Summary
Verticillium longisporum infects oilseed rape (Brassica napus) and Arabidopsis thaliana. To investigate the early response of oilseed rape to the fungal infection, we determined transcriptomic changes in oilseed rape roots at 6 days post‐inoculation (dpi) by RNA‐Seq analysis, in which non‐infected roots served as a control. Strikingly, a subset of genes involved in abscisic acid (ABA) biosynthesis was found to be down‐regulated and the ABA level was accordingly attenuated in 6 dpi oilseed rape as compared with the control. Gene expression analysis revealed that this was mainly attributed to the suppression of BnNCED3‐mediated ABA biosynthesis, involving, for example, BnWRKY57. However, this down‐regulation of ABA biosynthesis could not be observed in infected Arabidopsis roots. Arabidopsis ABA‐ defective mutants nced3 and aao3 displayed pronounced tolerance to the fungal infection with delayed and impeded symptom development, even though fungal colonization was not affected in both mutants. These data suggest that ABA appears to be required for full susceptibility of Arabidopsis to the fungal infection. Furthermore, we found that in both 6 dpi oilseed rape and the Arabidopsis nced3 mutant, the salicylic acid (SA) signalling pathway was induced while the jasmonic acid (JA)/ethylene (ET) signalling pathway was concomitantly mitigated. Following these data, we conclude that in oilseed rape the V. longisporum infection triggers a host‐specific suppression of the NCED3‐mediated ABA biosynthesis, consequently increasing plant tolerance to the fungal infection. We believe that this might be part of the virulence strategy of V. longisporum to initiate/establish a long‐lasting compatible interaction with oilseed rape (coexistence), which appears to be different from the infection process in Arabidopsis.
Keywords: ABA, Brassica napus, NCED3, RNA-Seq, Verticillium longisporum
Introduction
Brassica napus (oilseed rape) is an important oilseed crop grown worldwide and the cultivated area of oilseed rape has continuously increased in recent years (FAOSTAT, 2016). Lack of sufficient crop rotation and tillage operations results in an enrichment of soilborne pathogens, posing a real threat to crop cultivation (De Coninck et al., 2015). Verticillium longisporum is a hemibiotrophic soilborne pathogen causing verticillium stem striping in susceptible oilseed rape (De Coninck et al., 2015; Depotter et al., 2016). In this compatible interaction fungal hyphae enter the host through the roots without developing specific infection structures, and grow intra‐ and intercellularly through the root cortex towards the central cylinder. Fungal colonization of xylem vessels was thought to affect the xylem function and the transpiration stream, restricting water and nutrient transportation (Depotter et al., 2016; Eynck et al., 2007; Floerl et al., 2008), but a recent study showed that V. longisporum does not reduce xylem function in Brassica napus (Lopisso et al., 2017). In the field, symptoms on winter oilseed rape plants, including stem striping, appear in the late growing season, whereas stunted growth, leaf chlorosis as well as reduced leaf area are typically observable on root dip‐inoculated younger plants (Depotter et al., 2016; Eynck et al., 2009). This, together with the fungal growth within the plant, prevents an efficient disease management (Eynck et al., 2007; Happstadius et al., 2003). Efforts have been made to introduce V. longisporum resistance from B. rapa and B. oleracea genomes via resynthesis approaches, and several genotypes with increased resistance have been reported (Happstadius et al., 2003; Rygulla et al., 2007a, 2007b). However, so far no highly resistant germplasm of B. napus is available for cultivation.
Intriguingly, this plant–fungus interaction appears to involve post‐transcriptional gene silencing (PTGS) (Ellendorff et al., 2009). Argonaute (AGO) proteins and microRNAs directly modulate the infection process of Verticillium fungi and there is increasing evidence that noncoding RNAs play a pivotal role in cross‐kingdom RNA interference (Shen et al., 2014; Wang et al., 2016; Zhang et al., 2016). On the phytohormone level the plant–V. longisporum interaction has also been investigated in Arabidopsis thaliana (Floerl et al., 2012; Iven et al., 2012; Johansson et al., 2006; Ralhan et al., 2012), particularly regarding the involvement and role of phytohormone pathways relying on salicylic acid (SA) and jasmonic acid (JA) derivatives or ethylene (ET). It has been demonstrated that V. longisporum requires a COI1‐dependent but JA‐Ile‐independent mechanism to efficiently colonize Arabidopsis (Ralhan et al., 2012). However, since the Arabidopsis npr1‐1 mutant is more susceptible and shows decreased responses to the ethylene precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) or methyl jasmonate (MeJA) pretreatments, involvement of the SA pathway in the plant–fungus interaction was also postulated (Johansson et al., 2006). Another indication for the involvement of ET and SA in the Arabidopsis–V. longisporum interaction came from the observation that SA‐dependent PR1 and PR2 as well as the ET‐dependent PR4 expression was elevated at 7 days post‐inoculation (dpi) (Johansson et al., 2006).
The role of abscisic acid (ABA) in plant–pathogen interactions is somewhat controversial. ABA is generally believed to benefit plant resistance to abiotic stress, but it is also involved in stomatal closure and callose production and can therefore contribute to resistance against bacteria (Ton et al., 2009). An increased permeability of the cuticle and enhanced reactive oxygen species (ROS) production were observed in aba2 and aba3 mutants, which resulted in increased resistance to Botrytis cinerea (Cui et al., 2016; L’Haridon et al., 2011). ABA can contribute to plant defence against necrotrophic pathogens via enhancement of the JA response (Lorenzo et al., 2004). Hok et al. (2014) reported that the Arabidopsis ABA signalling mutant abi1‐1 was more susceptible to Hyaloperonospora arabidopsidis (Hpa), which is different from the observations made for V. longisporum by Johansson et al. (2006). Elevated ABA levels were found to be correlated with increased rice susceptibility to Xanthomonas oryzae pv. oryzae (Xu et al., 2013) and ABA was reported to negatively regulate the elicitor‐induced synthesis of the phytoalexin capsidiol in tobacco (Mialoundama et al., 2009) or to abolish the PTI‐induced chloroplastic reactive oxidative burst (de Torres‐Zabala et al., 2015) as well. Some bacterial pathogens are even able to manipulate the phytohormone homeostasis by increasing ABA levels in plants, thereby compromising plant defence reactions (de Torres‐Zabala et al., 2007, 2009). As demonstrated in the Ustilago maydis–Zea mays pathosystem, the fungus was capable of producing ABA, resulting in increased ABA levels within the plant cells at the early infection stages, which probably serves as part of the fungal virulence strategy (Morrison et al., 2015). Similar observations have been reported by works on B. cinerea, Cercospora spp., Fusarium spp., Rhizoctonia spp., Magnaporthe grisea and Magnaporthe oryzae (Cao et al., 2011; Siewers et al., 2006; Spence et al., 2015). On the other hand, ABA accumulation was suppressed during infection of Arabidopsis with Alternaria brassicicola (Flors et al., 2008). ABA promoted plant susceptibility to B. cinerea, while contributing to resistance against Sclerotinia sclerotiorum (Mbengue et al., 2016). An increase in SA was reported in xylem sap of oilseed rape after infection with V. longisporum (Ratzinger et al., 2009). Thus, depending on the plant–pathogen system, ABA can influence plant defence response in a positive or negative manner. Nevertheless, the role of ABA in the B. napus–V. longisporum interaction remains largely unsolved.
Although molecular understanding of plant–Verticillium interactions has been achieved with the model plant A. thaliana in recent years, less data is available for oilseed rape. The recently published genome sequence of B. napus (Chalhoub et al., 2014; Sun et al., 2017) and advanced parallel deep‐sequencing technology provide an opportunity to globally investigate the reprogramming of gene expression of oilseed rape in response to pathogen attacks. To get a deeper insight into the early transcriptional responses of oilseed rape to Verticillium infection, we conducted a comparative transcriptomic analysis between Verticillium‐infected and non‐infected oilseed rape at 6 dpi using RNA‐Seq experiments. Interestingly, a large fraction of ABA biosynthesis‐related genes was found to be significantly suppressed in the oilseed rape roots after infection with V. longisporum isolate 43 (Vl43), but not in Arabidopsis roots. This provoked us to hypothesize a possible role of ABA suppression in the early oilseed rape–fungus interaction, being distinct from Arabidopsis. Here, we report that the Vl43 infection triggers suppression of NCED3‐mediated ABA biosynthesis in the oilseed rape roots at the early infection stage. We demonstrate that ABA is required for full susceptibility of Arabidopsis to Vl43 infection. Thus, in oilseed rape the fungus‐induced suppression of ABA at the early infection stage may be part of the virulence mechanism, allowing the fungus to establish a long‐lasting compatible interaction with oilseed rape, which differs from the Arabidopsis–V. longisporum pathosystem.
Results
Transcriptomic analysis indicates suppression of ABA‐related gene expression in oilseed rape roots at the early infection stage
To investigate the early response of oilseed rape plants to the fungal infection, transcriptomic changes in oilseed rape roots in response to infection with Vl43 were investigated by RNA‐Seq at 6 dpi, in which non‐infected roots served as a control. The scatterplot in Fig. S1 contains all detected unigenes, being up‐regulated with a log2‐ratio ≥ 1 or down‐regulated (log2‐ratio ≤ −1). In total about 20% of all identified unigenes were differentially expressed at 6 dpi in oilseed rape as compared to the control (Fig. S1 and Table S1). Furthermore, we also analysed the Arabidopsis transcriptome after 6 dpi with Vl43 from root material by RNA‐Seq (Fig. S1 and Table S1). Differentially expressed genes (DEGs) were classified upon gene ontology (GO) analysis using KOBAS (Table S2; Wu et al., 2006) and the 25 most significantly affected biological processes are displayed in Table 1 (see Table S3 for the complete list). Intriguingly, GO terms for responses to ABA and ABA biosynthesis were one of the most affected biological processes in B. napus (Table 1); thus, we investigated the general biosynthesis of the phytohormones SA, JA, ET and ABA upon the GO terms in more detail (Fig. 1), revealing a significant difference between the plant species with respect to ABA biosynthesis. Following this, we searched for more ABA‐related genes in the whole oilseed rape RNA‐Seq dataset with the help of co‐expression analysis by an extensive ATTED‐II analysis with the database of closely related model plant Arabidopsis (Fig. S2; Obayashi et al., 2007). This led to the identification of 85 potentially ABA‐related DEGs (Fig. 2/Fig. S9). A meta‐analysis (Hruz et al., 2008) of the Arabidopsis homologues of these ABA‐related genes confirmed the involvement of the selected DEGs in diverse ABA‐mediated stress responses such as cold, drought, heat and salt treatments. Most (65) of the 85 ABA‐related genes that were suppressed at the early infection by Vl43 were found to be highly up‐regulated by ABA and abiotic stress treatments (Fig. 2/Fig. S9). The opposite can be observed only for several of the 20 Vl43 up‐regulated genes, which are displayed above the line in Fig. 2 (Fig. S9). Some of these genes have been described to function as negative regulators in ABA signalling, which could contribute to the overall down‐regulation of ABA responses.
Table 1.
GO enrichment analysis showing the 25 most significantly enriched GO terms for biological processes derived from all differentially expressed genes (DEGs) (log2 ≥ |1|) for Brassica napus and Arabidopsis thaliana
| Brassica napus | Arabidopsis thaliana | ||
|---|---|---|---|
| GO term | P‐value | GO term | P‐value |
| Response to abscisic acid | 0.00000019 | Cell wall organization | 0.00000039 |
| Response to hydrogen peroxide | 0.00002600 | Protein phosphorylation | 0.00002800 |
| Response to cadmium ion | 0.00009300 | Protein autophosphorylation | 0.00020000 |
| Response to water deprivation | 0.00012000 | TM tyrosine kinase signalling pathway | 0.00076000 |
| Response to high light intensity | 0.00033000 | Secondary metabolite biosynthesis | 0.00208000 |
| Response to salt stress | 0.00049000 | Peptidyl‐serine phosphorylation | 0.00289000 |
| Seed oilbody biogenesis | 0.00049000 | Signal transduction | 0.00349000 |
| Reductive pentose‐phosphate cycle | 0.00091000 | Chlorophyll biosynthetic process | 0.00436000 |
| Protein sumoylation | 0.00141000 | Regulation of monopolar cell growth | 0.00719000 |
| Photosynthesis, light harvesting in PS I | 0.00160000 | Root hair cell differentiation | 0.00840000 |
| Cell differentiation | 0.00183000 | Ion transport | 0.00965000 |
| Protein import into chloroplast stroma | 0.00240000 | Pectin catabolic process | 0.00987000 |
| Protein stabilization | 0.00261000 | Cell wall organization or biogenesis | 0.00992000 |
| Non‐photochemical quenching | 0.00274000 | Proteolysis/protein catabolic process | 0.01020000 |
| Vegetative/reproductive phase transition | 0.00315000 | Regulation of pollen tube growth | 0.01190000 |
| Abscisic acid biosynthetic process | 0.00360000 | Response to light stimulus | 0.01407000 |
| Lipid storage | 0.00373000 | Cellular response to iron ion starvation | 0.01564000 |
| Lignin biosynthetic process | 0.00376000 | DNA replication initiation | 0.01604000 |
| Response to heat | 0.00427000 | Cell wall modification | 0.01674000 |
| Protein–chromophore linkage | 0.00452000 | Base‐excision repair | 0.01699000 |
| Carbohydrate biosynthetic process | 0.00486000 | Intracellular signal transduction | 0.01797000 |
| Cotyledon vascular tissue formation | 0.00529000 | Chloroplast fission | 0.01859000 |
| Purine ribonucleoside metabolism | 0.00643000 | Cellular calcium ion homeostasis | 0.01931000 |
| Response to desiccation | 0.00684000 | Terpenoid biosynthetic process | 0.01967000 |
| Response to cytokinin | 0.00784000 | Lipid storage | 0.02079000 |
Figure 1.
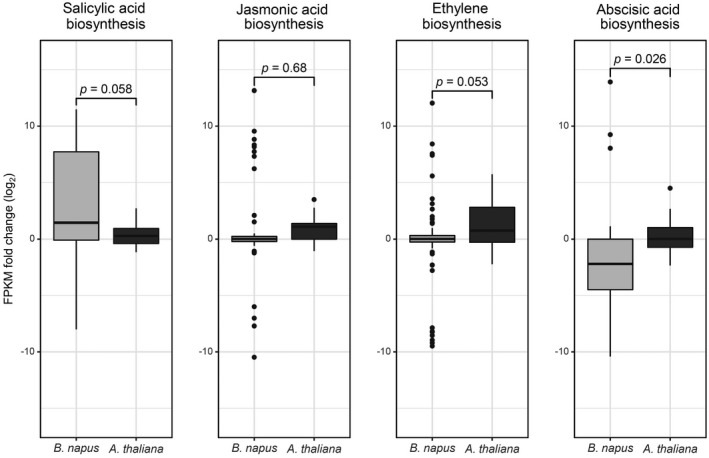
Verticillium longisporum‐induced differential expression of genes annotated to stress‐related phytohormone biosynthetic processes. For both plant species boxplots represent the log2 fold change expression of all genes annotated to the GO terms for salicylic acid (GO:0009697), jasmonic acid (GO:0009695), ethylene (GO:0009693) and abscisic acid (GO:0009688) biosynthetic processes that were differentially regulated in at least one species. P‐values calculated by Welch t‐tests indicate the significant difference between Brassica napus and Arabidopsis thaliana for each process.
Figure 2.
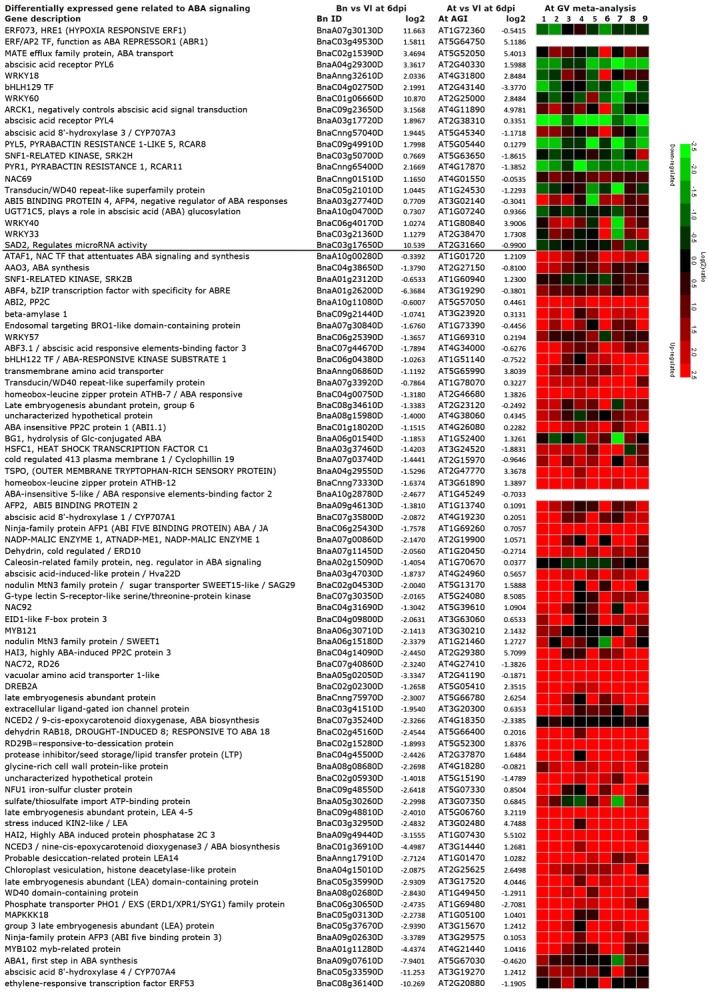
Differentially expressed genes related to abscisic acid (ABA) signalling as identified by RNA‐Seq of Verticillium longisporum (Vl43)‐infected oilseed rape (Bn) and Arabidopsis (At) roots at 6 days post‐inoculation. The line separates up‐and down‐regulated genes in response to the Vl43 infection of Brassica napus. The meta‐analysis of the orthologous Arabidopsis counterparts in response to various abiotic stress treatments focuses on ABA (1: AT‐00110, 2: AT‐00420), high light (3: AT‐00246), cold (4: AT‐00640), drought (5: AT‐00560, 6: AT‐00419), heat (7: AT‐00387), osmotic (8: AT‐00120) and salt stress (9: AT‐00534) treatments.
To verify the RNA‐Seq data, we selected some significantly expressed marker genes for several phytohormone pathways from the RNA‐Seq data and comparatively analysed their expression profiles in three independent biological replicates by RT‐qPCR. To enable an easier data comparison, we calculated the log2 values for the qPCR results and confronted them with the RNA‐Seq data. As shown in Table S4 the RNA‐Seq data could be generally confirmed.
ABA biosynthesis is attenuated in infected oilseed rape roots
The overall suppression of the ABA biosynthesis process at 6 dpi in oilseed rape roots implies an attenuated ABA production during the early interaction with V. longisporum. To facilitate the characterization of the early plant–fungus interactions, we established an ‘in vitro’ infection system, which allows a more accurate monitoring of the infection process and facilitated the generation of equally infected roots (Fig. S3). In Fig. S3A we show that Vl43 infection does not cause such severe symptoms in oilseed rape as observed in Arabidopsis (Fig. S4A), even though in both cases the fungus had successfully entered the plant after 3–4 dpi (Figs S3B and S4B). In a next step, we determined the ABA levels in leaf and root tissues at 3 and 6 dpi, while non‐infected plants served as controls. As shown in Fig. 3A, the basal ABA level remained relatively constant in the non‐infected roots and leaves (~2 ng ABA/100 mg fresh weight (FW) and ~10 ng ABA/100 mg FW, respectively) at 3 and 6 dpi. In response to the fungal infection ABA levels in the leaves were elevated approximately two‐fold. However, in root tissue we initially observed a slight increase at 3 dpi, but at 6 dpi the ABA level in oilseed rape was found to be strongly reduced (Fig. 3A), which is consistent with the RNA‐Seq data from 6 dpi. In addition, we analysed the expression profiles of the ABA biosynthesis gene NCED3 and one of its transcriptional activators, WRKY57, in response to the fungal infection (Fig. 3B). The expression patterns of the both genes behaved largely analogous to the dynamic changes in the ABA content in oilseed rape leaves. Notably, NCED3 and WRKY57 were already suppressed in roots at 3 dpi. These data suggest that in oilseed rape roots the ABA content was reduced at the early infection stage mainly via the suppression of ABA biosynthesis genes.
Figure 3.
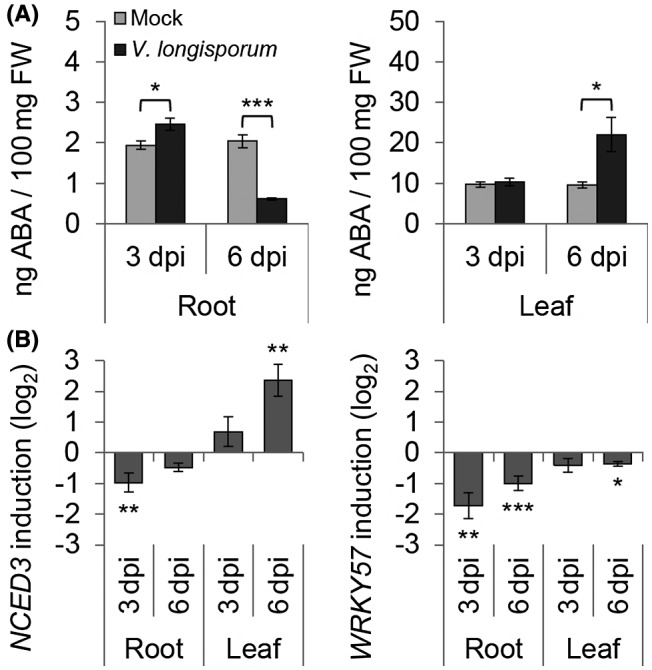
Abscisic acid (ABA) regulation on Verticillium longisporum (Vl43) infection in Brassica napus. Plant material of in vitro‐grown mock‐ and Vl43‐treated oilseed rape roots and leaves was sampled at 3 and 6 days post‐inoculation (dpi). (A) Competitive ELISA was applied to quantify the amount of ABA in 100 mg FW plant tissue. (B) RT‐qPCR analysis of the ABA biosynthesis key gene NCED3 and one of its transcriptional activators, WRKY57. Relative expression (log2) to PP2A/Actin2 was calculated with efficiency‐corrected Cq values and the induction (Vl43 vs mock) in response to the inoculation is displayed for each gene. Error bars indicate the standard error of the mean for three independent biological repetitions and statistics were performed using Student’s t‐test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001), comparing mock‐ and Vl43‐treated samples, respectively.
ABA biosythesis is not affected by Vl43 infection in Arabidopsis
In a similar experiment, transcriptomic changes in Vl43‐infected Arabidopsis were analysed by RNA‐Seq from root material obtained from the ‘in vitro’ infection system (Fig. S4). The results are summarized in Table S1. In total, about 30% of unigenes identified were differentially expressed in Arabidopsis roots at 6 dpi as compared to the non‐infected control (Fig. S1). By GO annotation these DEGs were analysed for their participation in biological processes (Fig. 1 and Table S2). Differing from oilseed rape, GO terms for responses to ABA and ABA biosynthesis were not significantly enriched in Arabidopsis. Furthermore, the comparison of the expression patterns of all DEGs assigned to the GO terms for biosynthesis of the phytohormones SA, JA, ET and ABA differed between oilseed rape and Arabidopsis roots (Fig. 1). While SA biosynthesis is predominantly induced in oilseed rape, JA/ET biosynthesis appears to be up‐regulated in Arabidopsis. Most notably, genes associated with ABA biosynthesis were not found to be collectively suppressed in Arabidopsis roots at 6 dpi (Figs 2 and 4B). By contrast to oilseed rape, ABA levels significantly increased in both roots and leaves of Arabidopsis on Vl43 infection (Fig. 4A), Accordingly, we did not observe down‐regulation of the ABA synthesis gene NCED3 and the transcriptional activator WRKY57 (Fig. 4B).
Figure 4.
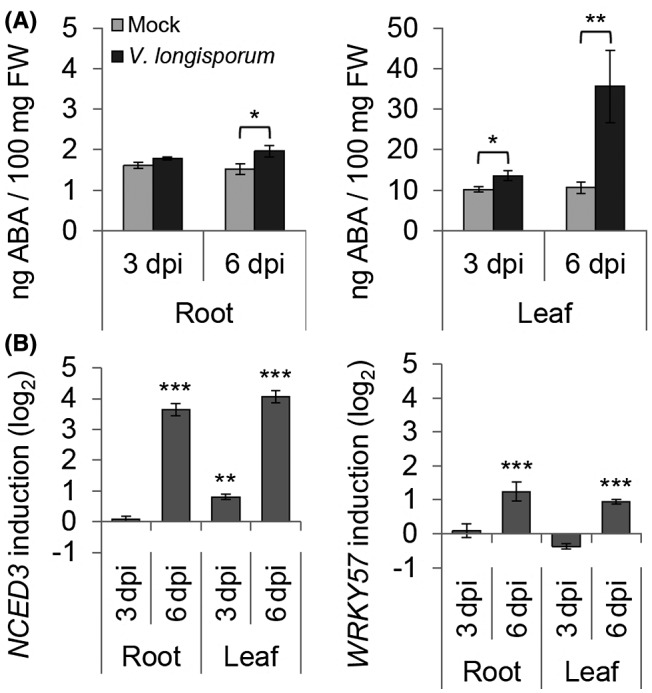
Abscisic acid (ABA) regulation on Verticillium longisporum (Vl43) infection in Arabidopsis thaliana. Plant material of in vitro‐grown mock‐ and Vl43‐treated oilseed rape roots and leaves was sampled at 3 and 6 days post‐inoculation (dpi). (A) Competitive ELISA was applied to quantify the amount of ABA in 100 mg FW plant tissue. (B) RT‐qPCR analysis of the ABA biosynthesis key gene NCED3, one of its transcriptional activators, WRKY57. Relative expression (log2) to PP2A/Actin2 was calculated with efficiency‐corrected Cq values and the induction (Vl43 vs mock) in response to the inoculation is displayed for each gene. Error bars indicate the standard error of the mean for three independent biological repetitions and statistics were performed using Student’s t‐test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001), comparing mock‐ and Vl43‐treated samples, respectively.
Suppression of ABA‐related genes goes along with an inhibition of the JA/ET pathway in oilseed rape roots
To investigate the impact of the ABA‐response suppression on other phytohormone pathways, we analysed the expression levels of three typical phytohormone marker genes PR1 (SA marker), PDF1.2 (JA marker) and ETR2 (ET marker) in response to the fungal infection (Fig. 5). At 3 dpi we observed only for ETR2 a slight induction (0.82‐fold log2 induction; P = 0.011) in oilseed rape roots, while at 6 dpi the expression of PR1 was strongly induced (8.97‐fold log2 induction; P < 0.001). However, the expression level of PDF1.2 was not significantly increased and no significant induction was observed for ETR2 at 6 dpi (Fig. 5A). These results suggest that V. longisporum might be able to interfere with the phytohormone crosstalk, e.g. via suppression of the NCED3‐mediated ABA biosynthesis accompanied with damped JA/ET responses at the early infection stage. We investigated the same phytohormone marker genes in infected Arabidopsis roots (Fig. 5B) and found PR1 already significantly induced at 3 dpi and further increased to a 7.3‐fold log2 induction at 6 dpi (P < 0.001). Additionally, PDF1.2 was strongly induced (7.59‐fold log2 induction; P < 0.001) at 6 dpi and the ethylene marker ETR2 was slightly (0.71‐fold log2 induction; P = 0.049) up‐regulated at the 6 dpi time point, which clearly differed from oilseed rape.
Figure 5.
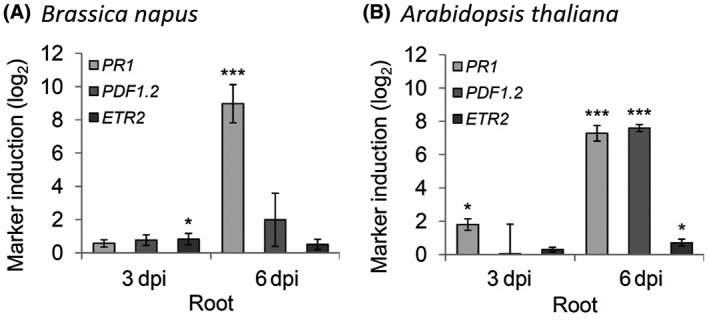
Effects on main defence hormone pathway marker gene expression. The expression of three typical phytohormone marker genes was investigated by RT‐qPCR in root material from Brassica napus (A) and Arabidopsis thaliana (B). PR1 for SA, PDF1.2 for JA and ETR2 for ET. Relative expression (log2) to PP2A/Actin2 was calculated with efficiency‐corrected Cq values and the induction (Vl43 vs mock) in response to the inoculation is displayed for each gene. Error bars indicate the standard error of the mean for three independent biological repetitions and statistics were performed using Student’s t‐test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001), comparing mock‐ and Vl43‐treated samples, respectively.
ABA‐deficient Arabidopsis mutants display reduced symptom development
To clarify the role of ABA in the plant–fungus interaction, we challenged the ABA biosynthesis mutants nced3 and aao3 with the Vl43 infection, and compared the results with Col‐0 wild‐type plants with respect to disease progression and symptom development. As compared to Col‐0, both nced3 and aao3 knockout mutant plants grown on soil displayed reduced symptoms as well. At 21 dpi, the stunted shoot growth and senescence‐like symptoms became obvious in the wild‐type plants, but were less pronounced in the nced3 and aao3 mutant. At 28 dpi, the wild‐type plants severely suffered from the infection and displayed strong growth depression as well as senescence‐like symptoms, while the nced3 and aao3 were much less affected, suggesting a delayed/impaired disease progression (Figs 6A and 7A). Comparing infected with uninfected plants, the relative rosette diameter and fresh weight were significantly enhanced in both mutants compared to Col‐0 (Fig. 6B,C). Although symptoms in the mutants were much less pronounced, the fungal biomass was not reduced as compared to the wild‐type (Figs 6D and 7B). These data demonstrate that in Arabidopsis the impaired ABA biosynthesis strongly affects symptom development, but does not negatively affect fungal colonization. In addition, we observed that external application of ABA on oilseed rape appears to impair the fungal infection progression, resulting in a reduced hypocotyl colonization as measured by determination of fungal DNA within hypocotyls (Fig. S8B,C), albeit data were not statistically significant due to a high variation in fungal infection and low case number in this preliminary experiment. Although ABA application at 6 dpi did not yet show altered symptoms, the observed tendency in decreased fungal colonization is consistent with observations that the Arabidopsis ABA mutants showed enhanced tolerance to Vl43 (Figs 6D and 7B).
Figure 6.
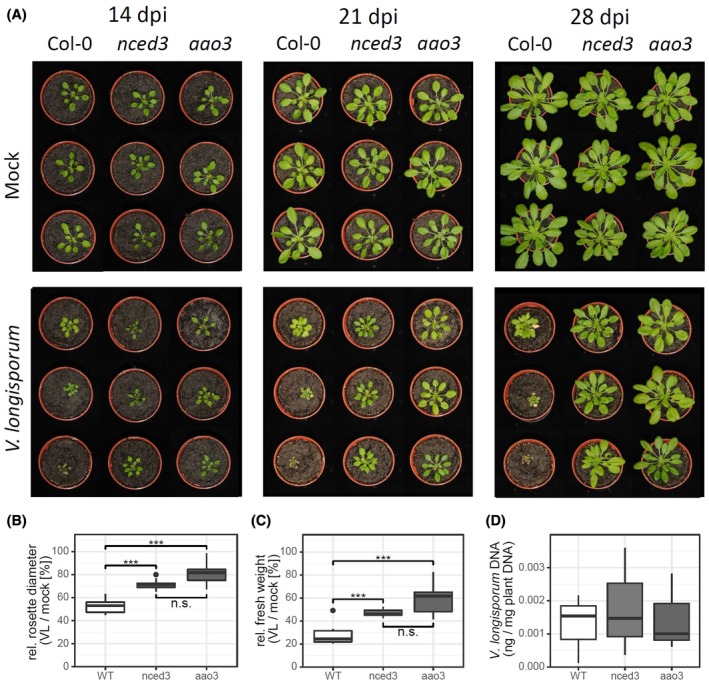
The Arabidopsis nced3 and aao3 mutants show delayed development of disease symptoms. Plants grown on sand/soil were photographed at the indicated time points (A). In an independent experiment plants were sampled at 24 days post‐inoculation (dpi) and the rosette diameter (B), the fresh weight (C) and relative amount of fungal DNA (D) were determined as described in the Experimental Procedures. Multiple comparison of means was performed by ANOVA and a post hoc Tukey test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Figure 7.
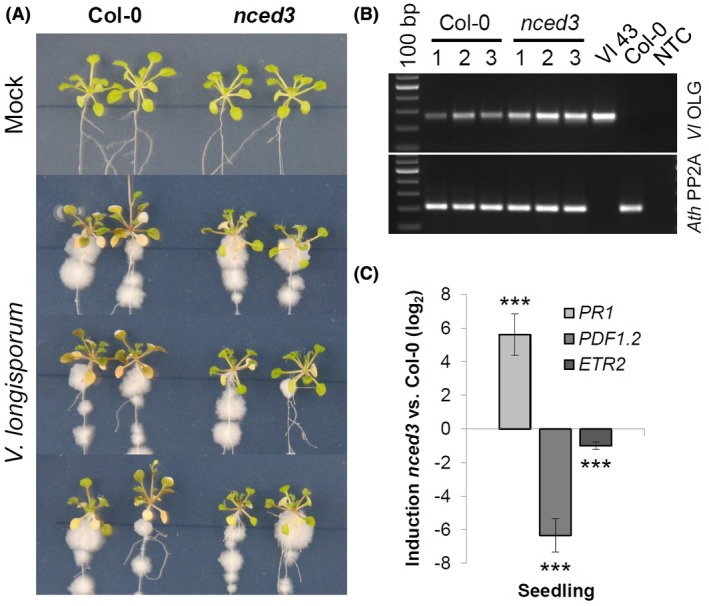
In vitro grown Arabidopsis nced3 plants show delayed disease symptom development and constitutively altered hormone marker expression. (A) Arabidopsis seedlings were grown on MS plates and photographed at 12 days post‐inoculation (dpi), indicating that nced3 knockout plants are more tolerant to Verticillium longisporum (Vl43) infection compared to Col‐0 wild‐type plants. (B) The infection progress was monitored by the amount of fungal DNA determined by semiquantitative PCR using leaf material harvested at 12 dpi. Comparing fungal amplicon signals to plant PP2A signals indicates that fungal growth was rather enhanced in the nced3 mutant. (C) Expression analysis of PR1 for salicylic acid, PDF1.2 for jasmonic acid and ETR2 for ethylene in the Arabidopsis nced3 mutant. Material of in vitro‐grown 3‐week‐old Arabidopsis seedlings of nced3 and Col‐0 was sampled. Relative expression (log2) to PP2A/Actin2 was calculated with efficiency‐corrected Cq values and the induction (nced3 vs Col‐0) in response to the nced3 knockout is displayed for each gene. Error bars indicate the standard error of the mean for three independent biological repetitions and statistics were performed using Student’s t‐test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001), comparing nced3 and Col‐0 samples, respectively.
To understand possible mechanisms underlying the nced3‐mediated tolerance, we compared the expression levels of the SA, JA and ET marker genes in the nced3 mutant and Col‐0 wild‐type plants. As shown in Fig. 7C, the nced3 mutant displayed constitutively higher PR1 levels, while PDF1.2 and ETR2 were both strongly suppressed as compared to Col‐0, suggesting that the suppression of ABA biosynthesis affects other phytohormone pathways in the nced3 mutant in a way similar to that observed in Vl43‐infected oilseed rape roots (Figs 5 and 7C). The lack of a functional NCED3 gene in the Arabidopsis mutant obviously promoted SA signalling while reducing JA/ET signalling. This could be a reason for the tolerance to fungal infection together with delayed disease symptoms and is in agreement with our observation that PDF1.2 expression was attenuated after 6 dpi in B. napus as compared to Arabidopsis (Fig. 5).
Discussion
To shed light on the early molecular events in the plant–V. longisporum interaction, we investigated transcriptomic changes at 6 dpi in B. napus and A. thaliana roots by RNA‐Seq experiments. The generated RNA‐Seq data provided us with a comprehensive view of molecular plant–Verticillium interactions. Surprisingly, most significantly enriched GO terms differed between oilseed rape and Arabidopsis, especially with respect to ABA signalling and biosynthesis (Table 1). ABA‐biosynthesis related genes were overall down‐regulated and ABA levels were accordingly decreased in oilseed rape roots at 6 dpi (Figs 1 and 2). Although interactions of Vl43 with both oilseed rape and Arabidopsis are compatible, we observed that infected B. napus plants display a less diseased phenotype (Figs S3A, S4A and S5), leading to the assumption that differential ABA signalling plays a pivotal role in the mediation of symptomless infection in oilseed rape within the early infection stage. While the ABA level more or less increased in leaves of oilseed rape, it was found to be significantly reduced in the roots at 6 dpi (Fig. 3A). These data underpin the RNA‐Seq results, reinforcing the hypothesis of a pivotal role for ABA in regulating the early plant–fungus interaction in oilseed rape. Interestingly, in accordance with the RNA‐Seq data, a reduction in ABA levels was not observed in the infected Arabidopsis roots, where the ABA levels by contrast appear to be gradually elevated during the infection progression (Fig. 4A). This observation is congruent with reports from Iven et al. (2012), who reported an increase in ABA levels in V. longisporum‐infected Arabidopsis roots at 6 and 8 dpi. It was furthermore shown that in Arabidopsis leaves ABA increased at 2 dpi (Roos et al., 2014) and also later at 15 dpi in Arabidopsis petioles in response to V. longisporum (Ralhan et al., 2012). The increase in ABA in the leaves of oilseed rape and Arabidopsis likely results from side effects, provoked by the fungal invasion into the roots and growth in the xylem, which could lead to an initially reduced water supply within the leaves. However, Reusche et al. (2012) reported that at 7 dpi V. longisporum triggers de novo xylem formation that can compensate for compromised water transport in brassicaceous host plants, which can explain why this vascular pathogen does not affect drought stress responses in oilseed rape (Lopisso et al., 2017). Furthermore, the increased ABA levels in Arabidopsis might contribute to an increased drought stress tolerance that has been observed for V. longisporum‐infected plants (Reusche et al., 2014).
In plants, the ABA level can be influenced by blocking ABA biosynthesis, by ABA inactivation or by degradation. In addition to suppressed ABA biosynthesis‐related genes in the RNA‐Seq data, some ABA catabolic genes described by Finkelstein (2013) have also been identified, e.g. CYP707A3 is up‐regulated, while CYP707A1 is down‐regulated (Fig. 2). Also, two genes involved in ABA inactivation (UGT71C5 and BG1; Liu et al., 2015b) were found in the RNA‐Seq data, which were up‐ and down‐regulated by the fungal infection, respectively (Fig. 2). Several ABA‐related genes up‐regulated in the B. napus response to V. longisporum infection have been described as negative regulators of the ABA response, e.g. WRKY18, WRKY33, ARCK1 and bHLH129. Others encode ABA receptor components (e.g. PYL4, 5 and 6), which might render the plant more sensitive to low ABA levels. Several ABA‐responsive genes containing the ABRE (C/TACGTGG/TC) and DRE (A/GCCGAC) cis‐elements in their promoters, including RD29B, NAC072, Hva22c, ERD10, LEA14 and RAB18 (Nakashima et al., 2014; Yoshida et al., 2014), were also found to be suppressed in oilseed rape (Fig. 2). Following this, we conclude that the infection resulted in a drop in ABA levels in the infected oilseed rape roots primarily by reducing ABA biosynthesis via suppression of gene expression of this process.
The RNA‐Seq data and qPCR experiments show that the expression levels of NCED3, a key gene involved in stress‐induced ABA biosynthesis (Jensen et al., 2013), and WRKY57, one positive regulator of NCED3 (Jiang et al., 2012), were both suppressed in the 6 dpi oilseed rape roots, supporting a crucial role of NCED3 for ABA biosynthesis (Fig. 3B). As the expression profile of NCED3 correlated mostly with that of WRKY57, we believe that suppression of ABA production is initiated upstream of this transcription factor in oilseed rape. Accordingly, this suppression is absent in the infected Arabidopsis roots in which both genes were up‐regulated (Fig. 4B), suggesting the involvement of the NCED3‐mediated ABA biosynthesis specifically in the early interaction of Vl43 and oilseed rape. Genetic evidence for a crucial role of NCED3‐mediated ABA biosynthesis in the plant–V. longisporum interaction came from the analysis of two Arabidopsis ABA‐deficient mutants, nced3 and aao3, both appearing to tolerate Vl43 colonization as compared to the wild‐type control, suggesting that ABA is required for full symptom development of Arabidopsis in response to Vl43 infection.
Since ABA is often reported to play a role in attenuating defence reactions towards biotic stress (Cao et al., 2011; Clay et al., 2009; Mialoundama et al., 2009), attempts to elevate ABA levels might be a genuine strategy for pathogens to manipulate their hosts. This can occur by producing ABA themselves (Cao et al., 2011; Morrison et al., 2015; Siewers et al., 2006; Spence et al., 2015; de Torres‐Zabala et al., 2015) or by activation of the plant ABA biosynthesis, for example via enhancing NCED3 gene expression as observed in the X. oryzae–rice interaction, resulting in increased susceptibility (Xu et al., 2013). Similar observations have been made for Pseudomonas syringae DC3000, which deploys type III effectors to induce the Arabidopsis NCED3 gene, resulting in elevation of ABA levels (de Torres‐Zabala et al., 2007, 2009). However, some Arabidopsis ABA mutants were more susceptible towards Pythium irregulare and A. brassicicola, thus ABA is believed to benefit plant responses against necrotrophic pathogens via enhancement of the JA response (Lorenzo et al., 2004). Accordingly, we found that the JA/ET response was compromised in oilseed rape when comparing the PDF1.2 and ETR2 expression level with that in Arabidopsis. Furthermore, we observed an elevated PR1 transcript accumulation that in Arabidopsis already started at 3 dpi was strongly induced at 6 dpi in both oilseed rape and Arabidopsis (Fig. 7). While SA, JA and ET can be negatively affected by ABA (Cao et al., 2011; Fan et al., 2009; Mauch‐Mani and Mauch, 2005; Ton et al., 2009), plants can attenuate ABA signalling through the action of SA/biotic stress‐inducible WRKY33 (Liu et al., 2015a) or via ET signalling (Anderson et al., 2004; Arc et al., 2013). Thus, the reciprocal antagonism between ABA and SA (Yasuda et al., 2008) might be the reason for ABA suppression in oilseed rape infected with V. longisporum. However, since the SA pathway appears to be activated in both plant species upon infection with V. longisporum, this phytohormone is likely not responsible for suppression of ABA in oilseed rape, although SA has been reported to exert a negative impact on ABA (Yasuda et al., 2008). On the contrary, the reduced ABA levels in B. napus could facilitate the relatively stronger induced expression of PR1 and SA biosynthetic genes in oilseed rape as compared to Arabidopsis at 6 dpi (Figs 1 and 5). It has been demonstrated that ABA can suppress PAL‐dependent SA biosynthesis (Cao et al., 2011) and promotes susceptibility in the rice–X.oryzae interaction by suppressing SA‐mediated defences (Xu et al., 2013). The SA increase in turn might then result in an attenuated expression of PDF1.2 (Thaler et al., 2012) and ETR2, which is consistent with our finding that in the nced3 mutant both PDF1.2 and ETR2 were suppressed while the PR1 was induced (Fig. 7C). It is therefore reasonable to speculate that this kind of phytohormone crosstalk at the early infection stage is fully employed in the decelerated and delayed disease symptom development in oilseed rape as compared to Arabidopsis. A similar pattern of differential marker expression for PR1 and PDF1.2 in B. napus roots was also reported by Kamble et al. (2013).
Interestingly, it appears that the fungal colonization is not impaired in A. thaliana ABA biosynthesis mutants as evidenced by similar amounts of fungal biomass in nced3 and aao3 plants as compared to Arabidopsis wild‐type plants. However, the external ABA application to oilseed rape reduced fungal colonization in hypocotyls as measured at 6 dpi, suggesting an impaired infection progression. This is in agreement with our hypothesis that the fungus may benefit from reduced ABA levels (Fig. S8). Whether ABA suppression is part of the plant defence response differing between oilseed rape and Arabidopsis or due to a host‐specific fungal influence is currently not clear and a highly intriguing question. As both plant species, B. napus and Arabidopsis, are susceptible to Vl43 infection, the observed up‐regulation of genes or accumulation of hormones might be caused by the pathogen infection process rather than by the plant defence response, which generally does not appear to be effective enough to restrict the disease progress. It has been demonstrated that in Arabidopsis the more resistant Bur ecotype showed a stronger induction of SA after V. longisporum infection than the more susceptible Ler ecotype, in which the level of ABA was higher (Häffner et al., 2014). Veronese et al (2003) showed that the Arabidopsis aba2‐1 mutant displayed reduced disease symptoms (anthocyanin accumulation) towards V. longisporum infection as compared to wild‐type plants (the strain Bob.70, 90‐02 was originally assigned to Verticillium dahliae; Inderbitzin et al., 2011, Table S2). These results are congruent with our findings that lower ABA levels go along with increased SA responses and result in reduced symptom development. This is further supported by analysis of the ABA mutants ahg2‐1 and aba1‐6, in which the SA pathway was constitutively activated, also conferring some resistance to the necrotrophic pathogens B. cinerea (Nishimura et al., 2009) and Plectosphaerella cucumerina (Sanchez Vallet et al., 2012), respectively.
Our data indicate that in oilseed rape SA biosynthesis is predominantly induced, while in Arabidopsis JA/ET biosynthesis is mainly up‐regulated on Vl43 infection. This discrepancy might be a hint for a specific evolutionary co‐existence of oilseed rape and V. longisporum as discussed by Eynck et al. (2009) and Inderbitzin et al. (2011). The fungus probably manipulates ABA signalling in oilseed rape roots in order to prevent a strong JA/ET resistance response and thereby establish a longer lasting compatible interaction with its host (coexistence). This would be in agreement with findings by Roos et al. (2015), who found for Arabidopsis that JA and MYC2 are required for full induction of TPS23/TPS27‐dependent production of monoterpenes, which stimulate germination and enhance V. longisporum invasion. Consequently, myc2‐1 mutants display higher resistance to V. longisporum. According to Anderson et al. (2004), MYC2 is responsive to both JA and ABA. In addition to down‐regulation of the positive regulator WRKY57, we observed differential expression of additional negative regulators of ABA signalling, including WRKY18 (Rushton et al., 2012; Shang et al., 2010; Xu et al., 2006), WRKY33 (Liu et al., 2015a) and bHLH129 (Tian et al., 2015), which were all up‐regulated at 6 dpi in oilseed rape roots. Furthermore, two NAC transcription factors, which were previously reported to be involved in ABA‐related responses, NAC72 and NAC69 (Fujita et al., 2004; Park et al., 2011), were down‐ and up‐regulated, respectively. Thus, we expect a much more complex network of transcription factors being involved in this regulation than presented in our summarizing model, which focuses for clarity on WRKY57‐ and NCED3‐dependent ABA biosynthesis (Fig. 8).
Figure 8.
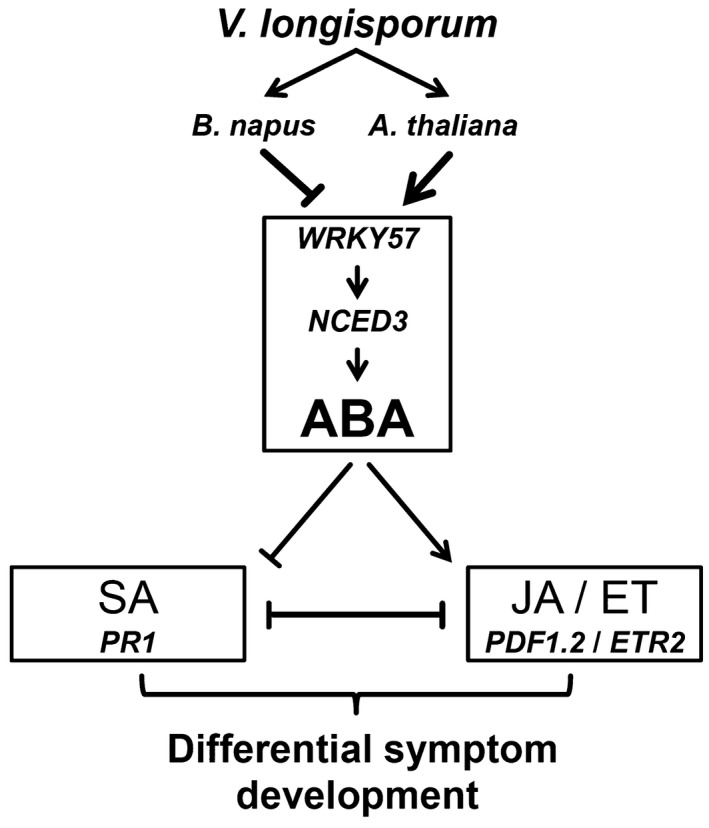
Simplified view of abscisic acid (ABA) signalling responses to the infection with Verticillium longisporum (Vl43) in oilseed rape and Arabidopsis roots at the early infection stages. ABA is functionally involved in plant–Vl43 interaction, which is different between oilseed rape and Arabidopsis at the early infection stages. Vl43 attenuates ABA signalling in oilseed rape roots by interference with NCED3‐mediated ABA biosynthesis triggering an enhanced salicylic acid (SA) and concomitantly impaired jasmonic acid (JA) and ethylene (ET) responses, which results in delayed symptom development. In Arabidopsis, JA/ET responses are not affected and full symptom development can be observed. The possible fungus‐induced suppression of ABA in oilseed rape may represent a specific virulence mechanism, allowing the fungus to successfully spread in the plant and coexist with its host in a longer lasting compatible interaction.
In conclusion, this study provides a molecular insight into early oilseed rape–V. longisporum interactions pointing to an important involvement of reduced ABA levels. In addition, our results constitute a unique platform for further characterizing and understanding this infection process, which may be useful for breeding of resistant oilseed rape varieties against V. longisporum infection.
Experimental Procedures
Fungal cultivation
Verticillium longisporum isolate Vl43 was provided by Dr Elke Diederichsen (FU Berlin, Germany) in the form of malt extract agar plates inoculated with the fungus. Glycerol stock cultures (22%) of Vl43 conidia were produced by transferring mycelium plugs into liquid Czapek–Dox medium. After 1 week growing in the dark on a rotating shaker, conidia were harvested with the help of a gauze filter (mesh 200 µm; Hydro‐Bios Kiel, Germany). Verticillium conidia from glycerol stocks were spread on potato dextrose agar (PDA) plates and grown for 2 weeks at room temperature (RT) in the dark. Plugs of these plates were again transferred to liquid Czapek–Dox medium and incubated on a rotary shaker for 3 days at RT in the dark to massively produce conidia. After 3 days, conidia were filtered through gauze, centrifuged for 8 min at 8000 rpm and resuspended in sterile Czapek–Dox medium to increase the concentration. Stocks for infection were supplemented with 86% glycerol to a final concentration of 22% and kept at −80 °C till the day of inoculation. At the day of inoculation stocks were thawed, centrifuged for 8 min at 8000 rpm and resuspended in tap water (‘in vivo’ infection) or sterile Czapek–Dox medium (‘in vitro’ infection) to a final concentration of 2 × 106 conidia/mL. Tap water and Czapek–Dox medium were used as mock treatments, respectively.
Plant material
Seeds of B. napus 'Express' (susceptible reference line #617 from NPZ Hans‐Georg Lembke KG, Holtsee, Germany), A. thaliana Col‐0 and the mutant lines (see below) were surface sterilized with 6% sodium hypochlorite/Tween 20 solution and 70% ethanol followed by washing (three times) with sterile distilled water.
Infection on soil of B. napus and A. thaliana
Sterilized seeds were first grown in vitro on ½ MS medium (0.5% sucrose) under short day conditions (8 h light, 22 °C). A. thaliana plants were grown in Petri dishes for 14 days whereas B. napus plants were grown in 250 mL salad cups for 7 days. At the day of inoculation plants were removed from the medium and root dipped in Vl43 conidia solution (or mock) for 15 min. B. napus plants were then transferred to plastic folding boxes (2 × 1.5 cm) filled with non‐sterile sand (Bauhaus hardware store, Belp, Switzerland) and fertilized with WUXAL® universal fertilizer‐treated tap water (Shen et al., 2014). A. thaliana plants were transferred to pots containing non‐sterile sand/soil (1:1) and also supplemented with fertilizer‐treated tap water. All the plants were placed in a climate cabinet (Vötsch VB 0714‐A, Reiskirchen, Germany) with short day conditions. For B. napus, roots of 15 infected and 15 non‐infected plants were sampled after 6 days and immediately flash‐frozen in liquid nitrogen. Photographs of A. thaliana plants were taken once per week to observe the infection process. At 24 dpi eight infected plants per line were deployed to measure and calculate fresh weight (FW) and diameter of rosettes in relation to the mean of mock‐treated controls. Statistics were calculated by a post hoc Tukey test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Infection on MS plates of B. napus and A. thaliana
Plants were pre‐grown as described for the in vivo infection. After 14 days A. thaliana plants were first transferred to 120 × 120 mm Petri dishes containing ½ MS (0.5% sucrose) and grown vertically for another 7 days before being inoculated. Seven‐day‐old B. napus plants were removed from the salad cups and placed on 120 × 120 mm Petri dishes containing ½ MS (0.5% sucrose) for direct inoculation and further cultivated under short day conditions. To inoculate the roots a brush was dipped in Vl43 conidia suspension (2 × 106/mL) or Czapek–Dox medium as mock treatment and used to distribute the conidia along the roots. Roots of B. napus plants were inoculated completely, whereas the upper 2 cm of A. thaliana roots were spared. Petri dishes were closed and plants were grown vertically in short day conditions. The progress of fungal growth and the infection was observed for the next 12 days (Figs 3A and 4A). Plant material was sampled at 3 and 6 dpi. Roots and leaves of B. napus and A. thaliana were sampled separately. All samples were immediately frozen in liquid nitrogen.
Verticillium genomic DNA (gDNA) detection in plant tissue
Brassica napus hypocotyls and Arabidopsis leaves from infected plants were sampled at the indicated time points. Each biological replicate consists of three individual hypocotyls, three in vitro Arabidopsis shoots or two complete Arabidopsis in vivo rosettes, respectively. For gDNA isolation the tissue was immediately flash‐frozen in liquid nitrogen and ground using a mortar and pestle. Total gDNA was isolated using cetyltrimethylammoniumbromide (CTAB) buffer (Rogers and Bendich, 1985) supplemented with β‐mercaptoethanol. For semiquantitative PCR amplification Dream Taq Polymerase (Thermo Scientific, Waltham, MA) was used according to the manufacturer's instructions (hypocotyls and in vitro shoots). For real‐time qPCR quantification of fungal DNA in rosettes the Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific) was used according to the manufacturer's instructions. Primers used for Vl43 detection (Eynck et al., 2007) are listed in Table S7. PCR was performed using the following conditions: 3 min at 95 °C; 35 cycles of 20 s at 95 °C, 20 s at 58 °C and 30 s at 72 °C. For semiquantitative PCR, products were run in a 1.7% agarose gel for 60 min at 75 V. Relative amounts of DNA were calculated by use of Image Lab software (Bio‐Rad, Hercules, CA). The amount of fungal DNA was normalized against PP2A signals from plant DNA. Error bars indicate the standard deviation (±SD) of three different biological repetitions and statistics were performed using Student’s t‐test (*P ≤ 0.05). For real‐time PCR, standard curves for the quantification of gDNA were calculated by dilution of a series of known concentrations of pure Arabidopsis and Vl43 DNA. For each sample the amount of fungal DNA in relation to plant DNA was calculated in ng/mg.
Vl43 hypocotyl assays
To detect Vl43 within the tissue of B. napus, hypocotyls were surface sterilized with a 6% sodium hypochlorite/Tween 20 solution and 70% ethanol for 3 min, followed by washing three times with sterile distilled water. The edges were cut and hypocotyls were placed on PDA plates. A possible fungal colonization of cotyledons and leaves at the observed time points was also checked, but not observed (Fig. S6). Pictures were taken with a stereomicroscope (Zeiss, Stereo Discovery.V20, Oberkochen, Germany) after 4 days.
Vl43 detection in A. thaliana roots
Roots of in vitro‐infected A. thaliana Col‐0 plants were sampled at 4 dpi and incubated in 10% KOH at 90 °C for 10 min. After that the samples were washed three times for 15 min in distilled water. Then the samples were stained in 10 µg/mL fluorescein isothiocyanate (FITC)‐wheat germ agglutinin (WGS) (Sigma‐Aldrich, St Louis, MO) and dissolved in 50 mM HEPES buffer, pH 7.5 (Roth GmbH + Co. KG, Karlsruhe, Germany), in the dark for 1 h. After staining the samples were washed three times for 15 min in 50 mM HEPES buffer in the dark. Samples were placed on microscope slides together with 50 mM HEPES buffer containing 2% DABCO and analysed using a confocal laser scanning microscope (Leica TCS SP1, Leica Microsystems, Wetzlar, Germany).
RNA isolation
Total RNA from different tissues was extracted by TRIzol® (Thermo Fisher Scientific Inc., Waltham, MA) reagent according to the manufacturer’s recommendation. The quality and concentration of RNA was controlled by gel electrophoresis and a NanoVue Plus Spectrophotometer (GE Healthcare Life Science, Chicago, IL).
RNA‐Seq data processing
Equal amounts of RNA from three biological replicates of mock‐ and Vl43‐inoculated soil‐grown oilseed rape roots as well as in vitro mock‐ and Vl43‐treated Arabidopsis roots were pooled for sequencing. Forward/reverse clean reads for B. napus and Arabidopsis were obtained by Illumina sequencing from Beijing Genomics Institute BGI (Hong‐Kong, China) and Novogene Co., Ltd (Hong‐Kong, China), respectively. Fragments per kilobase of transcript per million mapped reads (FPKM) values for all samples were calculated using the open‐source software tools HISAT and StringTie with default settings, as described by Pertea et al. (2016), with the B. napus genome (v. 4.1) and annotation (v. 5) (Chalhoub et al., 2014) and the TAIR10 reference genome/annotation (Lamesch et al., 2011) for Arabidopsis. GO terms for B. napus sequences were annotated by BLASTing the reference peptide library (v. 5) (Chalhoub et al., 2014) against A. thaliana reference peptides containing GO and KEGG annotations using KOBAS (Wu et al., 2006). The GO enrichment analysis was performed with R (R core Team, 2018) using the topGO (Alexa and Rahnenfuhrer, 2018) package. The bioinformatics pipeline is summarized in Fig. S7.
Data deposition
The data reported in this paper have been deposited in the National Center for Biotechnology Information BioProject archive, https://www.ncbi.nlm.nih.gov/bioproject, accession PRJNA419857 for B. napus and PRJNA563868 for A. thaliana).
Primer design and gene specificity
A list of the investigated genes and the primer pairs for amplification can be found in Tables S5 (B. napus) and S6 (A. thaliana). Primers were designed to detect all B. napus homeologues when applicable and checked with Vector NTI (Thermo Fisher Scientific) for their properties. Primers were synthesized by MWG Eurofins and the annealing temperature was set according to the respective data sheets. Primer specificity was checked on the NCBI homepage (www.ncbi.nlm.nih.gov/tools/primer-blast/) by PRIMER BLAST analysis and subsequent to the qPCR, the determination of a melting curve of the amplified PCR products was carried out, followed by gel electrophoresis of the qPCR products to confirm single product amplification.
Expression analysis by quantitative real‐time PCR
One microlitre of total RNA was treated with RNase‐free DNaseI (Fermentas) and then transcribed in a volume of 20 µL with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) into first‐strand cDNA according to supplier’s instructions. Two microlitres of a 1:10 diluted cDNA preparation was mixed with 18 µL Master Mix as described in the manual of the Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific). PCR was performed on a CFX96 Touch Real‐Time PCR Detection System (Bio‐Rad) using the following conditions: 3 min at 95 °C; 45 cycles of 10 s at 95 °C, 10 s at 59 °C, 10 s at 72 °C; and 10 s at 95 °C, melting curve from 65 °C to 95 °C. Gene expression was determined using the ΔCT method to calculate the relative expression according to Pfaffl (2001). Relative expression was calculated in relation to PP2A (At1G13320/BnaA06g33370D) and Actin2 (At3G18780/BnaA10g22340D) as reference genes, which have been found to be stably expressed in many plants under almost all conditions (Czechowski et al., 2005; Hruz et al., 2008; Liu et al., 2012) and with consideration of primer efficiency, which was estimated by standard curves for each gene generated by cDNA dilution series (1:10; 1:100; 1:1000). Each data point is based on three independent biological replicates measured with two technical replicates each. The relative expression to PP2A/Actin2 was log2 transformed and, to display the differential regulation, the induction was calculated as the difference between Vl43- and mock‐treated samples. Error bars indicate the standard error of the mean and statistics were performed using Student’s t‐test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001), comparing the relative log2 expression between mock‐ and Verticillium‐treated samples of each calculated induction.
ABA measurement from leaf and root material
Leaf and root tissue was groud under liquid nitrogen using mortar and pestle and 100 mg material was added to 1.5 mL 80% methanol and incubated overnight at 4 °C with shaking. Next day the 2 mL tubes were centrifuged for 10 min and 5000 rpm at 4 °C. One millilitre of the supernatant was transferred to a new tube and samples were dried in a speed vac until all liquid evaporated. The pellets were dissolved in 100 µL methanol by sonication for 15 min and afterwards 900 µL Tris‐buffered saline was added. One hundred microlitres of these samples was subjected to competitive ELISA from the Phytodetek® ABA Test Kit (AGDIA, Elkhart, IN) according to the manufacturer’s instructions and measured at 405 nm with an ELISA reader Model 680 (Bio‐Rad). The amount of ABA was calculated as ng/100 mg FW and each data point was based on three independent biological replicates measured with two technical replicates each. Error bars indicate the standard error of the mean and statistics were performed using the Student’s t‐test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001) comparing mock‐ and Vl43‐treated samples, respectively.
External application of ABA on oilseed rape seedlings
Seven‐day‐old oilseed rape seedlings were transferred on ½ MS medium without (mock) and with 10 µM ABA before inoculation. Seedlings were photographed at 6 dpi and hypocotyls were harvested for the quantification of fungal DNA as well as for Vl43‐hypocotyl assays.
Genotyping of A. thaliana T‐DNA lines
T‐DNA knockout lines were ordered from NASC and were genotyped according to the instructions of the SALK Institute (http://signal.salk.edu/tdnaprimers.2.html; Table S7). The nced3 knockout line has been selected on a T‐DNA insertion within exon 1, resulting in potential loss of function (At3G14440/GK‐129B08/N331021). The aao3 knockout line was selected on a T‐DNA insertion within exon 3, resulting in potential loss of function (At2G27150/SALK_072361C/N668480).
Statistical analyses
Graphs and plots were generated with Microsoft® Office 2007 and/or the ggplot2 package (Wickham, 2016) for R. All statistical calculations were performed with R (R core Team 2018). The comparison of means for two groups was performed using the Student’s or Welch t‐tests (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). Multiple comparison of means was performed by an analysis of variance (ANOVA) and a post hoc Tukey test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001) using SimComp (Hasler and Kluss, 2017).
Supporting information
Fig. S1 Scatterplot of all RNA‐Seq calculated FPKM values for all unigenes between mock‐ and Vl43‐infected samples for Brassica napus and Arabidopsis thaliana, respectively. Genes were considered to be differentially regulated when their log2 fold change was ≥1 (red) or ≤–1 (green).
Fig. S2 ATTED‐II search for ABA‐related co‐expressed genes with the database of closely related model plant Arabidopsis. The identified coregulatory networks are shown (A–E) and have been partly merged together. Genes that have also been found in the RNA‐Seq dataset are highlighted in yellow. Genes identified in the networks A–D are down‐regulated in the RNA‐Seq data, while genes from cluster E are up‐regulated.
Fig. S3 Characterization of the in vitro infection process of Brassica napus (Express 617) with Verticillium longisporum (Vl43). (A) Mock‐ and Vl43‐ inoculated B. napus seedlings were photographed at 3, 6 and 12 days post‐inoculation (dpi). (B) Hypocotyls were sampled and surface sterilized at different dpi and placed on potato dextrose agar to detect Vl43 in hypocotyls of infected B. napus seedlings. (C) Increment of fungal DNA in B. napus hypocotyls, measured by semiquantitative PCR using Verticillium‐ specific primers is displayed as the relative amount of fungal DNA at three different time points normalized by PP2A amplification from plant DNA. Error bars indicate the standard deviation (±SD) of three different biological repetitions and statistics were performed using the Student’s t‐test (*P ≤ 0.05).
Fig. S4 Characterization of the in vitro infection process of Arabidopsis thaliana (Col‐0) with Vertcillium longisporum (Vl43). Mock‐ and Vl43‐inoculated Col‐0 were photographed at 3, 6, 9 and 12 days post‐infection (A) and detection of WGA‐FITC stained hyphae at 4 dpi was documented using confocal microscopy (B).
Fig. S5 Disease symptom development in Verticillium longisporum (Vl43‐infected Arabidopsis and oilseed rape plants. Photos were taken at 36 days post‐inoculation.
Fig. S6 Fungal detection in the leaves. Oilseed rape leaves were cut and surface sterilized at 12 days post‐inoculation (dpi) and subsequently placed on potato dextrose agar for 4 days. As can be seen, no fungal growth could be observed at 12 dpi, indicating fungal growth within the plant was still restricted to the hypocotyls.
Fig. S7 Overview of the RNA‐Seq data procession pipeline for the Brassica napus experiment.
Fig. S8 Effects of external application of abscisic acid (ABA) on the infection progression of Verticillium longisporum (Vl43) by oilseed rape seedlings. Seedlings were transferred on ½ MS medium without (mock) or with 10 µM ABA before inoculation. (A) Brassica napus seedlings were photographed at 6 days post‐inoculation (dpi). (B) Fungal DNA in hypocotyls was measured by semiquantitative PCR using Verticillium‐specific primers as described above. Error bars indicate the standard deviation (±SD) of three different biological repetitions, consisting of three plants each. (C) At 6 dpi mock‐ and ABA‐treated hypocotyls were surface sterilized, cut into three segments and placed on potato dextrose agar to observe fungal growth. For each treatment six plants were employed. Two weeks after hypocotyl transfer, colonized hypocotyls were counted. In the mock control five out of six hypocotyls were colonized, while after ABA treatment colonization was only observed in one case.
Fig. S9 Differentially expressed genes related to abscisic acid (ABA) signalling as identified by RNA‐Seq of Verticillium longisporum (Vl43)‐infected oilseed rape (Bn) and Arabidopsis (At) roots at 6 days post‐inoculation. The line separates up‐and down‐regulated genes in response to the Vl43 infection of Brassica napus. The meta‐analysis of the orthologous Arabidopsis counterparts in response to various abiotic stress treatments focuses on ABA (1: AT‐00110, 2: AT‐00420), high light (3: AT‐00246), cold (4: AT‐00640), drought (5: AT‐00560, 6: AT‐00419), heat (7: AT‐00387), osmotic (8: AT‐00120) and salt stress (9: AT‐00534) treatments.
Table S1 Differentially expressed Brassica napus and Arabidopsis thaliana genes identified by RNA‐Seq.
Table S2 GO annotation table containing GO terms, KEGG pathways and RNA‐Seq expression data for all genes for which homologues were identified between Brassica napus and Arabidopsis thaliana.
Table S3 Complete list from the GO enrichment analysis of biological processes for Brassica napus and Arabidopsis thaliana.
Table S4 Verification of RNA‐Seq data by qPCR (comparison of log2 values), P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***).
Table S5 Brassica napus primers for qPCR.
Table S6 Arabidopsis thaliana primers for qPCR.
Table S7 Primer used for Arabidopsis T‐DNA genotyping and fungal quantification.
Acknowledgments
This work was financially supported by the Bundesministerium für Ernährung und Landwirtschaft (BMEL, grant no. 22006516) and the Bundesanstalt für Landwirtschaft and Ernährung (BLE, grant No. 2814IP004), Germany. The authors thank DAAD and BLE for travel grants (grant no. 57317839, 13/14‐15‐CHN), and the Chinese Scholarship Council (CSC) for providing a scholarship to Yan Zhao.
References
- Alexa, A. and Rahnenfuhrer, J. (2018) topGO: Enrichment Analysis for Gene Ontology. R package version 2.34.0. https://www.bioconductor.org/packages/release/bioc/html/topGO.html. Accessed 2 September 2019. [Google Scholar]
- Anderson, J.P. , Badruzsaufari, E. , Schenk, P.M. , Manners, J.M. , Desmond, O.J. , Ehlert, C. , Maclean, D.J. , Ebert, P.R. and Kazan, K. (2004) Antagonistic interaction between abscisic acid and jasmonate‐ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis . Plant Cell, 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arc, E. , Sechet, J. , Corbineau, F. , Rajjou, L. and Marion‐Poll, A. (2013) ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, F.Y. , Yoshioka, K. and Desveaux, D. (2011) The roles of ABA in plant–pathogen interactions. J. Plant Res. 124, 489–499. [DOI] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Wincker, P. (2014) Plant genetics. Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Clay, N.K. , Adio, A.M. , Denoux, C. , Jander, G. and Ausubel, F.M. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science, 323, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coninck, B. , Timmermans, P. , Vos, C. , Cammue, B.P. and Kazan, K. (2015) What lies beneath: belowground defense strategies in plants. Trends Plant Sci. 20, 91–101. [DOI] [PubMed] [Google Scholar]
- Cui, F. , Brosché, M. , Lehtonen, M.T. , Amiryousefi, A. , Xu, E. , Punkkinen, M. , Valkonen, J.P. , Fujii, H. and Overmyer, K. (2016) Dissecting abscisic acid signaling pathways involved in cuticle formation. Mol. Plant, 9, 926–938. [DOI] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depotter, J.R. , Deketelaere, S. , Inderbitzin, P. , von Tiedemann, A. , Höfte, M. , Subbarao, K.V. , Wood, T.A. and Thomma, B.P. (2016) Verticillium longisporum, the invisible threat to oilseed rape and other Brassicaceous plant hosts. Mol. Plant Pathol. 17, 1004–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellendorff, U. , Fradin, E.F. , de Jonge, R. and Thomma, B.P. (2009) RNA silencing is required for Arabidopsis defence against Verticillium wilt disease. J. Exp. Bot. 60, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eynck, C. , Koopmann, B. , Grunewaldt‐Stoecker, G. , Karlovsky, P. and von Tiedemann, A. (2007) Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. Eur. J. Plant Pathol. 118, 259–274. [Google Scholar]
- Eynck, C. , Koopmann, B. and von Tiedemann, A. (2009) Identification of Brassica accessions with enhanced resistance to Verticillium longisporum under controlled and field conditions. J. Plant Dis. Protect. 116, 63–72. [Google Scholar]
- Fan, J. , Hill, L. , Crooks, C. , Doerner, P. and Lamb, C. (2009) Abscisic acid has a key role in modulating diverse plant–pathogen interactions. Plant Physiol. 150, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAOSTAT . (2016) Statistical database. Food and Agriculture Organization of the United Nations: Statistic Division. Available at: http://www.fao.org/faostat/en/#data/QC/visualize. Accessed 9 December 2016.
- Finkelstein, R. (2013) Abscisic acid synthesis and response. Arabidopsis Book, 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floerl, S. , Druebert, C. , Majcherczyk, A. , Karlovsky, P. , Kües, U. and Polle, A. (2008) Defence reactions in the apoplastic proteome of oilseed rape (Brassica napus var. napus) attenuate Verticillium longisporum growth but not disease symptoms. BMC Plant Biol. 8, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floerl, S. , Majcherczyk, A. , Possienke, M. , Feussner, K. , Tappe, H. , Gatz, C. , Feussner, I. , Kües, U. and Polle, A. (2012) Verticillium longisporum infection affects the leaf apoplastic proteome, metabolome, and cell wall properties in Arabidopsis thaliana . PLoS One, 7, e31435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flors, V. , Ton, J. , van Doorn, R. , Jakab, G. , García‐Agustín, P. and Mauch‐Mani, B. (2008) Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola . Plant J. 54, 81–92. [DOI] [PubMed] [Google Scholar]
- Fujita, M. , Fujita, Y. , Maruyama, K. , Seki, M. , Hiratsu, K. , Ohme‐Takagi, M. , Tran, L.S. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2004) A dehydration‐induced NAC protein, RD26, is involved in a novel ABA‐dependent stress‐signaling pathway. Plant J. 39, 863–876. [DOI] [PubMed] [Google Scholar]
- Häffner, E. , Karlovsky, P. , Splivallo, R. , Traczewska, A. and Diederichsen, E. (2014) ERECTA, salicylic acid, abscisic acid, and jasmonic acid modulate quantitative disease resistance of Arabidopsis thaliana to Verticillium longisporum . BMC Plant Biol. 14, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happstadius, I. , Ljunberg, A. , Kristiansson, B. and Dixelius, C. (2003) Identification of Brassica oleracea germplasm with improved resistance to Verticillium wilt. Plant Breed. 122, 30–34. [Google Scholar]
- Hasler, M. and Kluss, C. (2017) SimComp: Simultaneous Comparisons for Multiple Endpoints. R package version, 3.2. https://CRAN.R-project.org/package=SimComp. Accessed 2 September 2019. [Google Scholar]
- Hok, S. , Allasia, V. , Andrio, E. , Naessens, E. , Ribes, E. , Panabières, F. , Attard, A. , Ris, N. , Clément, M. , Barlet, X. and Marco, Y. (2014) The receptor kinase IMPAIRED OOMYCETE SUSCEPTIBILITY1 attenuates abscisic acid responses in Arabidopsis . Plant Physiol. 166, 1506–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz, T. , Laule, O. , Szabo, G. , Wessendorp, F. , Bleuler, S. , Oertle, L. , Widmayer, P. , Gruissem, W. and Zimmermann, P. (2008) Genevestigator v3: a reference expression database for the meta‐analysis of transcriptomes. Adv. Bioinformatics, 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderbitzin, P. , Davis, R.M. , Bostock, R.M. and Subbarao, K.V. (2011) The ascomycete Verticillium longisporum is a hybrid and a plant pathogen with an expanded host range. PLoS One, 6, e18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iven, T. , König, S. , Singh, S. , Braus‐Stromeyer, S.A. , Bischoff, M. , Tietze, L.F. , Braus, G.H. , Lipka, V. , Feussner, I. and Dröge‐Laser, W. (2012) Transcriptional activation and production of tryptophan‐derived secondary metabolites in arabidopsis roots contributes to the defense against the fungal vascular pathogen Verticillium longisporum . Mol. Plant, 5, 1389–1402. [DOI] [PubMed] [Google Scholar]
- Jensen, M.K. , Lindemose, S. , de Masi, F. , Reimer, J.J. , Nielsen, M. , Perera, V. , Workman, C.T. , Turck, F. , Grant, M.R. , Mundy, J. , Petersen, M. and Skriver, K. (2013) ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana . FEBS Open Bio. 3, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. , Liang, G. and Yu, D. (2012) Activated expression of WRKY57 confers drought tolerance in Arabidopsis . Mol. Plant, 5, 1375–88. [DOI] [PubMed] [Google Scholar]
- Johansson, A. , Staal, J. and Dixelius, C. (2006) Early responses in the Arabidopsis–Verticillium longisporum pathosystem are dependent on NDR1, JA‐ and ET‐associated signals via cytosolic NPR1 and RFO1. Mol. Plant‐Microbe Interact. 19, 958–969. [DOI] [PubMed] [Google Scholar]
- Kamble, A. , Koopmann, B. and von Tiedemann, A. (2013) Induced resistance to Verticillium longisporum in Brassica napus by β‐aminobutyric acid. Plant Pathol. 62, 552–561. [Google Scholar]
- Lamesch, P. , Berardini, T.Z. , Li, D. , Swarbreck, D. , Wilks, C. , Sasidharan, R. , Karthikeyan, A.S. (2011) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40(D1), D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Haridon, F. , Besson‐Bard, A. , Binda, M. , Serrano, M. , Abou‐Mansour, E. , Balet, F. , Schoonbeek, H.J. , Hess, S. , Mir, R. , Léon, J. , Lamotte, O. and Métraux, J.P. (2011) A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog. 7, e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Shi, L. , Han, C. , Yu, J. , Li, D. and Zhang, Y. (2012) Validation of reference genes for gene expression studies in virus‐infected Nicotiana benthamiana using quantitative real‐time PCR. PLoS One, 7, e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kracher, B. , Ziegler, J. , Birkenbihl, R.P. and Somssich, I.E. (2015a) Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife, 4, e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Yan, J.P. , Li, D.K. , Luo, Q. , Yan, Q. , Liu, Z.B. , Ye, L.M. , Wang, J.M. , Li, X.F. and Yang, Y. (2015b) UDP‐glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis . Plant Physiol. 167, 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopisso, D.T. , Knüfer, J. , Koopmann, B. and von Tiedemann, A. (2017) The vascular pathogen Verticillium longisporum does not affect water relations and plant responses to drought stress of its host, Brassica napus . Phytopathology, 107, 444–454. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O. , Chico, J.M. , Sánchez‐Serrano, J.J. and Solano, R. (2004) JASMONATE‐INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate‐regulated defense responses in Arabidopsis . Plant Cell, 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch‐Mani, B. and Mauch, F. (2005) The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant Biol. 8, 409–414. [DOI] [PubMed] [Google Scholar]
- Mbengue, M. , Navaud, O. , Peyraud, R. , Barascud, M. , Badet, T. , Vincent, R. , Barbacci, A. and Raffaele, S. (2016) Emerging trends in molecular interactions between plants and the broad host range fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum . Front. Plant Sci. 31, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mialoundama, A.S. , Heintz, D. , Debayle, D. , Rahier, A. , Camara, B. and Bouvier, F. (2009) Abscisic acid negatively regulates elicitor‐induced synthesis of capsidiol in wild tobacco. Plant Physiol. 150, 1556–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, E.N. , Emery, R.J. and Saville, B.J. (2015) Phytohormone involvement in the Ustilago maydis–Zea mays pathosystem: relationships between abscisic acid and cytokinin levels and strain virulence in infected cob tissue. PLoS One, 10, e0130945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, K. , Yamaguchi‐Shinozaki, K. and Shinozaki, K. (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, N. , Okamoto, M. , Narusaka, M. , Yasuda, M. , Nakashita, H. , Shinozaki, K. , Narusaka, Y. and Hirayama, T. (2009) ABA hypersensitive germination2‐1 causes the activation of both abscisic acid and salicylic acid responses in Arabidopsis . Plant Cell Physiol. 50, 2112–2122. [DOI] [PubMed] [Google Scholar]
- Obayashi, T. , Kinoshita, K. , Nakai, K. , Shibaoka, M. , Hayashi, S. , Saeki, M. , Shibata, D. , Saito, K. and Ohta, H. (2007). ATTED‐II: a database of co‐expressed genes and cis elements for identifying co‐regulated gene groups in Arabidopsis. Nucleic Acids Res. 35(Database issue), D863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Kim, Y.S. , Kim, S.G. , Jung, J.H. , Woo, J.C. and Park, C.M. (2011) Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis . Plant Physiol. 156, 537–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M. , Kim, D. , Pertea, G.M. , Leek, J.T. and Salzberg, S.L. (2016) Transcript‐level expression analysis of RNA‐seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2018) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; http://www.R-project.org/. Accessed 2 September 2019. [Google Scholar]
- Ralhan, A. , Schöttle, S. , Thurow, C. , Iven, T. , Feussner, I. , Polle, A. and Gatz, C. (2012) The vascular pathogen Verticillium longisporum requires a jasmonic acid‐independent COI1 function in roots to elicit disease symptoms in Arabidopsis shoots. Plant Physiol. 159, 1192–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzinger, A. , Riediger, N. , von Tiedemann, A. and Karlovsky, P. (2009) Salicylic acid and salicylic acid glucoside in xylem sap of Brassica napus infected with Verticillium longisporum . J Plant Res. 122, 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusche, M. , Thole, K. , Janz, D. , Truskina, J. , Rindfleisch, S. , Drübert, C. , Polle, A. , Lipka, V. and Teichmann, T. (2012) Verticillium infection triggers VASCULAR‐RELATED NAC DOMAIN7‐dependent de novo xylem formation and enhances drought tolerance in Arabidopsis . Plant Cell, 24, 3823–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusche, M. , Truskina, J. , Thole, K. , Nagel, L. , Rindfleisch, S. , Tran, V.T. , Braus‐Stromeyer, S.A. , Braus, G.H. , Teichmann, T. and Lipka, V. (2014) Infections with the vascular pathogens Verticillium longisporum and Verticillium dahliae induce distinct disease symptoms and differentially affect drought stress tolerance of Arabidopsis thaliana . Environ. Exp. Bot. 108, 23–37. [Google Scholar]
- Rogers, S.O. and Bendich, A.J. (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76. [DOI] [PubMed] [Google Scholar]
- Roos, J. , Bejai, S. , Oide, S. and Dixelius, C. (2014) RabGAP22 is required for defense to the vascular pathogen Verticillium longisporum and contributes to stomata immunity. PLoS One, 9, e88187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos, J. , Bejai, S. , Mozūraitis, R. and Dixelius, C. (2015) Susceptibility to Verticillium longisporum is linked to monoterpene production by TPS23/27 in Arabidopsis . Plant J. 81, 572–85. [DOI] [PubMed] [Google Scholar]
- Rushton, D.L. , Tripathi, P. , Rabara, R.C. , Lin, J. , Ringler, P. , Boken, A.K. , Langum, T.J. , Smidt, L. , Boomsma, D.D. , Emme, N.J. , Chen, X. , Finer, J.J. , Shen, Q.J. and Rushton, P.J. (2012) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2. [DOI] [PubMed] [Google Scholar]
- Rygulla, W. , Friedt, W. , Seyis, F. , Lühs, W. , Eynck, C. , von Tiedemann, A. and Snowdon, R.J. (2007a) Combination of resistance to Verticillium longisporum from zero erucic acid Brassica oleracea and oilseed Brassica rapa genotypes in resynthesized rapeseed (Brassica napus) lines. Plant Breed. 126, 596–602. [Google Scholar]
- Rygulla, W. , Snowdon, R.J. , Eynck, C. , Koopmann, B. , Von Tiedemann, A. , Lühs, W. and Friedt, W. (2007b) Broadening the genetic basis of Verticillium longisporum resistance in Brassica napus by interspecific hybridisation. Phytopathology, 97, 1391–1396. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Vallet, A. , López, G. , Ramos, B. , Delgado‐Cerezo, M. , Riviere, M.P. , Llorente, F. , Fernández, P.V. , Miedes, E. , Estevez, J.M. , Grant, M. and Molina, A. (2012) Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina . Plant Physiol. 160, 2109–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, Y. , Yan, L. , Liu, Z.Q. , Cao, Z. , Mei, C. , Xin, Q. , Wu, F.Q. , Wang, X.F. , Du, S.Y. , Jiang, T. , Zhang, X.F. , Zhao, R. , Sun, H.L. , Liu, R. , Yu, Y.T. and Zhang, D.P. (2010) The Mg‐chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA‐responsive genes of inhibition. Plant Cell, 22, 1909–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, D. , Suhrkamp, I. , Wang, Y. , Liu, S. , Menkhaus, J. , Verreet, J.A. , Fan, L.J. and Cai, D. (2014) Identification and characterization of microRNAs in oilseed rape (Brassica napus) responsive to infection with the pathogenic fungus Verticillium longisporum using Brassica AA (Brassica rapa) and CC (Brassica oleracea) as reference genomes. New Phytol. 204, 577–594. [DOI] [PubMed] [Google Scholar]
- Siewers, V. , Kokkelink, L. , Smedsgaard, J. and Tudzynski, P. (2006) Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea . Appl. Environ. Microbiol. 72, 4619–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, C.A. , Lakshmanan, V. , Donofrio, N. and Bais, H.P. (2015) Crucial roles of abscisic acid biogenesis in virulence of rice blast fungus Magnaporthe oryzae . Front. Plant Sci. 6, 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, F. , Fan, G. , Hu, Q. , Zhou, Y. , Guan, M. , Wang, H. (2017) The high‐quality genome of Brassica napus cultivar 'ZS11' reveals the introgression history in semi‐winter morphotype. Plant J. 92, 452–468. [DOI] [PubMed] [Google Scholar]
- Thaler, J.S. , Humphrey, P.T. and Whiteman, N.K. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270. [DOI] [PubMed] [Google Scholar]
- Tian, H. , Guo, H. , Dai, X. , Cheng, Y. , Zheng, K. , Wang, X. and Wang, S. (2015) An ABA down‐regulated bHLH transcription repressor gene, bHLH129 regulates root elongation and ABA response when overexpressed in Arabidopsis . Sci. Rep. 5, 17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, J. , Flors, V. and Mauch‐Mani, B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. [DOI] [PubMed] [Google Scholar]
- de Torres‐Zabala, M. , Truman, W. , Bennett, M.H. , Lafforgue, G. , Mansfield, J.W. , Rodriguez Egea, P. , Bögre, L. and Grant, M. (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signaling pathway to cause disease. EMBO J. 26, 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala, M. , Bennett, M.H. , Truman, W.H. and Grant, M.R. (2009) Antagonism between salicylic and abscisic acid reflects early host‐pathogen conflict and moulds plant defence responses. Plant J. 59, 375–386. [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M., Littlejohn, G. , Jayaraman, S. , Studholme, D. , Bailey, T. , Lawson, T. , Tillich, M. , Licht, D. , Bölter, B. , Delfino, L. , Truman, W. , Mansfield, J. , Smirnoff, N. and Grant, M. (2015) Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants, 1, 15074. [DOI] [PubMed] [Google Scholar]
- Veronese, P. , Narasimhan, M.L. , Stevenson, R.A. , Zhu, J.K. , Weller, S.C. , Subbarao, K.V. and Bressan, R.A. (2003) Identification of a locus controlling Verticillium disease symptom response in Arabidopsis thaliana . Plant J. 35, 574–587. [DOI] [PubMed] [Google Scholar]
- Wang, M. , Weiberg, A. , Lin, F.M. , Thomma, B.P. , Huang, H.D. and Jin, H. (2016) Bidirectional cross‐kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants, 2, 16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2016) ggplot2: Elegant Graphics for Data Analysis. New York: Springer‐Verlag. [Google Scholar]
- Wu, J. , Mao, X. , Cai, T. , Luo, J. and Wei, L. (2006) KOBAS server: a web‐based platform for automated annotation and pathway identification. Nucleic Acids Res. 34, W720–W724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Chen, C. , Fan, B. and Chen, Z. (2006) Physical and functional interactions between pathogen‐induced Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors. Plant Cell, 18, 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Audenaert, K. , Hofte, M. and De Vleesschauwer, D. (2013) Abscisic acid promotes susceptibility to the rice leaf blight pathogen Xanthomonas oryzae pv. oryzae by suppressing salicylic acid‐mediated defenses. PLoS One, 8, e67413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, M. , Ishikawa, A. , Jikumaru, Y. , Seki, M. , Umezawa, T. , Asami, T. , Maruyama‐Nakashita, A. , Kudo, T. , Shinozaki, K. , Yoshida, S. and Nakashita, H. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid‐mediated abiotic stress response in Arabidopsis . Plant Cell, 20, 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T. , Mogami, J. and Yamaguchi‐Shinozaki, K. (2014) ABA‐dependent and ABA‐independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Zhao, Y.L. , Zhao, J.H. , Wang, S. , Jin, Y. , Chen, Z.Q. , Fang, Y.Y. , Hua, C.L. , Ding, S.W. and Guo, H.S. (2016) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants, 2, 16153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Scatterplot of all RNA‐Seq calculated FPKM values for all unigenes between mock‐ and Vl43‐infected samples for Brassica napus and Arabidopsis thaliana, respectively. Genes were considered to be differentially regulated when their log2 fold change was ≥1 (red) or ≤–1 (green).
Fig. S2 ATTED‐II search for ABA‐related co‐expressed genes with the database of closely related model plant Arabidopsis. The identified coregulatory networks are shown (A–E) and have been partly merged together. Genes that have also been found in the RNA‐Seq dataset are highlighted in yellow. Genes identified in the networks A–D are down‐regulated in the RNA‐Seq data, while genes from cluster E are up‐regulated.
Fig. S3 Characterization of the in vitro infection process of Brassica napus (Express 617) with Verticillium longisporum (Vl43). (A) Mock‐ and Vl43‐ inoculated B. napus seedlings were photographed at 3, 6 and 12 days post‐inoculation (dpi). (B) Hypocotyls were sampled and surface sterilized at different dpi and placed on potato dextrose agar to detect Vl43 in hypocotyls of infected B. napus seedlings. (C) Increment of fungal DNA in B. napus hypocotyls, measured by semiquantitative PCR using Verticillium‐ specific primers is displayed as the relative amount of fungal DNA at three different time points normalized by PP2A amplification from plant DNA. Error bars indicate the standard deviation (±SD) of three different biological repetitions and statistics were performed using the Student’s t‐test (*P ≤ 0.05).
Fig. S4 Characterization of the in vitro infection process of Arabidopsis thaliana (Col‐0) with Vertcillium longisporum (Vl43). Mock‐ and Vl43‐inoculated Col‐0 were photographed at 3, 6, 9 and 12 days post‐infection (A) and detection of WGA‐FITC stained hyphae at 4 dpi was documented using confocal microscopy (B).
Fig. S5 Disease symptom development in Verticillium longisporum (Vl43‐infected Arabidopsis and oilseed rape plants. Photos were taken at 36 days post‐inoculation.
Fig. S6 Fungal detection in the leaves. Oilseed rape leaves were cut and surface sterilized at 12 days post‐inoculation (dpi) and subsequently placed on potato dextrose agar for 4 days. As can be seen, no fungal growth could be observed at 12 dpi, indicating fungal growth within the plant was still restricted to the hypocotyls.
Fig. S7 Overview of the RNA‐Seq data procession pipeline for the Brassica napus experiment.
Fig. S8 Effects of external application of abscisic acid (ABA) on the infection progression of Verticillium longisporum (Vl43) by oilseed rape seedlings. Seedlings were transferred on ½ MS medium without (mock) or with 10 µM ABA before inoculation. (A) Brassica napus seedlings were photographed at 6 days post‐inoculation (dpi). (B) Fungal DNA in hypocotyls was measured by semiquantitative PCR using Verticillium‐specific primers as described above. Error bars indicate the standard deviation (±SD) of three different biological repetitions, consisting of three plants each. (C) At 6 dpi mock‐ and ABA‐treated hypocotyls were surface sterilized, cut into three segments and placed on potato dextrose agar to observe fungal growth. For each treatment six plants were employed. Two weeks after hypocotyl transfer, colonized hypocotyls were counted. In the mock control five out of six hypocotyls were colonized, while after ABA treatment colonization was only observed in one case.
Fig. S9 Differentially expressed genes related to abscisic acid (ABA) signalling as identified by RNA‐Seq of Verticillium longisporum (Vl43)‐infected oilseed rape (Bn) and Arabidopsis (At) roots at 6 days post‐inoculation. The line separates up‐and down‐regulated genes in response to the Vl43 infection of Brassica napus. The meta‐analysis of the orthologous Arabidopsis counterparts in response to various abiotic stress treatments focuses on ABA (1: AT‐00110, 2: AT‐00420), high light (3: AT‐00246), cold (4: AT‐00640), drought (5: AT‐00560, 6: AT‐00419), heat (7: AT‐00387), osmotic (8: AT‐00120) and salt stress (9: AT‐00534) treatments.
Table S1 Differentially expressed Brassica napus and Arabidopsis thaliana genes identified by RNA‐Seq.
Table S2 GO annotation table containing GO terms, KEGG pathways and RNA‐Seq expression data for all genes for which homologues were identified between Brassica napus and Arabidopsis thaliana.
Table S3 Complete list from the GO enrichment analysis of biological processes for Brassica napus and Arabidopsis thaliana.
Table S4 Verification of RNA‐Seq data by qPCR (comparison of log2 values), P ≤ 0.05 (*), P ≤ 0.01 (**), P ≤ 0.001 (***).
Table S5 Brassica napus primers for qPCR.
Table S6 Arabidopsis thaliana primers for qPCR.
Table S7 Primer used for Arabidopsis T‐DNA genotyping and fungal quantification.


