Abstract
Advanced Lipoxidation End-products (ALEs) are modified proteins that can act as pathogenic factors in several chronic diseases. Several molecular mechanisms have so far been considered to explain the damaging action of ALEs and among these a pathway involving the receptor for advanced glycation end products (RAGE) should be considered. The aim of the present work is to understand if ALEs formed from lipid peroxidation derived reactive carbonyl species (RCS) are able to act as RAGE binders and also to gain a deeper insight into the molecular mechanisms involved in the protein-protein engagement. ALEs were produced in vitro, by incubating human serum albumin (HSA) with 4-hydroxy-trans− 2-nonenal (HNE), acrolein (ACR) and malondialdehyde (MDA). The identification of ALEs was performed by MS. ALEs were then subjected to the VC1 Pull-Down assay (VC1 is the ligand binding domain of RAGE) and the enrichment factor (the difference between the relative abundance in the enriched sample minus the amount in the untreated one) as an index of affinity, was determined. Computation studies were then carried out to explain the factors governing the affinity of the adducted moieties and the site of interaction on adducted HSA for VC1-binding. The in silico analyses revealed the key role played by those adducts which strongly reduce the basicity of the modified residues and thus occur at their neutral state at physiological conditions (e.g. the MDA adducts, dihydropyridine-Lysine (DHPK) and N-2-pyrimidyl-ornithine (NPO), and acrolein derivatives, N-(3-formyl-3,4-dehydro-piperidinyl) lysine, FDPK). These neutral adducts become unable to stabilize ion-pairs with the surrounding negative residues which thus can contact the RAGE positive residues.
In conclusion, ALEs derived from lipid peroxidation-RCS are binders of RAGE and this affinity depends on the effect of the adduct moiety to reduce the basicity of the target amino acid and on the acid moieties surrounding the aminoacidic target.
Keywords: Advanced lipoxidation end products (ALEs), Human serum albumin (HSA), RAGE, Pull-down assay, VC1 domain, Reactive Carbonyl Species (RCS), 4-hydroxy-trans− 2-nonenal (HNE), Acrolein (ACR) and malondialdehyde (MDA)
Graphical abstract
Highlights
-
•
A wide set of ALEs-HSA was obtained by in vitro incubation of HSA with different RCS.
-
•
ALEs-HSA before and after VC1 enrichment were fully characterized by MS.
-
•
Retention efficiency of the identified ALEs-HSA by VC1 was determined.
-
•
Elucidation of structural requirements making an ALE a RAGE binder was obtained by computational studies.
-
•
The mechanism here proposed for ALEs can be considered as a general mechanism of protein-protein interaction.
1. Introduction
The oxidative degradation of lipids (lipid peroxidation) results in the formation of a wide variety of break-down products including small molecules containing a carbonyl moiety and characterized by chemical reactivity and for this reason called reactive carbonyl species (RCS) [1]. Lipid-derived reactive carbonyl species (RCS) are quite heterogeneous, belonging to different chemical classes including α,β-unsaturated aldehydes [4-hydroxynonenal (HNE), acrolein (ACR)], keto-aldehydes [methylglyoxal (MGO), 4-oxo-nonenal (ONE)] and di-aldehydes [malondialdehyde (MDA) and glyoxal (GO)] [2]. RCS react with different nucleophilic substrates and in particular with the nucleophilic amino acids of protein (arginine, lysine, cysteine and histidine) through a reaction called protein lipoxidation and involving the carbonyl moiety and the electrophilic center (e.g. the C3 of an α,β-unsaturated moiety). The reaction products between proteins and RCS, advanced lipoxidation products (ALEs), are now recognized not only as biomarker of RCS formation but also as bioactive/damaging biomolecules [3], [4]. Moreover, RCS are currently recognized as being involved in the onset and progression of several diseases including diabetic retinopathy [5], atherosclerosis [6], renal disease [7] and metabolic disorders [8]. Based on their pathogenetic role, ALEs are now considered as promising drug target and molecules effective in preventing ALE formation have been reported to have beneficial effects in some animal models [9], [10]. However, it should be considered that the effect of protein lipoxidation and of RCS can be double-sided, because besides a damaging mechanism as above mentioned, in some conditions and depending on their levels, they can exert protective effects associated with the induction of antioxidant defense mechanisms [11].
Several molecular mechanisms have so far been considered to explain the damaging action of ALEs, which can include one of the following mechanisms, depending on the damaging process and the target protein itself: protein dysfunction, protein oligomerization, signal transduction and immune response [3], [6]. Moreover, adduction of RCS to proteins can also lead to the formation of ALEs acting as binders and activators of some receptors as in the case of galectin-3, a glycan binding protein which has been suggested to aid in the removal of circulating AGEs and ALEs [12].
ALEs which are formed from RCS deriving either from lipid and reducing sugar oxidation, such as glyoxal and methylglyoxal, (in this paper called AGEs/ALEs) are known to act as binders and activators of the receptor RAGE [13], [14]. RAGE is a type I cell surface receptor that is expressed in several cells, such as endothelial cells, smooth muscle cells, but also dendritic cells and T-lymphocytes and is predominantly located in the lungs [15]. RAGE has been involved in many different pathologies with a marked oxidative base, such as diabetes, atherosclerosis, neurodegenerative diseases and many different ligands of RAGE have been identified, such as amyloid β peptide, S100/calgranulin protein, HMGB1 [16]. Two different pathways can be activated upon binding to the receptor: 1) the activation of the NADPH oxidase, resulting in the production of reactive oxygen species (ROS), which are detrimental to the cells and 2) the activation of the NF-κB pathway leading to a sustained pro-inflammatory and pro-fibrotic response [17].
However, there are some ALEs (in this paper called as ALEs-lipox), widely detected in several oxidative based and inflammatory diseases [6], [18], which are formed by RCS only deriving from a lipid peroxidation process, such as MDA, HNE and ACR. For these ALEs-lipox very few data are available on their RAGE interaction. Shanmugam et al. [19] reported that synthetic ALE (malondialdehyde-lysine [MDA-Lys]) induces oxidative stress and also activates the transcriptional factor nuclear factor-kB (NF-kB) in THP-1 monocytes partly via the receptor for AGEs (RAGE).
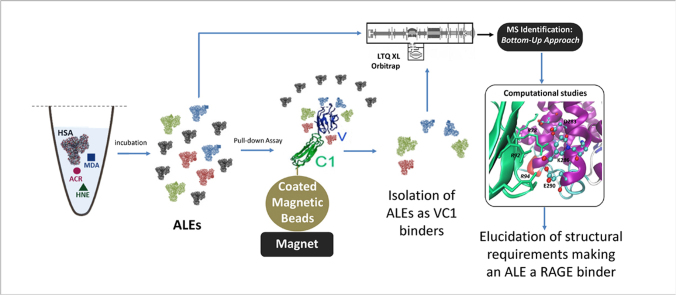
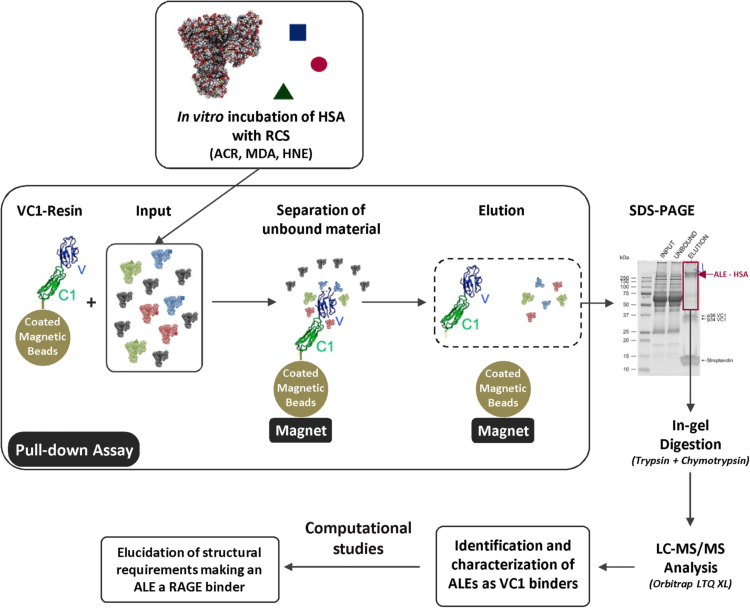
The aim of the present paper is to understand whether ALEs-lipox are able to act as RAGE binders as do AGEs and AGEs/ALEs and also to gain a deeper insight into the molecular mechanisms involved in the protein-protein engagement. Our starting hypothesis, the involvement of ALEs-lipox in RAGE activation, is supported by several facts such as that ALEs/AGEs and ALEs-lipox share some structural properties such as the covalent modifications by aldehydes of nucleophilic residues and that ALEs-lipox are pro-inflammatory and pro-fibrotic compounds activating the NF-kB pathway, a mechanism which could be addressed to a RAGE activation pathway. Fig. 1 shows the work flow of the study.
Fig. 1.
Work flow of the study. ALEs-lipox were firstly prepared by incubating HSA with lipid peroxidation derived RCS (HNE, ACR and MDA) and then fully characterized by MS. The RAGE binding ability of each identified ALE was then determined by using the VC1 assay as previously reported [20]. Computation studies were then carried out to explain the factors governing the affinity of the adducted moieties and the site of interaction on adducted HSA for VC1-binding.
To test this hypothesis, a set of ALEs was firstly prepared and fully characterized by MS. ALEs were formed using the well-known lipid peroxidation derived RCS and in particular HNE, ACR and MDA. Since ALEs are quite heterogeneous also when formed by a single attacking RCS, for each tested RCS the different ALEs were fully identified by a bottom-up MS approach in terms of adducted moiety and modification site. The RAGE binding ability of each identified ALE was then determined using the VC1 assay as previously reported [20]. For each identified ALE the VC1 binding ability was then related to the variation in the ionization state of the adducted residues as well as to the abundance of surrounding negative residues that, after the ALE generation, become available for RAGE binding. Computation studies were then carried out to explain the factors governing the affinity of the adducted moieties and the site of interaction on adducted HSA for VC1-binding.
The overall data permit the elucidation of the structural requirements for ALEs to become RAGE binders together with the molecular mechanisms involved in the protein-protein engagement.
2. Materials and methods
2.1. Reagents
Formic acid (FA), trifluoroacetic acid (TFA) and acetonitrile (ACN) were LC-MS grade; sodium dodecyl sulfate (SDS), ammonium bicarbonate, malondialdehyde tetrabutylammonium salt (MDA-TS), acrolein (ACR), HEPES, NaCl, sodium dihydrophosphate, disodium phosphate and all other chemicals were analytical grade and purchased from Sigma-Aldrich (Milan, Italy). 4-Hydroxy-2-trans-nonenal dimethylacetal (HNE-DMA, catalog Number H9538) was purchased from Sigma-Aldrich (Milan, Italy) and recombinant HSA expressed in P. pastoris were purchased from Sigma Aldrich (Milan, Italy). Streptavidin-coated magnetic beads (Streptavidin Mag Sepharose™) were purchased from GE Healthcare (Milan, Italy)
Ultrapure water was prepared by a Milli-Q purification system (Millipore, Bedford, MA, USA).
Any KD ™ Mini Protean® TGX ™ precast gel, Standard Precision Plus prestained protein standards, Laemmli sample buffer (2 ×/ 4 ×), Running buffer and Bio-Safe Coomassie, together with the threo-1,4-Dimercapto-2,3-butanediol (DTT) and iodoacetamide (IAA) were supplied by Bio-Rad Laboratories, Inc. Trypsin and Chymotrypsin sequencing grade were purchased from Roche Diagnostics SpA (Monza, Italy).
Digestion buffer was 50 mM ammonium bicarbonate; destaining solution was prepared mixing acetonitrile with digestion buffer (1:1 v/v); reducing solution was 10 mM DTT in digestion buffer; alkylating solution was 55 mM iodoacetamide in digestion buffer; extraction solution was prepared as follows: 3% TFA/30% ACN in H2O MilliQ.
2.2. In vitro generation of ALEs-HSA
4-hydroxy-trans− 2-nonenal (HNE) was prepared as previously described [21]. HSA modified with MDA was prepared dissolving HSA in 10 mM phosphate buffer pH 7.4 at a concentration equal to 100 μM (6.7 mg ml−1). HSA was incubated, as previously described [20] in the dark at 37 °C and 400 rpm and using the following molar ratios between protein and MDA: 1:6.3, 1:63, 1:630, 1:6300, 1: 12600. HSA modified with ACR or HNE was prepared dissolving HSA in 10 mM phosphate buffer pH 7.4 at a concentration equal to 75 μM (5 mg ml−1). HSA was incubated in the dark at 37 °C and at 400 rpm in molar ratios protein: ACR equal to 1:10, 1:100, 1:1000, 1:2500 and 1:5000 and in molar ration protein: HNE equal to 1:10, 1:100, 1:200, 1:1000 and 1:2000. HSA incubated without RCS was used as a control untreated sample. The reactions were stopped after 24, 48 or 72 h removing the excess of RCS by ultrafiltration using Amicon Ultra filter units 0.5 ml, cut-off 10 kDa (Millipore).
2.3. Intact protein analysis by MS
ALEs obtained by incubating HSA with RCSs were analyzed by direct infusion on a triple-quadrupole (TQ) mass spectrometer (Finnigan TSQ Quantum Ultra, ThermoQuest, Milan, Italy) equipped with an Electrospray Finnigan Ion Max source. For MS analyses, samples were desalted by using Amicon Ultra filter units 0.5 ml, cut-off 10 kDa (Millipore) and washed three times with water. Samples were then diluted to 1 mgml−1 with a final composition of CH3CN–H2O–HCOOH (30:70:0.1, v/v/v). Aliquots of 50 μl were injected into the mass spectrometer at a flow rate of 25 μl min−1 by using a ThermoQuest autosampler. Each sample was analyzed for 5 min under the following instrumental conditions: positive-ion mode; ESI voltage 3.5 kV, capillary temperature 350 °C, Q3 scan range 1200–1500 m/z, Q3 power 0.4 amu, scan time 1 s, Q2 gas pressure 1.5 Torr, skimmer offset 10 V, microscan set to 3. Full instrument control and ESI mass spectra acquisitions were carried out by Xcalibur software (version 2.0.7, Thermo Fisher Scientific, Rodano, MI, Italy). Mass spectra deconvolution was performed using MagTran software (version 1.02) [22].
2.4. VC1 pull-down assay
VC1-His-Strep was expressed and purified from Pichia pastoris culture supernatant as previously described [23]. The recombinant protein was immobilized on streptavidin-coated magnetic beads by exploiting the affinity of the Strep tag towards streptavidin. In order to obtain the VC1-resin, 50 μg of purified VC1-His-Strep in 170 μl of 20 mM HEPES pH 7.1, 100 mM NaCl were added to 5 μl of packed streptavidin coated-beads, previously equilibrated with the same buffer. A volume of 170 μl of 20 mM HEPES pH 7.1, 100 mM NaCl were added to the same amount of beads in a different tube in order to obtained the Control-resin. After 1 h of incubation at 4 °C on a rotary mixer, the unbound material was carefully removed and the magnetic beads were washed with 500 μl of buffer (20 mM HEPES pH 7.1, 100 mM NaCl). The VC1- and Control-resin were incubated for 1 h at 4 °C with 160 μl of ALE-HSA or untreated HSA at the concentration of 125 μg ml−1 in 20 mM HEPES pH 7.1, 100 mM NaCl. The unbound material was carefully removed and the beads were washed twice with 500 μl of Buffer (20 mM HEPES pH 7.1, 100 mM NaCl). The elution was performed by boiling the beads for 5 min in 15 μl of Laemmli Sample Buffer 4x mixed with 400 mM DTT, then with other 15 μl of buffer (20 mM HEPES pH 7.1, 100 mM NaCl). The two eluates were pooled.
2.5. Electrophoretic procedures
The fractions obtained from pull-down experiments were analyzed by SDS PAGE. To 20 μl of input and unbound fractions, 7 μl of Laemmli Sample Buffer 4x mixed with 400 mM DTT were added. The samples were denatured incubating for 5 min at 95 °C. Input, unbound and elution samples were separated by SDS-PAGE on Any KD ™ Mini Protean® TGX ™ precast gels and stained with Bio-Safe Coomassie blue (Bio-Rad). Images were acquired using the calibrated densitometer GS-800 and analyzed by the software Quantity one (Bio-Rad).
2.6. ALE-HSA in-gel digestion
Proteins bands corresponding to ALEs-HSA and obtained by incubating HSA with HNE, ACR and MDA (input), and those cut from the fraction retained by VC1, were excised from gels using a scalpel, finely chopped, transferred to a new eppendorf and washed with 200 μl of digestion buffer. An aliquot of 200 μl of destaining solution was added to each gel portion and heated at 37 °C for 10 min in the thermomixer (1400 rpm); the destaining solution was then discarded and this step was repeated until destaining was completed. Afterwards, gel pieces were incubated with 150 μl of reducing solution at 56 °C for 1 h and then with 150 μl of alkylating solution at room temperature for 45 min in the dark. In-gel digestion of ALE-HSA adducts was performed by overnight-incubation at 37 °C with 1 µg of sequencing-grade trypsin (Roche) dissolved in digestion buffer. ALE-HSA samples were also subjected to a second digestion by a sequencing-grade chymotrypsin (1 µg) for 7 h at 25 °C in the presence of calcium chloride (10 mM). The peptide mixtures were acidified with formic acid up to a final concentration of 1%.
To guarantee better protein detection, peptide mixtures were extracted by a 10 min-incubation with extraction solution and by an additional 10 min-incubation with pure acetonitrile. The two extracts were combined and dried in a vacuum concentrator (Martin Christ.). Digested peptide mixtures were then dissolved in an appropriate volume (20 μl) of 0.1% formic acid for mass spectrometry (MS) analysis.
2.7. Mass spectrometry analyses
Peptides from the in-gel digestion were separated by reversed-phase (RP) nanoscale capillary liquid chromatography (nanoLC) and analyzed by electrospray tandem mass spectrometry (ESI-MS/MS). For each analysis 5 μl of solubilized peptides were injected onto a C18HALO PicoFrit column (75 mM x 10 cm, 2.7 mM particles, pores 100 Å, New Objective, USA) by means of an autosampler. Samples were loaded onto the fused silica column at 400 nl/min of mobile phase consisting of 99% of phase A and 1% of phase B (0.1% HCOOH in CH3CN) for 15 min. Peptide separation was performed with a 55 min linear gradient of phase B (1–35%). The separative gradient was followed by 5 min at 80% of phase B to rinse the column, and 15 min of 99% of phase A and 1% of phase B served to re-equilibrate the column to the initial conditions. The nano-chromatographic system, an UltiMate 3000 RSLCnano System (Dionex), was connected to an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific Inc., Milan, Italy) equipped with a nanospray ion source (dynamic nanospray probe, Thermo Scientific Inc., Milan, Italy) set as follows: positive ion mode, spray voltage 1.8 Kv; capillary temperature 220 °C, capillary voltage 35 V; tube lens offset 120 V. The LTQ-Orbitrap XL mass spectrometer was operated in data-dependent acquisition mode (DDA) to acquire both the full MS spectra and the MS/MS spectra. Full MS spectra were acquired in "profile" mode, by the Orbitrap (FT) analyzer, in a scanning range between 300 and 1500 m/z, using a capillary temperature of 220 °C, AGC target = 5 × 105 and resolving power 60,000 (FWHM at 400 m/z). Tandem mass spectra MS/MS were acquired by the Linear Ion Trap (LTQ) in CID mode, automatically set to fragment the nine most intense ions in each full MS spectrum (exceeding 1 ×104 counts) under the following conditions: centroid mode, isolation width of the precursor ion of 2.5 m/z, AGC target 1 × 104 and normalized collision energy of 35 eV. Dynamic exclusion was enabled (exclusion dynamics for 45 s for those ions observed 3 times in 30 s). Charge state screening and monoisotopic precursor selection were enabled, singly and unassigned charged ions were not fragmented. Xcalibur software (version 2.0.7, Thermo Scientific Inc., Milan, Italy) was used to control the mass spectrometer.
2.8. Identification and localization of protein adducts
The software Proteome Discoverer (version 1.3.0.339, Thermo Scientific, USA), implemented with the algorithm SEQUEST, was used to compare the experimental full and tandem mass spectra with the theoretical ones obtained by the in silico digestion of the HSA sequence (Uniprot P02768). Trypsin and chymotrypsin were selected as the cleaving proteases, allowing a maximum of 2 missed cleavages. Peptide and fragment ion tolerances were set to 5 ppm and 10 mmu, respectively. Cysteine carbamidomethylation was set as fix modification (+57.02147); methionine oxidation was allowed as a variable modification in addition to the known HNE-, ACR- or MDA-derived modifications as listed in Table S1.
As a quality filter, only peptide with an Xcore value grater then 2.2 for doubly-charged peptides, 2.5 for triply-charged, 2.75 for quadruply-charged peptide ions, and 3 for charge states quintuple or higher were considered as genuine peptide identifications. To ensure the lowest number of false positives, the mass values experimentally recorded were further processed through a combined search with the Database Decoy, where the protein sequences are inverted and randomized. This operation allows the calculation of the false discovery rate (FDR) for each match, so that all the proteins out of range of FDR between to 0.01 (strict) and 0.05 (relaxed) were rejected.
For the localization of ALE-deriving modifications, the MS/MS spectra of modified peptides were manually inspected; for the confident mapping of the modification sites, spectra were requested to match the expected ions (b and/or y) neighboring the modified amino acid residue both at the N- and C-termini.
2.9. Semi-quantitative analysis of ALE-HSA adducts
The relative extent of each protein modification has been calculated by determining the amount of the modified peptide in respect to the native one, by assuming that the ionization efficiency of the native and the modified peptides are equal. In particular, the single ion traces (SIC) of the native and modified peptides were firstly reconstituted by setting as filter ion the m/z values of the corresponding precursor protonated peptides. The peak areas were then automatically calculated by the Qual Browser tool of the Xcalibur data system (version 2.0.7, Thermo Scientific Inc., Milan, Italy) and then the relative abundance calculated by using the Eq. (1).
| (1) |
The relative abundance of each modified peptide was determined in both the input and enriched samples. The retention efficiency of each identified ALEs towards VC1 was then determined by the Enrichment Factor value, calculated as the differences between the relative abundance in the enriched sample minus the amount in the untreated one (Enrichment factor= %VC1-%NoVC1).
2.10. Computational studies
The prediction of the pK values of the simulated adducts was performed by PM7-based semi-empirical calculations using MOPAC [24]. The simulations involved simplified model compounds by focusing on the adducted side chains and neglecting the backbone atoms. Specifically, the predictions involved the FDPK, mono and double N-propenal MA adducts of lysine. Indeed and as experimentally known, 2-amino pyrimidine and dihydropyridine (i.e., NPO and DHPK adducts) are neutral at physiological pH, while cyclic guanidines (see HTP0) retain the strong basicity of the guanidine group. With regard to HSA protein, the retrieved resolved structure (PDB Id 1A06) was used, after a preliminary optimization, to calculate the negative residues within a 5 Å radius sphere around each adducted residues and to generate the corresponding adducts as induced by MDA e ACR. As discussed in the Results section, two adducted HSA structures were manually generated using the VEGA suite of program [25]: the first carrying all the ACR-based adducts and the second structure with all the MDA-based adducts. The analysis was focused on Arg and Lys adducts only because they are numerous enough to derive a sort of structure-affinity relationships (SAR). Similarly, these analyses do not consider the HNE-induced modifications because they are too limited to develop clear relationships. In order to allow a suitable reorganization of the environments around all inserted adducts, the minimized adducted HSA structures underwent 1 ns of MD simulations by Namd [26] keeping the backbone atoms constrained to avoid excessive distortions of the resolved folding. With regard to RAGE structure, the NMR-derived V structure (PDB Id: 2mov) was prepared and utilized as done in previous studies. Protein-protein docking was performed using GRAMM [27] with the default parameters and generating 1000 complexes for each adducted HSA structure.
3. Results
3.1. Intact protein analysis of HSA and ALEs-HSA by MS
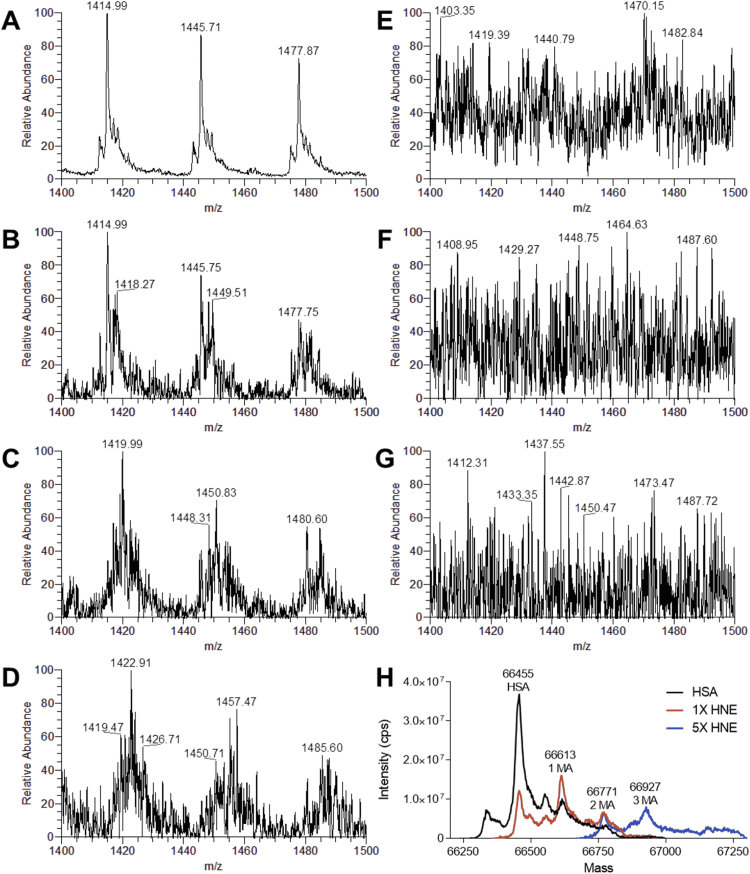
In order to investigate the interaction between RAGE and ALEs-lipox, different ALEs-lipox were produced in-vitro by incubating HSA with different concentrations of the well-known lipid derived RCS and in particular: acrolein (ACR), malondialdehyde (MDA) and 4-hydroxy-trans− 2-nonenal (HNE). After 24, 48 and 72 h, aliquots of the incubation mixtures were withdrawn, and the reaction was stopped by removing the excess of RCS by ultrafiltration. Intact protein analysis by direct infusion MS was used to evaluate the extent of HSA modifications. Fig. 2 shows the spectra of native HSA (panel A) and HSA incubated with increasing molar ratios of HNE [1:1 (B), 1:5 (C), 1:10 (D), 1:100 (E), 1:200 (F) and 1:1000 (G)]. Panel A) shows the MS-spectrum (mass range between m/z 1400 and 1500) of native and non-modified HSA, characterized by three sharp multicharged ions at m/z 1414.99, 1445.71 and 1477.87 relative to the three multicharged ions at 47 + , 46 + and 45 + . When HSA was incubated with HNE in a 1:1 M ratio, besides the three peaks relative to native HSA, a new set of peaks at m/z 1418.43, 1449.15, and 1481.40 is detectable and relative to the HNE Michael adduct of native HSA (MW shifted by 156 Da), as confirmed in the deconvoluted spectrum (panel H). The number of adducted HNE moieties per molecule of HSA increases proportionally with the increase of molar ratios reaching 3 and 5 HNE moles per mole of HSA at 1:5 and 1:10 HSA: HNE molar ratios, as also confirmed by the deconvoluted spectra (panel H shows the deconvoluted spectra HSA incubated in the presence of HNE at a 1:5 M ratio). At higher HNE molar ratios (1:100, 1:200 and 1:1000, panels E, F and G, respectively) the MS spectra do not show any detectable peaks due the formation of such a large number of adducts and consequently of so many ions which cover the m/z scan range thus eliminating the detection of sharp ions.
Fig. 2.
Direct infusion ESI-MS analysis of native and HNE-modified HSA. Mass spectra of HSA recorded in a mass range between m/z 1400 and 1500. A) Native HSA shows sharp intense peaks referred to the charge ions 47 + , 46 + and 45 + ; the deconvoluted spectrum reports a MW 66,455 Da (H). When HSA is reacted with HNE at increasing molar ratios 1:1 (B), 1:5 (C), 1:10 (D), additional peaks relative to HNE adducts appear. At higher molar ratios 1:100 (E), 1:200 (F) and 1:1000 (G) the MS spectra lose resolution and become flat due to the presence of multiple adducts. H) Deconvoluted spectra showing the MS of HSA and protein adducts. MS spectra relative to HSA incubated with HNE at 1:100 M ratio and higher cannot be deconvoluted due to the extent of modification.
Intact protein analysis was also performed on ALEs-lipox formed by incubating HSA with ACR and MDA, showing the same stepwise increase of modifications with the higher amount of RCS incubated with HSA (Supplementary Figs. 1 and 2).
Intact protein analysis well indicates that by using a wide range of molar ratios and different time-points a quite wide array of ALEs for each tested RCS was generated.
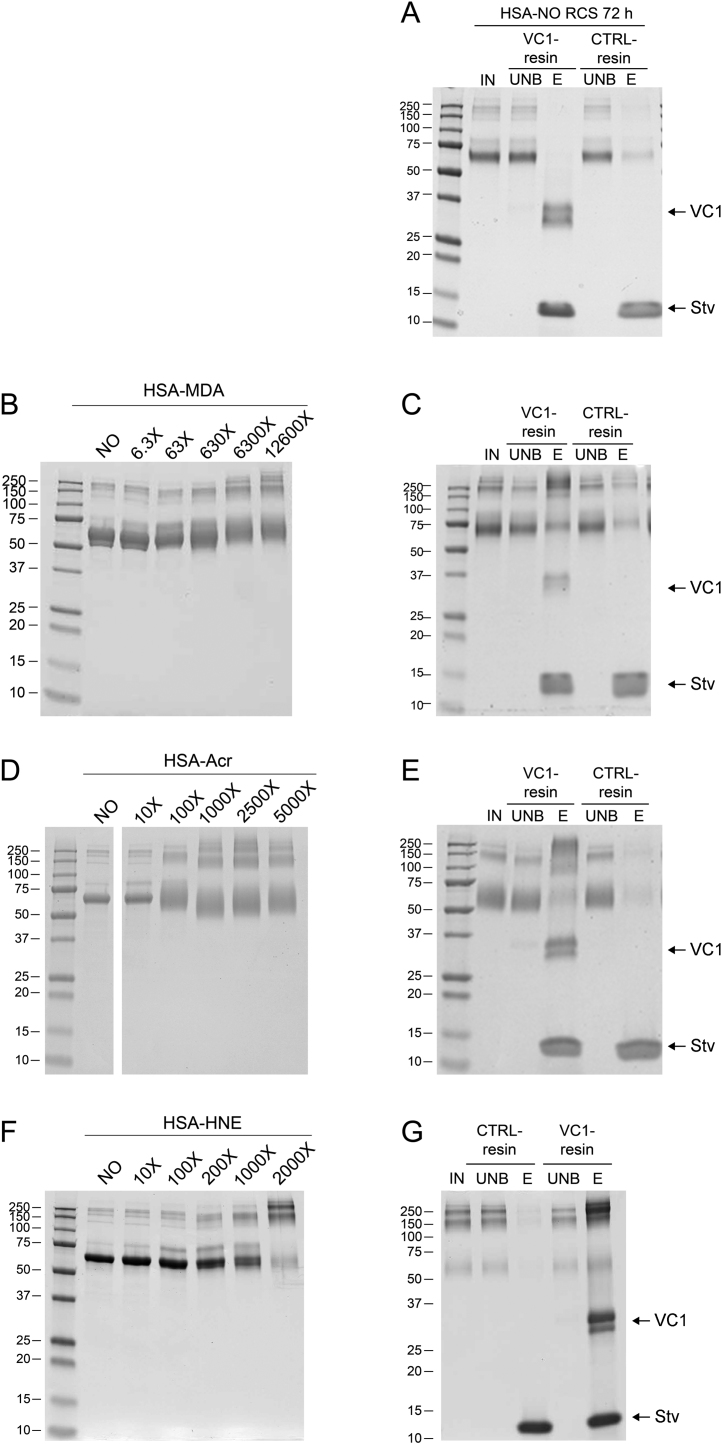
3.2. Pull-down assay with modified albumins
In order to characterize ALE-lipox modifications selectively enriched by RAGE, we performed a VC1 pull-down assay as previously described [20]. HSA and HSA treated with MDA, ACR or HNE were assayed for binding to VC1-resins and to control resin. As expected, unmodified HSA was not retained by the VC1-resin (Fig. 3, panel A). HSA modified by MDA and ACR are characterized by a different migration pattern on a SDS-PAGE analysis, with the appearance of oligomeric bands proportional to the increase of the HSA-RCS molar ratio (Fig. 3, panel B: HSA-MDA at 72 h, panel D: HSA-ACR 72 h, panel F: HSA-HNE 72 h). At increasing molar ratios and incubation time, higher amounts of albumin modified with MDA or ACR were eluted from the VC1 resin, with a predominance of the high molecular weight (HMW) species. The modified albumins were retained by the VC1-resin, but not by the control resin, as shown in Fig. 3 (panel C: HSA-MDA molar ratio 1:12,600, 72 h; panel: E HSA-ACR molar ratio 1:5000, 72 h, panel: G HSA-HNE molar ratio 1:2000, 72 h). The time course analysis and the pull-down experiments with HSA-MDA have previously been published [20]. The time course analysis and the pull-down experiments with HSA-ACR and HSA-HNE are reported in Supplementary Fig. 3 and Supplementary Fig. 4 respectively.
Fig. 3.
Modified albumin obtained by incubation of recombinant HSA in the presence of different molar ratio of RCS and VC1 pull-down assay. A) VC1 pull-down assay with untreated HSA. SDS PAGE analysis followed by Coomassie staining of the modified HSA obtained by 72 h incubation with the indicated molar ratio of RCS (panel B: MDA; panel D: ACR; panel F: HNE). The highest molar ratio of HSA-MDA (1:12,600), HSA-ACR (1:5000) or HSA-HNE (1:2000) were used as input (IN) in the pull-down assays with the VC1 and control (CTRL)-resins. The IN fractions, the unbound fractions (UNB) and eluates (E) were analyzed by SDS-PAGE followed by Coomassie staining. The gels show that untreated HSA does not bind VC1 (panel A), whereas high MW species of HSA-MDA (panel C), HSA-ACR (panel E) and HSA-HNE (panel G) are retained by the VC1-resin, but not by the CTRL-resin. Since the elution is performed in denaturing conditions, this step removes any associated molecule from the resin, including the two VC1 glycovariants (34 and 36 kDa) and streptavidin (Stv, 14 kDa), indicated by arrows.
3.3. Identification and localization of protein adducts by mass spectrometry
ALEs-lipox in the reaction mixtures and those enriched by VC1 were analyzed by bottom-up MS in order to identify the PTMs and to localize the amino acid residues involved in the protein adduct formation.
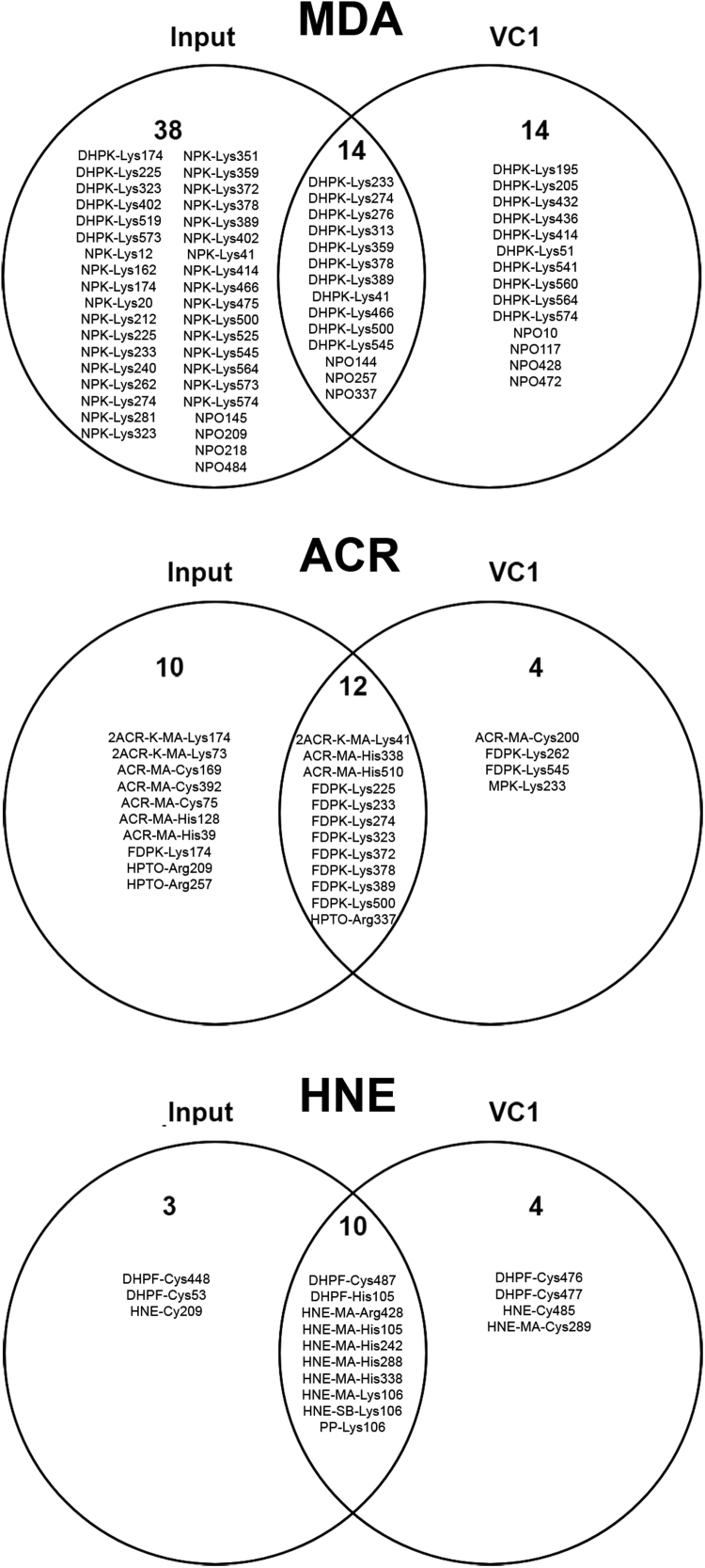
Table 1, Table 2 summarize the identified ALEs before and after VC1 enrichment, respectively. It should be noted that the tables summarize the overall PTMs identified at different molar ratios and incubation times. The ALEs not retained (identified only in the input samples), retained (identified after VC1 enrichment) and present in both input and elution samples are reported in the Venn diagrams of Fig. 4.
Table 1.
List of the PTMs identified on the indicated Amino acid residues in the ALEs samples obtained by in vitro incubation of HSA with RCS.
| PTMs | Amino Acid residue | |
|---|---|---|
| MDA | N-propenal-Lysine(NPK) | Lys12, Lys20, Lys41, Lys162, Lys174, Lys212, Lys225, Lys233, Lys140, Lys262, Lys274, Lys281, Lys323, Lys359, Lys351, Lys372, Lys378, Lys389, Lys402, Lys414, Lys466, Lys475, Lys500, Lys525, Lys545, Lys564, Lys573, Lys574 |
| dihydropyridine-lysine(DHPK) | Lys41, Lys174, Lys225, Lys233, Lys274, Lys276, Lys313, Lys323, Lys359, Lys378, Lys389, Lys402, Lys466, Lys500, Lys519, Lys545, Lys573 | |
| N-2-pyrimidyl-ornithine (NPO) | Arg209, Arg218, Arg144, Arg145, Arg257, Arg337, Arg484 | |
| ACR | Michael Adduct (N-propanal derivative)(ACR-MA) | His39, Cys75, His128, Cys169, His338, Cys392, His510 |
| N-2-(4 hydroxy-tetrahydro-pyrimidyl) ornitine | ||
| (propane-arginine, HTPO) | Arg209, Arg257, Arg337 | |
| N,N-bispropenal-Lysine(2ACR-K-MA) | Lys41, Lys73, Lys174 | |
| N-(3-formyl-3,4-dehydro-piperidinyl) lysine(FDPK) | Lys174, Lys225, Lys233, Lys274, Lys323, Lys372, Lys378, Lys389, Lys500 | |
| HNE | Michael adduct (HNE-MA) | His105, Lys106, His242, His288, His338, Arg428 |
| Schiff Base (HNE-SB) | Lys106 | |
| Novel cyclic adduct (HNE-CY) | Arg209 | |
| 2-pentyl-pyrrole (PP) | Lys106 | |
| 2,3 dihydro-pentyl-furan (DHPF) | Cys53, His105, Cys448, Cys487 |
Table 2.
List of the PTMs identified on the indicated Amino acid residues in the ALEs-HSA samples retained by VC1.
| PTMs | Amino Acid residue | |
|---|---|---|
| MDA | dihydropyridine-lysine(DHPK) | Lys41, Lys51, Lys195, Lys205, Lys233, Lys274, Lys276, Lys313, Lys359, Lys378, Lys389, Lys414, Lys432, Lys436, Lys466, Lys500, Lys541, Lys545, Lys560, Lys564, Lys574 |
| N − 2-pyrimidyl-ornithine (NPO) | Arg10, Arg117, Arg144, Arg257, Arg337, Arg428, Arg472 | |
| ACR | Michael Adduct (N-propanal derivative)(ACR-MA) | Cys200, His338, His510 |
| N − 2-(4 hydroxy-tetrahydro-pyrimidyl) ornitine (propano-arginine, HTPO) | Arg337 | |
| N,N-bispropenal-Lysine(2ACR-K-MA) | Lys41 | |
| N-(3-formyl−3,4-dehydro-piperidinyl) lysine(FDPK) | Lys225, Lys233, Lys262, Lys274, Lys323, Lys372, Lys378, Lys389, Lys500, Lys545 | |
| N-(3-Methylpyridinium)-lysine (MPK) | Lys 233 | |
| HNE | Michael adduct (HNE-MA) | His105, Lys106, His242, Cys289, His288, His338, Arg428 |
| Schiff Base (HNE-SB) | Lys106 | |
| Novel cyclic adduct (HNE-CY) | Arg485 | |
| 2-pentyl-pyrrole (PP) | Lys106 | |
| 2,3 dihydro-pentyl-furan (DHPF) | His105, Cys476, Cys477, Cys487 |
Fig. 4.
Venn diagrams of the identified ALEs as reported inTable 1and2. ALEs are reported as the modified amino acid residues. The upper diagram refers to ALEs obtained by treating HSA with MDA, the middle with ACR and the lower with HNE. The input reports ALEs not retained by VC1 and present only in the input samples; VC1 are the ALEs identified only after VC1-enrichment; the intersections report the ALEs found both in the input and VC1-enriched samples.
With regard to MDA (Table 2), only di-hydropyridine adducts on lysines (DHPK), and N-2-pyrimidyl-ornithine adducts on arginines (NPO) were retained by VC1-domain. The n-propenal modifications of lysine (NPK), largely identified before enrichment, were not identified after the enrichment.
ACR induced a set of modifications which were identified only after VC1 enrichment and in particular the N-(3-formyl-3,4-dehydro-piperidinyl) lysine (FDPK) modifications, the Michael adduct on cysteines, the double Michael adduct of lysines, the Michael adduct of histidine, the N-2-(4 hydroxy-tetrahydro-pyrimidyl) ornitine (propane-arginine, HTPO) and the Nε-(3-methylpyridinium)-lysine (MP-lysine) (Table 2).
Most of the ALEs generated by HNE were found both before or after enrichment, with the exception of few Michael adducts which were selectively retained by VC1 (not detected before enrichment) (Table 2). Interestingly, a novel + 138 Da adduct was detected on Arg209 and Arg485 with HNE (novel cyclic aduct, HNE-CY, Table 2). In agreement with the adducts which arginine yields with other RCS (as seen for ACR and MDA) and by considering the well-known reactivity of the guanidine function [see for example [28]) this adduct can be supposed to include a 2-amino pyrimidine scaffold even though its precise structural characterization would require additional studies.
3.4. Semi-quantitative analysis of ALE-HSA adducts
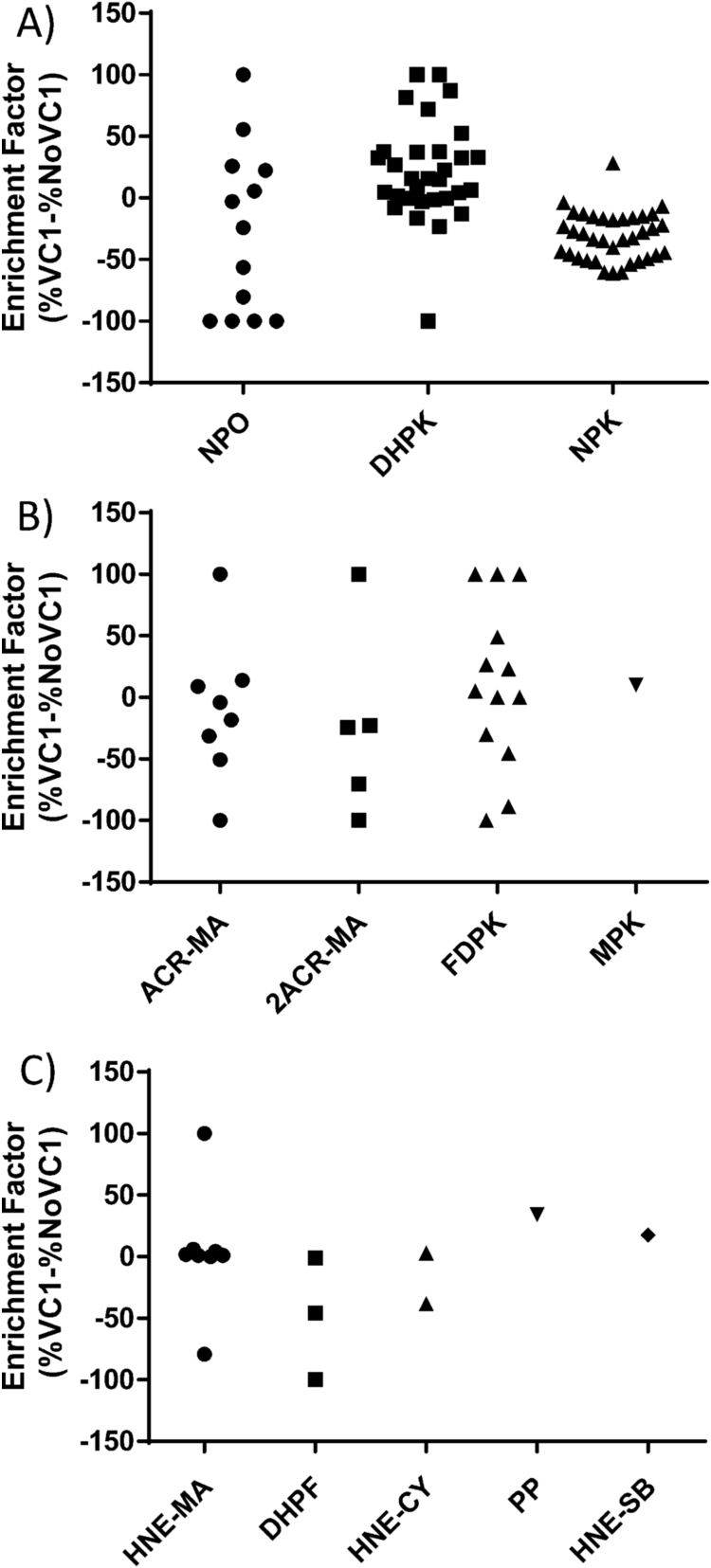
A semiquantitative analysis reporting the relative abundance of each modified peptide in respect of the native peptide was then carried out for each identified modification in both the input (% No VC1) and enriched samples (%VC1). The values were then used to calculate the enrichment factor (EF) calculated as reported in the method section. The values of the EF range between − 100 and + 100, where − 100 means that the modification has not been retained at all by the VC1 domain (it is only present in the input but not in the enriched sample), + 100 means that the modification has been identified because it is enriched by the VC1 (the modification is not detected in the input due to low abundance) and 0 means that they are equally distributed. Values between 0 and + 100 indicate that the modification is retained by VC1 and that the retention efficiency increases as the value increases above 0.
In Fig. 5, panels A, B and C show the EF for ALEs-lipox obtained by using MDA, ACR and HNE, respectively. Regarding MDA, an overview analysis indicates that most of the N-Propenal-Lys and NPO adducts are not retained, with a few exception, while DHPK modifications significantly increase the affinity of ALEs. As better described in the computational analysis paragraph the data indicate that for MDA, VC1 affinity is determined by the nature of the adduct.
Fig. 5.
Enrichment Factor (EF) graphical distribution. Enrichment Factor (EF) graphical distribution by means of vertical scatter plot overviewing the affinity of each identified ALEs towards VC1; all EF values spotted have been calculated as the differences between the relative abundance in the enriched sample minus the amount in the untreated one (Enrichment factor= %VC1-%NoVC1), and graphically grouped as: MDA-adducts (panel A), ACR-adducts (panel B) and HNE-adducts (panel C).
In the case of ACR, with some exceptions explained in the following paragraph, the FDPK adduct is more effective in respect to the mono and bis Michael adducts in increasing the binding towards VC1. In the case of HNE, most of the identified adducts were not found to be effective in increasing the retention affinity with the exception of few residues.
3.5. Computational results
With a view to rationalizing the key factors influencing the RAGE binding of the monitored adducts, in silico studies were performed. They were focused on the adducts on arginine and lysine residues as formed by ACR and MDA since they are numerous, with a very broad range of affinity, thus allowing the development of clear structure-affinity relationships.
As a preamble it should be remembered that the RAGE-ligand interacting regions (i.e. the VC1 domain) are characterized by a rich set of positively charged residues especially in the V portion: the resulting positive RAGE surface shows an understandable non-specific affinity for acidic proteins which can bind RAGE by stabilizing extended sets of salt bridges. On these grounds, one may figure out that a given covalent adduct can induce RAGE binding via two major mechanisms: it can introduce acid moieties which directly contact the RAGE residues (as seen for example with CML and CEL adducts) or it can reduce the basicity of the adducted residues. In this second indirect mechanism, the adducts can elicit RAGE binding mostly because they are able to destabilize ionic clusters on the protein surface and to liberate negatively charged residues which enhance their accessibility and they become available to stabilize ion-pairs with the positive RAGE residues.
The here monitored covalent adducts do not introduce negative functions and thus they should induce RAGE binding by the second indirect mechanism. Such a hypothesis implies that the monitored RAGE binding might be explained by considering two major factors: the basicity of the formed adducts and the number of the surrounding negatively charged residues. Indeed, when the generated adduct retains a marked basicity which renders the protonated form still predominant at physiological pH, the surrounding negative residues would remain shielded around it and unavailable to contact the RAGE residues. On the other hand, an adduct devoid of surrounding negative residues would be unable to induce strong ionic contacts with RAGE regardless of its basicity. On these grounds the here reported in silico analyses can be subdivided into three steps: in the first, the basicity of the considered adducts was predicted by semi-empirical calculations; in the second, the number of surrounding negative residues was calculated by using the refined HSA structure and by considering the number of aspartates and glutamates in a 5 Å radius sphere around each adducted residue. Finally, protein-protein docking simulations were performed by considering the V-domain of RAGE and the adducted HSA structures. For simplicity, the docking simulations involved the HSA structure including simultaneously all found adducts. Such a strategy clearly reduces the computational time and allows synergistic effects between adjacent adducts to be revealed.
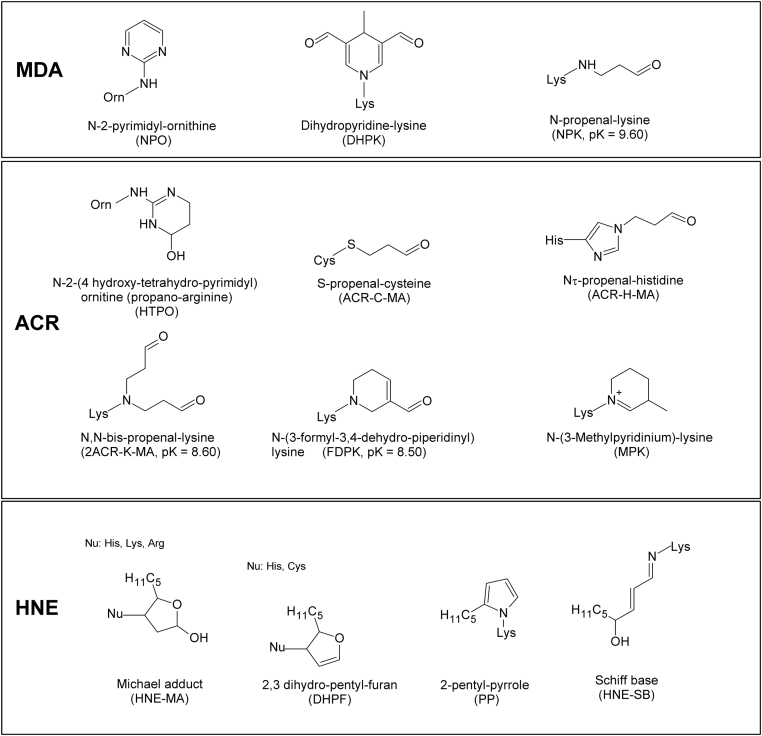
Fig. 6 shows the ALEs structures and for some of them reports the predicted basicity for the considered adducts and reveals that they can be subdivided into two main groups depending on whether they retain part of the basicity of the unmodified residues or become non-ionizable derivatives at physiological conditions. When focusing attention on arginine adducts, N-2-pyrimidyl-ornithine (NPO, from MDA) is completely neutral at physiological pH, while HTPO retains a large part of the arginines strong basicity. Similarly, and with regard to lysine adducts, the DHPK adduct (from MDA) is a neutral adduct, the single Michael adduct (N-Propenal lysine, from MDA) is slightly less basic than lysine (Δ = −0.6), while the double Michael adduct and the FDPK derivative (from ACR) show a similarly reduced basicity (Δ = −2.0) compared to the unmodified residue. In other words, and at physiological conditions, one may assume that (a) RP and DHPK adducts are always neutral, (b) HTPR and NP-lysine are still protonated, (c) double MA and FDPK are in equilibrium between the two forms and the neutral state can play a significant role.
Fig. 6.
Chemical structures of ALEs-lipox formed by incubating HSA with MDA, ACR and HNE and identified by MS. For some ALEs the pK values are reported in brackets.
This simple observation can explain: (a) why NPO is the only arginine adduct (considering both MDA and ACR) which shows some affinitive residues; (b) why the DHPK adducts show a retention efficiency markedly higher than N-propenal-lysines (among the MDA-induced lysine adducts) and (c) why double MA and FDPK show a comparable retention efficiency (among the ACR-induced lysine adducts). In general, one may conclude that the ionization properties of the monitored adducts play a pivotal role in determining their overall affinity towards RAGE, while the binding of each single adduct might be rationalized by considering its specific environment as discussed below.
Table S2 (supplementary material) compiles the number of negative residues included within a 5 Å radius sphere around each adducted residue and allows for some insightful considerations. With regard to neutral arginine adducts (NPO), Table S2 (supplementary materials) clearly evidences that the two highly affinitive adducts are surrounded by a markedly higher number of negative residues (5 for Arg10 and 4 for Arg472). Interestingly the other three NPO adducts showing a positive EF value (Arg117, Arg337 and Arg428) show 2 surrounding negative residues while all adducts with negative EF value show at most 1 residue.
The compiled results for lysine adducts appear to be less clear even though one may observe that almost all affinitive adducts possess more than 2 surrounding residues. The more difficult interpretability of the data concerning the lysine adducts can be explained by considering that (a) they include different adducts with different ionization properties and (b) some adducts (e.g. DHPK) can be directly involved in RAGE binding, thus requiring less surrounding negative residues. Moreover and due to the very high number of adducted lysines one may figure out synergistic effects between adjacent lysine adducts which may generate very extended ionic networks not conveniently described by considering 5 Å radius spheres.
Taken together, the obtained results seem to confirm that the MDA and ACR-induced adducts act as RAGE binders mostly when they occur in their neutral state at physiological pH and thus they are able to destabilize ionic clusters on protein surface and to liberate negatively charged residues which are no longer shielded and expose their carboxylate groups with a sort of “flowering effect” which promotes ion-pairing between adducted HSA and RAGE. With a view to confirming the supposed binding mechanism, the last part of these in silico studies involved protein-protein docking simulations which were performed to generate the corresponding RAGE-adducted HSA complexes. While avoiding a systematic analysis of all simulated HSA adducts, some representative putative complexes will be presented in order to further corroborate the proposed mechanism and to gain deeper insights on the involved interactions.
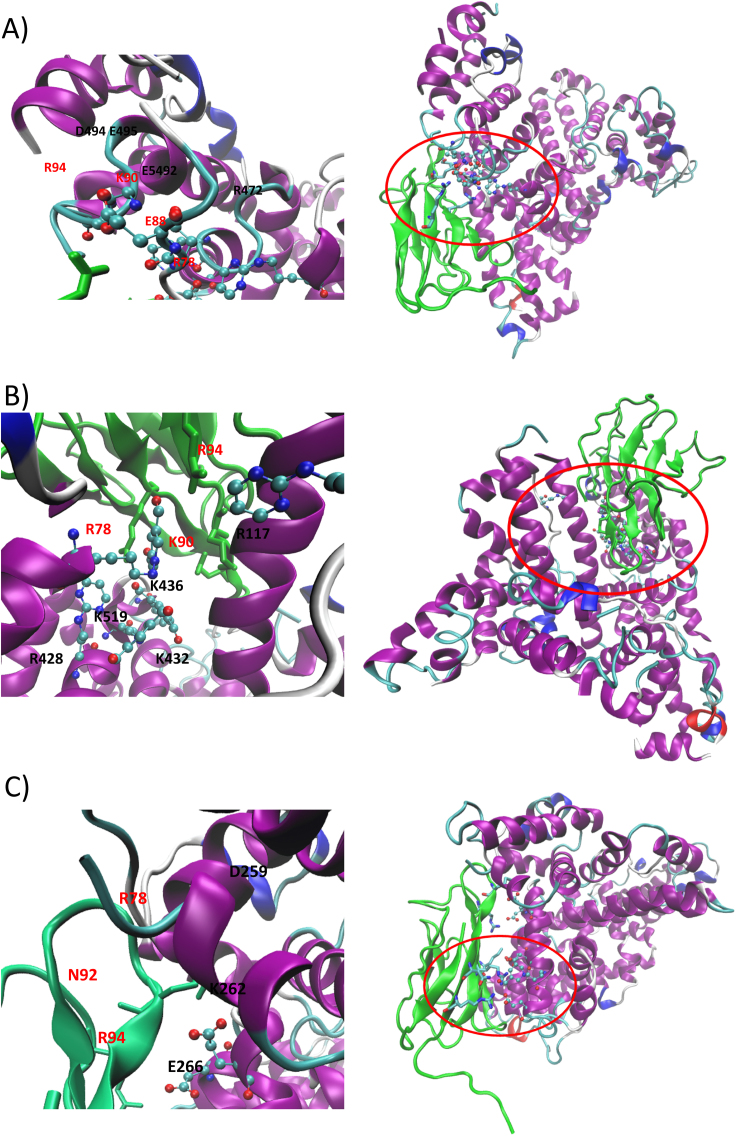
Fig. 7 (panel A) shows the putative complex as computed for the most affinitive NPO adduct (Arg472, from MDA) and reveals that the several negative residues surrounding the adduct are involved in an extended network of ion-pairs with the RAGE residues. In detail, Asp494 and Glu495 approach Arg70 of RAGE while Glu492 contacts RAGE-Lys90. Interestingly and although the RAGE surface is rich in positive residues, the complex also shows a salt bridge in which the positive residue comes from the HSA surface as seen in the salt bridge between RAGE-Glu88 and Arg410. It should be noted that also the simulated NPO adduct is found to elicit a charge transfer interaction with RAGE-Arg78. In this way, the interaction surface between HSA and RAGE is stabilized by a rich set of ionic interactions which are clearly promoted by the neutralization of Arg472 and which can explain the observed RAGE binding.
Fig. 7.
Putative RAGE-HSA complex as computed by protein-protein docking and induced by RP-Arg472 (6 A), DHPK-Lys436 (6B) and FDP-Lys262 (6 C). For each simulated adduct, the right panel shows the entire RAGE-HSA complex while the left panel focuses on key interactions stabilized by the adduct and its surrounding residues.
Fig. 7 (panel B) shows the computed complex for a very affinitive DHPK adduct (Lys436, from MDA) and highlights the potential synergistic role between adjacent adducts as supposed above. Indeed, the complex reveals that Lys433 is closely surrounded by several other MDA-based adducts (i.e., DHPK432 and DHPK519 plus NPO-Arg117 and NPO-Arg428) which contribute to the overall binding by directly interacting with the RAGE residues (see for example the reinforced H-bonds that DHPK436 elicits with Arg78 and Arg94), and through the surrounding negative residues (not reported in Fig. 7 for clarity). The found ion-pairs include Arg78-Glu400, Arg94-Asp183, Lys80-Glu188 and Lys32-Glu396. Notably, all the reported adducts show a positive EF value apart from DHPK519, whose poor results can be ascribed to the shielding effect played by the close Arg186 residue and which further confirms the key role of the ionizable surrounding residues.
Finally, Fig. 7 panel C reports the putative complex for a highly affinitive FPDK adduct (on Lys262, from ACR) and emphasizes the key role played by the three negative residues surrounding it and which are involved in clear ionic interactions with the positive RAGE residues. Interestingly and despite such a favorable environment Lys262 does not give affinitive adducts with MDA. This finding can be explained by considering that Lys262 generates only the ionized N-propenal adduct with MDA. Such a result suggests that the ionization properties play a largely dominant role which cannot be counteracted by the surrounding negative residues. Remarkably, the figure describing the entire RAGE-HSA complex shows that the ionic contacts are not limited to the region surrounding the FPD adduct but additional salt bridges are present in the entire contact surface. This suggests that the RAGE-HSA binding is initially promoted by the focused set of ionic contacts which can be stabilized around the adduct and then is reinforced by a more extended set of additional ion-pairs which characterize the entire contact surface. Notably, these additional contacts which do not involve adducted residues are per se unable to promote the RAGE-HSA binding but they play a clearly beneficial role in that the initial binding is triggered by the contacts stabilized by the residues surrounding the adduct with a sort of hierarchical mechanism which brings to mind that described for protein folding.
Although the affinity of the HNE-based adducts was not analyzed in silico, the observed EF values of the peptides including Lys106 are in clear agreement with the basicity of the detected adducts. Indeed, the retention efficiency is found to be quite high for those adducts which abrogate the lysine basicity (i.e. Schiff base and 2-pentylpyrrole) to markedly decrease when the adduct only slightly reduces the basicity as in the Michael adduct.
Even though HSA is not the ideal protein target to study in-depth the RAGE binding on cysteines since most HSA cysteines are involved in disulfide bridges, the general incapacity of the cysteine adducts to bind RAGE can be clearly rationalized in terms of the proposed flowering effect. Carbonylation of cysteines does not result in neutralization of positively charged residues but conversely it can neutralize particularly acid cysteines with an opposite and unfavorable electron-deficient effect on the protein surface
4. Discussion
Much evidence indicates that the pro-inflammatory and pro-fibrotic effects of AGEs and AGEs/ALEs are due, among other mechanisms, to RAGE binding and activation [18], [29], [30], [31]. The engagement mechanisms between AGEs and RAGE are still not fully elucidated and great scientific interest is now focused on understanding which are the structural moieties of AGEs that turn a protein to a RAGE binder and hence to a pro-inflammatory mediator. The study is further complicated by the fact that AGEs and AGEs/ALEs are characterized by a wide heterogeneity not only due to the variability of the adducted moieties which differ on the basis of the attacking RCS but also due to the variable target protein and sites of modification.
A rich set of positively charged residues represents a structural feature of RAGE which could explain its binding properties with ligands [32]. In particular such positive charges result in a positive RAGE surface, which drives the engagement with ligands characterized by acidic or in general negatively charge residues, able to stabilize the protein-protein interaction by an extended set of salt bridges [33]. While considering that the RAGE binding is clearly promoted by those AGEs/ALEs, which are characterized by the presence of carboxylic moieties such as CML and CEL [13], the mechanisms, by which a local and structurally limited modification, such as that induced by a AGE/ALE adduct, can trigger the RAGE affinity in large biomacromolecules, are still not precisely understood. This lack of understanding is even more pronounced when considering that many AGEs and almost all ALEs (see below) do not introduce acid functions but at most they modulate the basicity of the adducted residues.
While on one hand several studies have reported on the engagement between AGEs and AGEs/ALEs on VC1 and RAGE, on the other, little is known about the effect of ALEs from RCS formed only by lipid peroxidation such as HNE, MDA and ACR. These ALEs, here called ALEs-lipox, are formed abundantly in several oxidative based conditions and their damaging property has been widely reported. Several molecular mechanisms have been suggested to explain the ALEs-lipox damaging effect and among these are protein dysfunction, protein oligomerization, signal transduction and immune response [19]. Moreover, by considering a pro-inflammatory and pro-fibrotic response induced by ALEs-lipox, a RAGE dependent mechanism could also be considered.
In the present report we have evaluated the binding property of a wide set of ALEs-lipox formed by incubating HSA with the three most abundant lipid peroxidation RCS, namely HNE, ACR and MDA. The wide set of modifications were achieved by incubating the target protein with RCS at different molar ratios and time points as confirmed by MS intact protein analysis.
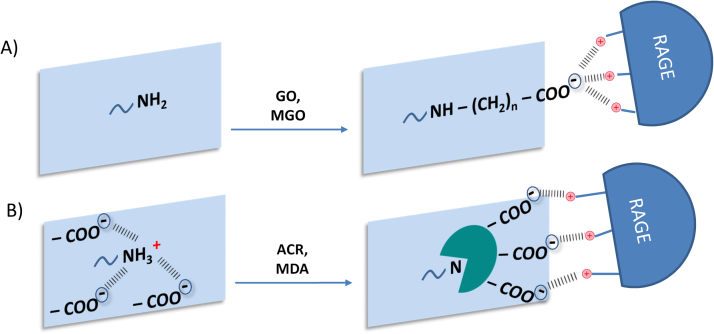
The high concentrations of the aldehydes and in particular their content in high molar excess in respect to the substrate, very far from biological conditions, were used in order to obtain the maximum number of adducts which can be generated from the reaction of RCS with the protein substrate. Although the resulting ALEs are not characterized by acid moieties, as reported in the literature and here confirmed by bottom-up analyses, we found using a VC1 pull-down assay that some of the generated ALEs bind to the VC1. This is a quite novel finding since to our knowledge no study has so far reported the ability of ALEs from lipid peroxidation RCS to interact with VC1. The VC1 binding assay coupled to a high resolution MS approach has also permitted the identification of which ALEs moieties are responsible for the VC1 binding and the sites of modification. The wide set of data and in particular the chemical diversity of the structures identified as binder and non binder, have also permitted a SAR study leading to the molecular explanation of why only some ALEs bind to VC1. As shown in Fig. 8, besides the well known VC1 engagement based on the acid residues of the protein ligand, we here found another potential mechanism which may involve adducts that do not introduce acid groups but only reduce the basicity of the modified residues. The mechanism, called the flowering effect, is based on a two-step process and involves exposed basic residues (mainly Lys and Arg) which in the non-adducted protein form a set of ionic bridges with the carboxylic groups of surrounding aspartate and glutamate residues. In the first step, the RCS form a covalent adduct with the positively charged residues, abolishing or greatly reducing their basicity. Consequently, the adducted residues are present in their neutral form at physiological conditions and such a change in their ionization state destabilizes the ionic bridges and renders the surrounding anionic residues more accessible and available to stabilize ion-pairs with the positive RAGE residues. Such a mechanism explains why only one set of the identified ALEs acts as RAGE binder. In other words we found the basic features that make an ALE a RAGE binder and which strictly depend on the type of the adduct and on the site of modification. In particular we found that only some of the several adducts formed by lipid peroxidation RCS abolish or greatly reduce the basicity of the target amino acids and among these RP (from MDA) and DHPK adducts are always neutral while HTPR, MA-Arg and NP-lysine do not affect the basic character. The second requirement is the presence of a cluster of negative side chains surrounding the target residue, which then becomes available to RAGE positive charges once the amino acid is modified, thus stabilizing the protein-protein complex.
Fig. 8.
Mechanism explaining AGEs/ALEs and ALEs-lipox binding to VC1. A) VC1 engagement based on the acid residues of the protein ligand. Such a mechanism occurs when RCS, such as GO and MGO react with the basic residue forming a carboxylated adduct (CML and CEL) which directly contacts the RAGE residues. B) Panel B summarizes the mechanism described in the present paper for explaining the binding of ALEs-lipox to VC1 and called the “flowering effect”. It is based on a two-step process and involves exposed basic residues (mainly Lys and Arg) which in the non-adducted protein form a set of ionic bridges with the carboxylic groups of surrounding aspartate and glutamate residues. Lipid peroxidation derived RCS react with the basic residue and abolish or greatly reduce their basicity. Consequently, the adducted residues shift in a neutral form and such a change in the ionization state destabilizes the ionic bridges and renders the surrounding anionic residues more accessible and available to stabilize ion-pairs with the positive RAGE residues.
We currently do not know whether ALEs-VC1 binding elicits RAGE activation and hence if they lead a RAGE-dependent biological response. Some studies such as that by Shanmugam et al. [34] reporting that ALEs from MDA can induce a RAGE dependent biological response, would suggest this. We are now working on this aspect by using cell models and ALEs enriched by the VC1 assay as potential RAGE activators.
5. Conclusions
In summary by using an integrated MS (intact protein and bottom-up approach) and computational approach we have found that some ALEs generated from lipid peroxidation RCS are RAGE binders. We have also found the basic features that ALEs from HNE, MDA and ACR must have to be a RAGE binder: 1) the covalent adducts should greatly reduce or abolish the basicity of the target amino acid, 2) the basic amino acid should be at the center of a set of carboxylic acids which, once the residue is modified, become available to freely contact the RAGE positive residues. Interestingly, the here proposed flowering effect can also be involved in the RAGE binding of the adducts which insert an acid function. Indeed and besides the direct contacts stabilized by the adduct, one may suppose that introduced anionic function exerts a similar effect on the surrounding negative residues thus promoting their ionic interactions with the positive residues on RAGE surface. Stated differently, the flowering effect might be a common mechanism triggering the affinity of adducted proteins towards RAGE even though it significantly increases its relevance for the adducts that do not add carboxylic functions such as those as seen here.
Moreover, one may expect that such a mechanism can modulate the affinity of carbonylated proteins towards a variety of target proteins not necessarily limited to RAGE. More generally and since a similar mechanism was proposed to explain the protein-protein interactions triggered by the acetylation of lysines, one may hypothesize that the ionic perturbation of the protein surface, that shifts from a polyzwitterionic situation to a polyanionic or polycationic condition, can be a general mechanism, by which post translational modifications involving ionizable side chains modulate the interacting capacity of the modified proteins.
Acknowledgements
This work has been funded by the European Union's Horizon 2020 research and innovation programme under the Marie Sklowdowska-Curie grant agreement number 675132 (http://cordis.europa.eu/project/rcn/198275_en.html).
Acknowledgments
Conflict of interest
GD, LP and GA are co-authors of a patent application (WO2016IB52391 20160427) entitled “Improved system for the expression of the receptor for the advanced glycation end products (AGEs) and the advanced lipid glycation end products (ALEs) and applications thereof”.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.101083.
Appendix A. Supplementary material
Supplementary material Supplementary Figure 1. Direct infusion ESI-MS analysis of native and ACR-modified HSA. Spectra of HSA with charge 47 + , 46 + and 45 + are shown. A) Native HSA shows a sharp intense peak with multiple low abundant peaks. When HSA is reacted with increasing concentrations of ACR, 1: 10 (B), 1: 100 (C), 1: 1000 (D), 1: 2500 (E) and 1: 5000 (F), the sharp peak diminishes and new peaks appear, confirming that HSA has been modified with different adducts of ACR. Supplementary Figure 2. Direct infusion ESI-MS analysis of native and MDA-modified HSA. Spectra of HSA with charge 47 + , 46 + and 45 + are shown. A) Native HSA shows a sharp intense peak with multiple low abundant peaks. When HSA is reacted with increasing concentrations of MDA, 1: 6.3 (B), 1: 63 (C), 1: 630 (D), 1: 6.300 (E) and 1: 12.600 (F), the sharp peak diminishes and new peaks appear, confirming that HSA has been modified with different adducts of MDA. Supplementary Figure 3: Time course analysis of the dose-dependent modification of HSA by ACR. Recombinant HSA was incubated with increasing concentrations of ACR (molar ration HSA-ACR from 1:10–1:5000) as reported in Materials and methods. At 24, 48 and 72 h of incubation, aliquots were withdrawn and after removal of free acrolein, pull-down assays with the VC1-resin or control (CTRL)-resin were performed. A specific binding of both the monomer of modified HSA and of the HMW-species to the VC1-resin was detected at increasing concentrations, starting from the molar ratio 1:1000. Supplementary Figure 4: Analysis of the dose-dependent modification of HSA by HNE. Recombinant HSA was incubated with increasing concentrations of HNE (molar ration HSA-HNE from 1:10–1:2000) as reported in Materials and methods. At 24, 48 and 72 h of incubation, aliquots were withdrawn and after removal of free HNE, pull-down assays with the VC1-resin or control (CTRL)-resin were performed. A specific binding of modified HSA to the VC1-resin was detected at increasing concentrations, starting from the molar ratio 1:200. At 1:1000 is evident the interaction with VC1 of both the monomer of modified HSA and the HMW-species.
Supplementary material Table S1 Known (literature based) covalent adducts induced by HNE, MDA and ACR and set as variable modifications within the PD parameters for the identification and localization of ALE-deriving adducts. Table S2 Number of negatively charged residues found within a 5 Å radius sphere around the adducted residues.
References
- 1.Gueraud F., Atalay M., Bresgen N., Cipak A., Eckl P.M., Huc L., Jouanin I., Siems W., Uchida K. Chemistry and biochemistry of lipid peroxidation products. Free Radic. Res. 2010;44(10):1098–1124. doi: 10.3109/10715762.2010.498477. [DOI] [PubMed] [Google Scholar]
- 2.Vistoli G., De Maddis D., Cipak A., Zarkovic N., Carini M., Aldini G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): an overview of their mechanisms of formation. Free Radic. Res. 2013;47(Suppl 1):3–27. doi: 10.3109/10715762.2013.815348. [DOI] [PubMed] [Google Scholar]
- 3.Aldini G., Vistoli G., Stefek M., Chondrogianni N., Grune T., Sereikaite J., Sadowska-Bartosz I., Bartosz G. Molecular strategies to prevent, inhibit, and degrade advanced glycoxidation and advanced lipoxidation end products. Free Radic. Res. 2013;47(Suppl 1):93–137. doi: 10.3109/10715762.2013.792926. [DOI] [PubMed] [Google Scholar]
- 4.Mol M., Regazzoni L., Altomare A., Degani G., Carini M., Vistoli G., Aldini G. Enzymatic and non-enzymatic detoxification of 4-hydroxynonenal: methodological aspects and biological consequences. Free Radic. Biol. Med. 2017;111:328–344. doi: 10.1016/j.freeradbiomed.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 5.Curtis T. The role lipid aldehydes and ALEs in the pathogenesis of diabetic retinopathy. Free Radic. Biol. Med. 2014;75(Suppl 1):S8. doi: 10.1016/j.freeradbiomed.2014.10.845. [DOI] [PubMed] [Google Scholar]
- 6.Negre-Salvayre A., Coatrieux C., Ingueneau C., Salvayre R. Advanced lipid peroxidation end products in oxidative damage to proteins. Potential role in diseases and therapeutic prospects for the inhibitors. Br. J. Pharmacol. 2008;153(1):6–20. doi: 10.1038/sj.bjp.0707395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacobini C., Menini S., Ricci C., Scipioni A., Sansoni V., Mazzitelli G., Cordone S., Pesce C., Pugliese F., Pricci F., Pugliese G. Advanced lipoxidation end-products mediate lipid-induced glomerular injury: role of receptor-mediated mechanisms. J. Pathol. 2009;218(3):360–369. doi: 10.1002/path.2536. [DOI] [PubMed] [Google Scholar]
- 8.Sasson S. Nutrient overload, lipid peroxidation and pancreatic beta cell function. Free Radic. Biol. Med. 2017;111:102–109. doi: 10.1016/j.freeradbiomed.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Anderson E.J., Vistoli G., Katunga L.A., Funai K., Regazzoni L., Monroe T.B., Gilardoni E., Cannizzaro L., Colzani M., De Maddis D., Rossoni G., Canevotti R., Gagliardi S., Carini M., Aldini G. A carnosine analog mitigates metabolic disorders of obesity by reducing carbonyl stress. J. Clin. Invest. 2018 doi: 10.1172/JCI94307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iacobini C., Menini S., Blasetti Fantauzzi C., Pesce C.M., Giaccari A., Salomone E., Lapolla A., Orioli M., Aldini G., Pugliese G. FL-926-16, a novel bioavailable carnosinase-resistant carnosine derivative, prevents onset and stops progression of diabetic nephropathy in db/db mice. Br. J. Pharmacol. 2018;175(1):53–66. doi: 10.1111/bph.14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Łuczaj W., Gęgotek A., Skrzydlewska E. Antioxidants and HNE in redox homeostasis. Free Radic. Biol. Med. 2017;111:87–101. doi: 10.1016/j.freeradbiomed.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 12.Vlassara H., Li Y.M., Imani F., Wojciechowicz D., Yang Z., Liu F.T., Cerami A. Identification of galectin−3 as a high-affinity binding protein for advanced glycation end products (age): a new member of the age-receptor complex. Mol. Med. 1995;1(6):634–646. [PMC free article] [PubMed] [Google Scholar]
- 13.Xue J., Rai V., Singer D., Chabierski S., Xie J., Reverdatto S., Burz D.S., Schmidt A.M., Hoffmann R., Shekhtman A. Advanced glycation end product recognition by the receptor for ages. Structure. 2011;19(5):722–732. doi: 10.1016/j.str.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossin N., Wautier M.P., Picot J., Stern D.M., Wautier J.L. Differential effect of plasma or erythrocyte AGE-ligands of RAGE on expression of transcripts for receptor isoforms. Diabetes Metab. 2009;35(5):410–417. doi: 10.1016/j.diabet.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Ott C., Jacobs K., Haucke E., Navarrete Santos A., Grune T., Simm A. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014;2:411–429. doi: 10.1016/j.redox.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González I., Romero J., Rodríguez B.L., Pérez-Castro R., Rojas A. The immunobiology of the receptor of advanced glycation end-products: trends and challenges. Immunobiology. 2013;218(5):790–797. doi: 10.1016/j.imbio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Kierdorf K., Fritz G. RAGE regulation and signaling in inflammation and beyond. J. Leukoc. Biol. 2013;94(1):55–68. doi: 10.1189/jlb.1012519. [DOI] [PubMed] [Google Scholar]
- 18.Baynes J.W. Chemical modification of proteins by lipids in diabetes. Clin. Chem. Lab. Med. 2003;41(9):1159–1165. doi: 10.1515/CCLM.2003.179. [DOI] [PubMed] [Google Scholar]
- 19.Aldini G., Dalle-Donne I., Facino R.M., Milzani A., Carini M. Intervention strategies to inhibit protein carbonylation by lipoxidation-derived reactive carbonyls. Med. Res. Rev. 2007;27(6):817–868. doi: 10.1002/med.20073. [DOI] [PubMed] [Google Scholar]
- 20.Degani G., Altomare A.A., Colzani M., Martino C., Mazzolari A., Fritz G., Vistoli G., Popolo L., Aldini G. A capture method based on the VC1 domain reveals new binding properties of the human receptor for advanced glycation end products (RAGE) Redox Biol. 2017;11:275–285. doi: 10.1016/j.redox.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colzani M., Criscuolo A., Casali G., Carini M., Aldini G. A method to produce fully characterized ubiquitin covalently modified by 4-hydroxy-nonenal, glyoxal, methylglyoxal, and malondialdehyde. Free Radic. Res. 2016;50(3):328–336. doi: 10.3109/10715762.2015.1118472. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z., Marshall A.G. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9(3):225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 23.Degani G., Colzani M., Tettamanzi A., Sorrentino L., Aliverti A., Fritz G., Aldini G., Popolo L. An improved expression system for the VC1 ligand binding domain of the receptor for advanced glycation end products in Pichia pastoris. Protein Expr. Purif. 2015;114:48–57. doi: 10.1016/j.pep.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Stewart J.J. Optimization of parameters for semiempirical methods VI: more modifications to the NDDO approximations and re-optimization of parameters. J. Mol. Model. 2013;19(1):1–32. doi: 10.1007/s00894-012-1667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedretti A., Villa L., Vistoli G. VEGA: a versatile program to convert, handle and visualize molecular structure on windows-based PCs. J. Mol. Graph. Model. 2002;21(1):47–49. doi: 10.1016/s1093-3263(02)00123-7. [DOI] [PubMed] [Google Scholar]
- 26.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katchalski-Katzir E., Shariv I., Eisenstein M., Friesem A.A., Aflalo C., Vakser I.A. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl. Acad. Sci. USA. 1992;89(6):2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo W. Base mediated direct C–H amination for pyrimidines synthesis from amidines and cinnamaldehydes using oxygen as green oxidants. Chin. Chem. Lett. 2016;27(1):4. [Google Scholar]
- 29.Del Turco S., Basta G. An update on advanced glycation endproducts and atherosclerosis. Biofactors. 2012;38(4):266–274. doi: 10.1002/biof.1018. [DOI] [PubMed] [Google Scholar]
- 30.Ramasamy R., Vannucci S.J., Yan S.S.D., Herold K., Yan S.F., Schmidt A.M. Advanced glycation end products and RAGE: a common thread in aging, diabetes, neurodegeneration, and inflammation. Glycobiology. 2005;15(7):16R–28R. doi: 10.1093/glycob/cwi053. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi M., Takino J., Yamagishi S. Involvement of the Toxic AGEs (TAGE)-RAGE System in the Pathogenesis of Diabetic Vascular Complications: a Novel Therapeutic Strategy. Curr. Drug Targets. 2010;11(11):1468–1482. doi: 10.2174/1389450111009011468. [DOI] [PubMed] [Google Scholar]
- 32.Park H., Adsit F.G., Boyington J.C. The 1.5 Å crystal structure of human receptor for advanced glycation endproducts (RAGE) ectodomains reveals unique features determining ligand binding. J. Biol. Chem. 2010;285(52):40762–40770. doi: 10.1074/jbc.M110.169276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch M., Chitayat S., Dattilo B.M., Schiefner A., Diez J., Chazin W.J., Fritz G. Structural basis for ligand recognition and activation of RAGE. Structure. 2010;18(10):1342–1352. doi: 10.1016/j.str.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shanmugam N., Figarola J.L., Li Y., Swiderski P.M., Rahbar S., Natarajan R. Proinflammatory effects of advanced lipoxidation end products in monocytes. Diabetes. 2008;57(4):879–888. doi: 10.2337/db07-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Supplementary Figure 1. Direct infusion ESI-MS analysis of native and ACR-modified HSA. Spectra of HSA with charge 47 + , 46 + and 45 + are shown. A) Native HSA shows a sharp intense peak with multiple low abundant peaks. When HSA is reacted with increasing concentrations of ACR, 1: 10 (B), 1: 100 (C), 1: 1000 (D), 1: 2500 (E) and 1: 5000 (F), the sharp peak diminishes and new peaks appear, confirming that HSA has been modified with different adducts of ACR. Supplementary Figure 2. Direct infusion ESI-MS analysis of native and MDA-modified HSA. Spectra of HSA with charge 47 + , 46 + and 45 + are shown. A) Native HSA shows a sharp intense peak with multiple low abundant peaks. When HSA is reacted with increasing concentrations of MDA, 1: 6.3 (B), 1: 63 (C), 1: 630 (D), 1: 6.300 (E) and 1: 12.600 (F), the sharp peak diminishes and new peaks appear, confirming that HSA has been modified with different adducts of MDA. Supplementary Figure 3: Time course analysis of the dose-dependent modification of HSA by ACR. Recombinant HSA was incubated with increasing concentrations of ACR (molar ration HSA-ACR from 1:10–1:5000) as reported in Materials and methods. At 24, 48 and 72 h of incubation, aliquots were withdrawn and after removal of free acrolein, pull-down assays with the VC1-resin or control (CTRL)-resin were performed. A specific binding of both the monomer of modified HSA and of the HMW-species to the VC1-resin was detected at increasing concentrations, starting from the molar ratio 1:1000. Supplementary Figure 4: Analysis of the dose-dependent modification of HSA by HNE. Recombinant HSA was incubated with increasing concentrations of HNE (molar ration HSA-HNE from 1:10–1:2000) as reported in Materials and methods. At 24, 48 and 72 h of incubation, aliquots were withdrawn and after removal of free HNE, pull-down assays with the VC1-resin or control (CTRL)-resin were performed. A specific binding of modified HSA to the VC1-resin was detected at increasing concentrations, starting from the molar ratio 1:200. At 1:1000 is evident the interaction with VC1 of both the monomer of modified HSA and the HMW-species.
Supplementary material Table S1 Known (literature based) covalent adducts induced by HNE, MDA and ACR and set as variable modifications within the PD parameters for the identification and localization of ALE-deriving adducts. Table S2 Number of negatively charged residues found within a 5 Å radius sphere around the adducted residues.