Summary
Astrocytes are particularly promising candidates for reprogramming into neurons, as they maintain some of the original patterning information from their radial glial ancestors. However, to which extent the position of astrocytes influences the fate of reprogrammed neurons remains unknown. To elucidate this, we performed stab wound injury covering an entire neocortical column, including the gray matter (GM) and white matter (WM), and targeted local reactive astrocytes via injecting FLEx switch (Cre-On) adeno-associated viral (AAV) vectors into mGFAP-Cre mice. Single proneural factors were not sufficient for adequate reprogramming, although their combination with the nuclear receptor-related 1 protein (Nurr1) improved reprogramming efficiency. Nurr1 and Neurogenin 2 (Ngn2) resulted in high-efficiency reprogramming of targeted astrocytes into neurons that develop lamina-specific hallmarks, including the appropriate long-distance axonal projections. Surprisingly, in the WM, we did not observe any reprogrammed neurons, thereby unveiling a crucial role of region- and layer-specific differences in astrocyte reprogramming.
Keywords: reprogramming, reactive gliosis, inflammation, AAV, lentivirus, cerebral cortex, cortical layers, axonal projection, electrophysiology, astrocytes
Graphical Abstract
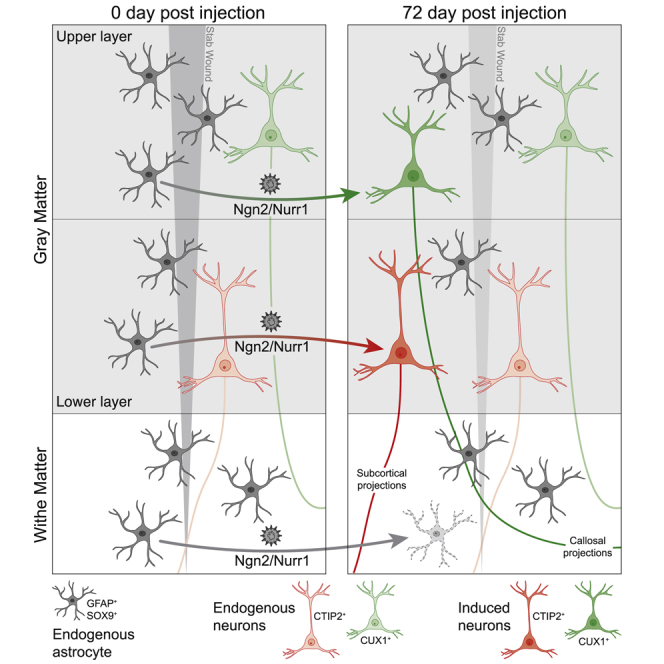
Highlights
-
•
AAV can be targeted to reactive astrocytes upon stab wound injury
-
•
Expression of Ngn2 and Nurr1 in these astrocytes induces pyramidal neurons
-
•
Induced pyramidal neurons acquire correct layer identity and axonal projections
-
•
Neurons cannot be induced in the white matter
Neurons dying after brain injury cannot be replaced. Mattugini, Bocchi, et al. show that local astrocytes can be converted into functional neurons acquiring appropriate layer identity and connectivity by expression of neurogenic factors in a mouse model of traumatic brain injury.
Introduction
The mammalian neocortex is a complex and highly organized structure containing many different types of neurons and glial cells. The intricate specificity of neuronal subtypes is established via a sequence of transcriptional regulators during development when neurons of different layer positions are generated in a sequential order from radial glial cells (Govindan and Jabaudon, 2017, Lodato and Arlotta, 2015, Greig et al., 2013). In the injured cerebral cortex, various combinations of transcription factors have been used to turn reactive glial cells (proliferating glia, NG2+ oligodendrocyte progenitors, or GFAP+ cells; for review, see Gascón et al., 2017 and Wang and Zhang, 2018) into neurons. Although much progress has been made since the first report describing in vivo glia-to-neuron reprogramming (Buffo et al., 2005), driving sustained and adequate reprogramming of layer-specific cerebral cortex neurons has still to be achieved.
Astrocytes display an amazing diversity in terms of positional identity, partly inherited from their radial glia ancestors (John Lin et al., 2017, Bayraktar et al., 2014) or instructed by surrounding neurons (Farmer et al., 2016, Lanjakornsiripan et al., 2018). As astrocytes differ in their morphology and gene expression at different laminar positions within the neocortical gray matter (GM; Lanjakornsiripan et al., 2018), we explored here whether they may be able to generate also different neuronal subtypes when converting to neurons in the adult brain. This is important, as laminar differences of neurons are key for cortical function. Excitatory projection neurons, the cortical pyramidal neurons, differ in their identity according to their laminar position, molecular hallmarks, morphology, and input-output connectivity (Molyneaux et al., 2007, Harris and Shepherd, 2015, Jabaudon, 2017). To reconstruct the circuitry of the cerebral cortex upon injury, it is therefore essential to obtain neurons with appropriate subtype identities and projections.
Results
Nurr1 Supports Proneural Factors to Achieve Highly Efficient Induction of Neurons from Astrocytes
Different viral vectors, such as lentivirus (LV), retrovirus (RV), and adeno-associated virus (AAV), have been used for in vivo reprogramming, achieving a broad range of reprogramming efficiency and neuronal survival (Gascón et al., 2017, Wang and Zhang, 2018). In order to directly compare these viral vectors, we analyzed their effects on infiltration of leukocytes (CD45+/Iba1− cells), microgliosis (Iba1+ cells), and astrogliosis (GFAP+ cells) after cortical stab wound (SW) injury extending through the GM into the white matter (WM), at a time point when acute inflammation and reactive gliosis have normally receded (13 days after injury; Mattugini et al., 2018). When LV or RV were injected 3 days after SW and analyzed 10 days post-injection (dpi), CD45+ leukocytes were abundant and reactive gliosis (Iba1+ and GFAP+ cells) was very strong at the site of injection (Figure S1A). Conversely, AAV injections showed very low levels of reactive gliosis and few immune cells at the injury and injection site (Figure S1A). The low reactivity upon AAV injection was independent of the number of vectors used (1–3) and of the factors included (data not shown).
Given their lower immunogenicity, we used AAVs to express the reprogramming factors and/or reporter proteins with inverted orientation and flanked by two pairs of loxP (Figure 1A; Atasoy et al., 2008). This allows the expression of the gene of interest specifically in astrocytes when injecting transgenic mice expressing Cre recombinase under the murine promoter of GFAP (mGFAP-Cre mice; Figures 1B and 1C; Gregorian et al., 2009), and injections into wild-type animals resulted in no GFP+ cells (data not shown). AAV-FLEx-GFP injected 3 days after SW resulted in GFP+ cells that were virtually all astrocytes, as detected by SOX9, GFAP, or morphology at 10, 24, and 72 dpi (Figures 1D–1I, S1B, and S1C). This is consistent with Cre protein expression detected mostly in astrocytes (100% ± 0%) and hardly in neurons (0.86% ± 1.5%). Accordingly, less than 10% were positive for neuronal markers, such as RBFOX3 (NeuN; Figures 1D–1I, S1B, and S1C), suggesting a small degree of possible leakiness of the mGFAP-Cre expression.
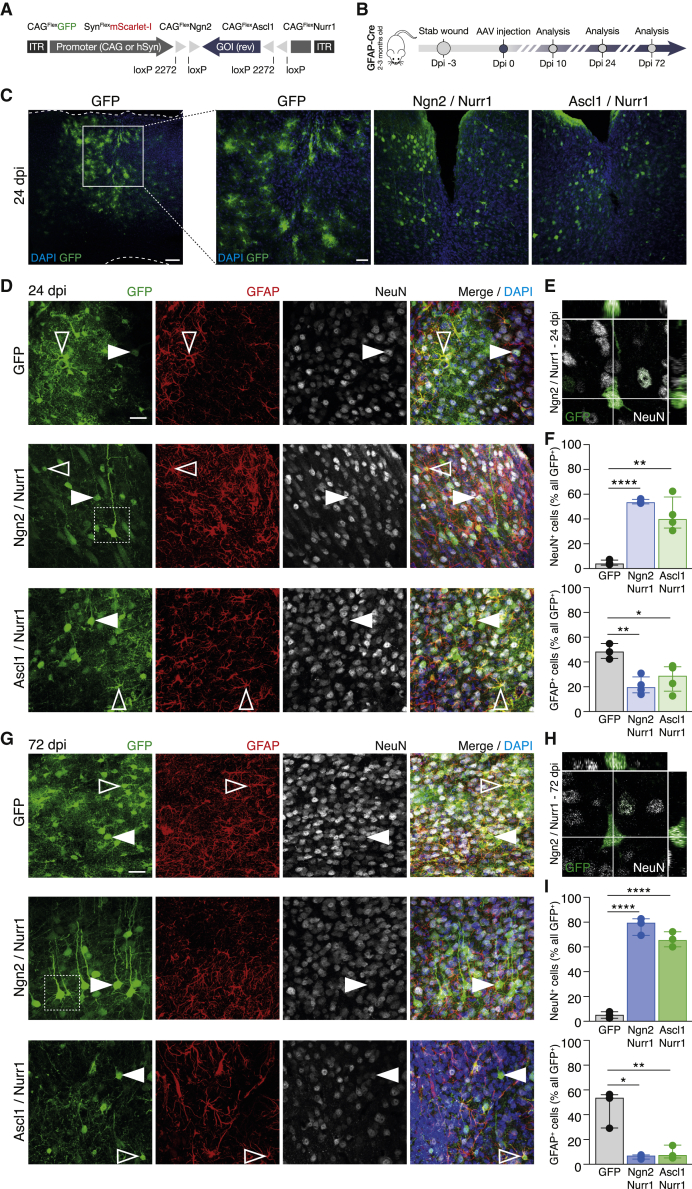
Figure 1.
Neurogenic Factors Reprogram Astrocytes into Neurons after Traumatic Brain Injury
(A and B) Scheme of the AAV-FLEx constructs (A) and experimental design (B).
(C) Photomicrographs showing an overview with GFP+ cells at 24 dpi of AAV encoding for GFP, GFP/Ngn2/Nurr1, and GFP/Ascl1/Nurr1.
(D–I) Photomicrographs showing GFP+/NeuN+ neurons (full arrowheads) and GFP+/GFAP+ astrocytes (empty arrowheads) at 24 (D) and 72 (G) dpi of GFP, Ngn2/Nurr1, and Ascl1/Nurr1. Example of Z-projection (E and H) of GFP/Ngn2/Nurr1 neurons (dashed square) used for the co-localization analysis (F and I). n = 3, 4, and 4 for GFP, Ngn2/Nurr1, and Ascl1/Nurr1, respectively.
Data are shown as median ± interquartile range (IQR). Student’s t test. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗∗p ≤ 0.0001. AAV, adeno-associated virus; dpi, days post-injection. Scale bars: 100 μm (C, left); 50 μm (C, right); 20 μm (D and G).
See also Figures S1 and S2.
In order to convert local reactive astrocytes into neurons, we used FLEx-switch AAVs (Figure 1A) containing either Neurogenin 2 (Ngn2) or achaete-scute homolog 1 (Ascl1; Figures 1B and 1C). The proneural transcription factor Ngn2 is sufficient to convert astrocytes into glutamatergic neurons in vitro (Heinrich et al., 2010) but rather inefficient on its own in proliferating reactive glia using RVs (Gascón et al., 2016). Given the high leukocyte invasion and reactive gliosis elicited by RVs, we first tested whether more neurons could be obtained using AAV-FLEx-Ngn2. However, only a small percentage of GFP+ cells was NeuN+ when AAV-FLEx-Ngn2 was co-injected with AAV-FLEx-GFP (Figure S2A) and/or AAV-FLEx-RFP (data not shown). We then combined Ngn2 with the transcription factor Nurr1 (Nr4a2) based on its increased expression in conditions favoring neuronal conversion (Gascón et al., 2016) and its neuroprotective role (Saijo et al., 2009, Sousa et al., 2007). Although the percentage of GFP+/NeuN+-induced neurons (iNs) was as low as in the control condition at 10 dpi (Figure S1B), it increased significantly to 53% at 24 dpi (with the remaining cells being mostly GFAP+ astrocytes; Figures 1D–1F). Interestingly, the combination of Ngn2/Nurr1 was also able to induce a significant increase in NeuN+ neurons in absence of SW, although less efficiently (Figure S2B). To determine whether Nurr1 is also effective with other proneural factors, we tested its combination with Ascl1, previously used in other reprogramming cocktails (Masserdotti et al., 2016). AAV-FLEx-Ascl1 injection resulted in only 20% NeuN+ iNs (Figure S2A), and its combination with Nurr1 doubled the proportion of iNs to about 40% (Figures 1D and 1F). Interestingly, injection of AAV-FLEx-Nurr1 alone showed low efficiency in neuronal induction (about 20% iNs; Figure S2A), indicating that the combination of a proneural factor, either Ngn2 or Ascl1, with Nurr1 is particularly efficient in astrocytes to neuron conversion.
Analysis of long-term survival at 72 dpi (or longer, 252 dpi; data not shown) showed even higher proportions of iNs with 80% NeuN+ upon Ngn2/Nurr1 and 70% NeuN+ upon Ascl1/Nurr1 (Figures 1G–1I). Importantly, the absolute number of GFP+/NeuN+ iNs also significantly increased, and GFP+/GFAP+ astrocytes decreased (Figure S2C), excluding a change in proportion due to selective cell death and rather suggestive of continued conversion of astrocytes into neurons.
Neurons Induced with Ngn2 and Nurr1 Originate from Both Proliferating and Quiescent Astrocytes
Although the above results are suggestive of astrocyte-to-neuron conversion, it is important to exclude neuronal labeling due to either exosome transfer of, e.g., Cre-containing particles or possibly cell fusion (Pesaresi et al., 2019, Ridder et al., 2014). To test this possibility, we labeled endogenous neurons by continuous EdU administration during embryonic neurogenesis (from E7.5 to E15.5; of note, EdU labeling is diluted in astrocytes during postnatal proliferation). After performing SW injury in the adult mice and injecting the reporter AAV-hSyn-mScarlet-I (non-flexed) to label endogenous neurons, we found that the majority of mScarlet-I+ neurons (about 80% in the deeper layers) were labeled by EdU at 24 dpi (Figures S3A and S3B). Conversely, when we performed the reprogramming protocol (AAV-FLEx-Ngn2 and AAV-FLEx-Nurr1, together with the reporter AAV-FLEx-hSyn-mScarlet-I), less than 20% of Ngn2/Nurr1 iNs were positive for EdU (Figures S3A and S3B). These results support the idea that Ngn2/Nurr1 iNs are not endogenous neurons generated at embryonic stages but rather come from astrocyte reprogramming. To further confirm this observation, we administered EdU directly after SW for 10 days, during the entire period of reactive astrocyte proliferation in this injury model (Mattugini et al., 2018, Buffo et al., 2008). About 20% of mScarlet-I+ iNs were EdU+ and hence derived from cells proliferating after the injury (i.e., glia and not neurons; Figures S3C and S3D). As previously reported, about 25%–30% of all astrocytes proliferate following SW injury (Buffo et al., 2008). Thus, this result is consistent with the astrocytic origin of iNs, which includes the proliferating astrocyte subpopulation.
Neurons Induced with Ngn2 and Nurr1 Acquire Mature Pyramidal Neuron Hallmarks and Molecular Identities according to Their Laminar Position
To examine the subtype identity of Ngn2/Nurr1 iNs, we focused on laminar, molecular, and morphological features. iNs distributed throughout upper and lower layers (Figure S3E). At 72 dpi, most CUX1+ iNs were located in upper layers, and virtually all CTIP2+ iNs were found at deeper positions similar to endogenous neurons (Figure 2A; Molyneaux et al., 2007). Notably, SW and viral vector injection did not alter CUX1 and CTIP2 expression patterns along the cortical column (Figure S4A) or the intensity of immunostaining or the soma size of iNs (Figure S4B). Single and combinatorial labeling for CUX1, SATB2, and BRN2 were mostly found in iNs located in upper layers, whereas FOXP2, CTIP2, TLE4, and TBR1 were mainly found in lower layer iNs (Figures S4C and S4D). Interestingly, immunoreactivity for these markers was only weakly detected in few iNs at 24 dpi, and a significantly higher fraction was strongly positive at 72 dpi (Figure S4E). Comparison to recent single-cell analysis (Zeisel et al., 2018) allowed us to molecularly define the subtypes of induced pyramidal cells (Figure S4F).
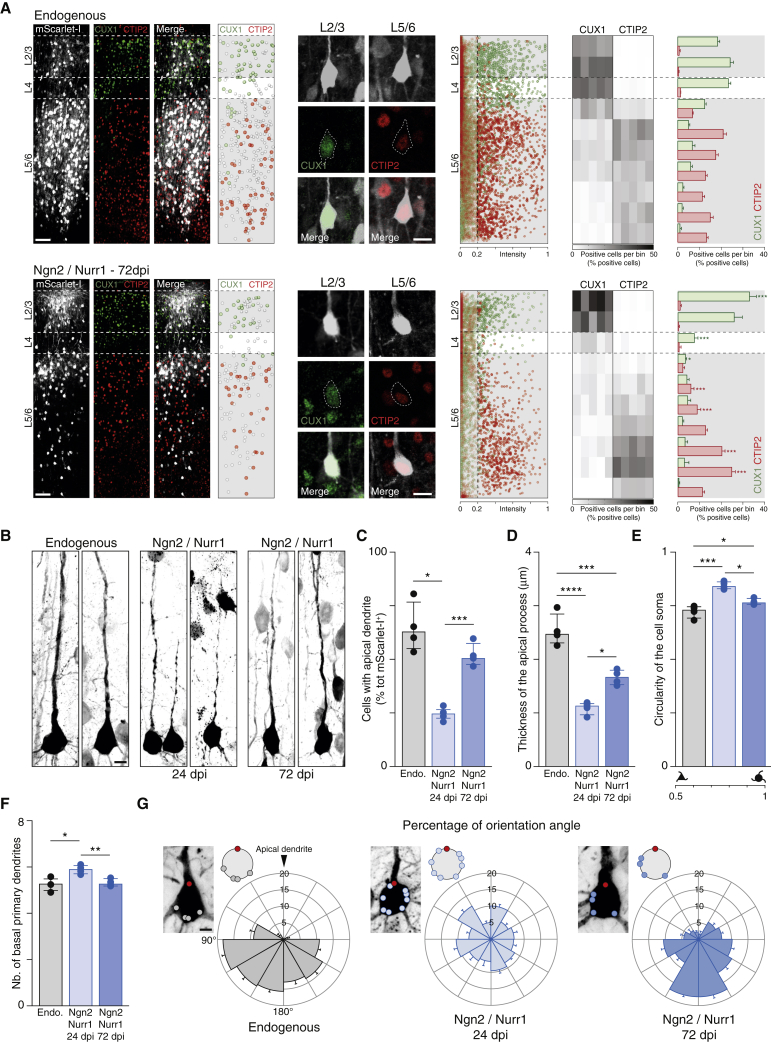
Figure 2.
Neurons Induced by Ngn2/Nurr1 Acquire Adequate Laminar Identity and Pyramidal Morphology
(A) Photomicrographs and quantitative analysis of endogenous and induced neurons at 72 dpi of AAV-mScarlet-I and AAV-FLEx-mScarlet-I/Ngn2/Nurr1. The vertical dashed line in the right panel represents the intensity threshold (intensity of 0.2) defining positive cells. The heatmap shows the cell distribution in ten bins. The color-coded columns represent the percentage of mScarlet-I+/CUX1+ or mScarlet-I+/CTIP2+ cells per bin of the total number of mScarlet-I+/CUX1+ or mScarlet-I+/CTIP2+, respectively, in an entire column. Histograms illustrate the average of percentage for each bin; n = 5 animals.
(B) Photomicrographs showing pyramidal morphology of mScarlet-I+ endogenous and induced neurons at 24 and 72 dpi.
(C–G) Histograms depicting the morphological analyses of endogenous and induced neurons at 24 and 72 dpi for the following parameters: percentage of cell with an apical dendrite (C), thickness of the apical dendrite (D), circularity of the cell soma (E), number of basal dendrites (F), and their orientation (G), demonstrating the gradual maturation process of iNs; n = 4 animals.
Data are shown as median ± IQR. One-way ANOVA. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001. L, layer. Scale bars: 100 μm (A, overview); 20 μm (A, close up); 10 μm (B); 5 μm (G).
See also Figures S3 and S4.
Remarkably, both CUX1+ and CTIP2+ iNs displayed a stereotypical pyramidal-shaped cell soma, a large apical dendrite oriented radially toward the pial surface, and an elaborated basal dendritic arbor, similar to endogenous neurons (Figure 2B; Harris and Shepherd, 2015). This was not the case when combining Ascl1 and Nurr1, which instead generated iNs of more variable morphologies (Figure 1G). Notably, Ngn2/Nurr1 iNs matured over time (Figure 2B). The number of Ngn2/Nurr1 iNs with an apical dendrite was significantly lower at 24 dpi compared to the endogenous neurons, and this difference was reduced and no longer significant at 72 dpi (Figure 2C). Likewise, the thickness of iN apical process grew between 24 and 72 dpi (Figure 2D), and the soma was initially significantly more circular at 24 dpi when compared to both iNs at 72 dpi or endogenous neurons (Figure 2E). Thus, iNs gradually acquired the shape of pyramidal neurons. Moreover, the number of primary basal dendrites was reduced between 24 and 72 dpi, indicative of potential pruning processes (Figure 2F). The mean number (Figure 2F) and orientation (Figure 2G) of iNs’ primary basal dendrites at 72 dpi was highly reminiscent of the basal dendritic arbor of endogenous pyramidal cells. Together, these data show that iNs develop over time a congruent assignment of laminar, molecular, and morphological features of mature pyramidal neurons.
Neurons Induced with Ngn2 and Nurr1 Develop Morphological and Functional Synaptic Connections and Long-Distance Axonal Projections
Given the morphological maturation of the Ngn2/Nurr1 iNs, we next examined parameters indicative of synaptic integration and connectivity. Because spines act as functional postsynaptic sites, we assessed their number on secondary dendrites and found a significant increase from 24 to 72 dpi in Ngn2/Nurr1 iNs, reaching numbers comparable to endogenous pyramidal neurons (Figure 3A). To probe the functional properties of these iNs and their synaptic inputs, we performed patch-clamp recordings on neurons induced by Ngn2/Nurr1 in acute slice preparations at 72 dpi. Their resting membrane potential and input resistance was well comparable to endogenous neurons (Figure S5A), and they generated action potentials similar to endogenous neurons (Figures 3B, 3C, S5B, and S5C). Most importantly, Ngn2/Nurr1 iNs received excitatory and inhibitory inputs (blocked by NBQX or bicuculline, respectively) at 72 dpi (Figure 3D), consistent with their functional integration.
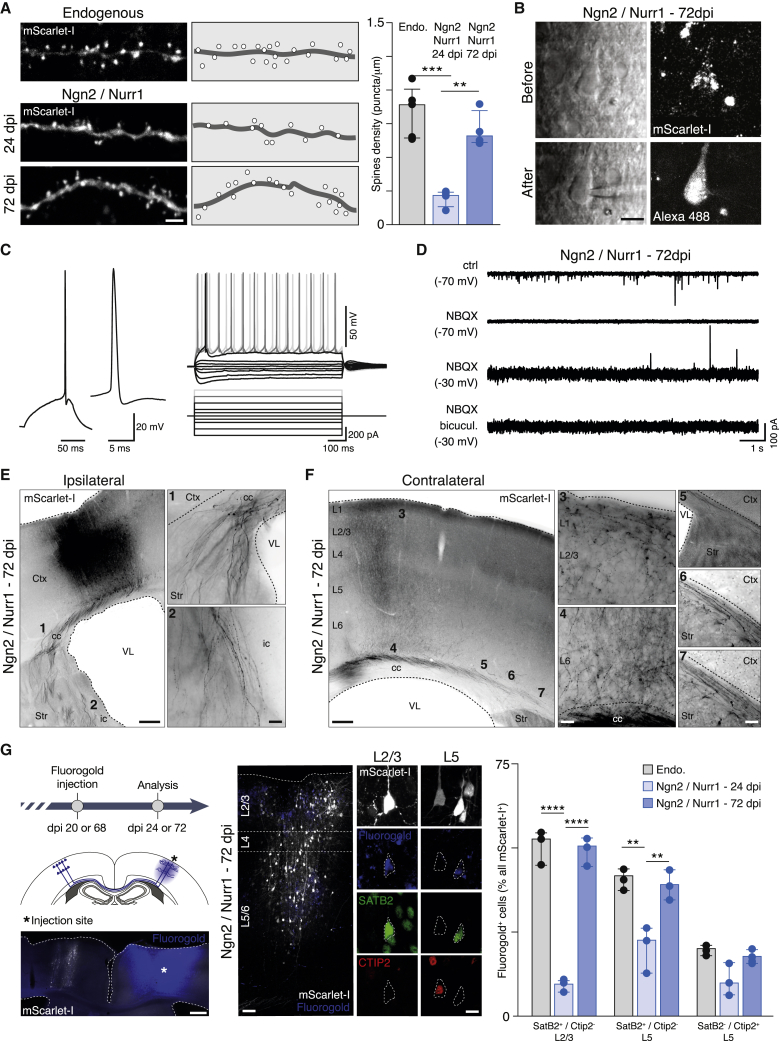
Figure 3.
Neurons Induced by Ngn2/Nurr1 Develop Spines and Project to the Appropriate Brain Regions
(A) Photomicrographs and quantification of spines on secondary dendrite segments of endogenous (top) and induced (bottom) neurons at 24 and 72 dpi of mScarlet-I/Ngn2/Nurr1; n = 4 animals.
(B) Patch-clamp recording of endogenous or induced neurons in acute slice preparations of the cortex at 72 dpi as shown in the laser-Dodt-contrast and fluorescence images of mScarlet-I+ cell, before (top) and after filling the recorded cell with Alexa 488 (bottom).
(C) Left: action potential in response to 100-ms current injection in 72 dpi iN shown in (B) (timescale expanded). Right: current-voltage responses recorded during 1-s current pulses from −300 to +400 pA are shown (of the same cell).
(D) Excitatory spontaneous synaptic currents recorded in voltage clamp at −70 mV in mScarlet-I+ iNs (top trace) are blocked by NBQX (2nd trace). Inhibitory spontaneous synaptic currents recorded at −30 mV (3rd trace from top) are blocked by bicuculline (bottom trace).
(E and F) Photomicrographs of mScarlet-I+ iN axons at 72 dpi in the ipsilateral (E) and contralateral (F) cortical hemisphere.
(G) Left: schematic drawing indicating the experimental schedule and positions of callosal neurons with a low-power overview photomicrograph depicting the Fluorogold injection site. Middle: retrogradely traced iNs at 72 dpi are mainly located in L2/3 and L5. Right: histograms illustrate the percentage of Fluorogold+ endogenous or induced neurons in L2/3 that are SATB2+/CTIP2− and in L5 that are SATB2+/CTIP2− or SATB2−/CTIP2+ neurons; n = 3 animals.
Data are shown as median ± IQR. One-way ANOVA. ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001. cc, corpus callosum; Crb, cerebellum; Ctx, cortex; ic, internal capsule; OB, olfactory bulb; SC: spinal cord; Str, striatum; Th, thalamus; VL, lateral ventricle. Scale bars: 100 μm (E, left; F, left; and G, overview); 50 μm (E, right and F, middle); 20 μm (B and G, close up); 10 μm (A and F, right).
See also Figure S5.
In order to monitor the efferent connections of Ngn2/Nurr1 iNs, we followed their axonal projections. At 24 dpi, only few mScarlet-I+ axons were found in the ipsilateral striatum and at the midline of the corpus callosum, but none reached the contralateral cortex (Figure S5D). At 72 dpi, however, subcortical axons reached the striatum (Figure 3E), thalamus, and midbrain (Figure S5E), and corticocortical axons crossed the corpus callosum into the contralateral cortex (Figures 3F and S5E). In order to investigate the subtype-specific origin of these projections, we performed retrograde labeling with Fluorogold injection into the contralateral cortex and stained for SATB2, which labels callosal projection neurons in layers 2/3 and 5. At 72 dpi, most Fluorogold+ iNs were SATB2+ located in L2/3 and L5, and fewer iNs were CTIP2+ in L5 (Figure 3G). In contrast, at 24 dpi, only a small fraction of iNs (located in L2/3 and L5) were Fluorogold+/SATB2+, supporting a maturational process occurring over time. Together, these data reveal for the first time a consistent assignment of axonal target selection and connectivity hallmarks by iNs after cortical injury in vivo.
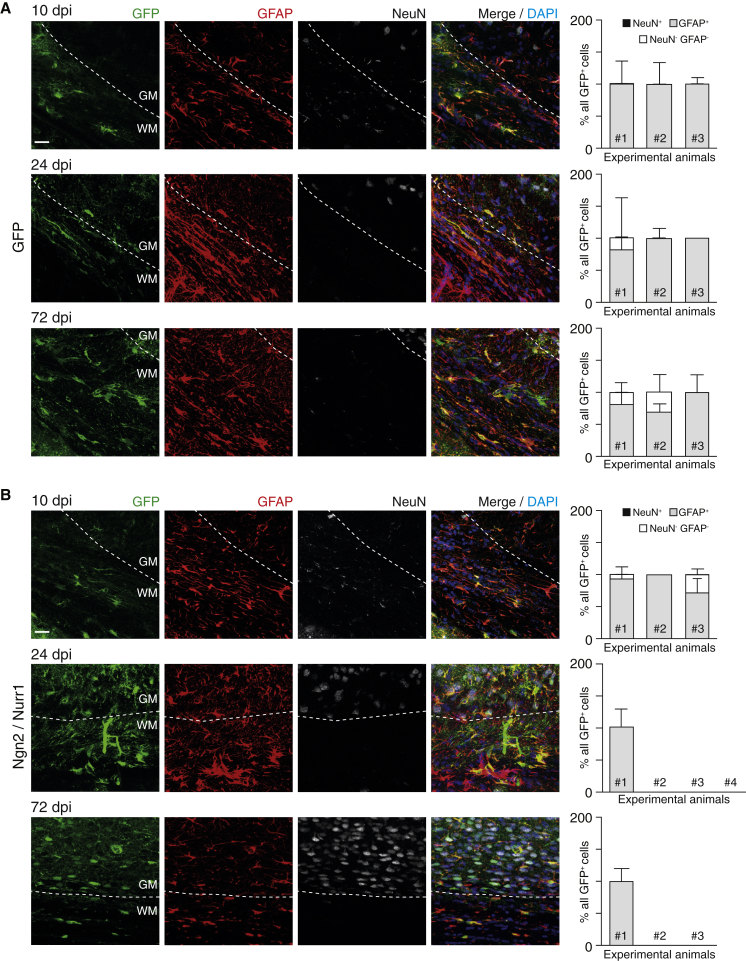
White Matter Astrocytes Fail to Undergo Neuronal Reprogramming
Given the amazing specificity and maturity of iNs seen in the GM, we were surprised not to see iNs in the WM. In control AAV-FLEx-GFP injections, GFP+ cells were detected at all time points (10, 24, and 72 dpi), indicating efficient targeting of WM astrocytes (Figure 4A). Surprisingly, however, when using the reprogramming factors Ngn2/Nurr1 (Figure 4B) or Ascl1/Nurr1 (data not shown), no GFP+/NeuN+ cells were detectable in the WM at 24 or 72 dpi. Instead, we found only GFAP+ cells, a few double-negative cells (GFAP−/NeuN−) and often no GFP+ cells at all, despite the fact that, at 10 dpi, GFP+ cells were always detectable (Figure 4B). Multiple mechanisms could explain these findings: WM astrocytes might be resistant to the reprogramming factors used here; the local environment might inhibit their maturation and integration; or they might reprogram into neurons and then migrate from the WM into the GM. In either case, these data reveal striking differences in astrocyte reprogramming between GM and WM.
Figure 4.
Absence of Induced Neurons after Transduction of White Matter Astrocytes with Ngn2/Nurr1 after Traumatic Brain Injury
(A and B) Photomicrographs showing GFP+ cells at 10, 24, and 72 dpi of GFP (A) and GFP/Ngn2/Nurr1 (B). Histograms show the percentage of marker+/− cells among the GFP+ cells for each experimental animal (3 or 4 per condition). Note that GFP+ cells were always detected in the control (all were astrocytes) but either not detectable or none were NeuN+ after Ngn2/Nurr1 injection. Data are shown as median ± IQR of different technical replicates, i.e., immunostainings of different sections. Scale bars: 20 μm.
Discussion
Direct neuronal reprogramming from local glial cells after injury represents a promising strategy for brain repair (Barker et al., 2018, Grade and Götz, 2017, Wang and Zhang, 2018). However, acquisition of adequate neuronal subtype identity and target region innervation is key for functional repair. Here, we describe a novel protocol using Ngn2 and Nurr1 to induce mature pyramidal neurons with different laminar and axonal projection identities from astrocytes after SW injury. Strikingly, Nurr1 is necessary for proneural factors to achieve the high conversion efficiency, and this is specific for the GM astrocytes, as no iNs are visible in the WM after applying the same reprogramming factors in the same lesion paradigm.
Surprisingly, we discovered an important role of Nurr1 in inducing cortical pyramidal neurons from local astrocytes. The combination of Nurr1 with the proneural factor Ngn2 achieved over 80% reprogramming efficiency and a significant increase in total number of iNs. Notably, efficiency and maturity of the converted neurons was much higher when Nurr1 was combined with Ngn2, compared to Ascl1. Nurr1 has also been previously used in reprogramming cocktails in vivo and in vitro toward generating dopaminergic neurons (Torper et al., 2015, Rivetti di Val Cervo et al., 2017), and although it is a well-known factor in specifying dopaminergic neurons in the ventral midbrain, it is also expressed in cortical and olfactory bulb neurons (Perlmann and Wallén-Mackenzie, 2004, Watakabe et al., 2007, Saino-Saito et al., 2004). Moreover, Nurr1 belongs to the orphan NR4A family exerting anti-inflammatory effects in different tissues (Rodríguez-Calvo et al., 2017). In the CNS, Nurr1 is recruited to the nuclear factor κB (NF-κB) promoter to regulate inflammatory genes and inhibit inflammatory signals in microglia and astrocytes (Saijo et al., 2009). This may be crucial for reprogramming, given the sensitivity of this process to reactive oxygen species (Gascón et al., 2016). Indeed, Nurr1 overexpression in neural stem cells upregulates trophic and survival signals to improve their response to oxidative stress, while downregulating cell cycle genes (Sousa et al., 2007). These multiple pathways controlled by Nurr1 may all contribute to its potent role in improving the efficiency and efficacy of cortical astrocyte-to-neuron reprogramming as uncovered here.
Strikingly, the combination of Ngn2 with Nurr1 not only achieved high efficiency in iN frequency and number, but also iNs displayed mature morphological and axonal projections adequate for their laminar position. iNs expressed CTIP2 and CUX1 combinations of characteristic TFs according to their laminar position. Both these iNs developed the typical morphology of pyramidal cells, with a prominent apical dendrite and an elaborated basal dendritic arbor, which gradually developed to resemble that of endogenous neurons. Finally, iNs acquired spine density and long-distance axonal projections to the striatum, midbrain, and thalamus as well as the contralateral cerebral cortex hemisphere. Most strikingly, these projections were not random, but rather iNs expressing SATB2 were found to project through the callosum, as is the case normally. Thus, these data show adequate neuronal subtype specification by direct in vivo reprogramming.
The precise identity reassignment according to the iN location prompted us to consider possible viral labeling artifacts. It is important to note that the AAVs used here also infect neurons, but as the flexed constructs are only recombined in Cre+ astrocytes, endogenous neurons do not express them. However, transfer of Cre to the endogenous neurons would drive the expression of the flexed constructs, as would the fusion of transduced astrocytes with endogenous neurons. Both cell fusion and Cre transfer would have to occur only in specific conditions, as we hardly see neurons labeled when we use only fluorescent reporter constructs, even when combined with only one proneural factor. Most importantly efficient labeling of endogenous neurons by embryonic EdU administration showed that mScarlet-I+ iNs were rarely EdU+, and the vast majority of mScarlet-I-labeled endogenous neurons (80%) were. This excludes the possibility of endogenous neurons as major contributor to the iNs. Together with the evidence that pyramidal neuron maturation was achieved gradually with, e.g., retrograde tracing iNs from the contralateral cortex only seen at 72, but not 24, dpi and the finding that 20% of iNs derive from cells proliferating after injury, these results lead us to favor the conclusion that the targeted astrocytes convert into neurons.
This then prompts the intriguing question of whether the layer-specific reprogramming is driven by cell-intrinsic mechanisms, specific migration, or environmental cues. Interestingly, astrocytes located in upper versus lower layers differ not only in their morphology but also in gene expression (Lanjakornsiripan et al., 2018). Surrounding neurons may play a key role in this process, as demonstrated in the cerebellum, where neuron-released Sonic hedgehog influenced local astrocyte transcriptional activity (Farmer et al., 2016). This supports the intriguing hypothesis that layer-dependent differences in cortical astrocytes might affect the outcome of reprogramming in terms of neuronal subtype identity. Such differences may also account for the apparent failure of WM astrocytes to turn into neurons, even though several possible mechanisms may contribute to the lack of iNs in the WM. However, regional specification may also pose hurdles to induce neurons that derive from other regions, such as cortical interneurons. Although the identity of the neurons reprogrammed by Ascl1 remains to be determined in detail, their phenotype was not typical of cortical GABAergic interneurons. Together, these results highlight the potential of astrocytes as promising starting cell population for direct neuronal reprogramming to generate neurons derived from the respective local brain region upon injury and highlight the fundamental importance of astrocyte subtypes not only to determine the efficiency of direct neuronal reprogramming but also the emerging neuronal subtype.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-GFAP | Dako | Cat. #Z0334; RRID: AB_10013382 |
| Mouse IgG1 anti-NeuN | Merk/Millipore | Cat. #MAB377; RRID: AB_2298772 |
| Rabbit anti-CUX1 | Santa Cruz | Cat. #Sc-13024; RRID: AB_2261231 |
| Mouse anti-SATB2 | Abcam | Cat. #Ab51502; RRID: AB_882455 |
| Guinea pig anti-SATB2 | SYSY | Cat. #327 004; RRID: AB_2620070 |
| Rat anti-CTIP2 | Abcam | Cat. #Ab18465; RRID: AB_2064130 |
| Chicken anti-GFP | Aves Labs | Cat. #GFP-1020; RRID: AB_10000240 |
| Rat anti-CD45 | BD/Bioscience | Cat. #550539; RRID: AB_2174426 |
| Rabbit anti-Iba1 | Wako | Cat. #019-19741; RRID: AB_839504 |
| Guinea pig anti-SOX9 | Lab. of Palle Serup | N/A |
| Rabbit anti-NeuN | Abcam | Cat. #Ab177487; RRID: AB_2532109 |
| Mouse anti-BRN2 | Santa Cruz | Cat. sc-393324; RRID: AB_2737347 |
| Rabbit anti-TBR1 | Abcam | Cat. ab31940; RRID: AB_2200219 |
| Rabbit anti-FOXP2 | Abcam | Cat. ab16046; RRID: AB_2107107 |
| Rabbit anti-TLE4 | Santa Cruz | Cat. sc-9125; RRID: AB_793141 |
| Bacterial and Virus Strains | ||
| Stable Competent E. coli (High Efficiency) | NEB | Cat.N: C3040 |
| rAAV2/5 CAG-FLEx-EGFP | SFB 870 Viral Vector Facility | https://www.sfb870.mcn.uni-muenchen.de/projects/subcluster-z/z04/index.html |
| rAAV2/5 CAG-FLEx-Ngn2 | SFB 870 Viral Vector Facility | https://www.sfb870.mcn.uni-muenchen.de/projects/subcluster-z/z04/index.html |
| rAAV2/5 CAG-FLEx-Nurr1 | SFB 870 Viral Vector Facility | https://www.sfb870.mcn.uni-muenchen.de/projects/subcluster-z/z04/index.html |
| rAAV2/5 CAG-FLEx-Ascl1 | SFB 870 Viral Vector Facility | https://www.sfb870.mcn.uni-muenchen.de/projects/subcluster-z/z04/index.html |
| rAAV2/1 hSyn-FLEx-pmScarletI-P2A-GCaMP6s | SFB 870 Viral Vector Facility | https://www.sfb870.mcn.uni-muenchen.de/projects/subcluster-z/z04/index.html |
| rAAV2/1 hSyn-pmScarletH-P2A-GCaMP6s | SFB 870 Viral Vector Facility | https://www.sfb870.mcn.uni-muenchen.de/projects/subcluster-z/z04/index.html |
| Chemicals, Peptides, and Recombinant Proteins | ||
| OptiPrep Density Gradient Medium | Sigma | Cat. #D1556 |
| DAPI | Sigma | Cat. #28718-90-3 |
| SYBR Green Real-Time PCR Master Mixes | Thermo Fisher Scientific | Cat. #4385612 |
| Fluorogold dye (Hydroxystilbamidine) | Sigma | Cat. #223769-64-0 |
| EdU (5-ethynyl-2′-deoxyuridine) | Invitrogen | Cat. #E10187 |
| Click-iT EdU Alexa Fluor 647 Imaging Kit | Invitrogen | Cat. #C10340 |
| Experimental Models: Cell Lines | ||
| 293T | ATCC | CRL-3216; RRID: CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6.Cg-Tg(Gfap-cre)77.6Mvs/2J | The Jackson Laboratory | Stock N: 024098; RRID: IMSR_JAX:024098 |
| Mouse: C57BL/6J | The Jackson Laboratory | Stock N: 000664; RRID: IMSR_JAX:000664 |
| Oligonucleotides | ||
| SV40 Forward primer (5′AGCAATAGCATCA CAAATTTCACAA3′) |
Laboratory of Eduard Ayuso (D’Costa et al., 2016) | N/A |
| SV40 reverse primer (5′CCAGACATGATAAGA TACATTGATGAGTT3′) |
Laboratory of Eduard Ayuso (D’Costa et al., 2016) | N/A |
| GFP Forward primer (5′GATGTTGTGGC GGATCTTGA3′) |
Laboratory of Eduard Ayuso (D’Costa et al., 2016) | N/A |
| GFP reverse primer (5′CAACAGCCACAACG TCTATATCATG3′) |
Laboratory of Eduard Ayuso (D’Costa et al., 2016) | N/A |
| Recombinant DNA | ||
| Plasmid: pAAV-CAG-FLEx-EGFP | Laboratory of Malin Parmar (Torper et al., 2015) | N/A |
| Plasmid: pAAV-hSyn-FLEx-pmScarletI-P2A-GCaMP6 | Laboratory of Tobias Rose | N/A |
| Plasmid: pAAV-hSyn-pmScarletH-P2A-GCaMP6s | Laboratory of Tobias Rose | N/A |
| Plasmid: pAAV-CAG-FLEx-Ngn2 | Laboratory of Malin Parmar (Torper et al., 2015) | N/A |
| Plasmid: pAAV-CAG-FLEx-Nurr1 | Laboratory of Malin Parmar (Torper et al., 2015) | N/A |
| Plasmid: pAAV-CAG-FLEx-Ascl1 | Laboratory of Malin Parmar (Torper et al., 2015) | N/A |
| Plasmid: pHelper | Cell Biolabs | Part N: 340202 |
| Plasmid: pAAV-RC5 | Cell Biolabs | Cat.N: VPK-425 |
| Plasmid: LV- PRSVLTR-psi-CAG-GFP-WPRE-LTRSIN | Laboratory of Magdalena Götz (Buffo et al., 2008) | N/A |
| Plasmid: RV- CAG-Ngn2-IRES-Dsred | Laboratory of Magdalena Götz (Heinrich et al., 2011) | N/A |
| Software and Algorithms | ||
| Zen software 2.3 lite | Carl Zeiss | RRID: SCR_013672; https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html |
| ImageJ | NIH | RRID: SCR_003070; https://imagej.nih.gov/ij/ |
| Photoshop | Adobe | RRID: SCR_014199; https://www.adobe.com/products/photoshop.html |
| Illustrator | Adobe | RRID: SCR_014199; https://www.adobe.com/products/illustrator.html |
| GraphPad Prism 7.0 | GraphPad Software | RRID: SCR_002798; https://www.graphpad.com |
| MATLAB | MathWorks | RRID: SCR_001622; https://www.mathworks.com |
| Other | ||
| FPLC | GE Healthcare | ÄKTA prime plus |
| HiTrap Q FF anion exchange chromatography column | GE Healthcare | Cat. 17505301 |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Magdalena Götz (magdalena.goetz@helmholtz-muenchen.de).
Experimental Model and Subject Details
Animals
Animal handling and experimental procedures were performed in accordance with German and European Union guidelines and were approved by the State of upper Bavaria. All efforts were made to minimize suffering and number of animals. Both male and female mice were used. mGFAP-cre mice were obtained from Jackson Laboratory (#024098). Mice were maintained in specific pathogen-free conditions in the animal facility, in 12:12 h light/dark cycles.
Method Details
Stab wound and viral injection
Adult (2-3 months) heterozygous mGFAP-cre mice (Jackson Laboratory #024098) were used for stab wound injury as described in Mattugini et al. (2018) with slight modifications. Briefly, animals were anesthetized and injured on one hemisphere from Bregma: RC: −0.4 to −1.4 mm; ML: −1.0 mm; DV: −1.2 mm. Further details on cortical stab wound injury and the neuronal loss associated with it are described in Frik et al. (2018) and Robel et al. (2011). Three days after stab wound injury mice were injected with AAV virus into the site of the lesion using a microinjector at the speed of 40-50 nl/min. For control experiments we injected 200-300 nL of -AAV-FLEx-GFP. To label endogenous neurons we injected 200-300 nL of AAV-mScarlet-I (non-FLEx). For reprogramming experiments, we injected 800-1000 nL of the following mixes containing 1/2 or 1/3 of each virus: AAV-FLEx-GFP / AAV-FLEx-Ngn2; AAV-FLEx-GFP / AAV-FLEx-Ascl1; AAV-FLEx-GFP / AAV-FLEx-Nurr1; AAV-FLEx-GFP/ AAV-FLEx-Ngn2 / AAV-FLEx-Nurr1; AAV-FLEx-GFP / AAV-FLEx-Ascl1 / AAV-FLEx-Nurr1; AAV-FLEx-mScarlet-I / AAV- FLEx-Ngn2/ AAV-FLEx-Nurr1. For retro- and lenti-viral vectors testing, wild-type animals were injured at the following coordinates from Bregma: RC: −0.4 to −1.4 mm; ML: −1.0 mm; DV: −0.6 mm. Three days after stab wound injury mice were injected with 300 nL of LV-GFP (VSV-G) or 800nl of RV-Ngn2-ires-dsRed. Where indicated, EdU (0,2 mg/ml) was administrated in 1% sucrose drinking water.
Immunohistochemistry
Mice were anesthetized and trans-cardially perfused with 4% paraformaldehyde (PFA) in PBS. Brains were post-fixed for 24 h in 4% PFA and then cryo-protected in 30% sucrose in PBS. Fifty-micrometer-thick brain sections were cut at the vibratome and incubated with blocking solution composed of PBS 2% bovine serum albumin (BSA Sigma Aldrich), 0.5% Triton X-100 (Sigma Aldrich) for 20 min and overnight with primary antibodies diluted in blocking solution. The day after, free-floating sections were washed in PBS and incubated with appropriate secondary antibodies, diluted in the blocking solution for 2 h. Finally, sections were washed and stained with 4’, 6-diamidino-2-phenylindole (DAPI; Sigma Aldrich) for 5 min.
Treatment with NaCitrate 10 mM pH 8.5/Tween 20 0.05% at 80°C for 30 min was performed before incubation with standard blocking solution for CTIP2, BRN2, and TLE4 immunohistochemistry. EdU was detected using the Click-iT EdU imaging kit (Invitrogen) following the protocol of the kit.
Viral vectors preparation
High-titer preparations of rAAV2/5 were produced based on protocol of Zolotukhin et al. (1999) with minor modifications. In brief, HEK293T cells were transfected with the CaPO4 precipitate method, pRC5, Ad helper and pAAV plasmid at a 1:1:1 molar ratio. After 72 h, cell pellets were harvested with AAV release solution, 50 U/ml benzonase was added, then incubated for 2 h at 37°C in a water bath. Cells were frozen and thawed in liquid nitrogen to allow rAAV release. Purification of rAAV vector was done with iodixanol density gradient (consisting of 15, 25, 40 and 56% iodixanol), followed by gradient spinning at 50,000 rpm for 2 h 17 min at 22°C in a Ti70 rotor (Beckman, Fullerton, CA, USA). rAAVs were collected at 40% iodixanol with 5-ml-syringe. rAAVs were dialyzed (Slide-A-Lyzer 10,000 MWCO 5ml) in buffer A overnight to remove iodixanol. Anion exchange chromatography column HiTrap QFF Sepharose column and superloop were connected with the ÄKTAprime plus chromatography system to collect the eluted fraction. To measure rAAV concentration, the eluted fraction was spun and washed once in PBS-MK Pluronic-F68 buffer with Millipore 30K MWCO 6ml filter unit. rAAVs were stored in glass vial tube in 4°C. rAAVs were titered by SYBR Green qPCR with GFP or SV40 primer (D’Costa et al., 2016). Usual titer was 3 x1014 to 5x1015 gc/ml. Retrovirus and Lentivirus were prepared as already described in Gascón et al., 2016, Hack et al., 2005, and Heinrich et al. (2011).
Retrograde labeling
Anesthetized animals were placed in a stereotaxic apparatus and injected with Fluoro-Gold (300 nl) into the contralateral homotypic cortex at 20 dpi or 68 dpi, in order to retrogradely label callosal projection neurons. Animal were scarified 4 days later (at 24 or 72 dpi, respectively) and analyzed.
Electrophysiology
Acute coronal slices (300 μm thick) were prepared from mGFAP-cre mice in chilled dissection solution based on either choline (in mM: 110 choline chloride, 25 NaHCO3, 25 D-glucose, 11.6 Na-ascorbate, 7 MgCl2, 3.1 Na-pyruvate, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2) or sucrose (in mM 85 NaCl, 75 sucrose, 2.5 KCL, 23 glucose, 24 mM NaHCO3, 1.25 NaH2PO4, 4 MgCl2, 0.5 CaCl2) as described previously (Scheuss et al., 2006, Weiler et al., 2018). When the cutting solution was choline based, slices were incubated in artificial CSF (ACSF) (in mM: 127 NaCl, 25 NaHCO3, 25 D-glucose, 2.5 KCl, 1 MgCl2, 2 CaCl2, 1.25 NaH2PO4) bubbled with carbogen (95% O2, 5% CO2) at 35°C until use. When the cutting solution was sucrose based, slices were first incubated in sucrose cutting solution bubbled with carbogen at 35°C for 30 - 60 min and then in bubbled ACSF at room temperature until use. In the recording chamber, the extracellular solution was ACSF bubbled with carbogen at room temperature. Somatic whole-cell recordings (pipette resistance, 3–4 MΩ) were performed on mScarlet-I expressing neurons, confirmed either by standard fluorescence or by 2-photon microscopy. The internal solution contained (in mM) 135 K-methylsulfonate, 10 HEPES, 10 Na-phosphocreatine, 4 MgCl2, 4 Na-ATP, 0.4 Na-GTP, 3 Na-L-ascorbate, and 30 μM Alexa 488. Data were acquired via Multiclamp 700B patch amplifier (Molecular Devices, Sunnyvale, CA) controlled with custom software written in Labview (National Instruments, Austin, TX). First, 100 ms long current pulses of various amplitudes were applied for eliciting a single action potential and determining the rheobase. Second, 1 s long current pulses were applied in steps of 100 pA from −300 pA to −100 pA, in steps of 10 pA from −50 pA to +50 pA and again in steps of 100 pA from 100 pA to 300 pA for recording spiking patterns. Third, spontaneous synaptic currents (SSCs) were recorded in voltage clamp at −70 mV (excitatory SSCs) and −30 mV (inhibitory SScs). In some cells, for confirming excitatory and inhibitory inputs, the recordings of spontaneous activity were repeated in 10 μM NMQX (2,3-Dioxo-6-nitro-1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide) and 10 μM bicuculline.
Quantification and Statistical Analysis
All images (30-40 μm thick z stacks) were acquired on a confocal laser scanning (Zeiss LSM710) microscope. At least three sections from each animal (at least 3) were analyzed for quantifications. All quantifications of confocal images were done using Zen or ImageJ software.
The proportions of neurons and astrocytes (Figures 1F, 1I, 4A, 4B, S1B, S1C, S2A, and S2B) were obtained by counting NeuN+, GFAP+, SOX9+, NeuN−/GFAP− and NeuN−/SOX9− cells among the transduced GFP+ population. Absolute numbers of neurons and astrocytes (Figure S2C) was calculated counting the numbers of NeuN+/GFP+ and GFAP+/GFP+ per mm3.
For CUX1/CTIP2 analysis (Figure 2A), the intensities of CUX1 and CTIP2 were measured on the somata of both endogenous and induced neurons (>2000 mScarlet-I+ cells per condition were measured) and plotted on the y axis according to their laminar position. The normalized intensity threshold defining cells positive for CUX1 or CTIP2 was arbitrary fixed at 0.2. The number of positive (CUX1+ or CTIP2+) cells was divided by the total number of m-Scarlet-I+ cells in each bin, representative of ten different cortical levels.
For CUX1/CTIP2 distribution (Figure S4A) the intensity level of CUX1 and CTIP2 immunostaining was measured along a vertical line with ImageJ software. The length of the line was then normalized as percentage distance from the ventricle and averaged values were represented divided in 135 bins covering the entire thickness cortex.
Intensity for CUX1 and CTIP2 immunostaining and size of the cell soma (Figure S4B) for mScarlet− and mScarlet+ neurons was calculated analyzing 50 cells per conditions. Both measurements were performed using DAPI staining as a reference using ImageJ software.
For molecular identity analysis (Figure S4D) proportion of SATB2+, CUX1+, BRN2+, CUX1+/SATB2+/BRN2+ (for upper layer) and TBR1+, FOXP2+, CTIP2+, TLE4+, CTIP2+/FOXP2+, CTIP2+/TLE4+ (for lower layer) were calculated among the mScarlet-I+ neurons present in upper (L2/3) and lower layers (L5/6) for both, endogenous and reprogrammed neurons. Our molecular analysis was then compared with the single cell RNAseq from Linnarsson laboratory (Figure S4F; http://mousebrain.org) in order to identify four cortical excitatory neurons subpopulation (TEGLU7: layer 2/3, TEGLU8: layer 4, TEGLU5: entorhinal superficial layer and TEGLU3: layer 6) generated by our reprogramming protocol.
For molecular identity maturation analysis (Figure S4E) the proportion of CUX1+ and CTIP2+ neurons was calculated among the mScarlet-I+ neurons present in the entire cortical column at 2 different time points (24 and 72 dpi) for induced neurons and compared to the endogenous proportion.
For EdU analysis (Figures S3A–S3D) the number of EdU+ cells was counted among the mScarlet-I+ neurons for both, endogenous and induced neurons located in the deep layers. For details on experimental time line see the respective figures.
For morphological analysis (Figures 2B–2G), 50 cells per conditions (endogenous and reprogrammed m-Scarlet-I+ neurons) were analyzed. The presence of the apical dendrite was defined by the presence of a prominent primary dendrite oriented to the pial surface. The thickness of the apical leading process was measured in its proximal part (5 μm from the cell soma). The circularity was measured using an ImageJ plugin drawing the cell soma. The number of primary processes was calculated as the number of processes leaving the cell soma in each neuron. The polarization was analyzed by measuring angles between the apical dendrite (red dots in Figure 2) and all the others primary dendrites (gray or blue dots) and plotted into a circular diagram divided in 12 different bins corresponding to the 360° orientation angles.
For spine density (Figure 3A), high power images were acquired and quantifications were done by counting only laterally emanating protrusions longer than 0.4 μm (Holtmaat et al., 2009).
For fluorogold analysis (Figure 3G) fluorogold+ neurons were counted among mScarlet-I+/SATB2+/CTIP2− (in layer 2/3 and in layer 5/6) and mScarlet-I+/SATB2−/CTIP2+ (in layer 5/6).
The analysis of electrophysiological data (Figures 3B, 3D, and S5A–S5C) was performed with custom routines in MATLAB (version R2018b, MathWorks, Natick, MA, USA).
All statistical tests were performed with GraphPad Prism 7.0 Software unless stated otherwise. Statistical significance was defined at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. All biological replicates (n) are derived from at least three independent experiments. All column graphs are expressed as median ± IQR. The normality of the distribution of data points was verified using Shapiro-Wilk test. For variables having normal distribution statistical significance was calculated using either two-sided, unpaired Student’s t test or one-way ANOVA followed by Tukey or Dunnett post-test, as indicated. When normality tests failed to show normal distribution, non-parametric Mann-Whitney or Kruskal-Wallis and Dunn’s multiple comparison tests were used, respectively. For electrophysiological analyses using MATLAB (version R2018b, MathWorks, Natick, MA, USA), statistical significance of the mean and variance was calculated using Wilcoxon rank-sum test or Levene’s test, respectively.
Data and Code Availability
This study did not generate datasets.
Acknowledgments
We are very grateful to Malin Parmar and Tobias Rose for providing the plasmids of the AAV-FLEx constructs; to Palle Serup for providing the SOX9 antibody; to Andreas Beyerlein from the Core Facility Statistical Consulting Helmholtz Zentrum München for statistical help; to the viral vector facility of the Collaboratives Research Center (SFB870); to Sabine Ulbricht, Manja Thorwirth, and Gabi Jaeger for the excellent technical help; to Alexandra Lepier and Ines Muehlhahn for establishing the AAV production in the lab; and to Adam O’Neill for excellent comments on the manuscript. This work was financed by the German Research Foundation (SFB 870, A08 to V.S. and A06 and Z04 to M.G.); Micronet (Era-Net, 01EW1705B); the Roger de Spoelberch Foundation and the advanced ERC grant ChroNeuroRepair (340793) to M.G.; and the SNF post-doctoral fellowship (P2GEP3_174900 and P400PB_183826) to R.B.
Author Contributions
M.G., N.M., and R.B. conceived the experiments, discussed the data, and wrote the manuscript. N.M. and R.B. performed all experiments except those specified below and analyzed the data. G.L.R. tested the immune response to viral vectors. V.S. performed and analyzed all electrophysiological experiments. O.T. and C.L.L. provided AAV expertise and scientific inputs. C.L.L. prepared all viral vectors.
Declaration of Interests
The authors declare no competing interests.
Published: September 2, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.neuron.2019.08.009.
Supplemental Information
References
- Atasoy D., Aponte Y., Su H.H., Sternson S.M. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker R.A., Götz M., Parmar M. New approaches for brain repair-from rescue to reprogramming. Nature. 2018;557:329–334. doi: 10.1038/s41586-018-0087-1. [DOI] [PubMed] [Google Scholar]
- Bayraktar O.A., Fuentealba L.C., Alvarez-Buylla A., Rowitch D.H. Astrocyte development and heterogeneity. Cold Spring Harb. Perspect. Biol. 2014;7:a020362. doi: 10.1101/cshperspect.a020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo A., Vosko M.R., Ertürk D., Hamann G.F., Jucker M., Rowitch D., Götz M. Expression pattern of the transcription factor Olig2 in response to brain injuries: implications for neuronal repair. Proc. Natl. Acad. Sci. USA. 2005;102:18183–18188. doi: 10.1073/pnas.0506535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo A., Rite I., Tripathi P., Lepier A., Colak D., Horn A.P., Mori T., Götz M. Origin and progeny of reactive gliosis: a source of multipotent cells in the injured brain. Proc. Natl. Acad. Sci. USA. 2008;105:3581–3586. doi: 10.1073/pnas.0709002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Costa S., Blouin V., Broucque F., Penaud-Budloo M., François A., Perez I.C., Le Bec C., Moullier P., Snyder R.O., Ayuso E. Practical utilization of recombinant AAV vector reference standards: focus on vector genomes titration by free ITR qPCR. Mol. Ther. Methods Clin. Dev. 2016;5:16019. doi: 10.1038/mtm.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer W.T., Abrahamsson T., Chierzi S., Lui C., Zaelzer C., Jones E.V., Bally B.P., Chen G.G., Théroux J.F., Peng J. Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science. 2016;351:849–854. doi: 10.1126/science.aab3103. [DOI] [PubMed] [Google Scholar]
- Frik J., Merl-Pham J., Plesnila N., Mattugini N., Kjell J., Kraska J., Gómez R.M., Hauck S.M., Sirko S., Götz M. Cross-talk between monocyte invasion and astrocyte proliferation regulates scarring in brain injury. EMBO Rep. 2018;19 doi: 10.15252/embr.201745294. e45294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascón S., Murenu E., Masserdotti G., Ortega F., Russo G.L., Petrik D., Deshpande A., Heinrich C., Karow M., Robertson S.P. Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell. 2016;18:396–409. doi: 10.1016/j.stem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Gascón S., Masserdotti G., Russo G.L., Götz M. Direct neuronal reprogramming: achievements, hurdles, and new roads to success. Cell Stem Cell. 2017;21:18–34. doi: 10.1016/j.stem.2017.06.011. [DOI] [PubMed] [Google Scholar]
- Govindan S., Jabaudon D. Coupling progenitor and neuronal diversity in the developing neocortex. FEBS Lett. 2017;591:3960–3977. doi: 10.1002/1873-3468.12846. [DOI] [PubMed] [Google Scholar]
- Grade S., Götz M. Neuronal replacement therapy: previous achievements and challenges ahead. NPJ Regen. Med. 2017;2:29. doi: 10.1038/s41536-017-0033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorian C., Nakashima J., Le Belle J., Ohab J., Kim R., Liu A., Smith K.B., Groszer M., Garcia A.D., Sofroniew M.V. Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J. Neurosci. 2009;29:1874–1886. doi: 10.1523/JNEUROSCI.3095-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig L.C., Woodworth M.B., Galazo M.J., Padmanabhan H., Macklis J.D. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack M.A., Saghatelyan A., de Chevigny A., Pfeifer A., Ashery-Padan R., Lledo P.M., Götz M. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat. Neurosci. 2005;8:865–872. doi: 10.1038/nn1479. [DOI] [PubMed] [Google Scholar]
- Harris K.D., Shepherd G.M. The neocortical circuit: themes and variations. Nat. Neurosci. 2015;18:170–181. doi: 10.1038/nn.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Blum R., Gascón S., Masserdotti G., Tripathi P., Sánchez R., Tiedt S., Schroeder T., Götz M., Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C., Gascón S., Masserdotti G., Lepier A., Sanchez R., Simon-Ebert T., Schroeder T., Götz M., Berninger B. Generation of subtype-specific neurons from postnatal astroglia of the mouse cerebral cortex. Nat. Protoc. 2011;6:214–228. doi: 10.1038/nprot.2010.188. [DOI] [PubMed] [Google Scholar]
- Holtmaat A., Bonhoeffer T., Chow D.K., Chuckowree J., De Paola V., Hofer S.B., Hübener M., Keck T., Knott G., Lee W.C. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabaudon D. Fate and freedom in developing neocortical circuits. Nat. Commun. 2017;8:16042. doi: 10.1038/ncomms16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Lin C.C., Yu K., Hatcher A., Huang T.W., Lee H.K., Carlson J., Weston M.C., Chen F., Zhang Y., Zhu W. Identification of diverse astrocyte populations and their malignant analogs. Nat. Neurosci. 2017;20:396–405. doi: 10.1038/nn.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanjakornsiripan D., Pior B.J., Kawaguchi D., Furutachi S., Tahara T., Katsuyama Y., Suzuki Y., Fukazawa Y., Gotoh Y. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun. 2018;9:1623. doi: 10.1038/s41467-018-03940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S., Arlotta P. Generating neuronal diversity in the mammalian cerebral cortex. Annu. Rev. Cell Dev. Biol. 2015;31:699–720. doi: 10.1146/annurev-cellbio-100814-125353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masserdotti G., Gascón S., Götz M. Direct neuronal reprogramming: learning from and for development. Development. 2016;143:2494–2510. doi: 10.1242/dev.092163. [DOI] [PubMed] [Google Scholar]
- Mattugini N., Merl-Pham J., Petrozziello E., Schindler L., Bernhagen J., Hauck S.M., Götz M. Influence of white matter injury on gray matter reactive gliosis upon stab wound in the adult murine cerebral cortex. Glia. 2018;66:1644–1662. doi: 10.1002/glia.23329. [DOI] [PubMed] [Google Scholar]
- Molyneaux B.J., Arlotta P., Menezes J.R., Macklis J.D. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Perlmann T., Wallén-Mackenzie A. Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res. 2004;318:45–52. doi: 10.1007/s00441-004-0974-7. [DOI] [PubMed] [Google Scholar]
- Pesaresi M., Sebastian-Perez R., Cosma M.P. Dedifferentiation, transdifferentiation and cell fusion: in vivo reprogramming strategies for regenerative medicine. FEBS J. 2019;286:1074–1093. doi: 10.1111/febs.14633. [DOI] [PubMed] [Google Scholar]
- Ridder K., Keller S., Dams M., Rupp A.K., Schlaudraff J., Del Turco D., Starmann J., Macas J., Karpova D., Devraj K. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLoS Biol. 2014;12:e1001874. doi: 10.1371/journal.pbio.1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P., Romanov R.A., Spigolon G., Masini D., Martín-Montañez E., Toledo E.M., La Manno G., Feyder M., Pifl C., Ng Y.H. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson’s disease model. Nat. Biotechnol. 2017;35:444–452. doi: 10.1038/nbt.3835. [DOI] [PubMed] [Google Scholar]
- Robel S., Bardehle S., Lepier A., Brakebusch C., Götz M. Genetic deletion of cdc42 reveals a crucial role for astrocyte recruitment to the injury site in vitro and in vivo. J. Neurosci. 2011;31:12471–12482. doi: 10.1523/JNEUROSCI.2696-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Calvo R., Tajes M., Vázquez-Carrera M. The NR4A subfamily of nuclear receptors: potential new therapeutic targets for the treatment of inflammatory diseases. Expert Opin. Ther. Targets. 2017;21:291–304. doi: 10.1080/14728222.2017.1279146. [DOI] [PubMed] [Google Scholar]
- Saijo K., Winner B., Carson C.T., Collier J.G., Boyer L., Rosenfeld M.G., Gage F.H., Glass C.K. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino-Saito S., Sasaki H., Volpe B.T., Kobayashi K., Berlin R., Baker H. Differentiation of the dopaminergic phenotype in the olfactory system of neonatal and adult mice. J. Comp. Neurol. 2004;479:389–398. doi: 10.1002/cne.20320. [DOI] [PubMed] [Google Scholar]
- Scheuss V., Yasuda R., Sobczyk A., Svoboda K. Nonlinear [Ca2+] signaling in dendrites and spines caused by activity-dependent depression of Ca2+ extrusion. J. Neurosci. 2006;26:8183–8194. doi: 10.1523/JNEUROSCI.1962-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa K.M., Mira H., Hall A.C., Jansson-Sjöstrand L., Kusakabe M., Arenas E. Microarray analyses support a role for Nurr1 in resistance to oxidative stress and neuronal differentiation in neural stem cells. Stem Cells. 2007;25:511–519. doi: 10.1634/stemcells.2006-0238. [DOI] [PubMed] [Google Scholar]
- Torper O., Ottosson D.R., Pereira M., Lau S., Cardoso T., Grealish S., Parmar M. In vivo Reprogramming of striatal NG2 glia into functional neurons that integrate into local host circuitry. Cell Rep. 2015;12:474–481. doi: 10.1016/j.celrep.2015.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.L., Zhang C.L. Engineering new neurons: in vivo reprogramming in mammalian brain and spinal cord. Cell Tissue Res. 2018;371:201–212. doi: 10.1007/s00441-017-2729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A., Ichinohe N., Ohsawa S., Hashikawa T., Komatsu Y., Rockland K.S., Yamamori T. Comparative analysis of layer-specific genes in mammalian neocortex. Cereb. Cortex. 2007;17:1918–1933. doi: 10.1093/cercor/bhl102. [DOI] [PubMed] [Google Scholar]
- Weiler S., Bauer J., Hübener M., Bonhoeffer T., Rose T., Scheuss V. High-yield in vitro recordings from neurons functionally characterized in vivo. Nat. Protoc. 2018;13:1275–1293. doi: 10.1038/nprot.2018.026. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets.