Abstract
Epididymitis is a commonly diagnosed disease associated with male infertility. However, little is known about the molecules that are involved in its development. This study was to identify critical genes associated with lipopolysaccharide-induced epididymitis and analyze the molecular mechanism of epididymitis through RNA sequencing. Experimental epididymitis models were generated by administering male Sprague–Dawley rats' lipopolysaccharide. A total of 1378 differentially expressed genes, including 531 upregulated and 847 downregulated genes, were identified in the epididymitis model rats compared with those in sham-operated rats by RNA sequencing. Functional enrichment analyses suggested that the upregulated genes were markedly enriched in inflammation-related biological processes, as well as in the tumor necrosis factor (TNF) signaling pathway, cytokine–cytokine receptor interactions, complement and coagulation cascades, and in the chemokine signaling pathway. Four downregulated genes (collagen type XXVIII alpha 1 chain [Col28α1], cyclin-dependent kinase-like 1 [Cdkl1], phosphoserine phosphatase [Psph], and fatty acid desaturase 2 [Fads2]) and ten upregulated genes (CCAAT/enhancer-binding protein beta [Cebpβ], C-X-C motif chemokine receptor 2 [Cxcr2], interleukin 11 [Il11], C-C motif chemokine ligand 20 [Ccl20], nuclear factor-kappa-B inhibitor alpha [Nfkbiα], claudin 4 [Cldn4], matrix metallopeptidase 9 [Mmp9], heat shock 70 kDa protein 8 [Hspa8], intercellular cell adhesion molecule-1 [Icam1], and Jun) were successfully confirmed by real-time polymerase chain reaction. Western blot demonstrated that CDKL1 was decreased, while MMP9 and NFKBIA were increased in the experimental model group compared with those in the sham-operated group. Our study sheds new light on the understanding of the early response of the epididymis during bacterial epididymitis.
Keywords: differentially expressed genes, epididymitis, inflammation, lipopolysaccharide
INTRODUCTION
Epididymitis is a disease characterized by the inflammation of the epididymis and is a commonly diagnosed factor in the investigation of male infertility. It frequently occurs in those aged 15–35 years who are sexually active.1,2 In the United States, epididymitis is estimated to be diagnosed in 0.7% of men aged 18–50 years who seek medical care.2 Epididymitis can be classified as either acute or chronic depending on the duration of the symptoms. Both acute and chronic epididymitis are treated with antibiotic and antiphlogistic pharmacotherapy, including fluoroquinolones, azithromycin, ciprofloxacin, piroxicam, ketorolac, and doxycycline. However, sperm counts cannot recover to normal in about 50% of men even after successful eradication of the pathogen by antibiotic treatment.3 Epididymitis can lead to male infertility; therefore, there is an urgent need to discover other therapies for epididymitis.
Bacterial infections are the most prevalent etiological factors of epididymitis.4 However, little is known about how the epididymis responds to invading bacteria. Lipopolysaccharides (LPS) are major components of the outer membrane of Gram-negative bacteria. Previous studies have established rat models of epididymitis by injecting LPS into the vas deferens.5,6 Cao et al.6 demonstrated that LPS-induced epididymitis decreases the expression of epididymal beta-defensins, and that the disruption of sperm associated antigen 11 (SPAG11E) expression is involved in the impairment of sperm motility. In another study, Silva et al.5 demonstrated that LPS stimulates an inflammatory response by upregulating the mRNA expression of proinflammatory cytokines such as interleukin 1 beta (Il1β), tumor necrosis factor (Tnf), interleukin 6 (Il6), and interferon gamma (Ifnγ). However, comprehensive studies on the response of epididymis caused by infections with Gram-negative bacteria are limited.
In this study, we analyzed the molecular mechanism of epididymitis in an animal model by RNA sequencing. The differentially expressed genes (DEGs) in response to the injected LPS were identified and analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Some DEGs were further studied by real-time polymerase chain reaction (PCR). We anticipate our results to be helpful in the development of therapies for epididymitis.
MATERIALS AND METHODS
Animals
Adult male Sprague–Dawley (SD) rats (aged 4 months old) were purchased from SLAC laboratory animal center (Shanghai, China) and were acclimatized in a room with controlled light (12 h light and 12 h dark), temperature of 26°C, and humidity of 50% for 1 week. Food and water were supplied ad libitum. All animal experiments were conducted according to the Guidelines for Animal Care and Use of Laboratory Animal and were approved by the Animal Ethics Committee of Renji Hospital (Shanghai, China).
Establishment of animal models of epididymitis
Twenty SD rats were randomly divided into two groups (n = 10 for each group). In the model group, rats were anesthetized with 3% (v/v) pentobarbital sodium (50 mg per kg body weight, intraperitoneally, DaTianFengTuo, Beijing, China). A 1.5–2-cm ventral midline incision was made to expose the testis and epididymis. The vas deferens was clamped with hemostats clamp, and 50 μl sterile saline containing 200 μg LPS (Yuanye Biotech Co., Ltd., Shanghai, China) was injected into the caput region of the epididymis with an insulin injection needle. Subsequently, the skin was closed. In the sham-operated group, rats underwent the same procedure, but were injected with sterile saline. Rats were sacrificed after 24 h of treatment, and the epididymal portion was dissected and washed with sterile saline. Tissues were stored at −80°C until use.
Examination of animal model by real-time PCR and enzyme-linked immunosorbent assay (ELISA)
The epididymitis model was examined by real-time PCR and ELISA. Frozen epididymal samples from rats in the model and sham-operated groups were homogenized in Trizol (Invitrogen, Carlsbad, CA, USA) in a tissue homogenizer (Tiangen Biotech Co., Ltd., Beijing, China). Total RNA was extracted and reverse-transcribed with a PrimeScript RT Master Mix (Takara Biotech Co., Ltd., Dalian, China) in a total volume of 20 μl. cDNA samples were amplified with SYBR Green PCR Master Mix (Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) in a 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) under the following cycle conditions: 50°C for 2 min, 95°C for 2 min, and 40 cycles of 95°C (15 s) and 60°C (60 s). Primers used for the PCR experiments are displayed in Supplementary Table 1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference gene to calculate the relative expression of each target gene.
Supplementary Table 1.
The primers used in real-time polymerase chain reaction
| Gene | Sequence (5’-3’) |
|---|---|
| Col28a1-ratF | GAAGACTCTGGCTGACCG |
| Col28a1-ratR | TCCTTGCTGGAGAACTGC |
| Cdkl1-ratF | AGCATCCCAACCTCGTCA |
| Cdkl1-ratR | CGTGCAAACCCAAAGTCA |
| Psph-ratF | GGGGATAAGGGAGCTGGTA |
| Psph-ratR | GGCTGCGTCTCGTCAAAA |
| Fads2-ratF | TTCCAGATTGAGCACCAC |
| Fads2-ratR | GTAGGCATCCAGCCACAG |
| Cebpb-rF | GCGCCATCGACTTCAG |
| Cebpb-rR | CGGACGGCTTCTTGCT |
| Cxcr2-rF | CATCCTGCCTCAGACCTA |
| Cxcr2-rR | AAGCCAAGAATCTCAGTAGC |
| Il11-rF | GCCAGATAGAGTCGTTGC |
| Il11-rR | AGGTAGGGAGTCCAGATTG |
| Cxcl6-rF | CAAGACATTACGGGCTATT |
| Cxcl6-rR | CTGGCACATCCTAACTCC |
| Ccl20-rF | CTGCTGGCTTACCTCTGC |
| Ccl20-rR | ATTTCCTCCTTGGGCTGT |
| Nfkbia-rF | CCTGGTCTCGCTCCTGTT |
| Nfkbia-rR | TCATCGTAGGGCAACTCA |
| Cldn4-rF | GTGGCTGGACAGGTTTGA |
| Cldn4-rR | GCAAGACCGTATGGGAAA |
| Mmp9-rF | GCATCTGTATGGTCGTGGCT |
| Mmp9-rR | CTGTAGGGGCCTCAGAAGGA |
| Hspa8-rF | GATTTGCTGCTCTTGGAT |
| Hspa8-rR | TGGTGAAAGTCTGGGTCT |
| Icam1-rF | AAACGGGAGATGAATGGT |
| Icam1-rR | TCTGGCGGTAATAGGTGTA |
| Jun-rF | GTGCCAACTCATGCTAACG |
| Jun-rR | TCTGTCGCAACCAGTCAAG |
Col28α1: collagen type XXVIII alpha 1 chain; Psph: phosphoserine phosphatase; Fads2: fatty acid desaturase; Cdkl1: cyclin-dependent kinase-like 1; Cxcl6: C-X-C motif chemokine ligand 6; Mmp9: matrix metallopeptidase 9; Nfκbiα: nuclear factor-kappa-B inhibitor alpha; Il11: interleukin 11; Icam1: intercellular cell adhesion molecule-1; Hspa8: heat shock 70 kDa protein 8; Cldn4: claudin 4; Cebpβ: CCAAT/enhancer-binding protein beta; Ccl20: C-C motif chemokine ligand 20; Cxcr2: C-X-C motif chemokine receptor 2
The protein concentrations of IL1β, TNF-α, and IL6 in epididymitis tissue samples were analyzed via ELISA kits from Thermo Fisher Scientific according to the manufacturer's instructions. A multimode reader (Thermo Fisher Scientific) was used to assess the absorbance at 450 nm. Each measurement was performed in triplicate.
RNA library construction, sequence, and data analysis
Total RNA from the epididymal samples was extracted using Trizol (Invitrogen), and the quality and concentration of the RNA were evaluated by NanoDrop 2000 (Thermo Fisher Scientific). RNA integrity was assessed using the RNA Nano 6000 Assay kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). RNA sequencing libraries were constructed with NEB NextUltra™ RNA Library Prep Kit for Illumina (New England BioLabs, Inc., Ipswich, MA, USA) following the manufacturer's recommendations. The amplified library fragments were purified, and the library quality was assessed on the Agilent Bioanalyzer 2100 system (Agilent Technologies). The quality-checked libraries were sequenced with the Illumina Hiseq2500 instrument (Illumina, Inc., San Diego, CA, USA), and paired-end reads were generated. Raw data in FASTQ format were first processed using in-house Perl scripts. Clean reads were obtained by removing reads containing adapter, ploy-N, and low-quality reads. The GC content and Q20 were calculated to evaluate the quality of sequencing. The clean reads were mapped to a reference genome with Hisat2 tools7 (http://ccb.jhu.edu/software/hisat2/index.shtml), and only reads with either a perfect match or one mismatch were further assembled into transcripts by String Tie (http://ccb.jhu.edu/software/stringtie/).8 The raw reads were uploaded to NCBI with the accession number SRP157380.
Identification of differentially expressed genes
Gene expression values were calculated by fragments per kilobase of exon per million fragments mapped (FPKM).9,10 DEGs between model and sham-operated groups were identified using DESeq11 with cutoff values of fold change ≥2 and false discovery rate (FDR) <0.01.
Functional enrichment analysis of DEGs
The functions of DEGs were annotated by GO, which included three subcategories: molecular function (MF), biological process (BP), and cellular component (CC).12 In addition, KEGG pathway analysis was performed to identify the signaling pathways in which the DEGs were enriched.13,14 GO and KEGG enrichment analyses were performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.7 with threshold of FDR <0.05.15
Confirmation of DEGs with real-time PCR
Fifteen DEGs were selected for confirmation through real-time PCR. The procedures were similar to those above, and the primers used are listed in Table 1.
Table 1.
The gene ontology enrichment results of upregulated genes
| Category | Term | Count | P | FDR |
|---|---|---|---|---|
| GOTERM_BP | GO:0006954~inflammatory response | 34 | 5.03×10−14 | 8.86×10−11 |
| GOTERM_BP | GO:0032496~response to lipopolysaccharide | 33 | 5.63×10−14 | 9.92×10−11 |
| GOTERM_BP | GO:0071222~cellular response to lipopolysaccharide | 23 | 3.27×10−11 | 5.76×10−8 |
| GOTERM_BP | GO:0042060~wound healing | 20 | 6.81×10−11 | 1.20×10−7 |
| GOTERM_BP | GO:0071347~cellular response to interleukin-1 | 18 | 7.11×10−11 | 1.25×10−7 |
| GOTERM_BP | GO:0045766~positive regulation of angiogenesis | 19 | 1.55×10−10 | 2.73×10−7 |
| GOTERM_BP | GO:0007155~cell adhesion | 27 | 2.98×10−10 | 5.24×10−7 |
| GOTERM_BP | GO:0030593~neutrophil chemotaxis | 14 | 4.50×10−10 | 7.94×10−7 |
| GOTERM_BP | GO:0070374~positive regulation of ERK1 and ERK2 cascade | 22 | 1.66×10−9 | 2.92×10−6 |
| GOTERM_BP | GO:1902042~negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 9 | 1.62×10−8 | 2.86×10−5 |
| GOTERM_BP | GO:0042127~regulation of cell proliferation | 23 | 2.06×10−8 | 3.62×10−5 |
| GOTERM_BP | GO:0010628~positive regulation of gene expression | 29 | 3.13×10−8 | 5.52×10−5 |
| GOTERM_BP | GO:0050900~leukocyte migration | 10 | 6.25×10−8 | 1.10×10−4 |
| GOTERM_BP | GO:0071356~cellular response to tumor necrosis factor | 16 | 3.69×10−7 | 6.49×10−4 |
| GOTERM_BP | GO:0007568~aging | 23 | 3.46×10−6 | 0.00609 |
| GOTERM_BP | GO:0042493~response to drug | 31 | 4.39×10−6 | 0.007737 |
| GOTERM_BP | GO:0030335~positive regulation of cell migration | 18 | 4.76×10−6 | 0.008396 |
| GOTERM_BP | GO:0043066~negative regulation of apoptotic process | 30 | 8.00×10−6 | 0.014103 |
| GOTERM_BP | GO:0045087~innate immune response | 20 | 8.87×10−6 | 0.015625 |
| GOTERM_BP | GO:0034097~response to cytokine | 12 | 1.55×10−5 | 0.027241 |
| GOTERM_BP | GO:0048661~positive regulation of smooth muscle cell proliferation | 11 | 1.84×10−5 | 0.032453 |
| GOTERM_BP | GO:0001525~angiogenesis | 16 | 2.10×10−5 | 0.037084 |
| GOTERM_MF | GO:0002020~protease binding | 14 | 5.26×10−6 | 0.007882 |
| GOTERM_MF | GO:0008009~chemokine activity | 8 | 1.62×10−5 | 0.02426 |
| GOTERM_MF | GO:0005125~cytokine activity | 15 | 1.83×10−5 | 0.027364 |
| GOTERM_CC | GO:0005615~extracellular space | 94 | 1.10×10−23 | 1.50×10−20 |
| GOTERM_CC | GO:0009986~cell surface | 47 | 8.98×10−13 | 1.22×10−9 |
| GOTERM_CC | GO:0005576~extracellular region | 49 | 1.53×10−11 | 2.08×10−8 |
| GOTERM_CC | GO:0070062~extracellular exosome | 109 | 2.86×10−10 | 3.89×10−7 |
| GOTERM_CC | GO:0031012~extracellular matrix | 24 | 1.84×10−8 | 2.50×10−5 |
| GOTERM_CC | GO:0009897~external side of plasma membrane | 23 | 1.99×10−7 | 2.72×10−4 |
| GOTERM_CC | GO:0045121~membrane raft | 21 | 2.97×10−6 | 0.00405 |
GO: Gene Ontology; BP: biological process; CC: cellular component; FDR: false discovery rate
Confirmation of DEGs by western blot
The epididymal tissues were lysed at 4°C with radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of Biotechnology, Shanghai, China) containing 1 mmol l−1 phenylmethanesulfonyl fluoride (Beyotime) and harvested for 30 min on ice. Following centrifugation (Eppendorf, Hamburg, Germany) at 12 000g for 10 min at 4°C, the supernatant was used for protein electrophoresis. The total protein concentrations were determined with a bicinchoninic acid (BCA) protein assay kit (Boster biological technology Co., Ltd., Wuhan, China) according to the manufacturer's protocol. A total of 20 μg of protein was separated on 12% (w/v) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were subsequently transferred electrophoretically onto polyvinylidene fluoride membranes (Millipore, Burlington, MA, USA). The membranes were immersed in blocking solution (0.05% [v/v] Tween 20 [Sigma, St. Louis, MO, USA] and 5% [w/v] bovine serum albumin [BSA; BBI Life Sciences Corp., Shanghai, China]) in Tris-buffered saline (Sangon Biotech, Shanghai, China) for 2 h at room temperature (25°C) and incubated with primary rabbit polyclonal antibodies against cyclin-dependent kinase-like 1 (CDKL1; Cat. No. PA5-45998; 1:1000; Thermo Fisher Scientific), nuclear factor-kappa-B inhibitor alpha (NFKBIA; Cat. No. 10268-1-AP; 1:500; Proteintech Group Inc., Rosemont, IL, USA), matrix metallopeptidase 9 (MMP-9; Cat. No. 10375-2-AP; 1:500; Proteintech), C-C motif chemokine ligand 20 (CCL20; Cat. No. 26527-1-AP; 1:2000; Proteintech), or GAPDH (Cat. No. 10494-1-AP; 1:2000; Proteintech) overnight at 4°C. The membranes were incubated with horseradish peroxidase-conjugated secondary antibodies against Cdkl1, Nfκbiα, Mmp-9, Ccl20, and GAPDH (anti-rabbit IgG; 1:10000; Cat. No. ab7090; Abcam) at 37°C for 2 h, which were detected using an enhanced chemiluminescence detection system (Amersham; GE Healthcare, Chicago, IL, USA). The intensity of the bands was analyzed by ImageJ software version 1.4.6 (National Institutes of Health, Bethesda, MD, USA).
Statistical analyses
Data except for those from RNA-seq were expressed as mean ± standard error of the mean (s.e.m.) and were processed in SPSS 21.0 (IBM, Chicago, CA, USA). Differences between the model and sham-operated groups were analyzed by Student's t-test. P < 0.05 was considered statistically significant.
RESULTS
LPS injection and the mRNA and protein levels of proinflammatory genes in the rat epididymitis model
On the basis of a previous study,5 we established a rat epididymitis model by injecting LPS into the head of the epididymis. The success of inducing epididymitis was demonstrated by real-time PCR and ELISA. After 24 h of LPS injection, the mRNA expression levels of pro-inflammatory factors Il1β, Il6, and Tnf-α were increased in the model group compared with those in the sham-operated group (P < 0.05; Figure 1a). Similarly, the protein levels of these proinflammatory factors were also markedly increased, as shown by the ELISA results (Figure 1b).
Figure 1.
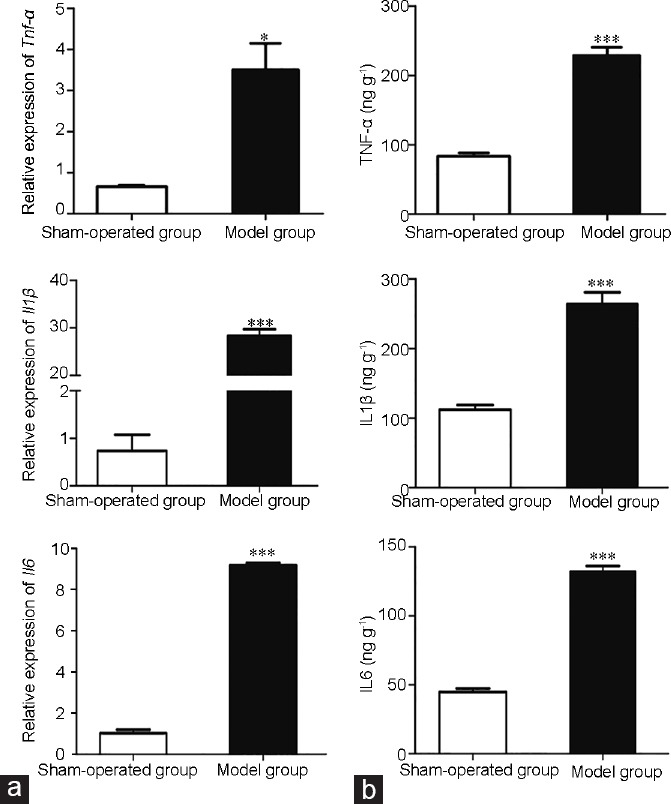
Demonstration of the epididymitis model by (a) real-time polymerase chain reaction and (b) enzyme-linked immunosorbent assay (ELISA). *P < 0.05, and ***P < 0.001 in model group compared to sham-operated group by Student's t-test. Error bars are standard error of the mean (s.e.m), n = 6 for each group. TNF-α: tumor necrosis factor alpha; IL1β: interleukin-1 beta; IL6: interleukin 6.
Quality control of the RNA sequence
The total RNA of epididymal portions of rats from the model and sham-operated groups was submitted for quality control and RNA library construction. After quality filtering, an average of 30 854 634 high-quality, clean reads were obtained for each sample. The average GC count was 50.3%, and the average Q30 was 94.4%. A large portion of clean reads (>93.5%) were mapped to the rat genome.
Identification of DEGs
A total of 1378 DEGs, including 531 upregulated and 847 downregulated, were identified in the epididymitis model compared with sham-operated rats (Figure 2a and Supplementary Table 2 (3MB, pdf) ). Hierarchical clustering of DEGs revealed that samples in the same group clustered closely, suggesting high reliability of the DEGs between groups, as well as high reproducibility in biological replicates (Figure 2b). Several members of the interleukin family (Il6, Il11, Il22ra2, interleukin 4 induced protein 1 [Il4i1], Il1b, and Il33), the NF-kappa-B family (Nfκbiα, Nfκbiζ, and Nfκb2), and the TNF family (TNF receptor superfamily member 12A [Tnfrsf12a], TNF-inducible gene 6 protein [Tnfaip6], Tnfaip3, Tnfrsf21, and Tnfrsf1a) were markedly upregulated in the model group.
Figure 2.
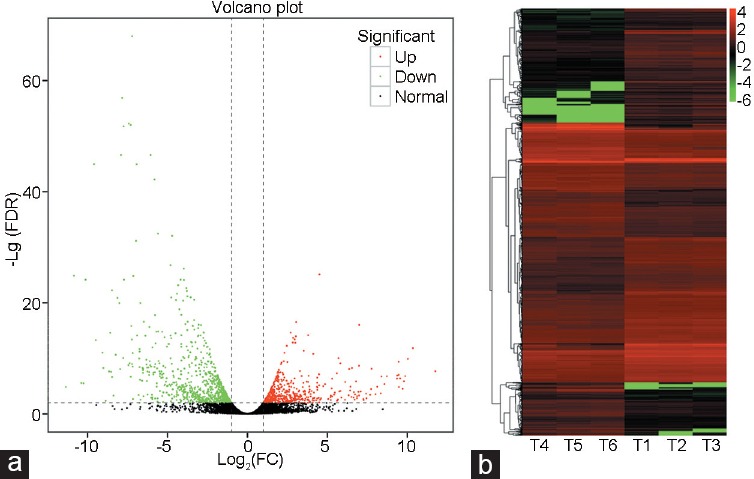
The (a) volcano plot and (b) heat map of differentially expressed genes. Red dots, green dots, and black dots in a signify the upregulated genes, the downregulated genes, and the nonchanged genes, respectively. (b) The X-axis is the sample number and the Y-axis is the differentially expressed genes. Red color means upregulation and green color means down-regulation. FC: fold change; FDR: false discovery rate.
Functional enrichment analysis of DEGs
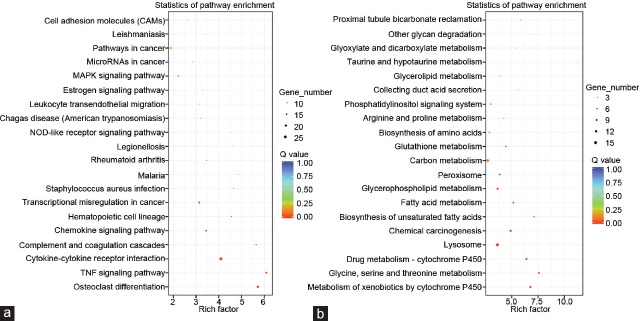
To investigate the functions of the DEGs further, GO and KEGG pathway enrichment analyses were performed for the upregulated and downregulated genes. At the cutoff value of FDR <0.05, the upregulated genes were markedly enriched in 22 GO-BP terms, including the inflammatory response (FDR = 8.86 × 10−11), the response to lipopolysaccharide (FDR = 9.92 × 10−11), and the cellular response to lipopolysaccharide (FDR= 5.76 × 10−8); three GO-MF terms, including the protease binding (FDR = 0.007882), the chemokine activity (FDR = 0.02426), and the cytokine activity (FDR = 0.027364); and seven GO-CC terms, including the extracellular space (FDR = 1.50 × 10−20), the cell surface (FDR = 1.22 × 10−9), and the extracellular region (FDR = 2.08 × 10−8) (Table 1). One hundred and nine upregulated genes (20.5%) were located on the extracellular exosome (FDR = 3.89 × 10−7). Moreover, these genes were markedly enriched in 10 pathways (Figure 3a), including the TNF signaling pathway (FDR = 3.74 × 10−8), cytokine–cytokine receptor interaction (FDR = 4.54 × 10−8), complement and coagulation cascades (FDR = 2.97 × 10−5), and the chemokine signaling pathway (FDR = 7.95 × 10−4). However, the downregulated genes were not enriched in any GO terms at the threshold of FDR <0.05, but were markedly enriched in nine pathways, including metabolism of xenobiotics by cytochrome P450 (FDR = 8.61 × 10−5); glycine, serine, and threonine metabolism (FDR = 1.00 × 10−4); and drug metabolism by cytochrome P450 (FDR = 1.54 × 10−4) (Figure 3b).
Figure 3.
The pathway enrichment analysis of (a) upregulated and (b) downregulated genes. MAPK: mitogen-activated protein kinase; TNF: tumor necrosis factor; NOD: nucleotide-binding oligomerization domain.
Confirmation of DEGs by real-time PCR
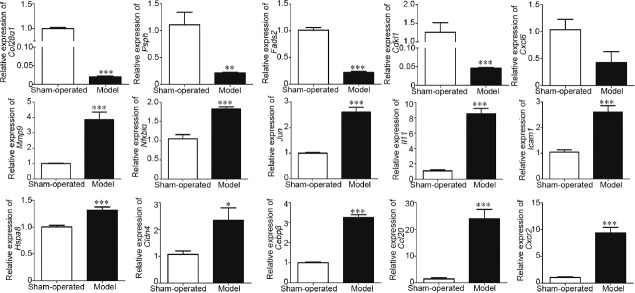
We selected 15 DEGs, including four downregulated genes (collagen type XXVIII alpha 1 chain [Col28α1], cyclin-dependent kinase-like 1[Cdkl1], phosphoserine phosphatase [Psph], and fatty acid desaturase 2 [Fads2]) and 11 upregulated genes (CCAAT/enhancer-binding protein beta [Cebpβ], C-X-C motif chemokine receptor 2[Cxcr2], interleukin 11 [Il11], C-X-C motif chemokine ligand 6 [Cxcl6], C-C motif chemokine ligand 20 [Ccl20], Nfκbiα, claudin 4[ Cldn4], matrix metallopeptidase 9 [Mmp9], heat shock 70 kDa protein 8 [Hspa8], intercellular cell adhesion molecule-1 [Icam1], and Jun) for analysis with real-time PCR. As shown in Figure 4, the expression of the four downregulated genes was consistent with the RNA-seq results. For the 11 upregulated genes, all except for Cxcl6 were consistent with the RNA-seq results.
Figure 4.
Relative expression of 15 differentially expressed genes measured by real-time PCR. *P < 0.05, **P < 0.01 and ***P < 0.001 in model group compared to sham-operated group by Student's t-test. Error bars are s.e.m., n = 3 for each group. Col28α1: collagen type XXVIII alpha 1 chain; Psph: phosphoserine phosphatase; Fads2: fatty acid desaturase; Cdkl1: cyclin-dependent kinase-like 1; Cxcl6: C-X-C motif chemokine ligand 6; Mmp9: matrix metallopeptidase 9; Nfkbiα: Nuclear factor-kappa-B inhibitor alpha; Il11: Interleukin 11; Icam1: intercellular cell adhesion molecule-1; Hspa8: heat shock 70 kDa protein 8; Cldn4: claudin 4; Cebpβ: CCAAT/enhancer-binding protein beta; Ccl20: C-C motif chemokine ligand 20; Cxcr2: C-X-C motif chemokine receptor 2; s.e.m.: standard error of the mean.
Western blot analysis
We selected four DEGs (Cdkl1, Mmp9, Ccl20, and Nfκbiα) for western blot analysis expression at the protein level. As shown in Figure 5, the protein expression of CDKL1 was decreased in the model group compared with that in the sham-operated group (P < 0.01). Moreover, the protein expression levels of MMP9 and NFKBIA were increased in the model group compared with those in the sham-operated group (P < 0.05). These results were consistent with the results of RNA-Seq. However, no significant difference was detected for CCL20 between these two groups.
Figure 5.
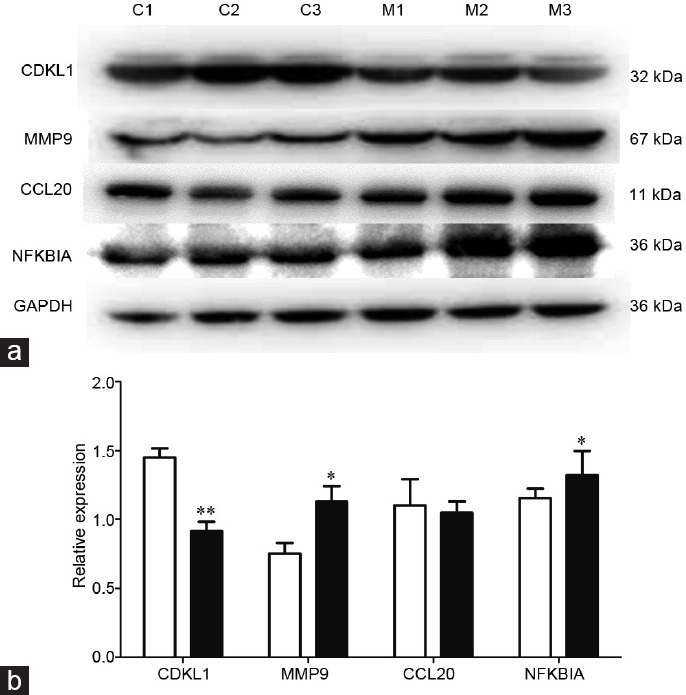
(a) Western blot analysis of CDKL1, MMP9, CCL20, and NFKBIA in model and sham-operated groups. C1–C3 are samples in the sham-operated group and M1–M3 are samples in the model group. (b) The intensity of the bands was analyzed by ImageJ. White column: sham-operated group; black column: model group. CDKL1: cyclin-dependent kinase-like 1; *P < 0.05 and **P < 0.01 in model group compared to sham-operated group by Student's t-test. Error bars are s.e.m., n = 3 for each group. MMP9: matrix metallopeptidase 9; CCL20: C-C motif chemokine ligand 20; NFKBIA: Nuclear factor-kappa-B inhibitor alpha; GAPDH: reduced glyceraldehyde-phosphate dehydrogenase; s.e.m.: standard error of the mean.
DISCUSSION
Epididymitis is a disease characterized by the inflammation of the epididymis and is frequently diagnosed in the investigation of male infertility.2,4 Although high-throughput sequence technology has been developed for several years, the molecular mechanism of epididymitis has not yet been comprehensively investigated. In this study, we established an epididymitis animal model and performed transcriptome analysis to identify the DEGs between the induced epididymitis and sham-operated rats. A total of 1378 DEGs, including 531 upregulated and 847 downregulated genes, were identified in the epididymitis model rats compared with sham-operated rats. Functional enrichment analyses by GO and KEGG suggested that the upregulated genes were markedly enriched in inflammation-related GO terms and in the TNF signaling pathway, cytokine–cytokine receptor interaction, complement and coagulation cascades, and the chemokine signaling pathway.
Bacterial infections from the urethra are the major causes of epididymitis. Therefore, it is critical to understand how the epididymis senses the presence of and responds to bacterial infection. LPS is the major component of the outer membrane of Gram-negative bacteria, contributing greatly to their structural integrity.16,17 LPS induces a strong response from normal animal immune systems. In previous studies, LPS has been used to establish epididymitis rat models5,6 by a unilateral and single injection into the caput region of the epididymis. To confirm that the animal model was established successfully, the expression levels of proinflammatory factors including Il1β, Il6, and Tnf-α were examined after 24 h of LPS injection. The results suggested that, at both the mRNA and protein expression levels, these proinflammatory factors were markedly increased in the model group compared with those in the sham-operated group. These changes are consistent with the signs of epididymitis reported in the literature,5,6 demonstrating successful establishment of the epididymitis model.
RNA-seq analysis identified a total of 1378 DEGs, including 531 upregulated and 847 downregulated, in the epididymitis model rats. Several members of the interleukin family, the NF-kappa-B family, and the TNF family were markedly upregulated in the model group. The NF-kappa-B family consists of transcription factors that regulate the expression of many proinflammatory genes.18 Our study demonstrated that LPS activated NF-kappa-B family members in the rat epididymis. Innate immunity is the first line of defense for organisms attacked by invading pathogens.19 NF-kappa-B can be activated by toll-like receptors, which are involved in recognition of infectious agents by macrophages.20 Our study demonstrated that LPS induced epididymitis by upregulating NF-kappa-B and the subsequent induction of proinflammatory cytokines. Our results are consistent with those in the study by Silva et al.5 Moreover, the high levels of TNF family and interleukin family members in semen impair sperm quality and are associated with male infertility.21
Apart from the proinflammatory genes, we also identified and confirmed other genes that might be associated with epididymitis. For example, Col28α1, Psph, Fads2, and Cdkl1 were markedly downregulated in the model group in our study. Col28α 1 belongs to a class of collagens containing von Willebrand factor type A-domains.22 It is reported that Col28α 1 is important in reducing inflammation and viral infections, as it contains a serine protease inhibitor domain.23 Fads2 is associated with Delta 6 desaturase.24 Schaeffer et al.25 demonstrated that gene polymorphisms and haplotypes in Fads2 are associated with polyunsaturated fatty acid membrane content and proinflammatory markers, suggesting that the activity of this enzyme modifies the effect of tissue inflammation. Vaittinen et al.26 reported that variations in the Fads1/2 genes were associated with the expression of genes in the Il-1β and NFκB pathway in adipose tissue after weight reduction. They demonstrated that polymorphism in Fads1/2 genes were associated with adipose tissue inflammation. We speculate that dysregulation of Fads2 results in the dysregulation of Delta 6 desaturase and subsequently the levels of serum arachidonic acid, which is a major substrate for eicosanoids that regulate inflammatory responses.27 Cdkl1 is a member of a large family of cell division cycle 2 (CDC2)-related serine/threonine protein kinases.28 However, there were no reports about the association of Cdkl1 and inflammation. In our study, the RNA-seq, real-time PCR, and western blot results all supported the marked downregulation of Cdkl1 in epididymitis tissues.
A total of 11 upregulated genes were confirmed by real-time PCR; among them, 10 were consistent with the RNA-seq data. The expression of NFKBIA, MMP9, and CCL20 at the protein level was confirmed by western blot; however, only the protein expression levels of NFKBIA and MMP9 were consistent with the RNA-seq data.
CONCLUSION
We identified 1378 DEGs, including 531 upregulated and 847 downregulated genes, in the epididymitis rat model compared with sham-operated rats. Functional enrichment analyses by GO and KEGG suggested that the upregulated genes were markedly enriched in inflammation-related GO terms and in the TNF signaling pathway, cytokine–cytokine receptor interaction, complement and coagulation cascades, and in the chemokine signaling pathway. Fourteen out of fifteen genes were confirmed by real-time PCR, and three out of four genes were demonstrated in western blots. Our study sheds new light on the understanding of the early response of the epididymis region during LPS epididymitis.
AUTHOR CONTRIBUTIONS
XS carried out the animal studies, participated in the bioinformatic analysis, and drafted the manuscript; NHL and YLW carried out the real-time PCR and performed the statistical analysis; and HXW and KH carried out the western blot; XS and BC conceived the study, participated in its design and coordination, and helped draft the manuscript. All authors read and approved the final manuscript and agreed with the order of presentation of the authors.
COMPETING INTERESTS
All authors declared no competing interests.
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation of China (No. 81471496).
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.McConaghy JR, Panchal B. Epididymitis: an overview. Am Fam Physician. 2016;94:723–6. [PubMed] [Google Scholar]
- 2.Trojian TH, Lishnak TS, Heiman D. Epididymitis and orchitis: an overview. Am Fam Physician. 2009;79:583–7. [PubMed] [Google Scholar]
- 3.Lu Y, Bhushan S, Tchatalbachev S, Marconi M, Bergmann M, et al. Necrosis is the dominant cell death pathway in uropathogenic Escherichia coli elicited epididymo-orchitis and is responsible for damage of rat testis. PLoS One. 2013;8:e52919. doi: 10.1371/journal.pone.0052919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michel V, Pilatz A, Hedger MP, Meinhardt A. Epididymitis: revelations at the convergence of clinical and basic sciences. Asian J Androl. 2015;17:756–63. doi: 10.4103/1008-682X.155770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva EJ, Ribeiro CM, Mirim AF, Silva AA, Romano RM, et al. Lipopolysaccharide and lipotheicoic acid differentially modulate epididymal cytokine and chemokine profiles and sperm parameters in experimental acute epididymitis. Sci Rep. 2018;8:103. doi: 10.1038/s41598-017-17944-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao D, Li Y, Yang R, Wang Y, Zhou Y, et al. Lipopolysaccharide-induced epididymitis disrupts epididymal beta-defensin expression and inhibits sperm motility in rats. Biol Reprod. 2010;83:1064–70. doi: 10.1095/biolreprod.109.082180. [DOI] [PubMed] [Google Scholar]
- 7.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Florea L, Song L, Salzberg SL. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res. 2013;2:188. doi: 10.12688/f1000research.2-188.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Feng Z, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–8. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang G, Meredith TC, Kahne D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol. 2013;16:779–85. doi: 10.1016/j.mib.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Quinn PJ. Lipopolysaccharide: biosynthetic pathway and structure modification. Prog Lipid Res. 2010;49:97–107. doi: 10.1016/j.plipres.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Osorio FG, Soria-Valles C, Santiago-Fernandez O, Freije JM, Lopez-Otin C. NF-kappaB signaling as a driver of ageing. Int Rev Cell Mol Biol. 2016;326:133–74. doi: 10.1016/bs.ircmb.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Hop HT, Arayan LT, Reyes AW, Huy TX, Min WG, et al. Heat-stress-modulated induction of NF-kappaB leads to brucellacidal pro-inflammatory defense against Brucella abortus infection in murine macrophages and in a mouse model. BMC Microbiol. 2018;18:44. doi: 10.1186/s12866-018-1185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira SC, de Oliveira FS, Macedo GC, de Almeida LA, Carvalho NB. The role of innate immune receptors in the control of Brucella abortus infection: toll-like receptors and beyond. Microbes Infect. 2008;10:1005–9. doi: 10.1016/j.micinf.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Kocak I, Yenisey C, Dundar M, Okyay P, Serter M. Relationship between seminal plasma interleukin-6 and tumor necrosis factor alpha levels with semen parameters in fertile and infertile men. Urol Res. 2002;30:263–7. doi: 10.1007/s00240-002-0269-y. [DOI] [PubMed] [Google Scholar]
- 22.Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, et al. Collagen XXVIII, a novel von Willebrand factor A domain-containing protein with many imperfections in the collagenous domain. J Biol Chem. 2006;281:3494–504. doi: 10.1074/jbc.M509333200. [DOI] [PubMed] [Google Scholar]
- 23.Gong D, Farley K, White M, Hartshorn KL, Benarafa C, et al. Critical role of serpinB1 in regulating inflammatory responses in pulmonary influenza infection. J Infect Dis. 2011;204:592–600. doi: 10.1093/infdis/jir352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinelli N, Girelli D, Malerba G, Guarini P, Illig T, et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am J Clin Nutr. 2008;88:941–9. doi: 10.1093/ajcn/88.4.941. [DOI] [PubMed] [Google Scholar]
- 25.Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–56. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 26.Vaittinen M, Walle P, Kuosmanen E, Mannisto V, Kakela P, et al. FADS2 genotype regulates delta-6 desaturase activity and inflammation in human adipose tissue. J Lipid Res. 2016;57:56–65. doi: 10.1194/jlr.M059113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng KT, Chang CY, Chang LF, Nesaretnam K. Modulation of obesity-induced inflammation by dietary fats: mechanisms and clinical evidence. Nutr J. 2014;13:12. doi: 10.1186/1475-2891-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyerson M, Enders GH, Wu CL, Su LK, Gorka C, et al. A family of human cdc2-related protein kinases. EMBO J. 1992;11:2909–17. doi: 10.1002/j.1460-2075.1992.tb05360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.