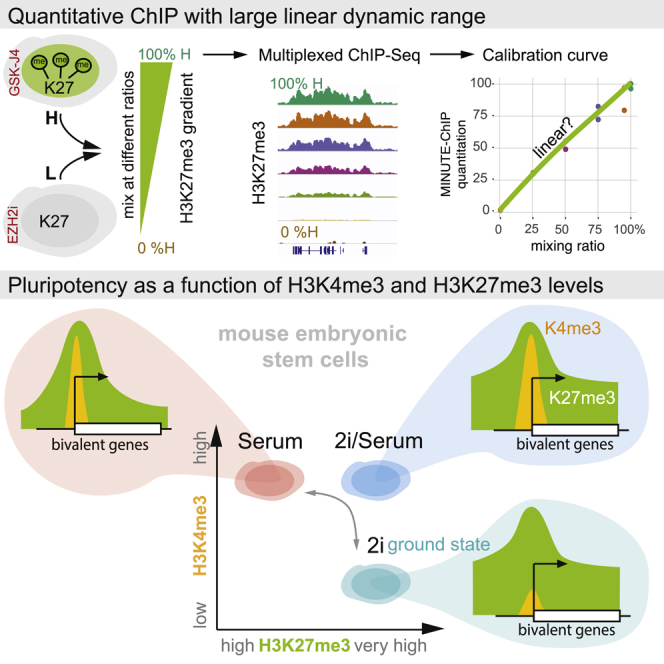
Summary
To understand the epigenomic foundation of naive pluripotency, we implement a quantitative multiplexed chromatin immunoprecipitation sequencing (ChIP-seq) method comparing mouse embryonic stem cells (ESCs) grown in 2i versus 2i/serum and serum conditions. MINUTE-ChIP has a large linear dynamic range for accurately quantifying relative differences in genome-wide histone modification patterns across multiple pooled samples. We find compelling evidence for a broad H3 lysine 27 trimethylation (H3K27me3) hypermethylation of the genome, while bivalent promoters stably retain high H3K27me3 levels in 2i. We show that DNA hypomethylation, as observed in 2i, is a contributor to genome-wide gain of H3K27me3, while active demethylation by JMJD3/UTX counteracts further accumulation of H3K27me3. In parallel, we find hypomethylation of H3 lysine 4 trimethylation (H3K4me3), particularly at bivalent promoters, to be a characteristic of the 2i ground state. Serum stimulates H3K4me3 independent of GSK-3b and ERK signaling, suggesting that low H3K4me3 and high H3K27me3 levels at bivalent promoters are a product of two independent mechanisms that safeguard naive pluripotency.
Keywords: pluripotency, ground state, naive stem cell, pluripotent stem cells, bivalency, bivalent promoters, polycomb, quantiative ChIP-seq, multiplexed ChIP-seq, epigenomics
Graphical Abstract
Highlights
-
•
MINUTE-ChIP enables accurate quantitative comparisons over a large linear range
-
•
High global H3K27me3 and low H3K4me3 characterize 2i pluripotent ground state
-
•
H3K27me3 levels at bivalent promoters are stably maintained between serum and 2i
-
•
Serum primes mESCs by stimulating H3K4me3 at bivalent promoters independent of 2i
Kumar et al. employ an improved multiplexed quantitative ChIP-seq methodology to characterize mESC pluripotency. They reveal that bivalent promoters are retained in the 2i ground state, which is characterized by abundant H3K27me3. H3K4me3 levels are high in serum-primed state and a stable 2i/serum intermediate but strikingly reduced in ground state.
Introduction
The mouse pluripotent ground state is attributed to naive epiblast cells of the inner cell mass and can be recapitulated ex vivo through inhibition of GSK-3b and mitogen-activated protein kinase (MAPK)/ extracellular signal-regulated kinase (ERK) signaling in mouse embryonic stem cells (mESCs) in serum-free conditions containing leukemia inhibitory factor (LIF) and two respective inhibitors (CHIR99021 and PD0325901, referred to as “2i”; Ying et al., 2008). Traditional serum-based culture in the presence of LIF (hereafter “serum”) maintains a more metastable pluripotent embryonic stem cell (ESC). Although rapidly interconvertible through change in media condition, significant differences in signaling, metabolism, transcriptional regulation, chromatin modifications, and nuclear organization distinguish the two conditions (Carey et al., 2015, Galonska et al., 2015, Habibi et al., 2013, Joshi et al., 2015, Leitch et al., 2013, Marks et al., 2012, van Mierlo et al., 2018, van Mierlo et al., 2019, Ying et al., 2008). ESCs maintain their pluripotent state until differentiation cues initiate a lineage commitment, and the exit from ground state pluripotency is considered as the first priming event for the differentiation cascade to follow.
A large number of chromatin regulators have been implicated in early embryonic developmental transitions. In particular, polycomb and trithorax group proteins have been implicated in the precise and rapid gene expression control. Methyltransferase subunits of polycomb repressive complex 2 (PRC2) and trithorax, EZH2, and MLL2 catalyze histone H3 lysine 27 trimethylation (H3K27me3) and H3 lysine 4 trimethylation (H3K4me3), respectively (Di Croce and Helin, 2013). Because of the co-occurence of H3K27me3 and H3K4me3, defined as “bivalency,” at developmentally regulated genes and their opposing functions (repressing and activating transcription, respectively), the two complexes are thought to set up a poised state, which can be resolved into an active or inactive state (Mikkelsen et al., 2007). Although PRC2 suppresses basal activity of developmentally regulated genes in mESCs, it is not essential for maintenance of pluripotency in culture. Nevertheless, changes in developmental stage, such as the transition between naive and primed pluripotency, often involve major changes in the epigenome of the cell. Capturing the true extent of epigenomic remodeling that takes place in such key transitions requires quantitative methodologies. Here, we implemented a quantitative, multiplexed chromatin immunoprecipitation sequencing (ChIP-seq) method to compare epigenomic profiles between 2i and serum conditions. We focused on the two histone modifications that establish promoter bivalency, H3K4me3 and H3K27me3 (Mikkelsen et al., 2007), and discover global changes in their stoichiometry that significantly affect local chromatin environments, such as bivalent domains, in unexpected ways.
Results
MINUTE-ChIP Allows Multiplexed Pooled ChIP with Unique Molecule Counting
The need for quantitative ChIP-seq has been widely appreciated, and a number of quantitative methods have been proposed. Barcoding-first ChIP techniques have gained popularity because of the benefit of multiplexing (Arrigoni et al., 2018, Chabbert et al., 2018, van Galen et al., 2016, Lara-Astiaso et al., 2014). Pooling barcoded samples greatly increases throughput without complicated automation, while effectively removing technical variability between samples. Challenges remain for fragmentation and ligation of crude chromatin in a manner that maximizes barcoding efficiency while avoiding any technical variability or sample loss that could confound the subsequent quantitative measurement. Mint-ChIP, developed by the Bernstein lab (van Galen et al., 2016), provides a formidable solution for these problems with a streamlined one-pot chromatin barcoding and a post-ChIP linear amplification that requires only one adaptor per chromatin fragment. In short, native chromatin is fragmented using micrococcal nuclease and subsequently blunted and ligated to double-stranded DNA adaptors that include a T7 RNA polymerase promoter and a sample barcode sequence. Finally, samples are combined and subsequent ChIP reactions are performed with the pooled samples. ChIP material is prepared into an Illumina-compatible library using linear amplification by virtue of T7 RNA polymerase, reverse transcription, and a low-cycle library PCR amplification (van Galen et al., 2016).
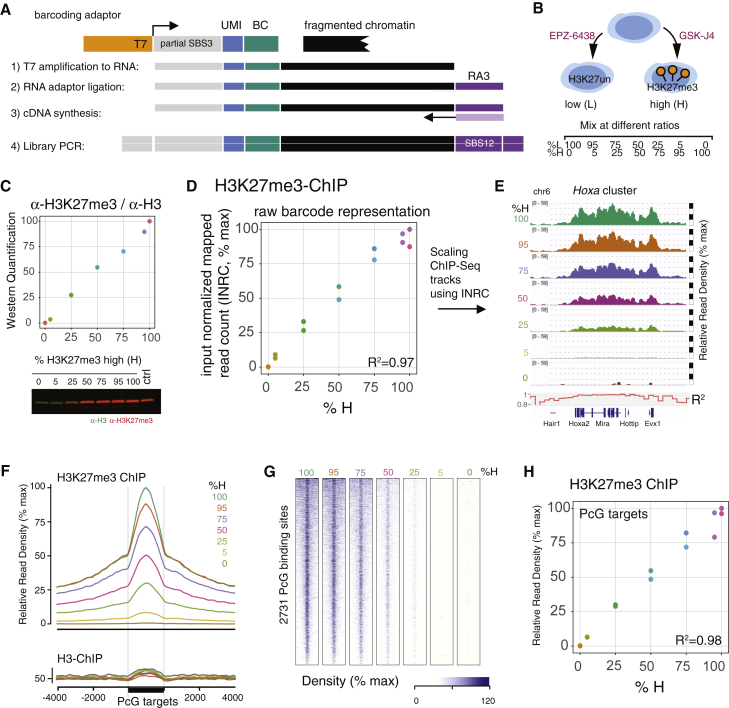
Here, we introduce unique molecule (unique molecular identifier [UMI]) counting and paired-end mapping of the chromatin fragments to this method, which we then termed MINUTE-ChIP for multiplexed indexed unique molecule T7 amplification end-to-end sequencing (Figure 1A; Table S1). Double-stranded DNA adaptors were used to barcode chromatin in a blunt-end ligation reaction. Pooling a human HeLa cell sample with the mouse ESC samples, adaptor cross-contamination was assessed to be very low, with a maximum of 1.5% mapping to the respective other species. (Figure S1A). As in the original Mint-ChIP design, adaptors carried a partial SBS3 for Illumina sequencing flanked by a T7 RNA polymerase for linear amplification. Between the SBS3 sequence and an 8-bp sample barcode at the 3′ end, a 6-bp randomized sequence was introduced, serving as a UMI. Using UMI information in addition to the ligated genomic sequence greatly increases the confidence in calling amplification duplicates and improves the quantitative representation of repetitive sequencing (Figure S1B).
Figure 1.
MINUTE-ChIP Method Accurately Reflects Global and Local Proportions
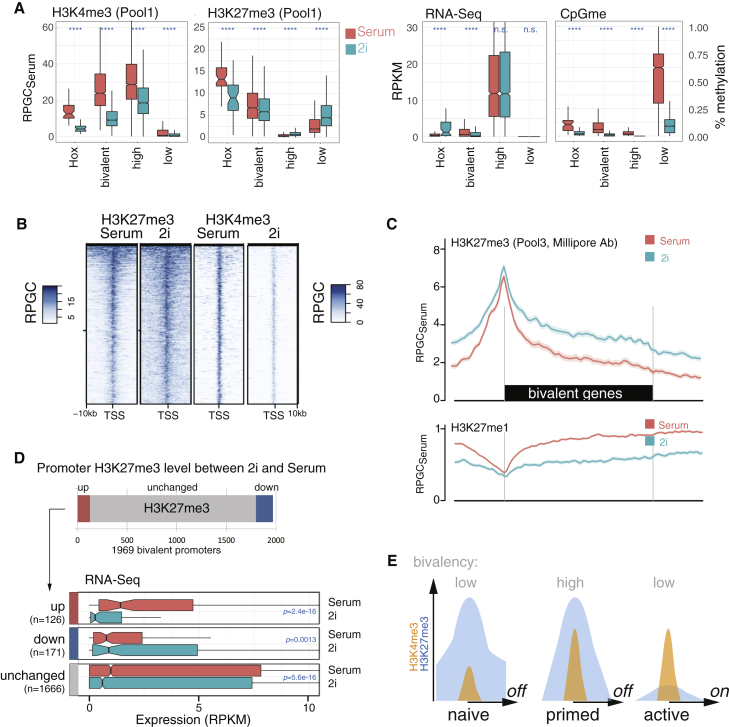
(A) Schematic of MINUTE adaptor design for barcoding chromatin. One-sided ligation of adaptor comprising T7 RNA polymerase promoter (T7), random 6-bp sequence (UMI), and 8-bp barcode (BC) is sufficient for subsequent linear amplification. cDNA is primed from a ligated RNA adaptor (RA3). SBS3 and SBS12 designate standard Illumina read1 and read2 sequencing primers.
(B) Setup for calibration experiment: mESC treated with EZH2 inhibitor EPZ-6438 (resulting in depletion of H3K27me3, “L”) or JMJD3/UTX inhibitor GSK-J4 (resulting in very high H3K27me3, “H”) were mixed in defined ratios to establish a gradient of H3K27me3 with seven artificial conditions (0%, 5%, 25%, 50%, 75%, 95%, and 100% H).
(C) Quantitative infrared fluorescent western blot to confirm mixing ratios. Control sample (ctrl) is an untreated mESC in 2i condition. Raw western blot signal (α-H3K27me3 fluorescence over α-H3 fluorescence) was arbitrarily scaled between 0 and 100. Representative blot of three technical replicates is shown.
(D) MINUTE-ChIP quantification after α-H3K27me3 immunoprecipitation: input-normalized raw mapped read counts (INRCs) for two technical replicates with different barcodes are plotted against mixing ratio. INRC is subsequently used to quantitatively scale ChIP-seq tracks. Coefficient of determination (R2) for a linear regression is given.
(E) Example genomic regions showing quantitatively scaled H3K27me3 ChIP-seq tracks (average of both replicates) arising from different mixing ratios. Scale bar on right side breaks at 25%, 50%, 75%, and 100% maximal signal. Coefficient of determination (R2) was calculated over 10-kb windows.
(F) H3K27me3 signal (average of two replicates) across 2,731 polycomb (PcG) target sites. For comparison, H3 MINUTE-ChIP data are shown below. SE is rendered as shaded area around lines.
(G) Heatmap of the same data across 2,731 PcG target regions.
(H) Quantification of mean H3K27me3 signal at PcG target sites by replicate. Coefficient of determination (R2) for a linear regression is given.
See also Figure S1.
For paired-end mapping, we modified the linear amplification strategy introduced in Mint-ChIP by priming the cDNA synthesis from a 3′ adaptor (RA3) ligated onto the amplified RNA, thus maintaining the original genomic fragment length, retrieving a typical mononucleosomal fragment length for histone ChIP (Figure S1C). As previously reported (van Galen et al., 2016), barcode representation in the pool may vary among samples, even at precisely matched adaptor DNA concentration. For accurate quantification, barcode representation after the ChIP has to be related back to the corresponding quantities in the input pool. A common issue of indexing-first ChIP protocols is that adaptors added in excess are carried over into the input pool. Amplification products from free adaptors, adaptor dimers, or other side reactions contribute contaminating sequences to an extent that sequencing of the input pool becomes infeasible. Thus, we have optimized the stoichiometry of adaptors to chromatin to enable sequencing of the input. Quantification against input representation is robust, as exemplified by an H3 ChIP from a pool containing two series of biological triplicate samples with varying input representation; all replicates lie within 10% variance (Figure S1D; Table S2).
MINUTE-ChIP Allows Accurate Quantitative Comparisons across a Large Dynamic Range
Quantitative ChIP-seq experiments report relative differences in occupancy of histone modifications. By definition, the measurements should be proportional to the true biological differences, over the dynamic range to be expected within the experiment. Because every experimental method has limitation as to linearity, sensitivity, and dynamic range, these parameters have not been established for most proposed quantitative ChIP-seq methods. Thus, we sought to benchmark MINUTE-ChIP using a calibration experiment. We aimed to generate a defined gradient of H3K27me3 that would span the entire physiologic range of the modification. To this end, we treated mESCs either with EZH2 inhibitor EPZ-6438, hereafter (EZH2i) to reduce H3K27me3 below detectable levels, or with an inhibitor to demethylases JMJD3/UTX, GSK-J4, to increase H3K27me3 above physiologic levels (Figure 1B). Mixing cells from these two treatments, H3K27me3 low (L) and H3K27me3 (H), in defined ratios (100:0, 95:5, 75:25, 50:50, 25:75 5:95, and 0:100 L:H), we created an artificial gradient of H3K27me3 (Figure 1B). Quantitative western blotting confirmed the mixing ratio (Figure 1C). Two replicates were prepared from each mixing ratio using two different barcodes and pooled for MINUTE-ChIP. From this pool, ChIP was performed in parallel using H3K27me3, H3K27me2, H3K27me1, and H3 antibodies. After demultiplexing reads by barcode (Figure 1A) and UMI deduplication, reads were mapped to the mm9 genome. Total mapped read counts per barcode were normalized to the respective input read counts, essentially correcting for uneven barcode representation in the input. The resulting input normalized mapped read count (INRC) is a quantitative measure of the abundance of ChIP epitope, e.g., H3K27me3, for each barcode (Figure 1D). Indeed, INRCs were proportional to the mixing ratios (R2 = 0.97). ChIP-seq tracks were scaled according to INRC to yielding quantitative genome-wide maps (Figure 1E). INCR scaling should result in a signal proportional to the true abundance of H3K27me3 nucleosomes across the genome. We assessed this by quantifying H3K27me3 at known polycomb targets, where the highest enrichment for this modification is expected (Figures 1F–1H). Indeed, the average H3K27me3 signal across 2731 PcG binding sites followed the mixing ratios proportionally (R2 = 0.98), over the entire gradient (Figures 1F–1H). Differences between the means of all calibration points were significant (Figure S1E). As expected, quantitative signal of H3 ChIP was essentially constant (Figures 1F and S1F).
In addition to inspecting highly enriched regions, we also sought to validate the sensitivity of our methods for genomic areas with much lower signal. To this end, we quantified H3K27me3 across highly expressed genes, where overall levels are ∼30-fold lower than at bivalent promoters (Figure S1G). Even at this low end, the MINUTE-ChIP quantification produced a proportional representation of the mixing ratios (Figure S1G). Further, we observed similar proportional representation in the H3K27me2 and H3K27me1 ChIPs and over a number of additional representative regions of the genome (Figures S1H–S1J). In conclusion, we have demonstrated that MINUTE-ChIP is a sensitive method with a large linear dynamic range for accurately quantifying relative differences in genome-wide histone modification patterns across multiple pooled samples.
High Levels of H3K27me3 and Low Levels of H3K4me3 Characterize 2i Ground State
Genome-wide epigenomic differences between naive and serum-primed mESCs have been extensively studied using qualitative methods. However, initial quantitative western blot experiments showed substantial differences in global levels of H3K27me3 and H3K4me3 (Figure S2A). As compared to serum condition, 2i-grown mESCs had approximately 1.7-fold more H3K27me3 and 2-fold less H3K4me3 (Figure S2A). This led us to investigate the H3K4me3 and H3K27me3 landscapes in a quantitative manner using our MINUTE-ChIP protocol.
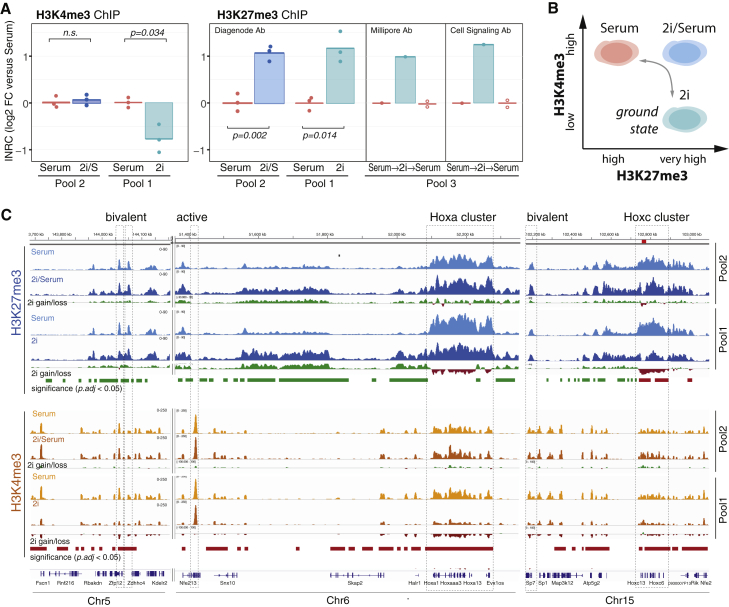
We performed three independent MINUTE-ChIP experiments (Figure 2A; Table S2): In “pool 1,” we compared biological triplicates of RW.4 mESCs grown in serum or serum-free 2i (2i) conditions using H3K27me3 (rabbit polyclonal from Diagenode) and H3K4me3 antibodies. In “pool 2,” we compared biological triplicates of RW.4 mESCs grown in serum or Serum with 2i inhibitors (“2i/serum”) conditions using H3K27me3 (rabbit polyclonal from Diagenode), H3K4me3, and H3 antibodies. In “pool 3,” we compared RW.4 mESCs sequentially grown in serum condition, three passages in 2i condition, and three passages in serum condition. We profiled H3K27me3 with two different antibodies (rabbit monoclonal from Cell Signaling Technology and rabbit polyclonal from Millipore) and H3K27me1. Pool 3 additionally included conditions with EZH2i and GSK-J4 inhibitors. MINUTE-ChIP quantification was performed as above, scaling tracks according to INRC. Common point of reference between the three independent MINUTE-ChIP experiments was the serum condition present in triplicates in each pool. Throughout the remainder of the manuscript, ChIP-seq tracks are consequently scaled relative to the serum condition (indicated as RPGCSerum), where serum condition is arbitrarily scaled to 1× coverage (reads per genomic content [RPGC]).
Figure 2.
MINUTE-ChIP Reveals High H3K27me3 and Low H3K4me3 Levels in Naive Mouse ESC
(A) Left: quantitative comparison of H3K4me3 levels in biological triplicates of Rw4 mESC grown in serum versus 2i condition (pool 1) or serum versus 2i/serum condition (pool 2). INRCs were calculated and plotted with respect to the serum condition in each pool. Right: quantitative comparison of H3K27me3 levels in biological triplicates of Rw4 mESC grown in serum versus 2i condition (pool 1 and pool 3) or serum versus 2i/serum condition (pool 2) is shown. Three different H3K27me3 antibodies were used as indicated. p values are reported from a two-sided t test.
(B) Schematic summary of pluripotent states defined by global H3K4me3 and H3K27me3 levels.
(C) Example H3K27me3 and H3K4me3 tracks from pool 1 and pool 2 (average of three biological replicates for each condition). Tracks are scaled as reads per genomic content relative to serum condition (RPGCSerum) as common reference point between the two pools. Additional tracks show gain (green) and losses (red) for each quantitative comparison, as well as 10-kb windows deemed to be significantly (p adj. < 0.05) increased (green) or decreased (red) among replicates.
See also Figure S2.
Across all MINUTE-ChIP experiments (including several full biological replicates and three different H3K27me3 antibodies; for details and primary data, see Table S2), we observed two-fold higher levels of H3K27me3 in the presence of 2i (Figure 2A). This difference was reversible within three passages (Figure 2A). An H3 ChIP control yielded no significant difference between the samples (Figure S1D). Mass spectrometry also supported a 1.5- (Figure S2B) to two-fold (van Mierlo et al., 2019) increase in H3K27me3 levels. It should be noted in this regard that MINUTE-ChIP measures H3K27me3 on the level of the nucleosome unit, which can carry zero, one, or two modified H3K27 residues, whereas quantitative mass spectrometry and western blot determine the abundance of H3K27me3 per H3 molecule. Thus, the different measurements must be compared under the caveat that the distribution of H3K27me3 histone molecules into singly and doubly modified mononucleosome is not known.
Nucleosomal H3K4me3 levels as quantified by MINUTE-ChIP were 1.8-fold decreased in 2i (Figure 2A), in agreement with the quantitative western blot (Figure S2A), representing a global difference that escaped observation in prior studies. Interestingly, H3K4me3 levels in 2i/serum were unchanged as compared to serum. 2i/serum condition thus appears to generate, at least on the level of the epigenome, an intermediate state between naive and serum-primed mESCs. This state is characterized by high H3K27me3 and H3K4me3, and we now more precisely define the ground state as H3K27me3 high and H3K4me3 low (Figure 2B). These results also suggest that independent mechanisms regulate H3K27me3 and H3K4me3 levels. The former is linked to the action of 2i inhibitors, whereas H3K4me3 levels are insensitive to 2i inhibitors but respond to removal of serum.
We generated quantitatively scaled genomic tracks for H3K27me3 and H3K4me3, combining the three biological replicate measurements, and further used the replicate information to determine regions with statistically significant gains or losses between conditions (Figure 2C; for replicate comparison, see Figure S2C). Qualitatively, our data resembled prior ChIP-seq data comparing serum and 2i conditions (Figures S2D and S2E). For example, a defined region in the Hoxc cluster (Hoxc12-c13) was fully depleted in H3K27me3 (Figure S2E). This is a characteristic feature of the 2i ground state, which has been previously described to coincide with strong transcriptional induction of the Hoxc12-c13 locus (Marks et al., 2012). The quantitative measurement, however, revealed more widespread differences between serum and 2i conditions for both H3K4me3 and H3K27me3, as evident from a DESeq statistical analysis (Anders and Huber, 2010) on 10-kb windows across the genome: 49.9% of regions significantly (adjusted p value [p adj.] < 0.05) gained and 0.1% lost H3K27me3 (p adj. < 0.05), while 0.3% gained and 21% lost H3K4me3 (Figure S2F). Demonstrating the robustness of the analysis, no significant differences were observed for H3 ChIP (Figure S2F). We thus sought to define how these global alterations of histone modification levels affect local chromatin environments and shape the pluripotent ground state.
Reduced H3K4me3 in 2i Ground State Particularly Affects Bivalent Promoters
As noted above, H3K4me3 showed a striking global loss in 2i, but not serum/2i, condition. As a consequence, we found that H3K4me3 was reduced across essentially all H3K4me3-enriched regions when grown in (serum-free) 2i condition (Figure 3A). In contrast, the H3K4me3 landscape showed virtually no significant differences between serum and 2i/serum (Figures 3A and S2F). This suggested that CHIR99021 and PD0325901 treatment per se did not affect H3K4me3 levels globally or locally, only when combined with serum withdrawal.
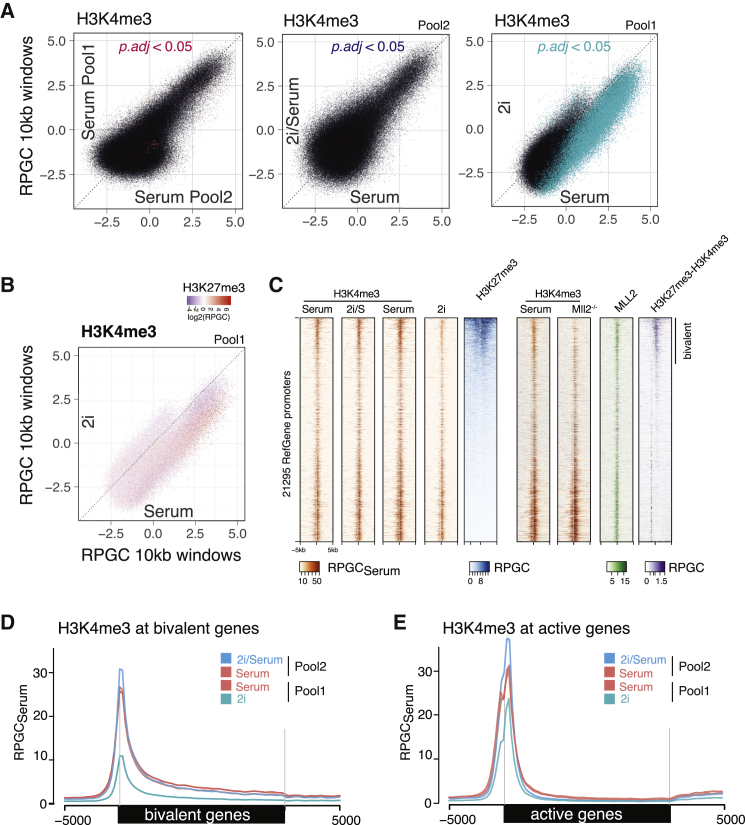
Figure 3.
Bivalent Promoters Are Depleted in H3K4me3 in 2i Ground State
(A) Genome-wide analysis of H3K4me3 across 10-kb windows. H3K4me3 density (RPGCSerum) is plotted as log2-fold enrichment over input read density on x and y axis. Mean of three biological replicates is plotted and significance based on three biological replicates each (adjusted p value [p adj.] < 0.05) was tested using DESeq. (Left) H3K4me3 comparison between serum conditions in pool 1 and pool 2 is shown. Middle: comparison of 2i/serum and serum condition (pool 2) is shown. (Right) Comparison of 2i and serum condition (pool 1) is shown.
(B) Genome-wide analysis of H3K4me3 across 10-kb windows as in (A), comparing 2i and serum condition (pool 1) with color scale representing H3K27me3 levels in the same bin.
(C) Heatmap of all RefSeq transcription start sites. Plotted are densities (RPGCSerum) of H3K4me3 tracks from pool 1 and pool 2, as well as H3K27me3 in serum condition. Additionally, published Mll2 ChIP, H3K4me3 ChIP in Mll2−/−, and H3K4me3-H3K27me3 Re-ChIP are shown (Mas et al., 2018). Entire heatmap is sorted by H3K27me3 levels.
(D) Average quantitative profile of H3K4me3 across 1,969 bivalent genes, comparing 2i/serum and 2i conditions quantitatively to their respective serum control.
(E) Average quantitative profile of H3K4me3 across bivalent genes, comparing 2i/serum and 2i conditions quantitatively to their respective serum control. y axis shows reads per genomic content relative to serum condition (RPGCSerum), where serum conditions are scaled to 1× coverage.
Interestingly, loss of H3K4me3 was particularly high in H3K27me3-enriched regions (Figures 3B and 3C). Thus, we compared the two major classes of H3K4me3 peaks in mouse ESCs at active and bivalent genes. Both showed a slight increase in H3K4me3 in 2i/serum condition (Figures 3D and 3E). The global reduction of H3K4me3 (Figure 2A) in 2i condition manifested in reduced peak sizes across all genes but most strikingly at bivalent promoters (Figures 3B and 3C; see Figure 2C for examples). Although active genes on average lost ∼35% of H3K4me3 (Figure 3E), bivalent promoters showed a more than 2-fold decrease in H3K4me3 in 2i (Figure 3D). This is in line with prior observations that H3K4me3 levels are reduced in 2i conditions relative to other promoters (Galonska et al., 2015), albeit our quantitative comparison now shows a much larger total effect size.
H3K4me3 at bivalent promoters is predominantly deposited by MLL2/COMPASS in mESCs, and several MLL methyltransferases redundantly target active genes (Denissov et al., 2014, Mas et al., 2018). MLL2−/− knockout in serum condition has a similar effect on H3K4me4 distribution as we observe in 2i condition (Figure 3C). Thus, we speculate that it is an attenuated MLL2 activity that stabilizes the 2i ground state. In line with this, MLL2 has been shown to be dispensable for pluripotency but to play a role in priming bivalent genes for activation during differentiation (Mas et al., 2018).
H3K27me3 Is Maintained at PcG Targets and Broadly Gained across Most of the Genome in 2i
Next, we turned to investigating the H3K27me3 landscape in light of the two-fold global increase in H3K27me3 (Figure 2A). Such increase in H3K27me3 was unexpected on the background of a widely accepted model that H3K27me3 is lost from polycomb targets, such as bivalent domains (Carey et al., 2015, Galonska et al., 2015, Joshi et al., 2015, Marks et al., 2012, Tee et al., 2014, Weiner et al., 2016). Thus, we revisited this hypothesis in the light of our quantitative datasets. We compared the level of H3K27me3 in serum and 2i at well-described polycomb (PcG) binding sites (Joshi et al., 2015; Figures 4A and S3A–S3D). In the same samples, we profiled H3K27me1, which is both an intermediate product of the EZH2 enzyme and the product of H3K27me3 demethylation (Figures 4A, 4B, and S3A–S3D). Conditions with EZH2i treatment provided an experimental baseline. Average H3K27me3 levels at the peak were unchanged in 2i (Figure 4A), and H3K27me3 was gained in the flanking regions, demonstrating that H3K27me3 is maintained quantitatively at PcG target sites in 2i ground state and not reduced as previously suggested by prior studies, including an alternative quantitative ChIP using Drosophila H2Av spike-in control (Figure S3E; Egan et al., 2016, van Mierlo et al., 2019). It should be noted that the difference between our and previous observations stems solely from the scaling factor, derived in our case from the total read counts in ChIP relative to input (Figures S3E–S3G). To our knowledge, dynamic range and proportionality have not been assessed for the H2Av spike-in method (Egan et al., 2016); thus, differences in scaling could arise if the measurement would not follow an ideal linear behavior.
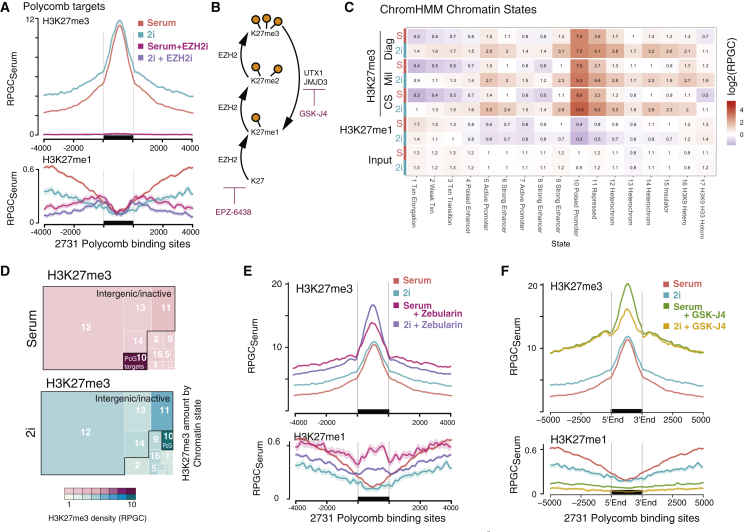
Figure 4.
H3K27me3 Is Maintained at PcG Targets and Gained Genome-wide
(A) Quantitative H3K27me3 and H3K27me1 profiles at 2,731 PcG target sites, in serum and 2i condition, as well as after treatment with EZH2 inhibitor EPZ-6438 (EZH2i) for 7 days. y axis shows reads per genomic content relative to serum condition (RPGCSerum), where serum condition is scaled to 1× coverage. SE is rendered as shaded area around lines.
(B) Schematic of enzymes involved in adding and removing H3K27 methylation and specific inhibitors used in pool 3.
(C) Heatmap of average H3K27me3, H3K27me1, and input read density across 17 functional chromatin states. Quantitative comparison between 2i and serum is shown using three different H3K27me3 antibodies. Color scale represents log2 of each pairwise 2i versus serum comparison.
(D) Treemap of total H3K27me3 amount (integral) across 17 functional chromatin states (as in C). Area is true to total proportions of H3K27me3. Color gradient shows average density (as in C).
(E) Average profile of H3K27me3 and H3K27me1 levels at PcG targets, in serum and 2i condition, as well as after treatment with DNMT inhibitor Zebularin. SE is rendered as shaded area around lines.
(F) Average profile of H3K27me3 and H3K27me1 levels at PcG targets, in serum and 2i condition, as well as after treatment with demethylase inhibitor GSK-J4.
See also Figures S3 and S4.
Strikingly, H3K27me3 levels were increased in the flanking regions of PcG target sites (Figure 4A) and virtually everywhere else in the genome, as exemplified by an analysis of 10-kb windows across the genome: 99% of the 10-kb bins exhibited a gain in H3K27me3 in 2i conditions (50% of those significant among three biological replicates; p adj. < 0.05), irrespective of their initial H3K27me3 density in serum condition (Figure S4A). Exceptions to this general increase were H3K4me3 co-enriched regions (Figure S4B) that maintained stable H3K27me3 levels. Few regions (including Hoxc and Hoxd clusters) significantly (p adj. < 0.05) lost H3K27me3 (Figures 2C, S4A, and S4B), albeit less pronounced than previously described (Marks et al., 2012, van Mierlo et al., 2019). Partitioning the genome in 17 functionally defined chromatin states (Ernst and Kellis, 2017), H3K27me3 was most enriched in ∼10,000 loci attributed to state 10 “poised promoter” (Figure 4C), which contained most of the well-defined PcG target sites (Figure S3C). H3K27me3 remained high or slightly increased in those regions (Figure 4C). In every other state, H3K27me3 was gained by a factor of 1.5- to 3-fold (Figure 4C). Such increase was even observed for state 1, corresponding to gene bodies of highly expressed genes (Figure S4C), which are known to carry high levels of H3K36me3 and have been thought to be devoid of any H3K27me3. However, our control experiments with EZH2i reveal that H3K27me3 is indeed present at considerable density, ∼3-fold (serum) and ∼2-fold (2i) below genome-wide average (Figure S3D). A broad, genome-wide gain in H3K27me3 thus accounted for the two-fold increase in global H3K7me3. Because H3K27me3 is a repressive modification known to also associate with repetitive sequences in certain circumstances, we wondered whether additional H3K27me3 would accumulate at interspersed or centromeric heterochromatin in 2i conditions. Because we were able to use UMI information to attribute accurate read counts to repetitive regions, we were able to quantitatively compare relative enrichment of histone modifications between the annotated genome and repetitive regions (Figure S4D): H3K27me3 was relatively depleted from major and minor satellite repeats, as well as intracisternal A particle (IAP) endogenous retroviruses (ERVs), in serum. Those regions, all known to be silenced predominantly by H3K9me3, showed almost no gain in H3K27me3 in 2i. Other interstitial repeats, such as SINE and LINE elements, gained H3K27me3 roughly two-fold (Figure S4D).
In summary, H3K27me3 was broadly gained in regions not typically targeted by PRC2. Comparison of our serum and 2i datasets with H3K27me3-depleted controls (EZH2i treatment) included in pool 3 (Figures 4A and S3D) demonstrates that only a small fraction (3.6% in serum and 2.0% in 2i) of H3K27me3-modified nucleosomes resides in PcG target regions (Figure 4D). This has important mechanistic implications, for example, that any effector protein (H3K27me3 “reader”) must be exquisitely sensitive to the density of H3K27me3 present on the chromatin fiber to selectively bind PcG target regions and not be titrated away by bulk H3K27me3 in the rest of the genome.
Gain in H3K27me3 Follows the Loss of CpG Methylation
Turning to the question of how ground state pluripotency invokes a broad increase in H3K27me3 at non-PcG targets, we considered that such global phenomenon could arise either from a shift in methylation or demethylation activity.
A known feature of ground state ESCs is a strong decrease in 5-methyl cytosine methylation at CpG dinucleotides (CpGme) (Galonska et al., 2015, Habibi et al., 2013, Leitch et al., 2013). Because a broadening of H3K27me3 in mESCs devoid of all CpGme has also been observed (Cooper et al., 2014, Habibi et al., 2013), it is conceivable that the gain in H3K27me3 is a consequence of low CpG methylation in 2i. Such mechanistic link could either involve a direct role of unmethylated CpGs in recruiting PRC2 (Oksuz et al., 2018, Perino et al., 2018), recruiting PRC1 (Moussa et al., 2019), or a more indirect effect, e.g., via regulating expression of PRC2 complex components. In a serum versus 2i comparison, H3K27me3 and CpGme appeared strongly anticorrelated (Figure S4E), as H3K27me3 was predominantly gained in regions that switch from high to low CpGme levels in 2i (Figure S4F). In contrast, constitutively unmethylated CpG islands underlying most of the above-mentioned PcG target sites and bivalent promoters stably maintain their high H3K27me3 levels (Figures S3A and S4F). We wondered whether we could recapitulate some or all features of H3K27me3 hypermethylation by artificially depleting CpG methylation. We treated mESCs with zebularin in serum condition and observed a broad increase in H3K27me3 (Figures 4E and S4G) similar to that described for DNMT knockout cell lines (Figure S4H; Brinkman et al., 2012). Zebularin appeared to have an additive effect with 2i (Figure S4G). This may be explained by the fact that remaining CpGme in 2i condition is removed by Zebularin (Galonska et al., 2015, Habibi et al., 2013, Leitch et al., 2013). Thus, our experiment supports the hypothesis that unmethylated CpGs are one determinant of the H3K27me3 landscape. Arguing against a strict requirement for unmethylated CpGs in acquiring H3K27me3, however, we found that even regions devoid of any CpG sites had H3K27me3 levels clearly above background in serum and gained H3K27me3 upon growth in 2i very similar to their CpG-containing flanking regions (Figure S4I). Although it is clear that DNMT knockout or otherwise reduced CpG levels per se do not induce the pluripotent ground state (Cooper et al., 2014, Galonska et al., 2015, Habibi et al., 2013, Leitch et al., 2013), it may be speculated that CpG demethylation is necessary albeit not sufficient for H3K27me3 hypermethylation and naive state. In fact, a recent bioRxiv preprint suggests that restoration of serum-like DNA methylation in the naive state also reverts back the H3K27me3 landscape, but the cell maintains a naive transcriptome and phenotype (McLaughlin et al., 2019). Vice versa, H3K27me3-deficient embryonic ectoderm development (EED)−/− protein mESCs have been shown to exhibit increased CpG methylation in 2i (van Mierlo et al., 2018) while also maintaining the naive transcriptome (Galonska et al., 2015).
Active Demethylation Limits H3K27me3 Levels Genome-wide
Finally, we wondered whether active demethylation contributed to maintaining a relatively lower H3K27me3 background outside of PcG target regions. GSK-J4 is a pan-demethylase inhibitor with specificity for H3K27me3 demethylases UTX and JMJD3 and, to a lesser extent, H3K4me3 demethylases (Heinemann et al., 2014). GSK-J4 treatment revealed that demethylases limit H3K27me3 levels in serum and 2i both at PcG targets and genome-wide (Figures 4F, S4J, and S4K). Gain in H3K27me3 was clearly at the expense of H3K27me1, the product of UTX/JMJD3 demethylases (Figure 4F). Analysis of the gain or loss of H3K27me3 according to initial H3K27me3 quantiles in serum revealed that GSK-J4 favors accumulation of H3K27me3 in regions exhibiting low to very low H3K27me3 while affecting H3K27me high regions to a lesser extent (Figure S4J). This would fit to a model in which UTX/JMJD3 acts relatively unspecific on H3K27me3 scattered across the genome, limiting H3K27me3 levels predominantly at non-polycomb targets. Such a model is also in agreement with genome-wide maps of UTX/JMJD3 that found a relatively flat distribution with weak enrichment at active gene promoters rather than polycomb targets (Banaszynski et al., 2013, Juan et al., 2017). Thus, it remains to be determined whether and how their localization and/or activity is modulated by the serum-to-2i transition. In summary, PRC2 and UTX/JMJD3 jointly balance H3K27me3 level in both serum and 2i conditions.
Maintenance of H3K27me3 and Loss of H3K4me3 at Bivalent Promoters
Compared to serum condition, the 2i ground state is characterized by low expression of particular lineage-specific genes that are thought to prime differentiation. Albeit dispensable for ground state pluripotency, PRC2 is thought to repress bivalent genes in mESCs, preventing premature activation of developmentally regulated genes. Deletion of the essential PRC2 component EED leads to a ∼2-fold increase in average expression of bivalent genes (Galonska et al., 2015). Previous studies report a loss or reduction of H3K27me3 at bivalent promoters as a hallmark of the 2i ground state (Carey et al., 2015, Galonska et al., 2015, Joshi et al., 2015, Marks et al., 2012, van Mierlo et al., 2019, Schlesinger and Meshorer, 2019, Tee et al., 2014, Weiner et al., 2016). This has been difficult to reconcile with the observation that bivalent genes were maintained silent or even further repressed in 2i conditions (Finley et al., 2018, Galonska et al., 2015, Joshi et al., 2015, Marks et al., 2012). Thus, it is an important question for understanding PRC2 function if the broad gain in H3K27me3 comes at the expense of maintaining PcG-mediated repression at targets such as bivalent domains.
To answer this question in a quantitative manner, we analyzed bivalent genes in serum versus 2i conditions. Overlapping largely with the PcG target sites discussed above, we find that H3K27me3 is quantitatively maintained at the ∼2,000 bivalent promoters (Figures 5A–5C, S5A, and S5B). Although individual datasets showed more variability in H3K27me3 peaks, 171 promoters lost and 126 gained H3K27me3 consistently more than 1.5-fold across our three independent MINUTE-ChIP experiments (Figure 5D; for examples, see Figure S5C). We wondered whether the quantitative changes in H3K27me3 levels we observe at these specific subsets of bivalent promoters could explain known gene expression changes. Indeed, bivalent genes gaining H3K27me3 in 2i condition were transcriptionally silenced (Figure 5D). Those losing H3K27me3 included a subset of Hox genes and regions previously shown to interact with Hox gene clusters (Joshi et al., 2015) and showed a variable degree of upregulation (Figures 5A and 5D). These data show that, with the exception of a small subset of bivalent genes, PRC2 is able to maintain its cognate activity in restricting bivalent gene transcription despite its broad gain in activity elsewhere. The pronounced reduction of H3K4me3 may additionally account for the small but consistent decrease in transcription from bivalent genes (Figures 5D, S5B, and S5C). Thus, our quantitative data reconcile apparent discrepancies arising from non-quantitative data and agree with the common notion that H3K27me3 antagonizes transcriptional activity.
Figure 5.
Bivalency Safeguarding Developmental Genes in Ground State Pluripotency Is Characterized by Low H3K4me3 and High H3K27me3
(A) Relative levels of H3K27me3 and H3K4me3 at bivalent, non-transcribed (low) and highly active (high) promoters, as well as Hox genes (Hox). y axis is normalized to RPGC in serum. RNA-seq expression levels and CpG methylation (% methylation of all CpGs in the region) levels at the same promoters (right) are shown. Wilcoxon signed rank test; ∗∗∗∗p < 0.0001; not significant (n.s.), p > 0.05.
(B) Heatmap across promoters of the bivalent genes as in (A) and Figure 3D.
(C) Average profiles of H3K27me3 and H3K27me1 across 1,969 bivalent genes. SE is rendered as shaded area around lines. Additional H3K27me3 antibody replicates are shown in Figure S5A.
(D) Number of bivalent promoters that changed H3K27me3 levels more than 1.5-fold between 2i and serum, consistent across the three different H3K27me3 antibody and biological replicates. 126 bivalent promoters gained H3K27me3 (“up”), 177 bivalent promoters lost H3K27me3 in 2i (“down”), and the remaining did not change consistently. Promoters that changed H3K27me3 more than two-fold are also shown individually in Figure S5C. For each of the three groups, up, down, and “unchanged,” the average expression level from triplicate RNA-seq in serum and 2i (Finley et al., 2018) is shown. Wilcoxon signed rank test.
(E) Hypothesis for transitions in bivalency: ground state bivalency (H3K4me4 low/H3K27me3 high) is refractory to activation (“naive”). For developmental genes to be activated upon lineage commitment, bivalent promoter needs to acquire H3K4me3 first (“primed”).
See also Figure S5.
Discussion
Here, we use a quantitative ChIP-seq approach to characterize the pluripotent ground state of mESCs. Our study provides a prime example for the need of an accurate quantitative ChIP-seq method in order to compare different developmental chromatin states: we find unexpected global shifts in histone modification levels that confounded prior studies. Our quantitative data allow us to mechanistically explain gene expression differences and provide the basis for substantially revising the model for bivalent promoter control in the ground state.
We show that the H3K27me3 is an abundant and extremely broad modification in the ground state of pluripotency that broadly covers the genome. Essentially no new H3K27me3 peaks are formed upon transition to a primed state; instead, the ambient H3K27me3 “background” is more and more suppressed, revealing existing H3K27me3 peaks at all cognate PcG targets (Figure 5E). Given the fact that neither H3K27me3 nor its antagonizing CpGme are strictly required for maintenance of the ground state (Galonska et al., 2015, McLaughlin et al., 2019, van Mierlo et al., 2019), it remains to be elucidated whether the broad coverage of the genome with H3K27me can be attributed to a specific function or rather represents a collateral effect of transcription factors networks overriding epigenomic control mechanisms to “lock in” a pluripotent state.
We further show that the ground state is characterized by particularly low levels of H3K4me3. This finding explains why bivalency, defined as co-occurrence of H3K27me3 and H3K4me3 on the same nucleosome, has been found to be dramatically reduced in 2i conditions (Weiner et al., 2016). Originally interpreted as loss of H3K27me3, our data provide an alternative explanation: bivalent domains start out in the ground state mainly covered by H3K27me3 and, only upon priming, accumulate H3K4me3 levels comparable to those at active genes (Figure 5E). Indeed, a recent report suggests that H3K4me3 specifies the robust and timely induction of bivalent promoters during differentiation (Mas et al., 2018). Given that H3K27me3 is per se dispensable for ground state pluripotency (Galonska et al., 2015, van Mierlo et al., 2019), we hypothesize that low H3K4me3 together with high H3K27me3 levels at bivalent promoters act to safeguard the ground state of pluripotency.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-H3 | Active motif | Cat# 39763; RRID:AB_2650522 |
| Rabbit monoclonal anti-H3K4me3 | Millipore | Cat# 04-745; RRID:AB_1163444 |
| Rabbit polyclonal anti-H3K27me3 | Millipore | Cat# 07-449; RRID:AB_310624 |
| Rabbit monoclonal anti-H3K27me3 | Cell Signaling Technology | Cat# 9733; RRID:AB_2616029 |
| Rabbit polyclonal anti-H3K27me3 | Diagenode | Cat# C15410195; RRID:AB_2753161 |
| Rabbit monoclonal anti-H3K27me2 | Cell Signaling Technology | Cat# 9728; RRID:AB_1281338 |
| Rabbit monoclonal anti-H3K27me1 | Cell Signaling Technology | Cat# 5326; RRID:AB_10695148 |
| IRDye 800CW Donkey anti-Mouse IgG (H + L) | LI-COR Bio-sciences | Cat# 926-32210; RRID:AB_621842 |
| IRDye 680RD Donkey anti-Rabbit IgG (H + L) | LI-COR Bio-sciences | Cat# 926-68073; RRID:AB_10954442 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Micrococcal Nuclease | NEB | Cat# M0247S |
| Proteinase K | Life technologies | Cat# 25530015 |
| RNase A | Life technologies | Cat# EN0531 |
| RNase H | NEB | Cat# M0297S |
| GSK-J4 | Sigma | Cat# SML0701 |
| Zebularin | Sigma | Cat# Z4775 |
| EZSolution EPZ-6438 | BioVision | Cat# 2428 |
| Accutase solution | Sigma | Cat# A6964 |
| PD0325901 | Sigma | Cat# PZ0162 |
| CHIR99021 | Sigma | Cat# SML1046 |
| Critical Commercial Assays | ||
| End-It DNA End-Repair kit | Lucigen | Cat# ER81050 |
| Fast-Link DNA Ligation kit | Lucigen | Cat# LK6201H |
| SureBeads Protein A/G Magnetic beads | Biorad | Cat# 161-4013/4023 |
| Silane beads | Life technologies | Cat# 37002D |
| Agencourt AMPure XP | Beckman Coulter | Cat# A63881 |
| High Sensitivity DNA Kit for Bioanalyzer | Agilent | Cat# 5067-4626 |
| Qubit DSDNA HS Assay Kit | Life technologies | Cat# Q32854 |
| Qubit RNA HS Assay Kit | Life technologies | Cat# Q32855 |
| HiScribe T7 Quick High Yield RNA Synthesis kit | NEB | Cat# E2050S |
| T4 RNA Ligase 2, truncated | NEB | Cat# M0242L |
| SuperScript III First-Strand Synthesis SuperMix | Life technologies | Cat# 18080-400 |
| NEBNext® High-Fidelity 2X PCR Master Mix | NEB | Cat# M0541L |
| NextSeq® 500/550 High Output Kit v2 (75 cycles) | Illumina | Cat# 20024906 |
| Deposited Data | ||
| All NGS data | This study | GEO: GSE126252 |
| Genomic files and Pseudocode | This study, Mendeley Data | https://doi.org/10.17632/s23bhg4xjv.1 |
| Experimental Models: Cell Lines | ||
| Mouse ESC: RW.4 (129/SvJ) | Karolinska Center for Transgene Technologies | N/A |
| Oligonucleotides | ||
| Primers | This study | See Table S1 |
| Software and Algorithms | ||
| bcl2fastq (v2.20) | Illumina | http://emea.support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| Bowtie 2 (v2.3.4.3) | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| SAMtools (v1.9) | Li et al., 2009 | http://www.htslib.org/ |
| BEDtools (v2.27.1) | Quinlan, 2014 | https://bedtools.readthedocs.io/en/latest/ |
| Je | Girardot et al., 2016 | https://gbcs.embl.de/portal/tiki-index.php?page=Je |
| deepTools (v3.1.0) | Ramírez et al., 2014 | https://deeptools.readthedocs.io/en/develop/ |
| Picard (v2.10.3) | https://broadinstitute.github.io/picard/ | |
| bwtool | Pohl and Beato, 2014 | https://github.com/CRG-Barcelona/bwtool/ |
| ngsplot | Shen et. al., 2014 | https://github.com/shenlab-sinai/ngsplot |
| SeqPlots | Stempor and Ahringer, 2016 | http://seqplots.ga |
| DESeq | Anders and Huber, 2010 | http://bioconductor.org/packages/release/bioc/html/DESeq.html |
| R (v) | R core team | https://www.r-project.org/ |
| Image Studio | LI-COR Bio-sciences | N/A |
| Other | ||
| Mod Spec® Service | Active Motif | Cat# 25085 |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Simon J Elsässer (simon.elsasser@scilifelab.se). This study did not generate new unique reagents.
Experimental Model and Subject Details
Rw4 murine (male,129X1/SvJ) embryonic stem cells (mESCs) were cultured feeder-free in 0.1% gelatin-coated dishes. Serum condition: Knockout DMEM (Life Technologies), 2 mM Glutamax (GIBCO), 0.1 mM non-essential amino acids (GIBCO), 15% ESC-grade fetal bovine serum (FBS) (GIBCO), 0.1 mM β-mercaptoethanol, and leukemia inhibitory factor (LIF) (Millipore), 2i/Serum condition: above medium supplemented with 2i; 1 μM MEK inhibitor PD0325901 (Sigma) and 3 μM GSK3 inhibitor CHIR99021 (Sigma). 2i condition: serum free ESGRO Complete Basal medium (Millipore) with 0.1mM LIF and 2i as described above. For drug treatment, mESCs were grown in respective medium supplemented with 10 μM GSK-J4 (Sigma) for 96 h, 50 μM Zebularin for 96 h or 10 μM EPZ-6438 (BioVision) for 7 days. For all conditions cells were passaged in 48 h intervals, using accutase (Sigma) for detachment. Cell line was tested for mycoplasma contamination.
Method Details
Immunoblot Analysis
1x106 cells were harvested for each growth condition, washed twice with phosphate buffered saline (PBS) and lysed in 500 μL of ice-cold radioimmunoprecipitation (RIPA) buffer (0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 10 mM HEPES [pH 7.6], 1 mM EDTA, 5% glycerol and 140 mM NaCl) supplemented with Protease Inhibitor Cocktail (PIC, Roche) on ice for 10 min. For the drug treated conditions, 10 μL of lysate from the ChIP sample was used for the analysis directly. Lysates were homogenized by sonication for 8-10 cycles at high power, 30 s on/off in a Bioruptor sonicator (Cosmo Bio Co. Ltd.). Samples were boiled at 95°C for 10 min with 6 × SDS sample buffer before loading onto 4%–20% Tris-glycine gels (BioRad). Resolved proteins were transferred to nitrocellulose membranes using the Trans-Blot® Turbo system (BioRad) according to manufacturer’s instructions. Membranes were then blocked for 1 h in 1% casein prepared in Tris-buffered saline and 0.1% Tween-20 (TBS-T) before blotting with respective primary antibodies diluted in TBST, overnight at 4°C. Blots were washed three times with TBST and incubated with secondary antibody in the same buffer for 1 h at room temperature (protect from light). Post three TBST washes, the membranes were imaged on a LI-COR Odyssey ® FC system. Quantitation of signal and analysis was performed using the LI-COR Image studio software. Primary antibodies included total H3 1:10,000 (Active motif 39763), H3K4me3 1: 5000 (Millipore 04-745), H3K27me3 1: 5000 (Millipore 07-449). The secondary antibodies were IRDye® 680RD anti-rabbit and IRDye® 800CW anti-mouse (LI-COR) at 1:5000 dilution.
Mass Spectrometry
5x106 mESCs per growth condition (Serum or 2i) were harvested, washed once with PBS, spun down at 800 g for 5 min. The cell pellets were flash frozen and sent to ActiveMotif for their Mod Spec® service. Briefly, histones were acid extracted, derivatized via propionylation, digested with trypsin, newly formed N-termini were propionylated, and then measured three separate times using the Thermo Scientific TSQ Quantum Ultra mass spectrometer coupled with an UltiMate 3000 Dionex nano-liquid chromatography system. The data was quantified using Skyline, and represents the percent of each modification within the total pool of that tryptic peptide.
MINUTE-ChIP
One-pot chromatin fragmentation and barcoding
MINUTE-ChIP and library preparation protocol is based on the Mint-ChIP protocol developed by the Bernstein lab (van Galen et al., 2016), with modifications as follows: 1x106 cells were harvested for each growth condition, washed twice with PBS and cell pellets were flash frozen at −80°C prior to use. Cells were resuspended in 50 μL PBS, lysed and digested to mono- to tri-nucleosomes fragments by adding 50 μL of 2x Lysis buffer (100 mM Tris-HCL [pH 8.0], 0.2% Triton X-100, 0.1% sodium deoxycholate, 10 mM CaCl2 and 1x PIC) containing 2U/μl of micrococcal nuclease (New England BioLabs, M0247S) and incubating on ice for 20 min and then 37°C for 10 min. Double-stranded DNA adaptors for barcoding and T7 amplification were generated by slow annealing of complementary single-stranded oligos (for sequences refer to Table S1), where the UMI bases were randomized. While resulting dsDNA adaptors may contain mismatched bases within the UMI, only the bottom strand UMI is amplified and sequenced. For each sample, 40 μL of the whole cell lysate containing the digested chromatin was taken forward into an overnight blunt end ligation reaction (End-It DNA repair kit and Fast-Link DNA ligation kit, Epicenter) with double stranded DNA adapters at 16°C. As in the original Mint-ChIP design, adaptors carried a partial SBS3 for Illumina sequencing flanked by a T7 RNA Polymerase for linear amplification. Between the SBS3 sequence and a 8bp sample barcode at the 3′ end, a 6bp randomized sequence was introduced, serving as a unique molecular identifier (UMI) (Figure 1A). UMI and sample barcode are ligated 5′ to the chromatin fragment and constitute the first 14 bases of read 1. The 4096 possible UMIs provide sufficient diversity to distinguish if two reads mapping to the exact same genomic location arose from a PCR amplification artifact or are indeed unique molecule. The adaptor concentration was optimized to 2.5 μM / reaction to reduce adaptor dimers. The ligation reaction was terminated with a lysis dilution buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 50 mM EGTA, 50 mM EDTA, 0.1% sodium deoxycholate and 1x PIC) and barcoded samples were combined into a single pool, spun down at 24,000 rpm for 10 min at 4°C.
Immunoprecipitation
50 μL Protein A/G magnetic beads (BioRad) were washed twice with PBS-T (PBS+ 0.1% Tween 20) and coupled to one of the following antibodies in the same buffer for 1 hr at room temperature with rotation: 3 μL H3 (Active motif 39763), 5 μL H3K4me3 (Millipore 04-745), 5 μL H3K27me1 (Cell signaling 5326), 5 μL H3K27me2 (Cell signaling 9728), 5 μL H3K27me3 (Cell signaling 9733 (rab mAb) or Diagenode C15410195 (rab pAb) or Millipore 07-449 (rab pAb)). Beads were then washed quickly with RIPA buffer. 200-400 μL of the cleared lysate pool was added to the pre-coupled magnetic beads and parallel ChIP assays were incubated further for 4 h at 4°C with rotation. 5% of the above volume was saved as the input pool and processed through the remaining protocol in a manner similar to the IPs. Next, the beads were washed (RIPA, RIPA high salt, LiCl and TE buffer) resuspended in ChIP elution buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.1% SDS, and 300 mM NaCl) containing 0.25 mg/mL Proteinase K (Thermo Fisher Scientific, 25530015) and eluted at 63°C for 1 h.
Linear amplification and library preparation
The native ChIP DNA (fragments longer than 100bp) was isolated using 1x SPRI beads (Beckman Coulter) and set up in an overnight in vitro transcription reaction (HiScribe T7 Quick High Yield RNA Synthesis kit, New England BioLabs). The resulting RNA was purified using Silane beads (Life Technologies) and an RNA 3′ adaptor was ligated onto it using T4 RNA Ligase 2, truncated (New England BioLabs) for 1 h at 25°C. The mixture was subsequently supplemented with components of the reverse transcription reaction (SuperScript III First-Strand Synthesis SuperMix, Life Technologies) to produce cDNA, primed using the ligated 3′ adaptor. Final libraries for each ChIP were produced using 150-200 ng of purified cDNA in a PCR reaction (High-Fidelity 2x master mix, New England BioLabs) for 8 cycles with 0.2 μM primers that carried a second 8bp barcode sequence. Quality assessment and concentration estimation of the purified DNA was done using the Qubit (Life Technologies) and BioAnalyzer (Agilent). Each library was then diluted to 4 nM and combined into a single pool before sequencing on the Illumina NextSeq500 platform.
Quantification and Statistical Analysis
Demultiplexing and deduplication
Sequencing was performed using 50:8:34 cycles (Read1:Index1:Read2) Illumina bcl2fastq was used to demultiplex paired-end sequencing reads by 8nt index1 read (PCR barcode). NextSeq lanes were merged into single fastq files, creating the primary fastq files. Read1 starts with 6nt UMI and 8nt barcode in the format NNNNNNABCDEFGH. A sample sheet was used with four columns corresponding to sample name, replicate name, barcode sequence, primary fastq name. For each line in the sample sheet, reads matching the barcode sequence were extracted from the primary fastq file, allowing up to one mismatch. Duplicate reads identified by identical first 24nt of read1 (spanning UMI, barcode and 10bp genomic sequence) were discarded. Read pairs matching parts of the adaptor sequence (SBS3 or T7 promoter) in either read1 or read2 were removed. Demultiplexed and deduplicated reads were written into sample-specific fastq file used for subsequent mapping.
Mapping
Sample-specific paired fastq files were mapped using bowtie2 v2.3.4.3 (Langmead and Salzberg, 2012) using–fast parameter to the mouse genome (mm9). Alignments were processed into sorted BAM files with samtools v1.9 (Li et al., 2009). Blacklisted regions were removed from BAM file using BEDTools v2.27.1 (Quinlan, 2014). Reads were also mapped to a metagenome of RepBase murine repetitive sequences using botwie2.
Estimating library diversity
For estimating library size, demultiplexed reads (without further filtering or deduplication) were pre-processed by moving the UMI sequence to the header and mapped to mm9 using bowtie2. The UMI-sensitive deduplication tool Je (Girardot et al., 2016) was then used to estimate the library diversity from the BAM file.
Generation of coverage tracks and quantitative scaling
Input coverage tracks with 1bp resolution in BigWig format were generated from BAM files using deepTools (Ramírez et al., 2014) bamCoverage (v3.1.0) and scaled to a reads-per-genome-coverage of one (1xRPGC, also referred to as ‘1x normalization’) using mm9 genome size 2654895218. ChIP coverage tracks were generated from BAM files using deepTools (v3.1.0) bamCoverage. Quantitative scaling of the ChIP-Seq tracks among conditions within each pool was based on their Input-Normalized Mapped Read Count (INRC). INRC was calculated by dividing the number of unique mm9-mapped reads by the respective number of Input reads: #mapped[ChIP] / #mapped[Input]. This essentially corrected for an uneven representation of barcodes in the Input and we could demonstrate that the INRC is proportional to the amount of epitope present in each condition.
One condition in the pool was chosen as the reference sample (in our case always Serum condition, no treatment), which was scaled to 1x coverage (also termed Reads per Genome Coverage, RPGC). All other conditions were scaled relative to the reference using the ratio of INRCs multiplied by the scaling factor determined for 1x normalization of the reference: (#mapped[ChIP] / #mapped[Input]) / (#mapped[ChIP_Reference] / #mapped[Input_Reference]) ∗ scaling factor.
Genome statistics
Statistics on genomic 10kb bins and custom intervals (gene sets, peak sets) were determined from scaled BigWig files using bwtool (Pohl and Beato, 2014) summary function. For calculation of enrichments in repetitive regions, repetitive read counts were scaled according to the INRC calculated from the mm9 genome. Summary statistics were visualized using R ggplot2 (https://ggplot2.tidyverse.org) and significance was calculated using ggpubr package (https://rpkgs.datanovia.com/ggpubr/).
For statistical comparison of conditions with replicates, nbinomTest from DESeq package (Anders and Huber, 2010) was applied to the summary tables generated with bwtools. Adjusted p value from nbinomTest output was written to a bedgraph file for visualization.
Track visualization
Density plots were generated directly BAM files using ngs.plot tool (Shen et al., 2014) with modifications: the in-built genome averaging was removed and replaced by a scaling factor calculated from the aligned read counts as above (#mapped[ChIP] / #mapped[Input]) / (#mapped[ChIP_Reference] / #mapped[Input_Reference]). Standard error is rendered as shaded area around lines. Equivalent plots can be generated using SeqPlots (Stempor and Ahringer, 2016) from the scaled BigWig files avaliable under GSE126252. Heatmaps and plots from published datasets were generated from BigWig files and generated with SeqPlots using a 50bp moving window average smoothing. For IGV visualization, a variable smoothing was applied depending on the size of the genomic region.
Quality control
Picard v2.10.3 (https://broadinstitute.github.io/picard/) was used to determine insert size distribution, duplication rate, estimated library size. For the latter, primary reads were demultiplexed separately from the procedure described above retaining all duplicate reads and mapped using bowtie2 before running Picard MarkDuplicates. For estimation of cross contamination, mm9 and hg19 fasta files were combined to build a hybrid botwie2 index and used for mapping the reads.
Data and Code Availability
The high-throughput data reported in this study have been deposited in GEO under the accession number GSE126252 and GSE133056, which includes demultiplexed and deduplicated reads and a quantitatively scaled bigwig track for each sample. Additional genomic files, pseudocode and supplemental data files are available on Mendeley Data (https://doi.org/10.17632/s23bhg4xjv.2).
Published datasets used in this study include: RNA-Seq Serum versus 2i (Finley et al., 2018, Marks et al., 2012) (GSM590124, GSM590125, GSM590126, GSM590127, GSM2345133, GSM2345139), CpG methylation (Habibi et al., 2013) (GSM1127953, GSM1127954), H3K37me3 (King et al., 2016, Marks et al., 2012) (GSM590114, GSM590113, GSM2229356, GSM2229359, GSM2229364), H3K4me3, H3K4me3-H3K27me3 Re-ChIP, H3K4me3 in MLL2KO (GSM2645495, GSM2645496), MLL2 ChIP (GSM2645501) (Mas et al., 2018), Ring1b and Suz12 ChIP (GSM1856437, GSM1856444), Spike-in controlled H3K27me3 ChIP (GSM3080981, GSM3080982, GSM3080988, GSM3080989), (van Mierlo et al., 2019).
Acknowledgments
We thank all members of the Elsässer lab for input into concept, experimental design, and manuscript. We thank Rozina Caridha for assistance with experiments and Laura Banaszynski for helpful discussions. Research was funded by Karolinska Institutet SFO Molecular Biosciences, Vetenskapsrådet (2015-04815), H2020 ERC-2016-StG (715024 RAPID), Ming Wai Lau Center for Reparative Medicine, Ragnar Söderbergs stiftelse, and Wissenschaftskolleg zu Berlin.
Author Contributions
B.K. and S.J.E. conceived experiments. B.K. performed experiments. B.K. and S.J.E. analyzed data. B.K. and S.J.E. wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: September 17, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.08.046.
Supporting Citations
The following reference appears in the Supplemental Information: Bao et al. (2015).
Supplemental Information
Sequences of oligos used for adaptors and PCR primers.
Detailed list and graphical overview of samples, barcodes, pools, antibodies, GEO accessions for three MINUTE-ChIP experiments comparing Serum and 2i condition.
References
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni L., Al-Hasani H., Ramírez F., Panzeri I., Ryan D.P., Santacruz D., Kress N., Pospisilik J.A., Bönisch U., Manke T. RELACS nuclei barcoding enables high-throughput ChIP-seq. Commun. Biol. 2018;1:214. doi: 10.1038/s42003-018-0219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L.A., Wen D., Dewell S., Whitcomb S.J., Lin M., Diaz N., Elsässer S.J., Chapgier A., Goldberg A.D., Canaani E. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W., Kojima K.K., Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman A.B., Gu H., Bartels S.J.J., Zhang Y., Matarese F., Simmer F., Marks H., Bock C., Gnirke A., Meissner A., Stunnenberg H.G. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22:1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Finley L.W.S., Cross J.R., Allis C.D., Thompson C.B. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabbert C.D., Adjalley S.H., Steinmetz L.M., Pelechano V. Multiplexed ChIP-seq using direct nucleosome barcoding: a tool for high-throughput chromatin analysis. Methods Mol. Biol. 2018;1689:177–194. doi: 10.1007/978-1-4939-7380-4_16. [DOI] [PubMed] [Google Scholar]
- Cooper S., Dienstbier M., Hassan R., Schermelleh L., Sharif J., Blackledge N.P., De Marco V., Elderkin S., Koseki H., Klose R. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S., Hofemeister H., Marks H., Kranz A., Ciotta G., Singh S., Anastassiadis K., Stunnenberg H.G., Stewart A.F. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141:526–537. doi: 10.1242/dev.102681. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Helin K. Transcriptional regulation by polycomb group proteins. Nat. Struct. Mol. Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Egan B., Yuan C.C., Craske M.L., Labhart P., Guler G.D., Arnott D., Maile T.M., Busby J., Henry C., Kelly T.K. An alternative approach to ChIP-seq normalization enables detection of genome-wide changes in histone H3 lysine 27 trimethylation upon EZH2 inhibition. PLoS ONE. 2016;11:e0166438. doi: 10.1371/journal.pone.0166438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nat. Protoc. 2017;12:2478–2492. doi: 10.1038/nprot.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley L.W.S., Vardhana S.A., Carey B.W., Alonso-Curbelo D., Koche R., Chen Y., Wen D., King B., Radler M.R., Rafii S. Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity. Nat. Cell Biol. 2018;20:565–574. doi: 10.1038/s41556-018-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galonska C., Ziller M.J., Karnik R., Meissner A. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell. 2015;17:462–470. doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardot C., Scholtalbers J., Sauer S., Su S.Y., Furlong E.E.M. Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinformatics. 2016;17:419. doi: 10.1186/s12859-016-1284-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi E., Brinkman A.B., Arand J., Kroeze L.I., Kerstens H.H.D., Matarese F., Lepikhov K., Gut M., Brun-Heath I., Hubner N.C. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Heinemann B., Nielsen J.M., Hudlebusch H.R., Lees M.J., Larsen D.V., Boesen T., Labelle M., Gerlach L.O., Birk P., Helin K. Inhibition of demethylases by GSK-J1/J4. Nature. 2014;514:E1–E2. doi: 10.1038/nature13688. [DOI] [PubMed] [Google Scholar]
- Joshi O., Wang S.Y., Kuznetsova T., Atlasi Y., Peng T., Fabre P.J., Habibi E., Shaik J., Saeed S., Handoko L. Dynamic reorganization of extremely long-range promoter-promoter interactions between two states of pluripotency. Cell Stem Cell. 2015;17:748–757. doi: 10.1016/j.stem.2015.11.010. [DOI] [PubMed] [Google Scholar]
- Juan A.H., Wang S., Ko K.D., Zare H., Tsai P.F., Feng X., Vivanco K.O., Ascoli A.M., Gutierrez-Cruz G., Krebs J. Roles of H3K27me2 and H3K27me3 examined during fate specification of embryonic stem cells. Cell Rep. 2017;18:297. doi: 10.1016/j.celrep.2016.12.036. [DOI] [PubMed] [Google Scholar]
- King A.D., Huang K., Rubbi L., Liu S., Wang C.Y., Wang Y., Pellegrini M., Fan G. Reversible regulation of promoter and enhancer histone landscape by DNA methylation in mouse embryonic stem cells. Cell Rep. 2016;17:289–302. doi: 10.1016/j.celrep.2016.08.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H.G., McEwen K.R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J.G., Smith A. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013;20:311–316. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A., Stunnenberg H.G. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas G., Blanco E., Ballaré C., Sansó M., Spill Y.G., Hu D., Aoi Y., Le Dily F., Shilatifard A., Marti-Renom M.A., Di Croce L. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat. Genet. 2018;50:1452–1462. doi: 10.1038/s41588-018-0218-5. [DOI] [PubMed] [Google Scholar]
- McLaughlin K.A., Flyamer I.M., Thomson J.P., Mjoseng H.K., Shukla R., Williamson I., Grimes G.R., Illingworth R.S., Adams I.R., Pennings S. DNA methylation directs polycomb-dependent 3D genome reorganisation in naive pluripotency: supplementary information. bioRxiv. 2019 doi: 10.1016/j.celrep.2019.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa H.F., Bsteh D., Yelagandula R., Pribitzer C., Stecher K., Bartalska K., Michetti L., Wang J., Zepeda-Martinez J.A., Elling U. Canonical PRC1 controls sequence-independent propagation of Polycomb-mediated gene silencing. Nat. Commun. 2019;10:1931. doi: 10.1038/s41467-019-09628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuz O., Narendra V., Lee C.H., Descostes N., LeRoy G., Raviram R., Blumenberg L., Karch K., Rocha P.P., Garcia B.A. Capturing the onset of PRC2-mediated repressive domain formation. Mol. Cell. 2018;70:1149–1162.e5. doi: 10.1016/j.molcel.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perino M., van Mierlo G., Karemaker I.D., van Genesen S., Vermeulen M., Marks H., van Heeringen S.J., Veenstra G.J.C. MTF2 recruits polycomb repressive complex 2 by helical-shape-selective DNA binding. Nat. Genet. 2018;50:1002–1010. doi: 10.1038/s41588-018-0134-8. [DOI] [PubMed] [Google Scholar]
- Pohl A., Beato M. bwtool: a tool for bigWig files. Bioinformatics. 2014;30:1618–1619. doi: 10.1093/bioinformatics/btu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R. BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014;47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger S., Meshorer E. Open chromatin, epigenetic plasticity, and nuclear organization in pluripotency. Dev. Cell. 2019;48:135–150. doi: 10.1016/j.devcel.2019.01.003. [DOI] [PubMed] [Google Scholar]
- Shen L., Shao N., Liu X., Nestler E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempor P., Ahringer J. SeqPlots - interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res. 2016;1:14. doi: 10.12688/wellcomeopenres.10004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee W.W., Shen S.S., Oksuz O., Narendra V., Reinberg D. Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell. 2014;156:678–690. doi: 10.1016/j.cell.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen P., Viny A.D., Ram O., Ryan R.J.H., Cotton M.J., Donohue L., Sievers C., Drier Y., Liau B.B., Gillespie S.M. A multiplexed system for quantitative comparisons of chromatin landscapes. Mol. Cell. 2016;61:170–180. doi: 10.1016/j.molcel.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mierlo G., Wester R.A., Marks H. Quantitative subcellular proteomics using SILAC reveals enhanced metabolic buffering in the pluripotent ground state. Stem Cell Res. (Amst.) 2018;33:135–145. doi: 10.1016/j.scr.2018.09.017. [DOI] [PubMed] [Google Scholar]
- van Mierlo G., Dirks R.A.M., De Clerck L., Brinkman A.B., Huth M., Kloet S.L., Saksouk N., Kroeze L.I., Willems S., Farlik M. Integrative proteomic profiling reveals PRC2-dependent epigenetic crosstalk maintains ground-state pluripotency. Cell Stem Cell. 2019;24:123–137.e8. doi: 10.1016/j.stem.2018.10.017. [DOI] [PubMed] [Google Scholar]
- Weiner A., Lara-Astiaso D., Krupalnik V., Gafni O., David E., Winter D.R., Hanna J.H., Amit I. Co-ChIP enables genome-wide mapping of histone mark co-occurrence at single-molecule resolution. Nat. Biotechnol. 2016;34:953–961. doi: 10.1038/nbt.3652. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of oligos used for adaptors and PCR primers.
Detailed list and graphical overview of samples, barcodes, pools, antibodies, GEO accessions for three MINUTE-ChIP experiments comparing Serum and 2i condition.
Data Availability Statement
The high-throughput data reported in this study have been deposited in GEO under the accession number GSE126252 and GSE133056, which includes demultiplexed and deduplicated reads and a quantitatively scaled bigwig track for each sample. Additional genomic files, pseudocode and supplemental data files are available on Mendeley Data (https://doi.org/10.17632/s23bhg4xjv.2).
Published datasets used in this study include: RNA-Seq Serum versus 2i (Finley et al., 2018, Marks et al., 2012) (GSM590124, GSM590125, GSM590126, GSM590127, GSM2345133, GSM2345139), CpG methylation (Habibi et al., 2013) (GSM1127953, GSM1127954), H3K37me3 (King et al., 2016, Marks et al., 2012) (GSM590114, GSM590113, GSM2229356, GSM2229359, GSM2229364), H3K4me3, H3K4me3-H3K27me3 Re-ChIP, H3K4me3 in MLL2KO (GSM2645495, GSM2645496), MLL2 ChIP (GSM2645501) (Mas et al., 2018), Ring1b and Suz12 ChIP (GSM1856437, GSM1856444), Spike-in controlled H3K27me3 ChIP (GSM3080981, GSM3080982, GSM3080988, GSM3080989), (van Mierlo et al., 2019).