22, 1207–1212 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Tsuyama T., et al. , Proton-dependent coniferin transport, a common major transport event in differentiating xylem tissue of woody plants. Plant Physiol. 162, 918–926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boija E., Johansson G., Interactions between model membranes and lignin-related compounds studied by immobilized liposome chromatography. Biochim. Biophys. Acta Biomembr. 1758, 620–626 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Boija E., Lundquist A., Edwards K., Johansson G., Evaluation of bilayer disks as plant cell membrane models in partition studies. Anal. Biochem. 364, 145–152 (2007). [DOI] [PubMed] [Google Scholar]
- 25.MacCallum J. L., Bennett W. F. D., Tieleman D. P., Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 94, 3393–3404 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohn M., Heinz E., Lüthje S., Lipid composition and fluidity of plasma membranes isolated from corn (Zea mays L.) roots. Arch. Biochem. Biophys. 387, 35–40 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Rühl J., Hein E. M., Hayen H., Schmid A., Blank L. M., The glycerophospholipid inventory of Pseudomonas putida is conserved between strains and enables growth condition-related alterations. Microb. Biotechnol. 5, 45–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen F., Tobimatsu Y., Havkin-Frenkel D., Dixon R. A., Ralph J., A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. U.S.A. 109, 1772–1777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Río J. C., et al. , Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem. 60, 5922–5935 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Jayakody L. N., et al. , Thermochemical wastewater valorization via enhanced microbial toxicity tolerance. Energy Environ. Sci. 11, 1625–1638 (2018). [Google Scholar]
- 31.Marrink S. J., Berendsen H. J. C., Simulation of water transport through a lipid membrane. J. Phys. Chem. 98, 4155–4168 (1994). [Google Scholar]
- 32.Lee C. T., et al. , Simulation-based approaches for determining membrane permeability of small compounds. J. Chem. Inf. Model. 56, 721–733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vermaas J. V., Beckham G. T., Crowley M. F., Membrane permeability of fatty acyl compounds studied via molecular simulation. J. Phys. Chem. B 121, 11311–11324 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Vermaas J. V., Bentley G. J., Beckham G. T., Crowley M. F., Membrane permeability of terpenoids explored with molecular simulation. J. Phys. Chem. B 122, 10349–10361 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Wennberg C. L., van der Spoel D., Hub J. S., Large influence of cholesterol on solute partitioning into lipid membranes. J. Am. Chem. Soc. 134, 5351–5361 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Barton N., et al. , Enabling the valorization of guaiacol-based lignin: Integrated chemical and biochemical production of cis,cis-muconic acid using metabolically engineered Amycolatopsis sp ATCC 39116. Metab. Eng. 45, 200–210 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Johnson C. W., et al. , Enhancing muconic acid production from glucose and lignin-derived aromatic compounds via increased protocatechuate decarboxylase activity. Metab. Eng. Commun. 3, 111–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salvachúa D., et al. , Bioprocess development for muconic acid production from aromatic compounds and lignin. Green Chem. 20, 5007–5019 (2018). [Google Scholar]
- 39.Lugo M. R., Sharom F. J., Kinetic validation of the models for P-glycoprotein ATP hydrolysis and vanadate-induced trapping. Proposal for additional steps. PLoS One 9, e98804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bársony O., et al. , A single active catalytic site is sufficient to promote transport in P-glycoprotein. Sci. Rep. 6, 24810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallinson S. J. B., et al. , A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion. Nat. Commun. 9, 2487 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cayley S., Lewis B. A., Guttman H. J., Record M., Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. J. Mol. Biol. 222, 281–300 (1991). [DOI] [PubMed] [Google Scholar]
- 43.Hearn E. M., Patel D. R., van den Berg B., Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proc. Natl. Acad. Sci. 105, 8601–8606 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu M., Kobayashi Y., Tanaka H., Wariishi H., Transportation mechanism for vanillin uptake through fungal plasma membrane. Appl. Microbiol. Biotechnol. 68, 673–679 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Yoneda A., et al. , Comparative transcriptomics elucidates adaptive phenol tolerance and utilization in lipid-accumulating Rhodococcus opacus PD630. Nucleic Acids Res. 44, 2240–2254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu W., Liu F., Singh S., Toward engineering E. coli with an autoregulatory system for lignin valorization. Proc. Natl. Acad. Sci. U.S.A. 115, 2970–2975 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Berg B., Going forward laterally: Transmembrane passage of hydrophobic molecules through protein channel walls. ChemBioChem 11, 1339–1343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gou M., Ran X., Martin D. W., Liu C. J., The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Nat. Plants 4, 299–310 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Bassard J. E., et al. , Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell 24, 4465–4482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbosa I. C. R., Rojas-Murcia N., Geldner N., The casparian strip—One ring to bring cell biology to lignification? Curr. Opin. Biotechnol. 56, 121–129 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Miao R., Lung S. C., Li X., Li X. D., Chye M. L., Thermodynamic insights into an interaction between ACYL-CoA–BINDING PROTEIN2 and LYSOPHOSPHOLIPASE2 in Arabidopsis. J. Biol. Chem. 294, 6214–6226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawasaki T., et al. , Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. 103, 230–235 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dima O., et al. , Small glycosylated lignin oligomers are stored in arabidopsis leaf vacuoles. Plant Cell 27, 695–710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chapelle A., et al. , Impact of the absence of stem-specific -glucosidases on lignin and monolignols. Plant Physiol. 160, 1204–1217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim E. K., Jackson R. G., Bowles D. J., Identification and characterisation of Arabidopsis glycosyltransferases capable of glucosylating coniferyl aldehyde and sinapyl aldehyde. FEBS Lett. 579, 2802–2806 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Tobimatsu Y., et al. , Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J. 76, 357–366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pandey J. L., et al. , Investigating biochemical and developmental dependencies of lignification with a click-compatible monolignol analog in Arabidopsis thaliana stems. Front. Plant Sci. 7, 1309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Q., et al. , Loss of function of cinnamyl alcohol dehydrogenase 1 leads to unconventional lignin and a temperature-sensitive growth defect in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 110, 13660–13665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan X., et al. , CAD1 and CCR2 protein complex formation in monolignol biosynthesis in Populus trichocarpa. New Phytol. 222, 244–260 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Jun S. Y., et al. , The enzyme activity and substrate specificity of two major cinnamyl alcohol dehydrogenases in sorghum (Sorghum bicolor), SbCAD2 and SbCAD4. Plant Physiol. 174, 2128–2145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park H., et al. , Biochemical characterization of the rice cinnamyl alcohol dehydrogenase gene family. Molecules 23, 2659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kunjapur A. M., Prather K. L. J., Microbial engineering for aldehyde synthesis. Appl. Environ. Microbiol. 81, 1892–1901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willis L., et al. , Cell size and growth regulation in the Arabidopsis thaliana apical stem cell niche. Proc. Natl. Acad. Sci. U.S.A. 113, E8238–E8246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Phillips J. C., et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klauda J. B., et al. , Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vermaas J. V., Petridis L., Ralph J., Crowley M. F., Beckham G. T., Systematic parameterization of lignin for the CHARMM force field. Green Chem. 21, 109–122 (2019). [Google Scholar]
- 67.Vanommeslaeghe K., et al. , CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abraham M. J., et al. , GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 69.Vermaas J. V., Hardy D. J., Stone J. E., Tajkhorshid E., Kohlmeyer A., TopoGromacs: Automated topology conversion from CHARMM to GROMACS within VMD. J. Chem. Inf. Model. 56, 1112–1116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Towns J., et al. , XSEDE: Accelerating scientific discovery. Comput. Sci. Eng. 16, 62–74 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.