Abstract
RAD51, is a key homologous recombination protein that repairs DNA damage and maintains gene diversity and stability. Previous studies have demonstrated that the over-expression of RAD51 is associated with chemotherapy resistance of tumor cells to chemotherapy, and enhanced activity of DNA damage repair (DDR) systems contributes to resistance of adult T-cell leukemia-lymphoma (ATL) resistance to chemotherapy. Thus, targeting RAD51 is a potential strategy for the sensitization of ATL cells to chemotherapeutic drugs by inducing DNA damage. In general, cells can repair minor DNA damage through DDR; however, serious DNA damage may cause cell toxicity in cells which cannot be restored. In the present, down regulation of RAD51 by shRNA and imatinib sensitized Jurkat cells to etoposide by decreasing the activity of homologous recombination (HR). We found that the suppression of RAD51 by shRNA inhibited tumor cells proliferation and enhanced apoptosis of Jurkat cells after etoposide treatment. Importantly, downregulation of RAD51 by imatinib obviously increased the apoptosis of Jurkat cell after etoposide treatment. These results demonstrated that RAD51 may be of great value to as a novel target for the clinical treatment of adult T-cell leukemia-lymphoma (ATL), and it may improve the survival of leukemia patients.
Keywords: DNA repair protein RAD51 homolog 1, adult T-cell leukemia-lymphoma, homologous recombination repair, imatinib, DNA damage
Introduction
A number of DNA-damaging chemotherapeutic agents used for the treatment of adult T-cell leukemia-lymphoma (ATL) result in cell death diametrically by inducing DNA damage. Examples include etoposide and anthracyclines, which are currently the most common treatments used in ATL (1,2). DNA damage causes cell cycle arrest and cell death. Therefore, drug resistance represents a challenge in the clinical treatment of leukemia, as strengthened DNA damage repair (DDR) serves a crucial role in the resistance of ATL cells to chemotherapy (3–5). Thus, targeting DNA repair pathways may be an effective strategy for eradicating leukemia cells resistant to chemotherapy.
An intact DDR system is critical for maintaining genomic stability and cell proliferation (6). Although in eukaryotes, DNA double-strand breaks (DSBs) are repaired by non-homologous end joining (NHEJ) mechanisms or homologous recombination (HR) (2,7), HR has high fidelity. Abnormal DSB repair induces various types of chromosomal aberrations, including deletions (loss of heterozygosity), aneuploidy and chromosomal translocations-events which are particularly relevant in carcinogenesis (6).
DNA repair protein RAD51 homolog 1 (RAD51) is a central protein in the homologous recombination (HR) repair pathway (8). The role of RAD51 is to recognize the homologous sequence and facilitate homologous pairing and DNA strand exchange, as well as to complete DNA replication using homologous sequences as a template (9,10). A previous study revealed that RAD51 is overexpressed in several tumors (11). The overexpression of RAD51 is also associated with increased tumor metastasis (12), high tumor grade (13), treatment resistance (14) and poor overall survival rate (15). Knockout of RAD51 disrupts the HR pathway and causes embryonic death in vertebrates, whereas high expression of RAD51 increases HR efficiency and results in tumor resistance (16). Therefore, the focus of the present study was RAD51 as a treatment target, in order to determine whether the recovery of its normal expression level would reduce tumor resistance, and even increase its sensitivity to DNA-damaging drugs.
Imatinib is a platelet-derived growth factor receptor tyrosine kinase, which is an inhibitor of proto-oncogenes c-KIT and c-ABL. Previous studies have demonstrated that imatinib (Gleevec) reduces RAD51 protein repression (17,18). Synthetic lethality via targeting the DDR pathways and HR defects has had clinical success with breast cancer type 1 susceptibility protein (BRCA1) functional loss and poly(ADP-ribose) polymerase inhibition (PARPi) (19). Direct inhibition of RAD51 with RNAi and small molecule inhibitors, such as halenaquinone (20), B02 (21), RI-1 (22) and IBR2 (23) aims to reduce the expression of RAD51. Indirect inhibition is predominantly achieved by functional damage to RAD51 protein recombinase activity or interference with RAD51 protein-protein interactions, such as with tyrosine receptor kinase inhibitors (TKIs) (24), histone deacetylase inhibitors (HDACis) (25) and methylamine Pterin (26). Novel treatments targeted at RAD51 combined with PARPi have been widely reported in combination with traditional cancer therapies (27,28). Thus, the present study aimed to expand this innovative treatment to other cancer types, particularly hematological malignancies.
In the present study, RAD51 was knocked down by small hairpin (sh)RNA in Jurkat cells, one of the highest RAD51-expressing ATL cell lines. Cell proliferation and apoptosis under etoposide treatment was evaluated in cell culture. In addition, DDR efficiency and apoptosis was examined following RAD51 silencing in response to etoposide in combination with imatinib treatment.
Materials and methods
Cell lines
293T/17, Namalwa (human Burkitt lymphoma cell), Karpas-299 (human anaplastic large cell lymphoma cells), Daudi (human Burkitt lymphoma cells), HL-60 (human myeloid leukemia cells), Su-DHL-4 (human diffuse large B-cell lymphoma cells), Kasumi-1 (human acute myeloid leukemia cells), HEL (human erythroleukemia leukemia cells), K562 (human chronic myeloid leukemia cells), THP1 (human monocyte leukemia cells) and Jurkat (human T lymphocytic leukemia cells) cell lines were purchased from the Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China). The lentivirus packaging cell line 293T/17 was maintained in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA). The remaining hematological tumor cells were cultured in Iscove's modified Dulbecco's medium (IMDM) or RPMI-1640 medium (Hyclone; GE Healthcare Life Sciences). Cell cultures were supplemented with 0.1 U/ml streptomycin, 0.1 U/µl penicillin and 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The cells were cultured at 37°C in a humidified atmosphere of 5% CO2.
Gene knockdown
shRNA targeting RAD51 was cloned into the pLVX-shRNA1 vector (Clontech Laboratories, Inc., Mountainview, CA, USA) to create shRAD51 and scrambled negative control (SCR) vectors. The RAD51 interference sequences were as follows: shRNA-RAD51-1, 5′-GAAGCTATGTTCGCCATTA-3′; shRNA-RAD51-2, 5′-GCCAACGATGTGAAGAAATT-3′; shRNA-RAD51-3, 5′-AAGCTATGTTCGCCATTAA-3′; shRNA-RAD51-4, 5′-GCAGTGATGTCCTGGATAA-3′; SCR, 5′-GTTCTCCGAACGTGTCACGT-3′. 293T/17 cells with shRAD51 or SCR and lentivirus packaging plasmids were co-transfected at a ratio of 4:3:2 using calcium phosphate precipitation to produce lentiviral particles. Transduction was performed in the presence of 5 µg/ml polybrene. Subsequent experiments were performed at 72 h post-transduction.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from cell lines using TaKaRa MiniBEST Universal RNA Extraction (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's instructions. First-strand cDNA was synthesized in a 10 µl reaction volume with 5X prime Script RT Master mix (Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions. Relative mRNA expression of RAD51 and GADPH was assessed by qPCR on an Applied Biosystems 7500 Fast Real-Time PCR system (Thermo Fisher Scientific, Inc.) with SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions. The following primers were used: GAPDH forward, 5′-CTCTGATTTGGTCGTATTGGG-3′ and reverse, 5′-TGGAAGATGGTGATGGGATT-3′; RAD51 forward, 5′-GCCACCGCCCTTTACAGAACA-3′ and reverse, 5′-TGGGATCAGCAGCAAACATCG-3′. Data were analyzed using the 2−ΔΔCq method (29), where ΔCq=(Cq target gene-Cq GAPDH).
Cell apoptosis analysis
Apoptosis assays were performed using a BD Bioscience Annexin V-allophycocyanin (APC) staining kit according to the manufacturer's protocols (BD Biosciences, Franklin Lakes, NJ, USA). Cells were seeded in 24-well plates at a density of 1×105 cells/ml and cultured in a medium with etoposide (20 µM) or PBS for 4 h at 37°C. Then, the cells were washed with PBS three times and incubated for 48 h at 37°C. Cells (5–10×105) were harvested, washed with PBS three times, and stained with Annexin V-APC and propidium iodide for 15 min at room temperature. Apoptotic cells were detected by flow cytometry.
Colony-forming ability assay and cytotoxicity test
For the clonogenic assay, soft agar culture was performed with 1.2% agarose. After 10 days, the number of colonies was counted using an inverted microscope, and the proliferation ability of the cells was observed. For the cytotoxicity test, cells were collected and the cell concentration was adjusted to 105/ml; 100 µl cell suspension was seeded in each well of a 96-well plate and different concentrations of imatinib were added and cultured at 37°C for 48 h; 10 µl Cell Counting Kit (CCK)-8 reagent was then added and incubated for a further 4 h. The absorbance at 450 nm was measured with a microplate reader.
Measurement of DNA repair capacity
HR, NHEJ reporter cassettes and pDsRed-N1 as internal controls were kindly provided by Dr Zhiyong Mao from the School of Life Science and Technology of Tongji University. NHEJ or HR reporter cassettes were linearized by I-SceI endonuclease and purified using Monarch PCR & DNA Cleanup kit (cat. no. T1030S; New England BioLabs, Inc., Ipswich, MA, USA). Cells were transfected with 0.5 µg NHEJ reporter construct or 2 µg HR reporter construct, and 0.2 µg of pDsRed-N1 as internal control. Transfections were performed using an Amaxa Nucleofector (Walkersville, MD, USA). Cells were analyzed by fluorescence-activated cell sorting at 72 h post-transfection, as previously described (30).
Immunoblotting and immunofluorescence
Cells were collected and lysed in radioimmunoprecipitation assay lysis buffer (EpiZyme Biotech, Shanghai, China) lysis buffer for protein extraction. Protein concentration was determined by a bicinchonic acid protein assay and the supernatant was used for western blotting. Proteins were separated by SDS-PAGE (10% gel) and transferred to polyvinylidene difluoride membranes, and membranes were blocked in 5% non-fat dry milk. Proteins were first incubated with anti-RAD1 (1:1,000; cat. no. ab133534; Abcam, Cambridge, MA, USA) overnight at 4°C with gentle rotation, and then incubated with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (cat. no. 7074S; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA) at room temperature. Signal development was performed using an enhanced chemiluminescence kit (EpiZyme Biotech).
For immunofluorescence analysis, cells grown on coverslips were fixed in 4% paraformaldehyde, permeabilized with PBS containing 0.1% Triton X-100 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and blocked with 5% bovine serum albumin (BSA) for 30 min at room temperature. Rabbit anti-human phosphorylated (γ-) H2A histone family member X (H2AX; 1:1,000; cat. no. ab11174; Abcam) was diluted in 5% BSA and applied at 4°C overnight. Following rinsing with PBS three times and incubating for 1 h with donkey anti-rabbit secondary antibody (cat. no. 20308-1, 1:1,000; Biotium, Hayward, CA, USA) at room temperature, the slides were washed three times in PBS and the cell nuclei were stained with DAPI (1:1,000; Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) for 10 min at room temperature. Images were captured with a Leica TCS SP2 confocal fluorescence microscope (×20; Leica Microsystems GmbH, Wetzlar, Germany).
Statistical analysis
All statistical analyses were performed using Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as the mean ± standard deviation. All quantitative experiments were conducted with a minimum of three independent experiments. For data analysis, two-tailed t-test or one-way analysis of variance followed by Tukey's post-hoc test were used. P<0.05 was considered to indicate a statistically significant difference. Flow cytometry data were analyzed with FCS Express 6 Flow software (De Novo Software, Glendale, CA, USA) and protein expression was quantified using the ImageQuant R 4.2A software (GE Healthcare Life Sciences).
Results
RAD51 is overexpressed in leukemia cells
To determine the expression of RAD51 in leukemia, the mRNA expression of RAD51 was measured in several cell lines by qPCR. Kasumi, Namalwa, Karpas 299, HL-60, Daudi, HEL, THP1, Su-4, K562 and Jurkat cells exhibited markedly higher RAD51 expression compared with the Kasumi cells, which were used as a control. The Jurkat cell line exhibited the highest level of RAD51 mRNA, which was ~7.25 times that of Kasumi cells (Fig. 1A). The Kasumi cell line was chosen as a control because it exhibited the lowest RAD51 expression of all tumor cell lines tested. Immunoblot analysis demonstrated that Jurkat cells exhibited markedly higher RAD51 protein expression compared with the other four cell lines (Fig. 1B and C). Consequently, Jurkat cells were selected for subsequent experiments.
Figure 1.
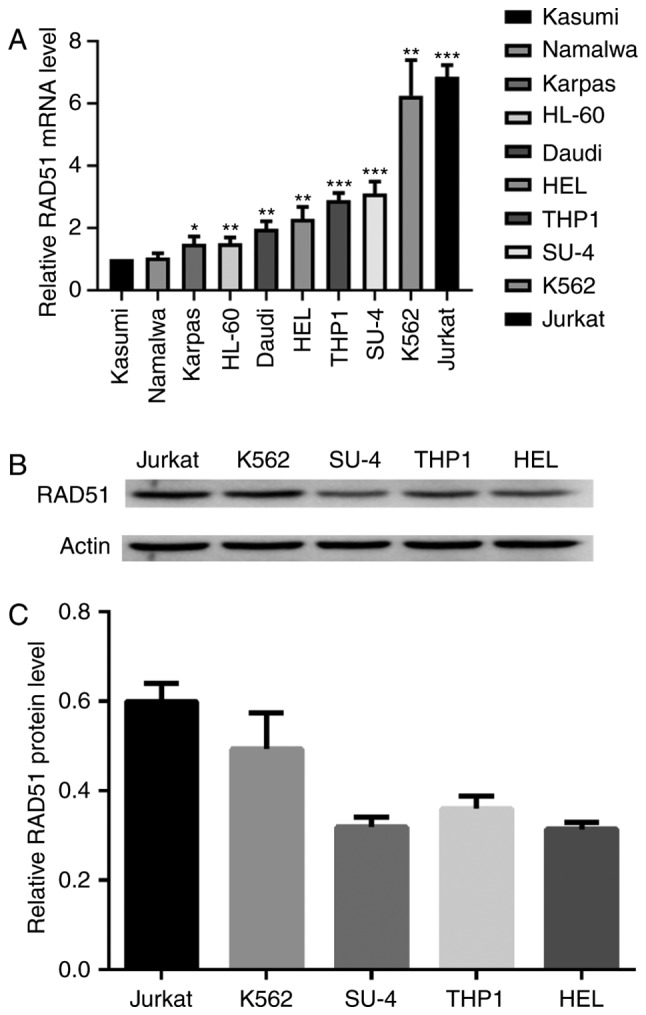
RAD51 expression is increased in leukemia cells. (A) RAD51 mRNA expression in Kasumi, Namalwa, Karpas, HL-60, Daudi, HEL, THP1, Su-DHL-4, K562, Jurkat cells was analyzed by reverse transcription-quantitative polymerase chain reaction relative to GAPDH as the internal control. *P<0.05, **P<0.01 ***P<0.001 vs. Kasumi. (B) Protein expression levels of RAD51 and β-actin in the cell lysates of Jurkat, K562, Su-DHL-4, THP1 and HEL cells was determined by immunoblotting. (C) RAD51 protein expression was quantified by densitometric analysis and normalized to GAPDH expression. The normalized RAD51 expression levels of Jurkat, K562, Su-DHL-4, THP1 and HEL are shown. Experiments were repeated at least three times. RAD51, DNA repair protein RAD51 homolog 1.
ShRNA-mediated down regulation of RAD51 in leukemia cells
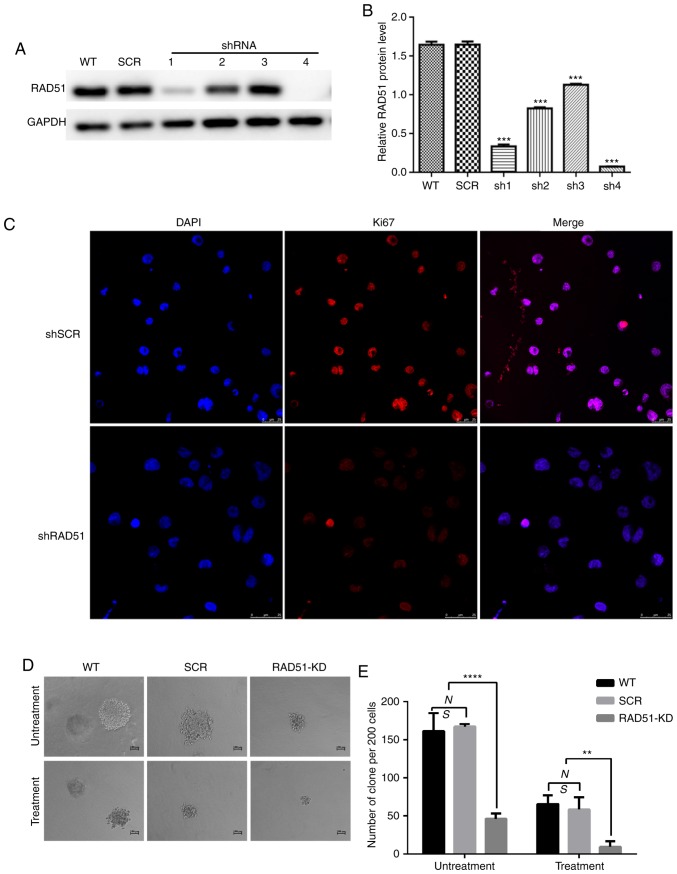
In order to determine whether the expression of RAD51 in infected Jurkat cells was decreased, and whether the four interferon fragments of RAD51 were designed to effectively degrade RAD51 mRNA, Jurkat cells were infected with lentivirus expressing shRNA targeting RAD51 (shRAD51-KD) or normal control (shRNA-SCR). As shown in Fig. 2A and B, RAD51-sh4 exerted a prominent interference effect. Consequently, RAD51-sh4 was selected for subsequent experiments.
Figure 2.
RAD51 silencing by shRNA sensitizes Jurkat cells to the DNA damage-inducing drug etoposide. (A) Protein expression of RAD51 and GAPDH in cell lysates of six different groups of Jurkat cells, including WT and Jurkat cells transduced with lentiviral vectors expressing SCR or with RAD51 shRNA1-4 clones. (B) Quantification of RAD51 interference efficiency. ***P<0.001 vs. WT. (C) Representative immunofluorescence staining for Ki67 (red) and DNA (blue) in SCR and RAD51 KD cells. (D and E) Colony formation assays were conducted using shRAD51- or SCR-transduced Jurkat cells with or without etoposide treatment. Representative images showing colony size are presented. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01, ****P<0.0001. RAD51, DNA repair protein RAD51 homolog 1; WT, wild-type; shRNA, small hairpin RNA; KD, knockdown; SCR, scramble.
RAD51 knockdown reduces cell proliferation and induces apoptosis of Jurkat cells
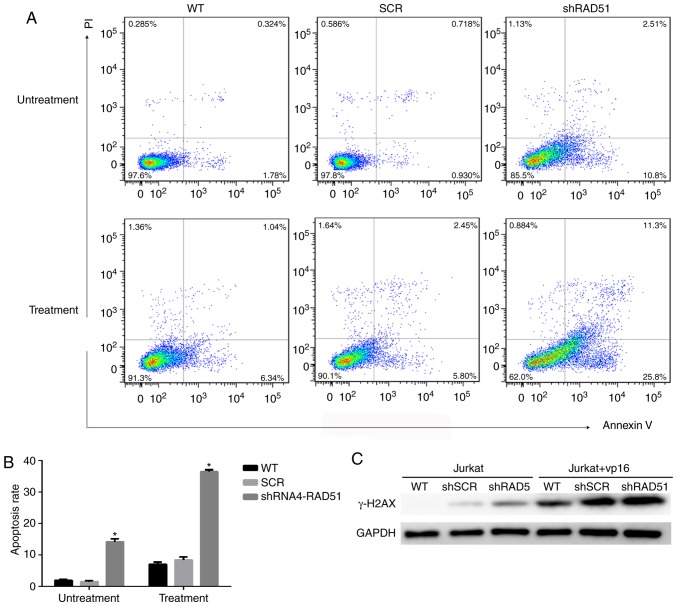
To elucidate the role of RAD51 in Jurkat cell DNA damage, RAD51 was downregulated with shRNA and the colony-forming ability and apoptotic rate was assessed following etoposide treatment. Ki67 is a well-known cell proliferation activity marker protein that reflects the proliferative activity of tumor cells (31). As shown in Fig. 2C, the results revealed that the expression of Ki67 in RAD51-KD cells was notably lower compared with that in SCR. Similarly, shRNA-mediated downregulation of RAD51 reduced the colony-forming ability of Jurkat cells by 3-fold under normal growth conditions, and further decreased the colony-forming ability of Jurkat cells by 4-fold following etoposide treatment (Fig. 2E). Furthermore, the colony size of Jurkat cells with RAD51 shRNA was markedly smaller compared with that of cells in the WT and control SCR groups (Fig. 2D). The apoptotic rate was examined by Annexin V staining coupled with flow cytometry. As shown in Fig. 3A and B, the mean apoptotic rate of Jurkat cells in the RAD51KD group was 14.12%, compared with 1.92 and 1.47% in the two negative control groups. Following etoposide treatment, the apoptotic rate of RAD51KD group was 36.42%, compared with 6.98 and 8.38% in the control groups, respectively (P<0.05). Taken together, these results demonstrated that RAD51 silencing results in reduced cell viability and an increased apoptotic rate under normal growth conditions with etoposide treatment.
Figure 3.
RAD51 knockdown induces apoptosis. (A) Effects of RAD51 knockdown on Jurkat cell apoptosis. After 3 days of lentivirus infection with negative control shRNA, WT or shRAD51, 20 µM of etoposide was added for 4 h and apoptosis was analyzed 48 h later by flow cytometry. (B) Quantification of the effects of RAD51 knockdown on Jurkat cell apoptosis. *P<0.05 vs. WT. (C) Western blot analysis of γ-H2AX expression. RAD51, DNA repair protein RAD51 homolog 1; WT, wild-type; sh, small hairpin RNA; SCR, scramble; γ-H2AX, phosphorylated H2A histone family member X.
Downregulation of RAD51 alters the efficiency of DDR in Jurkat cells
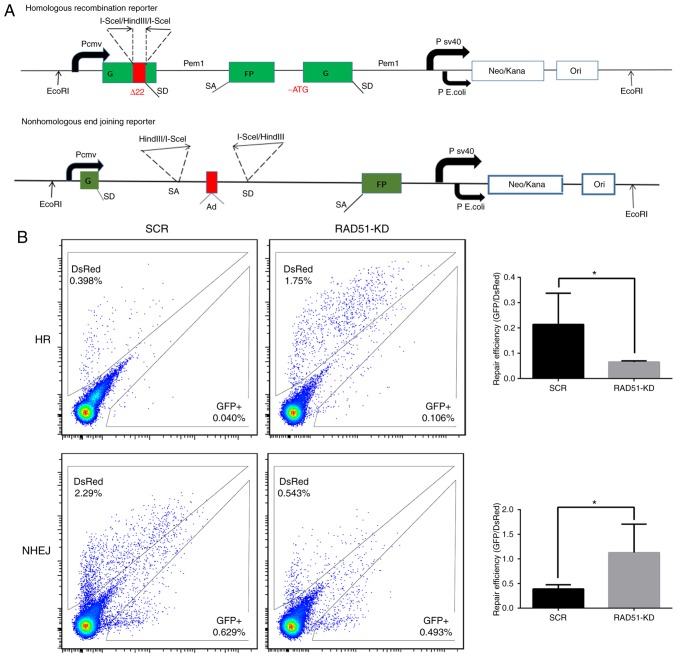
To investigate whether the increase in Jurkat cell apoptosis was associated with a decrease in DDR function, the expression of H2A histone family member X (H2AX) and the efficiency of DDR was measured in a quantitative manner in Jurkat cells. The phosphorylation of H2AX (γ-H2AX), is a sign of DNA DSBs (32). Therefore, γ-H2AX protein expression was detected in cells treated with or without etoposide. The expression of γ-H2AX was higher in Jurkat cells transduced with RAD51 shRNA compared with that with control shRNA, whereas γ-H2AX was notably higher in Jurkat cells transduced with RAD51 shRNA following etoposide treatment (Fig. 3C), indicating that RAD51 was indispensable for the repair of DSBs in Jurkat cells. Furthermore, fluorescent reporter constructs were used in which a functional GFP gene was reconstituted following an NHEJ or HR event (Fig. 4A). Notably, the results demonstrated that inhibition of RAD51 by shRNA reduced the efficiency of HR, but increased that of NHEJ (P<0.05; Fig. 4B).
Figure 4.
Inhibition of RAD51 impairs DNA repair in Jurkat cells. (A) Fluorescent reporter constructs were used to measure NHEJ or HR events. (B) Analysis of HR and NHEJ in SCR and shRAD51 Jurkat cells. Flow cytometric analysis results (left panel) show the gating for the analysis of GFP+ and DsRed+ cells using cells transfected with GFP or DsRed expression vectors, as well as cells transfected with a negative control plasmid to exclude auto-fluorescent cells. The ratio of GFP+ to DsRed+ cells (right panel), which was used as a measure of repair efficiency, was also presented. Experiments were repeated at least three times. *P<0.05. RAD51, DNA repair protein RAD51 homolog 1; NHEJ, non-homologous end joining; HR, homologous recombination; GFP, green fluorescent protein; sh, small hairpin RNA; SCR, scramble; KD, knockdown.
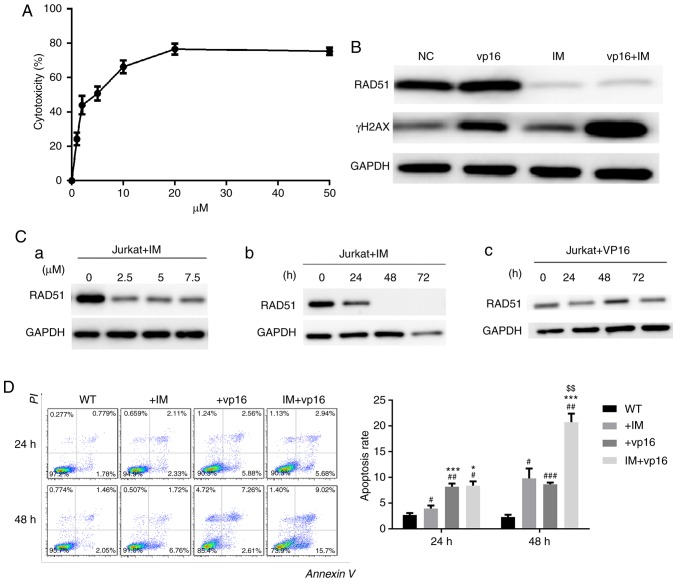
Imatinib inhibits the expression of RAD51 and enhances chemosensitivity in Jurkat cells
To test the toxicity of imatinib on Jurkat cells, CCK-8 assays were performed. Next, DSB levels in Jurkat cells following RAD51 knockdown were determined, as well as the apoptotic rate in cells treated with or without etoposide. The results demonstrated that the IC50 of imatinib toxicity to Jurkat cells was ~5 µM, and the toxic effect of imatinib on Jurkat cells at 20 µM stabilized (Fig. 5A).
Figure 5.
Imatinib inhibits the expression of RAD51 and enhances chemosensitivity in Jurkat cells. (A) The toxicity of imatinib to Jurkat cells was examined by Cell Counting Kit-8 assays. (B) Western blot analysis of RAD51 and γ-H2AX expression following the combinatory treatment of imatinib and etoposide. (C) IM reduced the expression of RAD51 in a (a) dose and (b) time-dependent manner. (c) Etoposide was used as a control. (D) Combined use of IM and etoposide increased the sensitivity of Jurkat cells to chemotherapy. *P<0.05, ***P<0.001 vs. IM group; $$P<0.01 vs. VP16 group; #P<0.05, ##P<0.01, ###P<0.001 vs. WT group. N=3. RAD51, DNA repair protein RAD51 homolog 1; γ-H2AX, phosphorylated H2A histone family member X; NC, negative control; WT, wild-type; IM, imatinib; PI, propidium iodide.
Using the IC50 of imatinib obtained from the experiment described above, imatinib reduced RAD51 protein expression, and this reduction in RAD51 was dose- and time-dependent (Fig. 5C-a and -b). In contrast, etoposide (VP16), which was used as a reference, did not reduce RAD51 expression, but increased it at 48 h, which may be associated with DDR initiation (Fig. 5C-c). The expression of γ-H2AX demonstrated that the combination therapy group had the most severe DSB injury, i.e., the weakest ability to repair DNA damage (Fig. 5B). Similarly, apoptosis evaluation indicated that combination therapy markedly increased Jurkat cell apoptosis. These results suggested that imatinib may specifically reduce RAD51 protein (Fig. 5D) in a dose- and time-dependent manner, and this downregulation of RAD51 by imatinib triggers apoptosis in Jurkat cells following etoposide treatment.
Discussion
In the present study, it was demonstrated that RAD51 was overexpressed in Jurkat cells, whereas RAD51 downregulation by shRNA and imatinib decreased cell viability and increased apoptosis in Jurkat cells treated with etoposide. Furthermore, RAD51 downregulation resulted in impaired DNA repair capacity, accompanied by decreased HR. Furthermore, downregulation of RAD51 by imatinib markedly increased the rate of apoptosis in combination with etoposide treatment. These results suggested that targeting RAD51 may be a potential strategy for the sensitization of ATL to DNA damage-based chemotherapy.
RAD51 is a central protein in HR repair pathways, which serves a vital role in the pathogenesis and drug resistance of leukemia (33,34). Although several tumor cells have been found to overexpress RAD51 (11,15), the underlying reason remains unclear. An increasing number of studies have demonstrated that mammalian RAD51 proteins are linked directly or indirectly to a number of other proteins, such as anti-tumor factors serine-protein kinase ATM (35), p53 (36), BRCA1/BRCA2 (27,28), proto-oncogene c-Abl (37) and SUMO-conjugating enzyme UBC9 (38). This prompted us to investigate the role of RAD51 in tumor development.
In the present study, downregulation of RAD51 by shRNA and imatinib reduced the efficiency of HR repair and increased chemosensitivity and apoptosis in ATL. These results suggested that RAD51 has the potential to become a novel target for the clinical treatment of acute leukemia, with the hope of increasing ATL patient survival rate.
Etoposide is a topoisomerase II (TOP2) inhibitor which is widely used as an anticancer drug. It increases the expression of the TOP2 cleavage complex and thus increases TOP2-mediated chromosome DNA breakage (39,40). In the present study, cells were treated with etoposide at a concentration of 20 µM for 4 h with etoposide to induce DNA damage (41), and all the analyses were performed 48 h later. The results demonstrated that downregulation of RAD51 reduced cell proliferation and increased the rate of apoptosis after etoposide treatment-induced DNA damage. This result may be related to DDR capacity, particularly HR abnormalities. The result of the subsequent experiments demonstrated showed that HR pathway repair in Jurkat cells was attenuated by RAD51 knockdown ed the rate of apoptosis following DNA damage. This result may be related to DDR capacity, particularly HR abnormalities. The result of the subsequent experiments demonstrated showed that HR pathway repair in Jurkat cells was attenuated by RAD51 knockdown and switched to the NHEJ pathway. This alteration resulted in an increase in apoptosis resulting from incomplete repair of Jurkat cells. A limitation of the present study was that whether key proteins in the NHEJ pathway were upregulated was not determined, so changes in the repair pathways could not be further confirmed.
Several small molecule RAD51 inhibitors have been developed (42,43). However, they lack specificity for RAD51 and are available only for in vitro studies of RAD51 activity. As shRNA is not currently applied in clinical treatment, inhibition of RAD51 with imatinib was also used in the current study (24). Imatinib is the first-line therapy for chronic myelocytic leukemia. It has been reported that imatinib treatment reduces the expression of RAD51 and is closely associated with reduced HR in tumor cell lines with different p53 states (18). Treatment of tumor cells with imatinib enhances sensitivity (24), but this effect does not occur in normal fibroblasts. In irradiated tumors, mitomycin, gemcitabine combined with imatinib decreases tumor cell proliferation. This synergistic effect was also demonstrated in vivo using a PC3 mouse tumor model: Combination of imatinib and radiotherapy alone significantly delayed tumor growth, at least partially due to a decrease in RAD51 expression (24). The results of the present study demonstrated that imatinib reduced RAD51 protein in ATL cells in a dose- and time-dependent manner. Therefore, the combined treatment of imatinib and chemotherapeutic drugs may be useful for the treatment of hematological tumors. Imatinib reduced the expression of RAD51, but the exact mechanism of how imatinib reduces RAD51 expression has not been fully elucidated; this requires further investigation in future experiments. For more far-reaching mechanisms, it will be necessary to determine the DNA damage response caused by RAD51 overexpression.
In conclusion, the RAD51 protein is key to HR repair pathways and was involved in the occurrence and drug resistance of leukemia. Increased expression of RAD51 recombination protein in various tumors is a common phenomenon (11). Acute leukemia is a malignancy with poor treatment outcomes (3). Although RNAi technology targets gene activity by silencing and has very high specificity, the clinical application of siRNA is currently limited by its off-target effects and short life span. The limitations of this study include the lack of data from peripheral blood samples and a non-cancerous cell line. In the present study, no normal peripheral blood samples or non-cancerous cell lines were used as negative controls; therefore, the experimental results can only indicate that RAD51 may serve an important role in blood cancer cell lines.
In the present experiment, RAD51 knockdown decreased the repair efficiency of Jurkat cells and increased their chemosensitivity, ultimately leading to cell apoptosis. Based on these results, RAD51 appears to be promising as a novel target for the clinical treatment of leukemia, and it may improve the survival of leukemia patients.
Acknowledgements
Not applicable.
Funding
This study was supported by Ministry of Science and Technology of China (grant no. 2016YFE0107200) and the National Natural Science Foundation of China (grant no. 81770151).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
MY, XT and ZF designed the research and performed experiments. WY, ZL, WZ, JZ and AL collected the samples, and participated in the collection and analysis of data. XT wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rich T, Allen R, Wyllie AH. Defying death after DNA damage. Nature. 2000;407:777–783. doi: 10.1038/35037717. [DOI] [PubMed] [Google Scholar]
- 2.Khanna KK, Jackson SP. DNA double-strand breaks: Signaling, repair and the cancer connection. Nature Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 3.Linker C, Damon L, Ries C, Navarro W. Intensified and shortened cyclical chemotherapy for adult acute lymphoblastic leukemia. J Clin Oncol. 2002;20:2464–2471. doi: 10.1200/JCO.2002.07.116. [DOI] [PubMed] [Google Scholar]
- 4.O'Grady S, Finn SP, Cuffe S, Richard DJ, O'Byrne KJ, Barr MP. The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat Rev. 2014;40:1161–1170. doi: 10.1016/j.ctrv.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Hijiya N, Thomson B, Isakoff MS, Silverman LB, Steinherz PG, Borowitz MJ, Kadota R, Cooper T, Shen V, Dahl G, et al. Phase 2 trial of clofarabine in combination with etoposide and cyclophosphamide in pediatric patients with refractory or relapsed acute lymphoblastic leukemia. Blood. 2011;118:6043–6049. doi: 10.1182/blood-2011-08-374710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 7.Kanaar R, Hoeijmakers JH, van Gent DC. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/S0962-8924(98)01383-X. [DOI] [PubMed] [Google Scholar]
- 8.Arnaudeau C, Helleday T, Jenssen D. The RAD51 protein supports homologous recombination by an exchange mechanism in mammalian cells. J Mol Biol. 1999;289:1231–1238. doi: 10.1006/jmbi.1999.2856. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 10.Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: Roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol. 2011;22:898–905. doi: 10.1016/j.semcdb.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 11.Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 2002;62:219–225. [PubMed] [Google Scholar]
- 12.Wiegmans AP, Al-Ejeh F, Chee N, Yap PY, Gorski JJ, Da Silva L, Bolderson E, Chenevix-Trench G, Anderson R, Simpson PT, et al. Rad51 supports triple negative breast cancer metastasis. Oncotarget. 2014;5:3261–3272. doi: 10.18632/oncotarget.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maacke H, Opitz S, Jost K, Hamdorf W, Henning W, Krüger S, Feller AC, Lopens A, Diedrich K, Schwinger E, Stürzbecher HW. Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int J Cancer. 2000;88:907–913. doi: 10.1002/1097-0215(20001215)88:6<907::AID-IJC11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 14.Short SC, Giampieri S, Worku M, Alcaide-German M, Sioftanos G, Bourne S, Lio KI, Shaked-Rabi M, Martindale C. Rad51 inhibition is an effective means of targeting DNA repair in glioma models and CD133+ tumor-derived cells. Neuro Oncol. 2011;13:487–499. doi: 10.1093/neuonc/nor010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbano R, Copetti M, Perrone G, Pazienza V, Muscarella LA, Balsamo T, Storlazzi CT, Ripoli M, Rinaldi M, Valori VM, et al. High RAD51 mRNA expression characterize estrogen receptor-positive/progesteron receptor-negative breast cancer and is associated with patient's outcome. Int J Cancer. 2011;129:536–545. doi: 10.1002/ijc.25736. [DOI] [PubMed] [Google Scholar]
- 16.Richardson C. RAD51, genomic stability, and tumorigenesis. Cancer Lett. 2005;218:127–139. doi: 10.1016/j.canlet.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Kubler HR, van Randenborgh H, Treiber U, Wutzler S, Battistel C, Lehmer A, Lehmer A, Wagenpfeil S, Hartung R, Paul R. In vitro cytotoxic effects of imatinib in combination with anticancer drugs in human prostate cancer cell lines. Prostate. 2005;63:385–394. doi: 10.1002/pros.20201. [DOI] [PubMed] [Google Scholar]
- 18.Ishida T, Takizawa Y, Kainuma T, Inoue J, Mikawa T, Shibata T, Suzuki H, Tashiro S, Kurumizaka H. DIDS, a chemical compound that inhibits RAD51-mediated homologous pairing and strand exchange. Nucleic Acids Res. 2009;37:3367–3376. doi: 10.1093/nar/gkp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 20.Takaku M, Kainuma T, Ishida-Takaku T, Ishigami S, Suzuki H, Tashiro S, van Soest RW, Nakao Y, Kurumizaka H. Halenaquinone, a chemical compound that specifically inhibits the secondary DNA binding of RAD51. Genes Cell. 2011;16:427–436. doi: 10.1111/j.1365-2443.2011.01494.x. [DOI] [PubMed] [Google Scholar]
- 21.Huang F, Motlekar NA, Burgwin CM, Napper AD, Diamond SL, Mazin AV. Identification of specific inhibitors of human RAD51 recombinase using high-throughput screening. ACS Chem Biol. 2011;6:628–635. doi: 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budke B, Kalin JH, Pawlowski M, Zelivianskaia AS, Wu M, Kozikowski AP, Connell PP. An optimized RAD51 inhibitor that disrupts homologous recombination without requiring Michael acceptor reactivity. J Med Chem. 2013;56:254–263. doi: 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Zhou L, Wu G, Konig H, Lin X, Li G, Qiu XL, Chen CF, Hu CM, Goldblatt E, et al. A novel small molecule RAD51 inactivator overcomes imatinib-resistance in chronic myeloid leukaemia. EMBO Mol Med. 2013;5:353–365. doi: 10.1002/emmm.201201760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhury A, Zhao H, Jalali F, Al Rashid S, Ran J, Supiot S, Kiltie AE, Bristow RG. Targeting homologous recombination using imatinib results in enhanced tumor cell chemosensitivity and radiosensitivity. Mol Cancer Ther. 2009;8:203–213. doi: 10.1158/1535-7163.MCT-08-0959. [DOI] [PubMed] [Google Scholar]
- 25.Kachhap SK, Rosmus N, Collis SJ, Kortenhorst MS, Wissing MD, Hedayati M, Shabbeer S, Mendonca J, Deangelis J, Marchionni L, et al. Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS One. 2010;5:e11208. doi: 10.1371/journal.pone.0011208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du LQ, Du XQ, Bai JQ, Wang Y, Yang QS, Wang XC, Zhao P, Wang H, Liu Q, Fan FY. Methotrexate-mediated inhibition of RAD51 expression and homologous recombination in cancer cells. J Cancer Res Clin Oncol. 2012;138:811–818. doi: 10.1007/s00432-011-1132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/S0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 28.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Seluanov A, Mao Z, Gorbunova V. Analysis of DNA double-strand break (DSB) repair in mammalian cells. J Vis Exp. 2010;8:2002. doi: 10.3791/2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholzen T, Gerdes J. The Ki-67 protein: From the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 32.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 33.Maacke H, Jost K, Opitz S, Miska S, Yuan Y, Hasselbach L, Lüttges J, Kalthoff H, Stürzbecher HW. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000;19:2791–2795. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- 34.Henning W, Stürzbecher HW. Homologous recombination and cell cycle checkpoints: Rad51 in tumour progression and therapy resistance. Toxicology. 2003;193:91–109. doi: 10.1016/S0300-483X(03)00291-9. [DOI] [PubMed] [Google Scholar]
- 35.Chen G, Yuan SS, Liu W, Xu Y, Trujillo K, Song B, Cong F, Goff SP, Wu Y, Arlinghaus R, et al. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J Biol Chem. 1999;274:12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 36.Stürzbecher HW, Donzelmann B, Henning W, Knippschild U, Buchhop S. P53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15:1992–2002. doi: 10.1002/j.1460-2075.1996.tb00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, Wang R, Sung P, Shinohara A, Weichselbaum R, Kufe D. Regulation of Rad51 function by c-Abl in response to DNA damage. J Biol Chem. 1998;273:3799–3802. doi: 10.1074/jbc.273.7.3799. [DOI] [PubMed] [Google Scholar]
- 38.Saitoh H, Pizzi MD, Wang J. Perturbation of SUMOlation enzyme Ubc9 by distinct domain within nucleoporin RanBP2/Nup358. J Biol Chem. 2002;277:4755–4763. doi: 10.1074/jbc.M104453200. [DOI] [PubMed] [Google Scholar]
- 39.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu CC, Li TK, Farh L, Lin LY, Lin TS, Yu YJ, Yen TJ, Chiang CW, Chan NL. Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science. 2011;333:459–462. doi: 10.1126/science.1204117. [DOI] [PubMed] [Google Scholar]
- 41.Muslimovic A, Nystrom S, Gao Y, Hammarsten O. Numerical analysis of etoposide induced DNA breaks. PLoS One. 2009;4:e5859. doi: 10.1371/journal.pone.0005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Z, Sun B, Clewell RA, Adeleye Y, Andersen ME, Zhang Q. Dose-response modeling of etoposide-induced DNA damage response. Toxicol Sci. 2014;137:371–384. doi: 10.1093/toxsci/kft259. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, He Y, Luo Y. Crystal structure of an archaeal Rad51 homologue in complex with a metatungstate inhibitor. Biochemistry. 2009;48:6805–6810. doi: 10.1021/bi900832t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.