ABSTRACT
Hypersaline aqueous environments at subzero temperatures are known to be inhabited by microorganisms, yet information on community structure in subzero brines is very limited. Near Utqiaġvik, Alaska, we sampled subzero brines (–6°C, 115–140 ppt) from cryopegs, i.e. unfrozen sediments within permafrost that contain relic (late Pleistocene) seawater brine, as well as nearby sea-ice brines to examine microbial community composition and diversity using 16S rRNA gene amplicon sequencing. We also quantified the communities microscopically and assessed environmental parameters as possible determinants of community structure. The cryopeg brines harbored surprisingly dense bacterial communities (up to 108 cells mL–1) and millimolar levels of dissolved and particulate organic matter, extracellular polysaccharides and ammonia. Community composition and diversity differed between the two brine environments by alpha- and beta-diversity indices, with cryopeg brine communities appearing less diverse and dominated by one strain of the genus Marinobacter, also detected in other cold, hypersaline environments, including sea ice. The higher density and trend toward lower diversity in the cryopeg communities suggest that long-term stability and other features of a subzero brine are more important selective forces than in situ temperature or salinity, even when the latter are extreme.
Keywords: cryopeg, sea ice, Arctic, Marinobacter, subsurface microbiology, bacterial diversity
An ice cave in Arctic permafrost yields subzero brines, likely ancient relic seawater, inhabited by robust microbial communities (dominated by a single strain of Marinobacter) distinctive from modern sea-ice brines.
INTRODUCTION
Microorganisms inhabit nearly all aqueous systems on the Earth, including those with extreme temperatures. In highly saline environments, the freezing point of water can be depressed well below 0°C, enabling liquid brine to exist within the system. Such subzero hypersaline brines serve as microbial habitats in many natural environments (Boetius et al. 2015) including sea ice (Brown and Bowman 2001; Junge et al. 2001; Eronen-Rasimus et al. 2016; Deming and Collins 2017; Yergeau et al. 2017; Rapp et al. 2018), cryopegs in permafrost (Gilichinsky et al. 2003, 2005; Spirina et al. 2017), ice-covered saline lakes (Ward and Priscu 1997; Murray et al. 2012), subglacial aquifers (Mikucki et al. 2009) and saline terrestrial ponds and springs (Meyer et al. 1962; Perrault et al. 2007; Niederberger et al. 2010). These brines are similar with respect to characteristic temperature and salinity but vary in other physical and chemical parameters. Two types of these subzero hypersaline habitats exist within kilometers of each other in the coastal region of the Alaskan Arctic: the brines of landfast sea ice (e.g. Ewert and Deming 2014) and the brines of cryopegs (Yoshikawa et al. 2004; Colangelo-Lillis et al. 2016). They serve as natural systems for studying the influence of extreme environmental conditions on microbial community structure and diversity.
Cryopegs are subsurface features within a stable permafrost matrix, where fluids exist in discrete layers and do not freeze at the subzero temperatures due to high concentrations of salts and other dissolved materials (Gilichinsky et al. 2003, 2005). Cryopegs remain at nearly the same temperature year-round, observed to range from –9 to –11°C in Siberian cryopegs (Gilichinsky et al. 2005) and –6 to –8°C in Alaskan cryopegs (Colangelo-Lillis et al. 2016; K. Yoshikawa, pers. comm.), with the minor variations related to season and distance from the surface. Several cryopegs have been categorized as thalassohaline (having a salt composition indicative of a seawater source) and are proposed to have formed from saturated marine sediments that were exposed to the atmosphere with falling sea level during glaciation periods, becoming desiccated and incorporated into permafrost along with surrounding material over thousands of years (Gilichinsky et al. 2003, 2005; Meyer et al. 2010a,b; Colangelo-Lillis et al. 2016). As thalassohaline permafrost features, cryopegs serve as natural systems for studying the effects of long-term exposure of marine microbial communities to subzero, hypersaline conditions in isolation from other aqueous environments and solar radiation.
The formation of cryopegs is analogous to the hypothesized scenario for recession of an ancient ocean on Mars. During desiccation of an ancient Martian ocean and cooling of the atmosphere, dissolved materials would have concentrated into highly saline brines that saturated surface materials. Hypersaline brines would have been forced underground as the Martian surface became cryotic, forming solid ice complexes and rejecting brines to the subsurface (Knauth and Burt 2002; Ehlmann et al. 2011; McEwen et al. 2011). These cryopeg-like systems could conceivably provide sanctuaries for life on Mars where the surface is too cold and irradiated to support life as known on the Earth (Jakosky et al. 2003; McKay et al. 2013; Westall et al. 2013; Hassler et al. 2014). Recent evidence suggests that subsurface brines of a more contemporary nature may exist on Mars (Ojha et al. 2015). Analog environments like cryopegs can thus serve as models for understanding the adaptations required and community structures that may result from long-term exposure to subzero, hypersaline conditions.
Microorganisms have been cultured from East Siberian and North Alaskan cryopegs (Gilichinsky et al. 2005; Kochkina et al. 2007; Pecheritsyna et al. 2007, 2012; Shcherbakova et al. 2009; Spirina et al. 2017), including isolates of Psychrobacter, Desulfovibrio and Brevibacterium, bacterial genera with cold-adapted and halotolerant species, as well as some eukaryotic yeast and mycelial fungi. Near Utqiaġvik, Alaska, cryopeg brines occur directly beneath a permafrost tunnel that was excavated to study freshwater ice wedge formation in the area (Yoshikawa et al. 2004; Meyer et al. 2010a,b). Paleoclimate and geological research date the sediment directly below the tunnel as deposited > 14 000 years ago and likely earlier (Meyer et al. 2010a,b). Today cryopeg brines exist roughly 8 m below the surface, and directly below a massive ice formation, previously characterized as a complex of ice wedges of meteoric origin [here we use the more general geological term ‘massive ice’, following more recent geological work; G. Iwahana (unpublished data)]. Measurements of 18O (water) from the cryopeg brines and the massive ice indicate separate origins, with the brines having originated most likely from seawater before the ice formed and been isolated for a minimum of 14 000 years (Colangelo-Lillis et al. 2016). Colangelo-Lillis et al. (2016) collected a limited volume of brine from below the tunnel floor in 2009 that was subjected to high-throughput DNA sequencing of the in situ viral community, which indicated low diversity relative to other aquatic environments, while counts of bacteria and virus-like particles (VLPs) suggested an active microbial community that likely included Marinobacter based on detection of Marinobacter genes in the virome. Another thalassohaline cryopeg system has also been identified near Utqiaġvik, at a different location and greater depth (40 m; Yoshikawa et al. 2004). From a brine sample recovered from this system, Spirina et al. (2017) cultured representatives of four bacterial phyla (seven genera), many of which were halotolerant, though no Marinobacter was reported. Information on the structure and diversity of the microbial community using non-cultivation methods is not available for this or any known cryopeg system.
During the formation of sea ice, the more extensively studied of the two subzero brine systems we examined (Bowman 2015), salts and other dissolved or suspended materials in seawater are concentrated into (sub)millimeter-scale brine channels and pockets that exist within the ice matrix (Petrich and Eicken 2017). The resulting brines increase in salinity and concentration of other materials as the brine volume shrinks with decreasing temperature during winter, allowing the brines to remain liquid well below –20°C (Cox and Weeks 1983). The presence of brine in sea ice takes the form of a liquid-filled network inhabited by microbial communities (Junge, Eicken and Deming 2004), where microbial motility along nutritional and salinity gradients may occur and nutrients can be exchanged with the underlying water column (Showalter and Deming 2018). Inhabiting this brine network over the lifetime of the ice comes with the stressors of fluctuating extremes in temperature and salinity. Temperatures in sea ice can range from ∼ –1 to below –20°C annually, with seasonal, diurnal and storm-induced fluctuations, while brine salinities vary as a function of temperature from near-fresh conditions (during melt season) to greater than 220 ppt in winter (Ewert and Deming 2014; Deming and Collins 2017). The nature of sea ice thus exposes the microbial community initially entrained within the ice to extreme physicochemical conditions, including stressful fluctuation events (Collins, Rocap and Deming 2010; Petrich and Eicken 2017). Specific adaptations to survive the combined extremes of subzero temperature and hypersalinity include exopolysaccharide production for cryoprotection (Krembs, Eicken and Deming 2011; Deming and Young 2017) and organic compatible solute uptake and metabolism for osmotolerance (Firth et al. 2016; Torstensson et al. 2019).
The classes Gammaproteobacteria, Alphaproteobacteria and Bacteroidia (including members of the former class Flavobacteriia, reassigned to class Bacteroidia in release 132 of the SILVA database) are abundant in sea ice and include members well adapted to cold and saline conditions (Bowman et al. 1997; Groudieva et al. 2004; Boetius et al. 2015). Adaptations to fluctuating extreme conditions are less well known (Ewert and Deming 2014), nor can we find community structure analyses of brines collected directly from sea ice without first melting the ice and thus altering brine salinity and composition. In 2018, Arctic sea ice covered 14.5 million km2 at its maximum extent and 4.6 million km2 at its minimum extent (Fetterer et al. 2017), demonstrating both the spatial scale of subzero hypersaline environments and their ephemeral and highly variable nature. These fluctuations induce a taxonomic shift, resulting in differently composed communities present in the ice as selective conditions change seasonally (Collins, Rocap and Deming 2010; Bowman et al. 2012; Eronen-Rasimus et al. 2015). As climate warming continues, the timing and extent of these seasonal fluctuations will likely change. Climate warming impacts can also be seen in permafrost, as the rate of permafrost thaw increases (Grosse et al. 2016). Reports of permafrost thaw and brine seepage into ice cellars across the North Slope of Alaska suggest a possible threat to food security for indigenous people of the region (Nyland et al. 2017). Cryopeg brines may also be affected by climate change and contribute to these threats. As the brines warm and migrate through the permafrost, their microbial communities may benefit from new conditions and food sources. Changes in the microbial communities of sea ice, permafrost and cryopegs will be important to monitor to understand the biogeochemical impacts of a warming climate.
In this study, we collected multiple cryopeg brines from below the permafrost tunnel near Utqiaġvik, Alaska, to investigate resident prokaryotic community structure and diversity between borehole locations and between years. We also sampled the massive ice deposit above the cryopeg brines, as well as nearby coastal sea-ice brines, to compare microbial community structure and diversity in cryopeg brines with that in adjacent environmental material and in another marine-derived subzero brine system. All samples were subjected to high-throughput amplicon sequencing of the V3–V4 hypervariable regions of the 16S rRNA gene, along with analyses of selected physicochemical conditions to consider possible selective forces acting in situ on the brine communities. The in situ composition and diversity of microbial communities in cryopeg brines are unknown to date—how such rarely sampled communities are structured in relation to those of surrounding and nearby extreme environments can help to reveal ecological mode and history. The influence of prolonged exposure of microbial communities to extremely low temperatures and high salinities, coupled with geophysical isolation, has not been fully explored. Here, we lay the foundation to explore these open questions, leading the way for future gene-based studies to uncover specific functions and interactions, including with viruses, that allow life to adapt and flourish in subzero hypersaline brines.
MATERIALS AND METHODS
Site descriptions
We collected samples near Utqiaġvik, Alaska, in May 2017 and May 2018 as part of a larger project to investigate gene exchange in subzero hypersaline environments. Cryopeg brines were obtained from the Barrow Permafrost Tunnel, which lies 6 m below the surface at 71.2944 °N, 156.7153 °W. The tunnel was excavated by the United States Army Cold Regions Research and Engineering Laboratory in the early 1960s, entirely within a massive ice formation in the permafrost (Meyer et al. 2010a). Working inside the tunnel at –6°C, we collected cryopeg brines and selected massive ice samples from discrete boreholes that generally penetrated ∼ 2 m below the tunnel floor, through the massive ice into permafrost and unfrozen (brine-saturated) cryopeg sediment (Fig. 1). Samples were obtained from both previously established boreholes (Colangelo-Lillis et al. 2016) and new boreholes, drilled with a cleaned and ethanol-rinsed ice auger or SIPRE corer (as in Colangelo-Lillis et al. 2016) until unfrozen sediment or liquid brine was encountered. This effort was time consuming (taking 4–6 hours to enter the tunnel, drill a borehole that yielded brine and collect a new sample) and challenging, given the low temperature and limited space in the tunnel (sharp ice crystals have been growing inward from the tunnel walls and ceiling since its excavation), but every precaution was taken to keep pristine any cryopeg brine encountered. We installed newly purchased, ethanol-rinsed PVC or ABS pipe in each new borehole, wearing ethanol-rinsed nitrile gloves for these and all proximal operations.
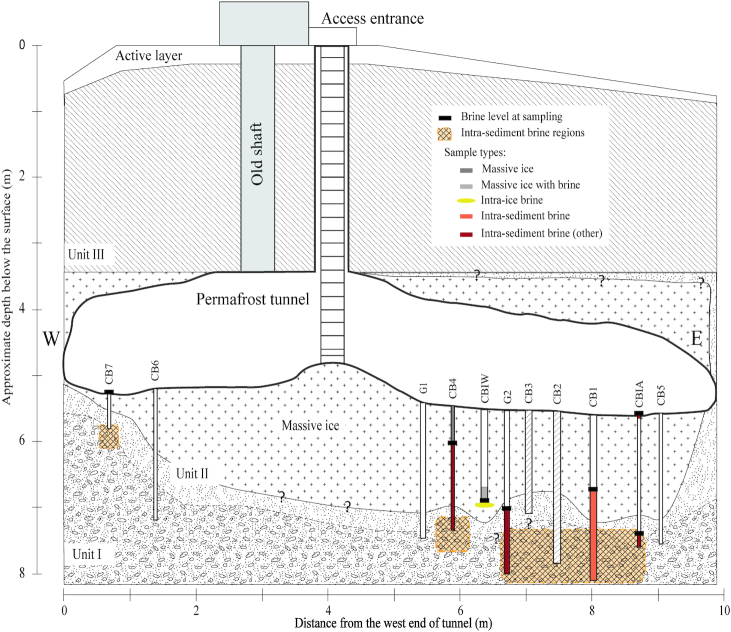
Figure 1.
North-facing cross-sectional diagram of the Barrow Permafrost Tunnel system. The tunnel is entered via the access ladder, and equipment and supplies are lowered through the same space. Cryopeg boreholes CB1, CB2, CB3 and the upper (dry) portion of CBIW were drilled prior to this study; CB2 ad CB3 were sealed and unavailable for this study. The other boreholes were drilled in 2017 and 2018, with pipes installed to preserve the holes and facilitate sampling. Crosshatched sections indicate confirmation of cryopeg sediments (intra-sediment brine regions). Units I–III refer to permafrost regions, following Meyer et al. (2010a). W and E indicate West and East directions, respectively. Question mark symbols ‘?’ indicate unresolved boundary position. See Table 1 for sample details, and Text S1 (Supporting Information) for borehole terminology and drilling details.
Landfast sea-ice sampling sites were located at 71.3730 °N, 156.5047 °W in May 2017 and 71.4730 °N, 156.7294 °W in May 2018 near the Barrow Sea Ice Mass Balance site operated by the University of Alaska Fairbanks (Druckenmiller et al. 2009). Air temperature during sampling in both years varied between –6 and –4°C. Sites were cleared of snow prior to drilling sackholes: snow depth was 16–19 cm in 2017 and 6–10 cm in 2018. Sea-ice brine (SB) was collected by drilling multiple sackholes, 1 m apart, into the sea ice, following Eicken et al. (2009). In May 2017, the sea ice was 117 cm thick, and sackholes were drilled to 75 cm; in May 2018, ice thickness was 110 cm, and sackholes were drilled to 55 cm. Vertical temperature profiles were measured from two sea-ice cores for each sampling year, following Eicken et al. (2009), to estimate sackhole brine temperature at relevant depths (and avoid inserting a thermoprobe into the pristine brine). Upper sea-ice temperatures (above 75 cm in 2017, above 55 cm in 2018) were –4°C in 2017 and –3°C in 2018.
Sample collection and processing
Cryopeg brine was collected using an apparatus composed of hand pump, acid-washed 2-L vacuum flask (initially autoclaved, then acid-washed/ethanol-rinsed between samples) and 1.5-m length of Masterflex and Teflon tubing (Cole-Parmer, Vernon Hills, IL, USA). Prior to the day of cryopeg sampling, multiple lengths of this tubing were acid-washed, rinsed with MilliQ water, rinsed with ethanol, dried to remove excess liquid that might freeze in the tunnel and sealed with sterile parafilm in the laboratory at the Barrow Arctic Research Center (BARC) for transport to the tunnel. Each cryopeg brine was collected from a discrete borehole (named accordingly; Fig. 1) and returned to BARC immediately for storage at –6°C until processing. Massive ice samples were collected by using an ethanol-cleaned spatula to gather ice shavings during ice auger drilling (CB4_IW1_17 and CB4_IW2_17) or as whole ice cores (CBIW_IW3_17 and CBIW_IW7_17) that were placed into sterile Whirl-Pak® bags. Massive ice samples were placed at –20°C upon return to BARC and kept frozen until later processing at the University of Washington. Further details on each borehole and sampling operation are provided in Text S1 (Supporting Information).
Sea-ice brines were collected in both years after allowing brine to drain from the surrounding sea ice into (covered) sackholes over a period of 3–5 hours. Brine was pooled in the field from multiple sackholes to obtain 20-L samples by hand-pumping the brines into an acid-washed cubitainer first rinsed with sample brine. Sea ice from the drained area was not collected. In 2018, we collected four additional (unpooled) 500-mL brine samples (SB1–4_18), each from a separate sackhole, to compare with the pooled sea-ice brine communities.
All brine samples were transported in insulated containers to subzero cold rooms at BARC (set to in situ brine temperatures) and processed as soon as possible after collection (within 2–8 hours). Samples collected from the permafrost tunnel and filtered for DNA extraction were limited in volume: 25–500 mL for cryopeg brine and 12.5–50 mL for melted massive ice. For sea-ice brines, a volume of 250 mL was filtered for each individual sackhole brine sample in 2018, while 2500 mL was filtered for the brine pooled from multiple sackholes each year. These samples were filtered through 0.22 µm filters (either Sterivex, without a prefilter, or 47-mm in-line GTTP Isopore filters with a 1.6-µm Whatman GF/A or a 3.0 µm polycarbonate prefilter [all filters from Merck-Millipore, Burlington, MA]; Table 1) in a BARC cold room at 4°C. The filters were stored immediately at –80°C until shipped (surrounded by –80°C ice packs) to the University of Washington in Seattle for similar storage until further processing.
Table 1.
Description of individual samples in this study by sample subtype.
| Sample subtype | # | Samplea | T(°C)b | S (ppt) | Description | Size fraction (µm)c |
|---|---|---|---|---|---|---|
| Massive ice | 1 | CB4_IW1_17 | –6 | 0 | Ice shavings, upper borehole | > 0.2 |
| 2 | CB4_IW2_17 | –6 | 0 | Ice shavings, lower borehole | > 0.2 | |
| Massive ice with brine | 3 | CBIW_IW3_17 | –6 | 7 | Ice core section, upper borehole | > 0.2 |
| 4 | CBIW_IW7_17 | –6 | 22 | Ice core section, lower borehole | > 0.2 | |
| Intra-ice brine | 5 | CBIW_17 | –6 | 140 | Brine, bottom of borehole | 0.2–1.6 |
| 6 | CBIW_18 | –6 | 121 | Brine, bottom of borehole | > 0.2 | |
| 7 | CBIW_re_18 | –6 | 120 | Recharged brine, 4 days later | > 0.2 | |
| Intra-sediment brine | 8 | CB1_0.2_09 | –6 | 115 | Brine, upper borehole | 0.2–3.0 |
| 9 | CB1_3.0_09 | –6 | 115 | Brine, upper borehole | > 3.0 | |
| 10 | CB1_18 | –6 | 122 | Brine, lower borehole | > 0.2 | |
| Intra-sediment brine (other) | 11 | CBIA_surf_18 | –6 | 120 | Brine, borehole surface seepage | > 0.2 |
| 12 | CBIA_18 | –6 | 112 | Brine, lower borehole, slurried with ice crystals, sulfidic odor | > 0.2 | |
| 13 | G2_18 | –6 | 109 | Brine, lower borehole, slurried with sediment | > 0.2 | |
| 14 | CB4_18 | –6 | 121 | Brine, lower borehole | > 0.2 | |
| Sackhole brine from sea ice | 15 | SB_17 | –4 | 78 | Brine, pooled from sackholes | 0.2–1.6 |
| 16 | SB_0.2_18 | –3 | 75 | Brine, pooled from sackholes | 0.2–3.0 | |
| 17 | SB_3.0_18 | –3 | 75 | Brine, pooled from sackholes | > 3.0 | |
| 18 | SB_18 | –3 | 75 | Brine, pooled from sackholes | > 0.2 | |
| 19 | SB1_18 | –3 | 75 | Brine, sackhole #1 | > 0.2 | |
| 20 | SB2_18 | –3 | 75 | Brine, sackhole #2 | > 0.2 | |
| 21 | SB3_18 | –3 | 75 | Brine, sackhole #3 | > 0.2 | |
| 22 | SB4_18 | –3 | 75 | Brine, sackhole #4 | > 0.2 |
CB indicates cryopeg borehole; G, borehole intended for geology; SB, sackhole brine; last two digits of sample name, sampling year (2009, 2017, 2018). See Text S1 (Supporting Information) for details of borehole terminology and drilling circumstances; Fig. 1 for tunnel locations of massive ice, permafrost, boreholes, and encountered brine and cryopeg sediment.
Temperature in tunnel recorded at tunnel floor; in sea ice, at sackhole depth.
Size fractionation indicates whether the sample was collected directly on a 0.2 µm filter (> 0.2) or first prefiltered with a 3.0 or 1.6 µm filter (0.2–1.6 or 0.2–3.0 µm); > 3.0 indicates the sample material collected on the 3.0 µm prefilter.
Designation of sample types
The samples collected for this study fell into three general categories: the two types of subzero brines, cryopeg brine and sea-ice brine; and the massive ice found above the cryopeg brine. The massive ice and cryopeg brine samples from below the permafrost tunnel were divided further into subtypes (Table 1), distinguished by salinity or locational source, as described below. Sea-ice brine samples were not separated further, as they were not similarly distinguishable. Table 1 lists all of the samples and whether or not the sample was prefiltered. Prefiltration was performed for selected samples, and processed for later metagenomic analyses, to separate eukaryotic and prokaryotic cells to the extent possible. Both filtered and prefiltered samples were included in this study, however, as the prokaryotic 16S rRNA gene was targeted for amplification and amplicons assigned to eukaryotes were removed in silico.
Massive ice
The massive ice samples were grouped by salinity (Table 1). Samples CB4_IW1_17 and CB4_IW2_17 were collected as auger shavings during the drilling of CB4 and had no detectable salinity (as measured on the melted sample); they were grouped as ‘massive ice’. Samples CBIW_IW3_17 and CBIW_IW7_17 were collected as core sections of the massive ice while drilling CBIW. They had detectable brine content (measurable salinity on the melted sample) and were grouped as ‘massive ice with some brine’.
Cryopeg brines
Where multiple samples were available from the same cryopeg borehole (CB), the cryopeg brine samples were grouped by locational source, i.e. ‘intra-ice brine’ encountered in situ within the massive ice and ‘intra-sediment brine’ encountered in contact with sediment. Remaining intra-sediment brines, represented by a single sample per location, were grouped as ‘intra-sediment brine (other)’. Although brine sampling in 2017 was limited (as in 2009; Colangelo-Lillis et al. 2016), collections from new boreholes drilled in 2018 increased the overall spatial coverage of this subsurface brine system. They also allowed consideration of potential changes over time, as intra-sediment brine was collected in 2018 from the same borehole sampled in 2009 [designated CB1 by Colangelo-Lillis et al. (2016)], while intra-ice brine was collected from another borehole [CBIW, originally designated IWC by Colangelo-Lillis et al. (2016)] in successive years, 2017 and 2018. CBIW also yielded brine samples 4 days apart in 2018 (Table 1). A cryopeg brine sample that had been collected and filtered in-line (3.0 µm prefilter, then 0.2 µm filter, with both filters stored at –70°C) in August 2009 by Colangelo-Lillis et al. (2016) was also included in this study. All individual samples with descriptors are listed in Table 1; further details about drilling circumstances and boreholes are provided in Text S1 (Supporting Information).
Enumeration of cells and VLPs
Samples were collected for bacterial and viral enumeration, fixed using 0.2-µm filtered 37% formaldehyde at a final concentration of 2%, and stored at 4°C until returned to the University of Washington. Total cell counts and the percentage of dividing cells were obtained by filtering the fixed samples onto 0.2 µm filters, cross-staining with 4′,6-diamidino-2-phenylindole (DAPI) and acridine orange, and analyzing by epifluorescence microscopy at 1562.5× (12.5× objective × 1.25× optivar × 100× oil immersion lens) on a 1953 Zeiss Universal microscope, following Sherr and Sherr (1983) and Ewert and Deming (2014). Total VLPs were enumerated at The Ohio State University using the wet-mount method described by Cunningham et al. (2016) using SYBR Gold (ThermoFisher Scientific, Waltham, MA). Counts below the detection limit of that method (1 × 106 VLP mL–1) were made at the University of Washington by filtering samples onto 0.02 µm anodisk filters and staining with SYBR Gold, following Noble and Fuhrman (1998).
Quantification of physicochemical conditions, nutrients and organic matter
We measured temperature in the field using a digital thermometer, brine salinity via refractometry and pH using pH paper. Where sample volume allowed, concentrations of particulate organic carbon and nitrogen (POC, PN), particulate and dissolved extracellular polysaccharides (pEPS, dEPS), dissolved organic carbon (DOC), and the nutrients PO4, NO3, NO2 and NH4 were determined. For POC (and PN), samples were collected on pre-combusted (500°C) Whatman GF/F filters (Merck-Millipore, Burlington, MA). Sample filters were fumed with HCL to remove inorganic carbon, and CHN was measured on a Leeman Labs Model CEC440 Elemental Analyzer in the Marine Chemistry Laboratory (MCL) at the University of Washington. DOC was measured on 0.2 µm sample filtrate using a Shimadzu TOC-VCSH DOC analyzer according to standard protocols in the MCL. For EPS measurements we used the phenol-sulfuric acid method following Krembs, Eicken, Deming (2011), with conversion from glucose-equivalents (standards based on glucose concentrations) to carbon-equivalents. Samples for pEPS were collected on 0.4 µm polycarbonate filters, with the filtrate (< 0.4 µm) representing dEPS. Nutrients were analyzed in the MCL using a Technicon AutoAnalyzer II (Knap et al. 1996).
DNA extraction and sequencing
We removed all Sterivex filters from their plastic housings using ethanol-rinsed, flame-sterilized pliers. The Sterivex and all 47-mm filters were cut into smaller pieces using similarly sterilized scalpels and forceps. DNA was extracted from cut filters using the DNeasy PowerSoil Extraction kit (QIAGEN, Hilden, Germany), quantified using Qubit dsDNA HS assay (Invitrogen, Carlsbad, CA) and checked for quality (A260/280 and A260/230) using a NanoDrop One spectrophotometer (ThermoFisher Scientific, Waltham, MA). Aliquots of extracted DNA containing at least 50 ng DNA were sent to GENEWIZ, Inc. (South Plainfield, NJ) for library preparation and sequencing.
The preparation of next generation sequencing libraries and Illumina MiSeq sequencing was conducted by GENEWIZ, Inc. (South Plainfield, NJ), where DNA samples were quantified using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) and DNA quality was checked on a 0.6% agarose gel. The sequencing library was constructed using a MetaVx Library Preparation kit (GENEWIZ, Inc., South Plainfield, NJ). Briefly, 50 ng of DNA were used to generate amplicons that cover the V3 and V4 hypervariable regions of the 16S rRNA gene of Bacteria and Archaea. The V3 and V4 regions were amplified using proprietary primer sets developed at GENEWIZ, Inc. (South Plainfield, NJ), where the forward primers contain the sequence ‘CCTACGGRRBGCASCAGKVRVGAAT’ (similar to 341F primers) and the reverse primers contain the sequence ‘GGACTACNVGGGTWTCTAATCC’ (similar to 806R primers; Huang et al. 2014). Indexed adapters were added to the ends of the amplicons by limited cycle PCR. Sequencing libraries were validated using an Agilent 4200 TapeStation (Agilent Technologies, Palo Alto, CA), and quantified by Qubit and real time PCR (Applied Biosystems, Carlsbad, CA). DNA libraries were multiplexed and loaded on an Illumina MiSeq instrument according to the manufacturer's instructions (Illumina, San Diego, CA). Sequencing was performed using a 2 × 250 paired-end (PE) configuration; image analysis and base calling were conducted by the MiSeq Control Software (MCS) on the MiSeq instrument.
Sequence processing
We received demultiplexed paired-end reads from GENEWIZ, Inc. (South Plainfield, NJ), which have been deposited in the Sequence Read Archive under BioProject accession number PRJNA540708 or individually under accession numbers SRR9003057–SRR9003078. Reads were assembled, quality-filtered and aligned following the MiSeq SOP for mothur v1.40.1 (Schloss et al. 2009; Kozich et al. 2013). To accommodate processing of the V3 and V4 regions of the 16S rRNA gene, we deviated from the SOP as follows: assembled reads shorter than 400 bp and longer than 480 bp were discarded; the SILVA release 132 non-redundant alignment reference provided through the mothur wiki (https://mothur.org/wiki/Silva_reference_files) was trimmed to accommodate the regions sequenced here by using the pcr.seqs command given the start and end positions of our sequences within the alignment; preclustering was set to allow up to four differences; classification of unique sequences was performed using the SILVA taxonomy file; and unique reads assigned to chloroplasts, mitochondria, Eukaryota and unknown domains were discarded. OTU clustering (at 97% similarity) was performed using the VSEARCH distance-based greedy clustering algorithm (Rognes et al. 2016) within mothur (updated here to v1.40.5 to fix an error in the mothur implementation of VSEARCH clustering) rather than the default Opticlust algorithm (Westcott and Schloss 2017) because of its lower memory requirement and higher speed on this size of dataset. We conducted taxonomic classification of OTUs using the SILVA (release 132) reference alignment provided in the mothur wiki. Statistical analyses and figures were generated using the R (version 3.4.4; R Core Team 2018) package Phyloseq (version 1.22.3; McMurdie and Holmes 2013). Singletons were removed from the OTU data before all analyses to avoid including sequencing error. A log of mothur commands used is available in Text S2 (Supporting Information).
Biodiversity and statistical calculations
The alpha diversity metrics calculated include the number of observed OTUs and Chao1 index to measure richness and the Shannon and Inverse Simpson indices to measure diversity. All reads but singletons (OTUs with a single read across samples) were kept to avoid creating statistical miscalculations that can propagate when rarefying reads. OTU abundances were log10(x + 1) transformed for beta-diversity analyses using base R to normalize read counts, stabilize variance and reduce error due to heteroscedasticity (McMurdie and Holmes 2014). Using the R package vegan (version 2.5.3; Oksanen et al. 2018), we performed an analysis of variance (ANOVA) along with a post hoc pairwise significance [Tukey's honestly significant difference (HSD)] test to assess the significance (P ≤ 0.05) of variance in the richness and diversity indices between the three main sample types: massive ice (n = 4), cryopeg brine (n = 10) and sea-ice brine (n = 8). To assess similarity of the communities, Bray–Curtis dissimilarity-based ordinations, based on transformed OTU abundances, were generated using non-metric multidimensional scaling (NMDS) with the ordinate function in Phyloseq set to iterate 1000 times to find the lowest stress ordination possible, along with an analysis of similarity (ANOSIM) using the ANOSIM function in vegan, on samples grouped further into subtypes (Table 1). NMDS ordinations were plotted in a split format showing sample similarities in one panel with OTU contributions, colored by genus, displayed adjacently. A principal component analysis (PCA) was performed in base R using the prcomp function for the subset of brine samples that were accompanied by the full suite of collected environmental data to assess which of those parameters might drive differences between cryopeg and sea-ice brines. Scripts used for biodiversity calculations and PCA are available in Text S2 (Supporting Information).
Minimal entropy decomposition
To assess variations in strain composition, we used the oligotyping program to conduct a minimal entropy decomposition (MED; version 2.0; Eren et al. 2015) analysis on sequences belonging to the genus Marinobacter from samples where they were the most relatively abundant taxa. We processed mothur taxonomy, count and fasta files generated after unique sequence classification to obtain only Marinobacter sequences. First, samples not dominated by Marinobacter were removed from the dataset in mothur using the remove.groups function. Then, the fasta file was screened using the screen.seqs function to select for sequences that were exactly 462-bp long (the median length of all sequences present after the initial sample removal), leaving a total of 529 228 sequences of equal length assigned to the genus Marinobacter. All alignment gaps were removed from the fasta file using the function degap.seqs. The processed mothur files were then used as input in the shell script mothur2oligo.sh, found at https://github.com/DenefLab/MicrobeMiseq/tree/master/mothur2oligo, which allows for sequences from a specific taxon to be extracted and reformatted for use in the MED analysis. Using the properly formatted Marinobacter sequence file, MED analysis was run with default parameters, where sequences that are dissimilar are iteratively split into nodes of similar sequences until the Shannon entropy of each base within all nodes reaches below a threshold of 0.0965.
RESULTS
Environmental conditions
During the May campaigns of both years, the temperature at the tunnel floor was –6°C. The temperature of cryopeg brine in situ was not measured directly to limit disturbance of such rarely accessed fluids; however, based on in situ thermistors deployed through a separate, dedicated borehole (CB3) in the tunnel between December 2010 and March 2012, temperature at cryopeg depth is relatively stable, oscillating between –8 and –6°C (K. Yoshikawa, pers. comm.). Sea-ice temperatures at the sackhole depth and time of sampling were slightly warmer, –4°C in 2017 and –3°C in 2018, though the ice (and its inhabitants) would have experienced much lower temperatures prior to May (e.g. upper sea ice temperatures near Utqiaġvik can fall below –20°C during winter; Ewert and Deming 2014, their Figure 1a; Petrich and Eicken 2017, their Figure 1.10). The sea-ice sackhole brines of this study had salinities of 78 ppt in 2017 and 75 ppt in 2018, as generally expected based on ice temperature (Cox and Weeks 1983), while the colder cryopeg brines had higher salinities of 140 ppt in 2017 and 112–122 ppt in 2018.
Additional environmental parameters measured on the subset of brine samples with sufficient volume showed that cryopeg brines were extremely rich in organic compounds, with concentrations of POC, DOC, pEPS and dEPS (in C equivalents) measured at the millimolar level, 2–4 orders of magnitude higher than the micromolar concentrations measured in sea-ice brine (precise values provided in Table 2). Most of the organic carbon in both types of brine was in the dissolved form: 30–102 mM DOC and 9–20 mM dEPS in cryopeg brines compared to 0.20–0.45 mM DOC and 0–0.20 mM dEPS in sea-ice brines. The particulate organic component was overall smaller but still much higher in the cryopeg brines: 2–12 mM POC and 0.09–14 mM pEPS in cryopeg brines compared to 0.020–0.022 mM POC and 0.001–0.002 mM pEPS in sea-ice brines. Cryopeg brines also contained much higher levels of NO2 and NH4+ relative to sea-ice brines, while NO3– and PO43– concentrations were similarly low in the two environments (Table 2). C:N ratios were generally low (6.7–7.4 in cryopeg and 8.2–10.4 in sea-ice brine), and pH values approached neutral (6.6 in cryopeg and 7.2 in sea-ice brine).
Table 2.
Concentrations of organic matter and nutrients for the subset of samples with sufficient volume.
| # | Sample | POC (µM) | DOC (μM) | pEPS (µM C) | dEPS (µM C) | PO4 (μM) | NO3 (μM) | NO2 (μM) | NH4 (μM) |
|---|---|---|---|---|---|---|---|---|---|
| 5 | CBIW_17 | 1.98 × 103 | 3.00 × 104 | 1.44 × 104 | 8.55 × 103 | 1.94 | 5.56 | 2.03 | 1.75 × 103 |
| 6 | CBIW_18 | 2.57 × 103 | 8.22 × 104 | 1.14 × 102 | 1.25 × 104 | 0.68 | 0.82 | 0.89 | 1.17 × 103 |
| 10 | CB1_18 | 1.24 × 104 | 1.02 × 105 | 1.63 × 103 | 1.94 × 104 | 1.22 | b.d.a | 16.2 | 3.35 × 103 |
| 14 | CB4_18 | 4.15 × 103 | 8.50 × 104 | 8.91 × 101 | 1.98 × 104 | 0.60 | 13.6 | 2.96 | 4.52 × 103 |
| 15 | SB_17 | 2.03 × 101 | 4.46 × 102 | 1.90 × 100 | 2.61 × 102 | 1.80 | 0.15 | 0.02 | 5.50 × 10–1 |
| 18 | SB_18 | 2.25 × 101 | 2.00 × 102 | 9.67 × 10–1 | b.d.a | 1.60 | 3.63 | 0.11 | 3.42 × 100 |
Below detection.
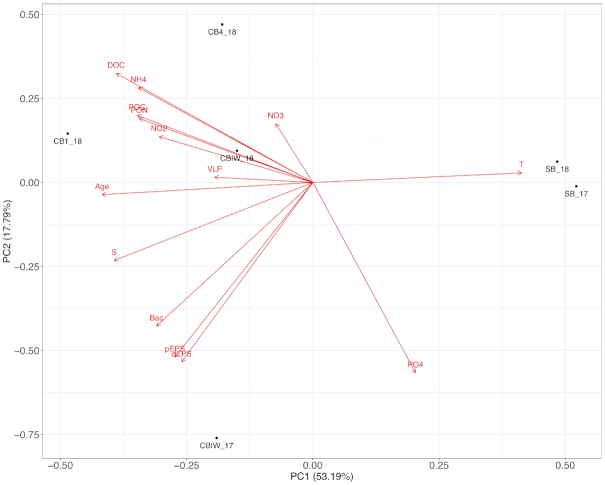
The PCA, based on physical and biogeochemical data (Table 2), resulted in a clear separation of cryopeg and sea-ice brines along PC1, which explained 53.2% of the variation between sample types (Fig. 2). Each of the factors enriched in cryopeg brines relative to sea-ice brines contributed nearly equally to explaining the differences between brine types. No single driving factor among those measured could be identified, though VLP abundance and concentrations of nitrate and nitrite appeared less influential than other factors.
Figure 2.
PCA of cryopeg and sea-ice brine samples that had a full suite of measured environmental factors. Vectors indicate contribution of each variable to each principal component.
DNA-containing cell and VLP abundance
The abundance of DAPI-stained cells in cryopeg brine was remarkably high (Table 3), ranging from 5.70 × 106 to 1.70 × 108 mL–1, compared to 1.11–2.22 × 105 mL–1 in sea-ice brine. In samples evaluated for the number of dividing cells (Table 3), 1.1–5.5% of the total were noted as dividing in cryopeg brine samples collected from boreholes (none were observed in CBIA_surf_18 from the tunnel floor), compared to 4.7% in sea-ice sackhole brine. A striking exception was cryopeg brine sample CB4_18, where 29% or 3.31 × 106 of the 1.14 × 107 cells mL–1 were observed as dividing. Although cellular abundance in this sample fell on the lower end of the range for cryopeg brines, the absolute number of dividing cells was twice that found in other cryopeg brines (1.30–1.82 × 106) and orders of magnitude higher than observed in sea-ice brine (5.22 × 103). VLP concentrations ranged from 3.54 × 106 to 3.89 × 108 mL–1 in cryopeg brine compared to 1.53–6.16 × 105 mL–1 in sea-ice brine (Table 3). Ratios of VLP to cells in cryopeg brines collected in 2017 and 2018 ranged from 0.31 to 3.3 [below the ratio of 10 reported by Colangelo-Lillis et al. (2016), for cryopeg brine sampled in 2009] but generally comparable to ratios in sea-ice brine, which ranged from 0.69 to 3.00 (Table 3). The lowest ratio was observed for cryopeg brine sample CB4_18.
Table 3.
Bacterial and viral abundances, % dividing cells and virus-to-bacteria ratio (VBR).
| # | Sample | Cells (mL–1) | Dividing cells (%) | VLP (mL–1) | VBR |
|---|---|---|---|---|---|
| 1 | CB4_IW1_17 | 4.04 × 105 | n.d.a | n.d. | n.d. |
| 2 | CB4_IW2_17 | 4.08 × 105 | n.d. | n.d. | n.d. |
| 4 | CBIW_IW7_17 | 1.40 × 107 | n.d. | n.d. | n.d. |
| 5 | CBIW_17 | 1.39 × 108 | n.d. | 1.22 × 108 | 0.9 |
| 6 | CBIW_18 | 1.22 × 108 | 1.3 | 3.52 × 108 | 2.9 |
| 7 | CBIW_re_18 | 1.18 × 108 | 1.1 | 3.89 × 108 | 3.3 |
| 8 | CB1_0.2_09b | 5.70 × 106 | 5.5c | 5.70 × 107 | 10 |
| 10 | CB1_18 | 9.57 × 107 | 1.9 | 1.24 × 108 | 1.3 |
| 11 | CBIA_surf_18 | 1.70 × 108 | 0.0 | n.d. | n.d. |
| 12 | CBIA_18 | 7.31 × 107 | 2.5 | 5.50 × 107 | 0.8 |
| 14 | CB4_18 | 1.14 × 107 | 29 | 3.54 × 106 | 0.3 |
| 15 | SB_17 | 2.22 × 105 | n.d. | 1.53 × 105,d | 0.7 |
| 18 | SB_18 | 1.11 × 105 | 4.7 | 6.16 × 105,d | 5.5 |
Subzero brine community structure
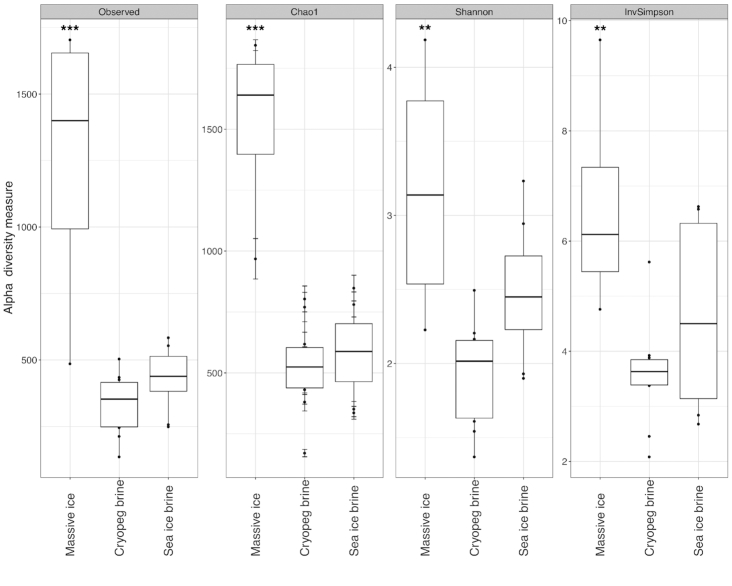
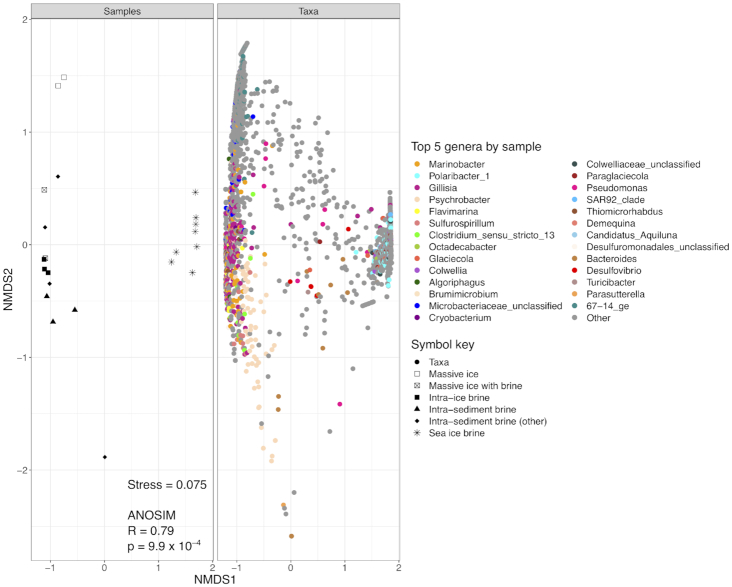
After quality control, sequence processing and removal of singletons, a total of 1 789 043 unique reads across the 22 sequenced samples could be assigned to the domains Bacteria and Archaea. Read totals for individual samples ranged from 29 543 to 143 814 (Table S1, Supporting Information). Rarefaction curves of OTUs versus sampling depth appear nearly saturated for all samples (Fig. S1, Supporting Information). The sampling blank (MilliQ rinse water from between-sample cleaning procedures for the cryopeg sampling tubing) contained a low number of reads (Fig. S1, Supporting Information). As these reads primarily matched the most abundant OTUs from the Marinobacter-dominated brine samples, with some exogenous members present exclusively in the blank (MilliQ source water was virtually devoid of cells by epifluorescence microscopy), we attribute them to low-abundance cell retention in the sample tubing, insufficient to influence the high-density brine samples. Samples were grouped according to sample type, massive ice (n = 4); cryopeg brine (n = 10); and sea-ice brine (n = 8), for richness and diversity analyses to compare between environments (Fig. 3). Average Shannon diversity (H′) values for the massive ice, cryopeg brine and sea-ice brine samples were 3.17, 1.92 and 2.49, respectively. ANOVA of all measures showed significant differences between sample types (P < 0.0001 for richness indices and P < 0.01 for diversity indices). Post hoc Tukey's HSD calculations were generated from the ANOVA for each measure to allow pairwise sample comparisons. Communities in massive ice samples (much lower in bacterial abundance) were significantly richer (P < 0.001, Observed/Chao1 index) and more diverse (P < 0.01, Shannon/Inverse Simpson index) than those in either cryopeg or sea-ice brines (Table S1, Supporting Information). Sea-ice brine communities appeared more diverse than those in cryopeg brines, but without statistical significance (P = 0.079, Shannon diversity index; P = 0.294, Inverse Simpson index). Cryopeg brines appeared less rich than sea-ice brines, but without statistical significance in number of observed OTUs (P = 0.679) or Chao1 richness index (P = 0.851). NMDS ordination (based on Bray–Curtis dissimilarity) of the samples by log10(x+1)-transformed OTU composition (Fig. 4) had a stress value of 0.075 and displayed a clear separation of cryopeg brine and massive ice samples from sea-ice brine samples, as also supported statistically by ANOSIM (R = 0.79, P = 9.9 × 10–4).
Figure 3.
Indices of microbial community richness (left panels) and diversity (right panels) for the three main sample types: massive ice, cryopeg brine and sea-ice brine. Massive ice samples (n = 4) differ significantly from cryopeg brines (n = 10) by every measure (** indicates P < 0.01 and *** indicates P < 0.001). Indices for sea-ice brines (n = 8) tend to be higher than cryopeg brines, also by every measure, but not significantly (e.g. P = 0.0796 for Shannon diversity index).
Figure 4.
Split NMDS ordination, using Bray–Curtis dissimilarities, showing relatedness of each sample and OTU contributions (colored by genus) to sample ordination. Left panel: black and white symbols indicate sample subtype; right panel: colors indicate taxonomy.
Taxonomic classifications
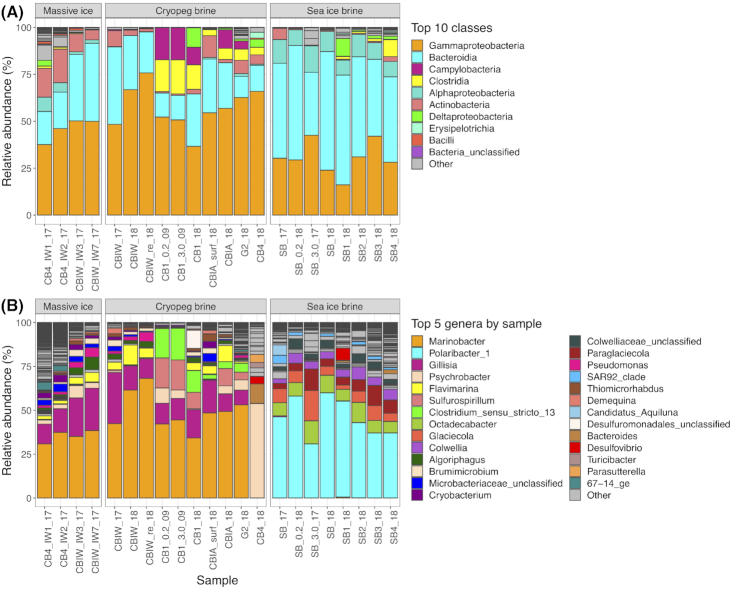
Across all samples, the classes Gammaproteobacteria and Bacteroidia dominated the microbial communities by relative abundance (Fig. 5A). Cryopeg brines were numerically dominated by Gammaproteobacteria (average relative abundance of 57%) followed by Bacteroidia (average relative abundance of 22%). Sea-ice brines were dominated by Bacteroidia (average relative abundance of 51%) followed by Gammaproteobacteria (average relative abundance of 30%). Archaea were rare, with all reads assigned to the domain making up only 0.094% of total reads across all samples. Only 22 OTUs (0.5%) were assigned to Archaea out of 3833 OTUs across all samples. The most abundant archaeal OTU (83% of all archaeal reads) belongs to the class Methanomicrobia genus Methanosaeata. Of the reads in this OTU, 89% came from a single massive ice sample (CB4_IW1_17). The class Methanomicrobia made up 10 of the 22 Archaeal OTUs, found primarily in massive ice samples; the class Nitrososphaeria made up 8 of the 22 Archaeal OTUs, found primarily in sea-ice brines. Few archaeal OTUs were identified in cryopeg brines, e.g. one in CB4_18 (0.063% of reads) and another in CBIW_18 (0.011% of reads), while others were even less abundant. Archaea were not considered further. No cyanobacterial reads, not attributed to chloroplasts, were detected in the sea-ice samples.
Figure 5.
Relative abundance plots of taxa in each sample, grouped by the three main sample types. (A) The top 10 classes across all samples are color coded; lower abundance classes are in gray. (B) The top five genera from each sample are color-coded and shown in each sample where they occur; lower abundance genera are in gray. Sample designations are further described in Table 1.
At the genus level, the differences in taxonomic composition increase between massive ice, cryopeg brines and sea-ice brines (Fig. 5B). All but one of the cryopeg brines were dominated by Marinobacter and Gillisia with average relative abundances of 49 and 15%, respectively.
The exception was the cryopeg brine community from CB4, which was dominated by Psychrobacter at 54% and Bacteroides at 11% (and also anomalous for its high percentage of dividing cells and some environmental properties). The general relative abundances of the cryopeg brine community members were validated by comparison to a shotgun metagenome of a 2017 sample from CBIW that depicts similar abundance values for the major taxa (Text S3 and Figure S2, Supporting Information). Massive ice samples, collected from boreholes during drilling to access cryopegs, were also dominated by Marinobacter and Gillisia with average relative abundances of 35 and 18%, respectively. A small number of reads (27–243) of the main OTU of Marinobacter from the cryopeg brines was also found in each of the sea-ice brine samples. In contrast, sea-ice brine communities were dominated by Polaribacter and Octadecabacter with average relative abundances of 46 and 8%, respectively (Fig. 5B). Although some minor differences were observed between individual sackhole brine communities that were spatially distinct, and pooling the brines appeared to favor the dominant genus (by de-emphasizing the more diverse rare members), the sea-ice brine communities were largely similar in composition.
Given the high relative abundances of Marinobacter in all but one of the cryopeg brine and massive ice samples at both genus and OTU levels, a MED analysis was performed to assess whether strain variation occurs between these samples. Approximately, 80% of Marinobacter sequences could be attributed to a single representative oligotype. BLAST analysis of this sequence against the NCBI 16S rRNA database showed 99.6% similarity with Marinobacter psychrophilus strain i20041 cultured from sea ice in the Canadian Basin (Zhang et al. 2008).
DISCUSSION
The cryopeg brine system underlying the Barrow Permafrost Tunnel provides a unique opportunity to consider microbial adaptation to subzero hypersaline conditions in a geophysically stable and isolated environment. Our efforts to characterize the cryopeg brines and their resident microbial communities and contrast them with the more variable and less isolated environment of sea-ice brines represent a first step in this direction. Based on radiocarbon dating, the cryopeg system we examined has been isolated for at least 14 000 years (Colangelo-Lillis et al. 2016) and, based on local stratigraphy, possibly from the last major transgression in 123 ka (Eisner et al. 2005). Cryopeg brines may thus share the selective pressures of subzero temperature and hypersalinity with contemporary sea-ice brines, but they diverge widely in age, physical environmental interactions, and the magnitude of seasonal fluctuations in temperature and salinity (Ewert and Deming 2014; K. Yoshikawa, pers. comm.).
They also diverge in organic content, as the cryopeg brines of this study were highly enriched in dissolved and particulate organic carbon and extracellular polysaccharides relative to sea-ice brines (millimolar versus micromolar levels; Table 2). The DOC levels in cryopeg brine (range of 30–102 mM) were also several orders of magnitude greater than those reported for other subzero brines from Blood Falls, Antarctica (80–450 µM DOC; Mikucki and Priscu 2007). Although more detailed chemical analyses are needed, much of the organic carbon in our cryopeg brine samples was in the form of dissolved carbohydrate (e.g. 9–20 mM dEPS), suggesting ample labile or semi-labile carbon to drive microbial heterotrophy. Millimolar levels of ammonia (1.2–4.5 mM NH4) in the brines and low C:N ratios (6.7–7.4) are also consistent with the remineralization of labile carbon by marine bacteria. The high concentrations of total and dividing cells and their viruses, and the organic-rich conditions of the cryopeg brines presented in this study, show that these subsurface thalassohaline environments can support remarkable microbial communities, despite continuously extreme temperature and salinity. The cell densities measured, which far exceeded those in the sampled sea-ice brines, resemble values for late-logarithmic growth phase in a nutrient-replete bacterial culture, suggesting that the cryopeg brines are near carrying capacity but that the organisms are not under growth-inhibiting nutrient limitation (Eiler et al. 2003). The in situ microbial communities in these subsurface brines thus appear to be well-adapted to the conditions of their subzero hypersaline habitat conventionally considered to be stressful.
Some evidence exists to suggest that cryopeg brines are anoxic environments (Gilichinsky et al. 2003; Colangelo-Lillis et al. 2016), which presented a sampling challenge in our study. We had no means to measure oxygen concentration in situ without risking contamination of the brines, nor were we able to prevent oxygen exposure during sampling despite best efforts; however, we collected and processed samples as rapidly as possible (2–8 hours from collection) to prevent alteration of the brine communities before filtration and freezing. Had oxygen exposure stimulated members of the community, our strict maintenance of in situ conditions of temperature and hypersalinity during the collection–processing period would have precluded significant growth. Cultured Marinobacter spp. have not been studied under cryopeg brine conditions of both subzero temperature and hypersalinity, but at –4.5°C and seawater salinity, Chua et al. (2018) estimated a doubling time of 9.9 days for Marinobacter gelidimuriae, a close relative of the dominant Marinobacter identified here. Using conditions very close to our cryopeg brines, however, Mykytczuk et al. (2013) reported much longer doubling times of 28 days at –5°C and 180 ppt (NaCl) and ∼ 144 days at –10°C and 180 ppt (NaCl) for their low-temperature, record-holding permafrost isolate, Planococcus halocryophilus OR1. We thus suggest that the taxonomic abundances and diversity indices presented here represent the best possible insight to the in situ community structure of these rarely sampled cryopeg brines.
The alpha-diversity indices of this study indicate that these dense cryopeg brine communities tend to be less diverse than those in sea-ice brines, as well as significantly less diverse than in the overlying massive ice. Higher diversity (H′ = 2.49) in sea-ice brines may be attributed to the broad fluctuations in environmental conditions that select for both phenotypic plasticity and different taxa during different seasons (Collins, Rocap and Deming 2010; Ewert and Deming 2014; Deming and Collins 2017). Lower diversity (H′ = 1.92) in cryopeg brines may be because the magnitude of fluctuations experienced is much lower, requiring less plasticity and favoring fewer taxa. Over the long period of geological isolation of cryopeg brines from the ocean, the extreme conditions may have forced taxa typically competitive under warmer, lower salinity conditions to become rare members of the community or be lost from it, lowering the overall diversity. An alternative (non-exclusive) explanation for lower diversity in cryopeg brines is that the measured high concentrations of organic substrates have allowed copiotrophic microbial taxa to outcompete others of oligotrophic mode. None of these explanations preclude the possibility that even relatively minor seasonal or annual fluctuations in environmental conditions over long periods of time may have played a role in structuring the microbial community. The observation of highest diversity (H′ = 3.17) in the low-abundance microbial assemblage in massive ice likely results from the depositional nature of this feature at the time of its formation. A very limited liquid phase and thus low biological activity in the massive ice would favor preservation of deposited cells, similar to what has been observed in other permafrost features (Jansson and Tas 2014). The presence of dominant cryopeg brine taxa in the massive ice samples may result from thermal migration of cryopeg fluids or interactions at the brine/ice boundary where partial melting and refreezing occurs as temperatures oscillate seasonally between –8 and –6°C. Massive-ice refreezing in cold seasons may entrain cryopeg brine taxa, causing major taxa to appear similar between these sample types while minor taxa differ.
Beta diversity analyses suggest that communities in sea-ice brines have distinct structures that differ from those present in the subterranean system of massive ice and cryopeg brines. Within that system, communities in massive ice samples with undetectable bulk ice salinity (measured by refractometry) separate farther from cryopeg brines than do those in massive ice samples that clearly contained some brine, implying some physical connectivity between massive ice and cryopeg brine. In line with this implication, communities in intra-ice brine (from CBIW) exhibit higher similarity to massive ice communities than to intra-sediment brine communities (from CB1; Fig. 4). Other samples of cryopeg brines (single samples per location) with confirmed or suspected connectivity with massive ice, i.e. by upward migration through the ice between sampling years (CBIA) or by drilling circumstances that may have partially mixed ice, sediment and brine (G2), also appear similar to the massive ice samples that contained some brine. The community in intra-sediment brine CB4_18, observed to be anomalous in other biological and chemical characteristics (Tables 2 and 3), appears unique in the NMDS plot relative to all other brine samples. When CB4 was drilled to a depth of 187 cm in 2017 (and left covered), no brine was encountered, but by 2018 brine had infiltrated into the void (and up the borehole to within 29 cm of the tunnel floor) created in the previous year (Text S1, Supporting Information). The history and location of this brine (the most westerly of other intra-sediment brines; Fig. 1) thus differ from the other cryopeg brines, potentially explaining the separation observed.
The common occurrence and high abundance of the classes Gammaproteobacteria and Bacteroidia in the landfast sea ice and cryopeg brines of this study, as well in other studies of Arctic and Antarctic sea ice (Brown and Bowman 2001; Bowman et al. 2012; Yergeau et al. 2017; Rapp et al. 2018), Baltic sea ice (Eronen-Rasimus et al. 2015), Arctic terrestrial springs (Niederberger et al. 2010), ice-covered saline lakes and aquifers (Ward and Priscu 1997; Mikucki and Priscu 2007; Murray et al. 2012) and other cryopeg systems in Siberia and Alaska (Gilichinsky et al. 2005; Pecheritsyna et al. 2007; Spirina et al. 2017), highlight the consistent and dominant occupation of subzero hypersaline niches by these classes. Genus-level differences between the brines studied here demonstrate the effects of local selective pressures on the specific adaptive capabilities found within each class.
The cryopeg brine communities of this study (with one exception) were dominated by members of the genus Marinobacter, a genus that appears to be cosmopolitan in cold, saline aqueous environments. For example, Marinobacter is abundant in a cold saline spring on Axel Heiberg Island in the Canadian High Arctic (Niederberger et al. 2010), a system that shares physicochemical characteristics with cryopeg brines, including temperature, hypersalinity, residence within or below permafrost, and isolation from light and the atmosphere (Perreault et al. 2007). Marinobacter is also abundant in Blood Falls, the saline subglacial outflow from Taylor Glacier, Antarctica (Mikucki and Priscu 2007; Campen et al. 2019). Cultivation studies have yielded isolates of Marinobacter from both the cold spring system on Axel Heiberg Island (Niederberger et al. 2010) and Blood Falls (Chua et al. 2018), as well as from a variety of other saline (if warmer) environments, including coastal sediments, oil-polluted seawater, low-temperature hydrothermal sediments and the phycospheres of diatoms and dinoflagellates (Handley and Lloyd 2013). Marinobacter isolates show metabolisms for utilizing a range of organic compounds, from simple amino acids and glucose to complex hydrocarbons, as well as for nitrate reduction, chemolithoautotrophic Fe(II) oxidation and other aerobic and anaerobic capabilities (Edwards et al. 2003; Kaye et al. 2011; Handley and Lloyd 2013). This wide variety of metabolic functions, along with the general traits of psychrotolerance, halotolerance, phenotypic plasticity, flexible heterotrophic metabolism and an r-strategist, copiotrophic lifestyle, make the Marinobacter genus highly successful across a variety of saline environments.
Two psychrophilic strains of Marinobacter have been isolated: M. psychrophilus strain i20041 from arctic sea ice (Zhang et al. 2008) and M. gelidimuriae from Blood Falls in Antarctica (Chua et al. 2018). Marinobacter gelidimuriae shares high 16S rRNA gene sequence similarity (97.8%) with M. psychrophilus strain i20041 (Chua et al. 2018), as does the one Marinobacter strain that dominates in our cryopeg brine samples (99.6%). If future work yields a cryopeg brine isolate with similar physiological characteristics, then together these organisms may represent a distinct clade of Marinobacter uniquely adapted to subzero, hypersaline brine environments. In the one cryopeg brine sample in our study where Marinobacter did not dominate (CB4_18), the genus was nevertheless present. Given the lower total bacterial density, higher percentage of dividing cells, lowest VBR and other anomalies of this sample, we speculate that its current Psychrobacter-dominated community was in a different stage of development, relative to those spatially distant from it (Fig. 1), and that Marinobacter may yet become dominant, a testable hypothesis with future sampling. The similarity in Marinobacter-dominated communities in other cryopeg locations over time, from 2009 to 2018 (Fig. 5B), lends support to this hypothesis.
The abundance and dominance of Marinobacter across the sampled cryopeg brines raises questions not only about the adaptability of this genus but also the potential interconnectivity of brine pockets within this subterranean massive ice-cryopeg system. Although individual brines varied in their chemical characteristics and composition of low-abundance members of the community, MED analysis indicates that a single Marinobacter strain dominated the samples. Several possible scenarios may explain this dominance across seemingly isolated borehole samples. Partial freeze–thaw cycles (between –8 and –6°C) at the brine/ice boundaries in a cryopeg system may separate the brine into distinct pools in colder seasons but allow subvolumes to connect in warmer seasons for repeat periods of homogenization. Alternatively, the sampled brines may have been physically isolated in situ yet contained communities that shared a common highly-adapted strain due to a common successional history driven by the same temperature and salinity conditions that favor this organism. The high level of relatedness of this Marinobacter strain to the contemporary sea-ice isolate, M. psychrophilus, suggests a selection for similar phenotypic traits important to competing successfully in both cryopeg and sea-ice brines, despite isolation of the cryopeg habitat over recent geologic time. The potential for phenotypic plasticity mediated by viral processes under the relevant in situ conditions remains an open question.
Subzero hypersaline aqueous environments are widespread at high latitudes on the Earth and long studied in sea ice given visible microbial colonization of its brine network. Despite the much smaller number of available studies for cryopegs, these systems are thought to be widespread in coastal permafrost settings (Yoshikawa et al. 2004; Gilichinsky et al. 2005). Local observations suggest that cryopeg brines may be seeping into traditional deep ice cellars, posing a possible threat to indigenous food security (Nyland et al. 2017). As the Earth's warming atmosphere and oceans continue to reduce the extent of polar sea ice and permafrost, microbial communities in subzero brines may be altered in ways that are difficult to predict yet may have strong influences not only on their immediate surroundings, but also on general ecosystem functions in the future. Uncovering microbial community composition and diversity in these extreme environments is thus urgent, and is now possible through interdisciplinary collaboration, modern sampling and DNA sequencing techniques, as exemplified here. Metagenomic, -viromic and -transcriptomic studies (ongoing from this study) will help to reveal details of metabolic function and adaptive mechanisms. Information on these systems may be valuable not only to understanding ecological dynamics during a changing climate on the Earth, but also in guiding considerations of potentially habitable environments beyond the Earth, including subsurface brine features that have been identified on Mars (Ojha et al. 2015) and icy moons, such as Europa and Enceladus (McCord et al. 2002; Zolotov 2007). The ecological patterns observed in terrestrial analog environments like cryopegs can inform expectations and features to anticipate when searching for life in similarly extreme environments throughout the Solar System.
CONCLUSIONS
The subzero hypersaline cryopeg and sea-ice brines sampled near Utqiaġvik, Alaska, harbored sizeable microbial communities, including dividing cells, despite being characterized by conditions considered biologically stressful. The cryopeg brines, isolated in a subsurface environment for at least 14 000 years, hosted particularly dense microbial assemblages. The lower diversity of the microbial communities in the cryopeg brines reflects the dominance of highly adapted bacterial members, particularly of the genus Marinobacter, that have thrived at the very low temperatures and hypersaline conditions of these brines. The more diverse sea-ice microbial communities, containing a canonical bacterial membership, are indicative of the seasonally fluctuating and ephemeral nature of this environment. Both brine environments contained highly abundant members from the same taxonomic classes but differed markedly at the genus level. We suggest that shared extreme temperature and salinity are sufficient to explain selection at the class level, but that genus-level distinctions are driven by other environmental parameters, including the magnitude and frequency of physical fluctuations experienced over time and the sources and levels of carbon and nitrogen, all of which remain to be fully explored. Ongoing research seeks to interrogate the function of these microbial ecosystems and the adaptive mechanisms used for exchanging genetic material when rapid adaptation is advantageous, in order to further the understanding of how life adapts to and thrives under extreme conditions.
Supplementary Material
ACKNOWLEDGEMENTS
Kenji Yoshikawa and Jerry Brown kindly supplied information and unpublished data regarding the Barrow Permafrost Tunnel and local geology. We thank Zhiping Zhong, in the laboratory of Matt Sullivan, for providing total VLP counts for this study along with other members of the larger team for support in the field: Max Showalter, Anders Torstensson, Hannah Dawson and Jodi Young. The Ukpeagvik Iñupiat Corporation science team provided essential logistical support and helped with access to the tunnel. Metagenome sequencing was kindly performed by Rachel Mackelprang and Christopher Chabot at California State University Northridge.
FUNDING
This work was supported by the Gordon and Betty Moore Foundation (grant number GBMF5488).
Conflicts of interest. There are no conflicts of interest to declare.
REFERENCES
- Boetius A, Anesio AM, Deming JW et al.. Microbial ecology of the cryosphere: sea ice and glacial habitats. Nat Rev Microbiol. 2015;13:677–90. [DOI] [PubMed] [Google Scholar]
- Bowman JP, McCammon SA, Brown M V et al.. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol. 1997;63:3068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JS, Rasmussen S, Blom N et al.. Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J. 2012;6:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JS. The relationship between sea ice bacterial community structure and biogeochemistry: a synthesis of current knowledge and known unknowns. Elem Sci Anth. 2015;3:000072. [Google Scholar]
- Brown MV, Bowman JP. A molecular phylogenetic survey of sea-ice microbial communities (SIMCO). FEMS Microbiol Ecol. 2001;35:267–75. [DOI] [PubMed] [Google Scholar]
- Campen R, Kowalski J, Lyons WB et al.. Microbial diversity of an Antarctic groundwater community and high resolution, replicate sampling inform hydrological connectivity in a polar desert. Environ Microbiol. 2019;21:2290–306. [DOI] [PubMed] [Google Scholar]
- Chua MJ, Campen RL, Wahl L et al.. Genomic and physiological characterization and description of Marinobacter gelidimuriae sp. nov., a psychrophilic, moderate halophile from Blood Falls, an Antarctic subglacial brine. FEMS Microbiol Ecol. 2018;94:1–15. [DOI] [PubMed] [Google Scholar]
- Colangelo-Lillis J, Eicken H, Carpenter SD et al.. Evidence for marine origin and microbial-viral habitability of sub-zero hypersaline aqueous inclusions within permafrost near Barrow, Alaska. FEMS Microbiol Ecol. 2016;92:1–15. [DOI] [PubMed] [Google Scholar]
- Collins RE, Rocap G, Deming JW. Persistence of bacterial and archaeal communities in sea ice through an Arctic winter. Environ Microbiol. 2010;12:1828–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GFN, Weeks WF. Equations for determining the gas and brine volumes in sea-ice samples. J Glaciol. 1983;29:306–16. [Google Scholar]
- Cunningham BR, Brum JR, Schwenck SM et al.. An inexpensive, accurate, and precise wet-mount method for enumerating aquatic viruses. Appl Environ Microbiol. 2015;81:2995-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming JW, Collins RE. Sea ice as a habitat for bacteria, archaea and viruses. In: Thomas DN.(ed). Sea Ice – An Introduction to its Physics, Chemistry, Biology and Geology. 3rd edn, West Sussex, UK: John Wiley and Sons, Ltd, 2017, 326–51. [Google Scholar]
- Deming JW, Young JN. The role of exopolysaccharides in microbial adaptation to cold habitats. In: Psychrophiles: From Biodiversity to Biotechnology, 2nd edn, Cham: Springer International Publishing, 2017, 259–84. [Google Scholar]
- Druckenmiller ML, Eicken H, Johnson M et al.. Towards an integrated coastal sea-ice observatory: system components and a case study at Barrow, Alaska. Cold Reg Sci Technol. 2009;56:61–72. [Google Scholar]
- Edwards KJ, Rogers DR, Wirsen CO et al.. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α- and γ-Proteobacteria from the deep sea. Appl Environ Microbiol. 2003;69:2906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlmann BL, Mustard JF, Murchie SL et al.. Subsurface water and clay mineral formation during the early history of Mars. Nature. 2011;479:53–60. [DOI] [PubMed] [Google Scholar]
- Eicken H, Gradinger R, Salganek M et al.. Field Technique for Sea Ice Research. Fairbanks, AK: University of Alaska Press, 2009. [Google Scholar]
- Eiler A, Langenheder S, Bertilsson S et al.. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl Environ Microbiol. 2003;69:3701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner WR, Bockheim JG, Hinkel KM et al.. Paleoenvironmental analyses of an organic deposit from an erosional landscape remnant, Arctic Coastal Plain of Alaska. Palaeogeogr Palaeocl. 2005;217:187–204. [Google Scholar]
- Eren AM, Morrison HG, Lescault PJ et al.. Minimum entropy decomposition: unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 2015;9:968–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eronen-Rasimus E, Lyra C, Rintala JM et al.. Ice formation and growth shape bacterial community structure in Baltic Sea drift ice. FEMS Microbiol Ecol. 2015;91:1–13. [DOI] [PubMed] [Google Scholar]
- Eronen-Rasimus E, Piiparinen J, Karkman A et al.. Bacterial communities in Arctic first-year drift ice during the winter/spring transition. Environ Microbiol Rep. 2016;8:527–35. [DOI] [PubMed] [Google Scholar]
- Ewert M, Deming JW.. Bacterial responses to fluctuations and extremes in temperature and brine salinity at the surface of Arctic winter sea ice. FEMS Microbiol Ecol. 2014;89:476–89. [DOI] [PubMed] [Google Scholar]
- Fetterer F., Knowles K, Meier W et al.. Sea Ice Index, Version 3.0. 2017. [Google Scholar]
- Firth E, Carpenter SD, Sørensen HL et al.. Bacterial use of choline to tolerate salinity shifts in sea-ice brines. Elem Sci Anthr. 2016;4:000120. [Google Scholar]
- Gilichinsky D, Rivkina E, Bakermans C et al.. Biodiversity of cryopegs in permafrost. FEMS Microbiol Ecol. 2005;53:117–28. [DOI] [PubMed] [Google Scholar]
- Gilichinsky D, Rivkina E, Shcherbakova V et al.. Supercooled water brines within permafrost—an unknown ecological niche for microorganisms: a model for astrobiology. Astrobiology. 2003;3:331–41. [DOI] [PubMed] [Google Scholar]
- Grosse G, Goetz S, McGuire AD et al.. Changing permafrost in a warming world and feedbacks to the Earth system. Environ Res Lett. 2016;11:040201. [Google Scholar]
- Groudieva T, Kambourova M, Yusef H et al.. Diversity and cold-active hydrolytic enzymes of culturable bacteria associated with Arctic sea ice, Spitzbergen. Extremophiles. 2004;8:475–88. [DOI] [PubMed] [Google Scholar]
- Handley KM, Lloyd JR. Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by Marinobacter species. Front Microbiol. 2013;4:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler DM, Zeitlin C, Wimmer-Schweingruber RF et al.. Mars’ surface radiation environment measured with the Mars Science Laboratory's Curiosity rover. Science. 2014;343:1244797. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhou Z, Duan H et al.. Methods and kits for identifying microorganisms in a sample. US9745611B2 Alexandria, VA: United States Patent and Trademark Office, 2014. [Google Scholar]
- Iwahana G, Eicken H, Cooper ZS et al.. A new perspective on the geological formation, evolution and isolation of subzero cryopeg sediments and brine in the coastl Arctic. (unpublished data) [Google Scholar]
- Jakosky BM, Nealson KH, Bakermans C et al.. Subfreezing activity of microorganisms and the potential habitability of Mars’ polar regions. Astrobiology. 2003;3:343–50. [DOI] [PubMed] [Google Scholar]
- Jansson JK, Taş N. The microbial ecology of permafrost. Nat Rev Microbiol. 2014;12:414–25. [DOI] [PubMed] [Google Scholar]
- Junge K, Eicken H, Deming JW. Bacterial activity at −2 to −20 degrees C in Arctic wintertime sea ice. Appl Environ Microbiol. 2004;70:550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junge K, Krembs C, Deming J et al.. A microscopic approach to investigate bacteria under in situ conditions in sea-ice samples. Ann Glaciol. 2001;33:304–10. [Google Scholar]
- Kaye JZ, Sylvan JB, Edwards KJ et al.. Halomonas and Marinobacter ecotypes from hydrothermal vent, subseafloor and deep-sea environments. FEMS Microbiol Ecol. 2011;75:123–33. [DOI] [PubMed] [Google Scholar]
- Knap A, Michaels A, Close A et al.. Protocols for the Joint Global Ocean Flux Study (JGOFS) Core Measurements, UNESCO, 1996. [Google Scholar]
- Knauth LP, Burt DM. Eutectic brines on Mars: origin and possible relation to young seepage features. Icarus. 2002;158:267–71. [Google Scholar]
- Kochkina GA, Ivanushkina NE, Akimov VN et al.. Halo-and psychrotolerant Geomyces fungi from arctic cryopegs and marine deposits. Microbiology. 2007;76:31–8. [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT et al.. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krembs C, Eicken H, Deming JW. Exopolymer alteration of physical properties of sea ice and implications for ice habitability and biogeochemistry in a warmer Arctic. Proc Natl Acad Sci USA. 2011;108:3653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord TB, Teeter G, Hansen GB et al.. Brines exposed to Europa surface conditions. J Geophys Res. 2002;107:5004. [Google Scholar]
- McEwen AS, Ojha L, Dundas CM et al.. Seasonal flows on warm Martian slopes. Science. 2011;333:740–3. [DOI] [PubMed] [Google Scholar]
- McKay CP, Stoker CR, Glass BJ et al.. The icebreaker life mission to Mars: a search for biomolecular evidence for life. Astrobiology. 2013;13:334–53. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer GH, Morrow MB, Wyss O et al.. Antarctica: the microbiology of an unfrozen saline pond. Science. 1962;138:1103–4. [DOI] [PubMed] [Google Scholar]
- Meyer H, Schirrmeister L, Andreev A et al.. Late glacial and Holocene isotopic and environmental history of northern coastal Alaska – results from a buried ice-wedge system at Barrow. Quat Sci Rev. 2010a;29:3720–35. [Google Scholar]
- Meyer H, Schirrmeister L, Yoshikawa K et al.. Permafrost evidence for severe winter cooling during the Younger Dryas in northern Alaska. Geophys Res Lett. 2010b;37:1–5. [Google Scholar]
- Mikucki JA, Pearson A, Johnston DT et al.. A contemporary microbially maintained subglacial ferrous “Ocean.” Science. 2009;324:397–400. [DOI] [PubMed] [Google Scholar]
- Mikucki JA, Priscu JC.. Bacterial diversity associated with Blood Falls, a subglacial outflow from the Taylor Glacier, Antarctica. Appl Environ Microbiol. 2007;73:4029–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AE, Kenig F, Fritsen CH et al.. Microbial life at –13 C in the brine of an ice-sealed Antarctic lake. Proc Natl Acad Sci USA. 2012;109:20626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytczuk NCS, Foote SJ, Omelon CR et al.. Bacterial growth at −15°C; molecular insights from the permafrost bacterium Planococcus halocryophilus OR1. ISME J. 2013;7:1211–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger TD, Perreault NN, Tille S et al.. Microbial characterization of a subzero, hypersaline methane seep in the Canadian high arctic. ISME J. 2010;4:1326–39. [DOI] [PubMed] [Google Scholar]
- Noble RT, Fuhrman JA. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–8. [Google Scholar]
- Nyland KE, Klene AE, Brown J et al.. Traditional Iñupiat ice cellars (SIĠḷUAQ) in Barrow, Alaska: characteristics, temperature monitoring, and distribution. Geogr Rev. 2017;107:143–58. [Google Scholar]
- Ojha L, Wilhelm MB, Murchie SL et al.. Spectral evidence for hydrated salts in recurring slope lineae on Mars. Nat Geosci. 2015;8:829–32. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R et al.. vegan: Community Ecology Package. R Package Version 2.5-3 2018. https://CRAN.R-project.org/package=vegan(25 July 2019, date last accessed). [Google Scholar]
- Pecheritsyna SA, Rivkina EM, Akimov VN et al.. Desulfovibrio arcticus sp. nov., a psychrotolerant sulfate-reducing bacterium from a cryopeg. Int J Syst Evol Microbiol. 2012;62:33–7. [DOI] [PubMed] [Google Scholar]
- Pecheritsyna SA, Shcherbakova VA, Kholodov AL et al.. Microbiological analysis of cryopegs from the Varandei Peninsula, Barents Sea. Microbiology. 2007;76:614–20. [PubMed] [Google Scholar]
- Perreault NN, Andersen DT, Pollard WH et al.. Characterization of the prokaryotic diversity in cold saline perennial springs of the Canadian high Arctic. Appl Environ Microbiol. 2007;73:1532–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrich C, Eicken H.. Overview of sea ice growth and properties. In: Sea Ice. Chichester, UK: John Wiley & Sons, Ltd, 2017, 1–41. [Google Scholar]
- Rapp JZ, Fernández-Méndez M, Bienhold C et al.. Effects of ice-algal aggregate export on the connectivity of bacterial communities in the central Arctic Ocean. Front Microbiol. 2018;9:1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing.2018. http://www.R-project.org/ (25 July 2019, date last accessed). [Google Scholar]
- Rognes T, Flouri T, Nichols B et al.. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T et al.. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova VA, Chuvil'skaya NA, Rivkina EM et al.. Novel halotolerant bacterium from cryopeg in permafrost: description of Psychrobacter muriicola sp. nov. Microbiology. 2009;78:84–91. [PubMed] [Google Scholar]
- Sherr EB, Sherr BF. Double-staining epifluorescence technique to assess frequency of dividing cells and bacteriovory in natural populations of heterotrophic microprotozoa. Appl Environ Microbiol. 1983;46:1388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter GM, Deming JW. Low-temperature chemotaxis, halotaxis and chemohalotaxis by the psychrophilic marine bacterium Colwellia psychrerythraea 34H. Environ Microbiol Rep. 2018;10:92–101. [DOI] [PubMed] [Google Scholar]
- Spirina EV, Durdenko EV, Demidov NE et al.. Halophilic-psychrotrophic bacteria of an Alaskan cryopeg—a model for astrobiology. Paleontol J. 2017;51:1440–52. [Google Scholar]
- Torstensson A, Young JN, Carlson LT et al.. Use of exogenous glycine betaine and its precursor choline as osmoprotectants in Antarctic sea-ice diatoms. J Phycol. 2019;53:663–75. [DOI] [PubMed] [Google Scholar]
- Ward BB, Priscu JC. Detection and characterization of denitrifying bacteria from a permanently ice-covered Antarctic Lake. Hydrobiologia. 1997;347:57–68. [Google Scholar]
- Westall F, Loizeau D, Foucher F et al.. Habitability on Mars from a microbial point of view. Astrobiology. 2013;13:887–97. [DOI] [PubMed] [Google Scholar]
- Westcott SL, Schloss PD. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere. 2017;2:e00073–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E, Michel C, Tremblay J et al.. Metagenomic survey of the taxonomic and functional microbial communities of seawater and sea ice from the Canadian Arctic. Sci Rep. 2017;7:42242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Romanovsky V, Duxbury N et al.. The use of geophysical methods to discriminate between brine layers and freshwater taliks in permafrost regions. J Glaciol Geocryol. 2004;26:301–9. [Google Scholar]
- Zhang DC, Li HR, Xin YH et al.. Marinobacter psychrophilus sp. nov., a psychrophilic bacterium isolated from the Arctic. Int J Syst Evol Microbiol. 2008;58:1463–6. [DOI] [PubMed] [Google Scholar]
- Zolotov MY. An oceanic composition on early and today's Enceladus. Geophys Res Lett. 2007;34:L23203. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.