Abstract
Increasing evidences support the notion that the impairment of intracellular degradative machinery is responsible for the accumulation of oxidized/misfolded proteins that ultimately results in the deposition of protein aggregates. These events are key pathological aspects of “protein misfolding diseases”, including Alzheimer disease (AD). Interestingly, Down syndrome (DS) neuropathology shares many features with AD, such as the deposition of both amyloid plaques and neurofibrillary tangles. Studies from our group and others demonstrated, in DS brain, the dysfunction of both proteasome and autophagy degradative systems, coupled with increased oxidative damage. Further, we observed the aberrant increase of mTOR signaling and of its down-stream pathways in both DS brain and in Ts65Dn mice.
Based on these findings, we support the ability of intranasal rapamycin treatment (InRapa) to restore mTOR pathway but also to restrain oxidative stress resulting in the decreased accumulation of lipoxidized proteins. By proteomics approach, we were able to identify specific proteins that showed decreased levels of HNE-modification after InRapa treatment compared with vehicle group. Among MS-identified proteins, we found that reduced oxidation of arginase-1 (ARG-1) and protein phosphatase 2A (PP2A) might play a key role in reducing brain damage associated with synaptic transmission failure and tau hyperphosphorylation. InRapa treatment, by reducing ARG-1 protein-bound HNE levels, rescues its enzyme activity and conceivably contribute to the recovery of arginase-regulated functions. Further, it was shown that PP2A inhibition induces tau hyperphosphorylation and spatial memory deficits. Our data suggest that InRapa was able to rescue PP2A activity as suggested by reduced p-tau levels.
In summary, considering that mTOR pathway is a central hub of multiple intracellular signaling, we propose that InRapa treatment is able to lower the lipoxidation-mediated damage to proteins, thus representing a valuable therapeutic strategy to reduce the early development of AD pathology in DS population.
Keywords: Down syndrome, mTOR, Rapamycin, Oxidative stress, Protein-bound HNE
Graphical abstract
1. Introduction
An increasing number of studies highlight the involvement of aberrant mTOR signaling in the pathogenesis and progression of neurodegenerative disorders, including Alzheimer Disease (AD) [1,2]. Indeed, mTOR pathway has a prominent role in the central nervous system through the regulation of several intracellular processes, such as, protein synthesis, transcription, autophagy, metabolism, and organelle biogenesis [3,4]. These functions are central for the maintenance of brain homeostasis and to regulate the proliferation of neural stem cells, the assembly of circuits, the plasticity of synapses and behavioral aspects like feeding, sleep and circadian rhythms [5,6]. As a consequence, mTOR pathway dysfunction is implicated in different types of brain disorders such as autism, epilepsy, Parkinson disease and AD. Noteworthy, experimental evidences showed that aberrant mTOR signaling is closely associated with the presence of the two key pathological features of AD: senile plaques, composed mainly of fibrillar β-amyloid (Aβ) peptide, and neurofibrillary tangles (NFT), composed of hyperphosphorylated tau protein [1,2,[7], [8], [9]].
Recently, we and others have investigated the role of mTOR in the development of AD [7,[9], [10], [11], [12], [13], [14], [15]]; we showed that in subjects with AD and AD-like dementia, as in Down syndrome (DS), the activation of mTOR signaling contributes to Aβ production and tau hyperphosphorylation [7,16,17]. DS, a genetic disorder characterized by the triplication of chromosome 21, is the most frequent genetic cause of intellectual disability associated with neurologic deficiencies, including early onset AD high incidence of clinical symptoms in the late 40ys of age [[18], [19], [20], [21]]. The early hyperactivation of mTOR signaling correlates with tau hyperphosphorylation in the brains of young DS subjects. In addition, hyperphosphorylation of mTOR is associated with increased activation of PI3K/Akt axis, decreased autophagosome formation, increased oxidative stress (OS) and accumulation of oxidized proteins [17,22]. Based on these evidences, pharmacological manipulation of mTOR signaling could represent a promising approach to reduce brain damage and so far, has entered clinical trials for several disorders [8,23].
Published data demonstrated the neuroprotective effects of oral administration of rapamycin in 3xTg-AD mice and J20 mouse models of AD, as indexed by reduced accumulation of plaques and tangles and improved cognitive function [[11], [12], [13],15,24,25]. We recently proved that administration of rapamycin by intranasal route in Ts65Dn mice (transgenic mouse model of DS) is able to rescue aberrant mTOR signaling by restoring autophagosome formation and by reducing Aβ production and tau hyperphosphorylation [26]. These effects were associated with the amelioration of cognitive function.
Based on these findings, we extended our study to investigate if intranasal administration of rapamycin (InRapa) was able to reduce lipoxidation-mediated damage to proteins in the frontal cortex of Ts65Dn mice. The age-dependent failure of degradative systems, including autophagy and proteasome results in the accumulation of damaged mitochondria and oxidized proteins [16,27,28]. It is likely that autophagy may provide protection against OS as supported by data showing that Atg4, an essential protease that controls the lipid modification of Atg8 and autophagosome formation, is a direct target for oxidation by hydrogen peroxide. On the other hand, it has also been suggested that low levels of ROS provide a signal to regulate autophagic survival and death processes [29,30]. Collectively, accumulated evidence suggests that free radicals are upstream and downstream of mTOR and numerous feedback and feed forward loops exist [31].
Considering the crosstalk between mTOR and OS, we have hypothesized that pharmacological rescue of mTOR hyperactivation might also be associated with reduced oxidative damage. Indeed, published studies from our group showed that oxidized proteins are significantly increased in the brain of DS subjects, prior and after development of AD [27,[32], [33], [34]]. In addition, a decrease of LC3 II/I levels ratio, an index of autophagosome formation, occurs early in DS brain (before development of AD neuropathology) and persists in DS brain with AD pathology, both compared to their respective age-matched controls [17,27]. Oxidative damage targets different components of the protein homeostasis control system such as GRP78, UCH-L1, V0-ATPase, cathepsin D and GFAP that couples, in DS brain, with decreased proteasome activity and reduced autophagosome formation [27,33]. Furthermore, a link between protein oxidative damage and aberrant mTOR/autophagy axis induction was proven in Ts65Dn mice [16].
In the present study, we provide evidences demonstrating the ability of InRapa treatment to restore, not only mTOR pathway, but also to modulate OS, resulting in decreased accumulation of lipoxidized (i.e. HNE-modified) proteins. Thus, the reduction of lipid peroxidation reactions has the potential to block/reduce the downstream attack, free radical-mediated, to multiple cellular targets, thus preserving redox homeostasis [35]. Among HNE-modified proteins, we found that reduced oxidation of arginase-1 (ARG-1) and protein phosphatase 2A (PP2A) might play a key role in lowering brain damage associated with synaptic transmission failure and tau hyperphosphorylation.
2. Materials and methods
2.1. Mouse colony
Ts65Dn (B6EiC3Sn a/A-Ts(1716)65Dn/J), a well-established mouse model of DS, carries a reciprocal translocation that is trisomic for approximately 104 genes (56%) on Mmu16, from Mrpl39 to the distal telomere, with homologues on HSA21. Mice were generated by repeatedly backcrossing Ts65Dn trisomic females with (B6EiC3SnF1/J) F1 hybrid males. The parental generations were purchased from Jackson Laboratories (Bar Harbour, ME, USA). These breeding pairs produce litters containing both trisomic (Ts65Dn) and euploid offsprings. The pups were genotyped to determine trisomy using standard PCR, as described by Reinoldth [36]. Mice were housed in clear Plexiglas cages (20 × 22 × 20 cm) under standard laboratory conditions with a temperature of 22 ± 2 °C and 70% humidity, a 12-h light/dark cycle and free access to food and water. Mice were sacrificed by cervical dislocation. Trunk blood was collected from the site where the animal was decapitated. Brain was dissected in two halves used, respectively, for immunochemical and immunofluorescence analysis. All the experiments were performed in strict compliance with the Italian National Laws (DL 116/92), the European Communities Council Directives (86/609/EEC). Experimental protocol was approved by Italian Ministry of Health authorization n◦ 1183/2016-PR. All efforts were made to minimize the number of animals used in the study and their suffering. All samples were flash-frozen and stored at −80 °C until utilization.
2.2. In Rapa treatment
6-month old Ts65Dn and euploid (Eu) mice were administered with intranasal rapamycin (In Rapa; Rapamune, Pfizer, New York, NY, USA) and Vehicle (Veh; saline with 1% DMSO) for 12 weeks. Mice were divided in 4 experimental groups Eu and Ts65Dn treated with vehicle or rapamycin (n = 10 per group) (Table 1). The treatment was conducted 3 times per week, with a dose of 0.1 μg/μl of rapamycin solution or vehicle in 10 μl (1 μg/mouse) as previously reported [26]. The treatment was well tolerated and no change in body weight or in the consumption of drinking water was observed. The rapamycin dose was chosen from a dose response pilot study performed before the treatment. In the dose–response treatment the animals were divided in three groups (n = 6 per group) and treated with 0.01, 0.05, 0.1 and 0.2 μg/μl of rapamycin. Our data demonstrated that the dose of rapamycin administered during the treatment, 0.1 μg/μl, was able to partially inhibit mTOR (Ser2448) phosphorylation in the frontal cortex (Suppl. Fig. 1A and B) and hippocampus [26].
Table 1.
Samples characteristics and experimental uses.
| Tissue | Treatment | Genotyping | n | Gender (m/f) | Experimental Use |
|||
|---|---|---|---|---|---|---|---|---|
| WB | IF | 2D electrophoresis | IP | |||||
| Frontal CX | Vehicle | Eu | 10 | 6/4 | 8 | 4 | 8 | 8 |
| Ts65Dn | 10 | 5/5 | 8 | 4 | 8 | 8 | ||
| InRapa | Eu | 10 | 6/4 | 8 | 4 | 8 | 8 | |
| Ts65Dn | 10 | 6/4 | 8 | 4 | 8 | 8 | ||
2.3. Sample preparation
Brain tissues of Ts65Dn and euploid mice (n = 8 per group) after treatment were sagittal divided in right and left hemispheres. The right portion was used for Immunofluorescence studies and the left portion was used for Biochemical and molecular biology studies. For Western blot, Slot blot and proteomics analysis the frontal cortex was homogenized in RIPA buffer (pH 7.4) containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 0,1% SDS, 1 mM PMSF, 1 mM NaF and 1 mM Na3VO4. Brains were homogenized by 20 strokes of a Wheaton tissue homogenizer. All the samples homogenate was centrifuged at 14,000×g for 10 min to remove cellular debris. The supernatant was extracted to determine the total protein concentration by the BCA method (Pierce, Rockford, IL, USA).
2.4. Measurement of total protein-bound 4-hydroxy-2-trans-nonenal (HNE-bound protein) and 3-nitrotyrosine (3-NT)
For the analysis of HNE-bound and 3-nitrotyrosine (3- NT) protein levels, 5 μl of the total protein extract from the frontal cortex of our groups of treatment were incubated with 5 μl of Laemmli buffer containing 0.125 M Tris base pH 6.8, 4% (v/v) SDS, and 20% (v/v) glycerol. The resulting samples (250 ng for each well) were loaded in each well on a nitrocellulose membrane under vacuum using a slot blot apparatus. The membranes were blocked in blocking buffer (3% bovine serum albumin) in TBS containing 0.01% Tween 20 for 1 h at room temperature and incubated with HNE polyclonal antibody (1:2000, Novus Biologicals, Abingdon, UK, #NB100-63093) or an anti-3-NT polyclonal antibody (1:1000, Santa Cruz, CA, USA, #sc-32757) in BSA 3% in TBS-T for 120 min. The membranes were washed in PBS following primary antibody incubation three times at intervals of 5 min each. The membranes were incubated respectively with an anti-goat and anti-mouse IgG alkaline phosphatase secondary antibody (1:5000, Sigma–Aldrich, St Louis, MO, USA) for 1 h. The membranes were washed three times in PBS for 5 min each and developed with Sigma fast tablets (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium substrate [BCIP/NBT substrate]). Blots were dried, acquired with Chemi-Doc MP (Bio-Rad, Hercules, CA, USA) and analyzed using Image Lab software (Bio-Rad, Hercules, CA, USA). No non-specific binding of antibody to the membrane was observed.
2.5. Two-dimensional (2D) electrophoresis
Frontal cortex homogenate from Eu (Veh and InRapa) and Ts65Dn (Veh and InRapa) (100 μg of proteins) were precipitated in cold absolute ethanol overnight. Each sample was than centrifuged at 10 000 g for 5 min. The pellet was dissolved in 200 μL of rehydration buffer: 8 M Urea, 20 mM Dithiothreitol, 2% (w/v) Chaps, 0,2% Bio-Lyte, 2 M Thiourea, and Bromophenol Blue. For the first-dimension electrophoresis, approximately 200 μl of sample were applied to 110-mm pH 3–10 IPG® ReadyStrip (Bio-Rad, Hercules, CA, USA). The strips were then actively rehydrated in the protean isoelectric focusing (IEF) cell (Bio-Rad, Hercules, CA, USA) at 50 V for 18 h. The isoelectric focusing was performed in increasing voltages as follows; 300 V for 1 h, then linear gradient to 8000 V for 5 h and finally 20,000 V/h. Strips were then stored at −80 °C until the 2D electrophoresis was to be performed. For the second dimension, the IPG® Strips, were thawed and equilibrated for 10 min in 50 mM Tris–HCl (pH 6.8) containing 6 M urea, 1% (w/v) sodium dodecyl sulfate (SDS), 30% (v/v) glycerol, and 0.5% dithiothreitol, and then re-equilibrated for 15 min in the same buffer containing 4.5% iodacetamide instead of dithiothreitol. Linear gradient precast criterion Bis-Tris gels (12%) (Bio-Rad, Hercules, CA, USA) were used to perform second dimension electrophoresis. Precision Protein™ Standards (Bio-Rad, Hercules, CA, USA) were run along with the samples at 200 V for 50 min. After running, the gels were incubated in fixing solution (10% acetic acid, 40% methanol) for 40 min and stained overnight at room temperature with 50 mL SYPRO Ruby gel stain (Bio-Rad, Hercules, CA, USA). The SYPRO ruby gel stain was then removed and gels stored in deionized water. For 2D blots, gels were blotted on nitrocellulose membranes (Bio-Rad, Hercules, CA, USA) and HNE proteins were detected on the membranes. Briefly, membranes were blocked for 1 h with 3% albumin in T-TBS, incubated with HNE polyclonal antibody (1:2000, Novus Biologicals, Abingdon, United Kingdom, #NB100-63093) overnight at 4 °C. After washing with T-TBS three times for 10 min, membranes were further incubated at room temperature for 1 h with the secondary antibody alkaline phosphatase conjugated anti-mouse IgG (1:5000; Sigma-Aldrich, St Louis, MO, USA). Membranes were then washed with T-TBS three times and developed using 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium solution (BCIP/NBT). In addition, we tested the specificity of anti-HNE antibody used in the proteomic approach. Thus, 1.0 mg of protein (BSA) was treated with 0, 1, 3 and 6 mM HNE (Cayman Chemical, Ann Arbor, MI, US) in 1 ml of sodium phosphate buffer and the samples were incubated in a 37 °C water bath for 2 h. The BSA-protein samples were loaded in SDS-page and transferred to nitrocellulose membrane, which was imaged using the Stain-Free Blot settings (Total Load) by the ChemiDoc MP imaging system. After transfer, the blot was incubated with HNE polyclonal antibody (1:2000) and developed using the Chemiluminescence settings in the Chemi Doc MP imaging system.
2.6. Western blot
Western blots were performed as previously described in Ref. [26]. Briefly proteins were separated via SDS-PAGE, transferred to nitrocellulose membrane. After transfer, the blot was imaged using the ChemiDoc MP imaging system using the Stain-Free Blot settings. This total protein signal was used for total protein normalization. Than the membrane was blocked with 3% of bovine serum albumin (SERVA Electrophoresis GmbH, Heidelberg, Germany) and incubated over night at 4 °C with primary antibody against: mTOR pSer2448 1:500 (Bio-Rad Laboratories, Hercules, CA, USA #AHP2657), mTOR 1:1000 (Cell Signaling, Danvers, MA, USA #2983S), Tau pSer404 1:2000 (Abcam, Cambridge, UK #ab92676), total tau 1:1000 (Santa Cruz, Santa Cruz CA, USA, #sc-5587), Atg12 (C-6) 1:500 (Santa Cruz, Santa Cruz CA, USA, #sc-271688), APG5 (C-1) 1:500 (Santa Cruz, Santa Cruz CA, USA, #sc-133158); Amyloid Precursor Protein 1:5000 (Sigma-Aldrich, St Louis, MO, USA # A8717); LC3 1:1000 (MBL International Corporation, Woburn, MA, USA #M152-3); PP2A 1:2000 (Genetex # GTX106334, Irvine, CA,USA); Arginase-1 1:2000 (Genetex # GTX109242, Irvine, CA,USA). All the membranes were incubated for 1 h at room temperature with secondary antibody horseradish peroxidase-conjugated anti-rabbit, anti-mouse or anti-goat IgG (1:20000, Bio-Rad Laboratories, Hercules, CA, USA). The blot was then imaged via the Chemi Doc MP imaging system using the Chemiluminescence settings. Subsequent determination of relative abundance via total protein normalization was calculated using Image Lab 6.0 software (Bio-Rad Laboratories, Hercules, CA, USA).
2.7. Image analysis
SYPRO ruby-stained gel and Blot images were obtained using a Chemidoc MP System (Bio-Rad, Hercules, CA, USA). All the images were saved in TIFF format. 2D gels and 2D blots images (8 gels and 8 blots for each group of study) were analyzed by PD-Quest 2D Analysis (7.2.0 version; Bio-Rad, Hercules, CA, USA). PD-Quest spot-detection software allows the comparison of 2D gels as well as 2D blots, from Eu (Veh and InRapa) and Ts65Dn (Veh and InRapa). Briefly, a master gel was selected followed by normalization of all gels and blots according to the total spot density. Gel-to-blot analysis was then initiated in two parts. First, manual matching of common spots was performed, that could be visualized among the differential 2D gels and 2D blots. After obtaining a significant number of spots the automated matching of all spots was then initiated. Automated matching is based on user-defined parameters for spot detection. These parameters are based on the faintest spot, the largest spot, and the largest spot cluster that occur in the master gel and are defined by the user. This process generates a large pool of data, approximately 400–800 spots. Only proteins showing computer-determined significant differential levels between the groups being analyzed were considered for identification. To determine significant differential levels of proteins, analysis sets were created using the analysis set manger software incorporated into the PD-Quest software. The numbers of pixels that occur in a protein spot were computed by the software corresponding to an increase/decrease in protein level. The image analysis was conducted first on blot and then on Sypro Ruby-stained expression gels. The two analyses were compared by software to normalize HNE-modified protein value to expression value for each spot matched.
2.8. In-gel trypsin digestion/peptide extraction
Protein spots identifies as significantly altered from our group comparison were excised from 2D-gels and transferred to individual Eppendorf microcentrifuge tubes for trypsin digestion as described previously [37]. In brief, DTT and IA were used to break, and cap disulfide bonds and the gel plug was incubated overnight at 37 °C with shaking in modified trypsin solution. Tryptic peptide solutions were reconstituted in water and stored at −80 °C until MS/MS analysis.
2.9. RP-HPLC-high resolution MS/MS characterization of tryptic peptides
High-resolution HPLC-ESI-MS/MS experiments were carried out by an Ultimate 3000 RSLC nano system coupled to an LTQ Orbitrap ELITE apparatus (Thermo Fisher Scientific, Waltham, MA, USA). Zorbax 300 SB-C18 (3.5 μm particle diameter; column dimension 1 mm × 150 mm) (Agilent Technologies, Santa Clara, CA, USA) was used as chromatographic column. The following eluents were used: (A) 0.1% (v/v) aqueous FA and (B) 0.1% (v/v) FA in ACN/water 80/20 v/v. The applied gradient was: 0–2 min 5% B, 2–40 min from 5 to 70% B (linear), 40–45 min from 70 to 99% B (linear), at a flow rate of 50 μl/min with a total run of 65 min. MS spectra were collected with 120,000 resolution and m/z range from 350 to 2000. In data-dependent acquisition mode the five most intense multiply-charged ions were selected and fragmented in ion trap by using CID 35% normalized collision energy. Tuning parameters were: capillary temperature 300 °C, source voltage 4.0 kV.
2.10. Immunofluorescence
Right brain hemispheres were removed and immersed in 4% paraformaldehyde for 24 h at 4 °C. Fixed brains were cryoprotected in successive 48 h with a solution of 20% sucrose and 0.02% NaN3 at 4 °C. Brains were frozen on a temperature-controlled freezing stage, coronal sectioned (20 μm) on a sliding cryostat (Leica Biosystems, Wetzlar, Germany), and stored in a solution of PBS containing 0.02% NaN3 at 4 °C. Brain sections were mounted on glass slide. Once dried, sections were blocked with a solution containing 10% normal goat serum, 0.02% NaN3, and 0.2% Triton X-100 in TBS. Slides were then incubated overnight at 4 °C with following antibodies: mTOR p-Ser2448 (mouse 1:500, #sc-293133 Santa Cruz, Santa Cruz CA, USA); Tau p-Ser404 (rabbit 1:500, #ab92676Abcam, Cambridge, UK). Slides were then washed with TBS and then incubated with Alexa Fluor 488 nm and-594nm secondary antibodies (Invitrogen Corporation, Carlsbad, CA, USA) at 1:1500 for 2h at room temperature. Slides were then washed again and incubated with DAPI (1:10.000). One slide per group was stained omitting primary antibodies to establish nonspecific background signal. Cover slips were placed using a drop of Fluorimount (Sigma-Aldrich, St Louis, MO, USA). All slides were imaged using Zeiss AXio (Carl Zeiss, Oberkochen, Germany). All immunolabeling acquisition intensities, field sizes, and microscopy settings were kept consistent across all images. Images were analyzed using ImageJ. Image montages for Figures were collated in Illustrator and Photoshop Cs6 (Adobe System, San Josè, CA, USA) software programs and were based upon cells that most closely approximated the group means.
2.11. Immunoprecipitation
Immunoprecipitation experiments were performed using SureBeads™ Protein A/G Magnetic Beads (BioRad SureBeads™ Protein G #161-4023). Briefly, 100 μl SureBeads were magnetized and washed with a solution of PBS/Tween20 0.1% v/v., then incubated for 30 min with 2 μg anti- PP2A (Genetex # GTX106334, Irvine, CA, USA) and Arginase-1 (Genetex # GTX109242, Irvine, CA, USA) at room temperature. After incubation, beads were washed 3 times with PBS/Tween20 0.1% v/v and the beads/antibody complexes were recovered and incubated with 100 μg of protein extracts for 1 h at room temperature. Proteins were separated by SDS-PAGE followed by immunoblotting on a nitrocellulose membrane (Bio-Rad, Hercules CA, USA). Membrane were incubated with the antibodies anti-HNE (1:2000, Novus Biologicals, Abingdon, UK), and then detected by the peroxidase-conjugated secondary antibody (1:20.000, Bio-Rad, Hercules, CA, USA) with Clarity™ Western ECL Blotting Substrates (Bio-Rad, Hercules, CA, US). Membranes were then acquired with ChemiDoc MP image system (Bio-Rad, Hercules, CA, USA) and analyzed using Image Lab software (Bio-Rad, Hercules, CA, USA).
2.12. ARG-1 activity
ARG-1 activity was measured by using the Arginase Activity Assay Kit (Sigma-Aldrich, St. Louis, Missouri, USA) according to the manufacturer instructions. Arginase activity was measured in the frontal cortex homogenate from Eu (Veh and InRapa) and Ts65Dn (Veh and InRapa). In details 50 μg of sample were incubated in a 96 well plate with 10 μL of Substrate buffer (l-Arginine and Mn solution) for 37 C for 2h. The reaction was stopped by adding 200 μl of the prepared Urea reagent to each well. The samples were analyzed with a spectrophotometer at 540 nm. The Arginase activity of our sample was determined by the following equation: Activity = (A430) sample – (A430) blank × (1mM × 50 x 103)/(A430) standard – (A430) water (V × T).
(T = Reaction time in minutes; V = sample volume (μL) added to well (1–40 μl); 1 mM = concentration of Urea Standard; 50 = reaction volume (μL); 103 = mM to μM conversion factor).
2.13. Statistical analysis
Each immunoblot experiment was performed three times using 8 samples per group. Immunohistochemistry analyzes were performed using at least 10 sections per brain of 4 mice per group. Details on sample size are summarized in Table 1. All statistical analyses were performed using a non-parametric one-way ANOVA with post hoc Bonferroni t-test. Data are expressed as mean ± SD per group. All statistical analyses were performed using Graph Pad Prism 7.0 software (GraphPad, La Jolla, CA, USA).
3. Results
3.1. InRapa effects on mTOR/autophagy axis, AD hallmarks and protein oxidative damage
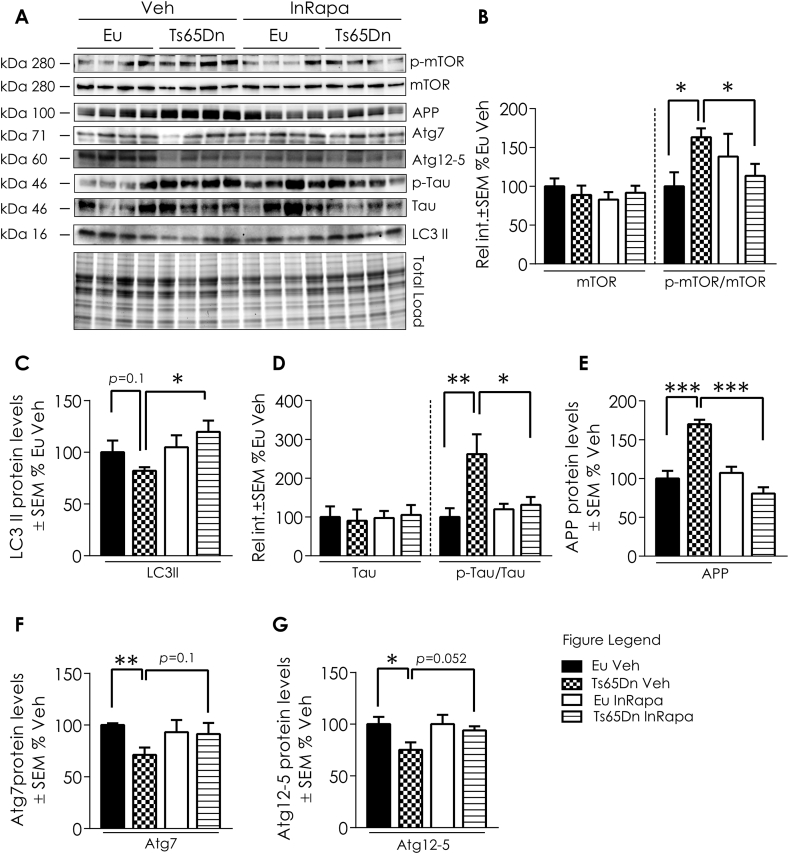
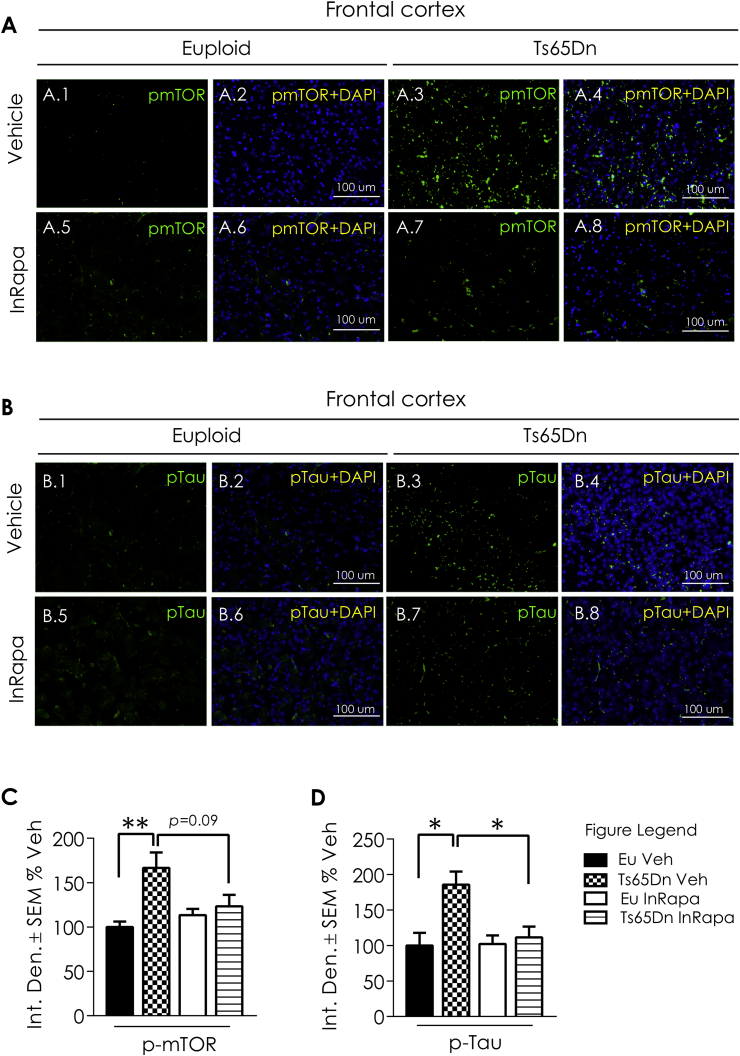
In order to evaluate the biochemical effects of InRapa treatment in the frontal cortex of Ts65Dn mice, we first analyzed the main target of rapamycin, mTOR. Collected data demonstrated that Ts65Dn mice compared with Eu Veh group show increased mTOR hyperphosphorylation at Ser2448, leading to mTORC1 over activation (63.1% increase; p = 0.014; Fig. 1A and B). The administration of InRapa in Ts65Dn mice was able to partially inhibit mTOR phosphorylation at Ser2448 leading to the restoration of mTORC1 to physiological levels, as demonstrated by the comparison with Eu mice. Indeed, as previously observed in the hippocampus [26], we found in the frontal cortex of Ts65Dn mice after InRapa treatment a significant reduction of mTOR phosphorylation compared to Veh (49.7% decrease; p = 0.027; Fig. 1A and B). These results were validated staining of mTOR, by immunofluorescence, in brain slices of mice cortex (Fig. 2A1-8). mTOR also plays a crucial role in regulating autophagy. Our published studies demonstrated an impairment of autophagy in the brain of Ts65Dn mice at different ages, and the ability of rapamycin to restore autophagy by dampening mTOR hyperactivation in the hippocampus of Ts65Dn mice [26]. To support these results, we measured by WB the levels of LC3 II, Atg7 and of the Atg12/Atg5 complex, which are well-recognized markers of autophagy induction, in mice in frontal cortex. Our results show that mTORC1 over activation in Ts65Dn Veh mice leads to decreased LC3II (about 30% reduction, p = 0.01; Fig. 1A and C), Atg7 (35% reduction, p = 0.01; Fig. 1A and F) and Atg12/Atg5 complex (25% reduction, p = 0.0024; Fig. 1A and G), while InRapa treatment by inhibiting mTOR phosphorylation led to the increase of LC3 II levels (about 40% increase compared to Ts65Dn Veh, p = 0.039; Fig. 1A and C) and show a trend of increase for Atg7 and Atg12/Atg5 complex (p = 0.1 and 0.052 respectively; Fig. 1A, F and G), therefore suggesting the increased induction of autophagy.
Fig. 1.
InRapa lowers tau hyper-phosphorylation and APP levels through mTOR inhibition and increased LC3 II levels. (A) Representative WB showing p-mTOR (Ser 2448), mTOR, LC3 II Atg 12, 7 and 5, p-tau (Ser404), tau, and APP total levels (B). Quantification of panel A showing mTOR protein levels, p mTOR (Ser2448)/mTOR ratio. (C) Quantification of panel A showing LC3 II protein levels. (D) Quantification of panel A showing Tau protein levels and Tau (Ser404)/Tau ratio. (E) Quantification of panel A showing APP protein levels. (F) Quantification of panel A showing At7 protein levels. (G) Quantification of panel A showing Atg12/Atg5 complex levels. Densitometry values shown in the bar graph are the mean of 8 (WB) samples per each group normalized per total load and are given as percentage of Eu Veh, set as 100%. Statistical significance was determined using one–way ANOVA and Student t-test analysis (*p < 0.05, **p < 0.01, ***p < 0.001).
Fig. 2.
IF image showing mTOR and Tau expression after In Rapa treatment. Panel A: Representative immunofluorescent images showing mTOR (Ser2448) signal in the CX of euploid treated with Veh (1,2) and InRapa (5,6) and Ts65Dn treated with Veh (3,4) and InRapa (7,8). Panel B: Representative immunofluorescent images showing Tau (Ser404) signal in the CX of euploid treated with Veh (1,2) and InRapa and (5,6) and Ts65Dn treated with Veh (3,4) and InRapa (7,8). DAPI (blue) was used to identify cell nuclei. Scale bar represent 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
As reported before the restoration of autophagy by rapamycin treatment ameliorates the aberrant regulation of APP and tau [26]. The overexpression of APP in DS was shown in humans and mouse samples and is confirmed in Ts65Dn Veh compared to Eu Veh, with an increase of about 70% (p = 0.0009; Fig. 1A and E). Interestingly, InRapa administration in Ts65Dn mice (Ts65Dn InRapa) is able to reduce total APP levels to euploid condition (about 50% decrease; p = 0.0001; Fig. 1A and E). To further investigate the efficacy of InRapa to reduce AD-related pathological features, we also examined tau hyperphosphorylation by WB (Fig. 1A) and immunofluorescence (Fig. 2B1-8) staining. Ts65Dn mice show increased phosphorylation of tau on Ser404 (about 162% increase, p = 0.008; Fig. 1A and D) compared with Eu mice. InRapa treatment on Ts65Dn mice demonstrated a robust decrease of tau phosphorylation, in Ser404, when compared to Veh Ts65Dn group (about 130% decrease, p = 0.022; Fig. 1A and D). A similar trend is observed in the IF images (Fig. 2B1-8).
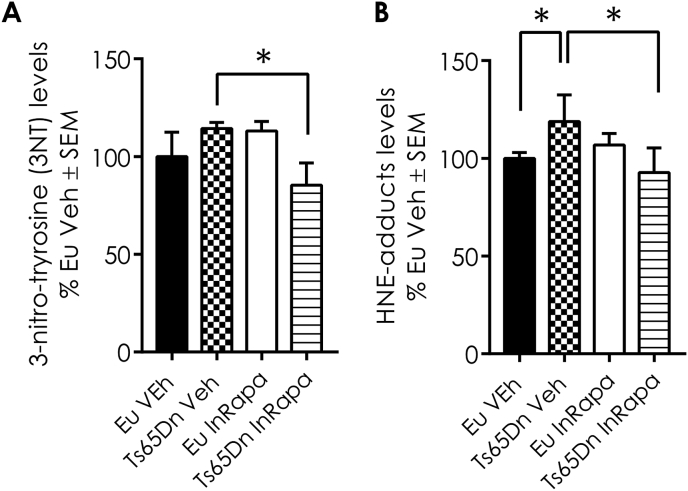
Previous studies by our group and others demonstrated that, in DS brain, alteration of autophagy is associated with increased OS [16,27,33,38,39]. We evaluated the levels of two different protein oxidation markers, 4-hydroxy-2-nonenal (HNE) protein adducts and protein-bound 3-nitrotyrosine (3-NT) in the frontal cortex of Ts65Dn and Eu mice treated with rapamycin or Vehicle. The increase of total 3-NT levels is not significant in Ts65Dn mice compared with Eu mice treated with Veh. However, InRapa treatment is able to reduce significantly the 3-NT total levels of about 30% (p = 0.045) in Ts65Dn (Fig. 3A). A significant increase of HNE adducts was observed (about 25%, p = 0.03) between Eu Veh and Ts65Dn Veh, which was is significantly reduced by the treatment with rapamycin (40%, p = 0.025, Fig. 3B).
Fig. 3.
InRapa reduces protein oxidative damage. In panel A) are shown the total protein-bound 3-NT and B) HNE of Ts65Dn mice and Eu treated with Veh and InRapa analyzed by slot blot assay. Densitometry values shown in the bar graph are the mean of 8 samples per each group normalized per total load and are given as percentage of Eu Veh, set as 100%. Statistical significance was determined using one–way ANOVA and Student t-test analysis (*p < 0.05, *p < 0.01).
3.2. Proteomics profile of HNE-modified proteins
Based on the analysis of total protein oxidation levels, we further analyzed protein-bound HNE profile by proteomics approach, in order to identify specific proteins showing increased levels of HNE-modification.
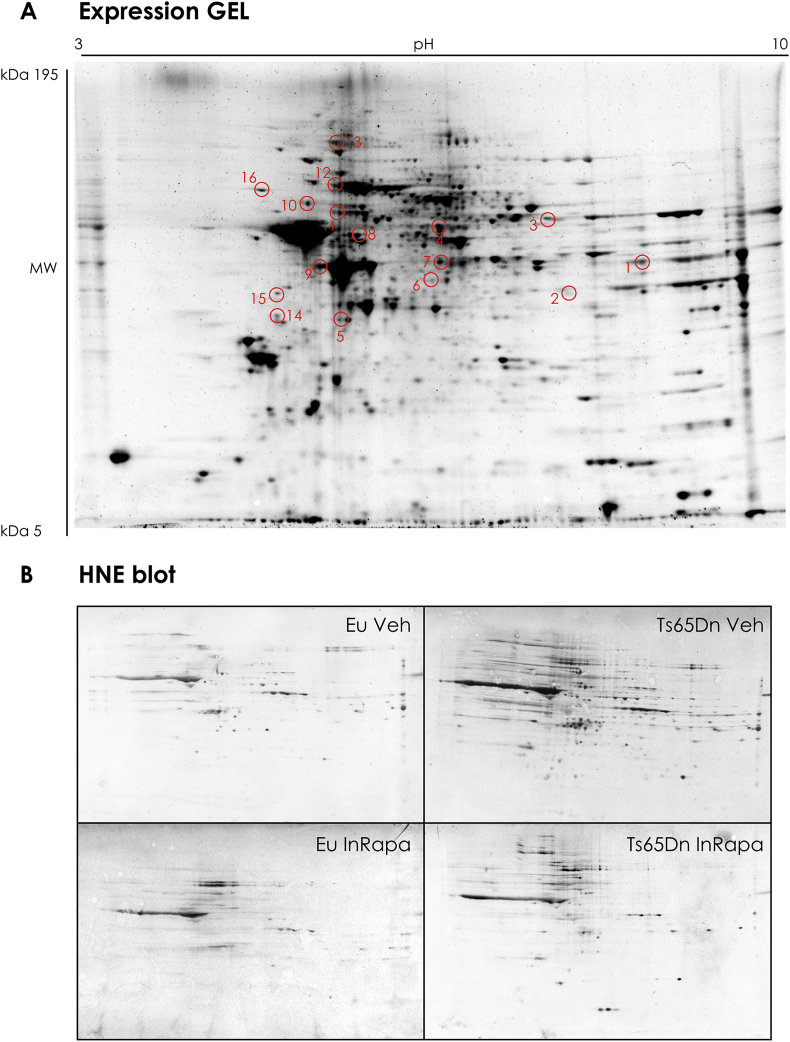
Subsequently, cortex homogenates were separated on two-dimensional (2D) gels and HNE modified-proteins were detected by immunoblotting (Fig. 4A). The overall intensities of protein spots that immunoreacted with HNE antibody appeared higher in Ts65Dn Veh group compared to Eu Veh and RAPA groups (Fig. 4B). On the other hand, the intensity of HNE-modified proteins is reduced in Ts65Dn treated with rapamycin compared with Ts65Dn Veh group (Fig. 4B), confirming a reduction of HNE adducts achieved by InRapa treatment. The 2D proteomics analyses identified a number of HNE-modified proteins (normalized to expression levels) in the cortex of the four groups of samples (Eu Veh, Ts65Dn Veh, Eu RAPA and Ts65Dn RAPA) and the following results were obtained (Table 2):
-
•
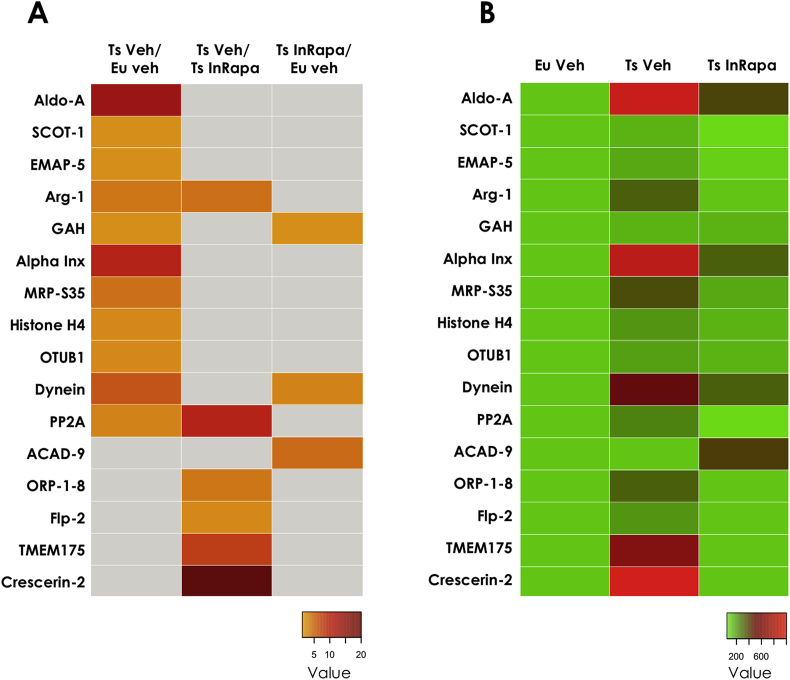
Increased HNE-bound protein levels in Ts65Dn mice compared with Eu (Ts65Dn Vehicle/Euploid Vehicle). To evaluate the genotype-dependent effect, we compared the Ts65Dn mice with Eu mice both treated with vehicle. Proteins showing increased levels of HNE-modification were identified as: Fructose-bisphosphate aldolase A with 12.6-fold increase; Succinyl-CoA:3-ketoacid coenzyme A transferase 1 and Guanine deaminase with 1.3-fold increase; Echinoderm microtubule-associated protein-like 5 with 1.6-fold increase; ARG-1 with 3.0-fold increase; α-internexin with 8.9-fold increase; 28S ribosomal protein S35 with 3.4-fold increase; Histone H4 with 1.9-fold increase; Ubiquitin thioesterase (OTUB1) with 1.8-fold increase; Dynein heavy chain 3 with 5.3-fold increase; Serine/threonine-protein phosphatase 2A (PP2A) regulatory subunit α isoform with 2.4-fold increase (Table 2). These data confirm the increase of OS in the frontal cortex of Ts65Dn mice compared with euploids, as displayed in the first column of the heat map (Fig. 5, panel A), and are noticeable comparing the first and the second column of the heat map (Fig. 5, panel B).
-
•
InRapa treatment reduces the levels of HNE-bound proteins in Ts65Dn mice (Ts65Dn Vehicle/Ts65Dn InRapa). To investigate the effects of rapamycin on Ts65Dn mice, we compared mice treated by InRapa with Tg mice treated with vehicle. Proteins that showed decreased HNE levels after InRapa treatment were identified as: oxysterol-binding protein-related protein 1-8 with 3-fold increase, formin-like protein 2 with 2-fold increase, endosomal/lysomomal potassium channel with 6.6-fold increase, crescerin-2 with 20-fold increase, ARG-1 with 3.3-fold increase and PP2A with 9.1-fold increase (Table 2). Intriguingly, ARG-1 and PP2A were also identified as HNE-modified in the comparison between Ts65Dn Veh and Eu Veh supporting that their oxidation is a common feature of Ts65Dn neurodegenerative process, while rapamycin is able to revert their oxidation and most-likely recover their function. Overall the analysis of this group of comparison confirm, as previously stated, that rapamycin is able to reduce OS levels in the frontal cortex of DS mice. These results are displayed in the second column of the heat map (Fig. 5, panel A) and are noticeable comparing the second and the third column of the heat map (Fig. 5, panel B).
-
•
InRapa treatment reduces lipoxidized proteins, to levels comparable with Eu mice (Ts65Dn InRapa/Euploid Vehicle). In this group of comparison, we found the increased HNE-modification of only three proteins in Ts65Dn InRapa compared with Eu Veh. Two proteins are in common with the first group of comparison: dynein heavy chain 3 (3.9-fold increase) and guanine deaminase (1.3-fold increase); while one protein, the Acyl-CoA dehydrogenase family member 9, is newly identified as increasingly HNE-modified (3.9-fold increase). Interestingly, these data suggest that InRapa is able to revert the oxidation of the majority of the proteins (see comparison Ts65Dn Veh and Eu Veh) with the exception of two, dynein heavy chain 3 (3.9-fold increase) and guanine deaminase (Table 2). Taken together, these findings indicate the efficacy of InRapa to significantly reduce the levels of HNE-modifications in the frontal cortex of Ts65Dn, although in the presence of only few oxidized proteins compared with Eu mice. Indeed, the third column of the heat map (Fig. 5, panel A) indicates few oxidized proteins in the comparison between Ts65Dn InRapa and euploids, as is also noticeable by observing the first and the third column of the heat map (Fig. 5, panel B).
Fig. 4.
Representative 2D gel and 2D-HNE blots from Ts65Dn mice and Eu treated with Veh and InRapa. Representative 2D gel and HNE blots from Ts65Dn mice and Eu treated with Veh and InRapa. The spots showing significantly HNE difference between the group of treatment are labeled. The spots numbers indicated on the maps are the same as those listed in Table 2.
Table 2.
Protein identified by MS/MS with significant different HNE-bound organized by groups of comparison. For each protein, full name and acronym, UNIPROT number, fold change between groups, sequence coverage and number of identified peptides are reported.
| Spot N. | Protein | Uniprot N. | Fold | Coverage | Peptides |
|---|---|---|---|---|---|
| Ts65Dn Veh/Eu veh | |||||
| 1 | Fructose-bisphosphate aldolase A (Aldo-A) | P05064 | 12.6 | 14.3 | 5 |
| 3 | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial (SCOT-1) | Q9D0K2 | 1.3 | 2.7 | 1 |
| 6 | Echinoderm microtubule-associated protein-like 5 (EMAP-5) | Q8BQM8 | 1.6 | 0.3 | 1 |
| 7 | Arginase-1 (Arg-1) | Q61176 | 3.0 | 5.2 | 1 |
| 8 | Guanine deaminase (GAH) | Q9R111 | 1,3 | 13,5 | 6 |
| 11 | Alpha-internexin (Alpha Inx) | P46660 | 8.9 | 5.4 | 2 |
| 12 | 28S ribosomal protein S35, mitochondrial (MRP-S35) | Q8BJZ4 | 3.4 | 4.3 | 1 |
| 13 | Histone H4 | P62806 | 1.9 | 9.7 | 1 |
| 14 | Ubiquitin thioesterase (OTUB1) | Q7TQI3 | 1.8 | 8.9 | 2 |
| 15 | Dynein heavy chain 3, axonemal | Q8BW94 | 5.3 | 1.25 | 4 |
| 16 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A) | Q76MZ3 | 2.4 | 5.43 | 3 |
| Ts65Dn Veh/Ts65Dn InRapa | |||||
| 4 | Oxysterol-binding protein-related protein 1-8 (ORP-1-8) | Q91XL9 | 3.0 | 0.85 | 1 |
| B9EJ86 | 0.8 | ||||
| 5 | Formin-like protein 2 (Flp-2) | A2APV2 | 2.0 | 1.1 | 1 |
| 7 | Arginase-1 | Q61176 | 3.3 | 5.2 | 1 |
| 9 | Endosomal/lysomomal potassium channel (TMEM175) | Q9CXY1 | 6.6 | 1.6 | 1 |
| 10 | Crescerin-2 | Q3TYG6 | 20 | 2.7 | 3 |
| 16 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A) | Q76MZ3 | 9.1 | 5.43 | 3 |
| Ts65Dn InRapa/Eu veh | |||||
| 2 | Acyl-CoA dehydrogenase family member 9, mitochondrial (ACAD-9) | Q8JZN5 | 3.9 | 4.0 | 2 |
| 8 | Guanine deaminase (GAH) | Q9R111 | 1.3 | 13.5 | 6 |
| 15 | Dynein heavy chain 3, axonemal | Q8BW94 | 2.3 | 1.25 | 4 |
Fig. 5.
Heat maps of protein-bound HNE levels. Panel A: Heat map showing, for all the proteins identified, the protein-bound HNE ratio between the groups of comparison. The gray square indicates no significant changes. Panel B: Heat maps showing, for all the proteins identified, the protein-bound HNE levels value, normalized on Eu Veh set as 100%, for each of the experimental group. Proteins acronyms are explained in Table 2 and in the list of abbreviations.
3.3. Validation experiments: ARG-1 and PP2A
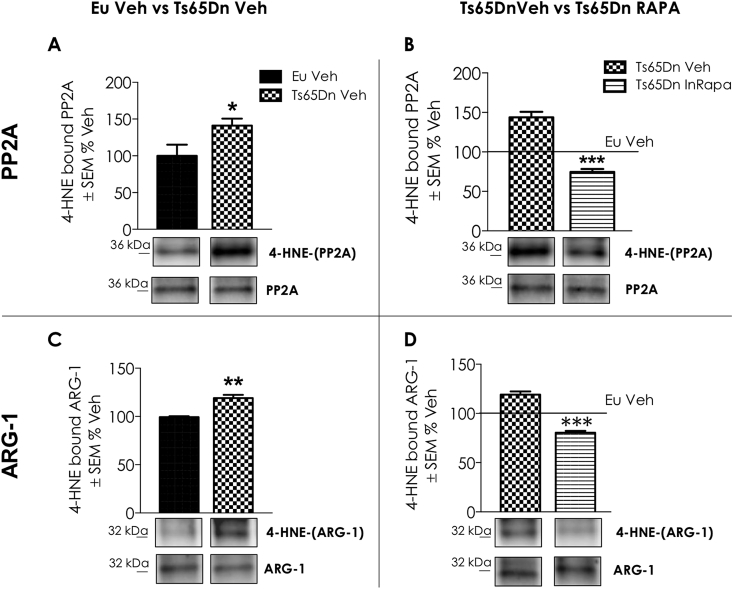
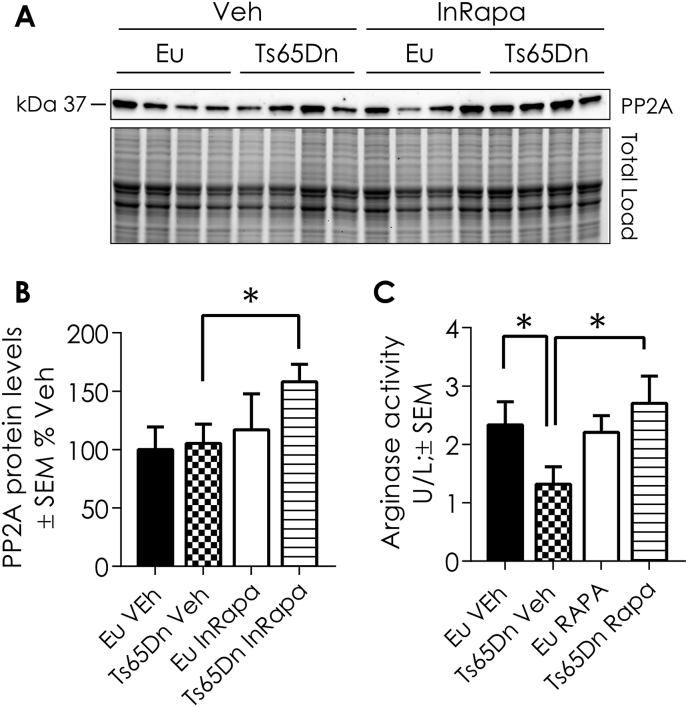
In light of these findings, ARG-1 and PP2A were selected for validation of the proteomics results. These proteins were immunoprecipitated and then probed by specific HNE antibody (Fig. 6A–D). In addition, the expression levels of PP2A and the expression levels and activity of ARG-1 were evaluated respectively by WB and enzyme assay (Fig. 7A–C). Proteomic data show that PP2A is significantly HNE-modified in Ts65Dn Veh group compared to Eu mice, suggesting the reduction of its phosphatase activity. Interestingly, rapamycin treatment is able to reduce the levels of HNE modifications on PP2A in Ts65Dn mice. The results obtained by immunoprecipitating PP2A protein confirmed proteomic data (Fig. 6A and B). Indeed, HNE-modifications is increased (about 40% increase, p = 0.04) on immunoprecipitated PP2A in Ts65Dn Veh group compared to Eu mice (Fig. 6A). Conversely, InRapa treatment was able to reduce the HNE modifications on PP2A (70% decrease, p = 0.001) in Ts65Dn RAPA compared with Ts65Dn Veh group (Fig. 6B). The analysis of PP2A expression levels show a significant increase, about 60% (p = 0.05), in InRapa Ts65Dn treated compared to Ts65Dn Veh (Fig. 7A and B). Therefore, these data demonstrate that InRapa treatment is able to recover PP2A status and conceivably its function by both decreasing HNE-adduct levels and increasing its expression levels.
Fig. 6.
Validation Experiments: immunoprecipitation of PP2A (Panel A and B) and ARG-1. HNE modifications levels detected by immunoprecipitation of PP2A and ARG-1 (n = 6/group) in Ts65Dn Veh vs Eu Veh and Ts65Dn InRapa vs Ts65Dn Veh. Panels A and B show the Immunoprecipitation blots and graph relative bar graph of HNE-modified PP2A between Ts65Dn Veh vs Eu Veh comparison group (A) and Ts65Dn InRapa vs Ts65Dn Veh comparison group (B). Panels C and D show the Immunoprecipitation blots and graph relative bar graph of HNE-modified ARG-1 between Ts65Dn Veh vs Eu Veh comparison group (C) and Ts65Dn InRapa vs Ts65Dn Veh comparison group (D). Densitometry values shown in the bar graph are the mean of 6 samples per each group, normalized on expression, and are given as percentage of Eu Veh, set as 100%. Statistical significance was determined using one–way ANOVA and Student t-test analysis (*p < 0.05, **p < 0.01, ***p < 0.001).
Fig. 7.
PP2A expression levels and ARG-1 enzyme activity. A) Representative WB showing PP2A protein levels. B) Quantification of panel A showing PP2A protein levels. C) Quantification of Arginase activity. Densitometry values shown in the bar graph are the mean of 8 (WB) samples per each group normalized per total load and are given as percentage of Eu Veh, set as 100%. ARG-1 assay was performed on 8 samples per group and is reported as U/L ± SEM. Statistical significance was determined using one–way ANOVA and Student t-test analysis (*p < 0.05, **p < 0.01, ***p < 0.001).
The proteomic results demonstrate the increased HNE-modification of ARG-1 in Ts65Dn mice treated with Vehicle compared to euploid mice, suggesting an impairment of ARG-1 activity. On the other hand, the same protein was less HNE-modified in InRapa Ts65Dn compared with Ts65Dn Veh group, thus suggesting the ability of rapamycin treatment to restore its activity. To confirm our hypothesis, we first immunoprecipitated ARG-1 and we evaluated its HNE-modification (Fig. 6C and D). The data obtained confirm the proteomics analysis showing an increase in HNE-modification on ARG-1 (20% increase, p = 0.01) in Ts65Dn Veh compared to Eu Veh (Fig. 6C). Interestingly, rapamycin treatment reduces the levels of HNE-modification on ARG-1 (20% decrease, p = 0.001) in Ts65Dn InRapa mice compared to Ts65Dn untreated mice (Fig. 6D). In addition, we measured the expression levels and the activity of ARG-1, by enzymatic assay, in the frontal cortex of the four groups of animals used in the present study. ARG-1 expression levels showed no differences between the groups of comparison (data not shown). However, in line with proteomics data, the ARG-1 activity (Fig. 7C) is reduced in Ts65Dn Veh group compared with euploid Veh (about 40% reduction, p = 0.05). However, InRapa treatment is able to recover the ARG-1 enzyme activity in Ts65Dn mice by a 45% increase (p = 0.04) compared to Veh group.
4. Discussion
Several studies demonstrated the involvement of OS in DS-related accelerated senescence and neuropathology [7,32,34,39]. Results from our group and from others have demonstrated that OS is an early event in the onset of pathological DS phenotype. It is supposed that the increase of OS during fetal stage can modify brain development [34,40], while in later life stages is mainly involved into the neurodegenerative phenomena contributing to the development of AD in DS patients [7,20,21]. Interestingly, the cause of increased OS lies on genes triplicated on Chr21 that are involved, directly or indirectly, in the overproduction of ROS and, in turn, in enhanced oxidative damage to proteins, nucleic acid and lipids [19,38,41].
Among triplicated genes, one of the most relevant OS inducer is the copper-zinc superoxide dismutase (SOD1), which when overexpressed demonstrates an imbalance in the ratio with CAT and GPX, resulting in the accumulation of H2O2. Among other proteins, encoded on Chr21, that are responsible for increased OS we can further mention APP, carbonyl reductase (CR), S100B and BACH1 [42].
Further, low levels of reducing agents and antioxidant enzymes, together with the impairment of the proteostasis network, contribute to exacerbation of brain pathology in DS [31]. mTOR is deeply involved in the regulation of the proteostasis network, through the on/off switch of translation and autophagy [4]. Autophagy is essential for preserving cellular homeostasis and physiological function in tissues and organs [43,44]. The disturbance of mTOR signaling leads to the abnormal accumulation of aggregated and unfolded/misfolded proteins, as demonstrated in DS brain [20]. The alteration of autophagy seems to be related with increased OS, however, this relationship is intricate, and increasing evidence suggests that the mTOR/autophagy axis plays a dual role in the cellular response to OS [31]. On one hand, increased levels of free radicals are involved in the regulation of autophagic survival signals and death processes and on the other hand, autophagy provides protection against oxidative damage [4]. Increased mTOR and reduced autophagy, could lead to the accumulation of ROS and to the increase of oxidized proteins [45]. Recent studies from our laboratory reported, in DS post-mortem brain, the early mTOR hyperactivation, within the brain, coupled with the impairment of its downstream and upstream pathways, increased amyloid-β load and hyperphosphorylated tau [17]. In addition, we demonstrated that the same DS cases reported increased oxidative damage to several components of autophagy and proteasome systems confirming a close connection between aberrant mTOR/autophagy, altered proteostasis network and increased OS [27,33]. Furthermore, the analysis of mTOR expression and phosphorylation in the hippocampus of Ts65Dn mice showed that the alteration of mTOR/autophagy axis could lead to the observed increase of oxidative damage [16]. Data from hippocampus [26] and cortex of Ts65Dn compared with Eu Veh confirm the aberrant mTOR phosphorylation in our DS model, coupled with reduced autophagy induction, increased APP/Aβ levels, increased tau phosphorylation and enhanced levels of lipoxidized proteins, indexed as protein-bound HNE. The further analysis of protein specific HNE-modification, by proteomics, revealed the involvement of key components in the development of neuronal damage in DS mice. Interestingly, the majority of proteins found to be lipoxidized in Ts65Dn mice compared to euploids, namely, fructose-bisphosphate aldolase A, succinyl-CoA:3-ketoacid coenzyme A transferase 1, alpha-internexin and ubiquitin thioesterase, have been previously identified as oxidatively modified in the frontal cortex from human DS sample [27,33]. In addition, few of those proteins, such as PP2A, were found poly-ubiquitinylated in DS human brain [46]. The overlap between proteomics data collected on mice and human DS support the strength of the Ts65Dn strain, which provides a valuable model to study the molecular mechanisms leading to brain damage and to test the efficacy of neuroprotective drug therapies [18,47,48].
Within this context, considering that i) oxidative damage impairs the proteostasis network and induces protein aggregation, which in turn favors further accumulation of oxidized cellular targets, starting a chain of harmful events for the neuron; ii) mTOR hyperactivation impairs protein degradative systems leading to accumulation of oxidized proteins, including HNE-modified; the ability to restore aberrant mTOR signaling and remove toxic aggregates could be essential to avoid neurodegeneration and ameliorate cognitive decline in DS [8]. Therefore, we aimed to treat Ts65Dn mice with InRapa for 3 months to rescue brain dysfunction. At the end of the treatment, beyond the increase of cognitive performances, as demonstrated by RAM and NOR tests, we observed the restoration of mTOR signaling along with increased autophagosome formation, reduced pathological markers (APP/Aβ and p-Tau) and reduced lipoxidative damage in both hippocampus and frontal cortex [26]. In particular, our data indicated that mTOR inhibition might lead to increased autophagosome formation (as suggested by increased LC3II, Atg7 and Atg12/5) [26,49,50], which is associated with the reduction of toxic aggregates and misfolded/lipoxidized proteins accumulation. Our results agree with previous studies demonstrating that rapamycin-induced activation of autophagy is one of the principal mechanisms by which the reduction of toxic protein aggregates is achieved in the brain of Tg-AD mice [43,44,[51], [52], [53]]. Indeed, we also supported that rapamycin is able to reduce oxidative damage by increasing autophagosome formation in SHSY-5Y neuroblastoma cells [16].
Proteomics data, by showing a reduction of total lipoxidized proteins in Ts65Dn InRapa compared with Eu, support the protective effect of InRapa towards OS. Moreover, this effect is evident if we consider the comparison between Ts65Dn Veh and Ts65Dn InRapa, which indicates that rapamycin led to reduced lipid peroxidation in InRapa Ts65Dn, as indexed by the number and the identity of HNE-bound proteins, compared with Ts65Dn Veh. Granted the property of rapamycin to reduce OS in Ts65Dn mice, we focused our attention on the specific targets of lipoxidation-mediated damage that could contribute to neurodegeneration and whose rescue by rapamycin could lead to the amelioration of brain pathology and to improved cognitive performance. Among these, we considered of great interest the lipoxidation and subsequent recovery of ARG-1 and PP2A.
PP2A is one of the most important serine/threonine phosphatases in mammalian brains. It accounts for as much as 1% of total cellular proteins in most tissue and cells. As a highly conserved enzyme, it has ubiquitous distribution and plays important roles in development, cell growth, and transformation [54,55]. Together with protein phosphatase 1 (PP1), PP2A accounts for more than 80% of the total serine/threonine phosphatase activity in mammalian cells. Tau phosphorylation levels are largely regulated by PP2A and, to a lesser degree, by PP1, PP2B and PP5 in the brain [56]. It is conceivable that the down-regulation of PP2A is partially responsible for the abnormal tau phosphorylation in AD and DS brain [[57], [58], [59], [60]]. In vivo, it was shown that PP2A inhibition induces tau hyperphosphorylation and spatial memory deficits [61], and these effects are reversed by the addition of acetyl-l- carnitine [62]. Moreover, PP2A inhibition by okadaic acid in rats leads to tau phosphorylation and impairment of spatial memory retention [63]. Interestingly, Specific PP2A inhibition has been also proven to affect many brain Ser/Thr kinases implicated in tau phosphorylation since PP2A inhibition can override the inhibition of these key tau kinases [64]. Thus, by deregulating the activity of tau protein kinases, PP2A dysfunction can promote aberrant stimulation of signaling cascades that contribute to neuronal and synaptic damage in AD and DS [56]. The regulation of PP2A is very complicated and until now, the mechanism for PP2A inactivation has not been fully understood [56]. However, among the potential ways of PP2A deregulation its increased lipoxidation might be responsible of the reduced phosphatase activity observed in DS.
Previous studies have demonstrated that the oxidation of PP2A lead to formation of disulfide bond in the catalytic subunit that is sufficient to inhibit its phosphatase activity [65]. In addition, it has also been shown that nitration of PP2A affects its function [66].
As reviewed in Di Domenico et al. [7] the increased activity of several kinases, including DYRK1A and RCAN1, encoded on Chr21, and GSK3β and CDK5, seems to have a prominent role in tau hyperphosphorylation occurring in the brain of DS mice. In addition, the reduction of autophagy, mediated by mTOR hyperactivation, could represent a further mechanism that lead to toxic tau deposition by avoiding its intracellular clearance [13]. In our previous publication [26], we discussed that the reduction of tau phosphorylation could be associated with the significantly decreased protein expression of RCAN1 and DYRK1A and with the improved degradation of hyperphosphorylated tau through the induction of autophagy. Our current data confirm the involvement of aberrantly regulated PP2A among the mechanism that lead to tau hyperphosphorylation. Ts65Dn mice after InRapa treatment demonstrate a robust and significant reduction of tau phosphorylation in the hippocampus and in the frontal cortex of DS mice. Therefore, here we support the involvement of a further mechanism causing tau hyperphosphorylation encompassing the reduced lipoxidation and the increased expression of PP2A. Our findings are in agreement with previous results obtained in AD by using memantine; indeed memantine, as well as rapamycin, was able to restore PP2A activity and reverse aberrant tau phosphorylation [67].
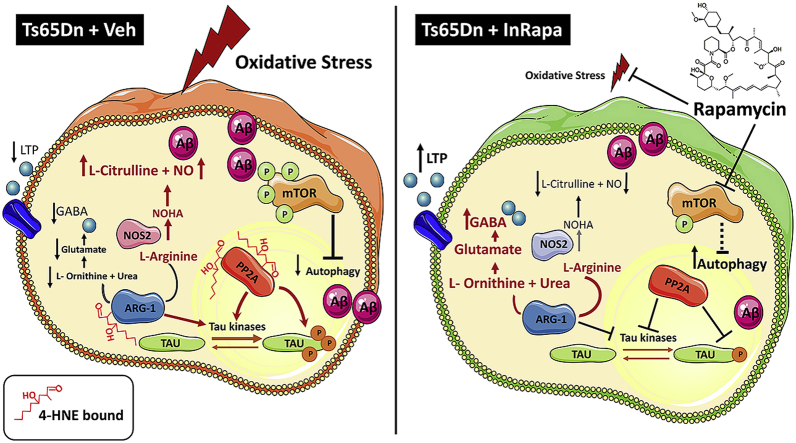
ARG-1 catalyzes the hydrolysis of l-arginine to urea and ornithine. It exists primarily in the liver but modest activity has been detected in extrahepatic tissues like the brain, where it produces ornithine that is then converted to glutamate and proline. Glutamate acts itself as neurotransmitter or can be converted to γ-amino butyric acid (GABA) [68]. To date, a large body of evidences has shown that synaptic deficits and memory impairment, in trisomic mouse models of DS, are largely determined by altered GABAergic signaling [69,70]. The Akt-mTOR pathway is involved in dendrite morphogenesis and synaptic plasticity, and it has been shown to modulate both glutamatergic and GABAergic synaptic transmission [71]. Montesinos and coworkers demonstrated that oral rapamycin treatment resulted in the fully restoration of hippocampal BDNF-LTP and basal synaptic transmission of Ts1Cje mouse model of DS [72]. Furthermore, many studies suggest that there is a competition between arginase and nitric oxide synthases (NOS), for l-arginine and that they control each other's levels [68]. Elevated NO levels, produced by increased NOS activity, drive nitration of Aβ and enhance aggregation and plaque formation [73], suggesting that the balance between ARG-1 and NOS determines the acceleration vs mitigation of amyloid deposition [74]. In addition, the NO-related nitration of tau has also been observed in different tauopathies [75,76]. A recent study by Hunt and colleagues [77] showed that overexpression of ARG-1 in the CNS of Tg4510 tau transgenic mice significantly reduced phospho-tau species and tangle pathology by decreasing several kinases capable of phosphorylating tau, decreasing inflammation, and modulating mTOR activity. Conversely, conditional deletion of ARG-1 in myeloid cells resulted in increased tau accumulation. Overall these studies suggest that the lipoxidation of ARG-1 contributes to its reduced activity thus leading to the development of brain neurodegeneration in DS by affecting different pathological aspects of the disease. Indeed, ARG-1 dysfunction might lead to altered synaptic transmission, aberrant mTOR activation, Aβ deposition and tau hyperphosphorylation. The mechanism by which HNE modification impairs ARG-1 activity is not known. We hypothesize that, similarly to PP2A, critical amino acid residues in the catalytic domain are oxidatively modified in the presence of increased levels of HNE. However, InRapa treatment, by reducing ARG-1 protein-bound HNE levels, rescues its enzyme activity and conceivably contributes to the recovery of the pathway above-mentioned as previously demonstrated by the increased levels of PSD95 and STX1 [26], and the reduced levels of mTOR phosphorylation, tau phosphorylation and Aβ deposition.
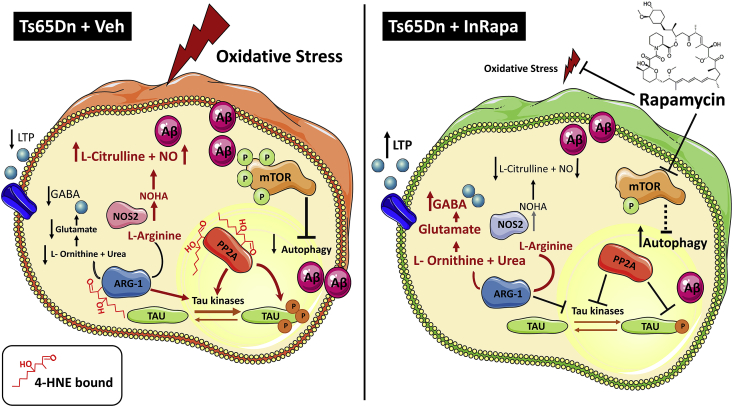
In conclusions, we previously demonstrated that InRapa treatment improves cognition in DS mice rescuing molecular pathways associated with aberrant mTOR phosphorylation. Here we added further pieces to the puzzle, by showing that the decrease of protein-bound HNE levels, by rapamycin treatment, is a crucial mechanism leading to reduced lipoxidative damage in DS mice associated with reduced AD neuropathology (Fig. 8). Considering that mTOR pathway is a central hub at the crossroad of multiple intracellular signaling, we propose that InRapa treatment is able to modulate lipoxidation and might represent a valuable therapeutic strategy to reduce the early development of AD pathology in DS population.
Fig. 8.
Implications of PP2A and ARG-1 oxidation and recovery in DS neuropathology. In the left panel an overview on the brain of Ts65Dn Veh group, where the PP2A and ARG-1 dysfunction might lead to altered synaptic transmission, aberrant mTOR activation, Aβ deposition and tau hyperphosphorylation. On the right panel, InRapa treatment, by reducing PP2A and ARG-1 protein-bound HNE levels, rescues their enzyme activity and reduces levels of mTOR phosphorylation, tau phosphorylation and Aβ deposition contributing to reduce AD-like brain oxidative damage in Ts65Dn mice.
Acknowledgments
This work was supported by the Ministry of Instruction, Universities and Research (MIUR) under the SIR program n° RBSI144MT to FDD, Jerome Lejeune Foundation with the grant # PERLUIGI/1484-PM2016A to MP and Fondi di Ateneo Progetti Grandi # RG116154C9214D1A from Sapienza University of Rome to FDD.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101162.
Abbreviations
- 3-NT
3-Nitrotyrosine
- Aβ
Amyloid-β
- ACAD-9
Acyl-CoA dehydrogenase family member 9
- AD
Alzheimer's Disease
- Aldo-A
Fructose-bisphosphate aldolase A
- Alpha Inx
Alpha-internexin
- APP
Amyloid Precursor Protein
- Arg-1
Arginase-1
- Atg
Autophagy related proteins
- CDK5
cyclin dependent kinase 5
- CNS
Central Nervous System
- CR
carbonyl reductase
- DS
Down Syndrome
- DYRK1A
Dual specificity tyrosine-phosphorylation-regulated kinase 1A
- EMAP-5
Echinoderm microtubule-associated protein-like 5
- Eu
Euploid
- Flp-2
Formin-like protein 2
- GABA
γ-amino butyric acid
- GAH
Guanine deaminase
- GSK3β
glycogen synthase kinase 3 isoforms β
- HNE
4-Hydroxynonenal
- Hsa21
human chromosome 21
- InRapa
Intranasal Rapamycin
- LC3
Microtubule-associated protein 1A/1B-light chain 3
- MRP-S35
28S ribosomal protein S35
- mTOR
mammalian Target of Rapamycin
- NOR
Novel Object Recognition
- NFT
Neurofibrillary Tangles
- ORP-1-8
Oxysterol-binding protein-related protein 1-8
- OS
Oxidative Stress
- OTUB1
Ubiquitin thioesterase
- PP1
protein phosphatase
- PP2A
protein phosphatase 2A
- PSD95
postsynaptic density protein 95
- Rapa
Rapamycin
- RAM
Radial Arm Maze
- RCAN
Regulator of Calcineurin 1
- RNS
Reactive Nitrogen Species
- ROS
Reactive Oxygen Species
- SCOT-1
Succinyl-CoA:3-ketoacid coenzyme A transferase 1
- Tg
Transgenic
- TMEM175
Endosomal/lysomomal potassium channel
- Veh
Vehicle
- GRP78
8 kDa glucose-regulated protein
- UCH-L1
Ubiquitin carboxy-terminal hydrolase L1
- V0-ATPase
Vacuolar-type H + -ATPase
- CatD
Cathepsin D
- GFAP
Glial fibrillary acidic protein
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Oddo S. The role of mTOR signaling in Alzheimer disease. Front. Biosci. 2012;4:941–952. doi: 10.2741/s310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai Z., Chen G., He W., Xiao M., Yan L.J. Activation of mTOR: a culprit of Alzheimer's disease? Neuropsychiatric Dis. Treat. 2015;11:1015–1030. doi: 10.2147/NDT.S75717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laplante M., Sabatini D.M. mTOR Signaling. Cold Spring Harb. Perspect. Biol. 2012;4(2) doi: 10.1101/cshperspect.a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perluigi M., Di Domenico F., Butterfield D.A. mTOR signaling in aging and neurodegeneration: at the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol. Dis. 2015;84:39–49. doi: 10.1016/j.nbd.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Hoeffer C.A., Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33(2):67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Troca-Marin J.A., Alves-Sampaio A., Montesinos M.L. Deregulated mTOR-mediated translation in intellectual disability. Prog. Neurobiol. 2012;96(2):268–282. doi: 10.1016/j.pneurobio.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Di Domenico F., Tramutola A., Foppoli C., Head E., Perluigi M., Butterfield D.A. mTOR in Down syndrome: role in Ass and tau neuropathology and transition to Alzheimer disease-like dementia. Free Radic. Biol. Med. 2018;114:94–101. doi: 10.1016/j.freeradbiomed.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tramutola A., Lanzillotta C., Di Domenico F. Targeting mTOR to reduce Alzheimer-related cognitive decline: from current hits to future therapies. Expert Rev. Neurother. 2017;17(1):33–45. doi: 10.1080/14737175.2017.1244482. [DOI] [PubMed] [Google Scholar]
- 9.Tramutola A., Triplett J.C., Di Domenico F., Niedowicz D.M., Murphy M.P., Coccia R., Perluigi M., Butterfield D.A. Alteration of mTOR signaling occurs early in the progression of Alzheimer disease (AD): analysis of brain from subjects with pre-clinical AD, amnestic mild cognitive impairment and late-stage AD. J. Neurochem. 2015;133(5):739–749. doi: 10.1111/jnc.13037. [DOI] [PubMed] [Google Scholar]
- 10.O' Neill C. PI3-kinase/Akt/mTOR signaling: impaired on/off switches in aging, cognitive decline and Alzheimer's disease. Exp. Gerontol. 2013;48(7):647–653. doi: 10.1016/j.exger.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 11.Caccamo A., Maldonado M.A., Majumder S., Medina D.X., Holbein W., Magri A., Oddo S. Naturally secreted amyloid-beta increases mammalian target of rapamycin (mTOR) activity via a PRAS40-mediated mechanism. J. Biol. Chem. 2011;286(11):8924–8932. doi: 10.1074/jbc.M110.180638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caccamo A., Majumder S., Richardson A., Strong R., Oddo S. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: effects on cognitive impairments. J. Biol. Chem. 2010;285(17):13107–13120. doi: 10.1074/jbc.M110.100420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caccamo A., Magri A., Medina D.X., Wisely E.V., Lopez-Aranda M.F., Silva A.J., Oddo S. mTOR regulates tau phosphorylation and degradation: implications for Alzheimer's disease and other tauopathies. Aging Cell. 2013;12(3):370–380. doi: 10.1111/acel.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siman R., Cocca R., Dong Y. The mTOR inhibitor rapamycin mitigates perforant pathway neurodegeneration and synapse loss in a mouse model of early-stage alzheimer-type tauopathy. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0142340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spilman P., Podlutskaya N., Hart M.J., Debnath J., Gorostiza O., Bredesen D., Richardson A., Strong R., Galvan V. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer's disease. PLoS One. 2010;5(4):e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tramutola A., Lanzillotta C., Arena A., Barone E., Perluigi M., Di Domenico F. Increased mammalian target of rapamycin signaling contributes to the accumulation of protein oxidative damage in a mouse model of down's syndrome. Neurodegener. Dis. 2016;16(1–2):62–68. doi: 10.1159/000441419. [DOI] [PubMed] [Google Scholar]
- 17.Perluigi M., Pupo G., Tramutola A., Cini C., Coccia R., Barone E., Head E., Butterfield D.A., Di Domenico F. Neuropathological role of PI3K/Akt/mTOR axis in Down syndrome brain. Biochim. Biophys. Acta. 2014;1842(7):1144–1153. doi: 10.1016/j.bbadis.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fillat C., Dierssen M., de Lagran M.M., Altafaj X. Insights from mouse models to understand neurodegeneration in Down syndrome. CNS Neurol. Disord. - Drug Targets. 2010;9(4):429–438. doi: 10.2174/187152710791556159. [DOI] [PubMed] [Google Scholar]
- 19.Wiseman F.K., Al-Janabi T., Hardy J., Karmiloff-Smith A., Nizetic D., Tybulewicz V.L., Fisher E.M., Strydom A. A genetic cause of Alzheimer disease: mechanistic insights from Down syndrome. Nat. Rev. Neurosci. 2015;16(9):564–574. doi: 10.1038/nrn3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Head E., Powell D., Gold B.T., Schmitt F.A. Alzheimer's disease in down syndrome. Eur. J. Neurodegener. Dis. 2012;1(3):353–364. [PMC free article] [PubMed] [Google Scholar]
- 21.Head E., Lott I.T., Wilcock D.M., Lemere C.A. Aging in down syndrome and the development of Alzheimer's disease neuropathology. Curr. Alzheimer Res. 2016;13(1):18–29. doi: 10.2174/1567205012666151020114607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer A.M., van Scheppingen J., Milenkovic I., Anink J.J., Adle-Biassette H., Kovacs G.G., Aronica E. mTOR Hyperactivation in down syndrome hippocampus appears early during development. J. Neuropathol. Exp. Neurol. 2014;73(7):671–683. doi: 10.1097/NEN.0000000000000083. [DOI] [PubMed] [Google Scholar]
- 23.Lipton J.O., Sahin M. The neurology of mTOR. Neuron. 2014;84(2):275–291. doi: 10.1016/j.neuron.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majumder S., Richardson A., Strong R., Oddo S. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0025416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin A.L., Zheng W., Halloran J.J., Burbank R.R., Hussong S.A., Hart M.J., Javors M., Shih Y.Y., Muir E., Solano Fonseca R., Strong R., Richardson A.G., Lechleiter J.D., Fox P.T., Galvan V. Chronic rapamycin restores brain vascular integrity and function through NO synthase activation and improves memory in symptomatic mice modeling Alzheimer's disease. J. Cereb. Blood Flow Metab. Offic. J. Int. Soc. Cereb. Blood Flow Metabol. 2013;33(9):1412–1421. doi: 10.1038/jcbfm.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tramutola A., Lanzillotta C., Barone E., Arena A., Zuliani I., Mosca L., Blarzino C., Butterfield D.A., Perluigi M., Di Domenico F. Intranasal rapamycin ameliorates Alzheimer-like cognitive decline in a mouse model of Down syndrome. Transl. Neurodegener. 2018;7:28. doi: 10.1186/s40035-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Domenico F., Coccia R., Cocciolo A., Murphy M.P., Cenini G., Head E., Butterfield D.A., Giorgi A., Schinina M.E., Mancuso C., Cini C., Perluigi M. Impairment of proteostasis network in Down syndrome prior to the development of Alzheimer's disease neuropathology: redox proteomics analysis of human brain. Biochim. Biophys. Acta. 2013;1832(8):1249–1259. doi: 10.1016/j.bbadis.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pajares M., Jimenez-Moreno N., Dias I.H., Debelec B., Vucetic M., Fladmark K.E., Basaga H., Ribaric S., Milisav I., Cuadrado A. Redox control of protein degradation. Redox Biol. 2015;6:409–420. doi: 10.1016/j.redox.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scherz-Shouval R., Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem. Sci. 2011;36(1):30–38. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Domenico F., Barone E., Perluigi M., Butterfield D.A. The triangle of death in Alzheimer's disease brain: the aberrant cross-talk among energy metabolism, mammalian target of rapamycin signaling, and protein homeostasis revealed by redox proteomics. Antioxidants Redox Signal. 2017;26(8):364–387. doi: 10.1089/ars.2016.6759. [DOI] [PubMed] [Google Scholar]
- 32.Butterfield D.A., Di Domenico F., Swomley A.M., Head E., Perluigi M. Redox proteomics analysis to decipher the neurobiology of Alzheimer-like neurodegeneration: overlaps in Down's syndrome and Alzheimer's disease brain. Biochem. J. 2014;463(2):177–189. doi: 10.1042/BJ20140772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Domenico F., Pupo G., Tramutola A., Giorgi A., Schinina M.E., Coccia R., Head E., Butterfield D.A., Perluigi M. Redox proteomics analysis of HNE-modified proteins in Down syndrome brain: clues for understanding the development of Alzheimer disease. Free Radic. Biol. Med. 2014;71:270–280. doi: 10.1016/j.freeradbiomed.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perluigi M., Di Domenico F., Buttterfield D.A. Unraveling the complexity of neurodegeneration in brains of subjects with Down syndrome: insights from proteomics. Proteonomics Clin. Appl. 2014;8(1–2):73–85. doi: 10.1002/prca.201300066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Domenico F., Tramutola A., Butterfield D.A. Role of 4-hydroxy-2-nonenal (HNE) in the pathogenesis of alzheimer disease and other selected age-related neurodegenerative disorders. Free Radic. Biol. Med. 2017;111:253–261. doi: 10.1016/j.freeradbiomed.2016.10.490. [DOI] [PubMed] [Google Scholar]
- 36.Reinholdt L.G., Ding Y., Gilbert G.J., Czechanski A., Solzak J.P., Roper R.J., Johnson M.T., Donahue L.R., Lutz C., Davisson M.T. Molecular characterization of the translocation breakpoints in the Down syndrome mouse model Ts65Dn. Mamm. Genome. 2011;22(11–12):685–691. doi: 10.1007/s00335-011-9357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Domenico F., Pupo G., Giraldo E., Badia M.C., Monllor P., Lloret A., Schinina M.E., Giorgi A., Cini C., Tramutola A., Butterfield D.A., Vina J., Perluigi M. Oxidative signature of cerebrospinal fluid from mild cognitive impairment and Alzheimer disease patients. Free Radic. Biol. Med. 2016;91:1–9. doi: 10.1016/j.freeradbiomed.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Di Domenico F., Pupo G., Mancuso C., Barone E., Paolini F., Arena A., Blarzino C., Schmitt F.A., Head E., Butterfield D.A., Perluigi M. Bach1 overexpression in Down syndrome correlates with the alteration of the HO-1/BVR-a system: insights for transition to Alzheimer's disease. J. Alzheimer's Dis. 2015;44(4):1107–1120. doi: 10.3233/JAD-141254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barone E., Head E., Butterfield D.A., Perluigi M. HNE-modified proteins in Down syndrome: involvement in development of Alzheimer disease neuropathology. Free Radic. Biol. Med. 2017;111:262–269. doi: 10.1016/j.freeradbiomed.2016.10.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perluigi M., di Domenico F., Fiorini A., Cocciolo A., Giorgi A., Foppoli C., Butterfield D.A., Giorlandino M., Giorlandino C., Schinina M.E., Coccia R. Oxidative stress occurs early in Down syndrome pregnancy: a redox proteomics analysis of amniotic fluid. Proteonomics Clin. Appl. 2011;5(3–4):167–178. doi: 10.1002/prca.201000121. [DOI] [PubMed] [Google Scholar]
- 41.Lockstone H.E., Harris L.W., Swatton J.E., Wayland M.T., Holland A.J., Bahn S. Gene expression profiling in the adult Down syndrome brain. Genomics. 2007;90(6):647–660. doi: 10.1016/j.ygeno.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 42.Perluigi M., Butterfield D.A. Oxidative stress and down syndrome: a route toward alzheimer-like dementia. Curr. Gerontol. Geriatr. Res. 2012;2012:724904. doi: 10.1155/2012/724904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Arencibia M., Hochfeld W.E., Toh P.P., Rubinsztein D.C. Autophagy, a guardian against neurodegeneration. Semin. Cell Dev. Biol. 2010;21(7):691–698. doi: 10.1016/j.semcdb.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuervo A.M. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24(12):604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Domenico F., Head E., Butterfield D.A., Perluigi M. Oxidative stress and proteostasis network: culprit and casualty of Alzheimer's-like neurodegeneration. Adv. Geriatr. 2014;2014:14. [Google Scholar]
- 46.Tramutola A., Di Domenico F., Barone E., Arena A., Giorgi A., di Francesco L., Schinina M.E., Coccia R., Head E., Butterfield D.A., Perluigi M. Polyubiquitinylation profile in down syndrome brain before and after the development of alzheimer neuropathology. Antioxidants Redox Signal. 2017;26(7):280–298. doi: 10.1089/ars.2016.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu T., Li Z.Y., Jia Z.P., Clapcote S.J., Liu C.H., Li S.M., Asrar S., Pao A., Chen R.Q., Fan N., Carattini-Rivera S., Bechard A.R., Spring S.S., Henkelman R.M., Stoica G., Matsui S.I., Nowak N.J., Roder J.C., Chen C., Bradley A., Yu Y.E. A mouse model of Down syndrome trisomic for all human chromosome 21 syntenic regions. Hum. Mol. Genet. 2010;19(14):2780–2791. doi: 10.1093/hmg/ddq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holtzman D.M., Santucci D., Kilbridge J., Chua-Couzens J., Fontana D.J., Daniels S.E., Johnson R.M., Chen K., Sun Y., Carlson E., Alleva E., Epstein C.J., Mobley W.C. Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc. Natl. Acad. Sci. U. S. A. 1996;93(23):13333–13338. doi: 10.1073/pnas.93.23.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morimoto N., Nagai M., Ohta Y., Miyazaki K., Kurata T., Morimoto M., Murakami T., Takehisa Y., Ikeda Y., Kamiya T., Abe K. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 50.Sheng R., Liu X.Q., Zhang L.S., Gao B., Han R., Wu Y.Q., Zhang X.Y., Qin Z.H. Autophagy regulates endoplasmic reticulum stress in ischemic preconditioning. Autophagy. 2012;8(3):310–325. doi: 10.4161/auto.18673. [DOI] [PubMed] [Google Scholar]
- 51.Nixon R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007;120(Pt 23):4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 52.Nixon R.A., Yang D.S. Autophagy failure in Alzheimer's disease--locating the primary defect. Neurobiol. Dis. 2011;43(1):38–45. doi: 10.1016/j.nbd.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai Z., Zhao B., Li K., Zhang L., Li C., Quazi S.H., Tan Y. Mammalian target of rapamycin: a valid therapeutic target through the autophagy pathway for Alzheimer's disease? J. Neurosci. Res. 2012;90(6):1105–1118. doi: 10.1002/jnr.23011. [DOI] [PubMed] [Google Scholar]
- 54.Janssens V., Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001;353(Pt 3):417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin L., Latypova X., Wilson C.M., Magnaudeix A., Perrin M.L., Terro F. Tau protein phosphatases in Alzheimer's disease: the leading role of PP2A. Ageing Res. Rev. 2013;12(1):39–49. doi: 10.1016/j.arr.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Sontag J.M., Sontag E. Protein phosphatase 2A dysfunction in Alzheimer's disease. Front. Mol. Neurosci. 2014;7:16. doi: 10.3389/fnmol.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu F., Grundke-Iqbal I., Iqbal K., Gong C.X. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur. J. Neurosci. 2005;22(8):1942–1950. doi: 10.1111/j.1460-9568.2005.04391.x. [DOI] [PubMed] [Google Scholar]
- 58.Gong C.X., Singh T.J., Grundkeiqbal I., Iqbal K. Phosphoprotein phosphatase-Activities in alzheimer-disease brain. J. Neurochem. 1993;61(3):921–927. doi: 10.1111/j.1471-4159.1993.tb03603.x. [DOI] [PubMed] [Google Scholar]
- 59.Gong C.X., Shaikh S., Wang J.Z., Zaidi T., Grundkeiqbal I., Iqbal K. Phosphatase-activity toward abnormally phosphorylated-tau - decrease in alzheimer-disease brain. J. Neurochem. 1995;65(2):732–738. doi: 10.1046/j.1471-4159.1995.65020732.x. [DOI] [PubMed] [Google Scholar]
- 60.Liang Z., Liu F., Iqbal K., Grundke-Iqbal I., Wegiel J., Gong C.X. Decrease of protein phosphatase 2A and its association with accumulation and hyperphosphorylation of tau in Down syndrome. J. Alzheimer's Dis. 2008;13(3):295–302. doi: 10.3233/jad-2008-13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian Q., Lin Z.Q., Wang X.C., Chen J., Wang Q., Gong C.X., Wang J.Z. Injection of okadaic acid into the Meynert nucleus basalis of rat brain induces decreased acetylcholine level and spatial memory deficit. Neuroscience. 2004;126(2):277–284. doi: 10.1016/j.neuroscience.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 62.Yin Y.Y., Liu H., Cong X.B., Liu Z., Wang Q., Wang J.Z., Zhu L.Q. Acetyl-L-carnitine attenuates okadaic acid induced tau hyperphosphorylation and spatial memory impairment in rats. J. Alzheimer's Dis. 2010;19(2):735–746. doi: 10.3233/JAD-2010-1272. [DOI] [PubMed] [Google Scholar]
- 63.Sun L., Liu S.Y., Zhou X.W., Wang X.C., Liu R., Wang Q., Wang J.Z. Inhibition of protein phosphatase 2A- and protein phosphatase 1-induced tau hyperphosphorylation and impairment of spatial memory retention in rats. Neuroscience. 2003;118(4):1175–1182. doi: 10.1016/s0306-4522(02)00697-8. [DOI] [PubMed] [Google Scholar]
- 64.Planel E., Yasutake K., Fujita S.C., Ishiguro K. Inhibition of protein phosphatase 2A overrides tau protein kinase I/glycogen synthase kinase 3 beta and cyclin-dependent kinase 5 inhibition and results in tau hyperphosphorylation in the hippocampus of starved mouse. J. Biol. Chem. 2001;276(36):34298–34306. doi: 10.1074/jbc.M102780200. [DOI] [PubMed] [Google Scholar]
- 65.Foley T.D., Petro L.A., Stredny C.M., Coppa T.M. Oxidative inhibition of protein phosphatase 2A activity: role of catalytic subunit disulfides. Neurochem. Res. 2007;32(11):1957–1964. doi: 10.1007/s11064-007-9394-x. [DOI] [PubMed] [Google Scholar]
- 66.Nakahata S., Morishita K. PP2A inactivation by ROS accumulation. Blood. 2014;124(14):2163–2165. doi: 10.1182/blood-2014-08-594093. [DOI] [PubMed] [Google Scholar]
- 67.Chohan M.O., Khatoon S., Iqbal I.G., Iqbal K. Involvement of I2PP2A in the abnormal hyperphosphorylation of tau and its reversal by Memantine. FEBS Lett. 2006;580(16):3973–3979. doi: 10.1016/j.febslet.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 68.Vural H., Sirin B., Yilmaz N., Eren I., Delibas N. The role of arginine-nitric oxide pathway in patients with Alzheimer disease. Biol. Trace Elem. Res. 2009;129(1–3):58–64. doi: 10.1007/s12011-008-8291-8. [DOI] [PubMed] [Google Scholar]
- 69.Contestabile A., Magara S., Cancedda L. The GABAergic hypothesis for cognitive disabilities in down syndrome. Front. Cell. Neurosci. 2017;11:54. doi: 10.3389/fncel.2017.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deidda G., Parrini M., Naskar S., Bozarth I.F., Contestabile A., Cancedda L. Reversing excitatory GABAAR signaling restores synaptic plasticity and memory in a mouse model of Down syndrome. Nat. Med. 2015;21(4):318–326. doi: 10.1038/nm.3827. [DOI] [PubMed] [Google Scholar]
- 71.Troca-Marin J.A., Casanas J.J., Benito I., Montesinos M.L. The Akt-mTOR pathway in Down's syndrome: the potential use of rapamycin/rapalogs for treating cognitive deficits. CNS Neurol. Disord. - Drug Targets. 2014;13(1):34–40. doi: 10.2174/18715273113126660184. [DOI] [PubMed] [Google Scholar]
- 72.Andrade-Talavera Y., Benito I., Casanas J.J., Rodriguez-Moreno A., Montesinos M.L. Rapamycin restores BDNF-LTP and the persistence of long-term memory in a model of Down's syndrome. Neurobiol. Dis. 2015;82:516–525. doi: 10.1016/j.nbd.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 73.Kummer M.P., Hermes M., Delekarte A., Hammerschmidt T., Kumar S., Terwel D., Walter J., Pape H.C., Konig S., Roeber S., Jessen F., Klockgether T., Korte M., Heneka M.T. Nitration of tyrosine 10 critically enhances amyloid beta aggregation and plaque formation. Neuron. 2011;71(5):833–844. doi: 10.1016/j.neuron.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 74.Colton C.A., Mott R.T., Sharpe H., Xu Q., Van Nostrand W.E., Vitek M.P. Expression profiles for macrophage alternative activation genes in AD and in mouse models of AD. J. Neuroinflammation. 2006;3:27. doi: 10.1186/1742-2094-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reyes J.F., Geula C., Vana L., Binder L.I. Selective tau tyrosine nitration in non-AD tauopathies. Acta Neuropathol. 2012;123(1):119–132. doi: 10.1007/s00401-011-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reynolds M.R., Reyes J.F., Fu Y.F., Bigio E.H., Guillozet-Bongaarts A.L., Berry R.W., Binder L.I. Tau nitration occurs at tyrosine 29 in the fibrillar lesions of Alzheimer's disease and other tauopathies. J. Neurosci. 2006;26(42):10636–10645. doi: 10.1523/JNEUROSCI.2143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hunt J.B., Jr., Nash K.R., Placides D., Moran P., Selenica M.L., Abuqalbeen F., Ratnasamy K., Slouha N., Rodriguez-Ospina S., Savlia M., Ranaweera Y., Reid P., Dickey C.A., Uricia R., Yang C.G., Sandusky L.A., Gordon M.N., Morgan D., Lee D.C. Sustained arginase 1 expression modulates pathological tau deposits in a mouse model of tauopathy. J. Neurosci. 2015;35(44):14842–14860. doi: 10.1523/JNEUROSCI.3959-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.