Abstract
The transcription factor hypoxia inducible factor-1α (HIF-1α) mediates adaptive responses to oxidative stress by nuclear translocation and regulation of gene expression. Mitochondrial changes are critical for the adaptive response to oxidative stress. However, the transcriptional and non-transcriptional mechanisms by which HIF-1α regulates mitochondria in response to oxidative stress are poorly understood. Here, we examined the subcellular localization of HIF-1α in human cells and identified a small fraction of HIF-1α that translocated to the mitochondria after exposure to hypoxia or H2O2 treatment. Moreover, the livers of mice with CCl4-induced fibrosis showed a progressive increase in HIF-1α association with the mitochondria, indicating the clinical relevance of this finding. To probe the function of this HIF-1α population, we ectopically expressed a mitochondrial-targeted form of HIF-1α (mito-HIF-1α). Expression of mito-HIF-1α was sufficient to attenuate apoptosis induced by exposure to hypoxia or H2O2-induced oxidative stress. Moreover, mito-HIF-1α expression reduced the production of reactive oxygen species, the collapse of mitochondrial membrane potential, and the expression of mitochondrial DNA-encoded mRNA in response to hypoxia or H2O2 treatment independently of nuclear pathways. These data suggested that mitochondrial HIF-1α protects against oxidative stress induced-apoptosis independently of its well-known role as a transcription factor.
Abbreviations: HIF-1, hypoxia inducible factor-1; PHD, prolyl hydroxylases; nDNA, nuclear DNA; mtDNA, mitochondrial DNA; HUVEC, human umbilical vein endothelial cell; ROS, reactive oxygen species; TOMM40, translocase of outer mitochondrial membrane 40; DMOG, dimethyloxaloylglycine; TIMM17A, translocase of inner mitochondrial membrane 17A; NTAD, N-terminal activation domain; CTAD, C-terminal activation domain; ID, inhibitory domain; PARP, cleaved poly (ADP-ribose) polymerase; AIF, apoptosis-inducing factor; ΔΨm, Mitochondrial membrane potential; WCL, whole cell lysates; HA, hemagglutinin; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HDAC, histone deacetylase; PCNA, proliferating cell nuclear antigen; TFAM, transcription factor A, mitochondrial; MT-ND1, mitochondrial-encoded NADH dehydrogenase I; MT-COII, mitochondrial-encoded cytochrome c oxidase II; MT-CYB, mitochondrial-encoded cytochrome b; SOD2, superoxide dismutase 2; CAT, catalase; TMRM, tetramethylrhodamine methyl ester; GSH, Glutathione; NFE2L2, Nuclear factor erythroid 2-related factor 2; HO-1, heme Oxygenase-1
Keywords: Mitochondria, HIF-1α, Oxidative stress, Apoptosis, ROS
Graphical abstract
Highlights
-
•
HIF-1α is recruited to mitochondria in response to oxidative stress.
-
•
Mitochondrial HIF-1α protects against oxidative stress induced apoptosis.
-
•
HIF-1α in mitochondria reduces ROS levels and reverses mitochondrial damage.
-
•
Mitochondrial HIF-1α reduces mtDNA encoded mRNA levels.
-
•
Mitochondrial HIF-1α may involve in liver fibrosis.
1. Introduction
Hypoxia is a pathological process that causes abnormal changes in metabolism, function and morphological structure of tissue because of insufficient oxygen supply. It's reported that hypoxia was accompanied with increased production of reactive oxygen species (ROS), thus provoking oxidative stress [1]. ROS is a double-edged sword: low level of ROS is a key signaling molecule in lots of pathophysiologic processes, while excessive ROS plays critical roles in damaging cellular components and initiating cell death [2].
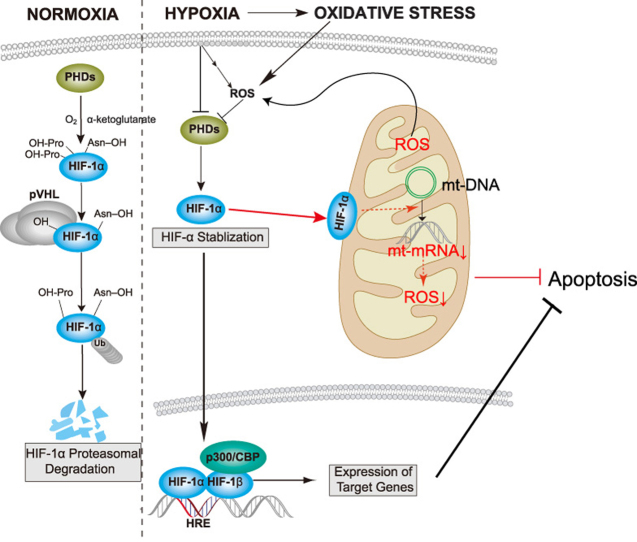
Hypoxia inducible factor-1 (HIF-1), a crucial transcription factor in the cellular response to hypoxia, is a heterodimer composed of a constitutively expressed β subunit and an O2-regulated α subunit. Under normoxic conditions, α subunit levels are regulated by ubiquitin-dependent proteasomal degradation. Conserved proline residues in the subunit are hydroxylated by O2-dependent prolyl hydroxylases (PHDs), and the modified residues are then ubiquitinated by the p-VHL-containing E3 ubiquitin ligase complex and degraded by the proteasome. However, under hypoxia, the HIF-1α subunit is stabilized due to inhibition of PHDs and thus accumulates in the nucleus [3]. HIF-1 binds to hypoxia-response elements and regulates the transcription of hundreds of genes involved in diverse processes such as erythropoiesis, angiogenesis, metabolic reprogramming, cell proliferation and apoptosis/survival, in response to hypoxia [4].
Mitochondria are the powerhouses of the cell and act as O2 sensors that not only generate ATP to fuel cellular processes but also represent a physical point of convergence for many oxidative stress-induced signals. Moreover, mitochondria are the main source of intracellular ROS in hypoxic cells [5]. As such, mitochondria play an important role in cell fate determination under hypoxia. When O2 levels drop, there is an imbalance between O2 and electron flow in the respiratory chain, resulting in excessive production of ROS in respiratory chain complexes, increased oxidation of macromolecules, and subsequent cellular dysfunction or death. Several studies have shown that HIF-1 reduces cellular ROS production by switching energy production from oxidative phosphorylation to glycolysis via multiple pathways [6]. For instance, HIF-1 represses mitochondrial respiration and electron transfer chain activity by activating transcription of the microRNA miR-201, which reduces expression of the iron–sulfur cluster assembly proteins ISCU1/2 and NDUFA4L2, thereby decreasing complex I activity [7]. HIF-1 also activates transcription of genes encoding glucose transporters and glycolytic enzymes, which increases flux from glucose to lactate [8]. In addition, HIF-1 activates the apoptotic protein BNIP3, which induces mitochondrial-selective autophagy under hypoxia [9].
Until recently, HIF-1-dependent regulation of mitochondrial function was thought to depend directly or indirectly on HIF-1 nuclear translocation. However, several studies have reported that HIF-1α localizes to the mitochondria after hypoxic exposure or preconditioning [10], [11]. Mylonis et al. also identified HIF-1α-mortalin-VDAC1-HK-II complex at mitochondrial outer membrane which inhibits hypoxia-induced apoptosis [12]. Here, we further investigated HIF-1α protein trafficking to mitochondria in more human cancerous and normal cell lines. We found that a small fraction of HIF-1α trafficked to the mitochondria after chemical or hypoxic stabilization in a highly reproducible manner. We also confirmed this phenomena in vivo. To study the specific functions of this subpopulation of HIF-1α (here referred to as mtHIF-1α), we expressed mutant proteins (termed mito-HIF-1α) that translocated only to the mitochondria. Our findings further elucidated the new mechanism for direct regulation of mitochondria by HIF-1α independently of its transcriptional activity.
2. Materials and methods
2.1. Animals
All animal experiments were approved by the Committee on Animal Research of Sichuan University. C57BL/6J male mice, 4 weeks of age, were obtained from Dashuo Biological Products (Chengdu, China). To induce liver fibrosis, the mice were injected intraperitoneally with 25% CCl4 (Sigma; 5 μL/g body weight in olive oil). Mice were randomly assigned to four groups of 5 mice to receive one injection every 3 days of olive oil (control group) or CCl4 which induces fibrosis for 2, 4, or 6 weeks. At the end of the treatment period, the mice were sacrificed, and samples were removed from the middle of right liver lobe. One sample was immediately fixed in 4% paraformaldehyde for morphological examination, and the remaining tissues were snap frozen and stored at −80 °C.
2.2. Cell lines and culture conditions
HK-2, HUVEC and HepG2 were purchased from the American Type Culture Collection (Manassas, VA, USA), HeLa and L02 cells were purchased from Shanghai Cell Bank (Chinese Academy of Sciences, Shanghai, China). All cell lines were cultured in the recommended medium purchased from Gibco (Grand Island, NY, USA,). For experiments, the cells were cultured under normoxia (21% O2) or hypoxia (1% O2) at 37 °C. When present, the PHD inhibitor dimethyloxaloylglycine (DMOG; Sigma-Aldrich, Milwaukee, Wis., USA) and HIF-1α inducer CoCl2 (Kelong, Shanghai, China) were added directly to the culture medium. For H2O2 treatment, cells were exposed to up to 1500 μM H2O2 in MEM or MEM alone for 8 h, after which the medium was replaced and the cells were cultured for an additional 24 h.
2.3. Antibodies and reagents
Antibodies to the following proteins were purchased as indicated: HIF-1α (MA5-16008, Thermo Scientific) for immunofluorescence of cells, HIF-1α (610959, BD Transduction Laboratories, Franklin L., New Jersey) for immunoblotting, HIF-1α (NB100-105, Novus Biologicals, Littleton, Colorado, USA) for co-immunoprecipitation and HIF-1α (NB100-449, Novus Biologicals) for immunofluorescence of tissues, Bcl-XL (ab32370, Abcam, Cambridge, UK), cytochrome c (ab76237, Abcam), Grp75 (ab2799, Abcam), translocase of outer mitochondrial membrane 40 (TOMM40; ab185543, Abcam), histone deacetylase (HDAC; ab19845, Abcam), hemagglutinin (HA; 3724S, Cell Signaling Technology, Danvers, MA, USA), TOMM34 (A4467, ABclonal, Wuhan, China), proliferating cell nuclear antigen (PCNA; sc-56, Santa Cruz Biotechnology, Santa Cruz, CA, USA), TOMM40 (ab185543, Abcam), TOMM70A (A4349, ABclonal), translocase of inner mitochondrial membrane 17A (TIMM17A; A6449, ABclonal), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 2118, Cell Signaling Technology), β-actin (8457, Cell Signaling Technology), cleaved poly (ADP-ribose) polymerase (PARP; 5625, Cell Signaling Technology), β-tubulin (CW0098, CWBio, Beijing, China), apoptosis-inducing factor (AIF; 5318, Cell Signaling Technology), transcription factor A, mitochondrial (TFAM; 8076, Cell Signaling Technology), mitochondrial-encoded NADH dehydrogenase I (MT-ND1; A5250, ABclonal), mitochondrial-encoded cytochrome c oxidase II (MT-CO2; A7702, ABclonal), mitochondrial-encoded cytochrome b (MT-CYB; A9762, ABclonal), ATP6 (A8193, ABclonal), HSP70 (4872, Cell Signaling Technology), HSP90 (4877, Cell Signaling Technology), Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) antibody (A-11001, Invitrogen, Carlsbad, Cal., USA), and Alexa Fluor 594-conjugated chicken anti-rabbit IgG (H+L) antibody (A-21442, Invitrogen). Proteinase K from Engyodontium album was purchased from Sigma-Aldrich, MitoTracker Green and Deep Red were purchased from Invitrogen, and complete, EDTA-free Protease Inhibitor Cocktail Tablets were purchased from Roche (Basel, Switzerland).
2.4. Plasmid construction and cell transfection
The vector for expression of mitochondrial-targeted HIF-1α (mito-HIF-1α) was constructed by fusing the mitochondrial targeting sequence (MTS) from Neurospora crassa F0-ATPase subunit 9 (Su9) to the upstream (5′) sequence of a hemagglutinin (HA)-tagged human HIF-1α gene by overlap extension PCR, followed by subcloning into pHBLV-CMVIE-IRES-ZsGreen (HanBio, Chengdu, China). A control vector encoding HA-tagged MTS (mito-HA) was also constructed. Vectors for expression of mutant mito-HIF-1α proteins lacking the HIF-1 N-terminal activation domain (NTAD), C-terminal transactivation domain (CTAD), or inhibitory domain (ID) were constructed by two rounds of PCR. All constructs were verified by sequencing before use. Plasmids were purified from bacteria using a Plasmid Miniprep Plus Purification Kit (GeneMark, Taiwan, China) and transfected into cell lines using a jetPRIME transfection kit (Polyplus Transfection, Illkirch, France) according to the manufacturers’ instructions. Briefly, HeLa cells (7.5 × 105) were seeded in T25 flasks, grown for 24 h, and then transfected with 2 μg of mito-HIF-1α, mito-HA, or mutant mito-HIF-1α plasmid DNA. Transfection efficiency was assessed by expression of the green fluorescent protein ZsGreen (GFP) by immunofluorescence and flow cytometry (Supplementary Fig. S1). The cells were cultured for 36 h before use in experiments.
2.5. qRT-PCR
RNA was isolated using TRIzol (Invitrogen), quantified using a NanoDrop2000 spectrophotometer (Bio-Tek, Winooski, VT, USA), and treated with gDNase from a FastQuant RT Kit (TIANGEN Biotech, Beijing, China) according to the manufacturer's instructions. Primers were designed using National Center for Biotechnology Information (NCBI) Primer-BLAST, and synthesized by Life Technologies (Shanghai, China). Sequences are listed in Supplementary Tables S1–S3. cDNA was prepared using an iScript cDNA synthesis kit (Bio-Rad, Hercules, California, USA). cDNA samples were diluted 1:10 and amplified by real-time PCR using SsoFast EvaGreen Supermix (Bio-Rad) and a CFX96 Real-time PCR Detection System (Bio-Rad). For each primer pair, the annealing temperature was optimized by gradient PCR. Data were analyzed using the comparative method (2-ΔΔCT) and the fold-change in mRNA expression was normalized to both 18S rRNA and β-actin mRNA.
2.6. Mitochondrial DNA amplification
Total cellular DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen, Dusseldorf, Germany) and quantified using a NanoDrop2000 spectrophotometer (Bio-Tek). Primers were designed using NCBI Primer-BLAST and synthesized by Life Technologies (Shanghai, China). Sequences are listed in Supplementary Table S4. qRT-PCR was performed using SsoFast EvaGreen Supermix (Bio-Rad) and a CFX96 Real-time PCR Detection System (Bio-Rad). For each primer pair, the annealing temperature was optimized by gradient PCR. DNA were amplified using 100 ng DNA/reaction. The target gene content was normalized to that of 18S and β-globin DNA.
2.7. Co-immunoprecipitation
Whole cell lysate was extracted from HeLa cells by IP lysis buffer (87787, Pierce, Thermo Fisher) after CoCl2 treatment for 4 h. Proteins were incubated with normal mouse IgG (sc-2025, Santa Cruz) and Protein A/G PLUS-Agarose (sc-2003, Santa Cruz) for 30 min at 4 °C. Afterwards, proteins were incubated with Protein A/G PLUS-Agarose and either normal mouse IgG of anti- HIF-1α antibody overnight at 4 °C. Collect the pellet after wash and centrifuge. Add 1X loading buffer and apply the samples to immunoblot analysis described below.
2.8. Immunoblotting
Proteins were extracted with mammalian protein extraction reagent (Thermo Fisher, Waltham, MA, USA), separated by 10–12% SDS-PAGE, and transferred to PVDF membranes. Membranes were blocked for 1 h with 5% skim milk in Tris-buffered saline-Tween 20 (TBST; 25 mM Tris·HCl [pH 7.5], 1 mM NaCl, and 0.1% Tween 20 [Sigma-Aldrich]) at 37 °C, and then incubated with primary antibodies diluted in blocking buffer overnight at 4 °C. The membranes were washed three times for 5 min each with TBST and then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at 37 °C. Finally, the membranes were washed three times for 5 min each with TBST and incubated with chemiluminescent reagents (4A Biotech, Beijing, China). Bands were detected with a FUSION FX5 Imaging System (VILBER Fusion, Paris, France).
2.9. Apoptosis assays using Alexa Fluor647-conjugated annexin V/PI staining
H2O2 or hypoxia treated cells were harvested using 0.25% EDTA free trypsin, pelleted, washed twice with cold 1 × annexin-binding buffer, and resuspended with 100ul 1 × annexin-binding buffer. Alexa Fluor647-conjugated annexin V/PI (Beijing 4 A Biotech, Beijing, China) were stained according to the manufacturer's indicated protocol and analyzed by flow cytometry. GFP positive cells were gated and analyzed.
2.10. Cell viability assay
Cells were harvested 24 h after transfection, resuspended at 5 × 104 cells/mL, aliquoted into 96-well plates at 200 μL/well, and incubated overnight at 37 °C to allow cell attachment. The cells were then treated with the indicated concentrations of H2O2 (or medium alone as a control) for 8 h, and then transferred to normal medium for an additional 24 h. An aliquot of 10 μL Cell Counting Kit-8 (CCK-8; 7sea, Shanghai, China) reagent was added to each well and the cells were incubated for 4 h at 37 °C. The optical density (OD) at 450 nm was measured using a microplate spectrophotometer. Cell viability was calculated as: % viability = (OD of the treatment group / OD of the control group) × 100%.
2.11. Subcellular fractionation
Intact mitochondria were isolated from cells and tissues using a Mitochondria Isolation Kit for Cultured Cells and a Mitochondria Isolation Kit for Tissue (both Thermo Fisher), respectively, according to the manufacturer's protocols. Mitochondrial proteins were extracted by lysis of mitochondria in mammalian protein extraction reagent (Thermo Fisher). Nuclei were isolated using a Nuclei Isolation Kit (Biohao, Shanghai, China) according to the manufacturer's instructions. To detect subcellular distribution of AIF, cells were separated into three distinct fractions: cytoplasmic, organelle (mitochondria), and nuclear/cytoskeletal, using a Cell Fractionation Kit (#9038, Cell Signaling Technology) according to the manufacturer's instructions.
2.12. Protease sensitivity analysis
Two aliquots of isolated intact mitochondria were treated with protease K with or without 1% Triton X-100 for 10 min at room temperature. A third aliquot remained untreated and served as a control. The reactions were stopped by addition of phenylmethylsulfonyl fluoride (2 mM), the samples were centrifuged, and the pelleted mitochondria were lysed. Protein samples were resolved by SDS-PAGE and immunoblotted as described above.
2.13. Caspase assay
The activity of generic caspases (caspase-1, -3, -4, -5, -6, -7, -8 and -9) was measured using the red fluorescent TF5-VAD-FMK probe (Abcam). Briefly, cells were harvested and resuspended in medium at 1 × 106 cells/mL. Aliquots of 0.5 mL cells were mixed with 1 μL of 500 × TF5-VAD-FMK and incubated for 1 h at 37 °C in the dark. The cells were washed twice with medium, resuspended in 0.5 mL medium, and analyzed by flow cytometry. Caspase activity was detected as fluorescence intensity in the APC channel (Ex/Em = 650/670 nm) after gating on GFP-positive cells.
2.14. Mitochondrial transmembrane potential assay
Mitochondrial membrane potential (ΔΨm) was measured by fluorescence microscopy and flow cytometry of cells incubated with tetramethylrhodamine methyl ester (TMRM; Invitrogen). TMRM is a red/orange cell-permeable fluorescent dye that is readily taken up and sequestered by healthy intact mitochondria, but the signal is lost as the ΔΨm is disrupted. For microscopy, 2 × 104 cells/well were seeded in 6-well plates, cultured overnight, and exposed to hypoxia or normoxia for 16 h. The cells were then incubated with 25 nM TMRM diluted in Hank's balanced salt solution (HBSS) for 30 min in the dark and analyzed with a fluorescent microscope (Olympus IX71, Tokyo, Japan). Fluorescence intensities on captured images were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA). For flow cytometry, the cells after hypoxia treatment were harvested, washed twice with HBSS, incubated for 30 min in the dark with 25 nM TMRM/HBSS, and analyzed by flow cytometry after gating on GFP-positive cells.
2.15. Intracellular ROS detection
ROS were quantified by flow cytometry using the CellROX Deep Red (Thermo Fisher) or Dihydroethidium (DHE; Beyotime Biotechnology, Nanjing, China) method. Briefly, cells were exposed to hypoxia or normoxia, harvested, and resuspended in medium at 1 × 106 cells/mL. Aliquots of 0.5 mL of cells were centrifuged and the pellet was resuspended in medium containing 500 nM CellROX Deep Red reagent or 5 μM DHE. The cells were gently mixed, incubated for 30 min at 37 °C in the dark, and analyzed by flow cytometry after gating on GFP-positive cells.
2.16. Measurement of cytochrome c release
Cytochrome c release was measured using a Human Cytochrome c ELISA Kit (ab119521, Abcam). Briefly, cells were exposed to hypoxia or normoxia, harvested, and resuspended in Lysis Buffer at 1.5 × 106 cells/mL. The lysate was incubated for 1 h at room temperature with gentle shaking, and the organelle-free supernatant was collected by centrifugation at 200 ×g for 15 min. The concentration of cytochrome c was determined by ELISA, according to the manufacturer's recommendations.
2.17. Measurement of GSH and GSSG
Cells were exposed to H2O2 and collected. Samples were prepared based on the manufacturer's instructions. The total GSH and GSSG levels were determined by GSH and GSSG Assay Kit (Beyotime Biotechnology, China).
2.18. Histology and fluorescence microscopy
For histology, liver samples were fixed for 24 h in 4% paraformaldehyde, embedded in paraffin, sliced into 5 μm-thick sections, and stained with hematoxylin and eosin (H&E) or Sirius Red. For fluorescence microscopy, frozen liver samples were sectioned and incubated with 100 nM MitoTracker Deep Red CMXRos (Thermo Fisher) at 37 °C for 20 min. Sections were washed with PBS (10 mM sodium phosphate, pH 7.4, 150 mM NaCl), fixed with 3.7% formaldehyde for 5 min, and washed again. The fixed sections were incubated with a 1:100 dilution of rabbit polyclonal anti-HIF-1α (NB100-449, Novus) in PBS at 37 °C for 2 h, washed, and then incubated with FITC-conjugated goat anti-rabbit secondary antibody at 37 °C for 30 min. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) at room temperature for 5 min. Sections were visualized and imaged using a fluorescence microscope (Zeiss, Imager Z2, Jena, Germany).
2.19. Statistical analysis
Data are shown as the mean ± standard error (SEM). Group differences were analyzed by unpaired Student's t-test or one-way or two-way analysis of variance (ANOVA) followed by Sidak's multiple comparisons test, as indicated, using GraphPad Prism version 7.02. A p value of ≤ 0.05 was considered significant.
3. Results
3.1. Oxidative stress induces mitochondrial translocation of endogenous HIF-1α
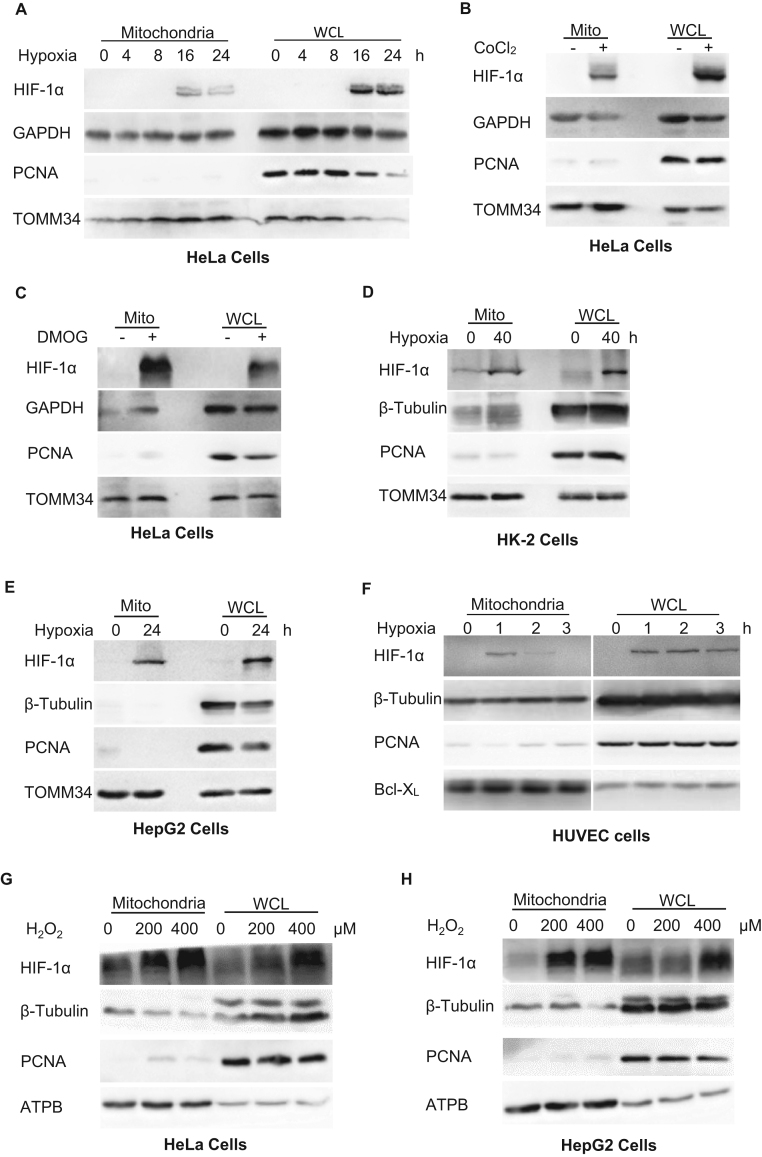
To determine the effects of hypoxia on HIF-1α localization, we analyzed whole cell lysates and mitochondria isolated from HeLa cells exposed to hypoxia for up to 24 h. HIF-1α expression was detected in the mitochondrial fractions after 16 h of hypoxia, as indicated by its co-localization with the mitochondrial marker protein TOMM34 (Fig. 1A). This observation suggested that HIF-1α translocates to the mitochondria under hypoxia, as reported for other cell lines [10], [11]. To verify this, we treated HeLa cells with CoCl2, which induces HIF-1α expression, or DMOG, which stabilizes HIF-1α by inhibition of PHDs, thereby mimicking the effects of hypoxia. These treatments also induced the appearance of HIF-1α in the mitochondrial fraction (Fig. 1B, C). Similar results were obtained when HIF-1α translocation was visualized by confocal fluorescence microscopy (Supplementary Fig. S2A). To further confirm direct association between HIF-1α and mitochondria, we conducted co-immunoprecipitation to detect mitochondrial proteins interacting with HIF-1α after CoCl2 treatment. The results suggested that HIF-1α interacts with Hsp70/Hsp90, chaperones escorting proteins targeting mitochondria and docking at the translocase of outer membrane (TOM) complex [13], [14], and TOMM40, a subunit of TOM complex [15], indicating the possibility that its importing pathway involving hsp70/hsp90-TOM interaction (Supplementary Fig. S3). Next, we confirmed the reproducibility of this phenomenon by analysis of HIF-1α expression in HK-2 (transformed human kidney cell line), HepG2 (human liver cancer cell line), HUVEC (human umbilical vein endothelial cell line) and L02 (normal human hepatic cell line) exposed to hypoxia (Fig. 1D–F, Supplementary Fig. S2B) and analysis of HK-2 cells treated with DMOG (Supplementary Fig. S2C).
Fig. 1.
Translocation of HIF-1α to the mitochondria during oxidative stress. A–H. Immunoblot analysis of isolated mitochondria or whole cell lysates (WCL). A. HeLa cells exposed to normoxia (0) or hypoxia for the indicated times. B, C. HeLa cells treated for 4 h with or without CoCl2 (B) or DMOG (C). D–F. Immunoblot analysis of HK-2 (D), HepG2 (E), and HUVEC (F) cells. G, H. HeLa (G) and HepG2 (H) cells exposed to H2O2 treatment at the indicated concentrations. GAPDH and β-tubulin were probed as cytosolic markers, PCNA as a nuclear marker, and Bcl-XL, TOMM34 and ATPB as mitochondrial markers.
We also confirmed this phenomenon by immunoblotting in HeLa and HepG2 cells exposed to H2O2 treatment, another classic oxidative stress inducer. The results indicated that H2O2 treatment induces mitochondrial translocation of HIF-1α in a dose-dependent way (Fig. 1G, H). Collectively, these experiments demonstrate that mitochondrial translocation of HIF-1α occurs in a highly reproducible manner across multiple cell lines exposed to oxidative stress.
3.2. Oxidative stress-induced apoptosis is inhibited by ectopic expression of mito-HIF-1α
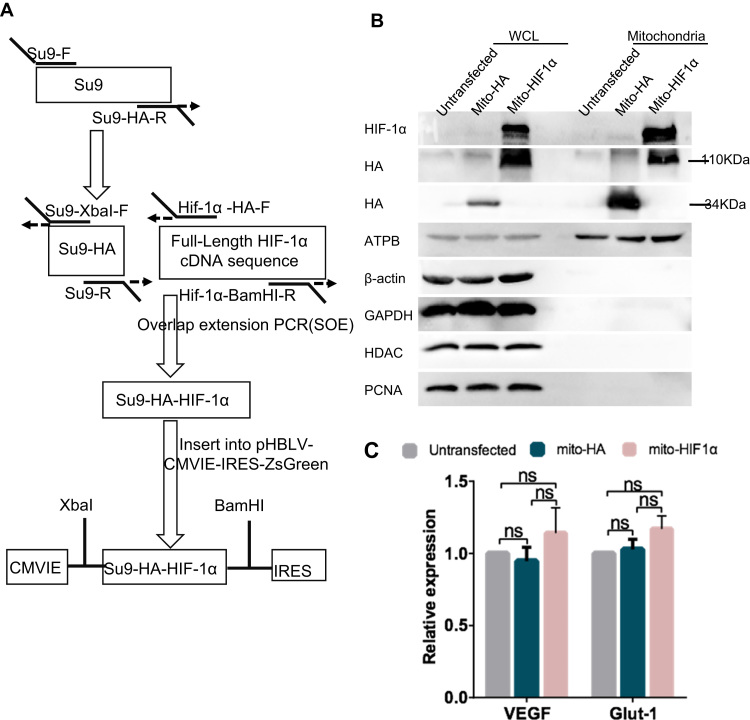
To investigate the functions of mitochondrial-targeted HIF-1α, we stably transfected HeLa cells with a vector encoding a GFP- and HA-tagged form of HIF-1α attached to an MTS, which we refer to as mito-HIF-1α. mito-HA, lacking only the HIF-1α sequence, was expressed as a control (Fig. 2A). Immunoblot analysis of untransfected and transfected HeLa cells confirmed that mito-HIF-1α was recruited to the mitochondria under normoxic conditions, indicating that the construct mimicked the behavior of endogenous mtHIF-1α during hypoxia (Fig. 2B). To determine whether mito-HIF1α functions as a classic nuclear transcription factor, we performed qRT-PCR analysis of several HIF-1α target genes, including vascular endothelial growth factor, and glucose transporter. However, we observed no significant differences in their transcription in untransfected or transfected cells (Fig. 2C), confirming that the ectopically expressed mitochondrial-targeted form of HIF1α did not act as a classical nuclear transcription factor.
Fig. 2.
Construction of mito-HIF-1α and mito-HA expression vectors. A. Construction of mito-HIF-1α overexpression vector. The MTS from N. crassa F0-ATPase subunit 9 (Su9) was fused to the 5′ sequence of the HA-tagged human HIF-1α gene and subcloned into the pHBLV-CMVIE-IRES-ZsGreen vector. The mito-HA control vector lacked the HIF-1α sequence. B. Immunoblot analysis of isolated mitochondria or whole cell lysates (WCL) of HeLa cells confirming the mitochondrial localization of mito-HIF1α and mito-HA. ATPB was probed as a mitochondrial marker, HDAC and PCNA as a nuclear marker, β-actin and GAPDH as a cytosolic marker. C. qRT-PCR analysis of vascular endothelial growth factor (VEGF) and glucose transporter 1(GLUT1) mRNA levels in untransfected or transfected HeLa cells. Data are normalized to RPS18 and β-actin mRNA levels and are expressed as the means ± SEM of n = 3 samples. ns, not significant.
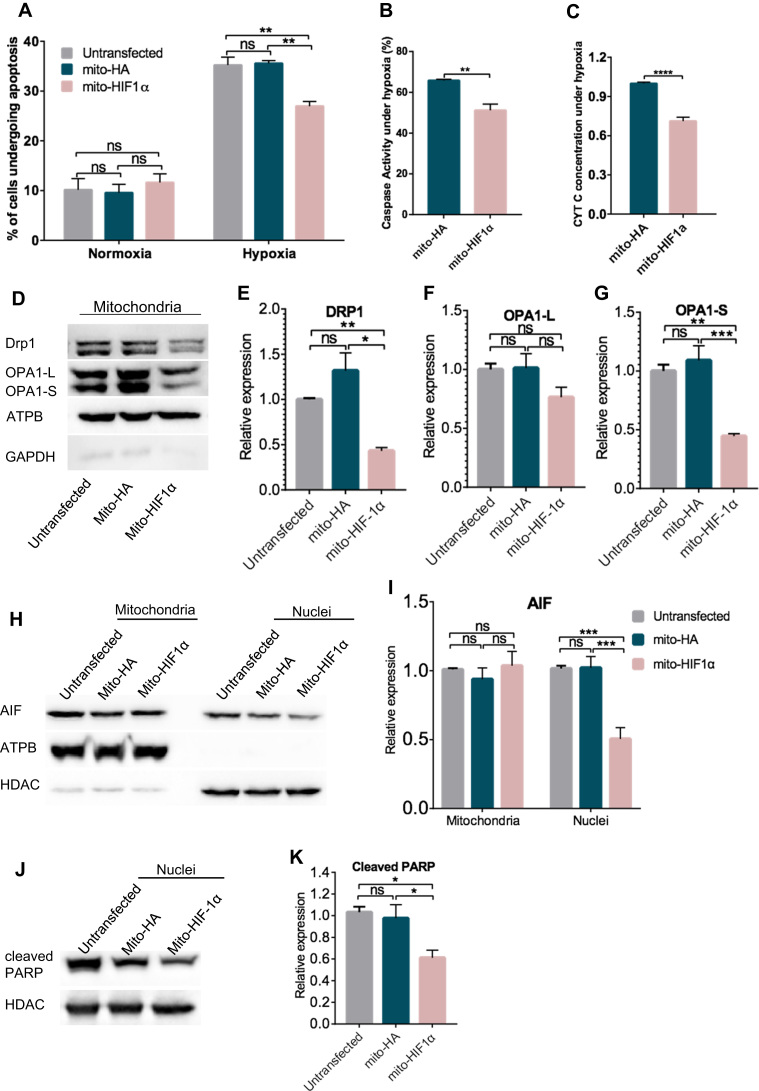
Next, we asked whether expression of mito-HIF-1α would affect apoptosis under hypoxia. Apoptosis was measured using fluorescent annexin V/PI staining and caspase assays after incubation of HeLa cells under normoxia or hypoxia for 16 h. We found that ~10% of untransfected and transfected cells underwent apoptosis under normoxia. Exposure to hypoxia increased the apoptosis rate of untransfected and mito-HA transfected cells to ~40%, whereas mito-HIF1α expression significantly reduced the apoptosis rate (Fig. 3A). Similarly, two additional markers of apoptosis—caspase activation (caspase-1, -3, -4, -5, -6, -7, -8, and -9) and cytochrome c release into the cytosol—both revealed that mito-HIF1α expression significantly reduced apoptosis compared with the control cells transfected with mito-HA (Fig. 3B, C).
Fig. 3.
Inhibition of hypoxia-induced apoptosis by expression of mito-HIF-1α. A, B. Flow cytometric analysis of apoptosis (annexin V/PI; A) and caspase activity (TF5-VAD-FMK; B) after exposure of HeLa cells to normoxia or hypoxia for 16 h. C. Cytochrome c concentration in the cytosol of transfected HeLa cells measured by ELISA. D–K. Immunoblot analysis of mitochondrial fusion/fission proteins in mitochondrial fractions(D-G), subcellular distribution of AIF in mitochondrial and nuclear fractions (H, I), and cleaved PARP in nuclear lysates (J, K) of untransfected and transfected HeLa cells exposed to hypoxia for 16 h. Signals were quantified with the Image J software and analyzed by GraphPad Prism software. Data are the means ± SEM (n ≥ 3). ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ATPB was probed as a mitochondrial marker, HDAC as a nuclear marker, and GAPDH as a cytosolic marker.
We also examined the effects of mitochondrial HIF-1α targeting on the recruitment of several additional mitochondrial proteins associated with apoptosis. Drp1 regulates mitochondrial outer membrane permeability, and a reduction in its expression is associated with the release of mitochondrial intermembrane space proteins such as cytochrome c, activation of caspases, and cell death [16]. OPA1 is involved in mitochondrial fusion and cristae remodeling, which is also associated with apoptotic cytochrome c release [17]. The tightness of cristae junctions correlates with oligomerization of membrane-bound OPA1L and soluble OPA1S. Proteolytic cleavage of OPA1L to OPA1S allows formation of the OPA1S complex, which, together with BNIP3, promotes mitochondrial outer membrane permeabilization, cytochrome c release, and cell death [18]. As shown in Fig. 3D–G, expression of mito-HIF-1α significantly reduced mitochondrial association of Drp1 and decreased the proportion of OPA1S, in accordance with reduction of cytochrome c release, in hypoxia-exposed HeLa cells compared with mito-HA expression, suggesting apoptosis was inhibited by expression of mito-HIF-1α.
Moreover, we verified the ability of mito-HIF-1α to reduce hypoxia-associated mitochondrial damage and cell death by examining the expression of AIF and cleaved PARP. AIF, released from mitochondria, accumulates in the nucleus of apoptotic cells and causes DNA degradation [19], while PARP is cleaved by caspase 3 during apoptosis [20]. Indeed, we observed that nuclear accumulation of AIF (Fig. 3H, I) and cleavage of PARP (Fig. 3J, K) was higher in untransfected and mito-HA-transfected cells compared with mito-HIF-1α-expressing cells upon exposure to hypoxia. Taken, together, these data demonstrate that expression of mitochondrial-targeted HIF-1α significantly inhibited hypoxia-induced apoptosis, as measured by multiple assays.
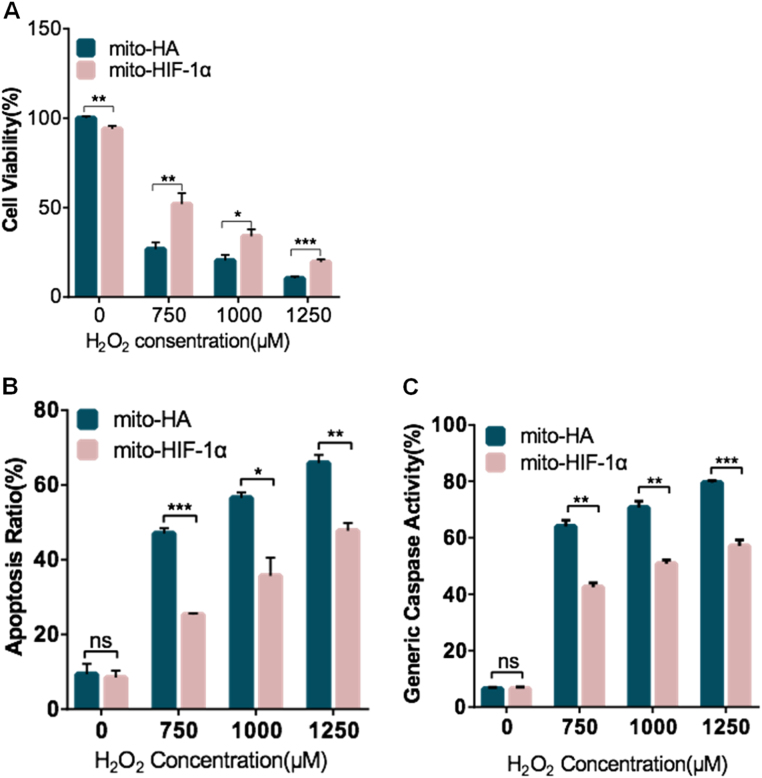
Finally, we asked whether mitochondrial-targeted HIF-1α can protect cells from oxidative stress induced by exposure to H2O2. For this, we examined cell viability (CCK-8 assay), apoptosis (annexin V/PI assay), and caspase activity (TF5-VAD-FMK assay) after cell treatment with different concentrations of H2O2. The results showed that, while H2O2 induced a dose-dependent decrease in cell viability in both the mito-HA- and mito-HIF-1α-expressing cells, the latter cells were significantly more resistant to cell death (Fig. 4A). Similarly, while treatment with moderate (750 μM) to high (1.25 mM) concentrations of H2O2 increased apoptosis, as measured by annexin V/PI staining (Fig. 4B) and caspase activity (Fig. 4C), mito-HIF-1α expression reduced cell death by ~50% compared with mito-HA. These data further demonstrate that mitochondrial-localized HIF-1α protects against oxidative stress-induced apoptosis.
Fig. 4.
Reduction of H2O2 -induced apoptosis by expression of mito-HIF-1α. A. Transfected HeLa cell viability was assessed after H2O2 treatment for 8 h using the CCK-8 assay. B, C. Flow cytometric analysis of apoptosis (annexin V/PI; B) and caspase activity (TF5-VAD-FMK; C) after H2O2 treatment of transfected HeLa cells for 24 h. Data are the means ± SEM (n ≥ 3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
3.3. Expression of mito-HIF-1α reverses oxidative stress-induced mitochondrial dysfunction
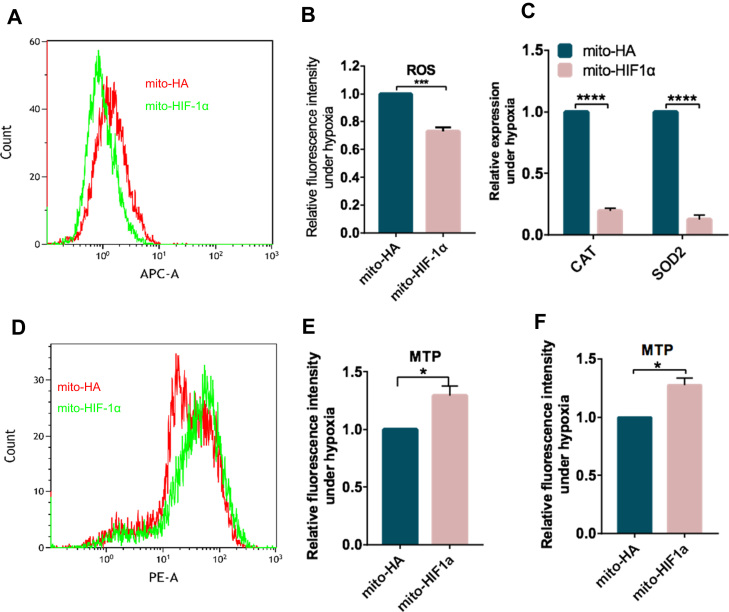
HIF-1α has been implicated in controlling ROS levels during hypoxia [6]. Based on our results thus far, we speculated that expression of mito-HIF-1α might reduce ROS production. To test this, we exposed transfected cells to hypoxia for 8 h and then labeled them with CellROX, a membrane-permeant redox-sensitive fluorescent dye. FACS analysis of the labeled cells indicated that ROS levels were reduced by ~30% in cells expressing mito-HIF-1α compared with the control cells (Fig. 5A, B). Superoxide anions are produced by mitochondrial complexes I and II and released into the mitochondrial matrix, where they can be converted to hydrogen peroxide and then to water by the activity of superoxide dismutase 2 (SOD2) and catalase (CAT) [21]. Notably, both SOD2 and CAT mRNA levels were markedly lower in mtHIF-1α-expressing compared with control cells, suggesting that mito-HIF-1α lowers ROS levels by inhibiting their production, rather than by promoting their detoxification (Fig. 5C).
Fig. 5.
Reduction of hypoxia-induced ΔΨm collapse and ROS production by mito-HIF-1α expression. A–F. Transfected HeLa cells were exposed to hypoxia for 8 h and then analyzed. A, B. Flow cytometry histograms (A) and quantification (B) of ROS production by the CellROX DeepRed assay. C. qRT-PCR analysis of superoxide dismutase 2 (SOD2) and catalase (CAT) mRNA levels. Data were normalized to RPS18. D, E. Flow cytometry histograms (D) and quantification (E) of ΔΨm in TMRM-labeled cells. F. Quantification of TMRM fluorescence intensity measured by microscopy. Data are the means ± SEM (n ≥ 3). *p < 0.5, ***p < 0.001, *** *p < 0.0001.
We next assessed the effects of mito-HIF-1α on mitochondrial transmembrane potential (ΔΨm), which plays a vital role in maintaining the physiological function of mitochondria. Cells were labeled with the membrane-permeant fluorescent dye TMRM, which is sequestered in the mitochondria of healthy cells [22]. Analysis of the cells by both fluorescence microscopy and flow cytometry showed that the ΔΨm of hypoxia-exposed cells was higher in mito-HIF-1α-expressing cells than the control cells (Fig. 5D–F).
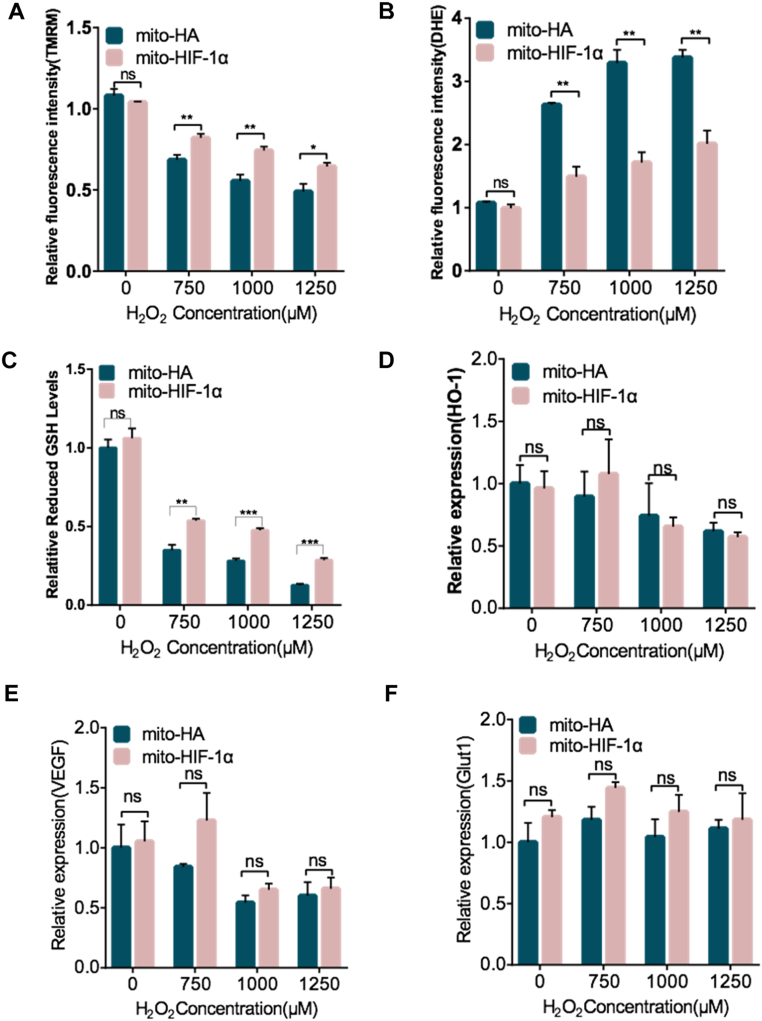
We also examined ROS levels and ΔΨm after H2O2 treatment for 8 h. Likewise, ΔΨm was higher (Fig. 6A) and ROS was lower (Fig. 6B) in cells expressing mito-HIF-1α than the control cells. Glutathione (GSH), the main non-protein thiol anti-oxidant, is required to scavenge ROS [23]. Therefore, we measured the GSH levels and found it significantly higher in mito-HIF-1α-expressing cells (Fig. 6C). Nuclear factor erythroid 2-related factor 2 (NFE2L2 or Nrf2)/heme Oxygenase-1 (HO-1) pathway is an important anti-oxidative and protective pathway for cells [24]. To exclude the possibility that mito-HIF-1α may affect nuclear pathways of HIF-1α, we detected mRNA levels of HO-1 and Glut 1 and VEGF, both of which are downstream genes of HIF-1α. The results indicated unchanged mRNA levels of these genes in both cell types (Fig. 6D–F), suggesting that anti-oxidative functions of HIF-1α in mitochondria is independent of its classic nuclear pathways. Taken together, these results demonstrate that mitochondrial-associated HIF-1α reduces ROS production and preserves mitochondrial function against oxidative stress.
Fig. 6.
Reduction of H2O2 -induced ΔΨm collapse and ROS production by mito-HIF-1α expression. A–F. Transfected HeLa cells were exposed to H2O2 treatment for 8 h and then analyzed. A, B. Quantification of ΔΨm in TMRM-labeled cells and ROS production after DHE staining by flow cytometry. C. Quantification of GSH levels by GSH/GSSG assay. D-F. qRT-PCR analysis of heme oxygenase-1 (HO-1), vascular endothelial growth factor (VEGF) and glucose transporter 1(GLUT1) mRNA levels. Data were normalized to RPS18 and β-actin mRNA. Data are the means ± SEM (n ≥ 3). *p < 0.5, ***p < 0.001, ****p < 0.0001, ns, not significant.
3.4. Mito-HIF-1α expression downregulates mitochondrial DNA-encoded mRNA expression
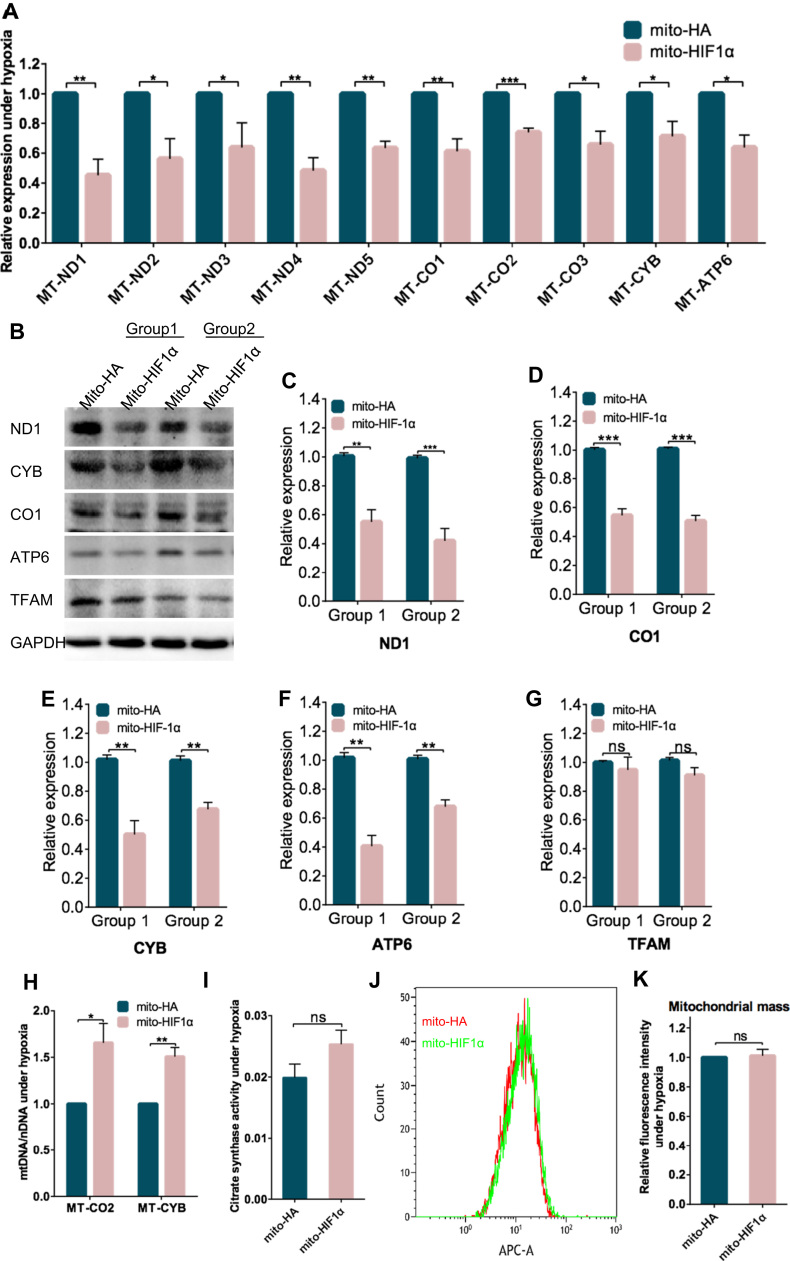
To further dissect the mechanisms underlying inhibition of apoptosis by mito-HIF-1α, we analyzed mRNA and protein levels of several mtDNA-encoded genes following cell exposure to hypoxia for 8 h. Both the mRNA levels (Fig. 7A) and protein levels (Fig. 7B–F) of all of the genes tested were lower in mito-HIF-1α-expressing cells than in control cells. Interestingly, the expression of TFAM, a key activator of mitochondrial transcription and regulator of mitochondrial genome replication [25], was unchanged (Fig. 7G). Next, to determine whether the reduction above could be explained by a reduction in mtDNA mass in the transfected cells, we measured the mtDNA copy number of mitochondrial-encoded cytochrome c oxidase subunit II (COII) and cytochrome b by qPCR. However, the copy number was significantly higher in the mito-HIF-1α-expressing cells than the control cells (Fig. 7H). Moreover, the activity of citrate synthase, which has been shown to reflect the overall volume of mitochondria and is thus a proxy for mitochondrial number [26] was not significantly different in mito-HA- and mito-HIF-1α-expressing cells (Fig. 7I). Finally, we also detected no significant difference between the two cell types in the fluorescence intensity of the mitochondrial marker MitoTracker Deep Red (Fig. 7J, K). Taken together, these data support the notion that mito-HIF-1α reduced the mRNA levels of mtDNA-encoded genes without affecting on mitochondrial mass.
Fig. 7.
Downregulation of mitochondrial DNA-encoded mRNA expression in mito-HIF-1α-transfected cells. Transfected HeLa cells were exposed to hypoxia for 8 h and then analyzed. A. qRT-PCR analysis of the indicated mitochondrial mRNAs. Data were normalized to RPS18. B-G. Immunoblot analysis (B) and corresponding quantification (C–G) of mitochondrial gene encoding proteins in whole cell lysates of transfected cells after 16 h of hypoxia. H. The ratio of mitochondrial DNA to nuclear DNA was determined by quantification of COII and Cyt B by qRT-PCR. Data were normalized to RPS18 and β-globin. I. Citrate synthase activity in whole cell lysates. J, K. Flow cytometry histograms (J) and quantification (K) of MitoTracker Deep Red fluorescence intensity. Data are the means ± SEM (n ≥ 3). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
3.5. The hypoxia-protective effects of mito-HIF-1α are independent of its transcriptional activity
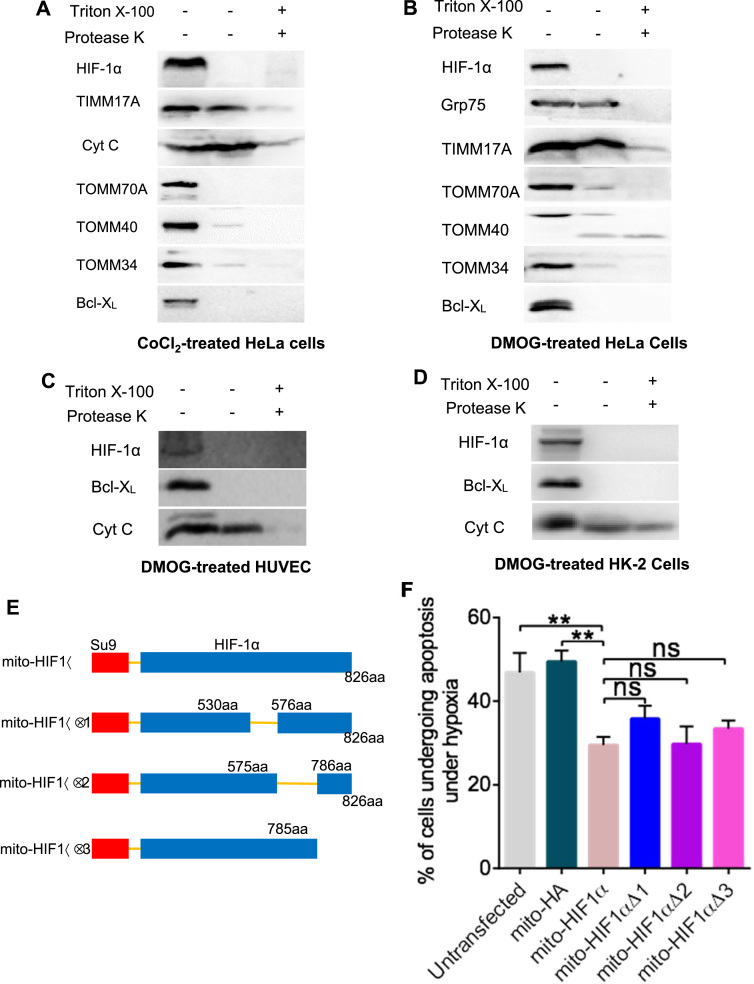
Although Mylonis et al. have reported that HIF-1α interacts with VDAC1 and hexokinase II, both locating in the mitochondrial outer membrane, mortalin, another component of this functional complex [12], also exists in the mitochondrial matrix [27], where mtDNA is located. Given that our results showed down regulated mitochondrial mRNA levels, it's of possibility that HIF-1α might exert functions in the matrix. Thus, we next investigated whether the reduction in mRNA of mtDNA-encoded target genes was mediated via the transcriptional activity of mitochondrial HIF-1α. Firstly, we examined the sub-mitochondrial localization of HIF-1α. Outer membrane proteins are protease sensitive, whereas internal proteins are accessible to proteases only after membrane disruption (e.g., experimentally by treatment with detergents) [28]. Therefore, we analyzed the protease sensitivity of HIF-1α and other mitochondrial proteins from CoCl2- and DMOG-treated cell lines. Intact mitochondria were isolated and incubated with protease K with or without detergent. HIF-1α and various mitochondrial marker proteins were then analyzed by immunoblotting. We found that HIF-1α and the outer membrane markers TOMM70A, TOMM40, TOMM34, and Bcl-XL were all digested by protease K alone, whereas the inner membrane marker TIMM17A and the intermembrane space marker cytochrome c were digested only after detergent treatment (Fig. 8A–D). These data indicate that HIF-1α is likely to be associated with the outer mitochondria membrane and thus inaccessible to mtDNA. This observation argues against the involvement of HIF-1α transcriptional activity in its ability to protect against hypoxia-induced damage.
Fig. 8.
The effects of mito-HIF-1α are independent of its transcriptional activity. A–D. Protease K sensitivity assay of the indicated mitochondrial proteins. Immunoblot analysis of intact mitochondria isolated from CoCl2-treated HeLa cells (A) or DMOG-treated HeLa (B), HUVEC (C), and HK-2 (D) cells. E. Construction of vectors for expression of mutant mito-HIF-1α proteins lacking the NTAD, ID, or CTAD. aa, amino acid. F. Flow cytometric analysis of apoptosis (annexin V/PI) in transfected HeLa cells after exposure to hypoxia for 16 h. Grp75 was probed as a matrix marker; TIMM17A as an inner membrane marker; cytochrome c as an intermembrane space marker; and TOMM70A, TOMM40, TOMM34, and Bcl-XL as outer membrane markers. Data are means ± SEM (n = 5). ns, not significant; **p < 0.01.
To probe this further, we transfected cells with vectors encoding mito-HIF-1α mutant proteins lacking the NTAD (transcription activation domain in N terminal) (mito-HIF1α∆1), ID (inhibitory domain which inhibits adjacent NTAD and CTAD) (mito-HIF1α∆2), or CTAD (transcription activation domain in C terminal) (mito-HIF1α∆3) (Fig. 8E). Notably, deletion of CTAD, NTAD, or ID had no effects on the ability of mito-HIF-1α to inhibit hypoxia-induced apoptosis (Fig. 8F), indicating that these domains are not required for this function of mito-HIF-1α. Therefore, although mitochondrial mRNA expression is reduced by mito-HIF-1α overexpression during hypoxia, the protective effect of mito-HIF-1α is independent of its conventional transcriptional activity.
3.6. Endogenous HIF-1α translocates to the mitochondria in fibrotic mouse liver
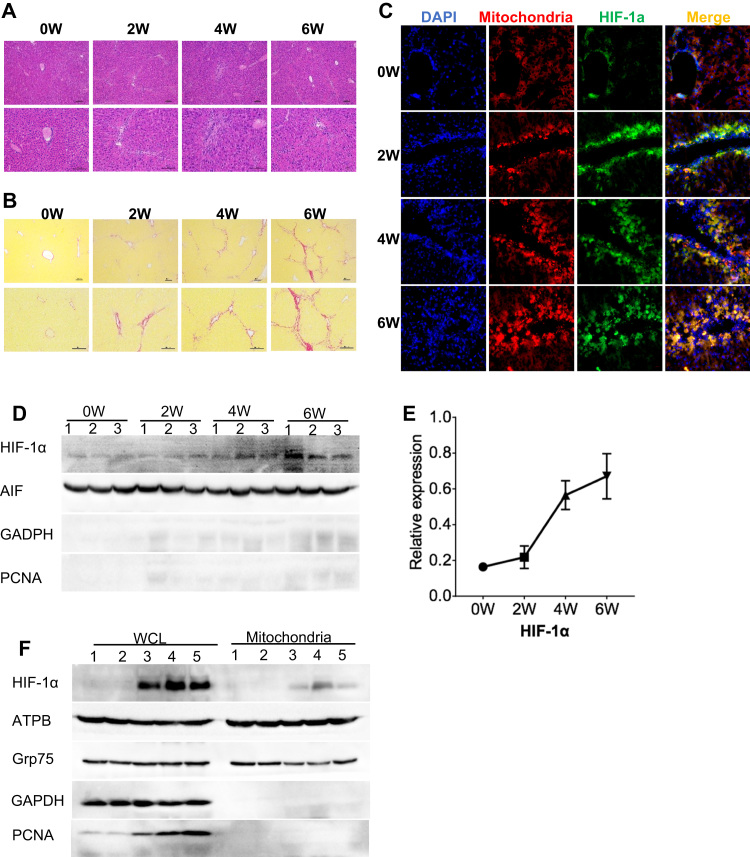
A growing body of work suggests that HIF-1α expression is significantly increased in hepatic fibrotic tissues [29] and oxidative stress drives the fibrogenic response in liver fibrosis [30]. Therefore, we asked whether HIF-1α translocates to the mitochondria during liver fibrosis. We treated mice with CCl4 for 2, 4, or 6 weeks to induce liver fibrosis, and then examined the localization of endogenous HIF-1α in liver cells. H&E staining of liver sections revealed that CCl4 induced changes associated with fibrosis. Levels of necrotic hepatocytes and inflammatory infiltrates gradually increased over the 6 weeks of CCl4 treatment (Fig. 9A), as was the level of collagen deposition, indicative of fibrosis, detected by Sirius red staining (Fig. 9B). Furthermore, immunofluorescence staining of HIF-1α showed that expression of HIF-1α was upregulated in parallel with the development of fibrosis (Fig. 9C). Notably, we also observed a progressive increase in the proportion of HIF-1α co-localized with MitoTracker DeepRed, indicating mitochondrial translocation (Fig. 9C). Finally, immunoblot analysis confirmed the specific translocation of HIF-1α to the mitochondrial fraction of fibrotic livers during CCl4 treatment (Fig. 9D–F).
Fig. 9.
Mitochondrial translocation of endogenous HIF-1α in mouse liver after induction of fibrosis. A, B. H&E (A) and Sirius red (B) staining of liver sections from mice administered CCl4 for 0, 2, 4, or 6 weeks. Scale bars, 100 µm. C. Immunofluorescence microscopy of liver sections stained for HIF-1α (green) and mitochondria (red, MitoTracker Deep Red) in fibrotic mouse liver. Nuclei were stained with DAPI (blue). D, E. Immunoblot analysis (D) and quantification (E) of mitochondrial proteins in liver extracts from mice administered CCl4 for 0, 2, 4, or 6 weeks. Lanes 1–3 indicate samples from three mice. F. Immunoblot analysis of the indicated proteins in whole cell lysates (WCL) and mitochondrial liver extracts from untreated mice (lanes 1 and 2) and mice treated with CCl4 for 6 weeks (lanes 3–5). GAPDH was probed as a cytosolic marker, PCNA as a nuclear marker, and AIF and ATPB as mitochondrial markers.
4. Discussion
Numerous studies have identified important roles for HIF-1α in cellular function and dysfunction through its transcriptional activity [4]. A few studies have reported that HIF-1α is translocated to the mitochondria in human and mouse cancer cell lines [10], [11]. Here, we show that a small fraction of HIF-1α associates with the mitochondria in a highly reproducible manner in human non-cancer as well as cancer cell lines after stabilization in response to oxidative stress, including hypoxia and H2O2 treatment. More importantly, we first detected expression of endogenous mtHIF-1α in mouse liver after induction of fibrosis. Besides, HIF-1α translocates to mitochondria in human liver cell line L02 and human liver carcinoma cell line HepG2, indicating that mitochondrial translocation of HIF-1α not only is broadly observed across cell lines but also is likely to be clinically relevant to the progress from liver fibrosis to liver carcinoma. Rane et al. reported that downregulation of miR-199a or hypoxic preconditioning induced HIF-1α association with the mitochondria of rat cardiac myocytes, where it contributed to the maintenance of mitochondrial membrane potential [11]. Given that mtHIF-1α has now been observed in many cell types in vitro and under pathological conditions in vivo, it is possible that mtHIF-1α may be responsible for some of the known functions of HIF-1α.
Our results showed that the kinetics of hypoxia-induced accumulation of HIF-1α at the mitochondria varied in the different cell lines investigated. In the human non-cancer cell lines L02 and HUVEC, HIF-1α was detected at the mitochondria within 1 h of hypoxia, but the effect was transient in HUVEC cells. In HeLa cells, mitochondrial HIF-1α was only detectable after 16 h hypoxia. However, the association was sustained for long periods in HeLa, Hep-G2, and HK-2 cells (up to 40 h). Briston et al. reported that HIF-1α could be detected in mitochondria within 2 h of hypoxia in the HCT116 cell line [10]. Therefore, although HIF-1α translocation to the mitochondria appears to be a common event, its kinetics and magnitude varies in both cancer and non-cancer cells. We speculate that the varying translocation patterns may partially reflect the individual cellular responses to hypoxia.
Previous studies indicated that HIF-1α reduces ROS levels via multiple pathways during hypoxia, including: improving the efficiency of complex IV by switching the cytochrome c oxidase subunit COX4-1 to COX4-2; induction of pyruvate dehydrogenase kinase 1 and lactate dehydrogenase A, the former shunting pyruvate away from the mitochondria and the latter converting pyruvate to lactate; induction of BNIP3, triggering mitochondrial-selective autophagy; and induction of microRNA-210, which blocks assembly of Fe/S clusters required for oxidative phosphorylation (reviewed in [31]). Our results here suggest the existence of a new pathway for decreasing ROS, which involves the association of HIF-1α with the mitochondrial outer membrane. We speculate that mtHIF-1α downregulates mitochondrial mRNA production, which inhibits respiratory chain activity (since some components are encoded by mtDNA) and decreases the demand for O2, thereby alleviating the degree of hypoxia and reducing ROS levels. Notably, previous study and ours have proved association of HIF-1α with outer mitochondrial membrane [12]; however, our results showed down regulation of mtDNA encoded mRNA levels in the matrix by ectopic expression of mtHIF-1α, which can’t be explained by the reported mechanism of HIF-1α/mortalin/VDAC1/HK-II [12]. We speculated that an unknown pathway involving downstream factors located in the matrix exists. Johnson et al. has reported that p53, another transcription factor translocating to mitochondria, inhibited mitochondrial translocation of RelA, which recruits to mitochondrial genome and represses mitochondrial gene expression, by binding to heat shock protein Mortalin which facilitates translocation of RelA to mitochondria, thus indirectly regulating mitochondrial transcription [32]. Therefore, it's possible that mtHIF-1α affects mitochondrial transcription through a similar mechanism.
Most studies of HIF-1α have focused on its transcriptional activity-dependent functions. However, our results indicate that the oxidative stress-related effects of mtHIF-1α occur independently of its translocation to the nucleus and do not require its transcriptional activity. In fact, several studies have identified functions for HIF-1α outside the nucleus. Khan et al. found that HIF-1α co-localized with peroxisomes, not the nucleus, in primary rat hepatocytes during hypoxia–reoxygenation [33]. Hubbi et al. showed that HIF-1α inhibits DNA replication during the cell cycle by direct binding to Cdc6, which promotes Cdc6 interaction with the minichromosome maintenance complex and results in reduced replication-origin firing under hypoxia [34]. Clearly, HIF-1α functions do not always involve its transcriptional activity and may occur at various subcellular locations outside the nucleus.
Intriguingly, other transcription factors have also been shown to translocate to the mitochondria; some of these affect mitochondrial gene expression while others interact with regulatory molecules. Ogita et al. revealed that the activator protein-1 (AP-1) complex binds to AP-1-like sites in non-coding regions of the mitochondrial genome [35], [36]. Other studies showed that NF-κB can move to the mitochondrial intermembrane space, where it negatively regulates mitochondrial mRNA expression [37], [38], [39]. CREB and p53 also relocate to the mitochondria. CREB binds to its cognate responsive elements in the D-loop of mtDNA, although the mechanism of p53 action is controversial [40], [41]. These studies demonstrate that many transcription factors have additional dynamic, non-canonical functions in mitochondria, which may partially reflect mitochondria-nucleus interaction.
In conclusion, we found that HIF-1α is recruited to the mitochondria in response to oxidative stress in vitro and in vivo, suggesting the possibility that mtHIF-1α might be involved in multiple oxidative stress-related diseases. Moreover, we further elucidated the mechanism underlying the direct regulation of mitochondria by HIF-1α independently of its nuclear activities, which sheds light on the regulatory role of HIF-1α under hypoxia.
Acknowledgements
This work was financially supported by 2013 program of Key Laboratory of National Health and Family Planning Commission and by the Natural Science Foundation of China (nos. 81270552 and 81273255).
Acknowledgments
Disclosure
The authors declare no competing interests.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2019.101109.
Contributor Information
Li Feng, Email: feng31@sohu.com.
You-Ping Li, Email: yzmylab@hotmail.com.
Appendix A. Supplementary material
Supplementary material
References
- 1.Smith K.A., Waypa G.B., Schumacker P.T. Redox signaling during hypoxia in mammalian cells. Redox Biol. 2017;13:228–234. doi: 10.1016/j.redox.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuhrmann D.C., Brune B. Mitochondrial composition and function under the control of hypoxia. Redox Biol. 2017;12:208–215. doi: 10.1016/j.redox.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schofield C.J., Ratcliffe P.J. Signalling hypoxia by HIF hydroxylases. Biochem. Biophys. Res. Commun. 2005;338(1):617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 4.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand M.D. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic. Biol. Med. 2004;37(6):755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 6.Semenza G.L. Hypoxia-inducible factor 1: regulator of mitochondrial metabolism and mediator of ischemic preconditioning. Biochim. Biophys. Acta. 2011;1813(7):1263–1268. doi: 10.1016/j.bbamcr.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan S.Y. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10(4):273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semenza G.L. HIF-1: upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008;283(16):10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Briston T., Yang J., Ashcroft M. HIF-1alpha localization with mitochondria: a new role for an old favorite? Cell Cycle. 2011;10(23):4170–4171. doi: 10.4161/cc.10.23.18565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rane S. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 2009;104(7):879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.I. Mylonis, et al., Mortalin-mediated and ERK-controlled targeting of HIF-1alpha to mitochondria confers resistance to apoptosis under hypoxia. 2017. 130(2), p. 466–479. [DOI] [PubMed]
- 13.Kriechbaumer V., von Loffelholz O., Abell B.M. Chaperone receptors: guiding proteins to intracellular compartments. Protoplasma. 2012;249(1):21–30. doi: 10.1007/s00709-011-0270-9. [DOI] [PubMed] [Google Scholar]
- 14.Li J. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 2009;284(35):23852–23859. doi: 10.1074/jbc.M109.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfanner N., Wiedemann N. Mitochondrial protein import: two membranes, three translocases. Curr. Opin. Cell Biol. 2002;14(4):400–411. doi: 10.1016/s0955-0674(02)00355-1. [DOI] [PubMed] [Google Scholar]
- 16.Xu W. Bax-PGAM5L-Drp1 complex is required for intrinsic apoptosis execution. Oncotarget. 2015;6(30):30017–30034. doi: 10.18632/oncotarget.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frezza C. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126(1):177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 18.Alavi M.V., Fuhrmann N. Dominant optic atrophy, OPA1, and mitochondrial quality control: understanding mitochondrial network dynamics. Mol. Neurodegener. 2013;8:32. doi: 10.1186/1750-1326-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daugas E. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J. 2000;14(5):729–739. [PubMed] [Google Scholar]
- 20.Yu S.W. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 21.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crowley L.C., Christensen M.E., Waterhouse N.J. Measuring mitochondrial transmembrane potential by TMRE staining. Cold Spring Harb. Protoc. 2016;2016(12) doi: 10.1101/pdb.prot087361. (p. pdb.prot087361) [DOI] [PubMed] [Google Scholar]
- 23.Mailloux R.J., McBride S.L., Harper M.E. Unearthing the secrets of mitochondrial ROS and glutathione in bioenergetics. Trends Biochem. Sci. 2013;38(12):592–602. doi: 10.1016/j.tibs.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Bao L. Role of heme Oxygenase-1 in low dose Radioadaptive response. Redox Biol. 2016;8:333–340. doi: 10.1016/j.redox.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell C.T., Kolesar J.E., Kaufman B.A. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim. Biophys. Acta. 2012;1819(9–10):921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 26.Neary M.T. Hypoxia signaling controls postnatal changes in cardiac mitochondrial morphology and function. J. Mol. Cell. Cardiol. 2014;74:340–352. doi: 10.1016/j.yjmcc.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Silva P. Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat. Struct. Mol. Biol. 2004;11(11):1084–1091. doi: 10.1038/nsmb846. [DOI] [PubMed] [Google Scholar]
- 28.Lorenz H., Hailey D.W., Lippincott-Schwartz J. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat. Methods. 2006;3(3):205–210. doi: 10.1038/nmeth857. [DOI] [PubMed] [Google Scholar]
- 29.Roth K.J., Copple B.L. Role of Hypoxia-Inducible Factors in the Development of Liver Fibrosis. Cell Mol. Gastroenterol. Hepatol. 2015;1(6):589–597. doi: 10.1016/j.jcmgh.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torok N.J. Dysregulation of redox pathways in liver fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;311(4):G667–G674. doi: 10.1152/ajpgi.00050.2016. (p. G667-g674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tormos K.V., Chandel N.S. Inter-connection between mitochondria and HIFs. J. Cell. Mol. Med. 2010;14(4):795–804. doi: 10.1111/j.1582-4934.2010.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson R.F., Witzel, Perkins N.D. p53-dependent regulation of mitochondrial energy production by the RelA subunit of NF-kappaB. Cancer Res. 2011;71(16):5588–5597. doi: 10.1158/0008-5472.CAN-10-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan Z., Michalopoulos G.K., Stolz D.B. Peroxisomal localization of hypoxia-inducible factors and hypoxia-inducible factor regulatory hydroxylases in primary rat hepatocytes exposed to hypoxia-reoxygenation. Am. J. Pathol. 2006;169(4):1251–1269. doi: 10.2353/ajpath.2006.060360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubbi M.E. A nontranscriptional role for HIF-1alpha as a direct inhibitor of DNA replication. Sci. Signal. 2013;6(262):ra10. doi: 10.1126/scisignal.2003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogita K. Transcription factor activator protein-1 expressed by kainate treatment can bind to the non-coding region of mitochondrial genome in murine hippocampus. J. Neurosci. Res. 2003;73(6):794–802. doi: 10.1002/jnr.10720. [DOI] [PubMed] [Google Scholar]
- 36.Ogita K. Localization of activator protein-1 complex with DNA binding activity in mitochondria of murine brain after In vivo treatment with kainate. J. Neurosci. 2002;22(7):2561. doi: 10.1523/JNEUROSCI.22-07-02561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cogswell P.C. NF-kappa B and I kappa B alpha are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-kappa B. J. Biol. Chem. 2003;278(5):2963–2968. doi: 10.1074/jbc.M209995200. [DOI] [PubMed] [Google Scholar]
- 38.Guseva N.V. Tumor necrosis factor-related apoptosis-inducing ligand-mediated activation of mitochondria-associated nuclear factor-kappaB in prostatic carcinoma cell lines. Mol. Cancer Res. 2004;2(10):574–584. [PubMed] [Google Scholar]
- 39.Zamora M. Recruitment of NF-kappaB into mitochondria is involved in adenine nucleotide translocase 1 (ANT1)-induced apoptosis. J. Biol. Chem. 2004;279(37):38415–38423. doi: 10.1074/jbc.M404928200. [DOI] [PubMed] [Google Scholar]
- 40.Ryu H. Antioxidants modulate mitochondrial PKA and increase CREB binding to D-loop DNA of the mitochondrial genome in neurons. Proc. Natl. Acad. Sci. USA. 2005;102(39):13915–13920. doi: 10.1073/pnas.0502878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuh R.A., Kristian T., Fiskum G. Calcium-dependent dephosphorylation of brain mitochondrial calcium/cAMP response element binding protein (CREB) J. Neurochem. 2005;92(2):388–394. doi: 10.1111/j.1471-4159.2004.02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material