Abstract
Cells employ multiple defence mechanisms to sustain a wide range of stress conditions associated with accumulation of modified self-biomolecules leading to lipo- and proteotoxicity. One of such mechanisms involves activation of the autophagy-lysosomal pathway for removal and degradation of modified lipids, proteins and even organelles. Biomolecules carbonylation, an irreversible oxidative modification, occurs in a variety of pathological conditions and is generally viewed as a marker of oxidative stress. Here, we used a model of rat primary cardiac cells to elucidate the role of autophagy-lysosomal pathway in the turnover of carbonylated biomolecules. Cells treated with inhibitors of autophagy-lysosomal degradation and primed with a short pulse of mild nitroxidative stress were studied using fluorescent microscopy and accumulation of carbonylated biomolecules in droplets- or vesicle-like structures was observed. Furthermore, systems-wide analysis of proteome regulation using relative label free quantification approach revealed the most significant alterations in cells treated with protease inhibitors. Interestingly, down-regulation of insulin signalling was among the most enriched pathway, as revealed by functional annotation of regulated proteins.
Keywords: Autophagy-lysosomal flux, Cellular stress, Protein and lipids carbonylation, Protein lipoxidation, Proteomics
Graphical abstract
Highlights
-
•
Starvation induced autophagy promotes cellular carbonylation.
-
•
Inhibition of autophagy-lysosomal flux leads to carbonyls accumulation.
-
•
Cellular carbonyls coincide with Nile Red positive structures.
-
•
Inhibition of autophagy-lysosomal flux induces proteome alterations.
-
•
Impairment of autophagy-lysosomal flux results in changes in metabolic and nutrient sensing pathways.
1. Introduction
Oxidation of cellular biomolecules with formation of carbonylated species occurs under numerous pathological conditions usually associated with overproduction or insufficient removal of reactive oxygen and nitrogen species (ROS and RNS). Carbonylation refers to a generic term describing oxidative modifications of proteins, lipids, carbohydrates and nucleic acids resulting in the introduction of aldehyde, ketone or lactam functional groups [1]. Formation of carbonylated biomolecules was linked to a wide range of pathological conditions including metabolic, cardiovascular and neurodegenerative disorders, and numerous studies showed positive correlation between accumulation of carbonylated species and development or progression of disease, establishing biomolecules carbonylation as a marker of oxidative stress related pathologies [2]. Thus, protein carbonyls are often used as a marker of redox dysregulation [3]. Lipid-derived carbonyls usually arise from the oxidation of free or phospholipid-esterified polyunsaturated fatty acids. Among the best known markers of lipid-derived carbonyls are α,β-unsaturated aldehydes such as hydroxy-nonenal and hydroxy-hexenal formed by lipid peroxidation of ω-6 and ω-3 polyunsaturated fatty acids, respectively [4]. Lipid-derived carbonyls represent a group of reactive electrophiles capable of modifying nucleophilic substrates including certain amino acid residues, leading to protein lipoxidation. Modified biomolecules often lose their functional activities and/or can change their sub-cellular localization [5]. Moreover, oxidized biomolecules can form neoantigens and thus participate in development of autoimmune diseases [6].
Since carbonylation is an irreversible modification which cannot be repaired by any known cellular enzymes, modified biomolecules need to be removed from the cells in order to prevent associated toxic effects. Cells have developed multiple ways of stress responses in conditions associated with accumulation of toxic products formed by modification of self-biomolecules. Carbonylated products of lipid-peroxidation can be removed via GSH conjugation, be reduced to alcohols or oxidized to less reactive carboxylic acids by variety of alcohol and aldehyde dehydrogenase enzymes present in the cell [7], [8]. Oxidized proteins are directed for proteasomal degradation, where oxidative stress activated 20S proteasomal system plays an important role providing fast ATP- and ubiquitin-independent removal of modified substrates [9]. Furthermore, the role of autophagy-lysosomal pathways in the degradation of modified biomolecules was demonstrated in a number of studies [10], [11], [12], [13]. Autophagy is usually subdivided in three types including macroautophagy (involving the formation of autophagosomes; hereafter referred as autophagy), microautophagy (direct engulfment of cytosolic material by lysosomes), and chaperon-mediated autophagy [14]. Autophagy, initially described to be activated by the lack of nutrients, is now recognized to be involved in a variety of cellular stress responses including hypoxia, DNA damage, viral infections, ER unfolded protein response, accumulation of damaged mitochondria and oxidative stress related pathologies [15]. Moreover, it was demonstrated that ROS are immediately produced upon nutrient deprivation [16], [17]. Previously, we demonstrated that mild nitroxidative stress induced autophagy in rat primary cardiac cells in a time dependent manner. Moreover, in this model, inhibition of lysosomal proteases prevented stress resolution and degradation of carbonylated substrates [18]. Here, we used primary cardiac cells in which autophagy was induced by 5 h starvation to monitor the effect of autophagy-lysosomal pathway inhibition on the accumulation of carbonylated species. Using specific inhibitors for each key step of the pathway (autophagosome formation, fusion with lysosome and its acidification, as well as inhibition of lipases and proteases) we have shown accumulation of carbonylated species in the form of droplet- or vesicle-like structures. To understand the effect of specific inhibitors on the regulation of the cellular proteome, we performed relative label free quantitative proteomics analysis and identified significant differences in proteome regulation by impairing each step of autophagy-lysosomal pathway.
2. Materials and methods
2.1. Materials
Rat primary cardiac cells were purchased from Innoprot (Elexalde Derio, Spain). Dulbecco's Modified Eagle Medium/Ham's F-12 (DMEM/F12), fetal bovine serum (FBS), horse serum, penicillin-streptomycin, L-glutamine, non-essential amino acids, sodium pyruvate, attachment factor and phosphate buffered saline (PBS) were obtained from Life Technologies GmbH (Darmstadt, Germany). Trypsin-EDTA solution, 2′,7′-dichlorofluorescin diacetate (DCFDA), thiazolyl blue tetrazolium bromide (MTT), triton-X-100, DMSO, 7-(diethylamino)-coumarin-3-carbohydrazide (CHH), paraformaldehyde (PFA), Nile Red, thiourea, β-mercaptoethanol, hydrochloric acid, formic acid, ammonium bicarbonate, 2,4-dinitrophenyl hydrazine (DNPH), 3-methyladenine, chloroquine diphosphate salt, goat anti-DNP antibody, ExtrAvidin peroxidase were purchased from Sigma-Aldrich GmbH (Taufkirchen, Germany). Goat anti-HNE antibody (ab46544), goat anti-MDA antibody (ab27644) were from Abcam (Berlin, Germany). Peroxidase-conjugated donkey anti-goat antibody was from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA) and mouse E06 biotinylated mAb from Avanti Polar Lipids, Inc. (Alabaster, USA). 3-Morpholinosydnonimine (SIN-1), E64d and Orlistat were purchased from Enzo Life Sciences GmbH (Lörrach, Germany). Methanol, ethanol, urea, SDS, SDC, glycerol, sucrose and dithiothreitol (DTT) were obtained from Carl Roth GmbH + Co. KG (Karlsruhe, Germany). Trypsin, glycine, bovine serum albumin (BSA), pepstatin A (pepA), Coomassie® Brilliant Blue G-250 and Tween® 20 were purchased from Serva Electrophoresis GmbH (Heidelberg, Germany). Iodoacetamide (IAA), Tris, HEPES and EDTA were purchased from Applichem GmbH (Darmstadt, Germany). Low fluorescence PVDF membranes, protein free blocking solution (AdvanBlock™), washing solution (AdvanWash™) and WesternBright™ Sirius HRP substrate were obtained from Advansta Inc. (Biozym Scientific GmbH, Hessisch Oldendorf, Germany). Acetonitrile (ULC-MS grade) and formic acid were purchased from Biosolve (Valkenswaard, Netherlands).
2.2. Cell culture
Primary rat cardiac cells (Innoprot, Elexalde Derio, Spain) were cultured on gelatine-coated 75 cm2 flasks or 96-well-plates (CELLSTAR®, Greiner Bio-One GmbH, Frickenhausen, Germany) in DMEM/F12 medium supplemented with 20% FBS, 5% horse serum, 2 mmol/L L-glutamine, 3 mmol/L sodium pyruvate, 0.1 mmol/L non-essential amino acids, 100 U/mL penicillin and 100 µg/mL streptomycin at 37 °C (humidified atmosphere of 5% CO2 and 95% air). When cells reached 80% confluence, they were washed with warm PBS, and cultured for 5 h in the absence (starved control) or presence of inhibitors (1 mmol/L 3-methyladenine, 10 µmol/L chloroquine, 25 µmol/L orlistat, or 5 µg/mL E64d with 5 µg/mL Pepstatin A) in non-supplemented DMEM/F12 medium. For mild nitroxidative stress induction in selected conditions, SIN-1 (50 μmol/L) was added for the last 30 min of incubation.
2.3. DCFDA assay
Rat cardiac cells were seeded on a black 96-well plate (20,000 cells per well), grown overnight, washed with PBS and incubated with 10 µmol/L DCFDA in transparent DMEM/F12 medium (1 h, 37 °C). After loading cells were washed with PBS, incubated in transparent DMEM/F12 medium (1 h, 37 °C), and treated with SIN-1 (50 μmol/L; in transparent DMEM/F12). Fluorescence (Ex = 485 nm, Em = 535 nm) was recorded every 3 min for 6 h at 37 °C using Paradigm™ Detection Platform (Beckman Coulter GmbH, Krefeld, Germany).
2.4. MTT assay
Rat cardiac cells were seeded on a 96-well plate (20,000 cells per well), grown overnight, washed with PBS and cultured in the absence or presence of inhibitors and SIN-1 as described above. After 5 h treatment medium was changed to 1.2 mmol/L MTT in transparent DMEM/F12 (4 h, 37 °C). After incubation SDS (5%, w/v) and HCl (0.005%, v/v) was added and incubated overnight (37 °C). Absorbance was measured at 562 nm (reference 690 nm) with a Paradigm™ Detection Platform (Beckman Coulter GmbH, Krefeld, Germany).
2.5. Fluorescence microscopy
Primary rat cardiac cells were grown on 96-well-plates till 80% confluence, medium was exchanged to non-supplemented DMED/F12 with or without inhibitors. After the treatment, cells were fixed with paraformaldehyde (4%, w/v, in PBS; 15 min), washed (PBS, two times), incubated with CHH (0.2 mmol/L, 2 h, RT), washed (PBS, three times), staining with Nile Red (1:1000, 1 mg/mL in DMSO, 10 min, 37 °C), washed (PBS, three times), and images were acquired using an inverted fluorescence microscope (DMI6000B, Leica Mikrosysteme Vertrieb GmbH, Wetzlar, Germany), equipped with 20 × (NA 0.40) and 40× objectives (NA 0.60), a 12 V/100 W halogen lamp as light source and a Leica DFC360FX camera. Images were acquired with Leica Application Suite (LAS AF) microscope software (version 2.3.0). For Nile Red and CHH images were acquired using λex= 552/λem= 578 and λex= 359/λem= 461, respectively. For confocal fluorescence microscopy experiments cells were cultured on glass-bottom 12-well dishes (Mattek Corporation, Ashland, MA). Images from single confocal sections were acquired every 0.5 µm using Leica SP5 microscope (63× magnification) and the corresponding overlays are shown.
2.6. Image analysis
CHH fluorescence intensity was determined by analysing the corrected total cell fluorescence (CTCF) with the Fiji implementation of imageJ [19]. CTCF is calculated by measuring the integrated density and subtracting the cell area multiplied for the background. CTCF = Integrated Density – (Cell Area × Mean Background Fluorescence). In total CTCF values were calculated for 100 cells for each condition selected from at least 9 images acquired from 3 independent replicates. Nile Red positive structures were quantified with ImageJ plugin Lipid Droplet Counter for at least 20 cells for each experimental condition. Mann-Whitney nonparametric t-test was performed using GraphPad Prism version 5.02 for Windows, GraphPad Software, San Diego California USA.
2.7. Protein extraction
For cell pellets collections, cells were grown in 75 cm2 flasks. Immediately after treatment cells were washed with PBS and collected by trypsinization (3 min). Cell pellets were washed in cold PBS and stored in −80 °C until analysis. For protein extraction, cell pellets were resuspended in 100 µL of lysis buffer (7 mol/L urea, 2 mol/L thiourea, 1% SDC in 50 mmol/L Tris-HCl, pH 7.5), sonicated on ice for 1 min using a Vibra-Cell™ tip sonicator (20 kHz, 1 min with on/off pulse, 5 s each, 30% amplitude; Sonics & Materials, Inc., Newtown, CT, USA), centrifuged at 10,000 g for 35 min and supernatant were collected. Bradford assay [20] was used to determine the protein concentration. Protein amount normalization was confirmed by SDS-PAGE stained with Coomassie Blue.
2.8. Western Blot
Proteins (10 μg) in lysis buffer were mixed with Laemmli sample buffer (62.5 mmol/L Tris-HCl pH 6.8, 20% v/v glycerol, 2% w/v SDS, 5% v/v ß-mercaptoethanol, 0.01% w/v bromophenol blue) and separated by SDS-PAGE (12% T, 0.75 mm). Proteins were blotted on a low-fluorescence polyvinylidene difluoride (PVDF) membrane using semidry Trans-Blot Turbo Transfer System (BioRad Laboratories GmbH, München, Germany). To visualize carbonyls (oxyblot) membranes were equilibrated (2 M HCl; 10 min), derivatized with DNPH (0.1 mg/mL in 2 M HCl, 5 min, RT), washed with 2 M HCl (10 min) and methanol (5 min, 5 times). Membranes were blocked overnight (4 °C, Immunoblot Blocking solution; AdvanBlock, Advansta), incubated with goat anti-DNP Ab (1:10,000 in blocking buffer, 1 h, RT), and washed (Immunoblot Washing solution, AdvanWash, Advansta). Membranes were incubated (1 h, RT) with peroxidase-conjugated donkey anti-goat Ab (1:10,000, in AdvanBlock, Advansta). To visualize HNE, MDA and oxidized phosphocholine adducts, protein were blotted on a PVDF membrane and blocked (AdvanBlock, Advansta) for 1 h at room temperature. Membranes were incubated with goat anti-HNE Ab (1:10,000; in blocking buffer, overnight, 4 °C), goat anti-MDA Ab (1:5 000; in blocking buffer, overnight, 4 °C), or mouse E06 mAb-TopFluor (1:500, in AdvanBlock, Advansta). Membranes were washed (Immunoblot Washing solution, AdvanWash, Advansta) and either probed with peroxidase-conjugated donkey anti-goat Ab (1:10,000, in AdvanBlock, Advansta) or ExtrAvidin - Peroxidase (1:5000). Membrane were visualized using WesternBright Sirius HRP substrate (Advansta) and images were acquired using Chemidoc™ MP (BioRad Laboratories GmbH, München, Germany).
2.9. Protein digestion
Proteins (50 μg) in lysis buffer were digested according to Filter Aided Sample Preparation (FASP) protocol [21]. Briefly, proteins were reduced with DTT (100 mmol/L, 1 h, RT), transferred onto Microcon centrifugal filters (30 kDa cut-off), centrifuged at 14,000 g, alkylated with iodoacetamide (50 mmol/L, 20 min in the dark), washed two times with urea buffer (8 mol/L Urea, 100 mmol/L TrisHCl, pH 8.5) and two times with ammonium bicarbonate buffer (50 mmol/L) before digestion with trypsin (1:25, enzyme to protein ratio, w/w; overnight at 37 °C in a humidified environment). Peptides were recovered in ammonium bicarbonate buffer (50 mmol/L), dissolved (250 ng/µL) in aqueous acetonitrile (3%, v/v) and used for LC-MS analysis.
2.10. Mass spectrometry analysis
A nano-Acquity UPLC (Waters GmbH, Eschborn, Germany) was coupled online to an LTQ Orbitrap XL ETD mass spectrometer equipped with a nano-ESI source (Thermo Fischer Scientific, Bremen, Germany). Eluent A was aqueous formic acid (0.1% v/v), and eluent B was formic acid (0.1% v/v) in acetonitrile. Samples (10 µL) were loaded onto the trap column (nanoAcquity symmetry C18, internal diameter 180 µm, length 20 mm, particle diameter 5 µm) at a flow rate of 10 µL/min. Peptides were separated on BEH 130 column (C18-phase, internal diameter 75 µm, length 100 mm, particle diameter 1.7 µm) with a flow rate of 0.4 µL/min. Peptides were separated using three steps gradients from 3% to 35% eluent B over 91 min, from 35% to 85% B over 4 min and then maintained at 85% of eluent B over 5 min. After an equilibration time of 20 min samples were injected every 120 min. The transfer capillary temperature was set to 200 °C and the tube lens voltage to 120 V. An ion spray voltage of 1.5 kV was applied to a PicoTip online nano-ESI emitter (New Objective, Berlin, Germany). The precursor ion survey scans were acquired at an orbitrap (resolution of 60,000 at m/z 400) for a m/z range from 400 to 2000. CID-tandem mass spectra (isolation width 2, activation Q 0.25, normalized collision energy 35%, activation time 30 ms) were recorded in the linear ion trap by data-dependent acquisition (DDA) for the top six most abundant ions in each survey scan with dynamic exclusion for 60 s using Xcalibur software (version 2.0.7).
For relative label free protein quantification.raw files were uploaded into Progenesis QI for proteomics (Waters GmbH, Eschborn, Germany) for feature detection, alignment, and quantification. Proteins were identified using Progenesis QI generated.mgf file by Sequest (Proteome Discoverer 2.2, Thermo Fisher Scientific) with the following parameter set: maximum of two missed cleavage sites, peptide mass tolerance of 10 ppm, peptide fragment tolerance of 0.8 Da, variable modifications for oxidation of Met and carbamidomethylation of Cys. Protein identification results (at least three rank 1 peptides per protein) were imported back into Progenesis QI and used for protein quantification using non-conflicting peptides. Functional annotation of regulated proteins (ANOVA p < 0.05) was performed using DAVID Bioinformatics Resources 6.8 [22].
3. Results and discussion
Previously, using a cardiac cell culture model of mild nitroxidative stress achieved by treatment with SIN-1, we demonstrated an increase in intracellular carbonylation associated with a strong perinuclear clustering [18]. Furthermore, cells were capable to remove accumulated carbonyls 16 h after stress exposure. Complementary analysis of protein and lipid carbonylation showed the dynamic nature of this oxidative modification. At the early time point upon stress induction we demonstrated an increase in carbonylated LPPs followed by a shift of the carbonyl specific signal to the protein fraction at later time points. Moreover, SIN-1-induced lipid peroxidation was associated with autophagy induction accompanied by the appearance of droplet-like structures. Here, we have extended our study to monitor biomolecules turnover under starvation and mild oxidative stress by inhibiting different stages of the autophagy-lysosomal degradation pathway.
3.1. Model characterization
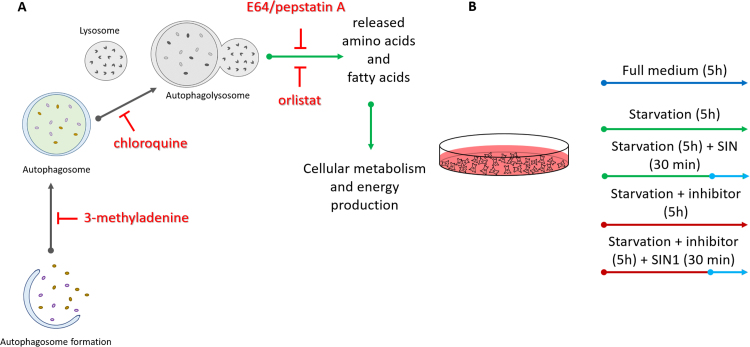
To monitor accumulation and subcellular distribution of carbonylated biomolecules upon starvation and to establish the role of the autophagy-lysosomal flux in biomolecules turnover, rat primary cardiac cells were cultured in serum free medium for 5 h in the absence or presence of specific inhibitors. Four different inhibitors along the autophagy-lysosomal degradation pathway were used in the study including 3-methyladenine, chloroquine, orlistat and a combination of E64 and pepstatin A (Fig. 1A). 3-Methyladenine (3-MA) is blocking autophagosome formation via the inhibition of type III phosphatidylinositol-3-kinases [23], [24]. Chloroquine blocks the autophagic flux by impairing fusion between autophagosome and lysosome [25], although it can inhibit lysosomal degradation by impairing lysosomal acidification and elicit the fusion of varied endolysosomal compartments [26]. Orlistat is a well know inhibitor of cellular lipases [27], [28]. In turn, E64 and pepstatin A inhibit cysteine and aspartic acid proteases, respectively [29], [30]. Finally, to investigate the effect of mild nitroxidative stress on the turnover of modified biomolecules, SIN-1 (50 mmol/L) was used to challenge the cells during the last 30 min of starvation and inhibitors treatment (Fig. 1B). A thirty min SIN-1 treatment was chosen based on the results of a DCFDA assay which demonstrated maximal free radicals production at this point, in good correspondence with the previously reported half-life for SIN-1 in aqueous medium (Fig. S1A) [31], [32]. Treatment time as well as inhibitors concentrations were optimized for rat primary cardiac cells to obtain a viable cell model without significant increase in cell death over the experimental time points (Fig. S1B).
Fig. 1.
Schematic representation of the treatment conditions used to study effects of autophagy-lysosomal pathway inhibitors (A) in rat primary cardiac cell culture model of starvation and nitroxidative stress (B).
3.2. Cellular carbonylation
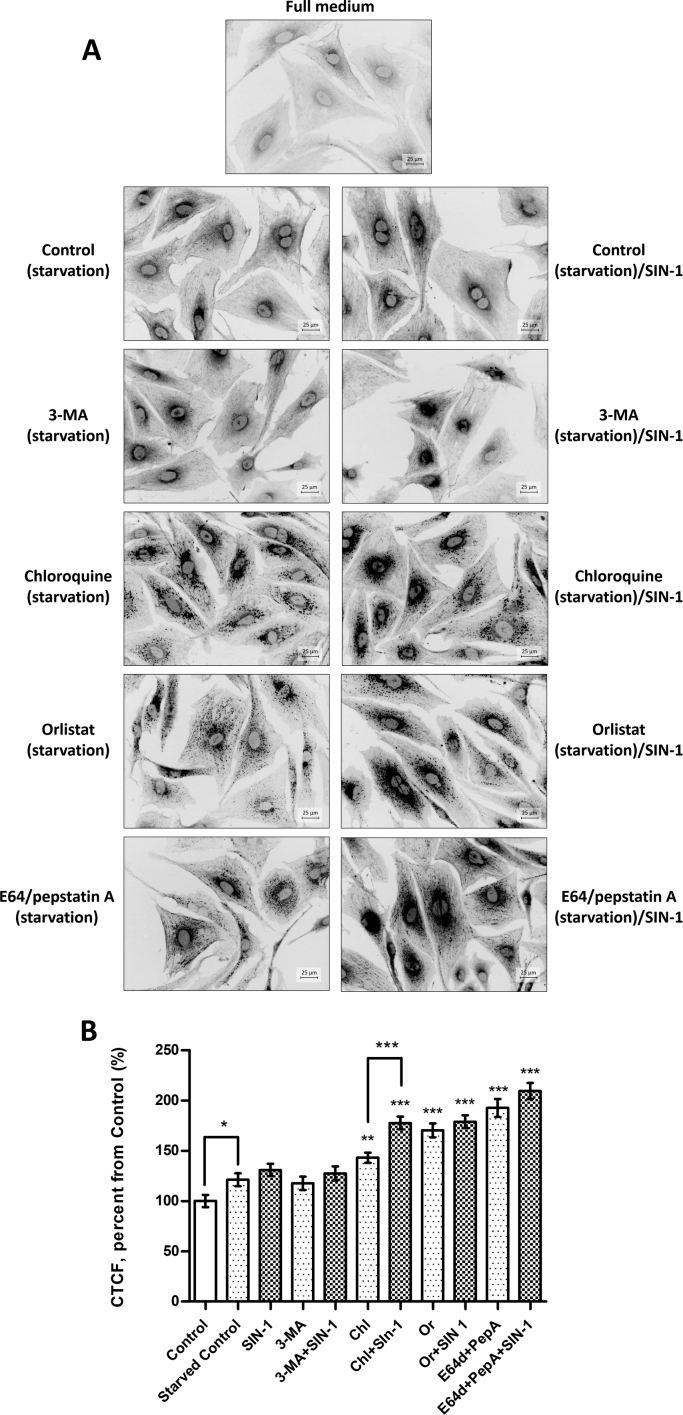
Cellular carbonylation was analysed by fluorescence microscopy using specific membrane permeable coumarin hydrazide derivative CHH (Fig. 2A) [33]. Starvation alone resulted in a mild increase of cellular carbonyls by 20% compared to the cells cultured in the full media (Fig. 2B). That is not surprising since starvation and autophagy induction are generally known to induce production of reactive oxygen species [34], [35]. A short pulse of SIN-1 treatment for the last 30 min of starvation further increased total cellular carbonylation by another 10%. Sub-cellular distribution of carbonylated species was similar to the previous reports on different cellular models of oxidative stress [18], [33], [36]. Thus, majority of the fluorescence signal was localized in the perinuclear area with a weaker distribution of carbonyl specific staining along filamentous structures distributed over the whole cell volume (Fig. 2A). Indeed, in our previous work focused on the identification of carbonylated proteins in cardiac cells, a significant number of oxidized proteins was attributed to the cell cytoskeleton and its regulation [18].
Fig. 2.
Fluorescence microscopy of cellular carbonyls labelled with CHH. Primary rat cardiac cells were cultured in serum-free medium in the absence or presence of inhibitors (for 5 h) and SIN-1 (50 µmol/L, 30 min), fixed with 4% PFA and labelled with CHH (0.2 mmol/L, 2 h). (A) Images are representatives of three independent experiments performed in triplicates. Scale bars 25 µm. (B) Carbonylation levels (given in corrected total cell fluorescence, CTCF) were evaluated by CHH fluorescence microscopy and images were quantified with ImageJ. Data are shown as means ± SEM (* p < 0.05, ** p < 0.001, *** p < 0.0001 vs. starved control; Mann-Whitney nonparametric t-test) of 100 cell images obtained from three independent experiments.
Treatment of cells with 3-MA during the 5 h starvation period did not result in significant changes in the total cellular carbonylation. Thus, 3-MA treatment alone or in combination with a short pulse of SIN-1 resulted in 18% and 27% increase in total cellular carbonylation respectively, in comparison to the cells cultured in the full medium.
Treatment with chloroquine resulted in much higher levels of total cellular carbonyls corresponding to the increase of 43% relative to the cells cultured in the full medium or 22% relative to the starved control cells (Fig. 2B). Multiple droplet or vesicle-like structures with high intensity of carbonyl specific signal were observed in chloroquine treated cells. Furthermore, association of carbonylated droplets with filamentous structures was even more evident (Fig. 2A). Pulse of SIN-1 treatment further increased total cellular carbonylation by another 30%, corresponding to the increase of 54% relative to the starved control cells. SIN-1 treatment also induced changes in sub-cellular distribution of carbonylated species leading to the denser distribution of carbonyl specific staining in the perinuclear area.
Exposure of starved cells to the lipase inhibitor orlistat resulted in droplet-like carbonyl positive staining distributed all over the cell volume without visible filamentous distribution. SIN-1 treatment of orlistat exposed cells induced perinuclear accumulation and closer association with filamentous structures. Increase in total cellular carbonylation corresponded to 70% and 79% for orlistat without and with SIN-1, respectively.
Finally, treatment with E64/pepstatin A provided even higher levels of total cellular carbonyls (74% and 100% increase for protease inhibitors without and with SIN-1 pulse, respectively, relative to starved control cells). In this case also droplet-like structures were observed with stronger accumulation around the nucleus and association with filamentous structure along the cell length. The final incubation with SIN-1 did not induce detectable changes in the distribution of carbonyl specific signals.
3.3. Accumulation of Nile Red-positive structures (NRS)
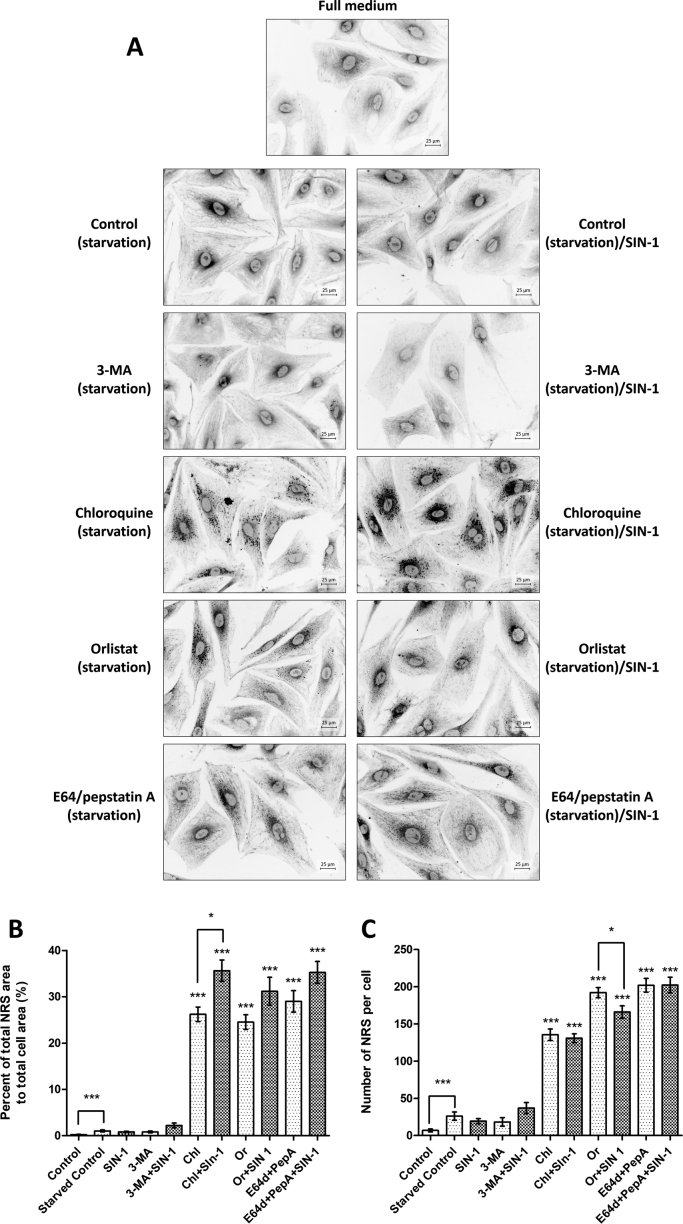
Since treatment with inhibitors resulted in accumulation of carbonyl specific signals mainly in droplet- or vesicle-like structures, we decided to the evaluate accumulation of lipids in starved cardiac cells using Nile Red staining specific for neutral lipids like triacyl glycerides (TGs) and cholesteryl esters (Fig. 3A). In cells starved for 5 h without and with SIN-1 pulse, minimal Nile Red staining was visible around the nucleus together with a few NRS present in the cytoplasm. 3-MA treatment did not result in significant changes in NRS numbers, intensity or distribution. However, chloroquine treatment resulted in significant accumulation of Nile Red positive structures. Large number of NRS accumulated upon 5 h chloroquine exposure with and without SIN-1 pulse. Interestingly, NRS often showed a longitudinal arrangement resembling the distribution of filamentous structures seen with CHH staining. Orlistat treatment also provoked high number of NRS distributed all over the cell volume with a higher density in the perinuclear space. Similar to CHH staining, SIN-1 pulse at last 30 min of orlistat treatment led to more longitudinal NRS distribution. For E64/pepstatin A treatment NRS accumulation was characteristic as well, with small perinuclear and filament-associated droplets. Quantification of the NRS surface area relative to the total cell area showed a significant increase of to 35% for chloroquine, orlistat and E64/pepstatin treatment with or without SIN-1 (Fig. 3B). Furthermore, for chloroquine, SIN-1 treatment resulted in a significant increase of NRS surface areas in relative to inhibitors alone.
Fig. 3.
Fluorescence microscopy of Nile Red-positive structures (NRS). Primary rat cardiac cells were cultured in serum-free medium in the absence or presence of inhibitors (for 5 h) and SIN-1 (50 µmol/L, 30 min), fixed with 4% PFA and labelled with Nile Red (1:1000, 1 mg/mL in DMSO, 10 min, 37 °C). (A) Images are representatives of three independent experiments performed in triplicates. Scale bars 25 µm. (B) NRS (given in percent of droplet surface area relative to the cell surface are) were quantified with ImageJ plugin Lipid Droplets Counter. Data are shown as means ± SEM (* p < 0.05, ** p < 0.001, *** p < 0.0001 vs. starved control; Mann-Whitney nonparametric t-test) of 20 cell images obtained from three independent experiments. (C) Number of NRS (provided relative to a cell area of 5000 µm2) quantified by ImageJ plugin Lipid Droplets Counter. Data are shown as means ± SEM (* p < 0.05, ** p < 0.001, *** p < 0.0001 vs. starved control; Mann-Whitney nonparametric t-test) of 20 cell images obtained from three independent experiments.
Comparison of NRS size (Fig. 3B) and number per cell (Fig. 3C) showed that relative to orlistat and E64/pepstatin A, exposure to chloroquine resulted in lower number of NRS, however their total area was comparable to the NRS areas of droplets formed after exposure to lipase and protease inhibitors, indicating formation of larger NRS upon chloroquine treatment. Furthermore, for all three inhibitors, SIN-1 treatment induced larger NRS although not increasing their number (Fig. 3B and C).
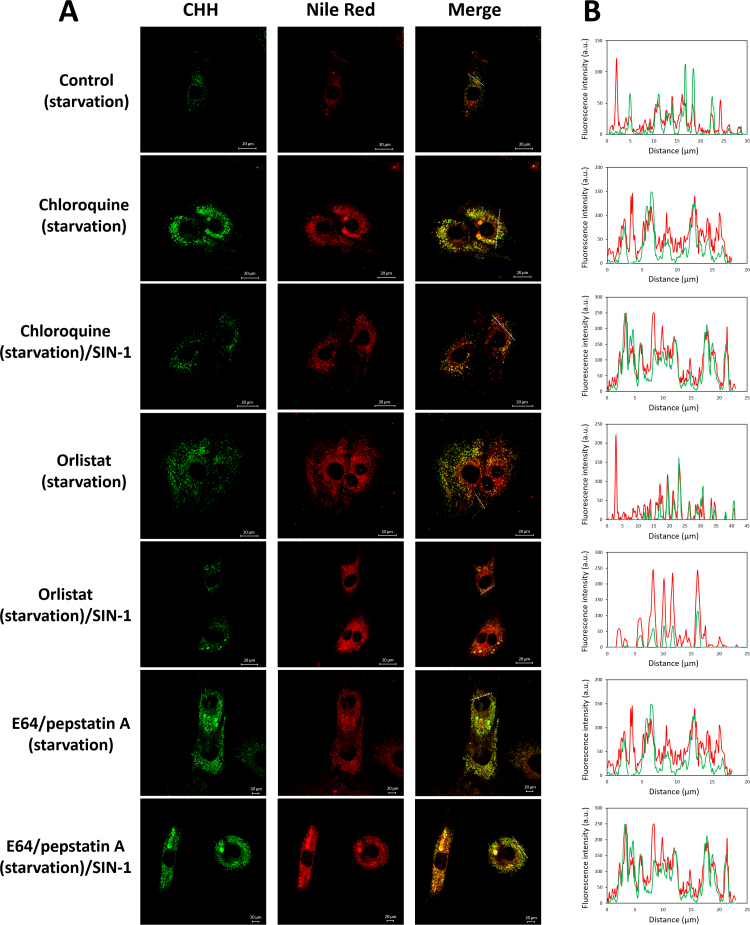
To illustrate the possible colocalization between carbonyl and NRS staining in chloroquine, orlistat and E64/pepstatin A treated cells, we performed confocal microscopy experiments (Fig. 4A and B). We observed a good coincidence of the CHH and Nile Red signals in chloroquine treated cells, whereas after SIN-1 pulse a portion of Nile Red staining closer to the nucleus did not show overlap with CHH staining. For orlistat exposure, NRS distributed throughout the whole cell volume showing a variable degree of coincidence with carbonyl-specific fluorescence. SIN-1 treatment resulted in perinuclear accumulation of overlapping CHH and Nile Red signals. In case of protease inhibitors good coincidence for carbonyl and NRS signals was visible without and with SIN-1 exposure.
Fig. 4.
Confocal microscopy of CHH and Nile Red-positive structures (NRS). Primary rat cardiac cells were cultured in serum-free medium in the absence or presence of inhibitors (for 5 h) and SIN-1 (50 µmol/L, 30 min), fixed with 4% PFA (w/v), labelled with CHH (0.2 mmol/L, 2 h) and labelled with Nile Red (1:1000, 1 mg/mL in DMSO, 10 min, 37 °C). (A) Single sections from the individual channels and the corresponding overlays (merged) are shown. Images are representative of two independent experiments. Scale bars 20 µm. (B) Fluorescence intensity profiles of the CHH (green) and Nile Red (red) signals along the dotted line marked in (A).
3.4. Western blot analysis
A significant increase in cellular carbonylation upon starvation in the presence of chloroquine, orlistat and E64/pepstatin A and SIN-1 can be attributed to both lipid and protein oxidation. Protein carbonylation is a generic term for products of different reactions resulting in formation of protein bound carbonyl groups like aldehydes, ketones and lactams [37]. These protein bound carbonyls can be very different in structure and be formed via several pathways including direct oxidation of amino acid residues (e.g. formation of aminoadipic semialdehyde via oxidative deamination on ε-amino group of lysine residues), Michael adducts formation between reactive α,β-unsaturated carbonyls formed by lipid peroxidation (e.g. 4-hydroxy-nonenal adducts on lysine, cysteine and histidine residues), and reaction with dicarbonyl compounds formed by lipid peroxidation, glucose autooxidation, or metabolic reactions (e.g. glyoxal and malondialdehyde adducts on lysine and arginine residues) [1], [38], [39].
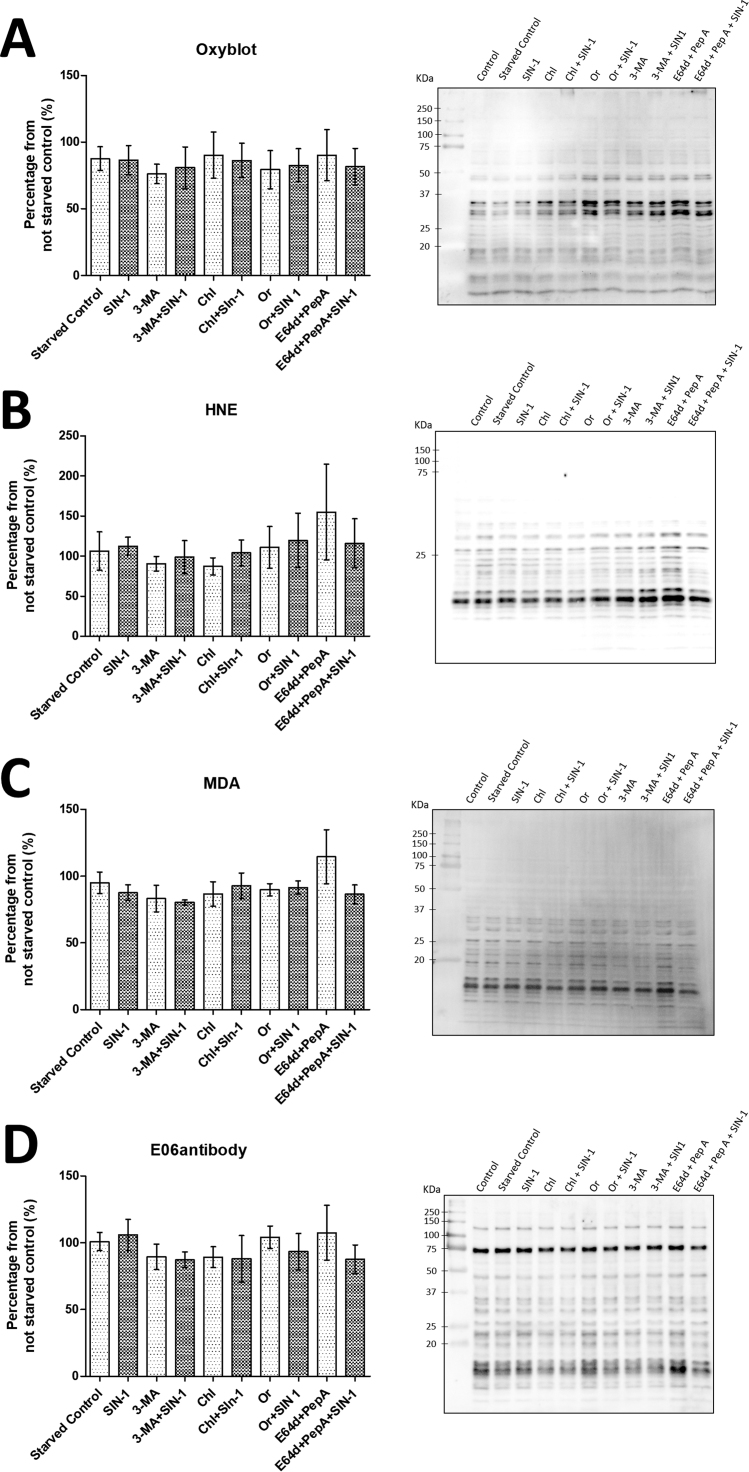
To evaluate the contribution of protein-derived carbonylation upon cell starvation, inhibitors treatments and SIN-1 priming, protein extracts were probed using oxyblot, anti-hydroxynonenal (anti-HNE), anti-malondialdehyde (anti-MDA), and EO6 antibody based immunodetection. Oxyblot analysis relays on the reaction of protein bound carbonyl groups with dinitrophenylhydrazine (DNPH), by a reaction mechanism similar to coumarine hydrazide-based derivatization used for the fluorescence microscopy experiments, and thus should address the total level of protein carbonylation in the system under the study. However, no significant differences in total protein bound carbonyls were observed between studied conditions by oxyblot analysis (Fig. 5A). A mild, however not statistically significant, increase in the intensity of the signals corresponding to the bands around 50 kDa and 35 kDa was evident for 3-MA, orlistat and E64/pepstatin treatment conditions. Indeed, in our previous model of SIN-1 treatment of rat cardiac cells, an increase in protein carbonylation upon different treatment time points was evident for the protein bands of the similar molecular weights [18].
Fig. 5.
Western Blot analysis of protein carbonylation using (A) oxyblot, (B) anti-MDA, (C) anti-HNE, and (D) anti-oxPC antibodies. Primary rat cardiac cells were cultured in serum-free medium in the absence or presence of inhibitors (for 5 h) and SIN-1 (50 µmol/L, 30 min), cell pellets were collected and used for protein extraction. 10 μg of proteins (normalized after measuring protein concertation and controlled by SDS-PAGE) from each experimental condition were separated by SDS-PAGE, blotted to PVDF membrane and probed with corresponding antibodies. Quantification of total protein carbonylation was performed from three independent experiments ± SEM. Image is representative of three independent experiments.
Then we used antibodies against protein adducts with HNE and MDA, a two well know products of lipid peroxidation, capable of forming carbonylated (via Michael addition) and non-carbonylated (via Schiff base formation) adduct with nucleophilic amino acid residues in proteins. Visualized protein band patterns were similar for both anti-HNE and anti-MDA western blot images (Fig. 5B and C), however both were quite different in comparison to oxyblot detection of total carbonylation. The most intense signals appeared for the bands with molecular weight between 10 and 20 kDa. For both immunoblots the only condition with noticeable, but still not statistically significant, difference versus starved control cells was E64/pepstatin A treatment. Interestingly, SIN-1 challenge did not result in increased detection of HNE/MDA-protein adducts.
Finally, we used EO6 natural antibodies, known to recognize the head group of oxidized phosphatidylcholine lipids, to visualize any oxPC-protein adducts (Fig. 5D) [40]. Lipid peroxidation can result in oxidative truncation of unsaturated fatty acyl residues in PC lipids with the formation of lipid-bound aldehydes. Such PC-bound aldehydes can react with accessible nucleophilic amino acid residues either via Schiff base or Michael addition mechanism depending on the structure of truncated fatty acyl residue. Similar to anti-MDA and anti-HNE western blots, E64/pepstatin A treated samples showed increased intensity for the bands between 10 and 20 kDa. Additionally, increased intensity was observed for the bands between 20 and 25 kDa.
Overall, western blot analysis was used to detect four different protein modifications associated with carbonylation, namely total carbonylation levels (oxyblot), Michael adducts with α,β-unsaturated aldehyde 4-HNE derived from lipid peroxidation, adducts with dicarbonyl compound malondialdehyde, and adducts with lipid aldehydes esterified to PC lipids. A detectable, but still not statistically significant accumulation of modified proteins was observed only in the case of cells treated with E64/pepstatin. Additional treatment with SIN-1 did not result in an increase of the carbonylation signal. It is not surprising that inhibition of cysteine and aspartic acid proteases results in attenuation in modified proteins turnover and accumulation of modified products. Interestingly, treatment with chloroquine and orlistat, although characterized by a significant accumulation of carbonylated products in the form of NRS, did not induce an increase in protein carbonylation and thus intense accumulation of carbonylated molecules under those conditions could be possibly attributed to lipid oxidation and/or any other protein-bound carbonyls not screened in this study.
Patterns of positively stained protein bands were quite similar for all three anti-LPP antibodies used in this study (anti-HNE, -MDA, and -oxPC), whereas oxyblot analysis resulted in quite different distribution of the signals. Interestingly, most of the LPP-protein adducts were detected for the proteins with a low molecular weight between 10 and 20 kDa.
3.5. Effect of autophagy-lysosomal flux inhibitors on proteome regulation in rat cardiac cells
To understand the effect of the different inhibitors on the regulation of cellular fate we performed analysis of proteome regulation using a bottom-up proteomics approach. First, we compared regulation of the proteomes between cells cultured in full medium or starved for 5 h to exclude the influence of starvation alone when comparing the effect of the inhibitors (Table S1). Overall, 35 and 25 proteins were up- or downregulated upon starvation (ANOVA p < 0.05), respectively. Analysis using DAVID functional annotation tool [22], [41] revealed enrichment of seven KEGG pathways (Table 1). Metabolic pathways showed the most significant enrichment. Indeed, starvation alone resulted in upregulation of several mitochondrial proteins involved in metabolism (aspartate and ornithine aminotransferases, subunit of pyruvate dehydrogenase complex) and energy production (beta and gamma subunits of ATP synthase). Cell starvation resulted in downregulation of several protein kinases including A-raf, Akt1, and PI3K-beta. On the other hand, adenylate cyclase 3 was shown to be upregulated by cell starvation. Regulation of these key signalling proteins resulted in enrichment of HIF-1, ErbB and adrenergic signalling pathways as well as lipolysis regulation. Among downregulated proteins (0.6-fold relative to full media control) was sequestosome-1 protein, also known as ubiquitin-binding protein p62, responsible for cargo targeting for autophagic degradation. Downregulation of sequestosome-1 is a well-known hallmark of autophagy activation leading to protein degradation upon delivering its cargo to the autophagosome [42]. Indeed, sequestosome-1 was downregulated in all studied conditions except E64/pepstatin treatment alone or in combination with SIN-1. Interestingly, oxysterol-binding protein-related protein 1, lipid binding protein associated with autophagy induction in yeasts [43], was slightly upregulated upon starvation.
Table 1.
Functional annotation analysis of differentially regulated proteins in rat primary cardiac cells upon 5 h starvation in the absence and presence of inhibitors of the autophagy-lysosomal pathway and SIN-1 treatment.
| KEGG pathway | # of proteins | p-value |
|---|---|---|
| Starvation alone (60 regulated proteins) | ||
| Metabolic pathways | 12 | 4.8E-3 |
| HIF-1 signalling pathway | 4 | 6.6E-3 |
| Adrenergic signalling in cardiomyocytes | 4 | 1.7E-2 |
| Regulation of lipolysis in adipocytes | 3 | 2.0E-2 |
| Central carbon metabolism in cancer | 3 | 2.3E-2 |
| Biosynthesis of amino acids | 3 | 3.8E-2 |
| ErbB signalling pathway | 3 | 4.9E-2 |
| 3-MA and SIN-1 (84 regulated proteins) | ||
| No significant enrichment | ||
| Chloroquine and SIN-1 (185 regulated proteins) | ||
| Parkinson's disease | 9 | 3.3E-4 |
| Insulin signalling pathway | 6 | 8.4E-3 |
| cGMP-PKG signalling pathway | 6 | 4.5E-2 |
| Starch and sucrose metabolism | 3 | 4.9E-2 |
| Orlistat and SIN-1 (143 regulated proteins) | ||
| Parkinson's disease | 5 | 3.1E-2 |
| cGMP-PKG signalling pathway | 5 | 4.4E-2 |
| E64/pepstatin and SIN-1 (350 regulated protein) | ||
| Tight junction | 13 | 1.6E-4 |
| Vascular smooth muscle contraction | 12 | 1.7E-4 |
| Alzheimer disease | 14 | 4.7E-4 |
| Oxytocin signalling pathway | 13 | 5.0E-4 |
| Sphingolipid signalling pathway | 11 | 8.4E-4 |
| Insulin signalling pathway | 11 | 2.1E-3 |
| cGMP-PKG signalling pathway | 12 | 2.8E-3 |
| Neurotrophin signalling pathway | 10 | 3.5E-3 |
| HIF-1 signalling pathway | 9 | 3.7E-3 |
| Ras signalling pathway | 14 | 4.6E-3 |
| Central carbon metabolism in cancer | 7 | 4.4E-3 |
| Fatty acid degradation | 6 | 5.4E-3 |
| Regulation of actin cytoskeleton | 13 | 6.7E-3 |
| Adherent junction | 7 | 8.4E-3 |
| Endocytosis | 15 | 1.0E-2 |
| Focal adhesion | 12 | 1.3E-2 |
| PI3K-Akt signalling pathway | 16 | 1,8E-2 |
| Phosphatidylinositol signalling system | 7 | 2.9E-2 |
Further we compared differences in proteome regulation between starved control and each inhibitor with or without SIN-1 challenge. 3-MA treatment resulted in significant regulation of 84 proteins (Table S2). All proteins upregulated for 3-MA treatment were upregulated upon combined 3-MA and SIN-1 exposure as well, indicating a leading role of inhibitor itself in the observed responses. However, functional annotation analysis of proteins regulated under these conditions did not show any significant pathways enrichment.
Strong upregulation of BAG5 protein was observed in SIN-1 (1.9-fold), 3-MA (2.1-fold) and 3-MA/SIN1 (2.0-fold relative to starved control) treated cells indicating increase in BAG protein level upon stress induction independent of the trigger. Indeed, BAG family proteins play an important role in cardiac function and their differential regulation was related to several pathologies. BAG5 expression in cardiomyocytes was significantly increased by ER stress. Furthermore, protein knockdown resulted in cell death. The role of BAG5 protein in regulation of GRP78 protein stability during ER stress was proposed [44]. Several other proteins were shown to be stabilized by BAG5 chaperon in different stress conditions. Thus, BAG5 was shown to stabilize PINK1 kinase during mitochondrial oxidative damage [45]. Interaction of BAG5 with DJ-1 protein during mitochondrial oxidative damage was shown to inhibit DJ-1 protective effects [46]. Another strongly upregulated protein (2.5- and 1.8-fold after 3-MA and 3-MS/SIN-1 treatments) was tumour protein 53-induced nuclear protein 1 (TP53INP1), a well-known stress protein with antioxidant associated tumour suppressive function. TP53INP1 is overexpressed during cellular stress response. In addition to its function in mediating p53 antioxidant activity, TP53INP1 acts as a positive regulator of autophagy through interactions with LC3 and ATG8-family proteins [47].
Chloroquine treatment alone or in combination with SIN-1 resulted in differential regulation of 185 proteins (Table S3). Here the direction of protein regulation (up or down) was driven by chloroquine treatment itself for the majority the proteins as well. Functional annotation analysis demonstrated enrichment of four KEGG pathways (Table 1), including Parkinson's disease, insulin and cGMP-PKG signalling pathway, and starch and sucrose metabolism. Enrichment of Parkinson's disease pathway was mostly driven by the presence of several mitochondrial proteins involved in oxidative phosphorylation and pro-apoptotic signalling, as well as protein kinase A. Interestingly, six proteins involved in insulin signalling pathway were shown to be regulated upon chloroquine and SIN-1 treatment, including insulin receptor substrate 1 (Irs1; 0.8- and 0.3-fold), protein tyrosine phosphatase PTPN1 (0.9- and 0.6-fold), two subunits of protein kinase A (0.7- and 0.3-fold for Prkacb; 1.0- and 0.5-fold for Prkar1a), and hexokinase I (1.0- and 0.6-fold for chloroquine alone or in combination with SIN-1, respectively), all of which were downregulated upon chloroquine or chloroquine/SIN-1 treatment. Furthermore, insulin receptor-related protein, the receptor involved in activation of IRS1 and PKT1/PKB pathway but not shown in the corresponding KEGG pathway map, was significantly downregulated as well (0.6- and 0.03-fold for chloroquine alone or in combination with SIN-1, respectively). The emerging role of the link between autophagy regulation, ER stress, insulin signalling, and development of insulin resistance is currently under investigation, especially in obesity related research [48], [49]. Similar to adipocytes, cardiomyocytes are insulin sensitive cells in which uptake of the circulating glucose from blood stream is tightly regulated via cross-talk between insulin signalling and GLUT4 transporter externalization on plasma membrane [50]. It is generally viewed that conditions leading to ER stress might result in the induction of autophagy which helps to maintain cellular homeostasis by degrading unfolded and damaged proteins. Impairment of ER stress-induced autophagy can result in insulin resistance via dysregulation of several signalling pathways. Interestingly, autophagy induced downregulation of insulin receptor was shown to contribute to ER stress mediated insulin resistance in adipocytes [48].
Diacylglycerol kinase gamma, an enzyme involved in phospholipid metabolism was already upregulated 1.5-fold upon 3-MA treatment and showed upregulation in the case of chloroquine (1.4-fold) and SIN-1 (2.2-fold) as well. Furthermore, treatment with chloroquine/SIN-1 also resulted in upregulation of diacylglycerol O-acyltransferase 2 (DGAT2; 1.1- and 1.8-fold for chloroquine alone or in combination with SIN-1, respectively). Interestingly, the DGAT1 isoform of this enzyme was shown to be responsible for the lipid droplets accumulation during starvation in a number of cell types. The DGAT1-mediated synthesis of triacylglycerides (TGs) and thus biogenesis of TG-rich LDs was shown to be an important mechanism in preventing cytotoxicity triggered by increased amount of intracellular free fatty acids and corresponding acylcarnitines released by autophagy-lysosomal degradation of cellular organelles [51], [52].
Treatment with orlistat and orlistat/SIN-1 resulted in regulation of 143 proteins and showed functional enrichment similar to chloroquine exposure based mostly on the same regulated proteins entries for Parkinson disease and cGMP-PKG KEGG pathways (Table 1 and S4). Interestingly, no upregulation for both diacylglycerol kinase and DGAT proteins was observed in cells treated with lipase inhibitors, indicating the stopping of the flux of autophagy-lysosomal degradation by inhibiting lipase activity results probably in reduced levels of released free fatty acids and thus no enzymes involved in their re-esterification into PL or TG are required under this condition.
The largest number of differentially regulated proteins (350) was detected upon E64/pepstatin A treatment (Table S5). Functional annotation resulted in enrichment of 18 KEGG pathways including some previously described for chloroquine and orlistat treatments as well as a number of new pathways including regulation of tight and adherent junctions, vascular smooth muscle contraction, regulation of actin cytoskeleton and focal adhesion indicating major reconfiguration of cardiac cell morphology and mobility upon proteases inhibition in starved cells (Table 1). Furthermore, several lipid metabolism and signalling pathways were shown to be regulated. Thus, E64/pepstatin treatment resulted in regulation of sphingolipid signalling pathway involving downregulation of sphingosine-1-phosphate lyase 1 (0.5- and 0.3-fold), and slight upregulation of ERK1 (1.2-fold in E64/pepstatin treatment together with SIN-1) and ERK-activating kinase MEK2 (1.6- and 1.8-fold for protease inhibitors alone or in combination with SIN-1, respectively). Proteins involved in fatty acid degradation pathway were generally downregulated including alcohol dehydrogenases 1 (0.3- and 0.1-fold) and 6 (0.1- and 0.04-fold), as well as acyl-CoA acetyltransferase (0.7- and 0.5-fold) and long-chain-fatty-acid-CoA ligase 3 (0.2- and 0.002-fold for protease inhibitors alone or in combination with SIN-1, respectively). Interestingly, insulin signalling pathway showed even higher enrichment than in chloroquine treated cells and was represented by 11 regulated proteins, the majority of which showed downregulation similar to chloroquine treatment condition however, few including tuberous sclerosis 1 protein homolog (TSC1) or hamartin were significantly upregulated (1.8- and 2.4-fold for protease inhibitors alone or in combination with SIN-1, respectively). Notably, treatment with protease inhibitors upon starvation resulted in significant upregulation of GLUT1 transporter (1.3- and 1.6-fold for protease inhibitors alone or in combination with SIN-1, respectively). Cardiac myocytes express mainly two types of glucose transporters – GLUT1 and GLUT4. GLUT1 on plasma membrane is mainly responsible for basal glucose uptake. GLUT4 is stored in the intracellular vesicles and translocated to the plasma membrane upon insulin stimulation. In adult hearts GLUT4 represents the main isoform and is the primary glucose transporter. GLUT1 increase in expression is a known hallmark of different pathological conditions including hypoxia where HIF-1 directly binds to the GLUT1 promoter. Both insulin and energy status (e.g. availability of free FAs) are important determinants of GLUT1 expression. GLUT1 expression was also shown to be stimulated by oxidative stress exposure [53]. In our cellular model of starvation accompanied by inhibition of proteolysis, deficiency in amino acid substrates might result in down regulation of insulin signalling and switch to insulin independent glucose uptake via upregulation of GLUT1 transporter.
4. Conclusions
Using a cardiac cell culture model of starvation-induced autophagy, we evaluated the effect of four inhibitors along the autophagy-lysosomal degradation pathway on the accumulation of carbonylated biomolecules. We demonstrated that inhibition of autophagosome-lysosomal fusion and lysosomal acidification as well as inhibition of cellular lipases and proteases resulted in significantly higher intensity of total cellular carbonylation independent from the short pulse of SIN-1 treatment. Carbonyl specific signals accumulated in the form of droplet- or vesicle-like structures coinciding with Nile Red positive staining in treated cells. Using western blot detection of four different types of protein modifications associated with introduction of carbonyl functional groups, we could demonstrate that only inhibition of cellular proteases resulted in slightly elevated, but still not statistically significant, signals of protein-bound carbonyls. In contrast, neither chloroquine nor orlistat exposure, although showing significantly higher total cellular carbonylation levels, revealed any increase in protein-bound carbonyls, at least for the set of protein modifications screened in the study. Analysis of systems-wide proteome regulation was performed using relative label free quantitative bottom-up proteomics which allowed the detection of the up- and downregulation of several well-known markers of the autophagic pathway including p62, BAG5, and TP53INP1 proteins. Furthermore, we observed upregulation of DGAT2 protein, isoform 1 of which was already reported as an important regulator of lipid metabolism in starved cells in association with the formation of lipid droplet responsible to reduce lipotoxicity. Interestingly, proteomics results indicated the existence of a cross-talk between autophagy, its inhibition and the insulin signalling pathway, a phenomenon previously reported in adipocytes and associated with development of insulin resistance. Furthermore, upregulation of GLUT1 transporter was detected in protease inhibitors-treated cells. Surprisingly, proteomics analysis did not reveal any significant functional enrichment in lipase inhibitor-treated cell, although this exposure resulted in high level of intracellular carbonyls associated with Nile Red positive droplet-like structures. Further investigations using systems-wide lipidomics analysis might facilitate understanding of cellular regulations in this condition.
Acknowledgment
We thank Prof. Ralf Hoffmann (Institute of Bioanalytical Chemistry, University of Leipzig) for providing access to his laboratory and mass spectrometers. The financial support from MASSTRPLAN project funded by the Marie Sklodowska-Curie EU Framework for Research and Innovation Horizon 2020 (Grant Agreement No. 675132; to MF and DPS), German Federal Ministry of Education and Research (BMBF) within the framework of the e: Med research and funding concept for SysMedOS project (to MF), as well as from the Spanish MINECO/FEDER, grant SAF2015-68590-R (to DPS), are gratefully acknowledged.
Acknowledgments
Disclosure
The authors declare that they have no conflicts of interest with the content of this article.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2019.101123.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Fedorova M. Diversity of protein carbonylation pathways: direct oxidation, glycoxidation and modifications by lipid peroxidation products. In: Ros J., editor. Protein Carbonylation: Principles, Analysis, and Biological Implications. 2017. pp. 48–84. [Google Scholar]
- 2.Frijhoff J. Clinical relevance of biomarkers of oxidative stress. Antioxid. Redox Signal. 2015;23(14):1144–1170. doi: 10.1089/ars.2015.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalle-Donne I. Protein carbonylation in human diseases. Trends Mol. Med. 2003;9(4):169–176. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 4.Cohen G. Signaling properties of 4-hydroxyalkenals formed by lipid peroxidation in diabetes. Free Radic. Biol. Med. 2013;65:978–987. doi: 10.1016/j.freeradbiomed.2013.08.163. [DOI] [PubMed] [Google Scholar]
- 5.Davies M.J. Protein oxidation and peroxidation. Biochem. J. 2016;473(7):805–825. doi: 10.1042/BJ20151227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scofield R.H. Modification of lupus-associated 60-kDa Ro protein with the lipid oxidation product 4-hydroxy-2-nonenal increases antigenicity and facilitates epitope spreading. Free Radic. Biol. Med. 2005;38(6):719–728. doi: 10.1016/j.freeradbiomed.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Sladek N.E. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J. Biochem. Mol. Toxicol. 2003;17(1):7–23. doi: 10.1002/jbt.10057. [DOI] [PubMed] [Google Scholar]
- 8.Mol M. Enzymatic and non-enzymatic detoxification of 4-hydroxynonenal: methodological aspects and biological consequences. Free Radic. Biol. Med. 2017;111:328–344. doi: 10.1016/j.freeradbiomed.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Nissan G., Sharon M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules. 2014;4(3):862–884. doi: 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Y. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143(1):291–299. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. J. Investig. Dermatol. 2013;133(6):1629–1637. doi: 10.1038/jid.2013.26. [DOI] [PubMed] [Google Scholar]
- 12.Pajares M. Redox control of protein degradation. Redox Biol. 2015;6:409–420. doi: 10.1016/j.redox.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y. Autophagy protects against oxidized low density lipoprotein-mediated inflammation associated with preeclampsia. Placenta. 2016;48:136–143. doi: 10.1016/j.placenta.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018;20(5):521–527. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 15.Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filomeni G. Under the ROS … Thiol network is the principal suspect for autophagy commitment. Autophagy. 2010;6(7):999–1005. doi: 10.4161/auto.6.7.12754. [DOI] [PubMed] [Google Scholar]
- 17.Scherz-Shouval R., Shvets E., Elazar I. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3(4):371–373. doi: 10.4161/auto.4214. [DOI] [PubMed] [Google Scholar]
- 18.Griesser E. Cross-talk between lipid and protein carbonylation in a dynamic cardiomyocyte model of mild nitroxidative stress. Redox Biol. 2017;11:438–455. doi: 10.1016/j.redox.2016.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schindelin J. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Wisniewski J.R. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 22.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y.T. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 2010;285(14):10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seglen P.O., Gordon P.B. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA. 1982;79(6):1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mauthe M. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Sala D. The C-terminal sequence of RhoB directs protein degradation through an endo-lysosomal pathway. PLoS One. 2009;4(12):e8117. doi: 10.1371/journal.pone.0008117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuohetahuntila M. Lysosome-mediated degradation of a distinct pool of lipid droplets during hepatic stellate cell activation. J. Biol. Chem. 2017;292(30):12436–12448. doi: 10.1074/jbc.M117.778472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bialecka-Florjanczyk E. Synthetic and natural lipase inhibitors. Mini-Rev. Med. Chem. 2018;18(8):672–683. doi: 10.2174/1389557516666160630123356. [DOI] [PubMed] [Google Scholar]
- 29.Muller S., Dennemarker J., Reinheckel T. Specific functions of lysosomal proteases in endocytic and autophagic pathways. Biochim. Biophys. Acta. 2012;1824(1):34–43. doi: 10.1016/j.bbapap.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung M. Cathepsin inhibition-induced lysosomal dysfunction enhances pancreatic beta-cell apoptosis in high glucose. PLoS One. 2015;10(1):e0116972. doi: 10.1371/journal.pone.0116972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konishi K., Watanabe N., Arai T. SIN-1 cytotoxicity to PC12 cells is mediated by thiol-sensitive short-lived substances generated through SIN-1 decomposition in culture medium. Nitric Oxide. 2009;20(4):270–278. doi: 10.1016/j.niox.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Schrammel A. Activation of soluble guanylyl cyclase by the nitrovasodilator 3-morpholinosydnonimine involves formation of S-nitrosoglutathione. Mol. Pharmacol. 1998;54(1):207–212. doi: 10.1124/mol.54.1.207. [DOI] [PubMed] [Google Scholar]
- 33.Vemula V., Ni Z.X., Fedorova M. Fluorescence labeling of carbonylated lipids and proteins in cells using coumarin-hydrazide. Redox Biol. 2015;5:195–204. doi: 10.1016/j.redox.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scherz-Shouval R., Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem. Sci. 2011;36(1):30–38. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Scherz-Shouval R., Elazar Z. Monitoring starvation-induced reactive oxygen species formation. Methods Enzymol. 2009;452:119–130. doi: 10.1016/S0076-6879(08)03608-2. [DOI] [PubMed] [Google Scholar]
- 36.Jung T. Intracellular distribution of oxidized proteins and proteasome in HT22 cells during oxidative stress. Free Radic. Biol. Med. 2006;40(8):1303–1312. doi: 10.1016/j.freeradbiomed.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 37.Dalle-Donne I. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 2006;10(2):389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domingues R.M. Lipoxidation adducts with peptides and proteins: deleterious modifications or signaling mechanisms? J. Proteom. 2013;92:110–131. doi: 10.1016/j.jprot.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Aldini G. Protein lipoxidation: detection strategies and challenges. Redox Biol. 2015;5:253–266. doi: 10.1016/j.redox.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman P. Correlation of antiphospholipid antibody recognition with the structure of synthetic oxidized phospholipids. Importance of Schiff base formation and aldol condensation. J. Biol. Chem. 2002;277(9):7010–7020. doi: 10.1074/jbc.M108860200. [DOI] [PubMed] [Google Scholar]
- 41.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahani M.H., Itakura E., Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy- derived amino acids. Autophagy. 2014;10(3):431–441. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villasmil M.L., Bankaitis V.A., Mousley C.J. The oxysterol-binding protein superfamily: new concepts and old proteins. Biochem. Soc. Trans. 2012;40(2):469–473. doi: 10.1042/BST20120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta M.K. GRP78 interacting partner Bag5 responds to ER stress and protects cardiomyocytes from ER stress-induced apoptosis. J. Cell. Biochem. 2016;117(8):1813–1821. doi: 10.1002/jcb.25481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X.J. BAG5 protects against mitochondrial oxidative damage through regulating PINK1 degradation. Plos One. 2014;9(1) doi: 10.1371/journal.pone.0086276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin L.X. BAG5 interacts with DJ-1 and inhibits the neuroprotective effects of DJ-1 to combat mitochondrial oxidative damage. Oxid. Med. Cell. Longev. 2017;2017:5094934. doi: 10.1155/2017/5094934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saadi H., Seillier M., Carrier A. The stress protein TP53INP1 plays a tumor suppressive role by regulating metabolic homeostasis. Biochimie. 2015;118:44–50. doi: 10.1016/j.biochi.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 48.Zhou L. Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol. Pharmacol. 2009;76(3):596–603. doi: 10.1124/mol.109.057067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang N. Autophagy regulates insulin resistance following endoplasmic reticulum stress in diabetes. J. Physiol. Biochem. 2015;71(2):319–327. doi: 10.1007/s13105-015-0384-1. [DOI] [PubMed] [Google Scholar]
- 50.Montessuit C., Lerch R. Regulation and dysregulation of glucose transport in cardiomyocytes. Biochim. Biophys. Acta. 2013;1833(4):848–856. doi: 10.1016/j.bbamcr.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen T.B., Olzmann J.A. Lipid droplets and lipotoxicity during autophagy. Autophagy. 2017;13(11):2002–2003. doi: 10.1080/15548627.2017.1359451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen T.B. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell. 2017;42(1):9–21. doi: 10.1016/j.devcel.2017.06.003. (e5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hrelia S. Doxorubicin induces early lipid peroxidation associated with changes in glucose transport in cultured cardiomyocytes. Biochim. Biophys. Acta. 2002;1567(1–2):150–156. doi: 10.1016/s0005-2736(02)00612-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material