Abstract
PIK3CD encodes the phosphoinositide 3-kinase (PI3K) catalytic subunit, p110δ, a lipid kinase linked to neurodevelopmental disorders, including schizophrenia (SZ). PIK3CD is regulated at the transcript level through alternate use of 5' untranslated exons (UTRs), promoters, and proinflammatory cytokines. Increases in global PIK3CD expression and downregulation by neuroleptics are observed in SZ, and preclinical efficacy of a p110δ-selective inhibitor is seen in rodent models of risk. Here, we cloned PIK3CD alternative transcripts in human brain and evaluated temporal- and tissue-specific expression. We quantified PIK3CD transcripts in B-lymphoblastoid cells from patients with SZ and examined 5' UTR transcriptional regulation by tumor necrosis factor α (TNFα) and interleukin-1β (IL1β) in patient-derived fibroblasts. We report that PIK3CD transcripts are differentially expressed in human brain in a developmental-specific manner. Transcripts encoding 5' UTRs -2A and alternative exon -1 (Alt1), P37 and AS1 and AS2 were increased in SZ. Alt1, P37, and AS2 were also preferentially expressed in fetal brain, and all transcripts were regulated by TNFα and IL1β. Our findings provide novel insight into the complexity of PIK3CD regulation in human brain, implicate PIK3CD in human neurodevelopment, and identify isoform-specific disruption in SZ.
Introduction
Abnormal PIK3CD signaling is implicated in diverse neurodevelopmental disorders, including schizophrenia (SZ), autism and developmental delay, whereby increased PIK3CD expression is observed in patients with SZ and autism (1,2), and gain-of-function genetic mutations are associated with developmental delay (3). In addition, PIK3CD has been identified as a candidate risk gene for SZ through familial-based genetic associations (1), and its expression is shown to be regulated by upstream variation in receptor tyrosine kinase function (1). The mechanisms underlying gene dysregulation and the transcripts involved are unknown. Like many genes implicated in SZ, the PIK3CD gene locus is highly complex. The gene encodes several 5' untranslated exons (5' UTRs; Fig. 1A) as well as an alternative splice isoform, P37 (JN190435), that results in a truncated protein product (Fig. 1C) (4–6). In addition to alternative transcript utilization, two novel antisense transcripts, PIK3CD AS1 (NR_027045.1) and PIK3CD AS2 (NR_126366.1) (Fig. 1B), have been identified and may function as an additional mechanism for transcriptional regulation of PIK3CD (7).
Figure 1.
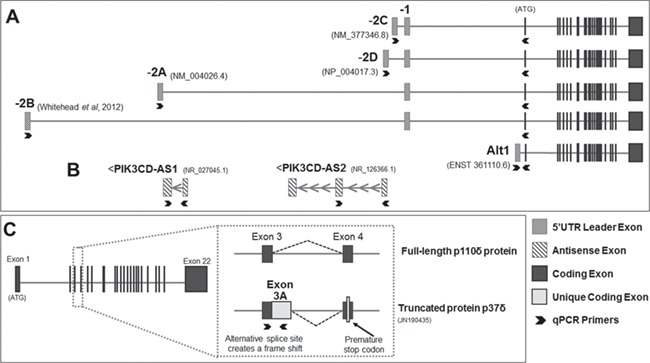
Human PIK3CD mRNA transcripts. (A) PIK3CD protein coding sequence (exons 1–22) are preceded by alternative 5' UTRs. mRNA sequences generally contain two 5' UTR exons (exon -2A, -2B, -2C, or -2D and exon -1; 2e not shown). Some transcripts contain an alternative exon -1 (Alt1, arrow), either as the sole leader exon or following exon -2A. Data derived from Whitehead et al (4), the Ensembl database, and cloning experiments described here. (B) PIK3CD-AS1 and PIK3CD-AS2 are recently identified lncRNAs located antisense to the upstream 5' UTR region of the PIK3CD gene. (C) The P37 transcript is formed by use of an alternative splice site between Exons 3 and 4 that includes intronic sequence immediately downstream of Exon 3 (termed Exon 3A), resulting in a frame shift that introduces a premature stop codon within Exon 4. The resulting product, p37δ, is a truncated protein lacking the kinase domain of full-length p110δ. The 5' UTR exon composition of P37 transcripts has not been investigated, although our preliminary data suggest transcripts contain -2A and Alt1. Transcript structure was derived from Refs. (4, 5). Block arrows denote location of PCR primers.
Importantly, emerging work in non-neuronal cells shows that PIK3CD is additionally regulated at the transcriptional level by immune genes that are linked to SZ, including the proinflammatory cytokines tumor necrosis factor α (TNFα) and interleukin-1β (IL1β) (8,9). These findings suggest that dysregulation of the immune system in SZ (10–17) could lead to secondary, transcriptional dysregulation of PIK3CD. Notably, the major histocompatibility complex (MHC) contains the most significant association of common variants in current genome-wide association studies of SZ and several of these variants reside in the TNFα gene (8). Additionally, environmental stimuli related to immunological activation, including infections during pregnancy and stress in early childhood, have been correlated with an increased risk of later SZ (18), and studies of peripheral cells and serum from patients with SZ have found increased levels of inflammatory molecules including TNFα and IL-1β (10–13,19). Nevertheless, despite strong genetic and environmental association to SZ, studies in postmortem human brain have been conflicting, with some reports failing to identify global expression changes in genes of the immune system in SZ (20) and others finding disease-associated increases in specific immune genes, such as the complement component 4, C4A (21). It is worth noting that such studies may not capture, or take into account, temporal dynamic changes in neuroimmune genes in response to the biological environment (i.e. immune challenge), nor the effects of antipsychotic agents on the neuroimmune system, or reflect the early effects of maternal or fetal immune activation on neuroimmune gene function during critical periods of brain development relevant to later risk for SZ (22).
In an original study in SZ, a pan-transcriptomic PIK3CD assay (1) revealed increased expression of PIK3CD in patient lymphoblastoid cell lines (LCLs). As emerging work highlights the clinical significance of disrupted alternative splicing and transcriptional dysregulation in neurological disease (23,24) and given that alternative splicing is especially prevalent in brain (23), understanding alternative transcription patterns of PIK3CD during normal brain development, as well as risk for SZ, is a pivotal next step.
The PIK3CD gene encodes for the p110δ protein, a Class I catalytic isoform of the phosphoinositide 3-kinase (PI3K) family that functions downstream of receptor tyrosine kinases to activate signaling cascades including the AKT and mTOR pathways (25). Originally identified as an immune system-specific isoform, PIK3CD expression has been reported in the Central Nervous System (CNS) and contributes to axonal growth, dendritic branching, and microglial activation (1,26–28). Importantly, inhibition of the p110δ protein in brain with the selective pharmacological inhibitor, IC87114, has been shown to ameliorate neurobehavioral and cognitive phenotypes in several rodent models of SZ (1,29), supporting PIK3CD as an important signaling molecule in the context of brain development, maturation, and disease. To date, it is unknown how or if alternative transcripts of PIK3CD, or its antisense RNAs, are utilized in brain tissue, or if specific transcripts contribute to increases previously observed in patients with SZ.
As noted, the complexity of the PIK3CD gene stems in part from the alternative use of one of several 5' UTR exons, termed -2A (NM_005026.4), -2B (from (5)), -2C (NM_377346.8), or -2D (NP_005017.3), which are spliced to a second 5' UTR exon, termed exon -1 (4,5). An additional non-coding leader exon, alternative exon -1 (Alt1; ENST 361110.6), appears to function as an alternative to utilization of exon -1 ((5) and Fig. 1A). The regulation of PIK3CD 5' UTRs has recently been investigated in non-neuronal cells, with evidence supporting that they are regulated by three separate promoters that are differentially sensitive to environmental regulation by proinflammatory cytokines, via activation of the downstream target NfκB (5). In addition to the complex 5' UTR exon usage, the recently isolated splice-variant of PIK3CD, P37, introduces a premature stop codon, which leads to a truncated form of the protein, p37δ. p37δ lacks a catalytic domain, yet remains functional through sequestration of regulatory molecules, allowing PI3K pathway activation to occur more rapidly (6,30). Finally, two novel antisense transcripts to PIK3CD, AS1 and AS2, have recently been identified (7). Long non-coding RNAs, including antisense RNAs, are abundantly expressed and utilized in the brain and have been associated with neurocognitive and degenerative disorders (31,32). The complexity of PIK3CD transcriptional regulation, in conjunction with evidence suggesting the importance of alternative and antisense RNAs in normal human brain development (31), led us to hypothesize that alternative PIK3CD transcripts, including AS1 and AS2 are differentially expressed across brain development and selectively altered in SZ.
Our study reveals alternate expression of multiple PIK3CD transcripts in human brain and identifies distinct 5' UTR exon usage that is temporally and spatially regulated across tissue and developmental stage. Furthermore, we show that specific PIK3CD transcripts, including AS1 and AS2, and the novel truncated variant, P37 are increased in patients with SZ. Finally, we show significant induction of PIK3CD transcript expression following acute cytokine stimulation but failed to detect any differential effects of diagnosis. Together, these findings further highlight the importance of alternative RNA transcript usage in brain and suggest that aberrant expression of specific PIK3CD transcripts may contribute to disease pathophysiology.
In summary, these data provide the first evidence to support alternate PIK3CD transcript expression in human brain, show that PIK3CD is developmentally regulated and altered in patients with SZ and suggest that dysregulation of proinflammatory cytokine signaling in SZ could be a mediator of altered PIK3CD transcription.
Results
Verification of multiple discrete PIK3CD transcripts with alternate 5' UTRs in fetal brain, and adult neocortex
Previous work has demonstrated that multiple distinct PIK3CD transcripts are expressed in leucocytes and non-neuronal peripheral cells ((4,5) and Fig. 1). Each transcript includes two untranslated exons: an alternate 5' UTR exon (termed -2A, -2B, -2C, -2D, and -2E) and a canonical exon -1, which span a 81 kb region upstream of the translational start codon in exon 1. Other transcripts with exon -1 spliced contain an alternative exon (termed Alt1; (5) and Fig. 1). Based on this complexity, we examined whether diversity in transcript expression was conserved in human brain and whether such variants were expressed differentially in fetal brain. Using isoform-selective primers, we identified full-length products of PIK3CD (~3.2 kb) containing 2A, -2B, -2C and -2D, and Alt 1 in fetal brain, adult neocortex (Fig. 2) and adult hippocampus (data not shown). -2E containing transcripts were not assessed due to below detectable quantities. All assayed isoforms were detectable in brain, with the exception of transcripts containing the -2B leader exon, which were minimally expressed in fetal brain and absent in adult brain. Such observations are consistent with previous findings demonstrating that expression of exon -2B is restricted to leucocytes (5). Cloning, sequencing and alignment of nucleotide sequences in NCBI BLAST verified that the cloned sequences match previously reported transcripts (4,5) and as presented in Figure 1. These data provide novel evidence that multiple PIK3CD transcripts are expressed in the developing and adult human brain and that alternate 5' UTR exon usage is conserved in human brain.
Figure 2.
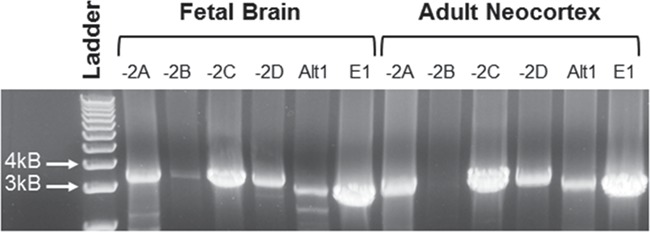
Expression of PIK3CD alternate 5' UTR transcripts in the adult and fetal human brain. Isoform specific primers were used (Table 1) to amplify cDNA transcripts from fetal and adult brain. PCR confirms a predicted amplicon size of ~3.2 kb for each PIK3CD 5' UTR isoform. 5' UTR -2B was poorly expressed in fetal brain and undetectable in adult neocortex.
Quantitative expression profiling of PIK3CD transcripts in fetal brain, adult neocortex, adult hippocampus and peripheral leucocytes
Previous evidence suggests that most non-leucocytic cell types express low levels of PIK3CD under basal conditions (4,5,25). After verifying the presence of PIK3CD alternative transcripts in brain, we sought to characterize quantitative expression of PIK3CD across tissues (leucocyte vs. brain) using isoform specific quantitative real-time polymerase chain reaction (qRT-PCR) (Fig. 3, Table 1). Consistent with previous data (4,5), we confirm that PIK3CD is more highly expressed in LCLs, with transcripts containing the 5' UTR exon -2A being 16-fold more abundant than in any brain region (Fig. 3D). Interestingly, in brain, transcripts containing -2C (t = −14.2, P < 0.0001), Alt1 (t = −51.53, P < 1 × 10−7) and transcript P37 (t = −14.5, P < 0.0001) were significantly more highly expressed in fetal brain compared to adult (Fig. 3). Transcripts containing 5’UTR exon -2B were undetectable in brain, but present at lower levels in LCLs (Fig. 3A–D). These results exemplify the tissue specificity of PIK3CD transcript usage and additionally imply that there is developmental-specific regulation of PIK3CD in human brain.
Figure 3.
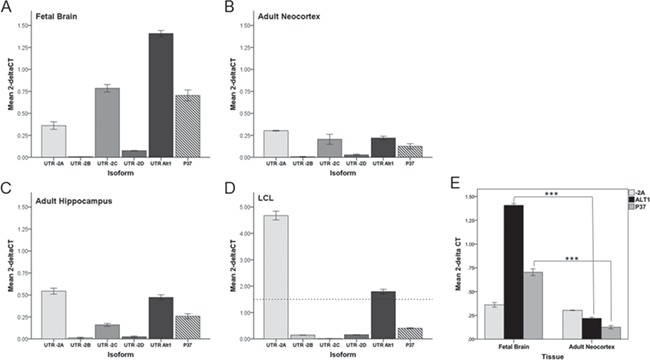
PIK3CD isoform expression is tissue- and development-stage specific. Quantitative analysis of PIK3CD isoform transcripts as measured by real-time qRT-PCR in fetal brain (A), adult neocortex (B), adult hippocampus (C), and LCL (D). (E) Statistical comparison of transcript expression between fetal brain and adult neocortex of variants differentially expressed in SZ (see Fig. 4), (***P < 0.00001). Transcript expression (cycle threshold (Ct) values) was normalized to PBGD (2-Δ CT). Error bars represent ±1 SEM for triplicate analysis from each tissue. Note difference in scale between neural tissues and LCL; dotted line in (D) denotes top of scale bar for (A)–(C) and (E).
Table 1.
PCR and qPCR primer sequences for PIK3CD transcripts
| Primer name | Sequence (5'-3') | Direction |
|---|---|---|
| PIK3CD -2A | TCCGAGCGGCCGCGAGCAGA | Forward |
| PIK3CD -2B | GTCTGTCTGGTGATACCAGG | Forward |
| PIK3CD -2C | TGGCGTCTTCCCGTCACT | Forward |
| PIK3CD -2D | TTCAAACCACCTTCTACCAC | Forward |
| PIK3CD Alt1 | GAGTAGTAGGAGCCACAAGCC | Forward |
| PIK3CD E1 | AACCTCAGCACCATCAAGCA | Forward |
| PIK3CD E22 | GGTTTTCCAGCTCTCACGGA | Reverse (PCR) |
| PIK3CD E1 | TGCTTGATGGTGCTGAGGTTG | Reverse (qPCR) |
| P37 E3 | CCCTTCTGGTCAACGTTAAG | Forward |
| P37 E3A | GAGTTTCACAGAGGAAGTGCT | Reverse |
| AS1 E1 | ACATCTGCGTGGTCGGGAGT | Forward |
| AS1 E2 | TATTACAGTGCCACTGCGCT | Reverse |
| AS2 E1 | ACAGAGGCTGGGAGGAGTTCT | Forward |
| AS2 E2 | TAGCATGGCCACTGGCTTAC | Reverse |
Altered expression of 5' UTR transcripts of PIK3CD and PIK3CD P37 in SZ
Given the findings that global PIK3CD transcript expression is elevated in peripheral LCLs of patients with SZ, that antipsychotic drugs attenuate transcriptional expression in the rodent and human brain (1), combined with new knowledge that differential utilization of promoters and 5' UTR exons occurs in human brain and at different stages of development (Fig. 3), we sought to determine the degree and specificity of transcript-specific dysregulation of PIK3CD in SZ. We used qRT-PCR with isoform specific custom primer and probes (Table 1) to measure all known PIK3CD transcripts in an independent cohort of LCLs derived from 30 SZ patients and 30 healthy control subjects. Transcriptional profiling revealed that PIK3CD “pan” (Ct range, 24–25), -2A (Ct range, 22–24), -2D (Ct range, 26–30), Alt1 (Ct range, 28–30) and P37 (Ct range, 23–25) transcripts were abundant in LCLs, while -2B and -2C were at levels below detection, confirming independent observations in experiments in Figure 3. PIK3CD transcripts containing the untranslated exon -2A (F(1, 59) = 6.82, P = 0.01; Fig. 4), as well as the novel truncated P37 transcript (F(1, 59) = 3.94, P < 0.05; Fig. 4), were significantly increased in SZ. Transcripts with the Alt1 leader exon also showed a trend for increased expression (F(1, 59) = 3.6, P = 0.06, Fig. 4). Pan-PIK3CD (assay spanning exons 8–9), as well as transcripts containing -2D, also showed trends toward being increased in SZ (P > 0.1; Supplemental Fig. 1); however, these failed to reach statistical significance. Synthesis of P37 specific cDNA, using a gene specific primer, indicated that both -2A and Alt1 can be utilized as part of the P37 transcript (data not shown), suggesting that increases in these three assays may represent a single transcript. These results further support dysregulation of PIK3CD transcriptional regulation in SZ and suggest involvement of specific alternative transcripts.
Figure 4.
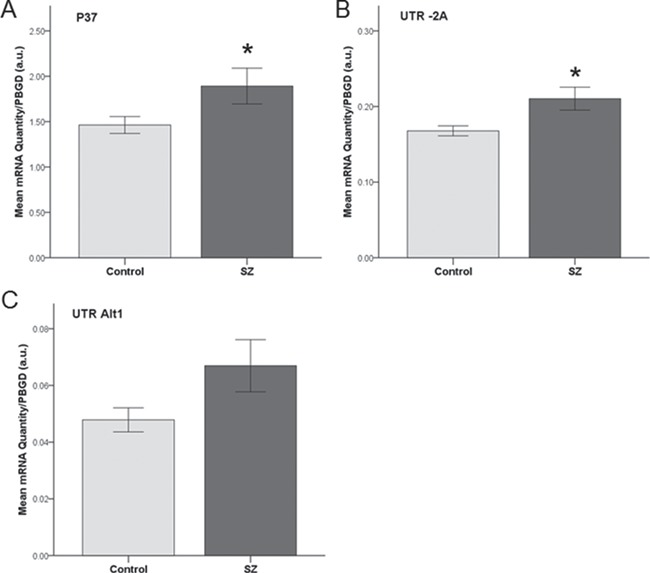
PIK3CD alternative transcript expression is increased in SZ. Quantitative analysis of PIK3CD isoform transcripts P37 (A), UTR-2A (B), and UTR Alt1 (C) as measured by qRT-PCR in LCLs from patients with SZ (n = 30) or normal controls (n = 30). All values were extrapolated using the standard curve method and normalized to PBGD. au, arbitrary units. Error bars represent ±1 SEM. *Represents P ≤ 0.05.
Expression of the novel antisense transcripts, PIK3CD AS1 and AS2 is increased in SZ
Bioinformatic study of the Ensembl database (GRCh38.p12) and the UCSC genome browser (GRCh38/hg38) inspected for annotated human PIK3CD transcripts, revealed two previously unstudied PIK3CD transcripts (AS1, NR_027045.1 and AS2 NR_126366.1) that are directly antisense to the 5' region upstream of the PIK3CD translational start site (7). This region contains the promoter regions 2 and 3 (5).
To investigate if alterations in expression of AS1 or AS2 are observed in SZ, we developed custom primers to span exons 1–2 of each antisense transcript. Both AS1 and AS2 were found to be highly expressed in LCLs (Ct ranges, 24–26) and additionally are significantly increased in SZ (AS1, F(1, 58) = 14.043, P < 0.001; AS2, F(1, 58) = 5.711, P = 0.02; Fig. 5B). These results suggest a possible positive relationship between AS1/AS2 expression regulation and other isoforms of PIK3CD. Current studies of the molecular role of AS1 and AS2 are underway. Second, we transcriptionally profiled AS1 and AS2 across developmental stage in human brain and compared to LCLs. While considerably lower in brain than in LCLs, AS1 expression was consistently expressed at comparable levels across fetal brain, adult neocortex, and adult hippocampus. Conversely, AS2 expression levels were 5-fold higher in fetal brain than in adult neocortex or hippocampus (Fig. 5A, right panel), comparable to levels seen in LCLs. This predominance of AS2 in fetal brain suggests a potential role for AS2 in early brain development.
Figure 6.
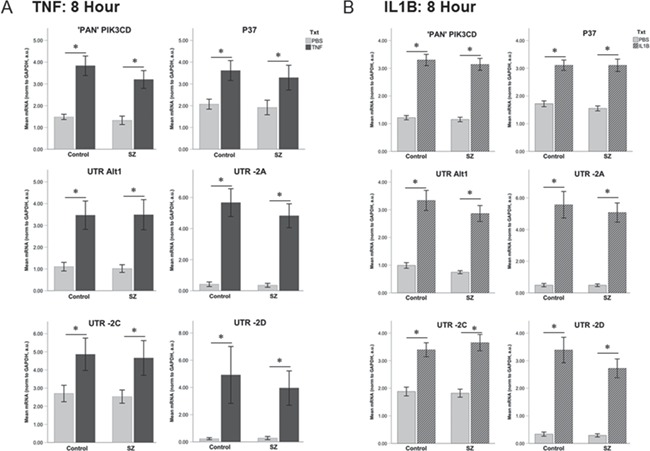
Proinflammatory cytokines induce PIK3CD transcript expression, in the absence of diagnosis effects. Acute cellular stimulation of human dermal fibroblasts from patients with SZ (N = 30) and normal controls (N = 30) with TNFα (A) of IL1β (B) for 8 hours induced transcription of all PIK3CD alternate transcripts as measured by qPCR. There was no effect of diagnosis for any assay in response to either cytokine stimulation. Note transcripts containing -2B are not expressed in human dermal fibroblasts. mRNA quantification was derived from a pooled-sample standard curve and normalized to β-actin. Error bars represent ±1 SEM for the average of experimental triplicate analysis for n = 30 vs. n = 30. a.u. = arbitrary units. *represents post hoc LSD P-value of <0.05 when compared to PBS control.
Figure 5.
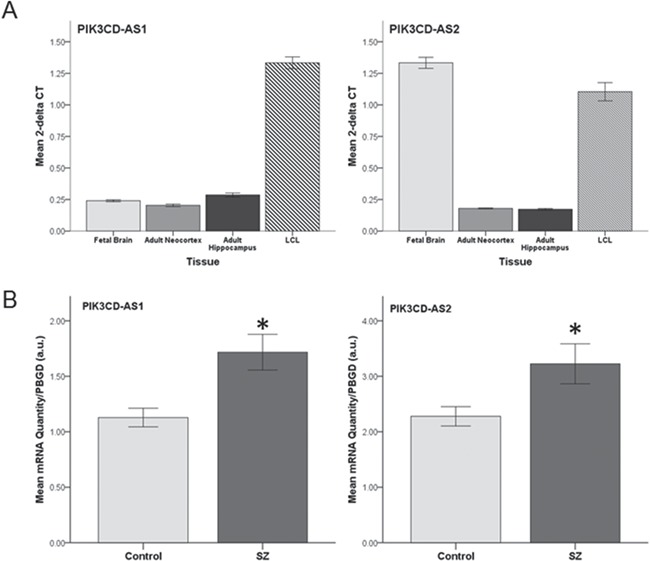
PIK3CD-AS1 and AS2 are increased in SZ and differentially expressed across tissue type. (A) Quantitative analysis of PIK3CD-AS1 and AS2 transcripts represented as mean 2-Δ CT normalized to PBGD across tissue types. Error bars represent ±1 SEM for triplicate analysis from each tissue. *Represents P ≤ 0.05. (B) Quantitative analysis of PIK3CD-AS1 and AS2 transcripts as measured by qPCR in LCLs from patients with SZ (n = 30) and normal controls (n = 30). mRNA quantification was derived from the standard curve method and normalized to PBGD. Error bars represent ±1 SEM for the average of triplicate analysis. au, arbitrary units.
Proinflammatory cytokines TNFα and IL1β induce PIK3CD transcript expression in primary human cells
Previous work in human endothelial cells, synovial fibroblasts and human leucocytes has reported isoform-selective induction of PIK3CD expression by TNFα and IL1β, mediated via inducible promoters and differential transcriptional regulation of PIK3CD 5' UTR exons (5). Given these data, we explored whether PIK3CD expression can be modulated by inflammatory cytokines in different cell types of human origin and examined whether human dermal fibroblasts represent a viable cell model system for studying the relationship between proinflammatory cytokines, PIK3CD regulation, and SZ.
HEK-293 cells and human primary fibroblast cell lines (N = 8) where stimulated with recombinant human TNFα, IL1β, or a PBS with 0.1% BSA control for 8 h. TNFα stimulation evoked the most robust transcriptional responses in both cell types, and while increases in PIK3CD in HEK293 cells were less in response to IL1β, dermal fibroblasts responded strongly to IL1β stimulation (Supplemental Fig. 2). In both HEK293 cells and fibroblasts, “pan” PIK3CD (analysis of variance [ANOVA], main effect of treatment: HEK293, F = 121.722, P = 0.001; Fibro, F = 5.888, P = 0.009) increased upon IL-1β and TNFα stimulation (Supplemental Fig. 2). Similar responses to IL1β and TNFα stimulation were observed for the P37 transcript (ANOVA, main effect of treatment: HEK293, F = 8.923, P = 0.05; Fibro, F = 3.265, P = 0.055), showing for the first time that P37 expression is regulated by proinflammatory cytokine stimulation. In HEK293 cells, TNFα stimulation increased transcript levels of UTRs -2A (F = 10.197, P = 0.046), -2C (F = 15.211, P = 0.027), and -2D (F = 55.417, P = 0.004). Similar results were observed in dermal fibroblasts with both TNFα and IL1β significantly increasing levels of transcripts containing UTRs -2A (F = 8.641, P = 0.002) and Alt1 (F = 5.568, P = 0.011). Transcripts containing -2C, while elevated under both conditions, however, failed to reach statistical significance (F = 2.542, P = 0.103). Together, these findings demonstrate that human primary non-immune cells can be utilized to examine the relationship between PIK3CD transcriptional regulation and cytokine stimulation and that proinflammatory cytokines increase expression of multiple alternative PIK3CD transcripts, in a cell-type specific manner (5).
Next, we utilized a larger cohort of patient-derived and control primary dermal fibroblasts (n = 30 vs. 30) to determine if there was a differential PIK3CD transcriptional response to cytokine stimulation in the context of SZ. In this larger cohort, expression of all PIK3CD transcripts was significantly elevated in response to both TNFα (Fig. 6A; ANOVA, main effect of treatment, “pan,” F = 169.977, P < 0.0001; P37, F = 34.024, P < 0.0001; Alt1, F = 64.892, P < 0.0001; UTR-2A, F = 264.916, P < 0.0001; UTR-2C, F = 24.75, P < 0.001; UTR-2D, F = 47.412, P < 0.001) and IL1β (Fig. 6B; ANOVA, main effect of treatment, “pan,” F = 121.351, P < 0.0001; P37, F = 57.048, P < 0.0001; Alt1, F = 63.862, P < 0.0001; UTR-2A, F = 86.212, P < 0.0001; UTR-2C, F = 39.879, P < 0.001; UTR-2D, F = 62.937, P < 0.001). No main effect of diagnosis or diagnosis–treatment interaction was observed (Fig. 6). Furthermore, baseline levels of PIK3CD transcript expression did not differ between SZ patients and controls, in contrast to that observed in LCLs (Fig. 4).
Discussion
Here, we present novel evidence that the leucocyte-enriched PI3K, PIK3CD, is expressed in human brain, with select transcripts containing alternate 5' UTR exons, being temporally and spatially regulated. We also show that alternative 5' UTR splicing is selectively affected in patients with SZ, with several isoforms showing increased expression in SZ. Notably, Alt1 containing and P37 transcripts are also predominantly expressed in fetal brain, suggesting a potential immature profile of expression related to disease. Furthermore, we demonstrate that two novel antisense RNAs, PIK3CD-AS1 and -AS2, are expressed in human brain and reveal increased expression in SZ. Taken together, these findings emphasize the importance of tight regulation of alternative RNA utilization in the PIK3CD gene and imply that dysregulation may contribute to disease pathophysiology in SZ. Finally, although we failed to find differential responses between patients and controls, we provide novel evidence to support a potential mechanism of disruption of PIK3CD transcriptional regulation in SZ, mediated via altered immune regulation of proinflammatory cytokine signaling.
Developmental programming and normal functioning of complex systems require stringent ontogenic- and cell-type-specific regulation of gene expression (33). Here, we provide novel evidence that cell-type- and tissue-, as well as developmental-, specific regulation of PIK3CD 5' UTR transcript expression occurs in the human brain and human LCLs. Confirming previous observations in human and mouse cell lines (4,5), we show that the PIK3CD -2A exon has the highest basal promoter activity and expression in leucocytes than in non-leucocytes and further demonstrate that, while expressed abundantly in human brain, transcripts containing -2A exon are not differentially expressed during development. These observations suggest that tight control of PIK3CD -2A RNA transcription in human brain is biologically important. In LCLs, we also show that basal levels of PIK3CD transcripts containing -2C and -2D are at low or undetectable levels, consistent with previous observations (5). We now reveal that -2D transcripts are also observed at low levels in human brain and that PIK3CD transcripts containing -2C exon are higher in brain and preferentially expressed during prenatal development, highlighting a novel roll for -2C containing PIK3CD transcripts in human brain.
Expanding upon previous findings of increased global transcript expression of PIK3CD in SZ (1), we reveal specific increases in expression of transcripts containing the 5' UTRs -2A or Alt1 in an independent cohort of SZ patients. While the precise function of alternative promoter usage and splicing of the PIK3CD 5' UTRs is unknown, 5' UTRs are often involved in the regulation of mRNA stability or translational efficiency (34,35), and such flexible use of exons and alternative promoters has been proposed to allow cells to dynamically respond to the biological environment (5). Investigation of the role of the PIK3CD 5' UTRs in cellular function is underway. Another finding of particular interest is the observation that P37, the transcript encoding the C-terminally truncated p37δ protein product, is increased in SZ. While p37δ lacks the catalytic domain of the full-length p110δ protein, it retains the p85-binding domain and has been shown to have growth-promoting properties (6,36). To date, the extent of our knowledge of p37δ has come from tumor biology. Here, we present novel evidence to support that P37 is not a tumor-specific variant (6,36) but is also expressed at high levels in human fetal brain and in SZ peripheral cells. These data warrant further investigation into the normal biological function of p37δ and the consequences of its overexpression.
The origins of dysregulated PIK3CD transcription in SZ are presently unclear; however, emerging evidence suggests that altered signaling in the PI3K pathway (1) may be attributable to altered upstream regulation by genes relevant to disease risk, such as the growth factor NRG1, or its receptor ErbB4 (1,29). Recently, Whitehead et al. (5) demonstrated that the proinflammatory cytokines, TNFα or IL1β, are key transcriptional regulators of full-length PIK3CD and determinants of alternative 5' UTRs usage. Importantly, increases in cytokine levels have been reported in patients with SZ and genome-wide association studies demonstrate consistent association with regions of the genome rich in immune genes, in particular MHC region (9–17,19). These findings implicate dysregulated immune gene or neuroimmune function in the disease as a potential mechanism of altered PIK3CD transcription. In support of this hypothesis, our data further confirm the relationship between cytokine stimulation and transcriptional regulation of PIK3CD (5).
In HEK293 cells and human dermal fibroblasts, TNFα and IL1β stimulation induced dramatic increases in total “pan” PIK3CD and transcripts containing each of the 5' UTR exons, consistent with prior observation in human endothelial cells and synovial fibroblasts (5). Our data in HEK293 cells are contrary to the absence of effects of TNFα stimulation previously reported in HEK293T cells (5). However, we note that our experiments were conducted on serum starved cells and at a 10-fold higher dose of TNFα, suggesting that higher doses of cytokine stimulation may be required in certain cell types to affect PIK3CD gene activity. We also report for the first time that the P37 transcript is sensitive to acute cytokine stimulation.
While robust increases in PIK3CD transcript expression were observed in response to cytokine stimulation, in a large cohort of patient (N = 30) and control-derived (N = 30) fibroblasts, we failed to detect effects of diagnosis on transcriptional response, or baseline levels of PIK3CD. These findings are potentially consistent with observations that pathological changes related to disease gene expression are highly tissue specific (37) and relevant to the pathology of the disease. Such findings would be consistent with observed changes in PIK3CD expression in patient brain and LCLs, but not in skin fibroblasts.
Finally, while our study focused specifically on investigation of the transcriptional landscape of PIK3CD, we acknowledge that insight into protein changes in response to cytokine stimulation is valuable. Indeed, although beyond the scope of our study, prior work characterizing PIK3CD transcript regulation by TNFα confirmed concomitant increases in p110δ protein following increases in PIK3CD transcript expression (5).
Alterations of PIK3CD transcriptional regulation in peripheral cells from SZ patients would have less impact on our understanding of neuropsychiatric disease without confirmation of conserved brain utilization of alternative PIK3CD transcripts. Indeed, our results demonstrate that multiple PIK3CD transcripts are expressed in human brain and that they are developmentally regulated. One of the prevailing theories in SZ is that the disorder is one of neurodevelopmental origins (38) and data support that genes implicated in genetic risk for SZ are predominantly expressed during prenatal brain development (38,39). The pronounced expression of -2C, Alt1, and P37 in fetal brain and differential expression in patients with SZ suggests that PIK3CD regulation is pertinent to normal human brain development and a contributor to disease risk.
Finally, we report that the putative PIK3CD antisense transcripts, PIK3CD-AS1 (NR_027045.1) and PIK3CD-AS2 (NR_126366.1) are increased in SZ. Non-coding RNAs have become of increasing interest in the field of neuroscience as they allow for additional levels of gene regulation and are abundantly expressed in the brain and differentially regulated during brain development (31,32,40,41). AS1 and AS2 are antisense to sequence within the intron between UTR -2A and -2D in an area with high epigenetic modifications (UCSC Genome browser) and known alternative promoter regions (5), suggesting the possibility that AS1 and AS2 can regulate transcription of PIK3CD. Importantly, PIK3CD-AS2 is expressed at a significantly higher level in fetal brain, signifying a potential novel role in neurodevelopment. Future studies utilizing knock-down and overexpression of AS1 and AS2 are underway to assess the functional role of the antisense transcripts and relationship between and regulation of full-length PIK3CD expression.
Taken together, our study shows that alternative promoter usage in the PIK3CD gene, which gives rise to expression of associated 5' UTR exons containing transcripts, is differentially regulated in both a tissue and developmental-specific manner in human brain and human peripheral cells and that this transcriptional landscape is altered in patients with SZ, in a cell-type specific manner and regulated by peripheral cytokines.
Materials and Methods
Molecular cloning and sequencing of PIK3CD cDNA transcripts in human brain
Commercial RNA from brain was obtained from Clontech (cortex, no. 636561; hippocampus, no. 636593; fetal total brain, no. 636526). Two micrograms of total RNA was reverse transcribed using Superscript III (Invitrogen, no. 18080-400) with oligo dT primers according to manufacturer's protocol. Polymerase chain reaction (PCR) amplification of full-length PIK3CD was performed using Platinum High Fidelity Taq polymerase (Invitrogen, no. 11304-011). PCR conditions were 94°C for 2 min for Taq activation, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 4 min with a final extension of 10 min at 68°C. All PCR reactions used a forward primer from a unique PIK3CD leader exon (Table 1, Fig. 1) in combination with a reverse primer located in exon 22 (Table 1). Amplified PCR products were separated by electrophoresis on 0.75% agarose gels stained with ethidium bromide and visualized under ultraviolet light. PCR products were cloned into the pCR 2.1 TOPO vector, and the cloning reaction was used to transform TOP10 competent cells (Invitrogen, no. K4500-02). Plasmid miniprep DNAs were prepared from bacterial cultures by using QIAprepSpin miniprep kit (Qiagen). Inserts in selected clones were sequenced by using M13 forward and reverse primers in the DNA sequencing Core at the University of Colorado, School of Medicine.
qRT-PCR analysis of PIK3CD transcript abundance in human fetal brain, adult brain and LCLs
Commercial RNA from brain was obtained from Clontech (cortex, no. 636561; hippocampus, no. 636593; fetal total brain, no. 636526). RNA derived from LCL lines from human subjects was extracted (see Human subjects below), as previously reported (1). cDNA was amplified using a Multi-Scribe kit with random primers (Life Technologies, no. 4304134). LCL cDNA from a subset of the control subjects (N = 6) was pooled for tissue-specific analysis. Gene expression levels were measured by qRT-PCR, using an ABI Prism 7900 sequence detection system with 384-well format (Applied Bio-systems), as previously described in detail (1). Assays were performed using SYBR green detection (Life Technologies, cat. no. 4367659) with PIK3CD leader forward primers (Table 1) and a reverse primer located in exon 1 (Table 1). A P37 specific assay was designed using a forward primer from exon 3 (Table 1) and a reverse primer from the included exon downstream from exon 3 (named exon 3A, Table 1). Isoform specific assays for AS1 and AS2 were designed to span exon 1 and 2 (Table 1). The “pan” PIK3CD was a stock Taqman Assay to Exons 7–8 of the PIK3CD transcript (Applied Bio-systems, Hs00908671). PCR conditions for each assay were 10 min at 95°C for Taq activation followed by 40 cycles of 95°C for 15 s, 60°C for 1 min. For cross-tissue comparisons (Fig. 3), each assay was performed on all tissues in a single 384 well plate, using a standard curve method. Mean quantities (triplicates) of 50 ng reactions were used for expression comparisons, and final quantities were calculated using the Δ-CT method, normalized to the endogenous control gene, porphobilinogen deaminase (PBGD, Applied Biosystems, Hs00609297). For LCL, case vs. control (N = 60) analysis, PCR data were acquired from the Sequence Detector Software (SDS version 2.0; Applied Bio-systems) and quantified by a standard curve method (1). In each experiment, the R2 of the curve was more than 0.99 and controls constituting no-template cDNA resulted in no detectable signal. The software plotted a standard curve of the cycles at threshold (Ct) vs. quantity of RNA. Primary data analysis is based on normalization of mRNA transcript quantity to PBGD.
Human subjects
Blood collection and Epstein-Barr virus lymphocyte transformation were approved by the Colorado Multiple Institutional Review Board, University of Colorado, School of Medicine and collected in accordance with the Declaration of Helsinki principles. All donors gave written informed consent. All subjects were drawn from individuals participating in the Department of Psychiatry “Genetic Research in Schizophrenia Study” [original PI, R.F.; current Principle Investigator (PI), A.J.L.], part of a larger original study of the Molecular Genetics of Schizophrenia, and as described previously (42). A consensus diagnosis of SZ based on Diagnostic and Statistical Manual of Mental Disorders, Third Edition, Revised or Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, criteria was made following a systematic and comprehensive examination of multiple sources of available information obtained from relatives, medical records, clinicians, and direct assessment using one or more diagnostic interviews including the Diagnostic Interview for Genetic Studies and the Structured Clinical Interview for Axis I DSM Disorders (SCID). Control subjects were evaluated using the SCID—Non-patient Edition. All subjects in the study were unrelated to each other. Ethnicities of case–control participants were recorded from self-report or family. Thirty unaffected controls (20 males and 10 females; 80% Caucasian, 10% African American, 3% Hispanic, non-Caucasian, 7% Asian) and 30 individuals with a diagnosis of SZ (21 males and 9 females; 83% Caucasian, 7% African American, 7% Hispanic, non-Caucasian, 3% Asian) were used for LCL studies.
Primary skin fibroblasts were obtained from the Schizophrenia Research Center Brain Bank at the University of Colorado, School of Medicine, all of which were donated through the Colorado Anatomical Gift Act. The Brain/Fibroblast Bank contains tissue collected postmortem from subjects with a diagnosis of SZ as well as from unaffected individuals and as described previously (43–45). Thirty unaffected controls (Caucasian; 22 males and 8 females; mean age at death 57.5+/− 12.6 (SD) years) and 30 individuals with a diagnosis of SZ (Caucasian; 22 males and 8 females; mean age at death 54.0+/− 16.3 (SD) years) were used for cytokine stimulation studies.
Cytokine cellular stimulation assay
HEK-293 cells (ATCC) were grown in DMEM (Gibco) with 10% fetal calf serum (Gibco), 100 mg/ml streptomycin, 100 units/ml penicillin (Gibco), and 2 mmol/L l-glutamine (Gibco) in an incubator (37°C/5% CO2). Cells were plated at a density of 1 × 106 cells/well in 6-well plates, serum-starved overnight, and stimulated with 100 ng/ml recombinant human TNFα (R&D Systems) or 20 ng/ml recombinant human IL1β (Peprotech) for 8 h. IL1β concentrations and stimulation times were selected based on established prior literature (5) and dose/time-response experiments presented in Supplemental Figure 3. One hundred nanograms per milliliter recombinant human TNFα/8 h was selected as producing the most consistent and robust response across all isoforms for each time point and dose in our cell lines of study (i.e. human fibroblasts; Supplemental Fig. 3). RNA was extracted using Qiagen Lysis Buffer. cDNA synthesis and expression assays were performed as described above. Human dermal fibroblasts were grown in DMEM/F12 (Gibco) with 15% fetal calf serum (Gibco), 100 mg/ml streptomycin, 100 units/ml penicillin (Gibco), and 2 mmol/L l-glutamine (Gibco) in an incubator (37°C/5% CO2). Cells were plated at a density of 0.5 × 106 cells/well in 6-well plates, serum-starved overnight, and stimulated as above. For fibroblast case vs. control (N = 60) analysis, qPCR data were acquired from the Sequence Detector Software (SDS version 2.0; Applied Biosystems) and quantified by a standard curve method (1). In each experiment, the R2 of the curve was more than 0.99 and controls constituting no-template cDNA resulted in no detectable signal. The software plotted a standard curve of the cycles at threshold (Ct) vs. quantity of RNA. Primary data analysis is based on normalization of average triplicate mRNA transcript quantity to GAPDH (Applied Biosystems, Hs99999905).
Statistical analysis
All statistical comparisons were performed using the SPSS statistics package version 24.0 Independent samples t tests were used to analyze the effect of age (fetal brain vs. adult brain) on measures of gene expression for the PIK3CD isoforms. ANOVA was used to compare gene expression data in patients vs. control subjects with diagnosis and sex as fixed factors. ANOVA with treatment, diagnosis, and sex as fixed factors was used to compare effects of TNFα and IL1β stimulation on PIK3CD isoform expression. Fisher's post hoc least significant difference (LSD) was used to investigate significant effects.
Conflict of Interest statement. The authors declare that they have no conflicts of interest with the contents of this article.
Funding
National Institutes of Mental Health [R01MH103716 to A.J.L.]; a scholarship from the University of Colorado RNA Biosciences Initiative (V.L.H); and funds from the Dr. Nancy Gary Endowed Chair held by A.J.L.
Supplementary Material
References
- 1. Law A.J., Wang Y., Sei Y., O'Donnell P., Piantadosi P., Papaleo F., Straub R.E., Huang W., Thomas C.J., Vakkalanka R. et al. (2012) Neuregulin 1-ErbB4-PI3K signaling in schizophrenia and phosphoinositide 3-kinase-p110 inhibition as a potential therapeutic strategy. Proc. Natl. Acad. Sci. U. S. A., 109, 12165–12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Poopal A. C., Schroeder L. M., Horn P. S., Bassell G. J., and Gross C. (2016) Increased expression of the PI3K catalytic subunit p110δ underlies elevated S6 phosphorylation and protein synthesis in an individual with autism from a multiplex family. Mol. Autism. doi: 10.1186/s13229-015-0066-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coulter T.I., Chandra A., Bacon C.M., Babar J., Curtis J., Screaton N., Goodlad J.R., Farmer G., Steele C.L., Leahy T.R. et al. (2017) Clinical spectrum and features of activated phosphoinositide 3-kinase δ syndrome: a large patient cohort study. J. Allergy Clin. Immunol., 139, e4, 597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kok K., Nock G.E., Verrall E.A.G., Mitchell M.P., Hommes D.W., Peppelenbosch M.P. and Vanhaesebroeck B. (2009) Regulation of p110delta PI 3-kinase gene expression. PLoS One., 4, e5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Whitehead M.A., Bombardieri M., Pitzalis C. and Vanhaesebroeck B. (2012) Isoform-selective induction of human p110δ PI3K expression by TNFα: identification of a new and inducible PIK3CDpromoter. Biochem. J., 443, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fransson S., Uv A., Eriksson H., Andersson M.K., Wettergren Y., Bergo M. and Ejeskär K. (2012) p37δ is a new isoform of PI3K p110δ that increases cell proliferation and is overexpressed in tumors. Oncogene., 31, 3277–3286. [DOI] [PubMed] [Google Scholar]
- 7. Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K. et al. (2004) Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet., 36, 40–45. [DOI] [PubMed] [Google Scholar]
- 8. Corvin A. and Morris D.W. (2014) Genome-wide association studies: findings at the major histocompatibility complex locus in psychosis. Biol. Psychiatry., 75, 276–283. [DOI] [PubMed] [Google Scholar]
- 9. Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014) Biological insights from 108 schizophrenia-associated genetic loci. Nature., 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fillman S.G., Weickert T.W., Lenroot R.K., Catts S.V., Bruggemann J.M., Catts V.S. and Weickert C.S. (2016) Elevated peripheral cytokines characterize a subgroup of people with schizophrenia displaying poor verbal fluency and reduced Broca's area volume. Mol. Psychiatry., 21, 1090–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pandey G.N., Ren X., Rizavi H.S. and Zhang H. (2015) Proinflammatory cytokines and their membrane-bound receptors are altered in the lymphocytes of schizophrenia patients. Schizophr. Res., 164, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Noto C., Maes M., Ota V.K., Teixeira A.L., Bressan R.A., Gadelha A. and Brietzke E. (2015) High predictive value of immune-inflammatory biomarkers for schizophrenia diagnosis and association with treatment resistance. World J. Biol. Psychiatry., 16, 422–429. [DOI] [PubMed] [Google Scholar]
- 13. Song X.-Q., Lv L.-X., Li W.-Q., Hao Y.-H. and Zhao J.-P. (2009) The interaction of nuclear factor-kappa B and cytokines is associated with schizophrenia. Biol. Psychiatry., 65, 481–488. [DOI] [PubMed] [Google Scholar]
- 14. Dickerson F., Stallings C., Origoni A., Schroeder J., Katsafanas E., Schweinfurth L., Savage C., Khushalani S. and Yolken R. (2016) Inflammatory markers in recent onset psychosis and chronic schizophrenia. Schizophr. Bull., 42, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. International Schizophrenia Consortium, Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F. and Sklar P. (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature., 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwab S.G., Mondabon S., Knapp M., Albus M., Hallmayer J., Borrmann-Hassenbach M., Trixler M., Gross M., Schulze T.G., Rietschel M. et al. (2003) Association of tumor necrosis factor alpha gene -G308A polymorphism with schizophrenia. Schizophr. Res., 65, 19–25. [DOI] [PubMed] [Google Scholar]
- 17. Shi J., Levinson D.F., Duan J., Sanders A.R., Zheng Y., Pe’er I., Dudbridge F., Holmans P.A., Whittemore A.S., Mowry B.J. et al. (2009) Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature., 460, 753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benros M.E., Nielsen P.R., Nordentoft M., Eaton W.W., Dalton S.O. and Mortensen P.B. (2011) Autoimmune diseases and severe infections as risk factors for schizophrenia: a 30-year population-based register study. Am. J. Psychiatry., 168, 1303–1310. [DOI] [PubMed] [Google Scholar]
- 19. Luo Y., He H., Zhang M., Huang X., Zhang J., Zhou Y., Liu X. and Fan N. (2014) Elevated serum levels of TNF-α, IL-6 and IL-18 in chronic schizophrenic patients. Schizophr. Res., 159, 556–557. [DOI] [PubMed] [Google Scholar]
- 20. Birnbaum R., Jaffe A.E., Chen Q., Shin J.H., BrainSeq Consortium, Kleinman J.E., Hyde T.M. and Weinberger D.R. (2018) Investigating the neuroimmunogenic architecture of schizophrenia. Mol. Psychiatry., 23, 1251–1260. [DOI] [PubMed] [Google Scholar]
- 21. Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V. et al. (2016) Schizophrenia risk from complex variation of complement component 4. Nature., 530, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown A. S., and Meyer U. (2018) Maternal immune activation and neuropsychiatric illness: a translational research perspective. Am. J. Psychiatry. doi: 10.1176/appi.ajp.2018.17121311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reble E., Dineen A. and Barr C.L. (2017) The contribution of alternative splicing to genetic risk for psychiatric disorders. Genes Brain Behav., 17, e12430–e12412. [DOI] [PubMed] [Google Scholar]
- 24. Lalonde E., Ha K.C.H., Wang Z., Bemmo A., Kleinman C.L., Kwan T., Pastinen T. and Majewski J. (2011) RNA sequencing reveals the role of splicing polymorphisms in regulating human gene expression. Genome Res., 21, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vanhaesebroeck B., Welham M.J., Kotani K., Stein R., Warne P.H., Zvelebil M.J., Higashi K., Volinia S., Downward J. and Waterfield M.D. (1997) P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. U. S. A., 94, 4330–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Low P.C., Manzanero S., Mohannak N., Narayana V.K., Nguyen T.H., Kvaskoff D., Brennan F.H., Ruitenberg M.J., Gelderblom M., Magnus T. et al. (2014) PI3Kδ inhibition reduces TNF secretion and neuroinflammation in a mouse cerebral stroke model. Nat. Comms., 5,–12. [DOI] [PubMed] [Google Scholar]
- 27. Eickholt B.J., Ahmed A.I., Davies M., Papakonstanti E.A., Pearce W., Starkey M.L., Bilancio A., Need A.C., Smith A.J.H., Hall S.M. et al. (2007) Control of axonal growth and regeneration of sensory neurons by the p110delta PI 3-kinase. PLoS One., 2, e869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmidt J.T., Mariconda L., Morillo F. and Apraku E. (2014) A role for the polarity complex and PI3 kinase in branch formation within retinotectal arbors of zebrafish. Dev. Neurobiol., 74, 591–601. [DOI] [PubMed] [Google Scholar]
- 29. Papaleo F., Yang F., Paterson C., Palumbo S., Carr G.V., Wang Y., Floyd K., Huang W., Thomas C.J., Chen J. et al. (2016) Behavioral, neurophysiological, and synaptic impairment in a transgenic Neuregulin1 (NRG1-IV) murine schizophrenia model. J. Neurosci., 36, 4859–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ejeskär K., Vickes O., Kuchipudi A., Wettergren Y., Uv A. and Rotter Sopasakis V. (2015) The unique non-catalytic C-terminus of P37delta-PI3K adds proliferative properties in vitro and in vivo. PLoS ONE., 10, e0127497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barry G. (2014) Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatry., 19, 410–416. [DOI] [PubMed] [Google Scholar]
- 32. Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F. and Mattick J.S. (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U. S. A., 105, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jasinska A.J., Zelaya I., Service S.K., Peterson C.B., Cantor R.M., Choi O.-W., DeYoung J., Eskin E., Fairbanks L.A., Fears S. et al. (2017) Genetic variation and gene expression across multiple tissues and developmental stages in a nonhuman primate. Nat. Genet., 49, 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kramer M., Sponholz C., Slaba M., Wissuwa B., Claus R.A., Menzel U., Huse K., Platzer M. and Bauer M. (2013) Alternative 5' untranslated regions are involved in expression regulation of human heme oxygenase-1. PLoS One., 8, e77224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang G., Guo X. and Floros J. (2005) Differences in the translation efficiency and mRNA stability mediated by 5'-UTR splice variants of human SP-A1 and SP-A2 genes. Am. J. Physiol. Lung Cell Mol. Physiol., 289, L497–L508. [DOI] [PubMed] [Google Scholar]
- 36. Fransson S. and Ejeskär K. (2013) High level of p37δ-mRNA relative to p110δ-mRNA in neuroblastoma tumors correlates with poor patient survival. Med. Oncol., 30, 724. [DOI] [PubMed] [Google Scholar]
- 37. Lage K., Hansen N.T., Karlberg E.O., Eklund A.C., Roque F.S., Donahoe P.K., Szallasi Z., Jensen T.S. and Brunak S. (2008) A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc. Natl. Acad. Sci. U. S. A., 105, 20870–20875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Birnbaum R. and Weinberger D.R. (2017) Genetic insights into the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci., 18, 727–740. [DOI] [PubMed] [Google Scholar]
- 39. Birnbaum R., Jaffe A.E., Chen Q., Hyde T.M., Kleinman J.E. and Weinberger D.R. (2015) Investigation of the prenatal expression patterns of 108 schizophrenia-associated genetic loci. Biol. Psychiatry., 77, e43–e51. [DOI] [PubMed] [Google Scholar]
- 40. Ravasi T. (2005) Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res., 16, 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Derrien T., Guigó R. and Johnson R. (2011) The long non-coding RNAs: a new (P) layer in the “Dark Matter”. Front. Genet., 2, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sinkus M.L., Lee M.J., Gault J., Logel J., Short M., Freedman R., Christian S.L., Lyon J. and Leonard S. (2009) A 2-base pair deletion polymorphism in the partial duplication of the alpha7 nicotinic acetylcholine gene (CHRFAM7A) on chromosome 15q14 is associated with schizophrenia. Brain Res., 1291,–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sinkus M.L., Adams C.E., Logel J., Freedman R. and Leonard S. (2013) Expression of immune genes on chromosome 6p21.3-22.1 in schizophrenia. Brain Behav. Immun., 32, 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bongiovanni R., Leonard S. and Jaskiw G.E. (2013) A simplified method to quantify dysregulated tyrosine transport in schizophrenia. Schizophr. Res., 150, 386–391. [DOI] [PubMed] [Google Scholar]
- 45. Canastar A., Logel J., Graw S., Finlay-Schultz J., Osborne C., Palionyte M., Drebing C., Plehaty M., Wilson L., Eyeson R. and Leonard S. (2012) Promoter methylation and tissue-specific transcription of the α7 nicotinic receptor gene, CHRNA7. J. Mol. Neurosci., 47, 389–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


