Summary
Botrytis cinerea is the causal agent of grey mould for more than 200 plant species, including economically important vegetables, fruits and crops, which leads to economic losses worldwide. Target of rapamycin (TOR) acts a master regulator to control cell growth and proliferation by integrating nutrient, energy and growth factors in eukaryotic species, but little is known about whether TOR can function as a practicable target in the control of plant fungal pathogens. Here, we characterize TOR signalling of B. cinerea in the regulation of growth and pathogenicity as well as its potential value in genetic engineering for crop protection by bioinformatics analysis, pharmacological assays, biochemistry and genetics approaches. The results show that conserved TOR signalling occurs, and a functional FK506‐binding protein 12 kD (FKBP12) mediates the interaction between rapamycin and B. cinerea TOR (BcTOR). RNA sequencing (RNA‐Seq) analysis revealed that BcTOR displayed conserved functions, particularly in controlling growth and metabolism. Furthermore, pathogenicity assay showed that BcTOR inhibition efficiently reduces the infection of B. cinerea in plant leaves of Arabidopsis and potato or tomato fruits. Additionally, transgenic plants expressing double‐stranded RNA of BcTOR through the host‐induced gene silencing method could produce abundant small RNAs targeting BcTOR, and significantly block the occurrence of grey mould in potato and tomato. Taken together, our results suggest that BcTOR is an efficient target for genetic engineering in control of grey mould, and also a potential and promising target applied in the biocontrol of plant fungal pathogens.
Keywords: Botrytis cinerea, host‐induced gene silencing, mycelial growth, pathogenicity, target of rapamycin
Introduction
Botrytis cinerea, a widespread necrotrophic fungal plant pathogen that can infect more than 200 plant species, including economically important vegetables, fruits and crops, and lead to grey mould rot or botrytis blight, causes widespread losses every year worldwide (Choquer et al., 2007; Dean et al., 2012; Williamson et al., 2007). More than 40% of these losses occur in greenhouse‐grown and field‐grown crops if chemical control is not used (Pedras et al., 2011; Villa‐Rojas et al., 2012). Thus, chemical pesticides are widely required for the control of B. cinerea. However, the widespread use of chemical pesticides results in serious water, soil, food and environmental pollution (Malhat et al., 2015; Oliveira et al., 2015; Tomenson and Matthews, 2009). To overcome this issue, biofungicides with low toxicity and high efficiency against key targets of pathogens have been developed and applied to control plant fungal disease. In addition, genetic engineering methods have also been extensively employed to improve resistance to plant pathogens in crop protection. For example, host‐induced gene silencing (HIGS), an RNA interference (RNAi)‐based approach in which small RNAs (sRNAs) are produced by the host plant to target invader transcripts, has emerged as an effective strategy for improving plant resistance against pathogens (Cai et al., 2018; Wang, Thomas et al., 2017).
HIGS has been widely used for crop protection and a wide range of transgenic crops acquire durable resistance against diseases by expressing double‐stranded RNA (dsRNA) that is subsequently processed into sRNAs targeting essential genes regulating growth or pathogenicity (Cheng et al., 2015; Ghag et al., 2014; Govindarajulu et al., 2015; Koch et al., 2013; Nowara et al., 2010; Nunes and Dean, 2012; Xu et al., 2018; Zhang et al., 2016; Zhu et al., 2017). For instance, engineered corn plants expressing a vacuolar ATPase (V‐ATPase) dsRNA can be protected from western corn rootworm feeding damage; durable resistance to Fusarium head blight and seedling blight can be acquired by wheat through HIGS targeting to an essential chitin synthase gene Chs3b; and the virulence genes of Verticillium dahliae can be silenced by plant‐mediated RNAi, compromising verticillium wilt in tomato and Arabidopsis (Baum et al., 2007; Cheng et al., 2015; Song and Thomma, 2018). In a study to control B. cinerea, Arabidopsis plants with hairpin RNAs simultaneously targeting dicer‐like genes of B. cinerea and V. dahliae exhibited enhanced resistance to both pathogens (Wang et al., 2016).
Target of rapamycin (TOR), a Ser/Thr protein kinase of large molecular weight with multiple functional domains, is a well‐known essential gene in eukaryotic species, but little is known about whether or not TOR can be developed and established as a key target for biological control of plant fungal pathogens. TOR was first discovered in yeast (Saccharomyces cerevisiae) by a genetic screen of mutants insensitive to rapamycin (Heitman et al., 1991). Rapamycin, a well‐known medicine produced by Streptomyces hygroscopicus, is able to efficiently repress TOR kinase activity. FK506 binding protein 12 kD (FKBP12), as a receptor of rapamycin, mediates inhibition of TOR by rapamycin and its defects confer resistance to rapamycin (Heitman et al., 1991). In the presence of rapamycin, FKBP12 forms a binary complex with rapamycin and further interacts with the FKBP12‐rapamycin binding domain (FRB) of TOR to form a ternary complex, resulting in abolishment of TOR activity (Chiu et al., 1994; Heitman et al., 1991; Koltin et al., 1991; Sabatini et al., 1994; Vezina et al., 1975). Based on the rapamycin‐FKBP12‐TOR system, the TOR signalling pathway has been extensively studied and gradually elucidated in yeast and mammals (Benjamin et al., 2011). The TOR protein consists of five highly conserved domains: HEAT repeats (Huntingtin, elongation factor 3 (EF3), a subunit of protein phosphatase 2A (PP2A) and TOR1), a FAT (FRAP, ATM and TRRAP) domain, a FRB (FKBP12‐rapamycin binding) domain, a kinase and a FATC (carboxy‐terminal FAT) domain, which reside in the TOR protein between the N‐terminal and the C‐terminal (Baretic and Williams., 2014; Sauer et al., 2013; Yang et al., 2013). Most eukaryotic organisms have one copy of the TOR gene, but two and three TOR genes have also been identified in yeast and Leishmania major, respectively (Heitman et al., 1991; Madeira da Silva and Beverley, 2010). In some eukaryotes, based on diverse components recruited by TOR, the TOR protein can form two different types of multiprotein complex: TOR complex 1 (TORC1) and TOR complex 2 (TORC2). The TOR protein recruits a regulatory‐associated protein of TOR (RAPTOR) and a lethal with SEC13 protein8 (LST8) to form TORC1, but combines LST8, stress‐activated map kinase‐interacting protein 1 (SIN1) and rapamycin‐insensitive companion of TOR (RICTOR) to form TORC2 in yeast and animals (Loewith et al., 2002; Wullschleger et al., 2006). Due to this difference, TORC1 is sensitive to rapamycin, but TORC2 shows insensitivity to rapamycin (Loewith et al., 2002). In addition, rapamycin‐sensitive TORC1 plays a major role in cell growth, development and proliferation in a temporal manner, while rapamycin‐resistant TORC2 seems to particularly regulate the development of the cell cytoskeleton (Feldman et al., 2009; Loewith et al., 2002; Takahara and Maeda, 2013; Wang and Proud, 2009; Wullschleger et al., 2006).
Various studies have shown that TOR is a central coordinator of energy, nutrient and stress signalling networks from yeast to mammals and plants (Dobrenel et al., 2016; Henriques et al., 2014; Loewith and Hall, 2011; Rexin et al., 2015; Schmelzle and Hall, 2000; Xiong and Sheen, 2015). However, little attention has been paid to TOR signaling study in phytopathogenic fungi are relatively less studied. Rapamycin was discovered in the early 1970s as an antifungal agent against the pathogenic yeast Candida albicans (Sehgal et al., 1975). Subsequent studies also found that rapamycin is effective against many phytopathogenic fungi, such as Botrytis cinerea, Mucor circinelloides, Fusarium fujikuroi, Fusarium oxysporum and Fusarium graminearum (Bastidas et al., 2012; Lopez‐Berges et al., 2010; Melendez et al., 2009; Teichert et al., 2006; Yu et al., 2014), indicating that a conserved TOR pathway also exists in plant pathogenic fungi. In addition, the research on TOR signalling in F. graminearum elucidated the TOR components and conserved functions in growth and development as well as its role in virulence (Yu et al., 2014). In B. cinerea, it was reported that a functional FKBP12 homologous protein existed with conserved function in bridging rapamycin, and the BcFKBP12 deletion relieved growth inhibition of rapamycin and reduced virulence of the strain T4 while not affecting the pathogenic development of the strain B05.10 (Gioti et al., 2006). Additionally, BcFKBP12 was reported to likely be involved in sulphur regulation and this regulation appears to be unrelated to TOR signalling (Melendez et al., 2009). To date, information on the TOR signalling pathway in B. cinerea is very limited, and TOR function remains to be determined.
In the present study, we functionally characterized B. cinerea TOR (BcTOR) in the regulation of vegetative growth and development as well as virulence of the B. cinerea strain B05.10 by bioinformatics analysis, pharmacological assays and chemical genetics approaches. We created transgenic potato and tomato plants expressing dsRNA specific to BcTOR based on the HIGS method, and found that transgenic plants could produce abundant sRNA molecules targeting BcTOR, significantly blocking the occurrence of grey mould caused by B. cinerea. Our results suggest that BcTOR can function as a potential and promising target in the control of grey mould disease.
Results
Molecular components of the TOR complex in B. cinerea
To investigate TOR signalling in B. cinerea, a yeast TOR protein sequence was used to search for the homologous sequences in the B. cinerea genome (http://fungi.ensembl.org/Botrytis_cinerea/Info/Index) and NCBI (http://www.ncbi.nlm.nih.gov/), respectively. The search results revealed a single TOR homologue gene (named BcTOR), located on the first chromosome of the B. cinerea genome (Fig. 1A). Further analysis showed that the full‐length genomic DNA of BcTOR spans about 8.12 kb and contains five exons and four introns in the B. cinerea genome (Fig. 1A), including a 7296 bp full‐length coding sequence encoding a protein of 2431 amino acid residues with a predicted molecular mass of 275 kDa. Alignment of BcTOR with TOR protein sequences from other species showed similar domain organization with significant identification and conservation of the FRB and kinase domains as well as the FATC domain at the C‐terminus (Fig. 1B), which are conserved in the phosphatidylinositol 3‐kinase related protein kinase family (Fruman and Rommel, 2014; Sauer et al., 2013; Schmelzle and Hall, 2000; Yang et al., 2013). Additionally, a conserved motif sequence named HEAT, reported to be involved in protein–protein interactions (Hara et al., 2002; Schmelzle and Hall, 2000), was found to be distributed throughout the N‐terminal region of BcTOR (Fig. 1B).
Figure 1.
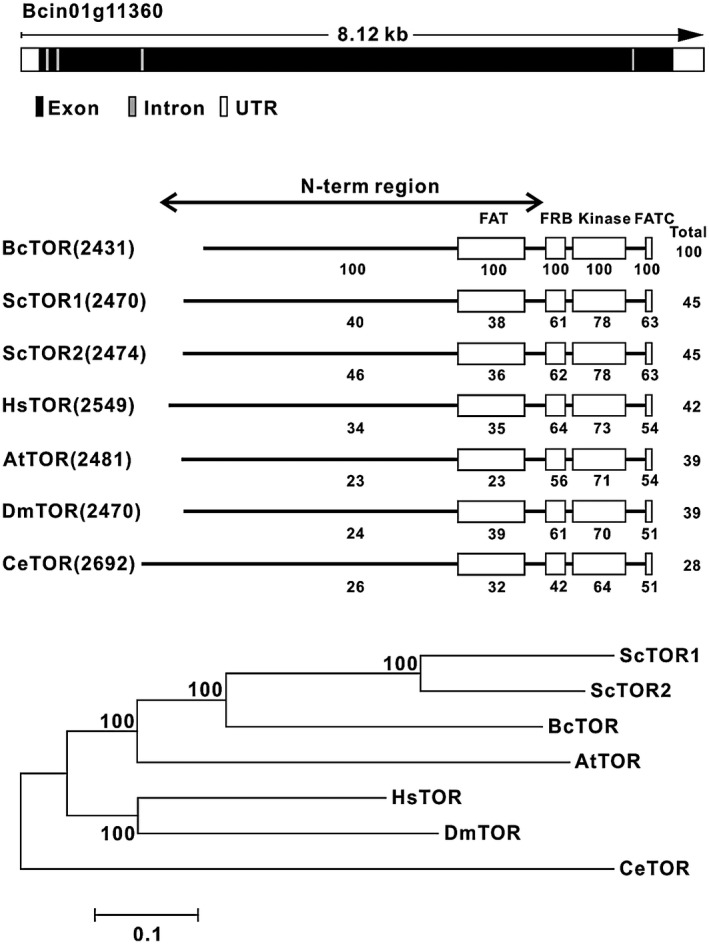
The information for the TOR homologue in Botrytis cinerea. (A) The gene locus and structure of the TOR homologue in B. cinerea. (B) Domain organization of BcTOR protein and comparison of the BcTOR amino acid sequence with those of TOR proteins from other organisms. (C) Phylogenetic analysis of BcTOR with that from other species. Bc, Botrytis cinerea; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens; At, Arabidopsis thaliana; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans. Protein domain diagram shows number of HEAT (Huntingtin, elongation factor 3 (EF3), a subunit of protein phosphatase 2A (PP2A) and TOR1) repeats; FAT (FRAP, ATM and TRRAP) domain; FKP12‐rapamycin binding (FRB) domain; and carboxy‐terminal FAT (FATC) domain.
Phylogenetic analysis revealed that BcTOR is evolutionarily conserved and depicts a closer evolutionary relationship with ScTOR1/2 in comparison with other TORs (Fig. 1C). In yeast or animals, TOR protein was reported to form two structurally and functionally distinct multiprotein complexes through recruiting shared and distinct TOR‐interacting components (Helliwell et al., 1994; Wullschleger et al., 2006). RAPTOR and LST8, as the core components of TORC1, each depicted one homologous gene through homologous comparison in the B. cinerea genome (Table 1). Additionally, RICTOR, a specific component of TORC2, also was found in the B. cinerea genome (Table 1), illustrating the existence of TORC2. These results indicate the existence of conserved TOR signalling in B. cinerea.
Table 1.
TORC1 and TORC2 homologues in Botrytis cinerea for various species
| Hs | Sc | Bc | |
|---|---|---|---|
| TORC1 | mTOR | TOR1/2 | TOR (Bcin01g11360) |
| RAPTOR | Kog1 | RAPTOR (Bcin06g02850) | |
| LST8 | Lst8 | LST8 (Bcin04g05980) | |
| ‐ | Toc89 | ‐ | |
| PRAS40 | ‐ | ‐ | |
| DEPTOR | ‐ | ‐ | |
| TORC2 | mTOR | TOR2 | TOR |
| SIN1 | Avo1 | SIN1 (Bcin16g01100) | |
| ‐ | Avo2 | ‐ | |
| RICTOR | Avo3 | RICTOR (Bcin03g06820) | |
| LST8 | Lst8 | LST8 | |
| PRR5 | Bit61 | ‐ | |
| DEPTOR | ‐ | ‐ |
Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Bc, Botrytis cinerea. TORC1, target of rapamycin complex 1; TORC2, target of rapamycin complex 2; mTOR, mammalian target of rapamycin; RAPTOR, regulatory‐associated protein of mTOR; LST8, lethal with SEC13 protein8; SIN, stress‐activated map kinase‐interacting protein 1; RICTOR, rapamycin‐insensitive companion of mTOR. (–) indicates that there are no shown/obvious homologues of the indicated proteins in the corresponding organisms.
BcFKBP12 rescues Arabidopsis sensitivity to rapamycin
Early studies demonstrated that the FKBP12 protein from yeast or humans could restore rapamycin sensitivity in Arabidopsis (Mahfouz et al., 2006; Ren et al., 2012; Sormani et al., 2007; Xiong et al., 2017). Our previous study also indicated that tomato FKBP12 could restore the inhibitory effect of rapamycin to TOR (Xiong et al., 2016). Likewise, in B. cinerea, FKBP12 homologous protein was found with conserved function in bridging rapamycin and FK506, and it was also reported that BcFKBP12 is involved in T4 strain virulence and sulphur regulation (Gioti et al., 2006; Melendez et al., 2009). Here, to further confirm BcFKBP12 function in TOR signalling, we cloned and expressed BcFKBP12 in Arabidopsis. As shown in Fig. 2A, there was no change in morphological phenotypes observed in these transgenic lines as compared with wild‐type (WT) plants. However, as expected, the transgenic lines expressing BcFKBP12 showed sensitivity to rapamycin and displayed a retarded growth phenotype with small cotyledons, reduced biomass (fresh weight) and shorter primary roots than that of WT under rapamycin treatment (Fig. 2A–E). In addition, this acquired response was dose‐independent (Fig. 2D,2). Similar results were observed in Arabidopsis expressing ScFKBP12, while no difference was found in WT under rapamycin treatment in comparison to DMSO treatment (Fig. 2D,E). These observations further confirm the conserved function of BcFKBP12 in mediating the inhibitory effect of rapamycin on TOR activity.
Figure 2.
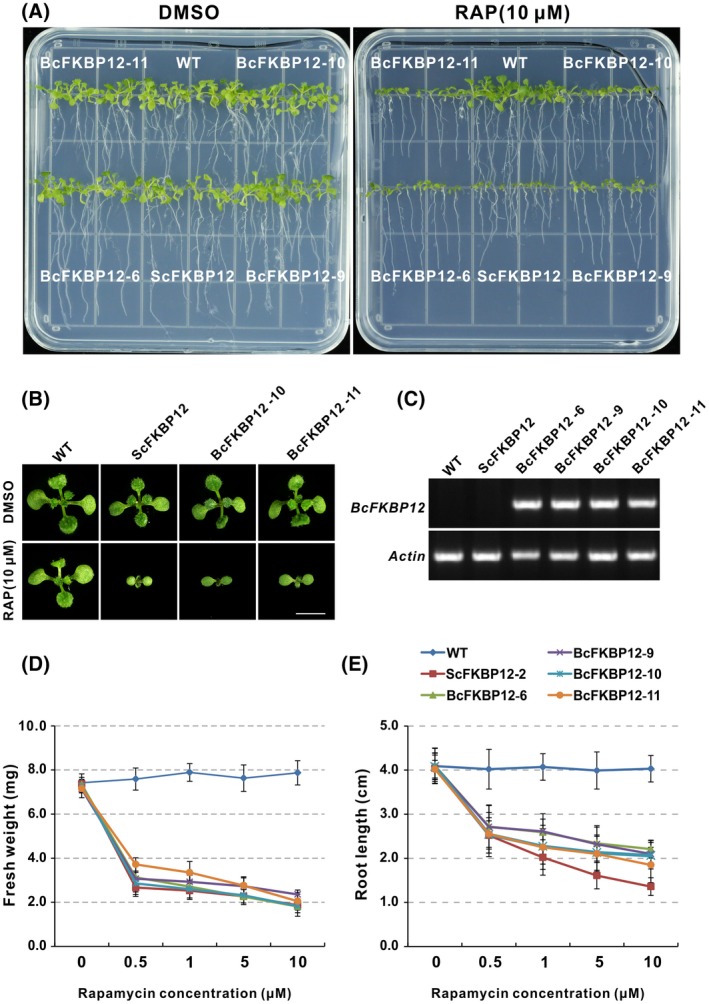
BcFKBP12 restores sensitivity of Arabidopsis to rapamycin. (A) Seedlings of BcFKBP12‐expressing lines showed growth retardation like ScFKBP12‐expressing plants under rapamycin treatment. (B) Cotyledon phenotype of wild‐type (WT), ScFKBP12‐ and BcFKBP12‐expressing plants under rapamycin treatment. Bar = 0.5 cm. (C) The detection of BcFKBP12 in transcript levels. (D, E) Dosage‐dependent curves of rapamycin for plant fresh weight and primary root length for different Arabidopsis plants measured at 10 days after germination. Error bars indicate SEM (n ≥ 40). The experiment was repeated three times for each treatment. SEM, standard error of the mean.
BcTOR regulates mycelial growth and conidiation, but not conidial germination of B. cinerea
Rapamycin is used to study the TOR signalling pathway in eukaryotic species. Previous reports indicated that rapamycin is able to suppress growth mediated by the FKBP12 orthologue in B. cinerea, but there is a lack of detailed description about TOR function in these studies. Here we set up a series of concentrations of rapamycin from 1 nM to 1 μM to observe the inhibition morphology of B. cinerea caused by rapamycin. The results show that radial growth of B. cinerea is severely inhibited on potato dextrose agar (PDA) amended with rapamycin even at a very low dose (1 nM) and nearly ceases growth with about 98% inhibitory rate at 100 nM (Fig. 3A,B,D). Microscopic observation of hyphal morphology showed that hyphae treated with rapamycin had more branches with increasing dose of rapamycin compared with the solvent control (Fig. 3C). Furthermore, twisted hyphal morphology was observed (Fig. 3C), similar to a previous study in F. graminearum (Yu et al., 2014). In accordance with retarded mycelial growth, there was a significant decline in biomass of B. cinerea when treated with rapamycin under liquid culture conditions (Fig. S1). These results suggest that TOR plays an important role in regulating the vegetative growth of B. cinerea.
Figure 3.
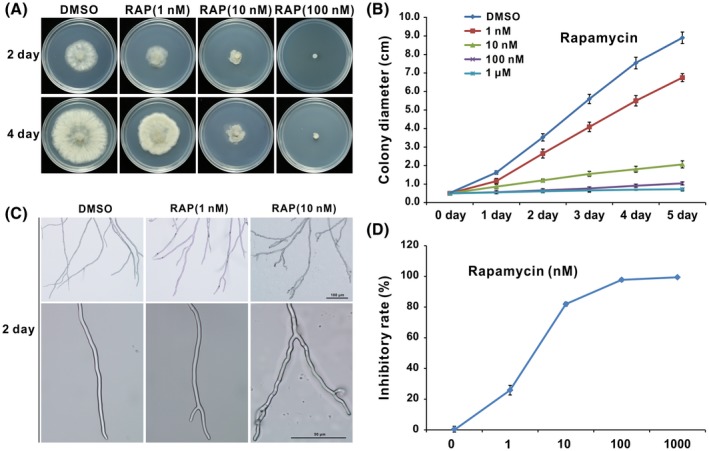
TOR inhibition under rapamycin retards mycelial growth of Botrytis cinerea. (A) Inhibitory effect of rapamycin against B. cinerea. (B) Dosage‐dependent curves of rapamycin for colony diameter of B. cinerea on potato dextrose agar plates. Error bars indicate the SEM (n ≥ 10). (C) Hyphal morphology of B. cinerea under rapamycin treatment. (D) Inhibitory rate of rapamycin on mycelial growth of B. cinerea. Error bars indicate the SEM (n ≥ 10). These experiments were repeated more than three times for each treatment.
In addition, rapamycin also exhibited a strong inhibitory effect on sporulation. Microscopic observation showed twisted hyphae with few conidiophores on 10 nM rapamycin plates compared to the control plate (Fig. 4A). Furthermore, spore number statistics showed that there is no difference in spore production between treatment of 1 nM rapamycin and DMSO solvent, while conidium production was dramatically decreased under 10 nM rapamycin treatment (Fig. 4B). The spores were not observed on PDA plates with 100 nM rapamycin (Fig. 4B). These results suggest that TOR activity is required for sporulation of B. cinerea. Subsequently, we investigated the effects of rapamycin on conidial germination through dropping spore suspensions on PDA plates supplemented with rapamycin or DMSO. As shown in Fig. 4C, after 3 days of incubation we observed smaller colonies on the rapamycin plates than on the DMSO plates. However, there was almost no difference in conidial germination at different doses of rapamycin compared with DMSO when observed at indicated time points (Fig. 4D), suggesting BcTOR is not involved in regulating conidial germination of B. cinerea.
Figure 4.
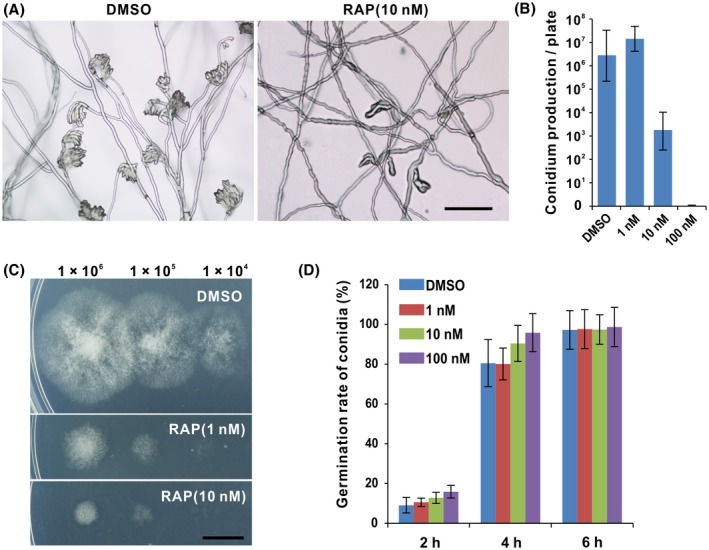
TOR inhibition represses conidiation but not conidial germination of Botrytis cinerea. (A) TOR affects sporulation of B. cinerea. (B) Conidium production under TOR inhibition. Error bars indicate the SEM (n = 10). The experiments were repeated three times. (C) Mycelial growth after germination under TOR inhibition. Scale bar represents 1 cm. (D) Conidial germination rate was calculated at the indicated time points. Data are represented as means ± SD (n = 3).
BcTOR inhibition reprograms gene expression to modulate growth and metabolism
To further investigate the role of BcTOR in the growth of B. cinerea, we performed whole genome expression profiling analysis on rapamycin and DMSO treatments through RNA sequencing (RNA‐Seq). RNA‐Seq data indicated that there were 3257 differentially expressed genes (DEGs), consisting of 1448 up‐regulated DEGs and 1809 down‐regulated DEGs (Fig. 5A and Table S1). Gene ontology (GO) enrichment analysis showed that these DEGs were categorized into 238 significantly enriched GO terms (q‐value < 0.01) (Table S2). In the top 30 of the most significantly enriched GO terms, most were associated with transcription, translation and ribosome biogenesis (Fig. 5B), which all belong to the conserved function of TOR signalling in regulation of growth (Loewith and Hall, 2011; Mahoney et al., 2009; Thoreen et al., 2012; Wullschleger et al., 2006). Furthermore, the DEGs classified in these top 30 terms displayed an almost unanimous down‐regulated trend (Fig. 5B and Table S2), suggesting that rapamycin represses BcTOR function in regulation of growth of B. cinerea. In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that many DEGs were mainly enriched on 169 KEGG pathways, including 91 down‐regulated pathways and 78 up‐regulated pathways (Tables [Link], [Link] and [Link], [Link]). In the top 20 most enriched pathways, the majority were involved in anabolism in down‐regulated groups, such as ‘ribosome’ and ‘ribosome biogenesis’, which were the two most significantly enriched pathways (Fig. 5C and Table S3). Conversely, in the up‐regulated top 20 enriched pathways, most participated in catabolism, such as ‘valine, leucine and isoleucine degradation’ and ‘fatty acid degradation’ (Fig. 5D and Table S4). These results show consistency with the conserved function of TOR in modulating metabolism (Robaglia et al., 2012; Wullschleger et al., 2006). Taken together, the above data indicate there is a conserved TOR function in the regulation of growth and metabolism in B. cinerea.
Figure 5.
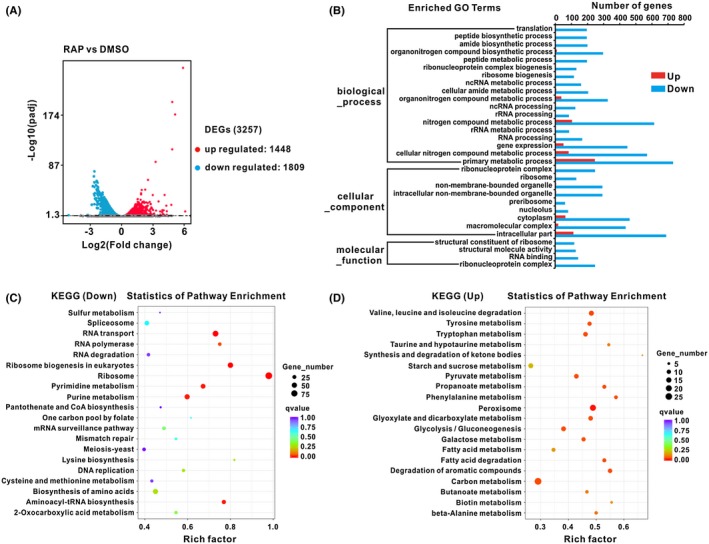
RNA‐Seq analysis of TOR function on growth of Botrytis cinerea. RNA sequencing was performed between rapamycin and DMSO treatment. Each treatment contained three biological replicates. (A) The number of DEGs. (B) Top 30 most significantly enriched GO terms. (C) Top 20 most enriched KEGG pathways for down‐regulated DEGs. (D) Top 20 most enriched KEGG pathways for up‐regulated DEGs.
BcTOR inhibition reduces infection by B. cinerea
To investigate whether TOR affects infection by B. cinerea, detached leaves of Arabidopsis and potato plants, as well as mature red tomato fruits from the supermarket, were drop‐inoculated with B. cinerea spore suspension mixed with indicated doses of rapamycin. After 3 days of incubation, larger water‐soaked infection lesions and more pathogen colonization were observed on leaves of both Arabidopsis and potato when infected with B. cinerea under DMSO treatment compared with that of rapamycin treatment (Fig. 6A–C). In particular, infected spots were hardly observed in the case of treatment with 1 μM of rapamycin (Fig. 6A). Similarly, tomato fruits showed smaller lesions and less pathogen biomass when infected with B. cinerea under rapamycin conditions at 7 days after incubation (Fig. 6A‐C). Furthermore, after 15 days of incubation, tomato fruits exhibited severe decay covered by mycelium under the solvent conditions, while rapamycin efficiently reduced infection by B. cinerea and even blocked the infection at high concentrations (Fig. S2). These findings suggest that BcTOR is required for pathogenicity of B. cinerea.
Figure 6.
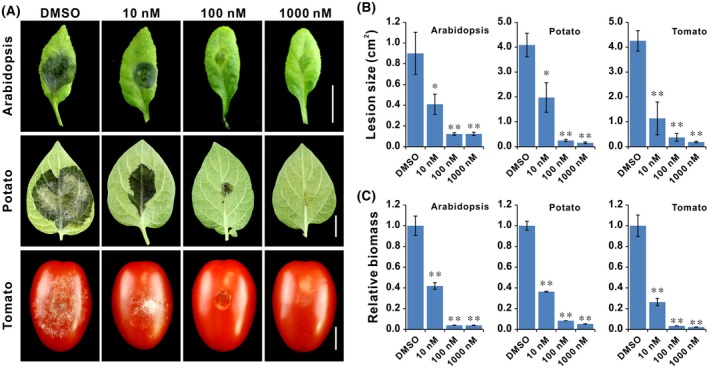
TOR inhibition reduces infection by Botrytis cinerea. Leaves of Arabidopsis and potato, as well as fruits of cherry tomato, were infected by B. cinerea by dropping a spore suspension mixed with various doses of rapamycin. (A) Infected phenotype of Arabidopsis and potato leaves (3 days post‐inoculation, dpi), and tomato fruits (7 dpi). (B, C) Lesion size and B. cinerea biomass were measured. Bar = 1 cm. Error bars indicate the SEM (n ≥ 20). The experiments were repeated more than three times for each treatment. *P < 0.05, **P < 0.01 (Student’s t‐test).
BcTOR silencing by plant‐mediated RNAi enhances resistance to B. cinerea in plants
HIGS has become a promising strategy in genetic engineering methods for crop protection. Transgenic plants and crops expressing dsRNAs that target essential growth and virulence genes of eukaryotic pathogens and pests are less prone to disease (Cai et al., 2018; Nunes and Dean, 2012; Wang, Thomas et al., 2017). Our previous results on BcTOR function showed that BcTOR could be a potential target in HIGS for crop protection. In order to test its potential, three fragments of BcTOR (BcRI1, BcRI2 and BcRI3) were selected to construct the RNA interference vector, and transgenic potato was first generated due to the ease of genetic transformation (Fig. 7A). A total of 25 independent transgenic potato lines were generated, including six BcRI1 lines (BcRI1‐1 to BcRI1‐9), nine BcRI2 lines (BcRI2‐1 to BcRI2‐6) and ten BcRI3 lines (BcRI3‐1 to BcRI3‐10), in which several lines were selected for pathogen infection experiment (Fig. S3). As shown in Fig. 7B, the selected transgenic potato lines all showed enhanced resistance to B. cinerea to different extents, manifested in smaller lesions and less pathogen biomass compared with WT or green fluorescent protein (GFP) dsRNA‐expressing plants (Fig. 7B‐D). In particular, BcRI3‐3 and BcRI3‐6 lines displayed near‐complete resistance to B. cinerea so water‐soaked lesions were hardly observed (Fig. 7A). In order to confirm that the enhanced resistance of transgenic potato is related to BcTOR transcript level, we examined the silencing efficiency of BcTOR by qRT‐PCR in these infected leaves. In accordance with the above observation, the transcript level of BcTOR was significantly decreased in the infected BcTOR‐silenced transgenic lines in comparison with WT or GFP dsRNA‐expressing plants (Fig. 7E). These results suggest that plants expressing dsRNA targeting BcTOR could efficiently reduce susceptibility to B. cinerea by interfering with the expression of BcTOR.
Figure 7.
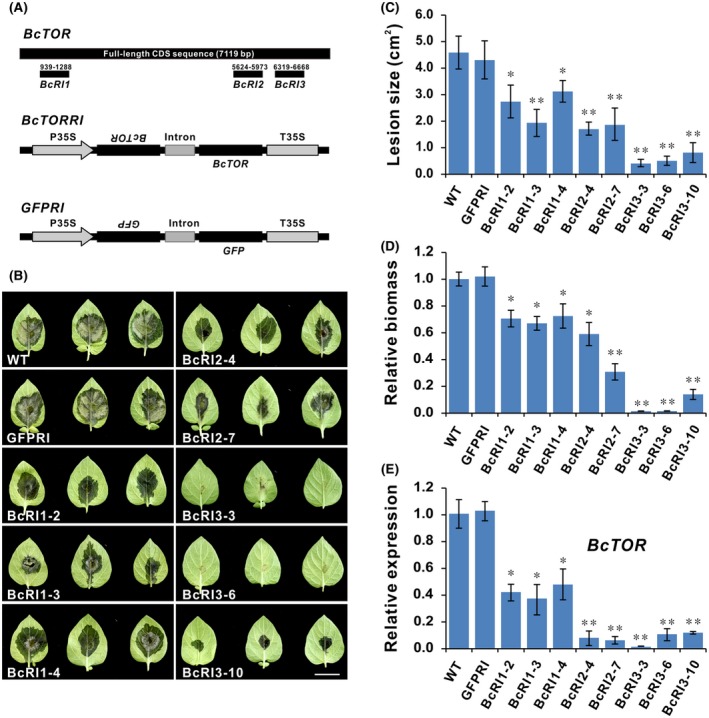
Transgenic potato expressing BcTOR dsRNA shows enhanced resistance to Botrytis cinerea. (A) Schematic diagram of RNA interference vector targeting BcTOR and GFP (control). (B) Infection phenotype of B. cinerea on detached leaves of transgenic potato lines and wild‐type (WT). Photographs were taken at 3 days post‐inoculation. Scale bar represents 2 cm. (C, D) Infection lesion size and relative biomass of B. cinerea. Error bars indicate the SEM (n ≥ 20). The experiments were repeated three times. (E) Relative expression level of BcTOR in B. cinerea during infection of transgenic and WT potato plants. Data are represented as means ± SD (n = 3). *P < 0.05, **P < 0.01 (Student’s t‐test).
The same constructs were used to generate transgenic tomato and a total of 27 independent transgenic lines were obtained (Fig. S4). Southern blotting results showed that among the selected lines, the BcRI1 and BcRI3 transgenic tomato lines contained two or more T‐DNA insertions, while both BcRI2 transgenic lines had a single copy (Fig. S5A–C). To demonstrate the specificity of the BcTOR sRNA molecules generated in these transgenic lines, we carried out sRNA‐Seq. The data showed that a large number of BcTOR‐specific sRNA molecules were generated in these transgenic lines (Fig. 8A–C and Tables [Link], [Link], [Link]). Corresponding to copy number, BcRI1 and BcRI3 lines had a higher abundance of sRNA molecules than BcRI2 lines. In addition, BcRI1 and BcRI3 lines also contained more species of BcTOR‐specific sRNAs compared with BcRI2 lines or WT plants (Fig. 8A–C and Tables [Link], [Link], [Link]). These results further confirm the presence of sRNA molecules derived from BcTOR RNAi constructs in transgenic tomato seedlings. Furthermore, pathogen infection experiments showed that, similar to the observation from the T1 generation, the T2 generation plants also displayed enhanced resistance against B. cinerea, suggesting that the trait is stable among generations (Figs 9A–E and S6A–D). Specifically, under normal growth conditions there was no difference between transgenic tomato plants expressing dsRNA targeting BcTOR and WT tomato plants; however, dsRNA‐expressing plants showed a distinct growth advantage after infection by B. cinerea compared with WT plants, where leaves showed serious decay (Fig. 9A). Additionally, consistent with BcTOR‐specific sRNA levels, tomato fruits from BcRI1 and BcRI3 lines had smaller lesions and less pathogen biomass than BcRI2 lines or WT fruits, as well as a decline in transcript levels of BcTOR during infection (Fig. 9B–E). Taken together, these results further demonstrate that BcTOR dsRNA‐expressing plants could constitutively produce BcTOR‐specific sRNAs and effectively inhibit the expression of BcTOR to resist B. cinerea, suggesting that BcTOR is an effective target for genetic engineering in the control of grey mould.
Figure 8.
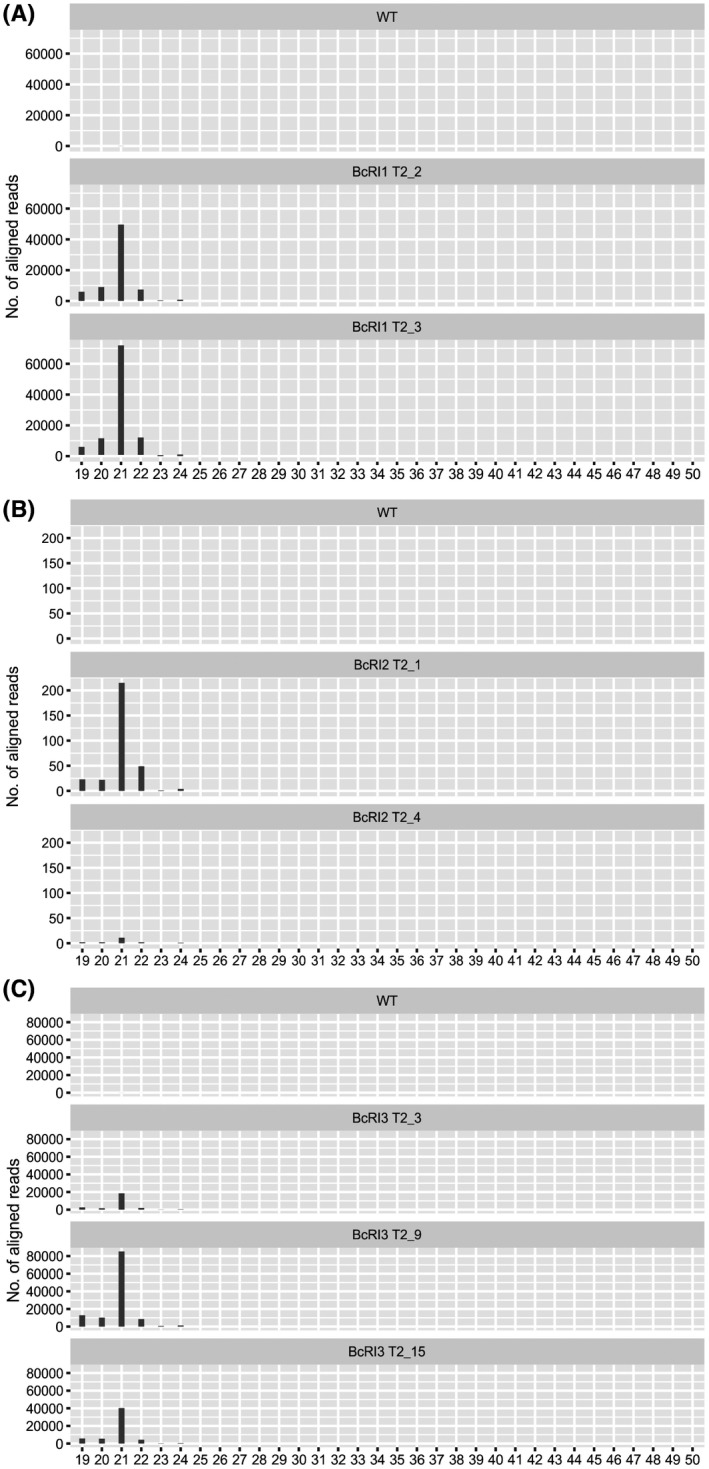
The abundance of sRNAs targeting BcTOR in transgenic tomato lines expressing BcTOR dsRNA. Two‐week‐old transgenic and wild‐type (WT) seedlings were harvested and used for small RNA (sRNA) sequencing. (A) sRNAs derived from BcRI1 construct lines. (B) sRNAs derived from BcRI2 construct lines. (C) sRNAs derived from BcRI3 construct lines.
Figure 9.
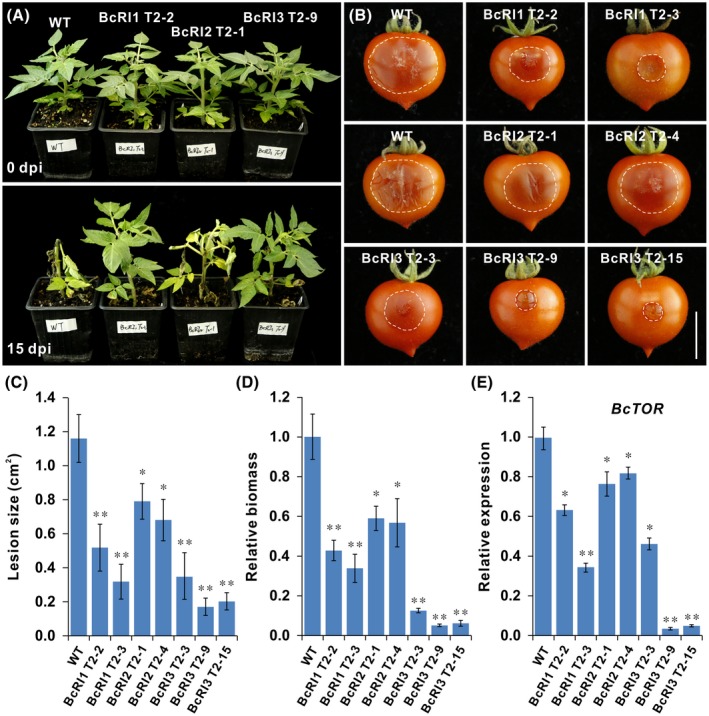
Transgenic tomato expressing BcTOR dsRNA shows enhanced resistance to Botrytis cinerea. (A) Phenotype of BcTOR dsRNA‐expressing tomato plants and wild‐type (WT) before and after infection by B. cinerea. Scale bar represents 1 cm. (B) Infection phenotype of B. cinerea on fruits from transgenic tomato and WT plants. Photographs were taken at 3 days post‐inoculation. Scale bar represents 1 cm. (C, D) Infection lesion size and relative biomass of B. cinerea. Error bars indicate the SEM (n ≥ 10). The experiments were repeated three times. (E) Relative expression level of BcTOR in B. cinerea during infection of transgenic and WT tomato fruits. Data are represented as means ± SD (n = 3). *P < 0.05, **P < 0.01 (Student’s t‐test).
Discussion
To control grey mould disease, the traditional strategy is to apply fungicides, but fungicide residues derived from the widespread use of chemical pesticides have caused serious environmental pollution issues (Malhat et al., 2015; Oliveira et al., 2015; Tomenson and Matthews, 2009). Biofungicides for key targets of pathogens are a promising application for controlling diseases. On the other hand, genetic engineering methods also are extensively applied to improve resistance to plant pathogens for crop protection. For instance, HIGS has been proved to be an efficient tool to unravel gene function as well as a promising approach to improve resistance to plant fungal pathogens by targeting key genes associated with growth and virulence (Cai et al., 2018; Wang, Thomas et al., 2017). However, selection of efficient targets is the key for HIGS.
TOR, a functionally and structurally conserved protein kinase, is a master regulator that controls cell growth by integrating nutrient, energy and growth factors in all eukaryotic species (van Dam et al., 2011; Takahara and Maeda, 2013; Wullschleger et al., 2006). TOR is extensively used as an excellent target in the control or therapy of diseases like Alzheimer's disease, Huntington's chorea and some cancers by using rapamycin or other active‐site TOR inhibitors (Blenis, 2017; Ciuffreda et al., 2010; Easton and Houghton, 2006; Jung et al., 2018; Santos et al., 2011; Wang, Valera et al., 2017), implying that TOR could also be a potential target in pathogens for disease control. To date, TOR signalling in phytopathogenic fungi such as B. cinerea, M. circinelloides and Fusarium spp. has been reported. TOR signalling was particularly well characterized in F. graminearum (Bastidas et al., 2012; Lopez‐Berges et al., 2010; Melendez et al., 2009; Teichert et al., 2006; Yu et al., 2014), suggesting that a conserved TOR pathway also exists in plant pathogenic fungi.
Genome scanning revealed the presence of conserved TOR signalling including conserved TOR protein and TOR components such as RAPTOR, LST8 of TORC1 and RICTOR of TORC2 in B. cinerea (Fig. 1A–C and Table 1), as well as the FKBP12 homologue, suggesting there is conserved TOR signalling in B. cinerea. Previous studies showed that the BcFKBP12 deletion relieved the growth inhibition of rapamycin, suggesting the role of BcFKBP12 in associating the conserved inhibitory effect of rapamycin to TOR in B. cinerea (Gioti et al., 2006). Based on the characteristics of this dependency, functional FKBP12 is usually used in studying TOR function in some rapamycin‐insensitive species, especially in the model plant Arabidopsis (Mahfouz et al., 2006; Ren et al., 2012; Sormani et al., 2007; Xiong et al., 2017). For example, Arabidopsis plants depicted susceptibility to rapamycin by expressing FKBP12 from yeast or human (Mahfouz et al., 2006; Ren et al., 2012; Sormani et al., 2007; Xiong et al., 2017). Consistent with the previous studies, BcFKBP12 also restored the sensitivity of Arabidopsis plants to rapamycin (Fig. 2A–E). This result further confirmed the conserved function of BcFKBP12 in bridging the inhibitory effect of rapamycin to TOR.
The TOR signalling pathway plays critical roles in controlling cell growth and proliferation, and in modulating downstream transcription, translation, autophagy and metabolism processes in a variety of eukaryotes (Dobrenel et al., 2016; Henriques et al., 2014; Mahoney et al., 2009; Rexin et al., 2015; Thoreen et al., 2012; Xiong and Sheen, 2015). Previous reports indicated that rapamycin suppresses growth of B. cinerea, but there is no detailed description of TOR function in these studies. Previously it has been reported that inhibition of TOR by rapamycin caused serious growth retardation, displaying twisted hyphal morphology, more branches and increased septation in F. graminearum (Yu et al., 2014). In this study, similar results were observed in B. cinerea when treated with rapamycin (Figs 3A–C and S1), suggesting that TOR plays a crucial role in regulating vegetative growth in B. cinerea. For mycelial growth, the retarded growth caused by rapamycin was supported by RNA‐Seq data that showed that many DEGs associated with ribosome, transcription and translation were down‐regulated under TOR inhibition (Fig. 5B–D and Tables [Link], [Link], [Link]). These data also reflect conserved functions of TOR signalling for ribosome biogenesis and assembly, transcription and translation (Takahara and Maeda, 2013; Wullschleger et al., 2006). In addition, similar to previous reports in F. graminearum, TOR inhibition also led to severe suppression of conidiation of B. cinerea when concentrations of more than 10 nM rapamycin were used (Fig. 4A,B). This implies that restricting the spread of B. cinerea by inhibiting TOR could be a promising approach. TOR inhibition reduces pathogenicity in F. graminearum associated with regulating mycelial growth and virulence (Yu et al., 2014), which offers an impressive example of TOR function on the virulence of plant fungal pathogens. Similarly, we observed reduction and even loss of infectivity in B. cinerea under TOR inhibition during infection (Fig. 6A–C), implying that BcTOR could be a potential target used to control B. cinerea.
HIGS is an effective strategy for crop protection and has also been proven to be effective in controlling necrotrophic fungal pathogens. For instance, Arabidopsis plants expressing dsRNA targeting BcDCL1 and BcDCL2 are more resistant to B. cinerea (Zhang et al., 2016). Here, we generated transgenic potato and tomato plants expressing dsRNA targeting BcTOR based on the HIGS method. In general, the transgenic potato and tomato plants displayed enhanced, or near‐complete resistance to B. cinerea, although there were some differences in resistance between transgenic lines (Figs 7B–E and 9A–E). sRNA‐Seq showed that a large number of BcTOR‐specific sRNA molecules were generated in these transgenic lines, and BcRI1 and BcRI3 lines had a higher abundance of sRNA molecules than BcRI2 lines (Fig. 8A–C and Tables [Link], [Link], [Link]). The data suggest that BcTOR dsRNA could be constitutively and efficiently processed into sRNA molecules (Fig. 8A–C and Tables [Link], [Link], [Link]). These data also explain the difference in resistance between transgenic tomato lines (Figs 8A–C and 9A–E, and Tables [Link], [Link], [Link]). In addition, the efficiency of dsRNA being processed into sRNAs may be responsible for differences among lines from different constructs. BcRI2 lines produced significantly fewer types of sRNA than BcRI1 and BcRI3 lines (Fig. 8A–C); an off‐target effect may also be the cause of the differences among these constructs. Through aligning BcTOR RNAi fragments with the cDNA database of tomato or B. cinerea, we found no possible off‐target genes in tomato plants, but in B. cinerea there were only two genes with more than 19 bases matched for each fragment (Table S9). Compared to BcRI2 lines, BcRI1 and BcRI3 contained more sRNA reads matching to non‐target genes, but these multiple target sRNA reads only account for very few of the total sRNA reads in BcRI1 and BcRI3 lines (Tables S8 and S9). In summary, our results demonstrate that BcTOR dsRNA‐expressing plants were able to effectively produce BcTOR‐specific sRNAs against B. cinerea, reducing infection. This study suggests that TOR is a potential and promising target in the biocontrol of plant fungal pathogens.
Experimental Procedures
Fungal strains and culture conditions
Botrytis cinerea strain B05.10 was incubated on PDA plates at 25 °C unless indicated otherwise. For the growth inhibition assay, 0.4 mm diameter agar plugs with fungal mycelia were placed on PDA plates with different concentrations of rapamycin (1, 10, 100, 1000 nM) or dimethylsulphoxide (DMSO, solvent control). The diameter of the colony was measured every day, and the hyphal tip growth and branching patterns were observed after 2 days' growth. For measurement of conidia production, spores were collected and counted after 10 days' cultivation. For the spore germination experiment, a spore suspension (1 × 106 spores/mL) was spread on PDA plates supplemented with rapamycin or DMSO for incubation, and spore germination was observed at different time points (2, 4 and 6 h).
In order to quantify the biomass of mycelia, potato dextrose broth (PDB) was used for liquid culture of mycelia. Five agar plugs with fungal mycelia were placed in a 100 mL triangular bottle with 50 mL PDB supplemented with the indicated concentrations of rapamycin or DMSO (solvent control), then the bottles were placed in an incubator at 25 °C. After 4 days' incubation, the mycelia were collected and freeze‐dried to constant weight before measuring the biomass of mycelia. To detect the expression level of related genes, mycelia cultivated in PDB for 4 days were treated by adding the indicated concentrations of rapamycin or DMSO (solvent control) for 12 h, and then the mycelia were collected for RNA extraction.
Plant material and growth conditions
The plant materials used in this work included A. thaliana ecotype Columbia (Col‐0), Solanum lycopersicum 'Micro‐Tom' and Solanum tuberosum 'Desirée'. All plants were grown in soil in an artificial climate culture room at 25/18 °C of day/night (except Arabidopsis plants were grown at 22 °C), 80% humidity and under a long‐day photoperiod consisting of a 16 h light regime. In addition, cherry tomato (Solanum lycopersicum var. cerasiforme) fruits purchased from the supermarket were also used in infection experiments.
Vector construction and plant transformation
Vector construction was based on a Gateway system according to previous reports (Earley et al., 2006; Xiong et al., 2017). Vector p35S‐IN4‐8GWN was used as the entry vector, and the plant binary vector KANA303, modified pEarleyGate303, was the destination vector (Xiong et al., 2017). For RNAi vector construction, three different segments of 350‐bp DNA sequences of BcTOR were selected, and the sense and antisense orientation fragments were then amplified with the primers (BcRI‐L‐Fu‐F/BcRI‐L‐Fu‐R) listed in Table S10a. The amplified products of antisense orientation fragments were then inserted into linearized p35S‐IN4‐8GWN (linearized by NotI/SbfI) to generate p35S‐BcRI(L)‐IN4‐8GWN by seamless cloning using a In‐Fusion HD Cloning Kit (Clontech, Palo Alto, CA, USA) following the user's manual. The sense fragments were amplified with primers (BcRI‐R‐Fu‐F/BcRI‐R‐Fu‐R) and linked into linearized p35S‐BcRI(L)‐IN4‐8GWN (digested by BbvCI/AscI) to obtain p35S‐BcRI(L)‐IN4‐BcRI(R)‐8GWN by the seamless cloning method. Finally, RNAi fragments were cloned into the destination vector KANA303 using Gateway LR clonase (Invitrogen, Carlsbad, CA, USA).
The BcTOR‐RNAi binary plasmid was transferred into Agrobacterium strain GV3101. Transgenic tomato was generated by Agrobacterium‐mediated transformation following the protocols described by Fillatti et al. (1987), and transgenic potato created by Agrobacterium‐mediated transformation according to the protocol described in Millam (2007). Transgenic plants were identified by PCR with primers of neomycin phosphotransferase II (NPTII) and BcTOR fragments (Table S10a). The positive transgenic plants were selected and used for subsequent experiments.
To create BcFKBP12‐expressing Arabidopsis, the coding sequence of BcFKBP12 (Bcin12g01360.1) was cloned in the plant binary vector KANA303 according to the experimental protocols described in previous reports (Xiong et al., 2016). The resulting destination vectors were transferred into Agrobacterium strain GV3101 for plant transformation. The floral dipping method was employed for generating transgenic Arabidopsis (Zhang et al., 2006). The transformation and screening of primary transformants were performed according to Zhang et al. (2006), and T3 generation transgenic plants were used for subsequent experiments.
Botrytis cinerea infection
For inoculation of plants, conidia were harvested from sporulating colonies in sterile water with 0.1% Tween‐20 and filtered with glass wool as described by Stefanato et al. (2009). For droplet inoculations, the concentration of spores was adjusted to 1 × 105 spores/mL and 5 μL of spore suspension with the indicated dose of rapamycin or DMSO was dropped onto the surface of single detached rosette leaves of 5‐week‐old Arabidopsis plants or 45‐day‐old potato plants starting from tissue culture plantlets or mature red cherry tomato fruits from supermarket and mature red tomatoes (cv. Micro‐Tom) harvested at 14 days after breaker stage from transgenic plants. Before inoculations, all detached leaves or fruits were surface cleaned or sterilized with 75% ethanol. The inoculated leaves or fruits were kept under a transparent plastic film cover to maintain high humidity and kept at 25 °C in the dark. Infection symptoms were observed and photographed at 3 days post‐incubation (dpi). For the incubation of intact tomato plants, the same density of spore suspension was applied for spray inoculations of 45‐day‐old intact tomato plants. Plants were kept prior to and during infection under sealed hoods at high humidity, and infection symptoms were observed and photographed at 15 dpi. The lesion area was measured using ImageJ software (http://imagej.nih.gov/ij/) from digitally computed images of leaves or tomato fruits.
DNA extraction and DNA quantification
To quantify B. cinerea growth by real‐time quantitative PCR (qPCR), 1 cm2 of leaf tissue or 1 cm3 of fruit tissue around the infected lesion area was collected at 3 dpi, and then ground into powder in liquid nitrogen. Total DNA was isolated using a plant genomic DNA extraction kit (Bioteke, Beijing, China) following the manufacturer’s user manual. To estimate the amount of fungal DNA in inoculated samples, purified DNAs were used for qPCR as described by Zhang et al. (2013). The primers listed in Table S10b were used.
Expression profiling sequencing and analysis
RNA‐Seq was carried out by Novogene Bioinformatics Technology Co. Ltd (Beijing, China). Hyphae of B. cinerea were grown for 4 days in PDB medium at 25 °C and then treated by adding 100 nM rapamycin or DMSO (as solvent control). For each treatment, three independent biological replicates were performed. After incubation for 12 h, the mycelia were collected and total RNA of B. cinerea mycelium was isolated using a RNAprep Pure Plant Kit (Tiangen, Beijing, China). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). The sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following manufacturer’s recommendations, the library preparations were sequenced on an Illumina Hiseq platform and 125 bp/150 bp paired‐end reads were generated. The clean reads were aligned to the reference B. cinerea genome using TopHat2 software (Kim et al., 2013). Transcript abundance was also normalized by transforming the data to reads per kilobase of exon model per million mapped reads (FPKM) method. Cufflinks and Cuffdiff were used to assemble the mapped reads and identify DEGs, respectively (Trapnell et al., 2012, 2013), and genes with an adjusted P‐value < 0.05 found by DESeq were assigned as differentially expressed. All the DEGs were annotated in the B. cinerea genome database website (http://fungi.ensembl.org/Botrytis_cinerea/Info/Index?db=core). Gene ontology enrichment (corrected P‐value < 0.05) of the DEGs was performed using GO sequencing software (Young et al., 2010). The enrichment of DEGs in KEGG pathways (corrected P‐value < 0.05) was obtained using KOBAS software (Kanehisa et al., 2008; Mao et al., 2005). RNA‐Seq data was validated by real‐time qRT‐PCR for the 20 DEGs showed on Fig. S7 and the corresponding primers are listed in Table S10c.
RNA isolation and qRT‐PCR analysis
Total RNA was isolated using the RNAprep Pure Plant Kit (TianGen, Beijing, China, DP441) and reverse transcription was performed using the PrimeScript RT Reagent Kit (Takara, Otsu, Japan) following the manufacturer’s instructions. Real‐time PCR was performed on a Bio‐Rad CFX96 System (Bio‐Rad, Hercules, CA, USA) using the TB Green Premix Ex Taq (Takara, Otsu, Japan).
DNA gel blotting
The genomic DNA samples were isolated from 2‐week‐old T2 generation seedlings of transgenic tomato lines. DNA gel blotting was performed using the DIG‐High prime DNA labelling and detection starter kit II (Roche, Mannheim, Germany) following the protocol provided with the kit. Genomic DNA was digested with the restriction enzyme EcoRI for DNA Southern blot analysis and about 10 μg of each was loaded on a 1% agarose gel. The probe location was a selected interference fragment of BcTOR, amplified and sequenced from corresponding plasmid with primers.
Small RNA sequencing
Total RNA extracted from 2‐week‐old transgenic and WT seedlings was used for sRNA sequencing. sRNA libraries were constructed using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs, Inc., Ipswich, MA, USA) according to the manufacturer’s instructions. Briefly, 1 μg of total RNA was ligated to a 3ʹ adapter and a 5ʹ adapter using Ligation Enzyme Mix. The resulting samples were reverse‐transcribed using Superscript II reverse transcriptase. Amplification was executed for the PCR products. All steps were performed according to the manufacturer's protocols. sRNA libraries were analysed for quality control and the average size of inserts was approximately 140–150 bp. The sequencing library was then sequenced on a Hiseq platform (Illumina) by Shanghai Personal Biotechnology Cp. Ltd (Shanghai, China). The quality information of raw data in FASTQ format was calculated and the raw data were filtered using the Personalbio self‐developed script. Clean data were obtained by removing adapter and low‐quality sequence. Filter Clean Reads from 18 to 36 nt in length and perform deduplication to obtain Unique Reads for subsequent analysis. The unique reads were aligned using BLAST with the corresponding target fragment of BcTOR. The reads count value of the sRNA was counted based on the number of sequences aligned to the corresponding target fragment of BcTOR.
Statistics
All experiments were repeated and yielded reproducible results. The most representative data are shown in this paper. Data are presented as means ± standard error of the mean, unless stated otherwise. Paired or unpaired, two‐tailed Student’s t‐tests were used to compare group differences. P‐values < 0.05 were considered significant.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Fig. S1 Biomass of B. cinerea under rapamycin and DMSO treatment in a liquid culture condition.
Fig. S2 Phenotype of tomato fruits infected by B. cinerea under TOR inhibition at 15 dpi.
Fig. S3 Identification of transgenic potato by PCR with primers of neomycin phosphotransferase II (NPTII) and BcTOR fragments.
Fig. S4 Identification of transgenic tomato by PCR with primers of neomycin phosphotransferase II (NPTII) and BcTOR fragments.
Fig. S5 DNA gel blotting analysis of tomato transgenic lines.
Fig. S6 Phenotype of BcTOR dsRNA expressing tomato in T1 generation.
Fig. S7 Validation of RNA‐seq data by real‐time PCR.
Table S1 DEGs between rapamycin and DMSO treatment
Table S2 Enriched GO terms
Table S3 Enriched KEGG pathway (down)
Table S4 Enriched KEGG pathway (up)
Table S5 Abundance statistics of sRNA target to BcTOR
Table S6 sRNA molecule sequence information
Table S7 sRNA blast information
Table S8 Potential off‐target genes in B. cinerea
Table S9 sRNAs matching to non‐targeting genes
Table S10 Primers used in this study
Acknowledgements
We thank Guozheng Qin (Chinese Academy of Sciences, Beijing, China) for kindly providing B. cinerea strain B05.10. This work was supported by the following grants: the National Natural Science Foundation of China (nos. 31801913, 31972469, 31672206 and 31801271), the China Postdoctoral Science Foundation (nos. 2017M622958 and 2018M633320), the Project of Chongqing Science and Technology Commission (nos. cstc2016jcyjA0822, cstc2019jcyj‐msxmX0127 and cstckjcxljrc15), Chengdu Agricultural Science and Technology Center local financial special fund project (NASC2019TI13), the Fundamental Research Funds for the Central Institutes and the Chinese Academy of Agricultural Sciences (19‐001‐09).
References
- Baretic, D. and Williams, R.L. (2014) The structural basis for mTOR function. Semin. Cell Dev. Biol. 36, 91–101. [DOI] [PubMed] [Google Scholar]
- Bastidas, R.J. , Shertz, C.A. , Lee, S.C. , Heitman, J. and Cardenas, M.E. (2012) Rapamycin exerts antifungal activity in vitro and in vivo against Mucor circinelloides via FKBP12‐dependent inhibition of Tor. Eukaryot Cell, 11, 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, J.A. , Bogaert, T. , Clinton, W. , Heck, G.R. , Feldmann, P. , Ilagan, O. , Johnson, S. , Plaetinck, G. , Munyikwa, T. , Pleau, M. , Vaughn, T. and Roberts, J. (2007) Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. [DOI] [PubMed] [Google Scholar]
- Benjamin, D. , Colombi, M. , Moroni, C. and Hall, M.N. (2011) Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 10, 868–880. [DOI] [PubMed] [Google Scholar]
- Blenis, J. (2017) TOR, the gateway to cellular metabolism, cell growth, and disease. Cell, 171, 10–13. [DOI] [PubMed] [Google Scholar]
- Cai, Q. , He, B. , Kogel, K.H. and Jin, H. (2018) Cross‐kingdom RNA trafficking and environmental RNAi‐nature's blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 46, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, W. , Song, X.S. , Li, H.P. , Cao, L.H. , Sun, K. , Qiu, X.L. , Xu, Y.B. , Yang, P. , Huang, T. , Zhang, J.B. , Qu, B. and Liao, Y.C. (2015) Host‐induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. [DOI] [PubMed] [Google Scholar]
- Chiu, M.I. , Katz, H. and Berlin, V. (1994) RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA, 91, 12574–12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquer, M. , Fournier, E. , Kunz, C. , Levis, C. , Pradier, J.M. , Simon, A. and Viaud, M. (2007) Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277, 1–10. [DOI] [PubMed] [Google Scholar]
- Ciuffreda, L. , Di Sanza, C. , Incani, U.C. and Milella, M. (2010) The mTOR pathway: a new target in cancer therapy. Curr. Cancer Drug Tar. 10, 484–495. [DOI] [PubMed] [Google Scholar]
- van Dam, T.J. , Zwartkruis, F.J. , Bos, J.L. and Snel, B. (2011) Evolution of the TOR pathway. J. Mol. Evol. 73, 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. , Rudd, J.J. , Dickman, M. , Kahmann, R. , Ellis, J. and Foster, G.D. (2012) The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel, T. , Caldana, C. , Hanson, J. , Robaglia, C. , Vincentz, M. , Veit, B. and Meyer, C. (2016) TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 67, 261–285. [DOI] [PubMed] [Google Scholar]
- Earley, K.W. , Haag, J.R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. and Pikaard, C.S. (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Easton, J.B. and Houghton, P.J. (2006) mTOR and cancer therapy. Oncogene, 25, 6436–6446. [DOI] [PubMed] [Google Scholar]
- Feldman, M.E. , Apsel, B. , Uotila, A. , Loewith, R. , Knight, Z.A. , Ruggero, D. and Shokat, K.M. (2009) Active‐site inhibitors of mTOR target rapamycin‐resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti, J.A.J. , Kiser, J. , Rose, R. and Comai, L. (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat. Biotechnol. 5, 726–730. [Google Scholar]
- Fruman, D.A. and Rommel, C. (2014) PI3K and cancer: lessons, challenges and opportunities. Nat. Rev. Drug Discov. 13, 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghag, S.B. , Shekhawat, U.K. and Ganapathi, T.R. (2014) Host‐induced post‐transcriptional hairpin RNA‐mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 12, 541–553. [DOI] [PubMed] [Google Scholar]
- Gioti, A. , Simon, A. , Le Pecheur, P. , Giraud, C. , Pradier, J.M. , Viaud, M. and Levis, C. (2006) Expression profiling of Botrytis cinerea genes identifies three patterns of up‐regulation in planta and an FKBP12 protein affecting pathogenicity. J. Mol. Biol. 358, 372–386. [DOI] [PubMed] [Google Scholar]
- Govindarajulu, M. , Epstein, L. , Wroblewski, T. and Michelmore, R.W. (2015) Host‐induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 13, 875–883. [DOI] [PubMed] [Google Scholar]
- Hara, K. , Maruki, Y. , Long, X. , Yoshino, K. , Oshiro, N. , Hidayat, S. , Tokunaga, C. , Avruch, J. and Yonezawa, K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell, 110, 177–189. [DOI] [PubMed] [Google Scholar]
- Heitman, J. , Movva, N.R. and Hall, M.N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science, 253, 905–909. [DOI] [PubMed] [Google Scholar]
- Helliwell, S.B. , Wagner, P. , Kunz, J. , Deuter‐Reinhard, M. , Henriquez, R. and Hall, M.N. (1994) TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Cell. Biol. 5, 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques, R. , Bogre, L. , Horvath, B. and Magyar, Z. (2014) Balancing act: matching growth with environment by the TOR signalling pathway. J. Exp. Bot. 65, 2691–2701. [DOI] [PubMed] [Google Scholar]
- Jung, S. , Gamez‐Diaz, L. , Proietti, M. and Grimbacher, B. (2018) “Immune TOR‐opathies,” a novel disease entity in clinical immunology. Front. Immunol. 9, 966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. , Araki, M. , Goto, S. , Hattori, M. , Hirakawa, M. , Itoh, M. , Katayama, T. , Kawashima, S. , Okuda, S. , Tokimatsu, T. and Yamanishi, Y. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Pertea, G. , Trapnell, C. , Pimentel, H. , Kelley, R. and Salzberg, S.L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A. , Kumar, N. , Weber, L. , Keller, H. , Imani, J. and Kogel, K.H. (2013) Host‐induced gene silencing of cytochrome P450 lanosterol C14alpha‐demethylase‐encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA, 110, 19324–19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltin, Y. , Faucette, L. , Bergsma, D.J. , Levy, M.A. , Cafferkey, R. , Koser, P.L. , Johnson, R.K. and Livi, G.P. (1991) Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl‐prolyl cis‐trans isomerase related to human FK506‐binding protein. Mol. Cell. Biol. 11, 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith, R. and Hall, M.N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics, 189, 1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith, R. , Jacinto, E. , Wullschleger, S. , Lorberg, A. , Crespo, J.L. , Bonenfant, D. , Oppliger, W. , Jenoe, P. and Hall, M.N. (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell, 10, 457–468. [DOI] [PubMed] [Google Scholar]
- Lopez‐Berges, M.S. , Rispail, N. , Prados‐Rosales, R.C. and Di Pietro, A. (2010) A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell, 22, 2459–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira da Silva, L. and Beverley, S.M. (2010) Expansion of the target of rapamycin (TOR) kinase family and function in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infectivity. Proc. Natl. Acad. Sci. USA, 107, 11965–11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz, M.M. , Kim, S. , Delauney, A.J. and Verma, D.P. (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell, 18, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney, S.J. , Dempsey, J.M. and Blenis, J. (2009) Cell signaling in protein synthesis ribosome biogenesis and translation initiation and elongation. Prog. Mol. Biol. Transl. 90, 53–107. [DOI] [PubMed] [Google Scholar]
- Malhat, F.M. , Haggag, M.N. , Loutfy, N.M. , Osman, M.A. and Ahmed, M.T. (2015) Residues of organochlorine and synthetic pyrethroid pesticides in honey, an indicator of ambient environment, a pilot study. Chemosphere, 120, 457–461. [DOI] [PubMed] [Google Scholar]
- Mao, X. , Cai, T. , Olyarchuk, J.G. and Wei, L. (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics, 21, 3787–3793. [DOI] [PubMed] [Google Scholar]
- Melendez, H.G. , Billon‐Grand, G. , Fevre, M. and Mey, G. (2009) Role of the Botrytis cinerea FKBP12 ortholog in pathogenic development and in sulfur regulation. Fungal Genet. Biol. 46, 308–320. [DOI] [PubMed] [Google Scholar]
- Millam, S. (2007) Potato (Solanum tuberosum L.) In Agrobacterium Protocols (Wang K. ed.), pp. 25–35. Totowa, NJ: Humana Press. [Google Scholar]
- Nowara, D. , Gay, A. , Lacomme, C. , Shaw, J. , Ridout, C. , Douchkov, D. , Hensel, G. , Kumlehn, J. and Schweizer, P. (2010) HIGS: host‐induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis . Plant Cell, 22, 3130–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes, C.C. and Dean, R.A. (2012) Host‐induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, B.R. , Penetra, A. , Cardoso, V.V. , Benoliel, M.J. , Barreto Crespo, M.T. , Samson, R.A. and Pereira, V.J. (2015) Biodegradation of pesticides using fungi species found in the aquatic environment. Environ. Sci. Pollut. R. 22, 11781–11791. [DOI] [PubMed] [Google Scholar]
- Pedras, M.S. , Hossain, S. and Snitynsky, R.B. (2011) Detoxification of cruciferous phytoalexins in Botrytis cinerea: spontaneous dimerization of a camalexin metabolite. Phytochemistry, 72, 199–206. [DOI] [PubMed] [Google Scholar]
- Ren, M. , Venglat, P. , Qiu, S. , Feng, L. , Cao, Y. , Wang, E. , Xiang, D. , Wang, J. , Alexander, D. , Chalivendra, S. , Logan, D. , Mattoo, A. , Selvaraj, G. and Datla, R. (2012) Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis . Plant Cell, 24, 4850–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexin, D. , Meyer, C. , Robaglia, C. and Veit, B. (2015) TOR signalling in plants. J. Biochem. 470, 1–14. [DOI] [PubMed] [Google Scholar]
- Robaglia, C. , Thomas, M. and Meyer, C. (2012) Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15, 301–307. [DOI] [PubMed] [Google Scholar]
- Sabatini, D.M. , Erdjument‐Bromage, H. , Lui, M. , Tempst, P. and Snyder, S.H. (1994) RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin‐dependent fashion and is homologous to yeast TORs. Cell, 78, 35–43. [DOI] [PubMed] [Google Scholar]
- Santos, R.X. , Correia, S.C. , Cardoso, S. , Carvalho, C. , Santos, M.S. and Moreira, P.I. (2011) Effects of rapamycin and TOR on aging and memory: implications for Alzheimer's disease. J. Neurochem. 117, 927–936. [DOI] [PubMed] [Google Scholar]
- Sehgal, S.N. , Baker, H. and Vézina, C. (1975) Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J. Antibiot., 28, 727–732. [DOI] [PubMed] [Google Scholar]
- Sauer, E. , Imseng, S. , Maier, T. and Hall, M.N. (2013) Conserved sequence motifs and the structure of the mTOR kinase domain. Biochem. Soc. 41, 889–895. [DOI] [PubMed] [Google Scholar]
- Schmelzle, T. and Hall, M.N. (2000) TOR, a central controller of cell growth. Cell, 103, 253–262. [DOI] [PubMed] [Google Scholar]
- Song, Y. and Thomma, B. (2018) Host‐induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis . Mol. Plant Pathol. 19, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormani, R. , Yao, L. , Menand, B. , Ennar, N. , Lecampion, C. , Meyer, C. and Robaglia, C. (2007) Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol. 7, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanato, F.L. , Abou‐Mansour, E. , Buchala, A. , Kretschmer, M. , Mosbach, A. , Hahn, M. , Bochet, C.G. , Metraux, J.P. and Schoonbeek, H.J. (2009) The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana . Plant J. 58, 499–510. [DOI] [PubMed] [Google Scholar]
- Takahara, T. and Maeda, T. (2013) Evolutionarily conserved regulation of TOR signalling. J. Biochem. 154, 1–10. [DOI] [PubMed] [Google Scholar]
- Teichert, S. , Wottawa, M. , Schonig, B. and Tudzynski, B. (2006) Role of the Fusarium fujikuroi TOR kinase in nitrogen regulation and secondary metabolism. Eukaryot. Cell, 5, 1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen, C.C. , Chantranupong, L. , Keys, H.R. , Wang, T. , Gray, N.S. and Sabatini, D.M. (2012) A unifying model for mTORC1‐mediated regulation of mRNA translation. Nature, 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomenson, J.A. and Matthews, G.A. (2009) Causes and types of health effects during the use of crop protection chemicals: data from a survey of over 6,300 smallholder applicators in 24 different countries. Int. Arch. Occ. Env. Health. 82, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D.R. , Pimentel, H. , Salzberg, S.L. , Rinn, J.L. and Pachter, L. (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Hendrickson, D.G. , Sauvageau, M. , Goff, L. , Rinn, J.L. and Pachter, L. (2013) Differential analysis of gene regulation at transcript resolution with RNA‐seq. Nat. Biotechnol. 31, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezina, C. , Kudelski, A. and Sehgal, S.N. (1975) Rapamycin (AY‐22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 28, 721–726. [DOI] [PubMed] [Google Scholar]
- Villa‐Rojas, R. , Sosa‐Morales, M.E. , Lopez‐Malo, A. and Tang, J. (2012) Thermal inactivation of Botrytis cinerea conidia in synthetic medium and strawberry puree. Int. J. Food Microbiol. 155, 269–272. [DOI] [PubMed] [Google Scholar]
- Wang, X. and Proud, C.G. (2009) Nutrient control of TORC1, a cell‐cycle regulator. Trends Cell Biol. 19, 260–267. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Weiberg, A. , Lin, F.M. , Thomma, B.P. , Huang, H.D. and Jin, H. (2016) Bidirectional cross‐kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants, 2, 16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Thomas, N. and Jin, H. (2017) Cross‐kingdom RNA trafficking and environmental RNAi for powerful innovative pre‐ and post‐harvest plant protection. Curr. Opin. Plant Biol. 38, 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Valera, J.C. , Zhao, X. , Chen, Q. and Silvio Gutkind, J. (2017) mTOR co‐targeting strategies for head and neck cancer therapy. Cancer Metast. Rev. 36, 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, B. , Tudzynski, B. , Tudzynski, P. and van Kan, J.A. (2007) Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. [DOI] [PubMed] [Google Scholar]
- Wullschleger, S. , Loewith, R. and Hall, M.N. (2006) TOR signaling in growth and metabolism. Cell, 124, 471–484. [DOI] [PubMed] [Google Scholar]
- Xiong, Y. and Sheen, J. (2015) Novel links in the plant TOR kinase signaling network. Curr. Opin. Plant Biol. 28, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, F. , Dong, P. , Liu, M. , Xie, G. , Wang, K. , Zhuo, F. , Feng, L. , Yang, L. , Li, Z. and Ren, M. (2016) Tomato FK506 binding protein 12KD (FKBP12) mediates the interaction between rapamycin and target of rapamycin (TOR). Front. Plant Sci. 7, 1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, F. , Zhang, R. , Meng, Z. , Deng, K. , Que, Y. , Zhuo, F. , Feng, L. , Guo, S. , Datla, R. and Ren, M. (2017) Brassinosteriod Insensitive 2 (BIN2) acts as a downstream effector of the Target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis . New Phytol. 213, 233–249. [DOI] [PubMed] [Google Scholar]
- Xu, J. , Wang, X. , Li, Y. , Zeng, J. , Wang, G. , Deng, C. and Guo, W. (2018) Host‐induced gene silencing of a regulator of G protein signalling gene (VdRGS1) confers resistance to Verticillium wilt in cotton. Plant Biotechnol. J. 16, 1629–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H. , Rudge, D.G. , Koos, J.D. , Vaidialingam, B. , Yang, H.J. and Pavletich, N.P. (2013) mTOR kinase structure, mechanism and regulation. Nature, 497, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M.D. , Wakefield, M.J. , Smyth, G.K. and Oshlack, A. (2010) Gene ontology analysis for RNA‐seq: accounting for selection bias. Genome Biol. 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. , Gu, Q. , Yun, Y. , Yin, Y. , Xu, J.R. , Shim, W.B. and Ma, Z. (2014) The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum . New Phytol. 203, 219–232. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S.S. , Niu, Q.W. and Chua, N.H. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Butelli, E. , De Stefano, R. , Schoonbeek, H.J. , Magusin, A. , Pagliarani, C. , Wellner, N. , Hill, L. , Orzaez, D. , Granell, A. , Jones, J.D. and Martin, C. (2013) Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 23, 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T. , Jin, Y. , Zhao, J.H. , Gao, F. , Zhou, B.J. , Fang, Y.Y. and Guo, H.S. (2016) Host‐induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae . Mol. Plant, 9, 939–942. [DOI] [PubMed] [Google Scholar]
- Zhu, X. , Qi, T. , Yang, Q. , He, F. , Tan, C. , Ma, W. , Voegele, R.T. , Kang, Z. and Guo, J. (2017) Host‐induced gene silencing of the MAPKK Gene PsFUZ7 confers stable resistance to wheat stripe rust. Plant Physiol. 175, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Biomass of B. cinerea under rapamycin and DMSO treatment in a liquid culture condition.
Fig. S2 Phenotype of tomato fruits infected by B. cinerea under TOR inhibition at 15 dpi.
Fig. S3 Identification of transgenic potato by PCR with primers of neomycin phosphotransferase II (NPTII) and BcTOR fragments.
Fig. S4 Identification of transgenic tomato by PCR with primers of neomycin phosphotransferase II (NPTII) and BcTOR fragments.
Fig. S5 DNA gel blotting analysis of tomato transgenic lines.
Fig. S6 Phenotype of BcTOR dsRNA expressing tomato in T1 generation.
Fig. S7 Validation of RNA‐seq data by real‐time PCR.
Table S1 DEGs between rapamycin and DMSO treatment
Table S2 Enriched GO terms
Table S3 Enriched KEGG pathway (down)
Table S4 Enriched KEGG pathway (up)
Table S5 Abundance statistics of sRNA target to BcTOR
Table S6 sRNA molecule sequence information
Table S7 sRNA blast information
Table S8 Potential off‐target genes in B. cinerea
Table S9 sRNAs matching to non‐targeting genes
Table S10 Primers used in this study


