Abstract
Overproduction of prostaglandin E2 (PGE2) has been linked to enhanced tumor cell proliferation, invasiveness and metastasis as well as resistance to apoptosis. 15-Keto prostaglandin E2 (15-keto PGE2), a product formed from 15-hydroxyprostaglandin dehydrogenase-catalyzed oxidation of PGE2, has recently been shown to have anti-inflammatory and anticarcinogenic activities. In this study, we observed that 15-keto PGE2 suppressed the phosphorylation, dimerization and nuclear translocation of signal transducer and activator of transcription 3 (STAT3) in human mammary epithelial cells transfected with H-ras (MCF10A-ras). 15-Keto PGE2 inhibited the migration and clonogenicity of MCF10A-ras cells. In addition, subcutaneous injection of 15-keto PGE2 attenuated xenograft tumor growth and phosphorylation of STAT3 induced by breast cancer MDA-MB-231 cells. However, a non-electrophilic analogue, 13,14-dihydro-15-keto PGE2 failed to inhibit STAT3 signaling and was unable to suppress the growth and transformation of MCF10A-ras cells. These findings suggest that the α,β-unsaturated carbonyl moiety of 15-keto PGE2 is essential for its suppression of STAT3 signaling. We observed that the thiol reducing agent, dithiothreitol abrogated 15-keto PGE2-induced STAT3 inactivation and disrupted the direct interaction between 15-keto PGE2 and STAT3. Furthermore, a molecular docking analysis suggested that Cys251 and Cys259 residues of STAT3 could be preferential binding sites for this lipid mediator. Mass spectral analysis revealed the covalent modification of recombinant STAT3 by 15-keto PGE2 at Cys259. Taken together, thiol modification of STAT3 by 15-keto PGE2 inactivates STAT3 which may account for its suppression of breast cancer cell proliferation and progression.
Keywords: Breast cancer, 15-Hydroxyprostaglandin dehydrogenase, 15-Keto-prostaglandin E2, STAT3, Thiol modification, MCF10A-ras cells, MDA-MB-231 xenografts
Highlights
-
•
15-Keto-prostaglandin E2 inhibits activation of STAT3 in MCF10A-ras cells.
-
•
15-Keto-prostaglandin E2 suppresses the growth and transformation of MCF10A-ras cells. .
-
•
15-Keto-prostaglandin E2 attenuated xenograft tumor growth and STAT3 phosphorylation. .
-
•
15-Keto PGE2 modulates STAT3 by directly binding to its Cys259 residue.
1. Introduction
An inflammatory lipid mediator prostaglandin E2 (PGE2) is formed from arachidonic acid by cyclooxygenase-2 (COX-2) [1]. Overproduction of PGE2 is constitutively elevated in various human malignancies, such as colon, gastric, lung and breast cancer [[2], [3], [4]]. COX-2-derived PGE2 plays an important role in inflammation and cancer progression through modulation of several intracellular signaling pathways [[5], [6], [7]].
The intracellular level of PGE2 is regulated not only by its biosynthesis but also by degradation. The key enzyme involved in catabolism of PGE2 is 15-hydroxyprostaglandin dehydrogenase (15-PGDH). Expression of 15-PGDH is low in various cancers, including those of colon, stomach, bladder, and breast [[8], [9], [10], [11]]. 15-PGDH knockout mice are susceptible to colon tumor induction [12]. Down regulation of 15-PGDH in breast cancer cells is associated with its gene silencing via hypermethylation of its promoter [11]. Overexpression and activity of 15-PGDH in various cancer cells have been shown to suppress their proliferation, invasiveness, metastatic potential, and growth [11,13,14]. Therefore, 15-PGDH is considered as a tumor suppressor [15].
15-Keto PGE2, an oxidized metabolite of PGE2 formed by 15-PGDH was initially considered biologically inactive. However, accumulating evidence supports that this prostaglandin modulates the diverse cellular signal transduction pathways. 15-Keto PGE2 containing the α,β-unsaturated ketone has been known as a ligand of peroxisome proliferator receptor γ (PPARγ) [16]. In line with this notion, 15-keto PGE2 inhibited the bacterial lipopolysaccharide (LPS)-induced cytokine production in Kupffer cells through activation of PPARγ [17]. Moreover, the mice treated by 15-keto PGE2 were shown to be resistant to sepsis by LPS [18]. Lu et al. reported that 15-PGDH overexpression resulted in elevated formation of 15-keto PGE2, whereas downregulation of 15-PGDH elevated the PGE2 level in hepatocellular carcinoma cells [19]. In addition, 15-keto PGE2 treatment increased p21 promoter activity via PPARγ activation in hepatocellular carcinoma cells [19]. 15-Keto PGE2 is metabolized to 13,14-dihydro-15-keto PGE2 by prostaglandin reductase 2 (PTGR2). Silencing of PTGR2 enhanced 15-keto PGE2 accumulation and stimulated apoptosis through generation of reactive oxygen species in pancreatic cancer cells [20]. Thus, it is likely that the anti-inflammatory and other cytoprotective activites of 15-PGDH are mediated by its product, 15-keto PGE2. However, whether 15-keto PGE2 can inhibit the tumor growth and progression remains largely unknown.
Signal transducer and activator of transcription 3 (STAT3) is a major transcription factor regulating cellular processes involved in proliferation, development, inflammation and cell survival [21]. In response to extracellular stimuli, such as cytokines and growth factors, STAT3 is recruited from the cytosol to their receptors and activated by receptor-associated Janus kinases (JAKs) [22]. Phosphorylation at the tyrosine 705 (Y705) residue facilitates the formation of a STAT3 dimer that translocates to the nucleus. This leads to transcription of target genes responsible for cell cycle progression, such as Cyclin D1 and c-Myc, as well as those involved in cell survival [23]. Aberrant overactivation of STAT3 is linked to tumorigenesis [24,25]. Notably, more than 40% of breast cancers exhibit constitutively activated STAT3 [26]. The increased phosphorylation of STAT3Y705 is associated with the metastasis of breast cancer and upregulation of genes involved in cell proliferation in breast cancer tissues [27]. Therefore, targeting the abnormally activated STAT3 signaling has been considered as an important cancer therapeutic strategy [24,28,29].
In this study, we investigated whether 15-keto PGE2 could inhibit STAT3 signaling and suppress the human breast cancer cell proliferation and tumor growth.
2. Materials and methods
2.1. Cell culture
MCF10A and MCF10A-ras cells were cultured in Dulbecco's Modified Eagle's Medium: Nutrient Mixture-F-12 (DMEM/F-12) supplemented with 5% heat-inactivated horse serum from Gibco (Grand Island, NY, USA), 10 μg/ml insulin, 100 ng/ml cholera toxin, 0.5 μg/ml hydrocortisone, 20 ng/ml human epidermal growth factor, 2 mmol/l L-glutamine and 100 units/ml penicillin/streptomycin. MDA-MB-231 and HeLa/P-STAT3-Luc cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) supplied from GenDEPOT (Barker, TX, USA) and 1% antibiotic-antimycotic mixture. PC3 cells were cultivated in RPMI 1640 containing 10% heat-inactivated FBS and 1% antibiotic-antimycotic mixture. These cell lines were grown at 37 °C in humidified atmosphere of 5% CO2. MCF10A, MDA-MB-231 and PC3 cells were obtained from American Type Culture Collection (ATCC). The MCF10A-ras was kindly provided by Prof. Aree Moon of Duksung Women's University, Seoul, South Korea.
2.2. Chemicals and biological reagents
15-Keto PGE2 (9,15-dioxo-11α-hydroxy-prosta-5Z,13E-dien-1-oic acid) and 13-14-dihydro-15-keto PGE2 (9,15-dioxo-11α-hydroxy-prosta-5Z-en-1-oic acid) were purchased from Cayman Chemicals (Ann Arbor, MI, USA). Dithiothreitol (DTT) was obtained from Sigma Chemical Co. (St. Louis, MO, USA). Primary antibodies for STAT3, P-STAT3Y705, JAK2 and P-JAK2 were products of Cell Signaling Technology (Danvers, MA, USA). Antibodies against β-Actin and Lamin B were supplied from Santa Cruz Biotechnology (Santa Cruz, CA, USA). 15-PGDH primary antibody was obtained from Cayman Chemicals. Anti-rabbit and anti-mouse horseradish peroxidase conjugated secondary antibodies were purchased from Zymed Laboratories Inc. (San Francisco, CA, USA). Human recombinant STAT3 protein (catalog number, ab43618) was obtained from Abcam, Cambridge, UK).
2.3. Biotinylation of 15-keto PGE2
To a solution of 15-keto PGE2 (4 mg) and N-(5-Aminopentyl) biotinamide trifluoroacetate salt (6 mg) in dry acetonitrile (1 ml) under Ar atmosphere was added N,N′-diisopropylcarbodiimide (4 μl) at room temperature. After completion of reaction monitored by thin-layer chromatography (TLC), the resulting mixture was concentrated under reduced pressure and purified by flash column chromatography (ethyl acetate: MeOH = 5 : 1). A white solid (3 mg): 1H NMR (800 MHz, DMSO) δ 7.72 (ddd, J = 14.5, 11.5, 5.4 Hz, 2H), 6.41 (s, 1H), 6.35 (s, 1H), 5.38–5.22 (m, 1H), 5.30 (d, J = 5.7 Hz, 1H), 4.30 (dd, J = 7.4, 5.4 Hz, 1H), 4.13–4.11 (m, 1H), 4.03 (q, J = 7.1 Hz, 1H), 3.51–2.80 (m, 4H), 2.64–2.34 (m, 6H), 2.28–1.89 (m, 13H), 1.63–1.15 (m, 18H), 0.86 (t, J = 7.2 Hz, 3H); 13C NMR (200 MHz, DMSO) δ 214.14, 199.68, 171.75, 162.66, 146.79, 143.09, 131.19, 130.86, 72.25, 70.25, 69.91, 61.01, 60.22, 59.15, 55.77, 55.44, 55.40, 53.27, 52.89, 46.54, 38.30, 38.27, 38.17, 35.18, 34.88, 31.23, 30.83, 28.85, 28.70, 28.20, 28.01, 27.89, 26.32, 25.30, 25.21, 24.71, 23.80, 23.54, 23.40, 22.08, 21.93, 20.74, 13.82; LR-MS (ESI) Calculated for C35H57N4O6S (M + H+) 661.4, Found 661.4.
2.4. Western blot analysis
2.4.1. Preparation of cell lysates
After treatment with 15-keto PGE2 or 13,14-dihydro-15-keto PGE2, MCF10A-ras cells were harvested at indicated time points. The cells were rinsed with cold phosphate-buffered saline (PBS) and then scraped in 1 ml PBS followed by centrifugation at 1700 g for 5 min at 4 °C. Whole cell lysates were prepared with 10× Cell Lysis Buffer purchased from Cell Signaling Technology (Danvers, MA, USA) diluted to 1× solution containing 1% of phenylmethylsulfonyl fluoride (PMSF). Cell pellets were resuspended and incubated for 1 h on ice followed by centrifugation at 18,000 g for 15 min. Supernatant was collected as whole cell lysates. For obtaining cytosolic and nuclear extracts, buffer A and buffer C were used, respectively. Pellets were resuspended in hypotonic buffer A [0.2% HEPES (pH 7.9), 0.01% MgCl2, 0.07% KCl, 0.007% DTT and 0.003% PMSF] for 15 min on ice, and 0.1% Nonidet P-40 (NP-40) was added for 4 min. The mixture was then centrifuged at 6000 g for 5 min at 4 °C. The supernatant contained the cytosolic protein. The pellets were rinsed twice with hypotonic buffer A and resuspended again in hypertonic buffer C [0.4% HEPES (pH 7.9), 0.01% MgCl2, 2.4% NaCl, 0.007% DTT, 0.003% PMSF, 0.005% EDTA and 20% glycerol]. After incubation for 1 h on ice, the mixtures were centrifuged at 18,000 g for 15 min at 4 °C. Supernatant was collected as nuclear extract.
2.4.2. Quantification of the protein concentration
The protein concentrations in whole cell lysates were determined by using the BCA protein assay kit supplied from ThermoFisher Scientific (Rockford, IL, USA). The protein concentrations in cytosolic and nuclear extracts were determined by using the Bradford Assay Reagents kit purchased from Bio-Rad (Hercules, CA, USA).
2.4.3. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE)
Equivalent amounts of protein from whole cell lysate or nuclear fraction were mixed with SDS sample loading dye and boiled for 5 min at 99 °C. Protein samples were resolved by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) microporous membrane supplied from PALL Corporation (Port Washington, NY, USA). Non-specific antibody binding sites were blocked by 3% skim milk in PBS containing 0.05% Tween 20 (PBST) for 1 h at room temperature. Membranes were washed and incubated with PBST containing specific primary antibodies. The membranes were rinsed three times again and incubated with respective horseradish peroxidase conjugated secondary antibodies for 1 h at room temperature. Peroxidase activity was detected by incubation with an ECL reagent for 5 min according to the manufacturer's instruction and visualized using the imagequant LAS-4000 purchased from Fujifilm Life Science (Stamford, CT, USA).
2.5. Reverse transcription
Total RNA was isolated from MCF10A-ras cells using TRizol® (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. To generate complementary DNA (cDNA), 1 μg of total RNA was reverse transcribed by using murine leukemia virus reverse transcriptase (Promega, Madison, WI, USA).
2.6. Quantitative real-time PCR
Quantitative real-time PCR was carried out on an Applied Biosystems (Foster City, CA). The primers used were as follows: CCND1-Foward (5′-TCTACACCGACAACTCCATCCG-3′), CCND1-Reverse (5′- TCTGGCATTTTGGAGAGGAAGTG-3′), BCL2-Forward (5′-ATCGCCCTGTGGATGACTGAGT-3′), BCL2-Reverse (5′-GCCAGGAGAAATCAAACAGAGGC-3′), β-Actin-Forward (5′-TGCTAGGAGCCAGAGCAGTA-3′), β-Actin-Reverse (5′-AGTGTGACGTTGACATCCGT-3′). The amplified transcript level of each specific gene was normalized to that of β-actin.
2.7. STAT3 promoter luciferase reporter gene assay
HeLa cells were transfected with P-STAT3-TA-luc vector supplied from Clontech (Mountain View, CA, USA) using FuGENE from Promega (Madison, WI, USA) according to manufacturer's instructions. The HeLa/P-STAT3-Luc cells were seeded at a density of 8 × 104 per well in six-well plates. Cells were pretreated with 15-keto PGE2 or 13,14-dihydro-15-keto PGE2 for 24 h and then stimulated with 10 ng/ml of Oncostatin M (OSM) for 6 h. The cells were washed with PBS and lysed in 1× reporter lysis buffer. Twenty μl of lysed cell extracts was mixed with 100 μl of the luciferase substrate, and the luciferase activity was determined using a luminometer (AutoLumat LB 953, EG&G Berthold). The β-galactosidase activity was measured to normalize the luciferase activity.
2.8. Wound migration assay
The Culture-Inserts (Ibidi) were transferred to 6-well plates. MCF10A and MCF10A-ras cells were seeded at a density of 5 × 104 cells per well in Culture-Inserts. After 24 h, the silicon inserts were removed, and the cells were photographed under a microscope. The separated walls were closed 24 h later, and the closed gap images were captured using a microscope.
2.9. Clonogenic assay
MCF10A-ras cells were plated in 6-well plates at a density of 200 cells per well. The DMEM/F-12 medium was changed every other day and treated with 15-keto PGE2 (20 μM) or 13,14-dihydro-15-keto PGE2 (20 μM). After 7 days of incubation, the colonies were fixed in cold methanol and stained by 0.5% crystal violet. The stained colonies were washed with PBS to remove excessive dye. Quantitative changes in clonogenicity were determined by extracting stained dye with 10% acetic acid, and the absorbance of the extracted dye at 570 nm was measured.
2.10. Anchorage independent growth assay
MCF10A-ras cells were plated on a 60 mm dish containing 0.5% (down) and 0.33% (up) double layer agar. The dishes were incubated at 37 °C in humidified incubator for 21 days. The cells were treated every day with DMEM/F-12 containing DMSO, 15-keto PGE2 (20 μM) or 13,14-dihydro-15-keto PGE2 (20 μM). The colonies were stained with crystal violet for 12 h and rinsed with PBS. The number of colonies was counted by the ECLIPSE Ti inverted microscope using the NIS-Elements AR (V.4.0) computer software program (NIKON Instruments Korea, Seoul, Korea).
2.11. Immunoprecipitation
MCF10A-ras cells were plated in a 100 mm dish and treated with 15-keto PGE2 (20 μM), 13,14-dihdro-15-keto PGE2 (20 μM) or biotinylated 15-keto PGE2 (40 μM). The cells were washed with ice-cold PBS and lysed in 1× lysis buffer for 1 h on ice followed by centrifugation at 18,000 g for 15 min. The protein concentration was determined by using the BCA protein assay kit (Pierce, Rockford, IL, USA). Cell lysates (500 μg) were subjected to immunoprecipitation by shaking the primary STAT3 antibody and protein A/G agarose bead suspension at 4 °C for 12 h. After centrifugation at 18,000 g for 1 min, immunoprecipitation beads were collected by pouring the supernatant and washed with cell lysis buffer. The immunoprecipitation beads were then mixed with 25 μl of 2× SDS electrophoresis sample buffer and boiled for 5 min at 99 °C. Supernatant (25 μl) from each sample was collected by centrifugation and loaded on SDS-polyacrylamide gel. For exogenous STAT3 homo-dimerization, PC3 cells were co-transfected with HA-tagged and Myc-tagged constitutively active STAT3s. The transfected PC3 cells were treated with 15-keto PGE2 (20 μM) or 13,14-dihydro-15-keto PGE2 (20 μM) for 12 h. Lysates from transfected PC3 cells were immunoprecipitated with indicated antibodies.
PC3 cells were transfected with GFP-tagged wild type (WT) STAT3, GFP-tagged C251A STAT3 or GFP-tagged C259A STAT3 vector. These cells were then treated with 40 μM biotinylated 15-keto PGE2 for 24 h and then lysed with a 1 × lysis buffer. Total protein was subjected to immunoprecipitation with STAT3 primary antibody at 4 °C for 12 h followed by the addition of protein A/G-agarose bead suspension (25% slurry, 20 mL) and additional shaking for 2 h at 4 °C. After centrifugation at 3000 rpm for 1 min, immunoprecipitated beads were collected by discarding the supernatant and washed with cell lysis buffer. The immunoprecipitate was then resuspended in 4 μl of 6 × SDS electrophoresis sample buffer and boiled for 5 min. Supernatant (24 μl) from each sample was collected by centrifugation and loaded on SDS-polyacrylamide gel. The binding of biotinylated 15-keto PGE2 to immunoprecipitated proteins was detected by streptavidin–horseradish peroxidase (HRP) conjugate (GE Healthcare).
2.12. Immunocytochemistry
MCF10A-ras cells were plated on the 8-well chamber slides at a density of 1 × 104 per well and treated with 15-keto PGE2 or 13,14-dihydro-15-keto PGE2. Cells were fixed in 4% formaldehyde for 15 min at 37 °C. After rinse with PBST, cells were treated with 0.1% Triton X-100 in PBS for 5 min and washed with PBST. Samples were blocked with 0.05% Tween-20 in PBS containing 5% bovine serum albumin (BSA) at room temperature for 30 min and washed with PBST and then incubated with diluted (1:200) primary antibody for overnight at 4 °C. After washing with PBST, samples were incubated with a diluted (1:1000) TRITC-conjugated anti-mouse secondary antibody in PBST containing 5% BSA at room temperature for 1 h. Samples were washed with 0.05% PBST containing 5% BSA, and then examined under a fluorescent microscope.
2.13. Docking study
A docking study was performed using covalent docking modules implemented in Maestro v9.5 (Schrödinger LLC, NY, USA). The STAT3 crystal structure (PDB code: 1BG1) was retrieved from PDB bank. The protein structure and the ligand were prepared according to the standard procedure of the Protein Preparation Wizard and Ligprep modules in Maestro v9.5. After adding hydrogens, STAT3 protein was neutralized and then energically minimized on only hydrogens. The structure of 15-keto PGE2 was drawn by ionization at pH 7.4 and energy minimization. 15-Keto PGE2 was docked to the molecular model of STAT3 in silico. In covalent docking module, the reaction type was set to Micheal addition between Cys251 or Cys259 of STAT3 protein and C13 atom of 15-keto PGE2. The grid box was automatically determined the residues within 5.0 Å of the Cys251 or Cys259 residue in STAT3. After minimization of residues within 3 Å of covalently bound ligand, the outputs 10 poses of ligand-receptor complex was scoring by Prime MM-GBSA to calculate a binding affinity. The best binding energy of 15-keto PGE2, low PrimeΔGbind, was chosen for the binding mode analysis.
2.14. Liquid chromatography –tandem mass spectrometry (LC-MS/MS) analysis
2.14.1. Sample preparation
Twenty μM of 15-keto PGE2 was reacted with recombinant STAT3 protein at room temperature for 3 h. The STAT3 protein treated with 15-keto PGE2 was digested into peptides by an in-solution method. Briefly, 6 M urea in 50 mM ammonium bicarbonate pH 8.0 (Sigma, St. Louis, MO, USA) was mixed with sample, and the mixture was incubated for 45 min at room temperature (RT). Then, 10 mM DTT and 30 mM iodoacetic acid were used to denature the proteins. The MS-grade trypsin protease (250 ng/ul) was added and incubated at 37 °C for 12 h. The resulting tryptic peptides were desalted using Pierce® C-18 spin columns (Pierce Biotechnology, Rockford, IL, USA) and were dried using Speed-Vac (Scanvac; LaboGene Aps, Lynge, Denmark). The tryptic peptides were reconstituted in mobile phase (Solvent A) for LC-MS analysis.
2.14.2. Direct-infusion mass spectrometry (DIMS) analysis of 15-keto PGE2
15-Keto PGE2 was reconstituted in 0.1% formic acid in 50% acetonitrile and loaded in a Hamilton syringe, injected by a syringe pump with a flow rate of 1 μl/min into the heated electrospray ionization (HESI) source and measured for 0.5 min on a Q-Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Fisher Scientific Inc., Germany). The operating source conditions for MS scan in positive ESI mode were optimized as follows: spray voltage, 3.6 KV; heated capillary temperature, 320 °C Nitrogen was used as damping gas. For higher-energy collision dissociation (HCD) experiments, keeping MS1 static, the precursor ion of interest was selected using the orbitrap analyzer, and the product ions were analyzed. The normalized collision energy (NCE) was set at 27. Resolutions of ions were set at 70,000 for MS1 and 17,500 for HCD experiments. The top 3 precursor ions in the MS scan were selected by orbitrap analyzer for subsequent MS/MS analysis.
2.14.3. LC-MS/MS analysis
For LC-MS analysis, Ultimate 3000 UHPLC system (Thermo Fisher Scientific Inc., Germany) was coupled to Q-Exactive™ Plus Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo Fisher Scientific Inc., Germany) equipped with a trap column (C18, 75 μm × 2 cm, 5 μm, Thermo Scientific Inc., Germany) for cleanup followed by an EASY-Spray column (C18, 75 μm × 50 cm, 2 μm, Thermo Scientific Inc., Germany). The peptides were separated by using the mobile phase comprising of 0.1% formic acid in water (Solvent A) and 0.1% formic acid in acetonitrile (Solvent B) in a gradient elution mode with a total run time of 100 min. The optimized linear gradient elution program was set as follows: (T min/% of solvent B): 0/5, 5/5, 69/40, 79/50, 80/80, 90/80, 91/5, 100/5. The column temperature was maintained at 60 °C, and the flow rate of the mobile phase was 300 nl/min throughout the run time.
The typical operating source conditions for MS scan in positive ESI mode were optimized as follows: spray voltage, 2.0 KV; heated capillary temperature, 250 °C; and nitrogen was used as damping gas. All the spectra were recorded under identical experimental conditions. The scan range was set from m/z 400–2000, and resolution of precursor ions was set at 70,000. The 10 most abundant ions were fragmented by data-dependent mode at a resolution of 17,500 with the exclusion duration of 30 s and the isolation window was performed with 2.0 m/z. The NCE were set at 27.
2.15. Xenograft assay
BALB/c (nu/nu) athymic mice were purchased from Charles River Laboratories (Skokie, IL, USA) and kept in a room at constant temperature (24 ± 2 °C) and humidity (50 ± 10%). The animal study was approved by Seoul National University Institutional Animal Care and Use Committees (IACUC). Seven-week old female BALB/c nude mice were subcutaneously injected with 2 × 106 MDA-MB-231 cells per mouse at both flanks. After 10 days of treatment, mice were randomly assigned to three groups (seven mice per group) and were treated with vehicle (5% DMSO in PBS), 15-keto PGE2 (70 μg/kg) or 15-keto PGE2 (140 μg/kg) daily by subcutaneous injection for 4 weeks. Tumor volumes were measured every other day with a digital caliper and calculated by formula 0.52 x length x width2. Mice were weighed three times a week.
2.16. Hematoxylin and eosin (H&E) staining and immunohistochemical analysis of xenograft tumors
The MDA-MB-231 xenograft tumors were removed and fixed in formalin solution (10% neutral buffered formaldehyde) at room temperature for 48 h. Slides containing 4 μm section of formalin-fixed and paraffin-embedded specimens of xenograft tumors were prepared for histopathological and immunohistochemical analyses. H&E staining was carried out as described previously [30]. The tumor sections were used to determine the expression of P-STAT3Y705 by immunohistochemical analysis. Slides were incubated with a primary antibodies for P-STAT3Y705 and visualized using the anti-rabbit horseradish peroxidase Envision System (DAKO). The counterstaining was done using Mayer's hematoxylin.
2.17. Statistical analysis
Statistical analysis for single comparison was performed using the Student's t-test. Data were expressed as means of ±SD from at least three independent experiments. p < 0.05 was considered a statistically significant difference.
3. Results
3.1. Comparison of enzymes involved in PGE2 metabolism in MCF10A-ras cells and non-oncogenic MCF10A cells
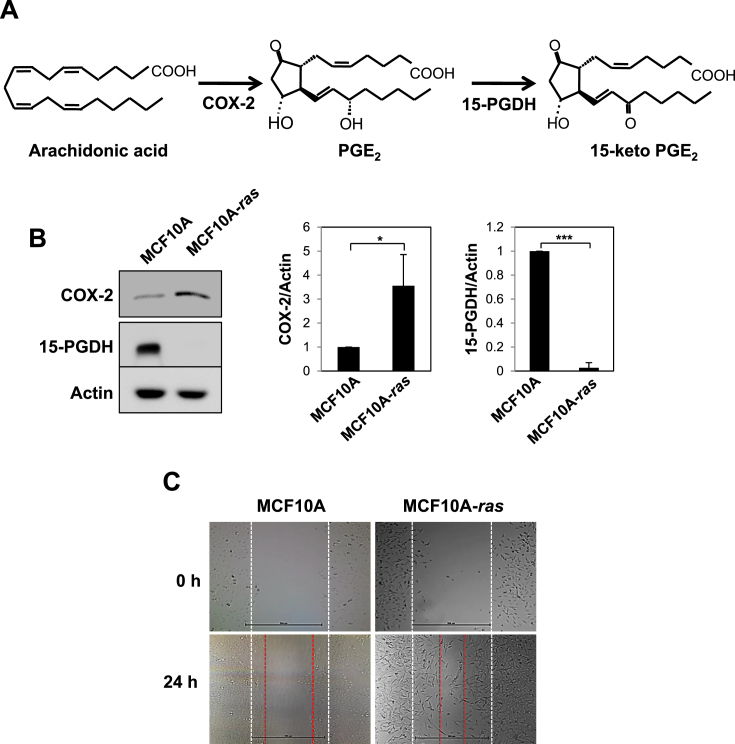
COX-2 converts arachidonic acid into PGE2, whereas 15-PGDH oxidizes the 15(S)-hydroxyl group of PGE2 to generate 15-keto PGE2 (Fig. 1A). In various tumors and transformed cells, down-regulation of 15-PGDH is frequently observed with concomitant overexpression of COX-2. Therefore, 15-PGDH has been speculated as an endogenous COX-2 antagonist [15]. In human breast cancer cells, PGE2 production has been shown to be markedly elevated compared to normal cells [31]. We analyzed total lysates of MCF10A and MCF10A-ras cells for the basal levels of COX-2 and 15-PGDH. The protein level of COX was higher in MCF10A-ras cells compared to normal breast MCF10A epithelial cells (Fig. 1B). However, the basal level of 15-PGDH was higher in MCF10A cells. Moreover, the migration of MCF10A-ras cells in terms of their wound healing ability was much higher than that in non-oncogenic MCF10A cells (Fig. 1C).
Fig. 1.
Comparison of enzymes involved in PGE2metabolism in MCF10A-ras cells and non-oncogenic MCF10A cells. A. PGE2 is synthesized by COX-2 and oxidized by 15-PGDH to form 15-keto PGE2. B. The protein levels of COX-2 and 15-PGDH were compared in MCF10A and MCF10A-ras cells by Western blot analysis. Actin was served as an equal loading control. The experiments were conducted at least 3 times and quantificated the band intensity of each protein compared to expression of actin. *p < 0.05, ***p < 0.001 (Two-sided t-test). C. The migration of MCF10A and MCF10A-ras cells was assessed by measuring the width of un-migrated space under a microscope.
3.2. 15-Keto PGE2 inhibits STAT3 signaling
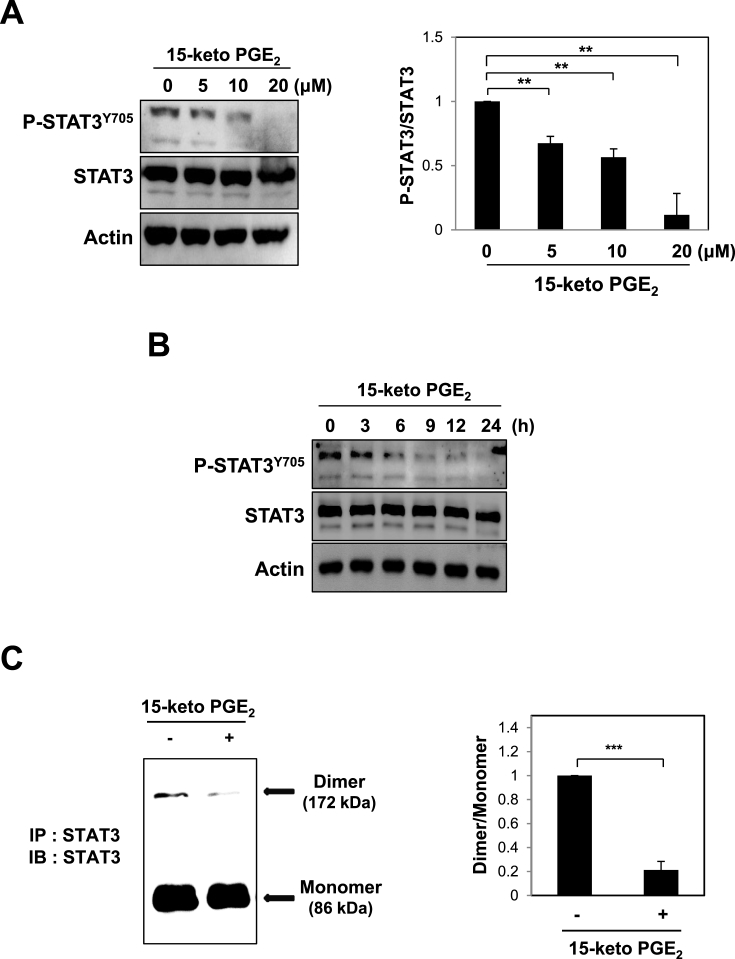
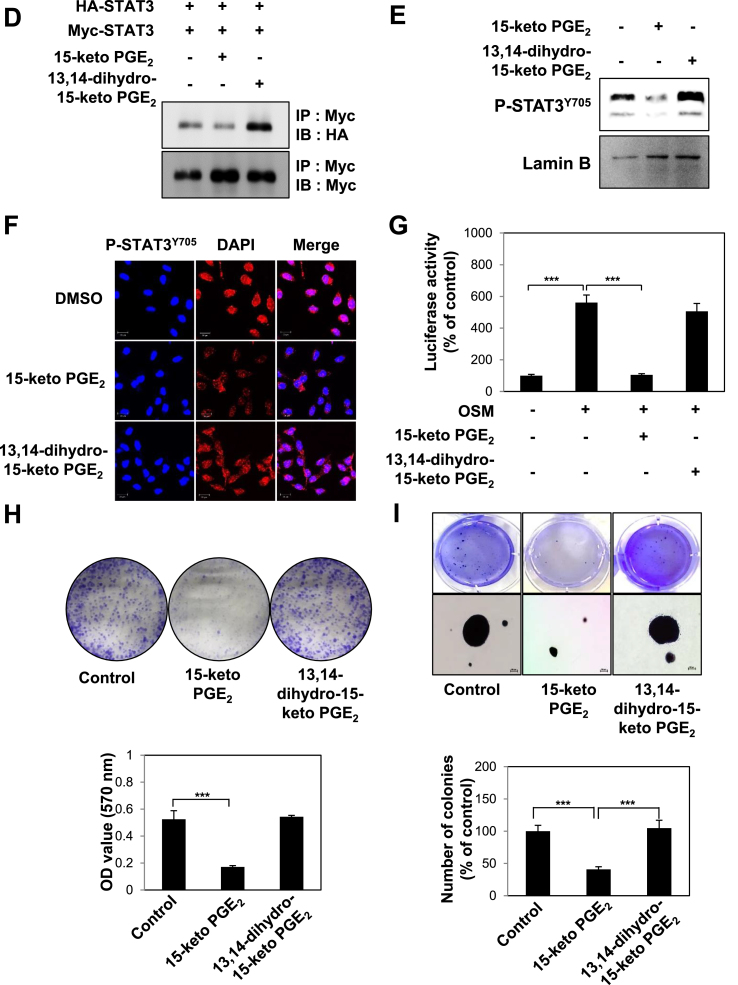
Ras gene mutation is found in certain types of human breast cancers, which associates with metastatic dissemination and poor prognosis [32]. STAT3 is constitutively activated in ras-transfected rat intestinal epithelial cells [33]. Phosphorylation of STAT3 at tyrosine 705 is a pivotal event for the activation of this transcription factor for cancer progression [34]. We investigated whether 15-keto PGE2 could affect the STAT3 signaling in MCF10A-ras cells. 15-Keto PGE2 significantly decreased STAT3 phosphorylation in a dose- (Fig. 2A) and time-dependent (Fig. 2B) manners. Phosphorylated STAT3 forms a homodimer and translocates to the nucleus where it binds to specific DNA sequences present in the promoter regions of target genes, thereby initiating their transcription [21,25]. Treatment of MCF10A-ras cells with 20 μM of 15-keto PGE2 resulted in interference of STAT3 homodimerization (Fig. 2C) and nuclear localization (Fig. 2D and E). To determine whether 15-keto PGE2 suppresses the transcriptional activity of STAT3, HeLa cells which show a lower level of STAT3 were transfected with P-STAT3-TA-luc vector and stimulated with Oncostatin M (OSM) for transactivation of STAT3. As illustrated in Fig. 2F, 15-keto PGE2 inhibited the transcriptional activity of STAT3 induced by OSM in HeLa/P-STAT3-luciferase reporter cells.
Fig. 2.
Effects of 15-keto PGE2on phosphorylation, dimerization, nuclear localization, and transcriptional activities of STAT3 in MCF10A-ras cells. A. MCF10A-ras cells were treated with indicated concentrations of 15-keto PGE2 for 24 h. The expression levels of STAT3 and P-STAT3Y705 were determined by Western blot analysis. **p < 0.01. B. MCF10A-ras cells were incubated with 15-keto PGE2 (20 μM) for indicated time periods. The protein level of phosphorylated STAT3 was measured by Western blotting. C. Whole lysates from MCF10A-ras cells treated with or without 15-keto PGE2 (20 μM) for 12 h were prepared for immunoprecipitation of STAT3 protein using protein A/G agarose conjugated C-terminus anti-STAT3 antibody. The STAT3 homo-dimerization was analyzed by Western blot analysis using N-terminus anti-STAT3 antibody. ***p < 0.001. D. MCF10A-ras cells were treated with 15-keto PGE2 (20 μM) for 24 h, and nuclear extracts were immunoblotted for detection of P-STAT3Y705. Lamin B was used as a nuclear protein marker. **p < 0.01. E. Immunocytochemical analysis was performed using antibodies against P-STAT3Y705. MCF10A-ras cells were treated with or without 15-keto PGE2 (20 μM) for 24 h. Nuclei were stained with DAPI and visualized under a confocal microscope. Two images were merged to verify co-localization. F. The luciferase assay was performed with HeLa/P-STAT3-luc reporter cells. The cells were treated with 15-keto PGE2 (10 or 20 μM) for 24 h and then stimulated with OSM (10 ng/ml) for another 6 h. Cells were then analyzed by a microplate luminometer. **p < 0.01, ***p < 0.001.
3.3. 15-Keto PGE2 inhibits transcription of STAT3 target genes and suppresses the proliferation and growth of MCF10A-ras cells
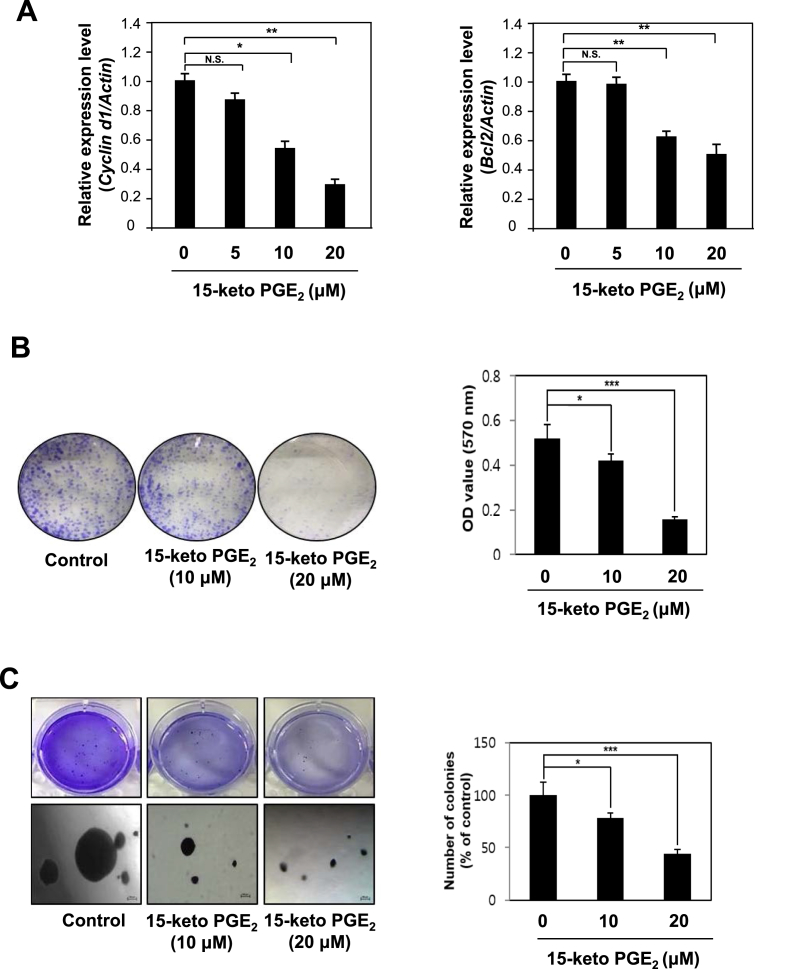
The transcription of genes induced by STAT3 encode proteins that belong to chemoattractants family (e.g., CCL5), MCP-1, and cell cycle regulators as well as those involved in antiapoptosis and angiogenesis [35]. 15-Keto PGE2 treatment inhibited the expression mRNA transcript of Cyclin D1 and Bcl-2 in a concentration-dependent fashion (Fig. 3A). Next, we conducted the clonogenic assay and the anchorage-independent growth assay to assess the anti-proliferative effects of 15-keto PGE2. 15-Keto PGE2 treatment markedly reduced the colony formation (Fig. 3B) and anchorage-independent growth (Fig. 3C) of MCF10A-ras cells.
Fig. 3.
Inhibitory effects of 15-keto PGE2on STAT3 target gene expression and neoplastic transformation of MCF10A-ras cells. A. MCF10A-ras cells were treated with 15-keto PGE2 (20 μM) or vehicle for 24 h. The mRNA levels of two representative STAT3 target genes Cyclin D1 and Bcl2 were determined by real-time PCR. β-Actin was measured to ensure equal amount of cDNA loaded. N.S.: not significant. *p < 0.05, **P < 0.01, N.S, not significant. B. MCF10A-ras cells were seeded in 6 well plates and treated with two different concentrations of 15-keto PGE2 (10 or 20 μM) or vehicle (DMSO) in DMEM/F12 medium containing 5% heat-inactivated horse serum and supplementary agents. The incubation conditions and other experimental details are described in Materials and methods. The colony size >100 μm was counted under a light microscope. Results are the means ± S.D. *p < 0.05, ***P < 0.001. C. MCF10A-ras cells were plated on a 60 mm dish containing 0.5% (down) and 0.33% (up) double layer agar. The cells were treated every day with DMEM/F-12 containing DMSO or 15-keto PGE2. After 3-week of incubation, the colonies were stained with crystal violet, and the number of colonies was counted as described in Materials and methods. Results are the means ± S.D. *p < 0.05, ***P < 0.001.
3.4. 15-Keto PGE2 did not inhibit the upstream signaling molecules of STAT3
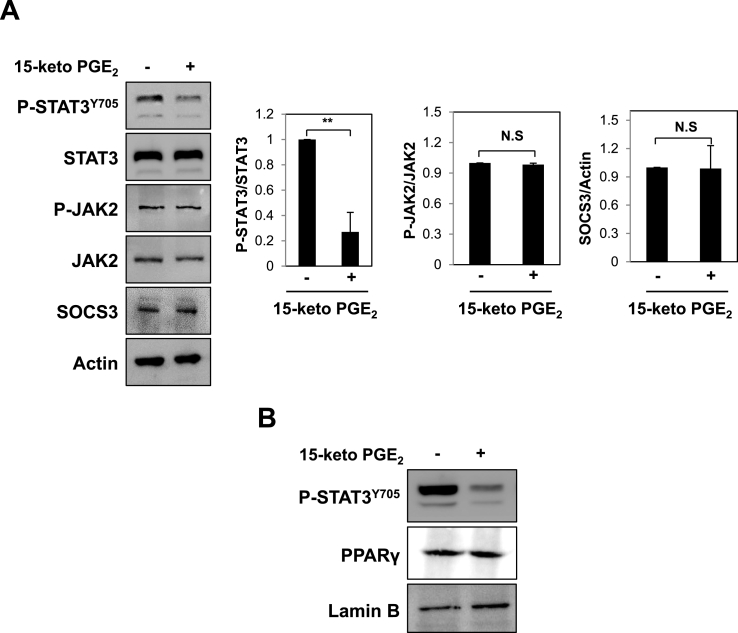
To elucidate the molecular mechanisms underlying inactivation of STAT3 by 15-keto PGE2, we examined its effect on signaling upstream of STAT3. Initially, we investigated whether 15-keto PGE2 could inhibit formation of phosphorylated STAT3 through regulation of JAK2, a well-known kinase responsible for STAT3 phosphorylation [36] and suppressor of cytokine signaling 3 (SOCS3), a negative regulator of STAT3 [37]. 15-Keto PGE2 (20 μM) affected neither JAK2 expression/phosphorylation nor SOCS3 expression while it suppressed STAT3 phosphorylation (Fig. 4A).
Fig. 4.
Effects of 15-keto PGE2on STAT3 regulating molecules and PPARγ in MCF10A-ras cells. A. MCF10A-ras cells were treated with 15-keto PGE2 (20 μM) for 24 h. The expression of JAK2, an upstream kinase of STAT3 and SOCS3, a negative regulator of STAT3, was assessed by Western blot analysis. **p < 0.01, N.S, not significant. B. Nuclear extracts were isolated from MCF10A-ras cells treated with 15-keto PGE2 (20 μM), and nuclear PPARγ levels were analyzed by Western blot analysis. Lamin B was used as a marker for nuclear protein.
It has been reported that the oxidized prostaglandin metabolites including 15-keto PGE2 modulate PPARγ activity as an endogenous ligand of this transcription factor [16]. Overexpression of 15-PGDH led to activation of PPARγ, and 15-keto PGE2 treatment increased transcriptional activity of p21 through PPARγ association in hepatocellular carcinoma cells [19]. However, we noticed that 15-keto PGE2 did not alter the expression levels of nuclear PPARγ (Fig. 4B).
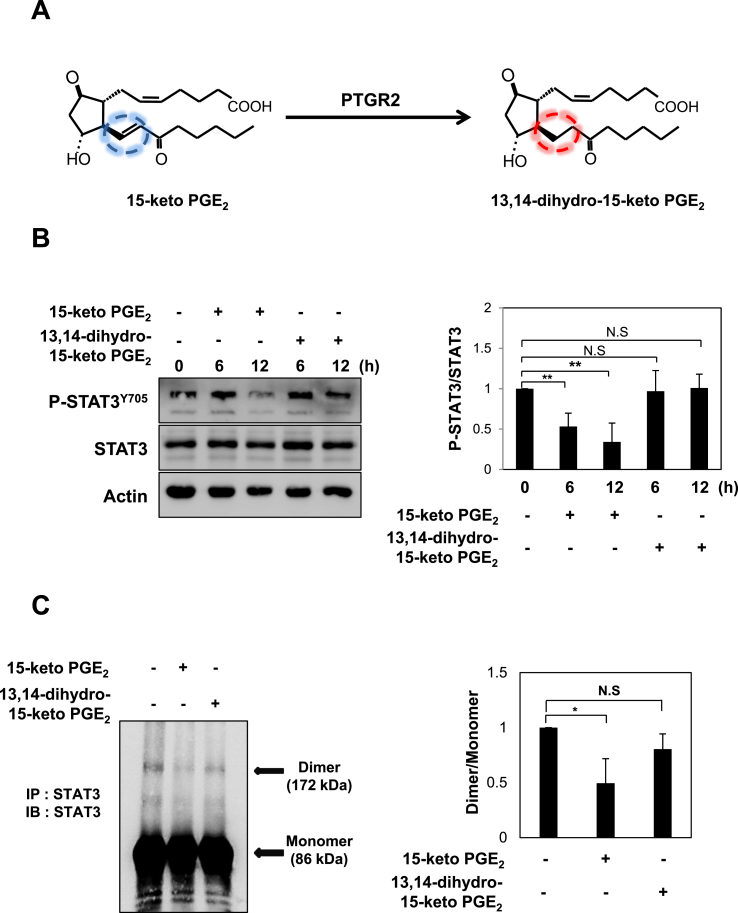
3.5. The α,β-unsaturated carbonyl moiety of 15-keto PGE2 is critical in STAT3 deactivation
15-Keto PGE2 has an α,β-unsaturated carbonyl moiety which is capable of interacting with nucleophilic cellular proteins [16]. 15-Keto PGE2 is reduced to 13,14-dihydro-15-keto PGE2 by PTGR2 [38] (Fig. 5A). To determine whether the α,β-unsaturated carbonyl group of 15-keto PGE2 plays an important role in the suppression of STAT3 signaling, the cells were treated with 13,14-dihydro-15-keto PGE2 lacking such electrophilic moiety. In contrast to 15-keto PGE2, 13,14-dihydro-15-keto PGE2 failed to inhibit STAT3 phosphorylation in MCF10A-ras cells (Fig. 5B). In addition, this non-electrophilic analogue did not exhibit a significant change in STAT3 homo-dimerization in the same cell line (Fig. 5C). In another experiment, we investigated the effect of 15-keto PGE2 and 13,14-dihydro-15-keto PGE2 on the dimer formation of exogenously introduced STAT3. For this purpose, PC3 cells were transfected with HA-tagged STAT3 and Myc-tagged STAT3 followed by treatment with 20 μM of 15-keto PGE2 or the same concentration of 13,14-dihydro-15-keto PGE2, and the immunoprecipitation assay was conducted. Again,15-keto PGE2, but not 13,14-dihydro-15-keto PGE2, inhibited the dimerization of exogenous STAT3 (Fig. 5D). Likewise, the nuclear translocation (Fig. 5E and F) and transcriptional activity (Fig. 5G) of STAT3 in the MCF10A-ras cells were inhibited by 15-keto PGE2 but not by 13,14-dihydro-15-keto PGE2.
Fig. 5.
Comparative effects of 15-keto PGE2and its non-electrophilic analogue, 13,14-dihydro-15-keto PGE2on STAT3 activation, and clonogenicity and anchorage-independent growth of MCF10A-ras cells. A. 15-Keto PGE2 is reduced to the 13,14-dihydro-15-keto PGE2 by PTGR2. 15-Keto PGE2 has an α,β-unsaturated carbon which is considered to target nucleophiles whereas 13,14-dihydro-15-keto PGE2 has no such electrophilic moiety. B. MCF10A-ras cells were treated with 20 μM each of 15-keto PGE2 or 13,14-dihydro-15-keto PGE2 for indicated time points. The expression levels of P-STAT3Y705 and STAT3 were measured by Western blot analysis. **p < 0.01 , N.S, not significant. C. MCF10A-ras cells were treated with 15-keto PGE2 (20 μM) or the same concentration of 13,14-dihydro-15-keto PGE2 for 12 h. Homo-dimerization of STAT3 was measured by the immunoprecipitation assay as described in Materials and methods. *p < 0.05, N.S., not significant. D. Human prostate cancer PC3 cells were co-transfected with HA-tagged STAT3 and Myc-tagged STAT3 and treated with 15-keto PGE2 or 13,14-dihydro-15-keto PGE2 (20 μM each) for 12 h. The total lysates obtained from the transfected cells were immunoprecipitated with anti-Myc antibody and analyzed by Western blotting with anti-HA antibody to measure the formation of a STAT3 dimer. E. MCF10A-ras cells were incubated with 15-keto PGE2 (20 μM) or 13,14-dihydro-15-keto PGE2 (20 μM) for 24 h. The nuclear extracts were separated and subjected to Western blot analysis. F. After MCF10A-ras cells were treated with 15-keto PGE2 (20 μM) or 13,14-dihydro-15-keto PGE2 (20 μM) for 24 h, the fixed cells were incubated with anti-P-STAT3Y705, which was detected using TRITC red fluorescence-labeled secondary antibody. G. The HeLa/P-STAT3-luc reporter cells were pretreated with 20 μM each of 15-keto PGE2 or 13,14-dihydro-15-keto PGE2 for 24 h followed by treatment with OSM for another 6 h. The cells were analyzed for the luciferase activity using a microplate luminometer. ***p < 0.001. H. MCF10A-ras cells treated with 15-keto PGE2 (20 μM) or 13,14-dihydro-15-keto PGE2 (20 μM) were subjected to the clonogenic assay. The colony size >100 μm was counted under light microscope. Results are the means ± S.D. ***p < 0.001. I. MCF10A-ras cells were plated on a 60 mm dish containing 0.5% (down) and 0.33% (up) double layer agar. The cells were treated every day with DMEM/F-12 containing DMSO, 15-keto PGE2 (20 μM) or 13,14-dihydro-15-keto PGE2 (20 μM). After 3-week of incubation, the colonies were stained with crystal violet, and the number of colonies was counted as described in Materials and methods. ***p < 0.001.
Further, the colony formation (Fig. 5H) and anchorage-independent MCF10A-ras cell growth (Fig. 5I) were not suppressed by 13,14-dihydro-15-keto PGE2. Above results suggest that the electrophilic α,β-unsaturated carbonyl group of 15-keto PGE2 is essential for its deactivating STAT3.
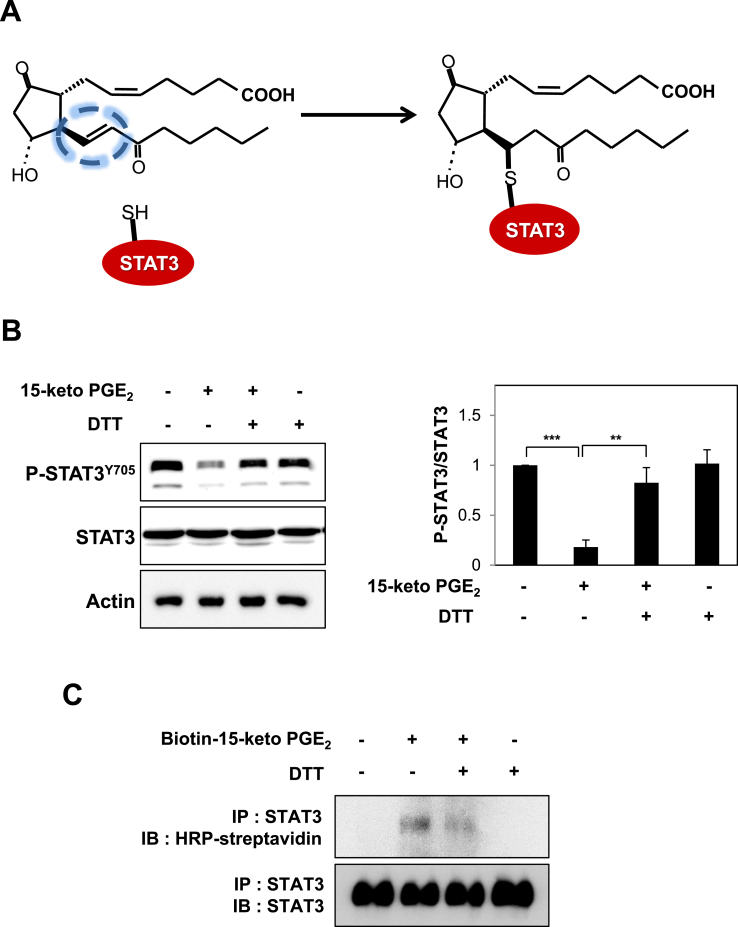
3.6. 15-Keto PGE2 covalently binds to STAT3 via Michael addition reaction
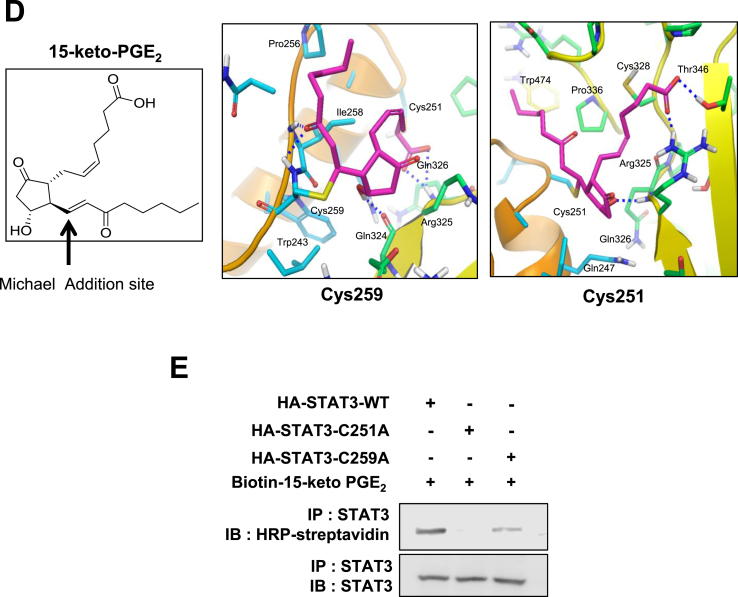
STAT3 possesses several cysteines [39] and many studies focus on targeting cysteine residues in the SH2 and DNA binding domains to inhibit the STAT3 signaling [[40], [41], [42], [43]]. We speculated that the electrophilic α,β-unsaturated carbonyl moiety of 15-keto PGE2 could modify the thiol group(s) of STAT3 (Fig. 6A) which might hamper its phosphorylation. Pretreatment of MCF10A-ras cells with the thiol reducing agent, DTT abolished the inhibitory effect of 15-keto PGE2 on STAT3 phosphorylation (Fig. 6B), lending support to the above supposition. To investigate direct interaction between 15-keto PGE2 and STAT3, MCF10A-ras cells were treated with biotinylated 15-keto PGE2, and the immunoprecipitation assay using the biotin-streptavidin system was performed. As shown in Fig. 6C, biotinylated 15-keto PGE2 directly bound to STAT3, and this was abrogated in the presence of DTT. A docking model predicted Cys251 and Cys259 of STAT3 as putative binding sites of 15-keto PGE2 (Fig. 6D).
Fig. 6.
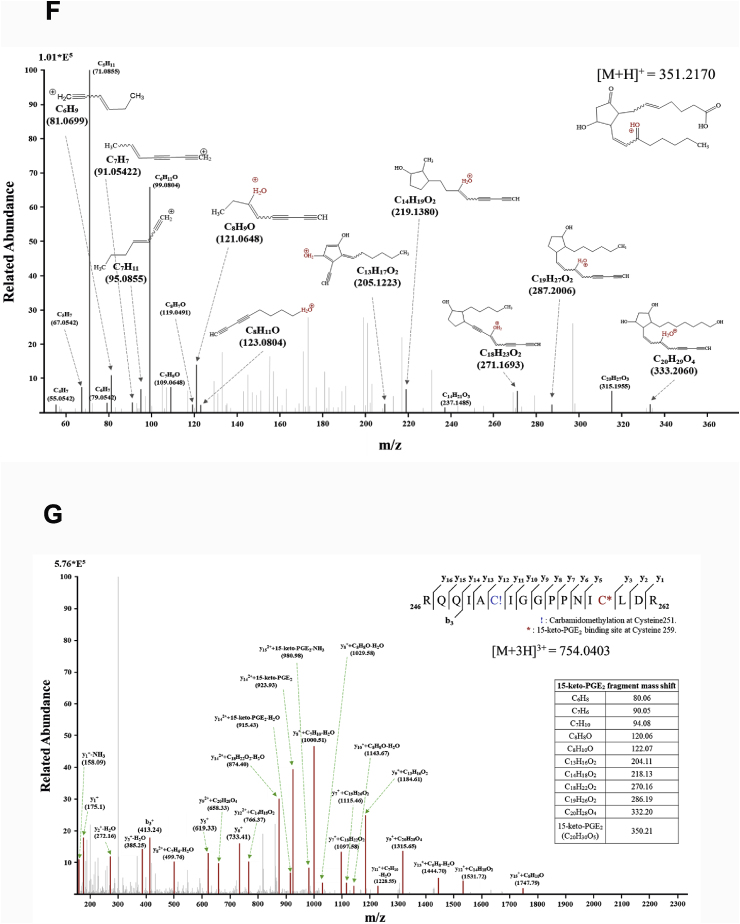
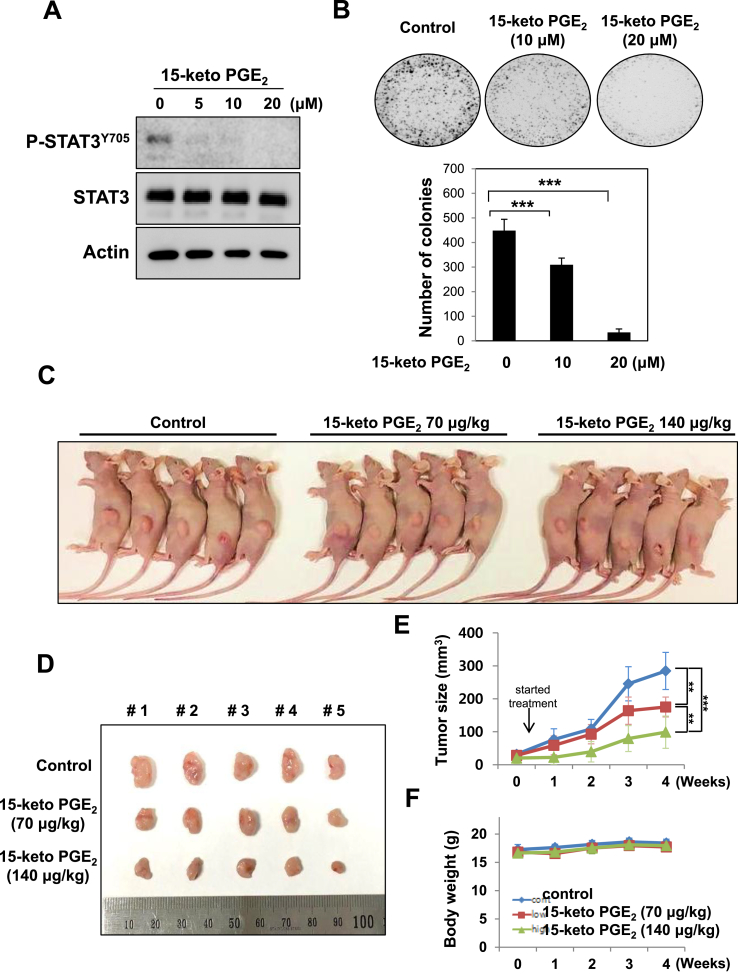
Covalent modification of STAT3 by15-keto PGE2. A. A proposed interaction between 15-keto PGE2 and a thiol group of STAT3. B. MCF10A-ras cells were pretreated with a thiol reducing agent, DTT (100 μM), for 1 h followed by exposure to 15-keto PGE2 (20 μM) for an additional 24 h. The protein levels of P-STAT3Y705 and STAT3 were determined by Western blot analysis. **p < 0.01, ***p < 0.001. C. The direct binding of 15-keto PGE2 to STAT3 was assessed using the avidin-biotin system. MCF10A-ras cells were pretreated with DTT (100 μM) for 1 h and then treated with biotinylated 15-keto PGE2 (40 μM) for an additional 12 h. The biotinylated 15-keto PGE2-STAT3 complex was detected by the immunoprecipitation and Western blot analyses. D. A putative model of 15-keto PGE2 covalently bound to Cys259 or Cys251 of STAT3 as predicted by a docking study. The blue dotted line represents a H-bond. E. PC3 cells were transfected with WT or Cys251 or Cys259-mutated STAT3 followed by treatment with 15-keto PGE2 (40 μM) for 24 h. WT STAT3 and STAT3 mutants were detected by anti-STAT3 antibody. F. Annotated fragment MS/MS spectrum of 15-keto PGE2 in positive ESI mode. The elucidation of fragment ion at m/z 333.2073, 315.1951 from the structure is given in the inset of the spectra. G. Mass spectrum of the STAT3 peptide (RQQIACIGGPPNICLDR) with 15-keto PGE2 binding. Annotated MS/MS spectrum illustrating the binding of 15-keto PGE2 to STAT3 protein at cysteine 259 residues was identified by LC-MS/MS. An exclamation mark indicates a carbamidomethylation on Cys251, and an asterisk indicates 15-keto PGE2 binding to Cys259. The precursor ion [M+3H]3 + for peptide with 15-keto PGE2 binding is m/z 754.0403. The 15-keto PGE2 fragment ions are inserted into the spectrum as a table. Each molecular formula present with b and y ions on the spectrum represents the 15-keto PGE2 fragment ion bound on Cys259. The sample preparation and other experimental details for mass spectral analysis are described in the Materials and methods. Sequence-informative fragmentation ions are summarized on the peptide sequence.
To further verify the direct interaction between 15-keto PGE2 and the cysteine residues of STAT3, we conducted site-directed mutagensis studies in which cysteine 251 and 259 were replaced by alanine. PC3 cells were transfected with WT or cysteine 251/259-mutated STAT3 followed by treatment with 40 μM of biotinylated 15-keto PGE2.. 15-Keto PGE2 barely bound to cysteine mutated STAT3 (Fig. 6E).
To further verify the binding site of 15-keto PGE2 on STAT3 protein, MS/MS analysis of recombinant STAT3 protein treated with 15-keto PGE2 was performed. First, the 15-keto PGE2 was analyzed by DIMS in the positive mode. A fragmentation pattern was obtained by HCD of the precursor ion corresponding to 15-keto PGE2. The MS/MS spectrum of 15-keto PGE2 (m/z 351.2170) shows the fragmentation information during HCD (Fig. 6F). Secondly, tryptic peptides of STAT3 protein treated with 15-keto PGE2 were analyzed by LC-MS/MS. Based on the fragmentation pattern, we were able to identify 15-keto PGE2 bound tryptic peptide derived from STAT3 protein as shown in Fig. 6G. From the MS scan, the molecular ion corresponding to [M+3H]3+ (m/z 754.0403) of 246-RQQIACIGGPPNICLDR-262 was identified as 15-keto PGE2 bound STAT3 peptide at the Cys259 residue. The fragment ions matched to the 15-keto PGE2 bound STAT3 peptide were assigned and confirmed manually. These data support the assignment of Cys259 as the primary binding site of 15-keto PGE2 on STAT3.
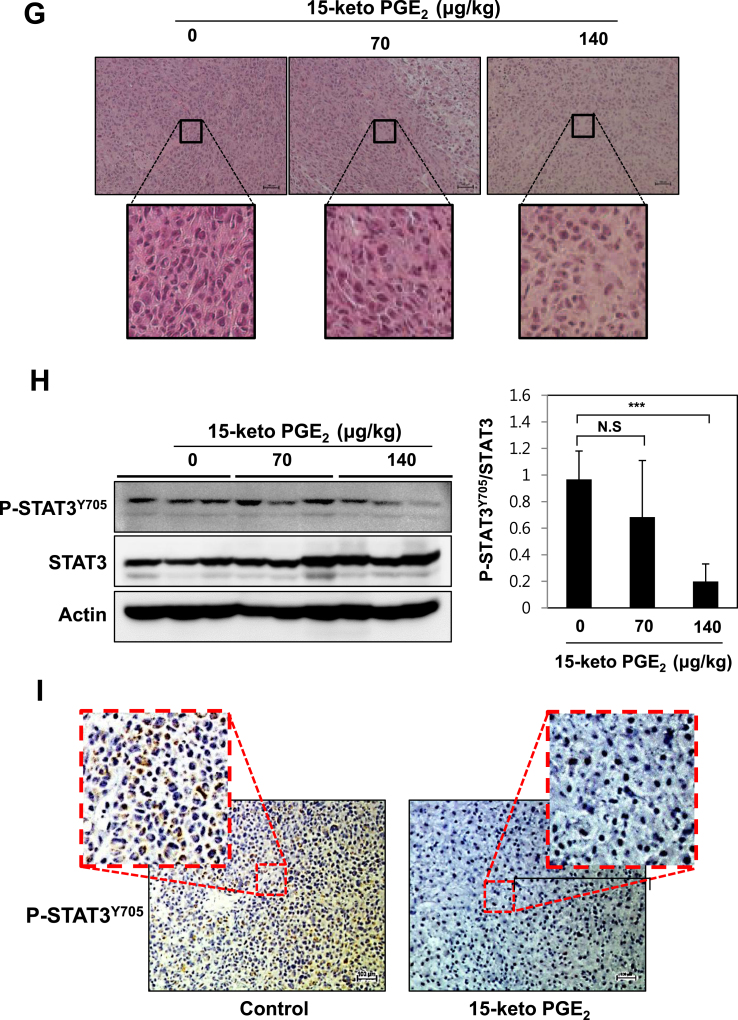
3.7. 15-Keto PGE2 attenuates tumor growth and suppresses STAT3 phosphorylation in a MDA-MB-231 xenograft model
Treatment of human breast cancer MDA-MB-231 cells with 10 or 20 μM of 15-keto PGE2 significantly decreased the phosphorylation of STAT3 (Fig. 7A). 15-Keto PGE2 also suppressed colony formation of MDA-MB-231 cells (Fig. 7B). We then examined whether 15-keto PGE2 could inhibit the growth of MDA-MB-231 cells transplanted to BALB/c nude mice. The mice xenografted with MDB-MB-231 cells were subcutaneously injected with two different doses of 15-keto PGE2 for 4 weeks. The tumor growth was significantly retarded in mice injected with 15-keto PGE2 at a dose of 70 μg/kg and 140 μg/kg (Fig. 7C and D). The average tumor volume in 15-keto PGE2-treated mice was dramatically reduced in a dose-dependent manner (Fig. 7E). However, there were no body weight loss and other signs of toxicity in mice treated with 15-keto PGE2 (Fig. 7F). Histopathological analysis of the tumor samples stained for H&E showed that tumor density was decreased in 15-keto PGE2-treated mice compared to the control animals (Fig. 7G). Furthermore, the phosphorylation of STAT3 was markedly decreased in the 15-keto PGE2 treated group (140 μg/kg) compared to the control group (Fig. 7H). Immunohistochemical analysis of tumor samples verified the significantly reduced expression of P-STAT3Y705 in the group treated with the higher dose (140 μg/kg) of 15-keto PGE2 (Fig. 7I), supporting the Western blotting data.
Fig. 7.
Inhibition of xenograft tumor growth and STAT3 phosphorylation in BALB/nude mice. A. MDA-MB-231 cells were treated with 15-keto PGE2 (20 μM) for 48 h. The expression levels of P-STAT3Y705 and STAT3 were measured by Western blot analysis. ***p < 0.001. B. MDA-MB-231 cells treated with 15-keto PGE2 (20 μM) for 24 h were subjected to the clonogenic assay. C. The representative shots of human mammary tumor (MDA-MB-231) xenografts in mice treated with vehicle, 15-keto PGE2 (70 μg/kg) and 15-keto PGE2 (140 μg/kg) for 4 weeks (n = 14/group). D, E. The size of tumors was measured with digital calipers. The calculated formula is 0.52 × length x width2. **p < 0.005, ***p < 0.001 (Two-sided t-test). F. The effect of 15-keto PGE2 treatment on body weight. G. H&E stained tumor tissue sections. Scale bar: 100 μM. Magnification:×4 and ×10. H. Total protein samples from the tumor specimens were subjected to Western blot analysis to measure the levels of P-STAT3Y705 and STAT3. Actin was used as the internal control. ***p < 0.001, N.S, not significant. I. The effect of the higher dose (140 μg/kg) of 15-keto PGE2 on the expression of P-STAT3Y705 was determined by immunohistochemical analysis (magnification, ×100).
4. Discussion
STAT3 as a transcription factor is involved in tumor formation, cancer cell proliferation and survival. Therefore, targeting STAT3 has been considered as a practical approach for anti-cancer therapy [22,24]. Many STAT3 signaling inhibitors developed so far target the upstream kinases of STAT3, particularly JAK2 [[44], [45], [46]]. However, a JAK2 inhibitor, cucurbitacin Ⅰ also induces a phenotypic change interfering connective tissue growth in non-tumor cells [46]. Therefore, specific inhibitors of STAT3, especially those targeting the down-stream events, are necessary for developing novel chemotherapeutic drugs. Eriocalyxin B [47], cryptotanshinone [48], S3I-201 [49], an NCI compound NSC-368,262 (C48) [42], and galiellalactone [43] have been reported as molecules targeting STAT3 directly. These compounds were found to suppress the phosphorylation, dimerization, nuclear translocation, and transcriptional activity of STAT3. For instance, eriocalyxin B covalently binds Cys712 of STAT3 and blocks phosphorylation and subsequent activation of STAT3 [47]. We found that 15-keto PGE2 directly inhibited STAT3 phosphorylation without affecting the JAK2 activity and the expression of SOCS3. As a result, the dimerization, nuclear translocation and transcriptional activity of STAT3 were repressed.
15-Keto PGE2 has an electrophilic α,β-unsaturated carbonyl group that can covalently modify nucleophilic cysteine residue(s) present in various proteins, regulating their functions/activities [16]. We observed that the α,β-unsaturated carbonyl moiety of 15-keto PGE2 plays an important role in directly blocking the STAT3 activation through covalent modification of Cys259 of STAT3. In support of this notion, STAT3 signaling was not inhibited by the non-electrophilic analogue, 13,14-dihydro-15-keto PGE2 lacking the α,β-unsaturated carbonyl group. The interaction between 15-keto PGE2 and STAT3 was disrupted by the thiol reducing agent, DTT, suggesting that the Michael addition reaction occurs between the α,β-unsaturated carbonyl group of 15-keto PGE2 and cysteine residue(s) in STAT3.
The STAT3 protein structurally consists of 6 domains [41]. These include N-terminal domain, a coiled-coil domain, a DNA binding domain, a linker domain, Src-homology 2 (SH2) domain and C-terminal transactivation domain. Of note, Cys418, Cys426, Cys468, Cys542, Cys550, Cys687, Cys712, and Cys718 located in the SH2 domain and the transactivation domain are known to be involved in STAT3 phosphorylation, dimerization, and activation [50]. Some compounds containing the α,β-unsaturated carbonyls group directly target the cysteine residues of STAT3, thereby deactivating this transcription factor. For example, eriocalyxin B having two α,β-unsaturated carbonyls directly interacts with STAT3 at the Cys712 of STAT3, which hampers the phosphorylation of STAT3 [47]. A small molecule compound, NSC-368,262 (C48) alkylates Cys468 of STAT3, resulting in suppression of STAT3 DNA binding [42]. Distinct from these compounds, 15-keto PGE2 interacts directly with Cys259 located in the coiled-coil domain of STAT3 to form a covalent linkage. In our previous study, curcumin, a natural polyphenolic compound present in the turmeric, induces apoptosis in MCF10A-ras cells through direct targeting Cys259 of STAT3 [51]. Zhang et al. reported that the coiled-coil domain also interacts directly with the gp130 peptide like SH2 domain which is required for STAT3 phosphorylation [52].
Other prostaglandin metabolites, especially A series of prostaglandins which also have an α,β-unsaturated carbonyl group, modulates the intracellular signaling involved cell proliferation and differentiation [53]. For instance, PGA2 acts as an electrophile inducing a conformation change of p53, thereby impairing p53-mediated apoptosis [54]. 15-Deoxy Δ12,14 prodtaglandin J2 (15d-PGJ2) undergoes nucleophilic addition to Cys136 on PTEN, thereby activating PI3K-Akt signaling [55]. 15d-PGJ2 inhibits IL-6-stimulated phosphorylation of STAT3 in endothelial cells which is attributable to its electrophilic nature [56]. This cyclopentenone prostaglandin also covalently modify the thiol groups of Keap1 [57] and other proteins including PPARγ [16,58].
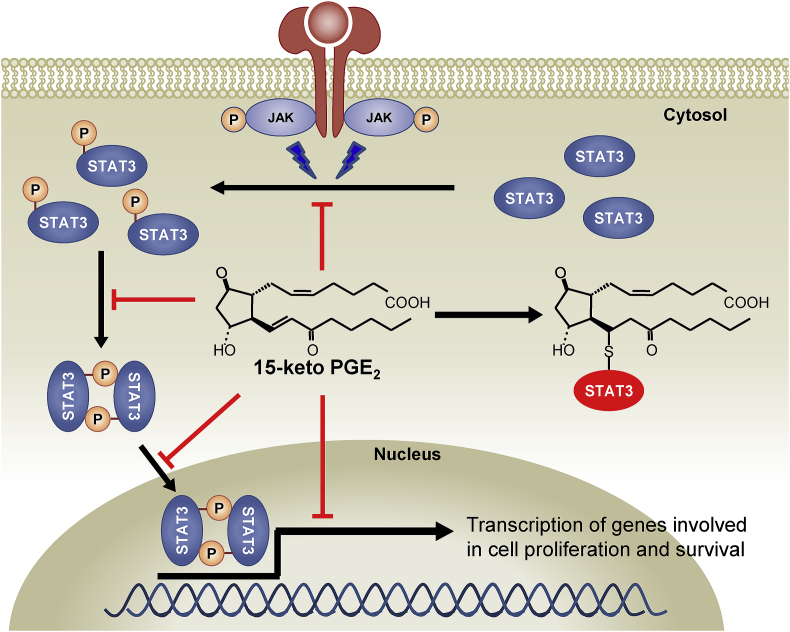
The compounds targeting STAT3 inhibit the growth of cells and induces apoptosis in cancers [22,29,48,52]. We observed that suppression of STAT3 signaling by 15-keto PGE2 accompanies proliferation of breast cancer cells and tumor growth. There has been no previous study evaluating the antitumor effect of 15-keto PGE2 in an animal model. Our present study demonstrates for the first time that the growth of xenograft tumors is suppressed by subcutaneous administration of 15-keto PGE2. In conclusion, 15-keto PGE2 inhibits STAT3 signaling through covalent modification, and suppresses breast cancer growth and progression (Fig. 8).
Fig. 8.
A proposed mechanism underlying suppression of STAT3 signaling and growth and progression of breast cancer by 15-keto PGE2. 15-Keto PGE2 covalently binds to STAT3, and this hampers the phosphorylation, dimerization, nuclear localization and transcriptional activity of STAT3.
Acknowledgements
This study was supported by Basic Science Research Program through the National Research Foundation, Ministry of Science and ICT (Grant No. 2012015106) and the Global Core Research Center (GCRC) grant (No. 2011-0030001) from the National Research Foundation Republic of Korea and the GRRC program of Gyeonggi province [GRRC-kyunghee2019(B03)], Republic of Korea.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101175.
Contributor Information
Hye-Kyung Na, Email: nhk1228@sungshin.ac.kr.
Young-Joon Surh, Email: surh@snu.ac.kr.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Nakanishi M., Rosenberg D.W. Multifaceted roles of PGE2 in inflammation and cancer. Semin. Immunopathol. 2013;35:123–137. doi: 10.1007/s00281-012-0342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rigas B., Goldman I.S., Levine L. Altered eicosanoid levels in human colon cancer. J. Lab. Clin. Med. 1993;122:518–523. [PubMed] [Google Scholar]
- 3.Schrey M.P., Patel K.V. Prostaglandin E2 production and metabolism in human breast cancer cells and breast fibroblasts. Regulation by inflammatory mediators. Br. J. Cancer. 1995;72:1412–1419. doi: 10.1038/bjc.1995.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., Dubois R.N. Eicosanoids and cancer. Nat. Rev. Canc. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenhough A., Smartt H.J., Moore A.E., Roberts H.R., Williams A.C., Paraskeva C., Kaidi A. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377–386. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 6.Legler D.F., Bruckner M., Uetz-von Allmen E., Krause P. Prostaglandin E2 at new glance: novel insights in functional diversity offer therapeutic chances. Int. J. Biochem. Cell Biol. 2010;42:198–201. doi: 10.1016/j.biocel.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Wang M.T., Honn K.V., Nie D. Cyclooxygenases, prostanoids, and tumor progression. Cancer Metastasis Rev. 2007;26:525–534. doi: 10.1007/s10555-007-9096-5. [DOI] [PubMed] [Google Scholar]
- 8.Backlund M.G., Mann J.R., Holla V.R., Buchanan F.G., Tai H.H., Musiek E.S., Milne G.L., Katkuri S., DuBois R.N. 15-Hydroxyprostaglandin dehydrogenase is down-regulated in colorectal cancer. J. Biol. Chem. 2005;280:3217–3223. doi: 10.1074/jbc.M411221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo S.H., Kang M.S., Kim K.H., An M.S., Ha T.K., Bae K.B., Oh M.K., Choi C.S., Oh S.H., Choi Y.K. Correlation of 15-prostagladin dehydrogenase expression with clinicopathological factors and survival rate in gastric adenocarcinoma. Int. J. Surg. (London, England) 2015;13:96–101. doi: 10.1016/j.ijsu.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Tseng-Rogenski S., Gee J., Ignatoski K.W., Kunju L.P., Bucheit A., Kintner H.J., Morris D., Tallman C., Evron J., Wood C.G., Grossman H.B., Lee C.T., Liebert M. Loss of 15-hydroxyprostaglandin dehydrogenase expression contributes to bladder cancer progression. Am. J. Pathol. 2010;176:1462–1468. doi: 10.2353/ajpath.2010.090875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf I., O'Kelly J., Rubinek T., Tong M., Nguyen A., Lin B.T., Tai H.H., Karlan B.Y., Koeffler H.P. 15-hydroxyprostaglandin dehydrogenase is a tumor suppressor of human breast cancer. Cancer Res. 2006;66:7818–7823. doi: 10.1158/0008-5472.CAN-05-4368. [DOI] [PubMed] [Google Scholar]
- 12.Myung S.J., Rerko R.M., Yan M., Platzer P., Guda K., Dotson A., Lawrence E., Dannenberg A.J., Lovgren A.K., Luo G., Pretlow T.P., Newman R.A., Willis J., Dawson D., Markowitz S.D. 15-Hydroxyprostaglandin dehydrogenase is an in vivo suppressor of colon tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12098–12102. doi: 10.1073/pnas.0603235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaliberova L.N., Kusmartsev S.A., Krendelchtchikova V., Stockard C.R., Grizzle W.E., Buchsbaum D.J., Kaliberov S.A. Experimental cancer therapy using restoration of NAD+-linked 15-hydroxyprostaglandin dehydrogenase expression. Mol. Canc. Therapeut. 2009;8:3130–3139. doi: 10.1158/1535-7163.MCT-09-0270. [DOI] [PubMed] [Google Scholar]
- 14.Kim H.R., Lee H.N., Lim K., Surh Y.J., Na H.K. 15-Deoxy-Δ12,14-prostaglandin J2 induces expression of 15-hydroxyprostaglandin dehydrogenase through Elk-1 activation in human breast cancer MDA-MB-231 cells. Mutat. Res. 2014;768:6–15. doi: 10.1016/j.mrfmmm.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Tai H.H. Prostaglandin catabolic enzymes as tumor suppressors. Cancer Metastasis Rev. 2011;30:409–417. doi: 10.1007/s10555-011-9314-z. [DOI] [PubMed] [Google Scholar]
- 16.Shiraki T., Kamiya N., Shiki S., Kodama T.S., Kakizuka A., Jingami H. Alpha,beta-unsaturated ketone is a core moiety of natural ligands for covalent binding to peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 2005;280:14145–14153. doi: 10.1074/jbc.M500901200. [DOI] [PubMed] [Google Scholar]
- 17.Yao L., Chen W., Song K., Han C., Gandhi C.R., Lim K., Wu T. 15-hydroxyprostaglandin dehydrogenase (15-PGDH) prevents lipopolysaccharide (LPS)-induced acute liver injury. PLoS One. 2017;12:e0176106. doi: 10.1371/journal.pone.0176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen I.J., Hee S.W., Liao C.H., Lin S.Y., Su L., Shun C.T., Chuang L.M. Targeting the 15-keto-PGE2-PTGR2 axis modulates systemic inflammation and survival in experimental sepsis. Free Radic. Biol. Med. 2018;115:113–126. doi: 10.1016/j.freeradbiomed.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Lu D., Han C., Wu T. 15-PGDH inhibits hepatocellular carcinoma growth through 15-keto-PGE2/PPARγ-mediated activation of p21WAF1/Cip1. Oncogene. 2014;33:1101–1112. doi: 10.1038/onc.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang E.Y., Chang Y.C., Shun C.T., Tien Y.W., Tsai S.H., Hee S.W., Chen I.J., Chuang L.M. Inhibition of prostaglandin reductase 2, a putative oncogene overexpressed in human pancreatic adenocarcinoma, induces oxidative stress-mediated cell death involving xCT and CTH gene expressions through 15-keto-PGE2. PLoS One. 2016;11:e0147390. doi: 10.1371/journal.pone.0147390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy D.E., Darnell J.E., Jr. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 22.Turkson J. STAT proteins as novel targets for cancer drug discovery. Expert Opin. Ther. Targets. 2004;8:409–422. doi: 10.1517/14728222.8.5.409. [DOI] [PubMed] [Google Scholar]
- 23.Darnell J.E., Jr. STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 24.Yue P., Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin. Investig. Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal B.B., Kunnumakkara A.B., Harikumar K.B., Gupta S.R., Tharakan S.T., Koca C., Dey S., Sung B. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Ann NY Acad Sci. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee K., Resat H. Constitutive activation of STAT3 in breast cancer cells: a review. Int. J. Cancer. 2016;138:2570–2578. doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh F.C., Cheng G., Lin J. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem. Biophys. Res. Commun. 2005;335:292–299. doi: 10.1016/j.bbrc.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 28.Inghirami G., Chiarle R., Simmons W.J., Piva R., Schlessinger K., Levy D.E. New and old functions of STAT3: a pivotal target for individualized treatment of cancer. Cell Cycle. 2005;4:1131–1133. doi: 10.4161/cc.4.9.1985. [DOI] [PubMed] [Google Scholar]
- 29.Liu D.B., Hu G.Y., Long G.X., Qiu H., Mei Q., Hu G.Q. Celecoxib induces apoptosis and cell-cycle arrest in nasopharyngeal carcinoma cell lines via inhibition of STAT3 phosphorylation. Acta Pharmacol. Sin. 2012;33:682–690. doi: 10.1038/aps.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato Y., Mukai K., Watanabe S., Goto M., Shimosato Y. The AMeX method. A simplified technique of tissue processing and paraffin embedding with improved preservation of antigens for immunostaining. Am. J. Pathol. 1986;125:431–435. [PMC free article] [PubMed] [Google Scholar]
- 31.Watson J., Chuah S.Y. Technique for the primary culture of human breast cancer cells and measurement of their prostaglandin secretion. Clin. Sci. (Lond.) 1992;83:347–352. doi: 10.1042/cs0830347. [DOI] [PubMed] [Google Scholar]
- 32.Wright K.L., Adams J.R., Liu J.C., Loch A.J., Wong R.G., Jo C.E., Beck L.A., Santhanam D.R., Weiss L., Mei X., Lane T.F., Koralov S.B., Done S.J., Woodgett J.R., Zacksenhaus E., Hu P., Egan S.E. Ras signaling is a key determinant for metastatic dissemination and poor survival of luminal breast cancer patients. Cancer Res. 2015;75:4960–4972. doi: 10.1158/0008-5472.CAN-14-2992. [DOI] [PubMed] [Google Scholar]
- 33.Zushi S., Shinomura Y., Kiyohara T., Miyazaki Y., Kondo S., Sugimachi M., Higashimoto Y., Kanayama S., Matsuzawa Y. STAT3 mediates the survival signal in oncogenic ras-transfected intestinal epithelial cells. Int. J. Cancer. 1998;78:326–330. doi: 10.1002/(SICI)1097-0215(19981029)78:3<326::AID-IJC12>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 34.Yu H., Lee H., Herrmann A., Buettner R., Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat. Rev. Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 35.Carpenter R.L., Lo H.W. STAT3 target genes relevant to human cancers. Cancers. 2014;6:897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sansone P., Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J. Clin. Oncol. 2012;30:1005–1014. doi: 10.1200/JCO.2010.31.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carow B., Rottenberg M.E. SOCS3, a major regulator of infection and inflammation. Front. Immunol. 2014;5:58. doi: 10.3389/fimmu.2014.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y.H., Ko T.P., Guo R.T., Hu S.M., Chuang L.M., Wang A.H. Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2. Structure. 2008;16:1714–1723. doi: 10.1016/j.str.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Li L., Shaw P.E. A STAT3 dimer formed by inter-chain disulphide bridging during oxidative stress. Biochem. Biophys. Res. Commun. 2004;322:1005–1011. doi: 10.1016/j.bbrc.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Kim J., Won J.S., Singh A.K., Sharma A.K., Singh I. STAT3 regulation by S-nitrosylation: implication for inflammatory disease. Antioxid Redox Signal. 2014;20:2514–2527. doi: 10.1089/ars.2013.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heidelberger S., Zinzalla G., Antonow D., Essex S., Basu B.P., Palmer J., Husby J., Jackson P.J., Rahman K.M., Wilderspin A.F., Zloh M., Thurston D.E. Investigation of the protein alkylation sites of the STAT3:STAT3 inhibitor Stattic by mass spectrometry. Bioorg. Med. Chem. Lett. 2013;23:4719–4722. doi: 10.1016/j.bmcl.2013.05.066. [DOI] [PubMed] [Google Scholar]
- 42.Buettner R., Corzano R., Rashid R., Lin J., Senthil M., Hedvat M., Schroeder A., Mao A., Herrmann A., Yim J., Li H., Yuan Y.C., Yakushijin K., Yakushijin F., Vaidehi N., Moore R., Gugiu G., Lee T.D., Yip R., Chen Y., Jove R., Horne D., Williams J.C. Alkylation of cysteine 468 in Stat3 defines a novel site for therapeutic development. ACS Chem. Biol. 2011;6:432–443. doi: 10.1021/cb100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Don-Doncow N., Escobar Z., Johansson M., Kjellstrom S., Garcia V., Munoz E., Sterner O., Bjartell A., Hellsten R. Galiellalactone is a direct inhibitor of the transcription factor STAT3 in prostate cancer cells. J. Biol. Chem. 2014;289:15969–15978. doi: 10.1074/jbc.M114.564252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad R., Raina D., Meyer C., Kufe D. Triterpenoid CDDO-methyl ester inhibits the Janus-activated kinase-1 (JAK1)-->signal transducer and activator of transcription-3 (STAT3) pathway by direct inhibition of JAK1 and STAT3. Cancer Res. 2008;68:2920–2926. doi: 10.1158/0008-5472.CAN-07-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miklossy G., Hilliard T.S., Turkson J. Therapeutic modulators of STAT signalling for human diseases. Nat. Rev. Drug Discov. 2013;12:611–629. doi: 10.1038/nrd4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Graness A., Poli V., Goppelt-Struebe M. STAT3-independent inhibition of lysophosphatidic acid-mediated upregulation of connective tissue growth factor (CTGF) by cucurbitacin I. Biochem. Pharmacol. 2006;72:32–41. doi: 10.1016/j.bcp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Yu X., He L., Cao P., Yu Q. Eriocalyxin B inhibits STAT3 signaling by covalently targeting STAT3 and blocking phosphorylation and activation of STAT3. PLoS One. 2015;10:e0128406. doi: 10.1371/journal.pone.0128406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin D.S., Kim H.N., Shin K.D., Yoon Y.J., Kim S.J., Han D.C., Kwon B.M. Cryptotanshinone inhibits constitutive signal transducer and activator of transcription 3 function through blocking the dimerization in DU145 prostate cancer cells. Cancer Res. 2009;69:193–202. doi: 10.1158/0008-5472.CAN-08-2575. [DOI] [PubMed] [Google Scholar]
- 49.Siddiquee K., Zhang S., Guida W.C., Blaskovich M.A., Greedy B., Lawrence H.R., Yip M.L., Jove R., McLaughlin M.M., Lawrence N.J., Sebti S.M., Turkson J. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu X., Sun Y.L., Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 51.Hahn Y.I., Kim S.J., Choi B.Y., Cho K.C., Bandu R., Kim K.P., Kim D.H., Kim W., Park J.S., Han B.W., Lee J., Na H.K., Cha Y.N., Surh Y.J. Curcumin interacts directly with the Cysteine 259 residue of STAT3 and induces apoptosis in H-Ras transformed human mammary epithelial cells. Sci. Rep. 2018;8:6409. doi: 10.1038/s41598-018-23840-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang T., Kee W.H., Seow K.T., Fung W., Cao X. The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol. Cell Biol. 2000;20:7132–7139. doi: 10.1128/mcb.20.19.7132-7139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honn K.V., Marnett L.J. Requirement of a reactive alpha, beta-unsaturated carbonyl for inhibition of tumor growth and induction of differentiation by “A” series prostaglandins. Biochem. Biophys. Res. Commun. 1985;129:34–40. doi: 10.1016/0006-291x(85)91398-1. [DOI] [PubMed] [Google Scholar]
- 54.Moos P.J., Edes K., Fitzpatrick F.A. Inactivation of wild-type p53 tumor suppressor by electrophilic prostaglandins. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9215–9220. doi: 10.1073/pnas.160241897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suh J., Kim D.H., Kim E.H., Park S.A., Park J.M., Jang J.H., Kim S.J., Na H.K., Kim N.D., Kim N.J., Suh Y.G., Surh Y.J. 15-deoxy-Δ12,14-prostaglandin J2 activates PI3K-Akt signaling in human breast cancer cells through covalent modification of the tumor suppressor PTEN at cysteine 136. Cancer Lett. 2018;424:30–45. doi: 10.1016/j.canlet.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 56.Wung B.S., Wu C.C., Hsu M.C., Hsieh C.W. 15-deoxy-Δ12,14-prostaglandin J2 suppresses IL-6-induced STAT3 phosphorylation via electrophilic reactivity in endothelial cells. Life Sci. 2006;78:3035–3042. doi: 10.1016/j.lfs.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 57.Oh J.Y., Giles N., Landar A., Darley-Usmar V. Accumulation of 15-deoxy-Δ12,14-prostaglandin J2 adduct formation with Keap1 over time: effects on potency for intracellular antioxidant defence induction. Biochem. J. 2008;411:297–306. doi: 10.1042/bj20071189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez-Gomez F.J., Cernuda-Morollon E., Stamatakis K., Perez-Sala D. Protein thiol modification by 15-deoxy-Δ12,14-prostaglandin J2 addition in mesangial cells: role in the inhibition of pro-inflammatory genes. Mol. Pharmacol. 2004;66:1349–1358. doi: 10.1124/mol.104.002824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.