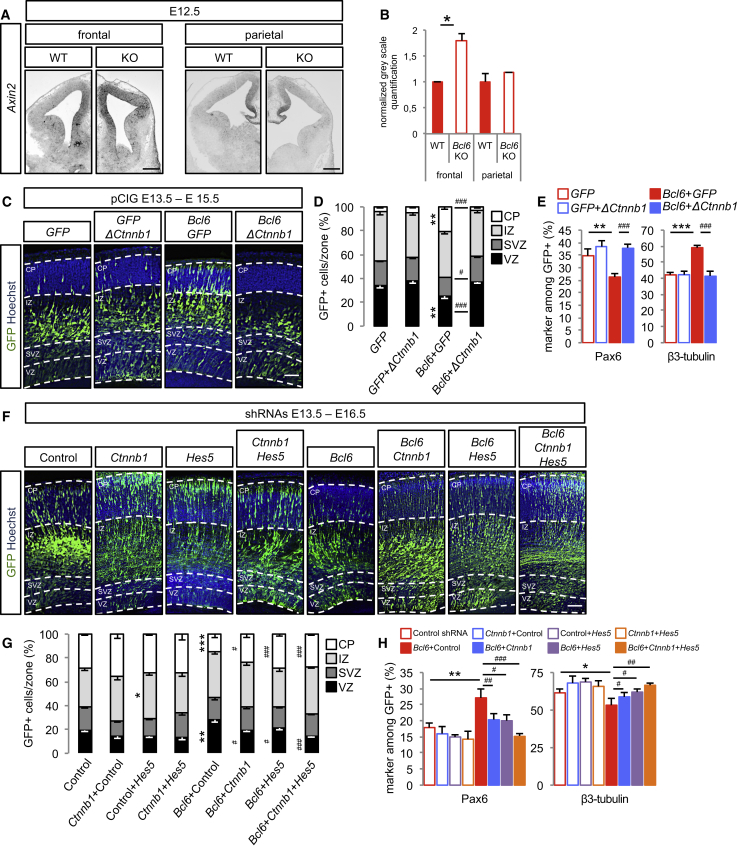
Figure 2.
Bcl6 Alters β-Catenin Signaling In Vivo to Promote Neurogenesis
(A) In situ hybridization of Axin2 Wnt reporter gene on coronal sections of E12.5 wild-type and Bcl6−/− frontal and parietal telencephalon. Scale bars, 500 μm.
(B) Normalized gray scale quantifications of Axin2 levels were performed using ImageJ software. Data are presented as mean + SEM. ∗p < 0.05 using Student’s t test.
(C–E) In utero electroporation of pCIG, pCIG+pCAG-Δ(1-90)Ctnnb1, pCIG-Bcl6+pCIG or pCIG-Bcl6+pCAG-Δ(1-90)Ctnnb1 at E13.5.
(C) Hoechst and GFP immunofluorescence was performed on coronal sections of E15.5 brains. Dashed lines mark the basal and apical margins of the VZ, SVZ, intermediate zone (IZ), and cortical plate (CP). Scale bar, 50 μm.
(D) Histograms show the percentage of GFP+ cells in VZ, SVZ, IZ, and CP. ∗∗p < 0.01 pCIG-Bcl6+pCIG versus pCIG and #p < 0.05, ###p < 0.001 pCIG-Bcl6+pCAG-Δ(1-90)Ctnnb1 versus pCIG-Bcl6+pCIG using two-way ANOVA followed by Tukey post hoc test.
(E) Histograms show the percentage of Pax6+ and Tuj1+ cells among the GFP+ cells. ∗∗p < 0.01, ∗∗∗p < 0.001 pCIG-Bcl6+pCIG versus pCIG+pCIG and ###p < 0.001 pCIG-Bcl6+pCAG-Δ(1-90)Ctnnb1 versus pCIG-Bcl6+pCIG using one-way ANOVA followed by Tukey post hoc test.
Data are presented as mean + SEM of n = 6 control embryos (1,856 cells), n = 10 embryos for stabilized β-catenin gain of function (2,727 cells), n = 9 embryos for Bcl6 gain of function (1,922 cells), and n = 9 embryos for Bcl6 and stabilized β-catenin double gain of function (1,588 cells).
(F–H) In utero electroporation of scramble (control), scramble+Ctnnb1, scramble+Hes5, scramble+Ctnnb1+Hes5, scramble+Bcl6, scramble+Ctnnb1+Bcl6, scramble+Hes5+Bcl6, and Ctnnb1+Hes5+Bcl6 shRNAs at E13.5.
(F) Representative images of Hoechst and GFP immunofluorescence performed on coronal sections of E16.5 brains. Dashed lines mark the basal and apical margins of the VZ, SVZ, IZ, and CP. Scale bar, 50 μm.
(G) Histograms show the percentage of GFP+ cells in VZ, SVZ, IZ, and CP. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus control and #p < 0.05, ###p < 0.001 versus Bcl6+control shRNAs using two-way ANOVA followed by Tukey post hoc test.
(H) Histograms show the percentage of Pax6+ and β3-tubulin+ cells among the GFP+ cells. ∗p < 0.05, ∗∗p < 0.01 versus control and #p < 0.05, ##p < 0.01, ###p < 0.001 versus Bcl6+control shRNAs using one-way ANOVA followed by Tukey post hoc test.
Data are presented as mean + SEM of n = 15 control embryos (4,476 cells), n = 7 embryos for Ctnnb1 shRNA (1,851 cells), n = 13 embryos for Hes5 shRNA (3,893 cells), n = 7 embryos for Ctnnb1+Hes5 shRNA (1,932 cells), n = 6 embryos for Bcl6 shRNA (2,093 cells), n = 9 embryos for Bcl6+Ctnnb1 shRNA (2,642 cells), n = 10 embryos for Bcl6+Hes5 shRNA (2,614 cells), and n = 9 embryos for Bcl6+Ctnnb1+Hes5 shRNA (2,993 cells).
See also Figures S2, S3, and S4.