ABSTRACT
The tick microbiota may influence the colonization of Ixodes scapularis by Borrelia burgdorferi, the Lyme disease bacterium. Using conserved and pathogen-specific primers we performed a cross-kingdom analysis of bacterial, fungal, protistan and archaeal communities of I. scapularis nymphs (N = 105) collected from southern Vermont, USA. The bacterial community was dominated by a Rickettsia and several environmental taxa commonly reported in I. scapularis, as well as the human pathogens B. burgdorferi and Anaplasma phagocytophilum, agent of human granulocytic anaplasmosis. With the fungal primer set we detected primarily plant- and litter-associated taxa and >18% of sequences were Malassezia, a fungal genus associated with mammalian skin. Two 18S rRNA gene primer sets, intended to target protistan communities, returned mostly Ixodes DNA as well as the wildlife pathogen Babesia odocoilei (7% of samples), a Gregarines species (14%) and a Spirurida nematode (18%). Data from pathogen-specific and conserved primers were consistent in terms of prevalence and identification. We measured B. burgdorferi presence/absence and load and found that bacterial beta diversity varied based on B. burgdorferi presence/absence. Load was weakly associated with bacterial community composition. We identified taxa associated with B. burgdorferi infection that should be evaluated for their role in vector colonization by pathogens.
Keywords: Borrelia burgdorferi, Ixodes scapularis, Lyme disease, tick microbiota, tick microbiome
This study describes the microbial communities of I. scapularis nymphs, in relation to B. burgdorferi presence/absence and identifies specific taxa associated with infection that should be further explored for their role in vector colonization by human pathogens.
INTRODUCTION
The microbiota of arthropod disease vectors are diverse and can be epidemiologically important because they may contain taxa that influence transmission of human pathogens (e.g. Dillon and Dillon 2004; Dong, Manfredini and Dimopoulos 2009; Weiss and Aksoy 2011; Engel and Moran 2013; Bourtzis et al. 2014; Hughes et al. 2014; Weiss et al. 2019). Tick microbiota, while still poorly understood in comparison to insect vectors of infectious disease, also may influence pathogen transmission (Narasimhan and Fikrig 2015; Bonnet et al. 2017; de la Fuente et al. 2017). Specific microbial taxa have been identified that are associated with both increased and decreased pathogen colonization in ticks (Clay et al. 2008; Narasimhan et al. 2014; Gall et al. 2016; Abraham et al. 2017), or that may indirectly affect pathogen transmission by altering tick fitness (Noda, Munderloh and Kurtti 1997; Zhong, Jasinskas and Barbour 2007; Hunter et al. 2015; Smith et al. 2015).
The role of the Ixodes scapularis microbiota in pathogen transmission is of particular interest because it is the vector of Lyme borreliosis, a tick-borne disease affecting an estimated 300 000 people per year in the United States (U.S.; CDC 2013; Mead et al. 2013), and several other important pathogens. Research into the I. scapularis microbiota could ultimately lead to management strategies to control either tick densities or infection prevalence with pathogens such as Borrelia burgdorferi (Narasimhan and Fikrig 2015), the disease-causing agent of Lyme borreliosis in North America. Experimental antibiotic treatments have demonstrated that the I. scapularis microbiota indirectly facilitates B. burgdorferi colonization by eliciting a tick immune response (Narasimhan et al. 2014). Additionally, some bacterial taxa occasionally present in I. scapularis are believed to reduce B. burgdorferi establishment through competitive interactions (Ross et al. 2018). Ixodes scapularis also contain intracellular bacteria that can potentially improve fitness by provisioning limiting nutrients (Hunter et al. 2015) or inducing production of an antifreeze glycoprotein (Neelakanta et al. 2010; Busby et al. 2012).
Despite this evidence, there are relatively few reports of microbiota-B. burgdorferi correlations from natural I. scapularis populations, and more information is needed on specific microbial taxa that may facilitate or inhibit pathogen transmission. In addition to B. burgdorferi infection prevalence the spirochete infectious dose, or load, is a potential risk factor in the transmission of B. burgdorferi (Piesman 1993; Ma et al. 1998). Furthermore, B. burgdorferi load may influence the expression pattern of Outer Surface Protein C, essential to B. burgdorferi colonization of hosts (Pal et al. 2004), due to density dependent interactions (De Silva et al. 1999; Yang et al. 2000). However, very little is known about the factors driving variation in B. burgdorferi loading (i.e. the number B. burgdorferi per tick) or whether load influences, or is influenced by, the I. scapularis microbiota. An additional challenge to tick microbiota research is that there are several sources of microbial community variation that could mask detection of such a relationship: I. scapularis microbiota vary by life stage, sex, geographic location, degree of engorgement and blood meal host identity (Moreno et al. 2006; Rynkiewicz et al. 2015; Van Treuren et al. 2015; Zolnik et al. 2016; Landesman et al. 2019), yet it is often difficult to control for all of these factors in a single study. Additionally, many studies into tick microbial communities focus on bacterial community, while data are generally lacking about communities of fungi, protista and archaea that may inhabit I. scapularis as well. Microbial community analyses are especially needed for nymphal stage I. scapularis, the life stage responsible for the majority of Lyme disease transmission to humans in the eastern United States (Barbour and Fish 1993).
To address these knowledge gaps, the objectives of this study were to (i) perform a cross-kingdom analysis of I. scapularis nymphs to characterize bacterial, archaeal, protistan and fungal inhabitants, (ii) determine if the I. scapularis microbial community is correlated with the B. burgdorferi presence/absence and/or spirochete load and (iii) identify specific microbial taxa that are correlated with B. burgdorferi presence/absence.
METHODS
Site description, tick collection and processing
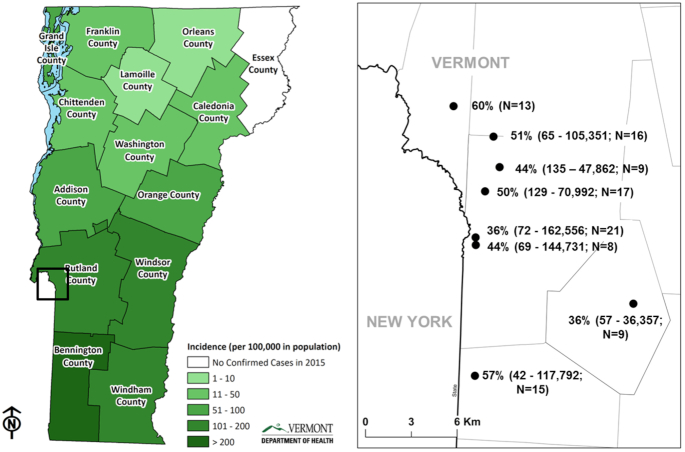
This research was conducted in Rutland County, Vermont, a region with one of the highest per capita rates of Lyme borreliosis reporting in the world (www.healthvermont.gov). Tick specimens for this study were collected from eight deciduous and mixed deciduous-coniferous forest sites from May 18 to July 13 2015, during peak nymphal activity (Fig. 1). Ticks were collected by dragging a white denim cloth (1 m2) across the forest floor on four separate 100 m transects per site. Ticks were removed from the denim cloth every 20 m, placed in 100% ethanol and stored in a −20°C freezer.
Figure 1.
County-level map of Vermont, USA, showing per capita human cases of Lyme disease in 2015, by county. Area of detail shows sample locations, B. burgdorferi infection prevalence and spirochete load range (min.—max.). Load was not calculated for one site.
Ticks were identified to genus by inspecting mouth parts and patterning of the anal groove. Over 99% of Ixodes species collected with the drag cloth method in the northeastern USA, including Vermont, are I. scapularis (David N. Allen, personal communication, May 5 2019; Piedmonte et al. 2018; Edwards et al. 2019). Individual nymphs were rinsed with 100% ethanol, air dried and added to 2 mL sample tubes, along with one sterile 2 mm steel bead and homogenized using a Tissue Lyser LT (Qiagen, Alameda, CA) at 55 Hz for 3 minutes. Genomic DNA was extracted using Qiagen's QIAamp DNA Micro Kit following the manufacturer's protocol. Whole nymph DNA was eluted in 60 µL of Qiagen's AE elution buffer solution. To test for laboratory contamination, we performed the same homogenization and DNA extraction procedure using a sterile bead but no tick (‘bead blanks’; n = 3). The DNA concentration of isolates was quantified on a Qbit® 2.0 fluorometer (Life Technologies, Carlsbad, CA) using the DS DNA (broad range) assay kit.
Borrelia burgdorferi testing
A total of 443 nymphs were tested for Borrelia burgdorferi infection and load—that is, both the presence/absence of B. burgdorferi and the number of spirochetes per tick—with quantitative real-time PCR (qPCR). We used a primer/probe system developed by Barbour et al. (2009). Forward and reverse primers (5′-GCTGTAAACGATGCACACTTGGT-3′ and 5′-GGCGGCACACTTAACACGTTAG-3′, respectively) targeted a 69bp region of the B. burgdorferi and B. miyamotoi 16S rRNA gene. To distinguish between these two species, we utilized a TaqMan™ probe (5′-TTCGGTACTAACTTTTAGTTAA-3′), including the FAM dye and MGBNFQ quencher, that was specific to B. burgdorferi. The PCR reaction was performed with a 1X QuantiNova mastermix (Qiagen) and primer and probe concentrations of 0.4 and 0.2 µM, respectively. Each sample and standard was run with 5 µL of template and a total reaction volume of 25 µL. The PCR cycle included a two-minute activation step at 95°C followed by 40 cycles of 95°C for 5 s (denaturation), and a combined annealing/extension step of 51°C for 5 s. PCR amplification was performed on a Qiagen Rotor Gene Q PCR thermal cycler. The qPCR run and qPCR reaction setup were performed in separate, temperature-regulated rooms. Each of the PCR runs included a blank comprised of sterile, RNase free water.
Each PCR run included amplicon standards of known concentration from which standard curves were generated. To prepare the standards, the 69bp target amplicon was synthesized and gel purified by Integrated DNA Technologies, Inc. (Coralville, Iowa). The amplicon was inserted into a StrataClone PCR cloning vector using the StrataClone Blunt PCR Cloning Kit by the Neuroscience Center of Biomedical Research Excellence of the University of Vermont (Burlington, VT). Plasmids were purified with a Qiagen plasmid maxi prep kit and sequenced by the Advanced Genome Technologies Core Facilities at the University of Vermont to confirm that they contained the correct amplicon. These plasmids were then linearized using BamH1, a high fidelity restriction enzyme (New England Biolabs, Ipswich, MA). Plasmid concentration was determined fluorometrically and 1:10 serial dilutions were prepared with molecular grade TE buffer. Standard concentrations were 41 040; 4104; 410; 41 and 10 plasmids/µL.
Spirochete load was calculated using the Rotor Gene Q Series software. Prior to the calculation, for each PCR run we applied a slope correction and removed data from the first five cycles, due to stochastic early amplification in some runs. To reduce the chance of false negatives, any amplification curve with an increase in fluorescence that was less than 5% of the maximum change found in any replicate was removed. Individual amplification curves were visually inspected for linear amplification, and removed if found. Ticks were considered B. burgdorferi positive if two out of the three technical replicates amplified and had a Ct value < 37. This threshold was justified by our observation that amplification of water blanks (one per qPCR run) and bead blanks (n = 3) was rare, and almost always at a Ct > 37 when it occurred. For B. burgdorferi-positive samples we ran three technical replicates to calculate the number of spirochetes per tick, based on the geometric mean of the replicates. Load data from any given run were included in the final data set if the standard curves had a PCR efficiency of 0.90–1.10 and r2 value > 0.99; load was only calculated if the technical replicates had similar Ct values at the 0.5 Ct threshold. Otherwise, these samples were considered B. burgdorferi-positive, but with an undetermined spirochete load.
Microbial community analyses
We used parallel PCR with universal primer sets and barcoding, combined with ultra-high throughput sequencing, to characterize the microbiota of B. burgdorferi-infected and uninfected nymphal-stage I. scapularis. Among the 443 nymphs tested for B. burgdorferi presence/absence and load, we selected 105 for microbial community analysis, including 20 B. burgdorferi-negative and 85 B. burgdorferi-positive nymphs with a range of spirochete loads (e.g. tens to >100 000). Universal primer sets targeted prokaryotes (515F/806R, v4 region of 16S rRNA gene; Bergmann et al. 2011; Apprill et al. 2015), archaea (Arch349F/806R, 16S rRNA gene; Takai and Horikoshi 2000), fungi (ITS1F/ITS2R, ITS gene; Schoch et al. 2012) and protista (Euk-1391F/EukBr-7R and Euk566F/1200R, targeting the V9 and V4-V5 regions of the 18S rRNA gene, respectively; Amaral-Zettler et al. 2009; Hadziavdic et al. 2014; Table 1).
Table 1.
Summary of primer sets used and the number of reads obtained. ITS = Internal transcribed spacer region of rDNA; IGS = Intergenic spacer region of rRNA; mito = mitochondria; *Only the forward read was analyzed.
| Primer Pair (Forward/Reverse) | Gene Region | Target Group | Expected size (nt) | # paired reads | Reference |
|---|---|---|---|---|---|
| 515F/806R | 16S | Prokaryotes | 292 | 23 407 998 | 1,2 |
| Arch349F/806R | 16S | Archaea | 457 | 200 676 | 3 |
| 566F/1200R | 18S | Protists | >660* | 12 220 122 | 4 |
| Euk-1391F/EukBr-7R | 18S | Protists | 95–175 | 6852 490 | 5 |
| ITS1F/ITS2R | ITS | Fungi | >580 | 34 221 582 | 6 |
| APdsb298F_APdsb670R | dsb | Anaplasma | 392 | 1827 983 | 7 |
| APdsb365F_APdsb715R | dsb | Anaplasma | 370 | 1985 614 | 7 |
| BT18SF_BT18SR | 18S | Babesia | 399–449* | 863 988 | 8 |
| FBbslF_FBbslR2 | 5S-23S IGS | B. burgdorferi | 250 | 391 485 | 9 |
| FBLBM16_23F3_FBLBM16_23R3 | 16S-23S IGS | B. miyamotoi/lonestari | 295–350 | 260 668 | 9 |
| 16Splus1_16Sminus1 | 16S (mito) | Ixodes | 450–454 | 51 862 | 10 |
We used primer sets for species-level detection of known pathogens, including B. burgdorferi (FBbslF_FBbslR2 5′-CGAGTTCGCGGGAGAGTA and 5′-TCCTAGGCATTMACCATAGMCT-3′; Table 1), Borrelia miyamotoi/Borrelia lonestari (FBLBM16_23F3_FBLBM16_23R3; 5′-GGTCAAGGGTTCGARTCCCT-3′ and 5′-GTTCAACTCCTCCTGATCCCA-3′; Fredericks et al., in prep), various strains of Anaplasma phagocytophilum (APdsb298F_APdsb670R: 5′-AGGGTTGATAAAATGCACGGC-3′ and 5′-TAAGTCGCTGGGTCTCTGGA-3′; APdsb365F_APdsb715R: 5′-AGGTCCCTAAGCATCACTCCT-3′ and 5′-TCTGCCTGTTGAGTCTGGTG-3′; Keesing et al. 2018) and species within the Babesia and Theileria genera (BT18SF_BT18SR; 5′-GACACAGGGAGGTAGTGACAAG-3′ and 5′-CTAAGAATTTCACCTCTGACAGT-3′; Georges et al. 2001). Finally, we tested a 16S rRNA mitochondrial primer pair (16Splus1_16Sminus1; 5′-CTGCTCAATGATTTTTTAAATTGCTGT-3′ and 5′-GTCTGAACTCAGATCAAGT-3′; Nadolny et al. 2011) for potential species-level identification of Ixodes species.
Prior to sequencing, PCR amplification was carried out using the Fluidigm Access Array system, which automates the generation of amplicon libraries within an enclosed chamber, reducing preparation time and minimizing potential sources of contamination (Yue et al. 2012; Brown et al. 2016). Briefly, DNA samples were diluted to 2 ng/µL and a mastermix for amplification was prepared using the Roche High Fidelity Fast Start Kit and 20x Access Array loading reagent. Mastermix (4 µL) and target DNA (1 µL) were added to 48 wells of a PCR plate. In a separate plate, 20X primer solutions were prepared by adding 2 µL of each forward and reverse primer (50 µM each), 5 µL of 20X Access Array Loading Reagent and water to a final volume of 100 µL. Mastermix and primer (4 µL each) were added to their respective inlets of a primed Fluidigm LP48.48 IFC, which was placed in a Fluidigm Juno for loading, PCR amplification and harvesting (see Table S1, Supporting Information for PCR conditions). Harvested product was transferred to a new 96 well plate and diluted 1:100 in water, and 1 µL of diluted product was used for a second round of amplification with Illumina linkers and barcodes. Products were quantified on a Qubit fluorometer (Life Technologies, Carlsbad, CA) and stored at −20°C. All samples were run on a Fragment Analyzer (Advanced Analytics, Ames, IA) to confirm amplicon regions and expected sizes. Samples were then pooled in equal concentrations and these pooled products were size selected on a 2% agarose E-gel and extracted from the isolated gel slice with a Qiagen gel extraction kit. Cleaned size selected products were run on an Agilent Bioanalyzer to confirm appropriate profile and determination of average size. Products were then added to an Illumina HiSeq 2500 for amplicon sequencing of 2 × 250 nt paired end reads. PCR amplification and sequencing were performed at the Roy J. Carver Center for Biotechnology at the University of Illinois (Urbana, IL).
Bioinformatics
We performed a sub-OTU analysis with DADA2 denoising, which generated 100% amplicon sequence variants (ASVs; Callahan et al. 2016). For each primer set, we truncated the sequence at a position where the overall quality generally remained above Q20. All reads for which quality values led to an expectation that more than two errors would occur over that length (max-ee) were removed. Paired reads were joined for all but the Euk566F/1200R and BT18SF_BT18SR primer pairs, for which the analysis was restricted to the forward read, due to non-overlapping amplicons. For the 16S rRNA gene and 18S rRNA gene data sets, taxonomic classification was performed using 99% OTU sequences from the SILVA database (release 128; Quast et al. 2013). Primer sequences were used to extract relevant subsets of the database, allowing up to 30% mismatch across both primers, and a special naive Bayesian classifier was trained for each primer set. ITS sequences were classified against the UNITE database (v. 01/12/2017; Kõljalg et al. 2013). Initial processing, quality filtering and taxonomy steps were performed at the Center for Bioinformatics and Computational Biology of the University of Delaware (Newark, DE) using Qiime2–2018.2, and additional filtering steps were performed at Green Mountain College (Poultney, VT) with Qiime2–2018-11 (Caporaso et al. 2010; Bolyen et al. 2018).
For pathogen-specific primers, paired or single end (for non-overlapping amplicons) reads were matched to sequences in a custom database of tick-borne pathogens, using vsearch38 (Rognes et al. 2016). Sequences were filtered when percent identity fell below 90% and alignment lengths below 50%. A maximum of 50 hits were retained for each sample-amplicon pair. A pathogen was determined to be present in a sample if there was a greater than 98% match to the database and a minimum of 10 reads present. Representative sequences from the Ixodes-specific primer set were manually searched against the NCBI database.
Statistics
Statistical analyses were performed using PRIMER 7 (v. 7.0.13) and R (v 3.4.1). Microbial community data analyses focused on alpha and beta diversity indices derived from the prokaryote (515F/806R) and fungal (ITS1F/ITS2R) data sets. To test the hypothesis that microbial alpha diversity was significantly different between B. burgdorferi-infected and -uninfected ticks, we modeled (i) the total number of bacterial or fungal ASVs per sample and (ii) Shannon's diversity index as linear functions of site, sample day and B. burgdorferi presence/absence. Sampling day was recorded categorically as one of three time periods of nymphal I. scapularis activity: ‘early’ (May 18 to June 4), ‘peak’ (June 5–22) and ‘late’ (June 23 to July 13). Models were fit using maximum likelihood optimization and competing models were compared and weighted using Akaike's Information Criterion (Burnham and Anderson 2002). For beta diversity, we calculated separate log transformed Bray–Curtis similarity matrices from fungal and bacterial ASV tables, and weighted and unweighted UniFrac distance matrices from the bacterial ASV table. Prior to calculating these matrices, we removed ASVs that had fewer than 10 sequences in the data set and rarefied the tables to 10 700 and 11 400 sequences per sample for bacteria and fungi, respectively. We performed three-way Analysis of Similarity (ANOSIM; Clarke 1993) separately on bacterial and fungal ASV tables with site, sample day and B. burgdorferi presence/absence as predictors. For the bacterial ASV table, we removed ASVs classified as Archaea, Mitochondria or Chloroplast. Prior to calculating diversity indices, we also removed all Borrelia from the bacterial ASV table, because we (i) had an independent measure of B. burgdorferi infection and load as our dependent variables and (ii) wanted to explore any correlation between B. burgdorferi presence/absence and the composition of the rest of the microbiota. We used the BEST analysis in Primer to look for correlations between Bray–Curtis similarities and the differences in B. burgdorferi load among samples. BEST performs a permutation test for Spearman's rank correlation between sample distances in the Bray–Curtis matrix and pairwise differences in log-transformed B. burgdorferi loading.
RESULTS
We obtained 6.8 to 34.2 million sequence reads from the 515F/806R, ITS1F/ITS2R, Euk566F/1200R and Euk-1391F/EukBr-7R primer pairs, and 200 000 reads from Arch349F/806R (Table 1). For two of the three bead blanks there were few (e.g. <20) to no sequences. However, for one bead blank there were 654 sequences obtained from the Arch349F/806R primer pair, 172 705 sequences from the Euk-1391F/EukBr-7R primer pair, 38 sequences from the Euk566F/1200R primer pair and 4087 sequences from the 515F/806R primer pair. These sequences were classified as unidentified bacteria, unidentified Mammalia (for both 18S rRNA gene primers) and Lactococcus, respectively.
We based our detection limit (Ct = 37) for qPCR on the behavior of sample blanks, since we consistently observed amplification below our lowest standard, in which case amplification was rarely observed and almost always above Ct 37 when found. Quantitative PCR runs used for load analysis had efficiencies that ranged from 0.93 to 1.11 and technical replicates with < 0.50 Ct difference. The overall B. burgdorferi infection prevalence, determined by qPCR, was 47% (208/443 positive), with a range of 36%–60% by site. The spirochete load among all B. burgdorferi-positive nymphs tested ranged from 42 to 162 556 (Fig. 1).
Prokaryotes (515F/806R)
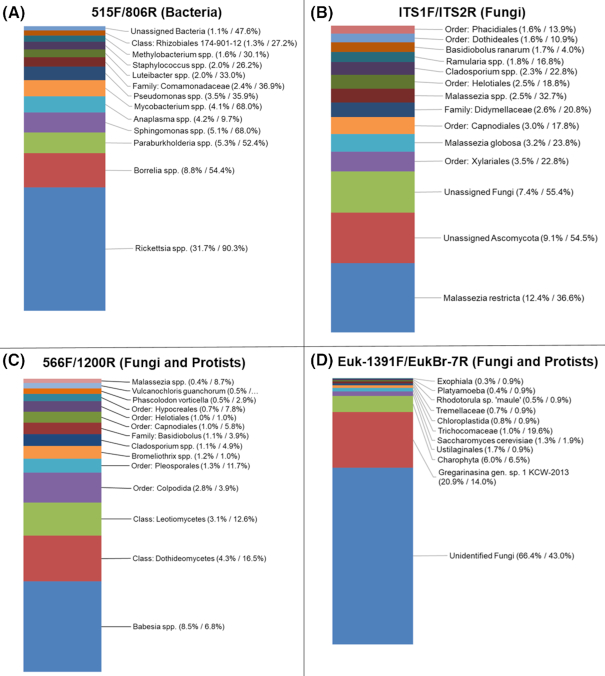
With the 515F/806R primer pair we obtained 2128 bacterial ASVs, of which 1852 were present in individual samples (hereafter referred to as ‘rare ASVs’). The most abundant bacterial genus was Rickettsia, which represented 31.7% of all bacterial sequences and was present in 90.3% of samples. There were 258 ASVs identified as Rickettsia, with a single dominant ASV that was found in all Rickettsia-positive samples. The other Rickettsia ASVs were rare, and all but four had >97% similarity to the most common Rickettsia ASV (the other four were present in single samples with <10 copies). A nucleotide BLAST search (blastn algorithm) of the most common, and several of the next most common ASVs, indicated Rickettsia buchneri as the most likely match. Other abundant taxa were Borrelia (8.8% of sequences and 54% of samples, according to the HiSeq data), Burkholderia-Paraburkholderia (5.3% of sequences and 53% of samples), Sphingomonas (5.1% of sequences and 68% of samples), Mycobacterium (4.1% of sequences and 68% of samples) and Anaplasma (4.2% of sequences and 9.7% of samples; Fig. 2 and Table S2, Supporting Information). Additional taxa that comprised >1% of all sequences, and were found in at least 10% of samples, were Pseudomonas, Comamonadaceae, Luteibacter, Staphylococcus and Methylobacterium. We found two archaeal ASVs in a single sample that were from the Woesearchaeota DHVEG-6 phylum (data not shown).
Figure 2.
Taxonomy summaries for the 515F/806R, ITS1F/ITS2R, 566F/1200R and Euk-1391F/EukBr-7R primer sets. In parentheses: number of sequences as a percentage of total, and percentage of samples in which the taxon was found.
All samples determined to be B. burgdorferi-negative with the 515F/806R primer set were also determined to be B. burgdorferi negative by qPCR analysis. An additional 13 samples were B. burgdorferi negative with 515F/806R; these corresponded to qPCR samples that were B. burgdorferi-positive but with the lowest loads in the data set (Table S3, Supporting Information).
Fungi (ITS1F/ITS2R)
We obtained 1712 fungal ASVs, of which 1537 were present in individual samples. The most abundant fungal taxon was Malassezia restricta, which comprised 12.4% of fungal sequences and was found in 36.6% of samples (Fig. 2 and Table S2, Supporting Information). Malassezia globosa and unidentified Malassezia were found in 23.8% and 32.7% of samples and comprised 3.2% and 2.5% of sequences, respectively. Other abundant taxa were from the orders Xylariales (3.5% of sequences and 22.8% of samples), Capnodiales (3.0% and 17.8%), Helotiales (2.5% and 18.8%), the family Didymellaceae (2.6% and 20.8%) and the genera Cladosporium (2.3% and 22.8%) and Ramularia (8% and 16.8%). We found the entomopathogenic fungi Beauveria bassiana, Lecanicillium fusisporum and unidentified Lecanicillium, each in three different tick samples. Unidentified Ascomycota and unidentified fungi comprised 9.1% and 7.4% of sequences, respectively; for each taxon, 93% of these unidentified taxa were rare ASVs. A blastn search of a subset of these rare ASVs indicated that they represented a diversity of taxa, rather than a small number of dominant ASVs.
Eukaryotes (Euk566F-1200R and Euk-1391F/EukBr-7R)
The Euk566F-1200R primer pair returned > 99.5% of sequences classified as Ixodes and an additional 0.3% of the sequences were other non-target taxa (e.g. unidentified Eukaryota and unidentified Opisthokonta). There were 36 fungal and 12 protistan taxa which comprised 73% and 17.5% of the sequences, respectively, after removing the Ixodes sequences. The most abundant fungi were Dothideomycetes (4.3% of sequences in 16.5% of samples, after filtering Ixodes sequences), Leotiomycetes (3.1% of sequences in 12.6% of samples) and Pleosporales (1.3% of sequences in 11.7% of samples; Fig. 2 and Table S2, Supporting Information). The most abundant protists were two Babesia ASVs, which comprised 8.5% of the non-Ixodes sequences and were found in 7/103 (7%) of samples. A blastn search revealed Babesia odocoilei as the closest match for both of these ASVs. Other relatively abundant protists were an unidentified Colpodida (2.8% of sequences in four samples), Bromeliothrix (1.2% of sequences in one sample), Phascolodon vorticella (0.5% of sequences in three samples) and Vulcanochloris guanchorum (0.5% of sequences in one sample; Table S2, Supporting Information).
With the Euk566F-1200R primer pair we also found an unidentified Spirurida (Phylum: Nematoda) in 18% of samples, and an unidentified Tylenchida (Phylum: Nematoda) in one sample. A blastn search of the Spirurida taxa indicated Dirofilaria repens as the best match (208/210 nt matched) for all ASVs. Other top matches were Onchocercidae, Loa loa, Rumenfilaria andersoni, Elaeophora schneideri, Filarioidea and Cercopithifilaria bainae (206–207 nt matched).
With the Euk-1391F/EukBr-7R primer-pair we found > 95% of sequences were unidentified Acari and an additional 4.2% of sequences were unidentified Animalia or unidentified Mammalia. The most abundant protists were an unidentified Charophyta, found in seven samples, and Gregarinasina gen. sp. 1 KCW-2013, found in 15 samples. There were seven fungal taxa, each found in 1–2 samples and unidentified fungi found in 46 samples (Fig. 2 and Table S2, Supporting Information).
Archaea (Arch349F/806R)
With the Arch349F/806R primer pair we found an unidentified Archaea in the same sample in which we found Woesearchaeota DHVEG-6 with the 515F/806R primer set. Additionally, we found the same dominant bacterial taxa identified with the 515F/806R primer set (e.g. Rickettsia, Borrelia, Anaplasma, Burkholderia-Paraburkholderia, etc.; Table S2, Supporting Information). Unidentified bacteria comprised 40% of the sequences; many of these were revealed by a blastn search to be from the 18S rRNA gene region of Ixodes.
Pathogen- and tick-specific primer sets
Using pathogen-specific primers we found Borrelia burgdorferi in 62/103 samples and no B. miyamotoi (Table S2, Supporting Information). We found A. phagocytophilum in 11/103 samples with both Anaplasma-specific primer sets and Babesia odocoilei/ Babesia spp. RD1 (equal e scores) in 6/103 samples. For each sample-primer pair these species hits almost always comprised >97% of reads. The 16Splus1_16Sminus1 primer set returned sequences that matched the 18S region of Ixodes species, which was not the intended target, and thus no confirmation of identifications could be made.
Potential drivers of alpha diversity
AIC analysis indicated that B. burgdorferi presence/absence was very weakly predictive of the number of bacterial ASVs (ΔAIC = 1.28; data not shown) and not a significant predictor of Shannon's Diversity Index. Neither site nor sampling day was predictive of bacterial alpha diversity. AIC analysis indicated that fungal alpha diversity was affected by both site and sampling day but not B. burgdorferi presence/absence; models that combined both site and day as predictors of the number of ASVs as well as Shannon's Diversity Index had the greatest model support (Table 2).
Table 2.
Summary of fungal alpha diversity modeling using Akaike's Information Criterion and Shannon's Index and Number of ASVs as response variables.
| Shannon's Index | ||||
|---|---|---|---|---|
| Model | Null | Day | Site | Site + Day |
| # parameters | 2 | 4 | 8 | 10 |
| Negative Log Likelihood | 414.9 | 408.3 | 406.3 | 400.6 |
| AIC | 833.7 | 824.6 | 828.7 | 821.2 |
| ΔAIC | 12.5 | 3.4 | 7.5 | 0 |
| AIC weight | 0 | 0.18 | 0.02 | 1 |
| P(Best Model) | 0 | 0.15 | 0.02 | 0.83 |
| No. of ASVs | ||||
| Model | Null | Day | Site | Site + Day |
| # parameters | 2 | 4 | 8 | 10 |
| Negative Log Likelihood | 158.5 | 148.8 | 149.3 | 140.6 |
| AIC | 321 | 305.6 | 314.6 | 301.1 |
| ΔAIC | 19.9 | 4.4 | 13.5 | 0 |
| AIC weight | 0 | 0.11 | 0 | 1 |
| P(Best Model) | 0 | 0.098 | 0.001 | 0.901 |
Drivers of bacterial and fungal beta diversity
After quality filtering and rarefaction, 103 samples, of which 83 were B. burgdorferi-positive, were retained for bacterial beta diversity analysis. The BEST analysis provided evidence of a weak correlation between the pairwise Bray–Curtis distance between samples and the difference in their B. burgdorferi loads (ρ = 0.052, P = 0.066). Using a three-way ANOSIM, we found a significant correlation between B. burgdorferi presence/absence and bacterial community similarity as estimated with the Bray–Curtis similarity matrix (R = 0.188, p = 0.014) and the weighted UniFrac distance matrix (R = 0.246, P = 0.012). Similarity estimated by the unweighted UniFrac distance matrix was also correlated with B. burgdorferi presence/absence (R = 0.142, P = 0.055; Table 3). After removing rare ASVs, the effect was significant with the weighted UniFrac Matrix (P = 0.036), but weaker with the Bray–Curtis matrix ( P = 0.072) and not significant with the unweighted UniFrac matrix ( P = 0.405, Table S4, Supporting Information). Differences in site and sample day were both significant factors of beta diversity, with and without the rare ASVs (Table 3; Table S4, Supporting Information).
Table 3.
Three-way ANOSIMs of bacterial and fungal community similarity with site, sample day (three equal time periods from May 18 to July 13, 2015) and B. burgdorferi presence/absence as factors. Borrelia spp. were filtered from the data set and Bray–Curtis distance matrices were log (x+1) transformed. Significance was determined by comparing the ANOSIM R to the R values obtained from 999 999 random permutations. For negative R values, the P-value was calculated as (100-P) *2 (two-tail test).
| Site | Sample Day | B. burgdorferi | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Kingdom | Data | Seqs/sample | N | R | P | R | P | R | P |
| Bacteria | Bray–Curtis | 10 700 | 102 | 0.123 | 0.002 | 0.209 | 0.0006 | 0.188 | 0.014 |
| Weighted Unifrac | 0.069 | 0.051 | 0.046 | 0.213 | 0.246 | 0.012 | |||
| Unweighted Unifrac | 0.043 | 0.144 | 0.096 | 0.05 | 0.142 | 0.055 | |||
| Fungi | Bray–Curtis | 11 400 | 101 | 0.063 | 0.026 | 0.05 | 0.169 | −0.145 | 0.028 |
We hypothesized that Spirurida presence may also influence the I. scapularis microbial community, either due to the presence of its own unique microbiota or other interactions with I. scapularis. Using a two-way ANOSIM of the Bray-Curtis matrix, with B. burgdorferi and Spirurida presence/absence as factors, we found that bacterial assemblages varied significantly between Spirurida-positive and Spirurida-negative ticks (R = 0.153; p = 0.022; Table 4). This was also the case with B. burgdorferi presence/absence (R = 0.162, P = 0.012). After removing rare ASVs, significance for B. burgdorferi presence/absence was similar, and the results for Spirurida presence/absence showed a higher R value and stronger significance (R = 0.176, P = 0.007; Table S5, Supporting Information). There was a significant effect of B. burgdorferi infection with the weighted UniFrac metric (R = 0.144, P = 0.039). For Spirurida presence/absence, we found a negative correlation with the unweighted UniFrac metric (R = −0.12; P = 0.036), implying greater average similarity between groups than within groups. However, these results with the UniFrac matrices were not significant after removing the rare ASVs (Table S5, Supporting Information).
Table 4.
Two-way ANOSIMs of bacterial and fungal community similarity with B. burgdorferi presence/absence and Spirurida infection status as factors. Borrelia spp. were filtered from the dataset and Bray–Curtis distance matrices were log (x+1) transformed. Significance was determined by comparing the ANOSIM R to the R values obtained from 999 999 random permutations. For negative R values, the P-value was calculated as (100-P) *2.
| B. burgdorferi | Spirurida | ||||||
|---|---|---|---|---|---|---|---|
| Kingdom | Data | Seqs/sample | N | R | P | R | P |
| Bacteria | Bray–Curtis | 10 700 | 102 | 0.162 | 0.012 | 0.153 | 0.022 |
| Weighted Unifrac | 0.144 | 0.039 | 0.028 | 0.346 | |||
| Unweighted Unifrac | 0.042 | 0.209 | −0.12 | 0.036 | |||
| Fungi | Bray–Curtis | 11 400 | 101 | −0.067 | 0.072 | 0.047 | 0.126 |
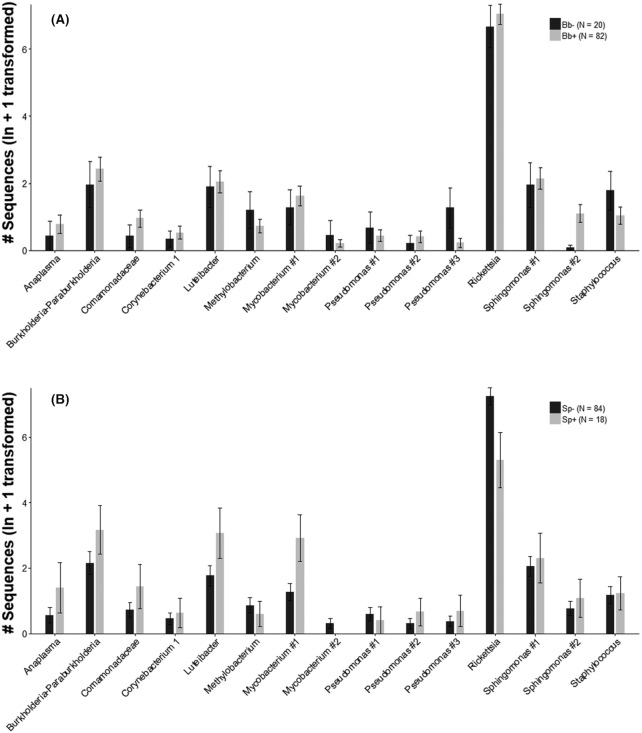
Using a two-way Similarity Percentages (SIMPER) analysis, we found that a total of 466 ASVs were needed to explain 70% of the community dissimilarity based on B. burgdorferi presence/absence, and a total of 390 ASVs were needed to explain 70% of the dissimilarity based on Spirurida presence/absence. ASVs making the largest contribution to community dissimilarity, and comprising at least 0.5% of sequences and at least 5% of samples, are plotted in Fig. 3. ASVs showing the largest differences based on B. burgdorferi presence/absence were a Pseudomonas ASV (#3 in Fig. 3A) and a Staphylococcus ASV, which were more abundant in B. burgdorferi-negative nymphs, and a Sphingomonas ASV (#2 in Fig. 3A), which was more abundant in B. burgdorferi-positive nymphs. ASVs showing the largest differences based on Spirurida presence/absence were a Luteibacter and a Mycobacterium ASV (#1 in Fig. 3B), which were more abundant in Spirurida-positive nymphs, and a Rickettsia ASV, which was more abundant in Spirurida-negative nymphs (Fig. 3B). These qualitative comparisons may inform the development of more targeted PCR analyses using species presence/absence by B. burgdorferi infection status.
Figure 3.
(A), SIMPER Plot of ln+1 transformed ASV sequence counts (±SE) by (A) B. burgdorferi presence/absence (Bb+/Bb−) and (B), Spirurida presence/absence (Sp+/Sp-) for individual ASVs with an overall frequency >0.5% and contribution to community similarity of at least 0.5%, as determined by two-way SIMPER. The taxonomic identity of the ASVs appear in the X-axis. Y-axis values are low due in part to the large number of samples with zeroes, representing samples not infected by B. burgdorferi.
After quality filtering and rarefaction, 101 samples were retained for fungal beta diversity analysis. With the three-way ANOSIM we found a negative correlation measure between B. burgdorferi-infected and -uninfected nymphs (R = −0.145, P = 0.028, Table 3). PERMDISP indicated that average deviations from the centroid were 69.2 (±SE 0.19) and 67.1 (±SE 0.53) for B. burgdorferi-infected and -uninfected ticks respectively (p perm = 0.037), suggesting the negative correlation was due to greater dispersion in B. burgdorferi-positive ticks. When removing the rare ASVs, the negative correlation due to B. burgdorferi presence/absence was weaker (R = −0.115, P = 0.146), as was evidence for greater dispersion ( p perm = 0.113). Fungal communities were significantly different by site (R = 0.063, P = 0.026), but not by sampling day ( P = 0.169), and these results for site and day were similar after removing the rare ASVs (Table S4, Supporting Information).
Due to the large number of non-target taxa (e.g. Acari and Ixodes), statistical analyses were not performed on data obtained from the 18S rRNA gene and Arch349F/806R primer sets.
DISCUSSION
Using parallel PCR with universal primer sets, followed by high-throughput sequencing, we performed a cross-kingdom analysis of the I. scapularis microbiota of field-collected ticks. This analysis uncovered several pathogens of human and wildlife significance and insights into the potential source of colonizing microbes. Below we discuss our taxonomic findings from each of the five universal primer sets tested, and identify specific ASVs correlated with B. burgdorferi presence/absence that should be further studied for potential positive or negative roles in vector colonization by human pathogens. Our analysis provides insight into how tick microbiota may be affected by the presence of pathogens, information that may ultimately inform management strategies that reduce pathogen exposure risk (Narasimhan and Fikrig 2015).
We found that B. burgdorferi presence/absence was correlated with bacterial beta diversity, more strongly due to differences in the relative abundance of taxa, rather than differences in species occurrence (Table 3). Fungal beta diversity also varied by B. burgdorferi presence/absence, due to differences in the dispersion of samples (Anderson et al. 2006), and this pattern with fungi appeared to have been driven by the rare ASVs (Table 3; Table S3, Supporting Information). Importantly, our analysis controlled for a number of sources of microbial community variation, including life stage, degree of engorgement (all were questing ticks) and geographic location (Moreno et al. 2006; Rynkiewicz et al. 2015; Van Treuren et al. 2015; Zolnik et al. 2016), thereby enabling us to focus primarily on microbial taxonomic correlations with B. burgdorferi infection. Sex is also a driver of microbial community variation—but one that is difficult to determine in nymphs—and our large sample size was expected to reduce any potential sample bias due to sex.
The 20 B. burgdorferi-negative samples, as determined by qPCR, were also negative by Illumina sequencing (both the FBbslF_FBbslR2 and 515F/806R primer sets; Table S3, Supporting Information). The 12 samples that were positive by qPCR, but negative by Illumina sequencing, had the 12 lowest B. burgdorferi load estimates in the data set, accounting for most of the discrepancies among primer sets. These results suggest that qPCR was the most sensitive test for B. burgdorferi presence/absence. The FBbslF_FBbslR2 primer set detected seven more B. burgdorferi-positive samples than 515F-806R; this somewhat lower sensitivity of 515F/806R may be due to greater competition among targets for this conserved primer set. The number of Illumina reads were lower than qPCR loads because (i) Illumina read counts, unlike qPCR samples, were based upon 1 µL of sample and not adjusted for DNA extraction elution volume, (ii) there could have been a loss of sample during gel extraction and (iii) samples for Illumina sequencing were diluted to 2 ng/µL prior to sequencing, in order to reduce PCR inhibition. Because the dilution was based on whole tick DNA, and not microbial DNA specifically, quantitative comparisons of the primer sets were not performed. Two samples exhibited large discrepancies among primer sets: samples with loads of 31 352 (T1194) and 82 985 (T1168) had no hits with the FBbslF_FBbslR2 primer set (for T1194 the 515F-806R primer set detected 142 reads). The cause for these large differences between qPCR and the FBbslF_FBbslR2 primer set is unknown.
Although our data concerning microbiome-B. burgdorferi interactions are correlative, several other studies also indicate that I. scapularis microbiota may influence pathogen colonization of ticks. Ross et al. (2018) found that colonization by B. burgdorferi may be impeded by antagonistic interactions with species such as Pseudomonas, which possess genes for a Type VI secretion system. Consistent with their findings, we identified a relatively abundant and influential Pseudomonas ASV (Fig. 3A) that was present in 20% of B. burgdorferi-negative (4/20) but only 3.7% of B. burgdorferi-positive nymphs (3/82; P = 0.026 based on a test with 20 000 permutations). In addition to a Pseudomonas ASV, we found several other genera that should be evaluated for their potential to either inhibit (e.g. Staphylococcus) or facilitate (e.g. Sphingomonas) B. burgdorferi colonization (Fig. 3A). The tick microbiota is also believed to trigger an immune response in I. scapularis that facilitates B. burgdorferi colonization of the midgut (Narasimhan et al. 2014). Similarly, the I. scapularis microbiota can be altered by presence of A. phagocytophilum, which appears to facilitate colonization of this pathogen, potentially at the expense of B. burgdorferi colonization (Abraham et al. 2017). The I. scapularis microbiota may also indirectly affect pathogen transmission by altering tick fitness, due to the presence of R. buchneri, a nutritional endosymbiont that has the genetic machinery for folate synthesis (Hunter et al. 2015).
Both the I. scapularis microbiota, as well as B. burgdorferi colonization of I. scapularis, are correlated with the identity of the blood meal host (LoGiudice et al. 2003; Ostfeld and Keesing 2012; Landesman et al. 2019). Therefore, our observed microbiota-B. burgdorferi correlations may also be an indicator of host identity, rather than, or in addition to, species interactions. Furthermore, parasitic nematodes from the Spirurida order also infect a limited range of hosts (e.g. Joseph et al. 2011) and our SIMPER analysis of Spirurida-microbiota correlations uncovered several influential taxa (Luteibacter, a Mycobacterium, A. phagocytophilum and Rickettsia; Fig. 3B) that are from orders shown to be correlated with host species identity (Landesman et al. 2019). We therefore suggest that knowledge of the host blood meal, which is difficult to discern from questing ticks (Allan et al. 2010), is a critical missing factor in this and other analyses of tick microbiota.
Spirochete load is an important but under-studied risk factor for transmission of B. burgdorferi (e.g. De Silva et al. 1999; Yang et al. 2000), and large variation in B. burgdorferi loads in I. scapularis were reported here and by Wang et al. (2003). Our sampling design included a large proportion (80%) of infected nymphs, allowing us to study microbial community composition across orders of magnitude of spirochete loads for detection of potential microbiota-B. burgdorferi load correlations. BEST analysis suggested that nymphs with a higher spirochete load tend to have more distinct microbiota than nymphs with a lower spirochete load, although the correlation was weak. However, there was a considerable amount of noise in the data set, as evidenced by the large number of rare ASVs, and our analysis is limited by the fact that sequence counts, while useful for comparing relative abundances between groups, are not a reliable estimate of absolute abundances (Krehenwinkel et al. 2017). Thus, our evidence for a weak correlation between microbial community composition and spirochete load, combined with data showing differences in presence/absence by B. burgdorferi infection status (e.g. a Pseudomonas, a Staphylococcus and a Sphingomonas), indicate taxa which should be the focus of continued analyses to determine their potential relationships to B. burgdorferi infection and transmission. Such an analysis would aid in the search for microbial taxa that may be interacting with B. burgdorferi within I. scapularis.
Borrelia burgdorferi load values should be interpreted with caution, as these estimates are prone to pipetting error, especially at low load values, as well as error due to variation in qPCR efficiency among separate runs (efficiency ranged from 0.93 to 1.11 among runs used for load estimates). Additionally, the presence of B. miyamotoi, which also binds to our primer set, could reduce the reaction efficiency, although we found little evidence that this pathogen was present in samples.
The most commonly encountered bacteria was a Rickettsia—most likely Rickettsia buchneri—an obligate intracellular bacterium that is transmitted transovarially and trans-stadially (Kurtti et al. 2015) and that is believed to be one of the few stable members of the I. scapularis microbiota (Ross et al. 2018). Anaplasma phagocytophilum, found in 9.7% of samples, is the agent of human granulocytic anaplasmosis and the second most common tick-borne disease in Vermont (www.healthvermont.gov). In 2015, there were reported 55 cases per 100 000 individuals of anaplasmosis in Vermont, and this increased to 189 cases per 100 000 in 2017 (Natalie A. Kwit, Vermont Department of Health, Personal Communication).
Many of the abundant bacterial taxa found (e.g. Sphingomonas, Mycobacterium, Pseudomonas, Comamonadaceae, Luteibacter, Staphylococcus, Methylobacterium and Rhizobiales; Fig. 2) have been previously reported in I. scapularis (e.g. Benson et al. 2004; Moreno et al. 2006; Narasimhan et al. 2014; Rynkiewicz et al. 2015; Van Treuren et al. 2015) and in other Ixodes species (Heise, Elshahed and Little 2010; Carpi et al. 2011; Kwan et al. 2017; Diaz-Sanchez et al. 2019), suggesting that they may be members of the I. scapularis internal microbiota. Many of these abundant genera contain species that inhabit plants and soil, suggesting that some may have colonized the tick surface (e.g. Ross et al. 2018; Zolnik et al. 2018) where they are less likely to interact with ticks or their internal microbiota. However, Luteibacter, Sphingomonas, Pseudomonas and Staphylococcus have been found in salivary glands of Ixodes ovatus and Ixodes persulcatus (Qiu et al. 2014). Furthermore, Pseudomonas was found on both the surface and within the viscera of I. scapularis (Ross et al. 2018). Surface washing with bleach may be more effective than ethanol at removing surface-dwelling organisms, thereby allowing for an analysis of primarily internal microbes (Binetruy et al. 2019). However, for this study we used an ethanol wash because we were also interested in surface-associated taxa, such as entomopathogenic fungi (see below), that may play a role in reducing tick fitness (e.g. Kirkland, Westwood and Keyhani 2004). Furthermore, it has been suggested that surface microbes may potentially enter internal structures via surface openings (Hynes 2014; Ross et al. 2018), although this mechanism remains untested.
Environmental microbes may also be acquired from the forest (Zolnik et al. 2018) and other tick environments. Many of the dominant fungi in our data set, including the Xylariales, Capnodiales (including Cladosporium and Ramularia), Pleosporales and Helotiales (Fig. 2) are plant-associated fungi (Webster and Weber 2007; Hyde et al. 2013), providing additional evidence that the tick microbiota contains many environmental taxa. The most commonly encountered fungi were from the genus Malassezia, which contains species that frequently colonize mammalian skin (Galuppi and Tampieri 2008; Boekhout et al. 2010). The absence of Malassezia from sample blanks suggests that they may have a non-human origin, and raises the possibility that I. scapularis acquire Malassezia, as well as other environmental microbes, due to physical contact with their vertebrate host during blood feeding (Hynes 2014). We found Beauveria bassiana, an entomopathogenic fungus shown to reduce fitness of several non-Ixodes tick species (Samish and Rehacek 1999; Kaaya and Hassan 2000; Gindin et al. 2002), in three samples. Lecanicillium fusisporum, an entomopathogenic fungus of aphids (Vu, Hong and Kim 2007), and unidentified Lecanicillium were each found in three other samples. Although we found no evidence that any of these taxa affected tick densities (data not shown), the ability to screen large numbers of ticks for entomopathogenic fungi may aid in the collection of natural specimens to be evaluated as biological control agents.
Only five protistan taxa were found with the Euk566F/1200R primer set, despite obtaining over 12 million reads (Table S2). This may be due to competition for primer binding by Ixodes DNA, which comprised the vast majority of sequence reads, and/or low abundance of protists inhabiting I. scapularis. The protists detected in this study were likely a mixture of environmental species as well as true residents of the I. scapularis microbiota. The Euk-1391F/EukBr-7R primer set was also dominated by Ixodes taxa, and returned even fewer protistan species, with less taxonomic resolution than the Euk566F/1200R primer set.
With the Euk-1391F/EukBr-7R primer set we detected a Gregarine species (Gregarinasina gen. sp. 1 KCW-2013), which are frequently found associated with invertebrates, including arthropods (Rueckert, Betts and Tsaousis 2019). The most abundant protist was a Babesia spp., with Babesia odocoilei as the most likely species (Fig. 2 and Table S2, Supporting Information). This classification is supported by a blastn search of amplicons from the Euk566F/1200R primer set, for which the two Babesia ASVs had greater than 99% identity to B. odocoilei in 7/103 samples. We also found evidence for B. odocoilei with the Babesia-specific primer set in 6/103 samples (Table S2, Supporting Information). B. odocoilei has established populations in I. scapularis in New England (Armstrong et al. 1998; Steiner et al. 2014) and nearby regions of Canada (e.g. Milnes et al. 2019). Babesia odocoilei is transmitted between I. scapularis and the white-tailed deer (Odocoileus virginianus; Emerson and Wright 1968; Emerson and Wright 1970; Waldrup et al. 1990) and is the agent of cervid babesiosis, affecting captive animals and, potentially, wild populations of Elk and Caribou (Mathieu et al. 2018; Milnes et al. 2019). Interestingly, 5 of 7 Babesia odocoilei positive samples were also B. burgdorferi positive, suggesting that an unexpectedly high proportion of larva fed upon, and were infected with B. burgdorferi, by white-tailed deer. Alternatively, this finding may indicate that there are additional unknown hosts for B. odocoilei or that B. odocoilei is transmitted transovarially. To the best of our knowledge the latter mechanism has not been evaluated for this species. Babesia microti, the causative agent of human Babesiosis, is endemic in the northeastern United States but was not detected in our samples. In 2015 there were approximately two cases (3.4 per 100 000 people) of human Babesiosis in Rutland County, Vermont (Natalie A. Kwit, Vermont Department of Health, Personal Communication), suggesting that B. microti prevalence in nymphs was low during the study period.
With the Euk566F/1200R primer set (Table S2, Supporting Information) we also found a Spirurida nematode. Dirofilaria repens (Table S2, Supporting Information), one of the causative agents of heartworm and human dirofilariasis (Joseph et al. 2011; Kartashev et al. 2011), was the closest match in NCBI. However, using longer sequencing reads from samples collected in New York and Connecticut, USA Tokarz et al. (2019) found Spirurida more closely matched to Acanthocheilonema viteae. A comparison of our shorter (210 nt) amplicons revealed 99%–100% similarity to the corresponding sequence in the Tokarz et al. sequences (Rafal Tokarz, personal communication, May 3, 2019). It remains to be determined if nematode DNA found in I. scapularis is from living organisms and whether I. scapularis is a competent vector for transmission.
The archaea primer set returned bacteria, but no archaea, and the 515F/806R primer pair returned one archaeon (Table S2, Supporting Information), suggesting that archaea are largely absent from these ticks. However, Nakao et al. (2013) identified archaea in their tick samples, which may indicate that our primer sets lack the sensitivity to detect additional archaeal species.
CONCLUSIONS
Advances in PCR and DNA sequencing technologies now allow for the cost-effective analysis of large numbers of tick specimens, enabling new insights into the composition and function of tick microbiota. Using the Fluidigm Access Array system, combined with Illumina's HiSeq platform, we performed a cross-kingdom analysis of the microbiota of I. scapularis nymphs in southern Vermont, a region with one of the highest rates of Lyme disease reported in the USA. In addition to commonly studied human pathogens (B. burgdorferi and A. phagocytophilum), we identified several taxa of potential veterinary and wildlife significance in I. scapularis, including B. odocoilei and a Spirurida nematode. Entomopathogenic fungi were found in nine samples. With the fungal primer set, we also found an abundance of Malassezia, suggesting that animal skin and fur may be an important source of environmental microbes within the whole tick microbial community. Finally, we found differences in the relative abundances of bacteria inhabiting I. scapularis based on B. burgdorferi presence/absence. Most notably a Pseudomonas ASV, a potential competitor with B. burgdorferi in the I. scapularis midgut, was more common in B. burgdorferi-negative nymphs. However, despite testing >80 B. burgdorferi-positive ticks across a large range of spirochete loads, we found only a weak association between microbial community composition and spirochete load. Nevertheless, these findings offer considerable promise for the discovery of microbial taxa that may influence the transmission of tick-borne pathogens and future research should seek to identify mechanisms by which microbial interactions may be harnessed to disrupt disease transmission pathways.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Robert Cluss, David Allen (Middlebury College), Tim Hunter (BioTek Instruments), Tabitha Finch, Judith van Houten and Rex Forehand (Vermont Genetics Network) for their thoughtful advice and support throughout the project, Brian Haggerty-Perrault, Alex Stephanson and Madison Mercer for tick collections and preparation of DNA extracts, and several land owners in Fair Haven, Middletown Springs, Poultney and Wells, Vermont, who granted access to their properties for this project. We thank Sheryl White of the Neuroscience COBRE Molecular Core of the University of Vermont for preparing the qPCR standards, Mark Band and Chris Wright of the Carver Biotechnology Center for DNA sequencing services and Shawn Polson of the Center for Bioinformatics and Computational Biology of the University of Delaware for bioinformatics analyses. Finally, we thank two anonymous reviewers whose comments have significantly improved this manuscript. This research was funded by an Institutional Development Award (IDeA) to W.J.L. from the National Institute of General Medical Sciences of the National Institutes of Health under Grant number P20GM103449.
Conflict of interest. None declared.
REFERENCES
- Abraham NM, Liu L, Jutras BL et al.. Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc Nat Acad Sci, USA. 2017;114:E781–E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan BF, Goessling LS, Storch GA et al.. Blood meal analysis to identify reservoir hosts for Amblyomma americanum ticks. Emerg Infect Dis. 2010;16:433–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral-Zettler LA, McCliment EA, Ducklow HW et al.. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One. 2009;4:e6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Ellingsen KE, McArdle BH. Multivariate dispersion as a measure of beta diversity. Ecol Lett. 2006;9:683–93. [DOI] [PubMed] [Google Scholar]
- Apprill A, McNally S, Parsons R et al.. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–37. [Google Scholar]
- Armstrong PM, Katavolos P, Caporale DA et al.. Diversity of Babesia infecting deer ticks (Ixodes dammini). Am J Trop Med Hyg. 1998;58:739–42. [DOI] [PubMed] [Google Scholar]
- Barbour AG, Bunikis J, Travinsky B et al.. Niche partitioning of Borrelia burgdorferi and Borrelia miyamotoi in the same tick vector and mammalian reservoir species. Am J Trop Med Hyg. 2009;81:1120–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Fish D. The biological and social phenomenon of lyme disease. Science. 1993;260:1610–6. [DOI] [PubMed] [Google Scholar]
- Benson MJ, Gawronski JD, Eveleigh DE et al.. Intracellular symbionts and other bacteria associated with deer ticks (Ixodes scapularis) from Nantucket and Wellfleet, Cape Cod, Massachusetts. Appl Environ Microbiol. 2004;70:616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann GT, Bates ST, Eilers KG et al.. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem. 2011;43:1450–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binetruy F, Dupraz M, Buysse M et al.. Surface sterilization methods impact measures of internal microbial diversity in ticks. Parasites Vectors. 2019;12:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T, Guého-Kellermann E, Mayser P et al.. Malassezia and the Skin: Science and Clinical Practice. Berlin: Springer Verlag, 2010. [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR et al.. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints, 2018. [DOI] [PMC free article] [PubMed]
- Bonnet SI, Binetruy F, Hernandez-Jarguin AM et al.. The tick microbiome: why non-pathogenic microorganisms matter in tick biology and pathogen transmission. Front Cell Infect Microbiol. 2017;7:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis K, Dobson SL, Xi Z et al.. Harnessing mosquito–Wolbachia symbiosis for vector and disease control. Acta Trop. 2014;132:S150–S63. [DOI] [PubMed] [Google Scholar]
- Brown SP, Ferrer A, Dalling JW et al.. Don't put all your eggs in one basket: a cost-effective and powerful method to optimize primer choice for rRNA environmental community analyses using the Fluidigm Access Array. Mol Ecol Resour. 2016;16:946–56. [DOI] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-theoretic Approach. New York: Springer Verlag, 2002. [Google Scholar]
- Busby AT, Ayllon N, Kocan KM et al.. Expression of heat shock proteins and subolesin affects stress responses, Anaplasma phagocytophilum infection and questing behaviour in the tick, IxodesScapularis. Med Vet Entomol. 2012;26:92–102. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ et al.. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al.. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpi G, Cagnacci F, Wittekindt NE et al.. Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One. 2011;6:e25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Reported cases of Lyme disease by state or locality, 2002–2011. 2013.
- Clarke KR. Non-parametric multivariate analyses of changes in community structure. Austral Ecology. 1993;18:117–43. [Google Scholar]
- Clay K, Klyachko O, Grindle N et al.. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol Ecol. 2008;17:4371–81. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Antunes S, Bonnet S et al.. Tick-Pathogen interactions and vector competence: identification of molecular drivers for Tick-Borne diseases. Front Cell Infect Microbiol. 2017;7:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva AM, Zeidner NS, Zhang Y et al.. Influence of outer surface protein A antibody on Borrelia burgdorferi within feeding ticks. Infect Immun. 1999;67:30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Sanchez S, Hernandez-Jarguin A, Torina A et al.. Characterization of the bacterial microbiota in wild-caught Ixodes ventalloi. Ticks Tick Borne Dis. 2019;10:336–43. [DOI] [PubMed] [Google Scholar]
- Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49:71–92. [DOI] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MJ, Russell JC, Davidson EN et al.. A 4-Yr survey of the range of ticks and Tick-Borne pathogens in the lehigh valley region of eastern pennsylvania. J Med Entomol. 2019;56:1122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson H, Wright W. Correction. J Wildl Dis. 1970;6:519. [Google Scholar]
- Emerson H, Wright W. The isolation of a Babesia in white-tailed deer. J Wildl Dis. 1968;4:142–4. [DOI] [PubMed] [Google Scholar]
- Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. [DOI] [PubMed] [Google Scholar]
- Gall CA, Reif KE, Scoles GA et al.. The bacterial microbiome of Dermacentor andersoni ticks influences pathogen susceptibility. ISME J. 2016;10:1846–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galuppi R, Tampieri M. Epidemiology and variability of Malassezia spp. Parassitologia. 2008;50:73–6. [PubMed] [Google Scholar]
- Georges K, Loria G, Riili S et al.. Detection of haemoparasites in cattle by reverse line blot hybridisation with a note on the distribution of ticks in Sicily. Vet Parasitol. 2001;99:273–86. [DOI] [PubMed] [Google Scholar]
- Gindin G, Samish M, Zangi G et al.. The susceptibility of different species and stages of ticks to entomopathogenic fungi. Experim Appl Acarol. 2002;28:283–8. [DOI] [PubMed] [Google Scholar]
- Hadziavdic K, Lekang K, Lanzen A et al.. Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS One. 2014;9:e87624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise SR, Elshahed MS, Little SE. Bacterial diversity in Amblyomma americanum (Acari: Ixodidae) with a focus on members of the genus Rickettsia. J Med Entomol. 2010;47:258–68. [DOI] [PubMed] [Google Scholar]
- Hughes GL, Dodson BL, Johnson RM et al.. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci U S A. 2014;111:12498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Torkelson JL, Bodnar J et al.. The Rickettsia endosymbiont of Ixodes pacificus contains all the genes of de novo folate biosynthesis. PLoS One. 2015;10:e0144552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde KD, Jones EBG, Liu J-K et al.. Families of Dothideomycetes. Fungal Diversity. 2013;63:1–313. [Google Scholar]
- Hynes WL. How ticks control microbes: innate immune responses. In: Sonenshine DE, Roe RM(eds). Biology of Ticks. New York: Oxford University Press, 2014, 129–46. [Google Scholar]
- Joseph E, Matthai A, Abraham LK et al.. Subcutaneous human dirofilariasis. J Parasit Dis. 2011;35:140–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaya GP, Hassan S. Entomogenous fungi as promising biopesticides for tick control. Experim Appl Acaro. 2000;24:913–26. [DOI] [PubMed] [Google Scholar]
- Kartashev V, Batashova I, Kartashov S et al.. Canine and human dirofilariosis in the rostov region (southern Russia). Vet Med Int. 2011;2011:685713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F, Ostfeld RS, Okanga S et al.. Consequences of integrating livestock and wildlife in an African savanna. Nature Sustainability. 2018;1:566–73. [Google Scholar]
- Kirkland BH, Westwood GS, Keyhani NO. Pathogenicity of entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae to Ixodidae tick species Dermacentor variabilis, Rhipicephalus sanguineus, and Ixodes scapularis. J Med Entomol. 2004;41:705–11. [DOI] [PubMed] [Google Scholar]
- Krehenwinkel H, Wolf M, Lim JY et al.. Estimating and mitigating amplification bias in qualitative and quantitative arthropod metabarcoding. Sci Rep. 2017;7:17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti TJ, Felsheim RF, Burkhardt NY et al.. Rickettsia buchneri sp. nov., a rickettsial endosymbiont of the blacklegged tick Ixodes scapularis. Int J Syst Evol Microbiol. 2015;65:965–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JY, Griggs R, Chicana B et al.. Vertical vs. horizontal transmission of the microbiome in a key disease vector, Ixodes pacificus. Mol Ecol. 2017;26:6578–89. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K et al.. Towards a unified paradigm for sequence‐based identification of fungi. Mol Ecol. 2013;22:5271–7. [DOI] [PubMed] [Google Scholar]
- Landesman WJ, Mulder K, Allan BF et al.. Potential effects of blood meal host on bacterial community composition in Ixodes scapularis nymphs. Ticks Tick Borne Dis. 2019;10:523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGiudice K, Ostfeld RS, Schmidt KA et al.. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proc Nat Acad Sci, USA. 2003;100:567–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu A, Pastor AR, Berkvens CN et al.. Babesia odocoilei as a cause of mortality in captive cervids in Canada. Can Vet J. 2018;59:52. [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Seiler KP, Eichwald EJ et al.. Distinct Characteristics of Resistance to Borrelia burgdorferi-Induced Arthritis in C57BL/6N Mice. Infect Immun. 1998;66:161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS, Nelson C, Hinckley A et al.. Estimating the public health burden of Lyme disease in the United States. International Conference on Lyme Borreliosis and Other Tick-Borne Diseases2013.
- Milnes EL, Thornton G, Leveille AN et al.. Babesia odocoilei and zoonotic pathogens identified from Ixodes scapularis ticks in southern Ontario, Canada. Ticks Tick Borne Dis. 2019;10:670–6. [DOI] [PubMed] [Google Scholar]
- Moreno CX, Moy F, Daniels TJ et al.. Molecular analysis of microbial communities identified in different developmental stages of Ixodes scapularis ticks from Westchester and Dutchess Counties, New York. Environ Microbiol. 2006;8:761–72. [DOI] [PubMed] [Google Scholar]
- Nadolny RM, Wright CL, Hynes WL et al.. Ixodes affinis (Acari: Ixodidae) in southeastern Virginia and implications for the spread of Borrelia burgdorferi, the agent of Lyme disease. J Vector Ecol. 2011;36:464–7. [DOI] [PubMed] [Google Scholar]
- Nakao R, Abe T, Nijhof AM et al.. A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks. The ISME journal. 2013;7:1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S, Fikrig E. Tick microbiome: the force within. Trends Parasitol. 2015;31:315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S, Rajeevan N, Liu L et al.. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host & Microbe. 2014;15:58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelakanta G, Sultana H, Fish D et al.. Anaplasma phagocytophilum induces Ixodes scapularis ticks to express an antifreeze glycoprotein gene that enhances their survival in the cold. J Clin Invest. 2010;120:3179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda H, Munderloh UG, Kurtti TJ. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. 1997;63:3926–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostfeld RS, Keesing F. Effects of host diversity on infectious disease. Annu Rev Ecol Evol Syst. 2012;43:157–82. [Google Scholar]
- Pal U, Yang X, Chen M et al.. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedmonte NP, Shaw SB, Prusinski MA et al.. Landscape features associated with blacklegged tick (Acari: Ixodidae) density and Tick-Borne pathogen prevalence at multiple spatial scales in central New York State. J Med Entomol. 2018;55:1496–508. [DOI] [PubMed] [Google Scholar]
- Piesman J. Dynamics of Borrelia burgdorferi transmission by nymphal Ixodes dammini ticks. J Infect Dis. 1993;167:1082–5. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Nakao R, Ohnuma A et al.. Microbial population analysis of the salivary glands of ticks; a possible strategy for the surveillance of bacterial pathogens. PLoS One. 2014;9: e103961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P et al.. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B et al.. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross BD, Hayes B, Radey MC et al.. Ixodes scapularis does not harbor a stable midgut microbiome. ISME J. 2018;12:2596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert S, Betts EL, Tsaousis AD. The symbiotic spectrum: where do the Gregarines fit? Trends Parasitol. 2019;35:687–94. [DOI] [PubMed] [Google Scholar]
- Rynkiewicz EC, Hemmerich C, Rusch DB et al.. Concordance of bacterial communities of two tick species and blood of their shared rodent host. Mol Ecol. 2015;24:2566–79. [DOI] [PubMed] [Google Scholar]
- Samish M, Rehacek J. Pathogens and predators of ticks and their potential in biological control. Annu Rev Entomol. 1999;44:159–82. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S et al.. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Nat Acad Sci, USA. 2012;109:6241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TA, Driscoll T, Gillespie JJ et al.. A Coxiella-like endosymbiont is a potential vitamin source for the Lone Star tick. Genome Biol Evol. 2015;7:831–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner FE, Pinger RR, Vann CN et al.. Infection and co-infection rates of Anaplasma phagocytophilum variants, Babesia spp., Borrelia burgdorferi, and the rickettsial endosymbiont in Ixodes scapularis (Acari: Ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J Med Entomol. 2014;45:289–97. [DOI] [PubMed] [Google Scholar]
- Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol. 2000;66:5066–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Tagliafierro T, Sameroff S et al.. Microbiome analysis of Ixodes scapularis ticks from New York and Connecticut. Ticks Tick Borne Dis. 2019;10:894–900. [DOI] [PubMed] [Google Scholar]
- Van Treuren W, Ponnusamy L, Brinkerhoff RJ et al.. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl Environ Microbiol. 2015;81:6200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu VH, Hong SI, Kim K. Selection of entomopathogenic fungi for aphid control. J Biosci Bioeng. 2007;104:498–505. [DOI] [PubMed] [Google Scholar]
- Waldrup K, Kocan A, Barker R et al.. Transmission of Babesia odocoilei in white-tailed deer (Odocoileus virginianus) by Ixodes scapularis (Acari: Ixodidae). J Wildl Dis. 1990;26:390–1. [DOI] [PubMed] [Google Scholar]
- Wang G, Liveris D, Brei B et al.. Real-Time PCR for simultaneous detection and quantification of borrelia burgdorferi in Field-Collected Ixodes scapularis Ticks from the Northeastern United States. Appl Environ Microbiol. 2003;69:4561–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster J, Weber RWS. Introduction to Fungi. Third Edition Cambridge: Cambridge University Press, 2007. [Google Scholar]
- Weiss B, Aksoy S. Microbiome influences on insect host vector competence. Trends Parasitol. 2011;27:514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss BL, Maltz MA, Vigneron A et al.. Colonization of the tsetse fly midgut with commensal Kosakonia cowanii Zambiae inhibits trypanosome infection establishment. PLoS Pathog. 2019;15:e1007470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Goldberg MS, Popova TG et al.. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol Microbiol. 2000;37:1470–9. [DOI] [PubMed] [Google Scholar]
- Yue M, Schmieder R, Edwards RA et al.. Microfluidic PCR combined with pyrosequencing for identification of allelic variants with phenotypic associations among targeted Salmonella genes. Appl Environ Microbiol. 2012;78:7480–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Jasinskas A, Barbour AG. Antibiotic treatment of the tick vector Amblyomma americanum reduced reproductive fitness. PLoS One. 2007;2:e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnik CP, Falco RC, Daniels TJ et al.. Transient influence of blood meal and natural environment on blacklegged tick bacterial communities. Ticks Tick Borne Dis. 2018;9:563–72. [DOI] [PubMed] [Google Scholar]
- Zolnik CP, Prill RJ, Falco RC et al.. Microbiome changes through ontogeny of a tick pathogen vector. Mol Ecol. 2016;25:4963–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.