Sex-determination in Mercurialis annua is not related to chromatin conformation or DNA methylation of floral homeotic genes but might be regulated upstream of these genes by one or more unknown gender-specific factors that affect hormonal homeostasis.
Keywords: Chromatin, cytokinin, dioecy, DNA methylation, epigenetics, feminization, floral homeotic gene, Mercurialis annua, nuclear proteome, sex-determination
Abstract
In plants, dioecy characterizes species that carry male and female flowers on separate plants and it occurs in about 6% of angiosperms; however, the molecular mechanisms that underlie dioecy are essentially unknown. The ability for sex-reversal by hormone application raises the hypothesis that the genes required for the expression of both sexes are potentially functional but are regulated by epigenetic means. In this study, proteomic analysis of nuclear proteins isolated from flower buds of females, males, and feminized males of the dioecious plant Mercurialis annua revealed differential expression of nuclear proteins that are implicated in chromatin structure and function, including floral homeotic proteins. Focusing on floral genes, we found that class B genes were mainly expressed in male flowers, while class D genes, as well as SUPERMAN-like genes, were mainly expressed in female flowers. Cytokinin-induced feminization of male plants was associated with down-regulation of male-specific genes concomitantly with up-regulation of female-specific genes. No correlation was found between the expression of class B and D genes and the changes in DNA methylation or chromatin conformation of these genes. Thus, we could not confirm DNA methylation or chromatin conformation of floral genes to be the major determinant regulating sexual dimorphisms. Instead, determination of sex in M. annua might be controlled upstream of floral genes by one or more sex-specific factors that affect hormonal homeostasis. A comprehensive model is proposed for sex-determination in M. annua.
Introduction
The majority of angiosperms are hermaphrodites and monoecious (sexually monomorphic), whereby both male and female organs are found on the same individual plant. In contrast, only about 6% of the angiosperms are dioecious (sexually dimorphic), where male and female flowers are carried on separate individual plants (Renner and Ricklefs, 1995; Charlesworth, 2002). Obviously, the question of cost of sexual reproduction in dioecious species has been considered by evolutionary biologists, since there is a greater cost when two individuals are required for production of offspring in contrast to hermaphrodites and monoecious plants, where one individual is sufficient. This question has puzzled botanists for generations, including Darwin (1877). Various theoretical considerations, definitions, and models have been proposed over the years to explain dioecy, but mechanistic studies to understand the regulation of sex-determination in dioecious species at the molecular level have failed to provide a comprehensive model of the process (Obeso, 2002). In this study we do not ask the question of why dioecy exists, but rather of how? Genetic aspects related to sex-determination in dioecious species have been studied quite intensively, including our own search for molecular markers in dimorphic species (Golan-Goldhirsh et al., 2001; Khadka et al., 2002, 2005; Yakubov et al., 2005). Dioecious plants show diversity in sex-determination systems that range from a single locus to heteromorphic chromosomes, indicating the independent origin of dioecy in various plant families (Charlesworth and Charlesworth, 1978; Akagi et al., 2014; Harkess et al., 2017; Puterova et al., 2018).
The annual dioecious Mercurialis annua has been used as a model plant for dioecy because it has a short life cycle that makes it amenable to molecular-genetic studies, in contrast to most dioecious plants that are woody perennials. In addition, M. annua is also amenable to sex conversion by hormonal treatment, which allows a myriad of experimental designs for particular biological questions of sex determination. Phytohormones play a role in sex determination in plants, often acting in a species-specific manner to specify gender (reviewed in Golenberg and West, 2013). Ethylene and gibberellins contribute to sex differentiation in cucumber and maize, respectively (Hansen et al., 1976; Trebitsh et al., 1997). Hormone-induced sex change has also been shown in Spinacia oleracea (West and Golenberg, 2018). In M. annua, exogenous application of auxins has been shown to induce masculinization while cytokinins induce feminization of male plants (Delaigue et al., 1984; Durand and Durand, 1991). Identification of male-specific molecular markers and recent genetic analyses have revealed that male M. annua possess homomorphic XY chromosomes, but which genetic components are responsible for sex determination and floral dimorphism is not yet fully known (Khadka et al., 2002, 2005; Russell and Pannell, 2015; Veltsos et al., 2018). The genome of M. annua has recently been assembled and contains over 34 000 genes, of which about third have been assigned to linkage groups, with the sex chromosome appearing as the largest group. Based on genetic mapping and exome resequencing, it has been estimated that about one-third of the Y chromosome has lost recombination capacity, which might facilitate divergence between the sexes in M. annua (Veltsos et al., 2019). Furthermore, transcriptome analysis of males and females has revealed differential gene expression between them at the first leaf stage, while expression of sex-biased genes peaks just prior to, and after, flowering (Cossard et al., 2019).
Most studies related to the regulation of flower development have been performed in hermaphroditic model species such as Arabidopsis thaliana that have four whorled flowers. Mercurialis annua belongs to type II dioecious species and has three apparent whorls, without the rudiment whorl of the opposite sex (Mitchell and Diggle, 2005). Dioecious species are considered to be amenable for identification of the genetic and epigenetic components involved in dimorphic flowers and sex determination. Therefore, in this study we have used this species to examine several functional classes of floral homeotic, MADS box-containing transcription factors (TFs) that regulate organ identity in various whorls, which are described by the ABCDE model (Coen and Meyerowitz, 1991; Krizek and Fletcher, 2005; Theißen et al., 2016). Thus, the class A proteins APETALA1 (AP1) and AP2 together with the class E proteins SEPALLATA1 (SEP1) to SEP4 specify sepals, class B proteins such as AP3 and PISTILLATA (PI) together with class A and class E proteins specify petals, class C AGAMOUS (AG) together with class B and class E proteins specify stamens, class C and class E proteins specify carpels, and class D proteins such as SHATTERPROOF1 (SHP1)/AGAMOUS-LIKE1 (AGL1) together with class E proteins specify ovules (Theißen and Saedler, 2001; Soltis et al., 2007). The class B–E proteins play the key role in the development of reproductive whorls, i.e. the stamens and carpels, whilst the SUPERMAN (SUP) transcription factor has been proposed to act as a negative regulator of class B genes to maintain boundaries between the two whorls (Bowman et al., 1992; Yun et al., 2002; Wuest et al., 2012; Ó’Maoiléidigh et al., 2013; Stewart et al., 2016; Prunet et al., 2017).
Multiple studies that have examined the expression patterns of MADS-box floral genes in type I dioecious plants such as Silene latifolia and Rumex acetosa (Hardenack et al., 1994; Ainsworth et al., 1995) as well as in type II dioecious plants such as Thalictrum dioicum and S. oleracea (Di Stilio et al., 2005; Pfent et al., 2005) have shown that their expression in dioecious plants essentially follows the classical ABC model of flowering (Coen and Meyerowitz, 1991). Consistent with these findings, Sather et al. (2010) showed that silencing of class B genes in S. oleracea is sufficient to alter the floral gender of males into hermaphrodites or females due to transformation of stamens into carpels. However, the genetic and/or epigenetic regulation of the sexually dimorphic expression of floral genes is poorly understood. The ability of hormone application to cause sex-reversal suggests sexual bi-potency in M. annua, and that the genes required for the development of both sexes are present in both genders but they may be restrained by various factors, including epigenetics in the floral primordia, to bring about dioecy. Epigenetics refers to changes in heritable phenotypes that do not involve changes in the DNA sequence but instead involve changes in the regulation of gene expression. This is brought about by multiple mechanisms that control chromatin structure and function, including DNA methylation and histone modification, which are often controlled by sRNA-based mechanisms (Gibney and Nolan, 2010).
The capacity for hormonal sex-reversal in M. annua prompted us to examine the expression pattern its floral homeotic genes and to determine whether their epigenetic regulation represents the major constituent in sex determination. We found that differential expression of floral homeotic genes was associated with sexual dimorphism in M. annua and that cytokinin was involved in their transcriptional control. Furthermore, cytokinin-induced feminization of males was accompanied by extensive changes in nuclear proteins that are involved in chromatin structure and function. However, the relationship between sexual dimorphism and epigenetic regulation of floral homeotic genes could not be confirmed in the present work. Based on our results, a model is proposed for sex determination in M. annua.
Materials and methods
Plant growth conditions
Dioecious Mercurialis annua (Euphorbiaceae) of Belgian origin was used in this study. Seeds were sown in trays containing standard gardening soil and seedlings were transplanted into 2.5-l pots and grown in a controlled climate growth chamber at 27 °C with photoperiod of 14/10 h light/dark and light intensity of ~400 µmol m−2 s−1.
Feminization of male plants by treatment with 6-benzylaminopurine
At the onset of flowering (plants ~25 d old), male and female plants were separated. Feminization of the isolated male plants was done by spraying 1 mg l−1 6-benzylaminopurine (BAP) three times daily as described previously (Durand and Durand, 1991; Khadka et al., 2005). Samples from three biological replicates were collected and either used immediately for isolation of nuclei or stored at –80 °C until analysis. Each biological replicate consisted of a pool of flower buds from 3–5 plants with >20 flower buds from each plant.
Isolation of nuclei
Flower buds were cut into small pieces in ice-cold nuclei isolation buffer (NIB; Saxena et al., 1985) supplemented with protease inhibitor cocktail (Sigma). The homogenates were gently rotated at 4 °C for 1 h and then filtered through a 100-μm nylon mesh followed by passing through a 30-μm nylon mesh. The filtered homogenate was then centrifuged 300 g at 4 °C for 8 min, the supernatant was discarded, and the nuclei pellet was gently washed to remove the upper chloroplast layer. The pellets were washed twice with NIB and inspected under a confocal microscope to ensure that they were of high quality.
Proteomic analysis
The nuclei isolated from the flower buds were subjected to analysis by the proteomic services of The Smoler Protein Research Center at the Technion, Israel. Briefly, the samples were digested with trypsin, analysed by LC-MS/MS on a Q-Exactive (ThermoFisher Scientific), and identified using the Discoverer1.4 software against the Ricinus communis, Jatropha curcas, and Arabidopsis protein databases (http://uniprot.org). All the identified peptides were filtered with high confidence, top rank, and mass accuracy. High-confidence peptides were passed the 1% false discovery rate (FDR) threshold. The peak area on the chromatogram of a protein was calculated from the average of the peptides from each protein. The PANTHER classification tool was used for categorization of differentially expressed proteins (Mi et al., 2013). The proteomics analysis was repeated, and the two datasets were compared and showed ~60% repeatability; low repeatability and reproducibility is often seen in proteomics (Tabb et al., 2010).
Nucleic acid extraction and cDNA synthesis
Genomic DNA was extracted using a PureLink Genomic DNA Mini Kit (ThermoFisher Scientific) according to the manufacturer’s protocol. Total RNA was extracted using a RNeasy Mini Kit (Qiagen). The first strand of cDNA was synthesized from 1 µg total RNA treated with DNase (Epicentre) using a Verso cDNA Synthesis Kit (ThermoFisher Scientific).
Isolation of genes and partial promoter sequences
Floral homeotic cDNA clones were prepared by PCR using M. annua flower cDNA as the template and appropriate degenerate primers (based on conserved regions of A. thaliana, R. communis, and J. curcas; Supplementary Table S1) for the recovery of class B (AP3, PI, TM6), class C/D (AG, AGL5, AGL11), and two SUPERMAN-like (SUP-like) gene products. Touchdown PCR conditions were as follows: 95 °C for 2 min; 40 cycles of 95 °C for 30 s; 65–45 °C for 30 s; 72 °C for 60 s; 72 °C for 5 min. The PCR products were cloned into the pJET1.2 plasmid vector (ThermoFisher Scientific) and sequenced at the Biotechnology Center, Ben-Gurion University of the Negev, Beer-Sheva, Israel. To obtain the full cDNA sequence, 3´- RACE was performed as described by Yadav et al. (2012) and 5′-RACE was performed using a 5´-Full RACE Core Set kit (Takara). The class B orthologs were designated as MaPI (for PISTILLATA), MaAP3 (for APETALA3), MaTM6 (for TOMATO MADS-box 6). The AGAMOUS-like orthologs were designated as MaAG1 (for AGAMOUS, class C), MaAGL1 (for AGL11/STK, class D), and MaAGL3 (for SHP2/AGL5, class D). The SUP-like genes were designated as MaSL1 and MaSL2.
The upstream promoters of MaAP3, MaAGL1, MaPI, MaSL1, and MaSL2 were isolated by a semi-random sequence walking strategy modified from Aquino and Figueiredo (2004). Briefly, a gene-specific primer was used for linear amplification of the specific DNA segment for 20 high-stringency cycles (95 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min). The random walking primer was then added and a low-stringency cycle (95 °C for 30 s, 35 °C for 30 s, 72 °C for 2 min) was used for unspecific binding and amplification. Following this, 30 high-stringency cycles were used for exponential amplification. The desired fragments were screened by semi-nested PCR using an asymmetrical ratio (1:5) of walking primer and nested gene-specific primer. The products of interest were purified, cloned, and sequenced as described above. The sequences are available under NCBI GenBank accession numbers KR781112-KR781116 and MN068012-MN068021.
For reference, 135 bp of the Actin gene was amplified using primers designed from conserved region of mRNA of J. curcas, R. communis, and Populus trichocarpa. The amplified product of the M. annua ACTIN (Act) gene was confirmed by direct sequencing from both ends.
Gene expression analysis
Quantification of gene expression was done by quantitative or semi-quantitative RT-PCR analysis using gene-specific primers. qPCR reactions were carried out using Perfecta SYBR green supermix (Quanta Biosciences) on an Applied Biosystems 7500 Real-Time PCR System. All reactions were performed for three biological samples, each with three technical replicates. The PCR conditions were as follows: 94 °C for 15 s, 40 cycles of 94 °C for 5 s, 60 °C for 30 s. Each reaction was normalized against the expression of the Actin gene. The relative gene expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
MNase assays
Nuclei prepared from male and feminized M. annua flower buds were used for micrococcal nuclease (MNase) assays essentially as described by Zhao et al. (2001). MNase assays were performed for three biological replicates, each consisting of nuclei derived from flower buds from at least three individual plants. The recovery of DNA after MNase treatment was checked by PCR and separated on agarose gel containing ethidium bromide.
DNA methylation analysis
For cytosine methylation analysis, chop-PCR (methylation-sensitive enzyme digestion followed by PCR) and bisulfite sequencing were performed as described previously (Yadav et al., 2018). For the chop-PCR, genomic DNA was treated with the methylation-sensitive restriction enzymes HpaII or MspI and subjected to PCR to amplify various gene fragments containing the restriction site CCGG. Bisulfite conversion was done by adding a mixture of sodium bisulfite, hydroquinone, and urea and incubating at 55 °C for 16 h. The samples were desalted using a PCR purification kit and desulfonated by adding NaOH to a final concentration of 0.3 M. The DNA was then purified using a QIAquick PCR purification kit (Qiagen). The bisulfite-converted DNA was used for PCR amplification of promoter and gene-body fragments. The PCR products were cloned into the pJET1.2 vector. At least 10 individual clones from each region were sequenced by Macrogen, Netherlands. The sequences were analysed and scored using the Kismeth online service (Gruntman et al., 2008).
Results
Feminization of male Mercurialis annua: setting-up the experimental system
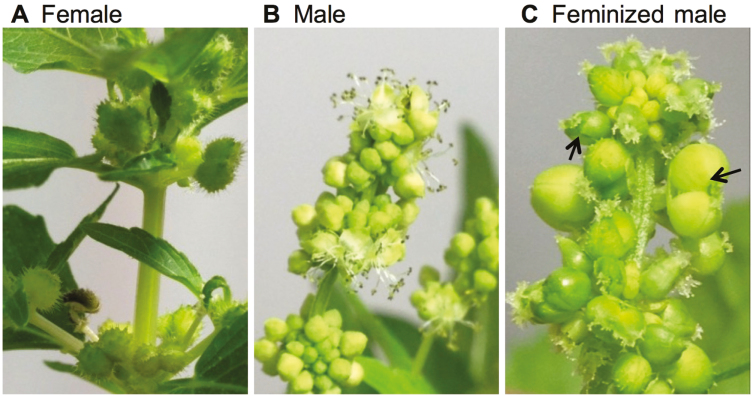
Female and male M. annua plants have distinct inflorescence morphologies (Fig. 1A, B). In female plants, flowers develop directly at the leaf axils with short pedicels, while in male plants clusters of flowers develop on long pedunculated inflorescences. Feminization of male flowers by the cytokinin BAP caused development of female flowers that yielded fertile seeds on male inflorescences (Fig. 1C; see also Khadka et al., 2005).
Fig. 1.
Morphology of dioecious Mercurialis annua. (A) Female plant, (B) male inflorescence, and (C) feminized male inflorescence, induced by spraying plants with 6-benzylaminopurine three times daily for 4 weeks. Feminized males produced bi-carpellet flowers, some of which are indicated by arrows.
Proteome analysis of flower-bud nuclei
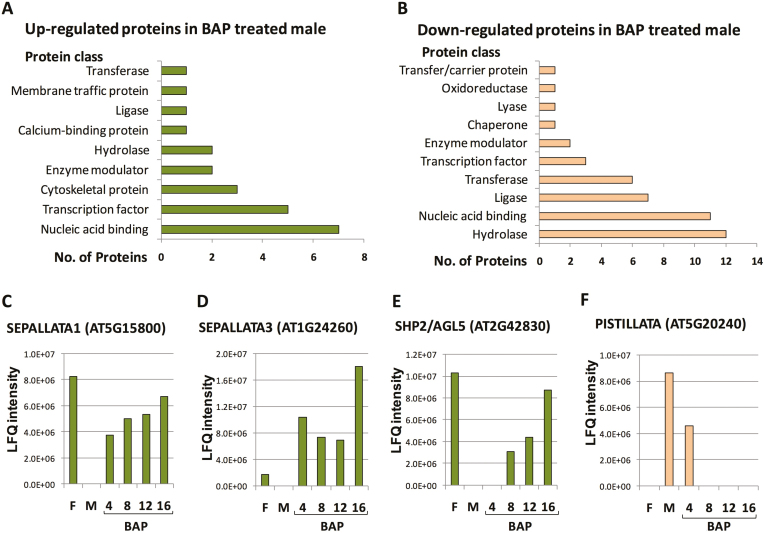
To identify the regulatory genes involved in BAP-induced sex alteration of M. annua, we performed proteome analysis of nuclear proteins derived from the flower buds of females, males, and males treated with BAP for 4, 8, 12, and 16 d. The proteome data indicated that the core histone proteins H2A, H2B, H3, and H4 showed the highest intensities among the proteins identified (Supplementary Tables S2, S3).The occurrence of cytoplasmic proteins such as rbcS resulted from contamination of nuclei during preparation. The major difference between the genders was that 52 proteins present in female flowers were absent in males, while 244 proteins present in male flowers were absent in females. This was consistent with a recent transcriptome analysis in which a higher number of male-biased genes (1385) were found compared to female-biased genes (325) (Veltsos et al., 2019). Among the 52 proteins expressed only in female flowers, 49 were up-regulated in feminized males (Supplementary Table S4). Out of the 244 male-specific proteins, 84 were disappeared from the proteome during the course of feminization (Supplementary Table S5), as follows: 39 proteins were absent after 4 d of BAP treatment, a further 15 after 8 d, a further 12 after 12 d, and a further 18 after 16 d. Multiple classes of proteins were identified by categorization analysis of the differentially expressed proteins in the feminized males. The major up-regulated protein classes were nucleic acid-binding proteins, transcription factors, and cytoskeleton proteins (Fig. 2A), and the major down-regulated classes were hydrolases, nucleic acid-binding proteins, ligases, and transferases (Fig. 2B). Among the differentially expressed proteins, four floral organ identity MADS-box transcription factors were identified. The class E proteins SEP1 and SEP3 and the class D protein SHP2/AGL5 were up-regulated during feminization, reaching their highest levels at day 16 (Fig. 2C–E). In contrast, the class B protein PI was down- regulated within 4 d of BAP treatment and could not be detected thereafter (Fig. 2F). The proteome data for floral homeotic proteins were confirmed by RNA analysis (see below). In addition to floral homeotic proteins, other chromatin and transcription regulatory proteins were up-regulated in BAP-treated males. These included DNA TOPOISOMERASE 2 (TOP2), HISTONE H1, ATP-dependent DNA helicase 2 subunit KU80, ZINC-FINGER HOMEODOMAIN 9 (ZHD9), WRKY39.1, RINGLET2 (RLT2), and the bromodomain-containing protein, GTE4. The bZIP transcription factor related to Arabidopsis BZIP21 as well as the protein related to Arabidopsis DEFECTIVE IN EXINE FORMATION 1 (DEX1) were down-regulated in BAP-treated males. The proteome data also revealed 132 proteins not found in male and female flowers that were present during the course of feminization (Supplementary Table S6). Among them were the histone acetyltransferase GCN5, which is related to the Arabidopsis histone acetyltransferase of the GNAT family that is involved in transcriptional activation, and MINICHROMOSOME MAINTENANCE (MCM) proteins, which function as components of the MCM2-7 complex that is implicated in seed development in Arabidopsis (Herridge et al., 2014). There were also 70 proteins present in male and female flowers that were gradually down-regulated during the course of feminization (Supplementary Table S7), including a histone H2B variant related to Arabidopsis HTB5 and HTB6, which are presumed to be involved in chromatin compaction, and COMPASS component SWD2, a homolog of the Arabidopsis ANTHESIS POMOTING FACTOR 1 (APRF1) that is implicated in promoting flowering under long-day conditions (Kapolas et al., 2016).
Fig. 2.
Proteome analysis of nuclei of Mercurialis annua isolated from flower buds of female, male, and feminized male plants (treated with 6-benzylaminopurine, BAP). (A) Down-regulated and (B) up-regulated proteins following male feminization. (C-F) The label-free quantification (LFQ) intensity reflecting the relative amounts of the indicated proteins, which were calculated using peptide intensities normalized between the samples (the corresponding Arabidopsis homolog gene ID is given in brackets). F, female; M, male. (This figure is available in colour at JXB online.)
Differential expression of floral homeotic genes
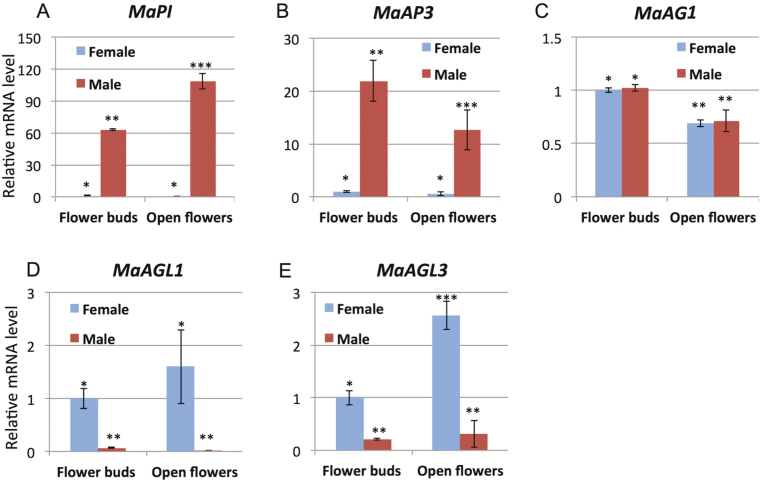
Based on the results of the proteome analysis, we cloned and examined the expression of M. annua orthologs of floral homeotic MADS-box genes. The homology of the isolated genes was confirmed by phylogenetic analyses (Supplementary Figs. S1–S3). We examined the patterns of RNA expression of the isolated genes in female and male flowers at the bud and opened-flower stages. The class B genes MaPI and MaAP3 were highly expressed in male flowers and poorly expressed in female flowers (Fig. 3A, B) whilst the class C gene MaAG1 was highly expressed at similar levels in both female and male flowers (Fig. 3C). In contrast to the class B genes, the class D genes MaAGL1 and MaAGL3 were highly expressed in female flowers and poorly expressed in male flowers (Fig. 3D, E). The expression levels of most of floral homeotic genes were significantly different between the floral bud and open-flower stages in a sex-specific manner. There was higher expression in open flowers of MaPI in males and of MaAGL3 in females, while there was higher expression in flower buds of MaAP3 in males and of MaAG1 in both females and males (Fig. 3A–C, E).
Fig. 3.
Expression of MADS-box genes in flower buds and open flowers of female and male plants of Mercurialis annua. Relative expression of (A) MaPI, (B) MaAP3, (C) MaAG1, (D) MaAGL1, and (E) MaAGL3 determined using RT-qPCR. The relative transcript levels are normalization to the Actin gene. Data are means (±SE) of three biological replicates. Significant differences between means are indicated by different numbers of asterisks as determined by Tukey’s HSD test (P<0.05). (This figure is available in colour at JXB online.)
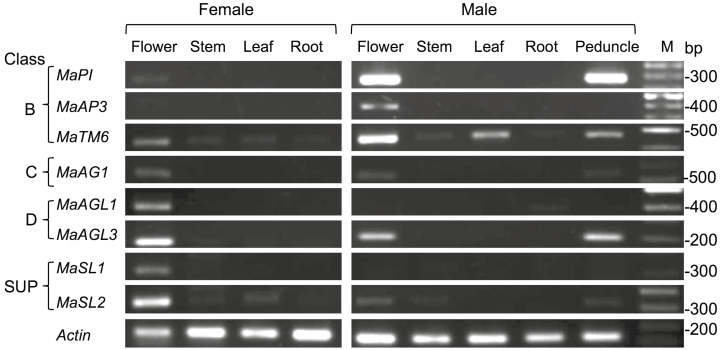
Examination of the organ specificity of gene expression showed that MaPI and MaAP3 were almost exclusively expressed in male flowers (Fig. 4), although MaPI was also strongly expressed in male peduncles. Expression of MaTM6 was evident in flowers of both females and males, but its expression in vegetative organs was very low in female plants. In the males, MaTM6 expression was high in flowers, moderate in leaves and peduncles, and very low in stems and roots. MaAGL1 and MaSL1 were exclusively expressed in flowers of female plants. MaAG1 was expressed at moderate level in flowers of both sexes, and at a lower level in the peduncle of male plants. MaAGL3 was highly expressed in flowers of females and had slightly lower expression in flowers and peduncles of male plants.
Fig. 4.
Expression patterns of floral homeotic genes in different organs of female and male plants of Mercurialis annua. Expression of class B, C, D, and SUPERMAN-like (SUP) genes was determined using semi-quantitative PCR using cDNAs derived from RNA extracted from the various plant organs. Actin was used as a ubiquitously expressed reference gene. M, molecular size markers.
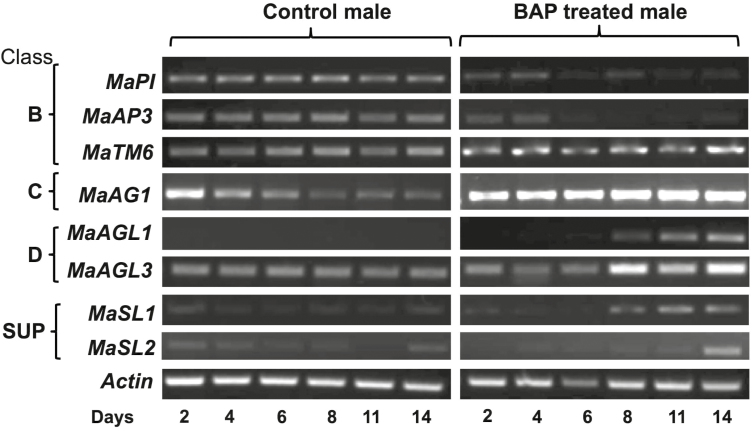
BAP-induced feminization of male plants resulted in changes in the expression patterns of floral genes (Fig. 5). The expression of the class B identity gene MaTM6 was not significantly affected by feminization, while MaPI and MaAP3 were down-regulated. In contrast, the expression of the class C/D floral genes MaAG1, MaAGL1, MaAGL3, and MaSL1 was up-regulated by feminization. A significant up-regulation of class C/D genes was observed at 8–11 d of BAP treatment. Thus, the RNA results confirmed the proteome data with respect to the down- and up-regulation of male and female identity proteins, respectively, during the course of feminization.
Fig. 5.
Time-course of the expression of floral genes in Mercurialis annua during feminization induced by treatment with 6-benzylaminopurine (BAP). Plants at 25 d old were sprayed three times daily with water (Control) or with BAP and newly emerging inflorescences were collected on the days indicated. RNA was extracted and subjected to cDNA synthesis, which was then used to determine expression using semi-quantitative PCR. Class B, C, D, and SUPERMAN-like (SUP) genes are indicated. Actin was used as the reference gene.
Epigenetic regulation of floral genes
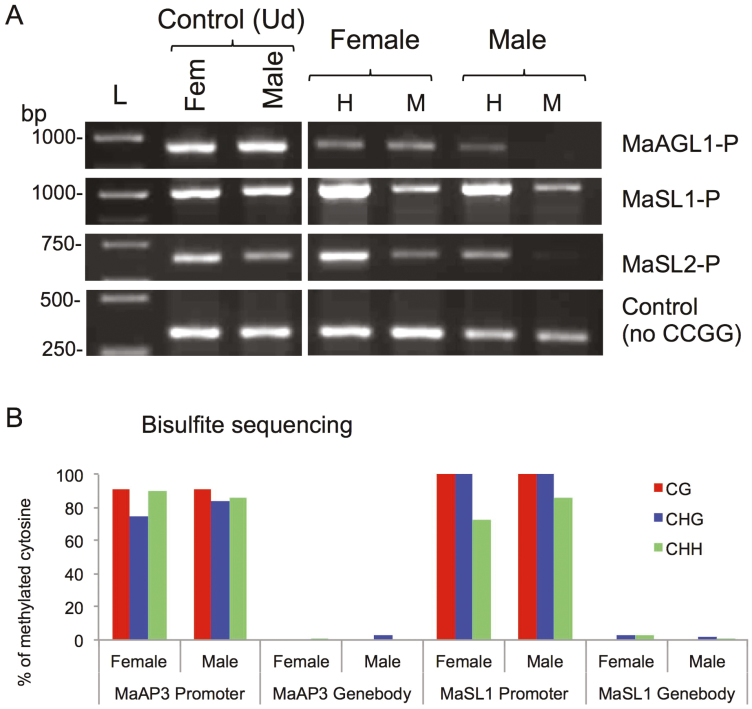
Epigenetics has often been implicated in sex-determination in dioecious plants (Janousek et al., 1996; Bräutigam et al., 2017) and therefore we investigated the involvement of epigenetic mechanisms (i.e. DNA methylation) in the regulation of floral gene expression. We examined the status of cytosine methylation at the promoter regions of the differentially expressed genes MaSL1, MaSL2, and MaAGL1 using chop-PCR assays with the methylation-sensitive enzymes HpaII and MspI. Both enzymes recognize the CCGG site but differ in their sensitivity to cytosine methylation: HpaII is sensitive when either of the cytosines is methylated while MspI is sensitive only when the external one is methylated, thus allowing CG and CHG methylation to be distinguished. The assays indicated that there were no differences in the CpG methylation status of the genes between female and male flowers; however, CHG methylation appeared to be absent from the promoter regions of MaAGL1 and MaSL2 in male flowers in so far as no recovery of the PCR fragment could be detected in the MspI digest (Fig. 6A). We also perform bisulfite sequencing of the MaAP3 and MaSL1 promoter and gene body regions that showed no differences in DNA methylation status between male and female flowers. The promoter regions of both genes were highly methylated in all cytosine contexts (CG, CHG, and CHH) while their gene bodies were essentially unmethylated (Fig. 6B).
Fig. 6.
Transcriptionally active floral genes are methylated in both females and males of Mercurialis annua. (A) Analysis of DNA methylation at the promoters (-P) of MaAGL1, MaSL1, and MaSL2 by as determined by chop-PCR. A fragment of MaSL1 lacking the CCGG site (no CCGG) was used as an internal control. Left panel is a control of undigested DNA (Ud). Ud, Undigested DNA; H, HpaII; M, MspI; L, molecular size markers in base pairs. (B) Analysis of methylation at the promoter and in the gene-body of MaAP3 and MaSL1 as determined by bisulfite sequencing. The percentage of cytosine methylation for each fragment was determined from at least 10 different clones. (This figure is available in colour at JXB online.)
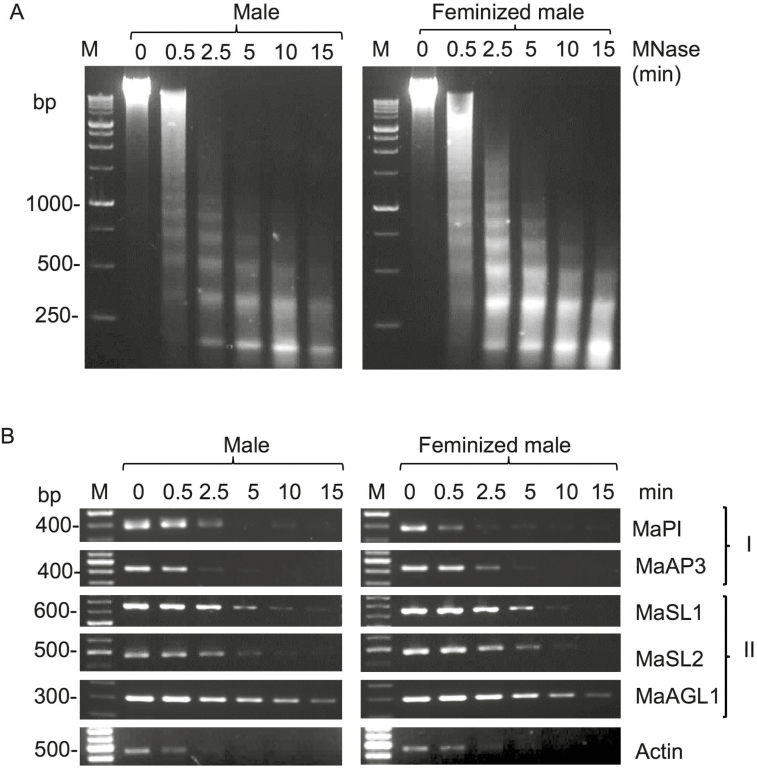
Since no differences in DNA methylation were found between male and female flowers, we used micrococcal nuclease (MNase) assays to investigate the chromatin configuration of the promoters of several floral genes during the course of feminization. After 14 d of BAP treatment, the MNase-treated nuclei from male and feminized male flower buds showed similar progressive digestion of genomic DNA with notable nucleosomal ladders (Fig. 7A). The MNase-digested DNAs were used as templates for PCR analysis of the promoter regions of several genes, and no notable differences in digestion patterns were found between male and feminized male flowers (Fig. 7B). However, two groups of major digestion patterns reflecting open and relatively closed chromatin configuration were observed. Group I consisted of the promoter regions of the class B genes MaPI and MaAP3, and showed higher sensitivity to MNase digestion that was similar to actin, a constitutively expressed gene. Group II consisted of the class D gene MaAGL1 together with MaSL1 and MaSL2, and were more resistant to MNase digestion (Fig. 7B). Thus, it appeared that class B genes that assumed an open chromatin conformation in male flowers remained open upon feminization, while class D genes remained in a relatively closed configuration in feminized male flowers.
Fig. 7.
Analysis of chromatin configuration of selected floral genes in Mercurialis annua as determined by micrococcal nuclease assays. (A) Nuclei prepared from male and feminized male flower buds (treated with 6-benzylaminopurine for 14 d, before female flowers were visible) were treated with MNase for the time periods indicated. DNA was extracted from the treated nuclei and resolved on 1.5% agarose gel. M, molecular size markers, in base pairs. (B) Assessment of chromatin configuration of promoters as determined by PCR using DNA recovered from the MNase-treated nuclei shown in (A). Group I refers to male-related identity genes and Group II refers to female-related identity genes. Actin was used as the reference for open chromatin configuration. M, molecular size markers.
Discussion
The data presented here regarding the expression of floral identity genes are consistent with their known functions in determining sexual identity of floral organs in various plant species. It has been shown previously that class B genes are highly expressed in male flowers of the type-II dioecious plants T. dioicum and S. oleracea (Di Stilio et al., 2005; Pfent et al., 2005) and our results showed that male flowers were characterized by a strong expression of the class B genes MaPI and MaAP3, which was concomitant with suppression of female identity genes such as MaAGL1 (class D) and MaSL1 (Figs. 2–4). The involvement of cytokinin in sex-determination has been reported in a variety of plant species including Arabidopsis (Lindsay et al., 2006) and the oilseed crops Plukenetia volubilis and Jatropha curcas (Pan and Xu, 2011; Fu et al., 2014). We found that BAP-induced feminization was accompanied by increased expression of the class C gene MaAG1 (Fig. 5) and this was concomitant with suppression and activation, respectively, of class B male-specific (e.g. MaPI) and class D female-specific (MaAGL1, MaAGL3) genes thus specifying female flower development. In Arabidopsis, an increase in the number of carpels following BAP treatment is correlated with an increase in the expression of WUSCHEL (WUS) (Lindsay et al., 2006; Gordon et al., 2009), the protein product of which specifies stem cell identity in both the floral and the shoot apical meristems (Laux et al., 1996; Mayer et al., 1998). WUS also activates the class C homeotic gene AGAMOUS, which is required for specifying both stamen and carpels (Theißen et al., 2016). Thus, it is possible that cytokinin may act in re-specifying the male floral meristem toward female-producing flowers by activation of WUS-like genes in M. annua concurrently with activation of MaAG1 and suppression and activation, respectively, of male and female floral genes to bring about female flower formation.
Sex conversion: proteomic data
Proteome analysis of BAP-feminized males showed differential expression of several protein families, including nucleic acid-binding proteins and transcription factors (Fig. 2). Some of the proteins up-regulated following BAP treatment were involved in chromatin structure and function, suggesting that sex conversion is an intricate process that requires substantial genome reorganization to allow transcriptional activation and repression of genes to bring about feminization. Among these proteins were topoisomerase 2 (TOP2) that can relieve superhelical DNA (a characteristic of heterochromatin) by introducing transient double-strand DNA breaks (reviewed in Nitiss, 2009) and an ATP-dependent DNA helicase 2 subunit KU80, which is involved in DNA non-homologous end-joining (NHEJ) that is required for the repair of double-strand breaks (West et al., 2002). In addition, the proteomic data indicated that BAP treatment resulted in up-regulation of a linker histone H1.1, which is involved in heterochromatin formation and regulation of gene expression (Hergeth and Schneider, 2015), as well as up-regulation of a structural maintenance of chromosomes (SMC) protein (Supplementary Table S4). SMC proteins function in a range of nuclear processes, including chromosome condensation, DNA repair, and epigenetic transcriptional silencing of genes (Harvey et al., 2002). The proteomic data also indicated that multiple transcription factors were up-regulated in response to BAP treatment including WRKY transcription factor 39.1, a group II WRKY protein with a C2H2 zinc finger-like motif (Agarwal et al., 2014) and the basic leucine zipper (bZIP) class transcription factors EMBP-1 the homolog of which in wheat has been implicated in the abscisic acid signaling pathway (Guiltinan et al., 1990) and in histone gene expression (Mikami et al., 1994). Other transcription factors up-regulated in feminized males included a member of the zinc-finger homeodomain protein sub-family (ZF-HD) related to Arabidopsis ZHD9/ATHB34, the expression of which, in common with other members in this group, is elevated during floral development (Tan and Irish, 2006). Another homeodomain protein up-regulated in feminized males was a homeodomain-like transcriptional regulator RLT2, which is implicated in activation of expression of seed storage genes in Arabidopsis (Sundaram et al., 2013).
Consistent with the conversion of male flowers into females, some proteins involved in male reproductive organs were down-regulated in the course of feminization (Supplementary Table S5). Among these was the bZIP transcription factor related to Arabidopsis bZIP21/TGA9, which is implicated in male gametogenesis. Plants lacking bZIP21/TGA9 and bZIP65/TGA10 are defective in anther development (Murmu et al., 2010). In addition, the proteomic data showed down-regulation of the DEFECTIVE IN EXINE FORMATION protein that is related to Arabidopsis DEX1, which is required for exine pattern formation during pollen development (Paxson-Sowders et al., 2001). Overall, the proteome data suggested that both the up-regulation of sex-specific proteins as well as the suppression of proteins for the opposite sex function were important for sexual dimorphism of M. annua.
Expression pattern of floral genes
The Mercurialis orthologs of the SHP2/AGL5, SEP1, and SEP3 proteins were up-regulated in feminized males (Fig. 2C–E), suggesting that they play roles in female flower specification. This is consistent with the role of SHP in carpel development in Arabidopsis where constitutive expression results in a partial conversion of the first whorl sepals into carpel-like structures as demonstrated by extensive proliferation of stigmatic papillae (Favaro et al., 2003; Pinyopich et al., 2003). Similarly, in Gerbera hybrida two duplicated orthologs of a SEP-like gene, GRCD1 and GRCD2, are sub-functionalized for stamen and carpel identity, respectively (Zhang et al., 2017). The PI protein, which was down-regulated in feminized males, is involved in controlling the development of whorls 2 and 3 in Arabidopsis, Antirhinum, and tomato (Tröbner et al., 1992; Goto and Mayerowitz, 1994; Guo et al., 2016). This suggest that cytokinin switched off male control genes (e.g. PISTILLATA) concomitantly with up-regulation of female identity genes, thus leading to the replacement of stamen by carpels, as in the development of normal dioecious female flowers.
Using a cell-free translation system with RNAs derived from M. annua male and female flowers, Delaigue et al. (1984) found variation in peptides between the two sexes and that cytokinin-induced feminization of male flowers led to the expression of female-specific peptides. Similarly, we found that cytokinin-induced feminization of M. annua male flowers was associated with up-regulation of female-specific floral genes concomitantly with down-regulation of male-specific genes (Fig. 5). In Arabidopsis, exogenous application of BAP has been reported to promote differentiation of carpeloid tissue and to suppress stamen development. This is similar to the effect obtained by overexpressing SUP in tobacco plants, leading to the proposition that SUP may regulate sex-determination pathways by promoting female organ differentiation via its effect on cytokinin signaling (Nibau et al., 2011). Alternatively, cytokinin may affect male and female flower development via controlling SUP expression. Indeed, in M. annua the SUP-like genes exhibited female flower-specific expression (Fig. 4), as previously seen in the dioecious Popolus tomentosa and Silene latifolia (Kazama et al., 2009; Song et al., 2013). The sup mutant in Arabidopsis is associated with the ectopic expression of AP3 in the fourth whorl (Bowman et al., 1992), and therefore SUP has been proposed to function as a negative regulator of AP3. The concomitant expression of class B and SUP-like genes in male flower buds suggests that SUP-like gene(s) might not be a transcriptional regulator of class B genes in M. annua. An alternative possibility is that the expression of SUP in male flower buds is negatively regulated post-transcriptionally.
The expression of the class B gene MaAP3 was restricted to male flowers, while MaTM6 (AP3-related) and MaPI were expressed in flowers as well as in peduncles (Fig. 4). It is notable that TM6, which is absent in Arabidopsis, was also weakly expressed in the leaves of M. annua female plants and in other vegetative organs. The TM6 ortholog CpTM6-2 in Carica papaya is expressed at a low level in sepals and at a high level in leaves (Ackerman et al., 2008), while the ortholog VvTM6 in Vitis vinifera is expressed throughout the plant but at higher levels in flowers and berries (Poupin et al., 2007). It has been proposed that a gene duplication event of the paleoAP3 genes resulted in two types, namely the euAP3 and TM6 lineages that are distinguished by their C-terminal regions (Kramer et al., 1998). These duplicated genes probably to some extent adopted different functions (sub-functionalization), as demonstrated by their tissue-specific patterns of expression and the differing effects of their loss-of-function on flower development (Eckardt, 2006).
The expression of the class C gene MaAG1 was similar in male and female flowers of M. annua (Fig. 4), suggesting that it may not be involved in gender determination. This is consistent with a previous study that showed that the C class AG genes are involved in the floral quartet that specifies both stamens and carpels (reviewed in Theißen et al., 2016). The class D genes MaAGL1 and MaAGL3 were highly expressed in female flowers, and MaAGL3 was also expressed in male flowers and peduncles. Our results suggested that the class B genes MaAP3 and MaPI together with the class C gene MaAG1 have a role in determining the identity of male floral organs. The proteins that they encode may participate in the floral quartet that controls gene expression and the identity of the male reproductive organs (Theißen et al., 2016). On the other hand, the class D genes MaAGL1 and MaAGL3 together with class C and class E genes may form a floral quartet that specifies female floral organs, carpels, and ovules. Notably, in seed plants the class B genes have been suggested to have a primary role in sex determination (Winter et al., 1999), with expression of both class B and class C genes specifying male reproductive organs while the expression of only class C genes specifies female reproductive organs. Thus, switching from male to female and vice versa can be solely driven by changes in the spatio-temporal expression of class B genes (Winter et al., 1999; Theissen and Melzer, 2007). However, our data showed that induction of feminization was associated not only with the up-regulation of female-related class C and class D genes, but also with the turning off of the expression of male-related class B genes, which might be crucial for the development of female flowers in otherwise male plants of M. annua.
Epigenetics and sex-determination
Our data showed that there was no clear relationship between floral homeotic genes and their epigenetic make-up (Table 1). Gene expression was primarily regulated at the chromatin level, where gene transcription requires open chromatin to allow the transcription machinery to approach the gene locus. Analysis of chromatin accessibility using MNase assays revealed that in male flowers the class B genes MaPI and MaAP3 assumed an open chromatin conformation similar to the constitutively expressed gene Actin (Fig. 7). On the other hand, the class D gene MaAGL1 and the SUP-like genes MaSL1 and MaSL2 appeared to acquire a relatively closed conformation compared with Actin, which was consistent with the lack of expression in male flowers. Surprisingly, however, no apparent change in accessibility of chromatin to MNase was evident upon feminization and up-regulation of MaAGL1 and MaSL1. This suggests that chromatin can assume different levels of open conformation as reflected by variable sensitivity to MNase, including hyper-accessible DNA sites (Schwartz et al., 2019) that which probably provide another regulatory layer for control of gene expression (Ishihara et al., 2010; Kotomura et al., 2015). Similarly, no change in chromatin accessibility was observed for the down-regulated class B genes MaPI and MaAP3, the transcription of which was possibly halted in an open chromatin environment by other means (e.g. suppressor proteins).
Table 1.
Summary of the expression levels of floral genes in Mercurialis annua in relation to their epigenetic constraints
| Gene | Expression | DNA methylation | Sensitivity to MNase | Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | Male | Feminized male | Female | Male | Feminized male | Female | Male | Feminized male | Female | Male | Feminized male | |
| MaPI | – | ++ | – | High | High | S | O E | O S | ||||
| MaAP3 | – | +++ | – | mALL | mALL | High | High | m S | O m E | O S | ||
| MaActin | +++ | +++ | +++ | High | High | E | E | O E | ||||
| MaAGL3 | +++ | + | +++ | E | e | E | ||||||
| MaAGL1 | +++ | – | ++ | mALL | mCG | Low | Low | m E | PO m S | PO E | ||
| MaSL1 | ++ | – | ++ | mALL | mALL | Low | Low | m E | PO m S | PO E | ||
| MaSL2 | ++ | – | – | mALL | mCG | Low | Low | m E | PO S | PO S | ||
| MaAG1 | +++ | ++ | +++ | E | E | E |
Expression: –, no expression; +, low expression; ++/+++, high expression.
DNA methylation: mAll, methylated at all C contexts; mCG, methylated at the CG context only.
Score: S, silent; E, expressed; e, low expression; O, open chromatin; PO, partially open chromatin.
The nature of gene regulation by DNA methylation is not fully understood, but it has been generally implicated in regulating chromatin structure and function (Niederhuth and Schmitz, 2017). DNA methylation was detected at promoters but not in gene-bodies of the floral genes that we examined (Fig. 6). Interestingly, the methylation status of all the genes was similar in both sexes despite their differential expression. In Arabidopsis, gene methylation has been reported to correlate with the level of gene expression: gene-body methylation is correlated with constitutively and highly expressed genes, while promoter methylation is correlated with weakly expressed genes that are usually tissue-specific (Zhang et al., 2006; Zilberman et al., 2007). However, we did not find a consistent correlation between DNA methylation and expression of the floral genes in M. annua (Table 1, Fig. 7), with MaAP3, MaAGL1, MaSL1, and MaSL2 being normally transcribed in spite of being heavily methylated at their promoters. Thus, it appears that DNA methylation at the promoter regions of M. annua floral genes had a positive effect on their expression, in contrast to the commonly observed effect of suppression of expression by methylation, particularly when transposable elements are concerned (Lisch, 2009). This finding may possibly be explained by a lowering of the affinity of repressors to their binding sites as a result of DNA methylation. Indeed, there are studies that have similarly shown that DNA methylation at promoters contributes to transcriptional activation of certain tissue-specific genes (Niesen et al., 2005; Weber et al., 2007; Rishi et al., 2010; Bahar Halpern et al., 2014).
Most studies that have addressed epigenetic regulation of sex determination have highlighted various genes that are not related to floral homeotic genes. Bräutigam et al. (2017) examined the sex-determining region of Populus balsamifera and identified PbRR9 as showing a clear pattern of gender-specific methylation. PbRR9 encodes a protein that is a member of the two-component response regulator (type-A) gene family involved in cytokinin signaling. A detailed study of the occurrence of androecy in the Cucurbitaceae species Cucumis melo and C. sativus implicated the ethylene biosynthetic enzymes CmACS-7 and CmACS-11 in sex determination (Boualem et al., 2009, 2015), and they are required for epigenetic repression of the male-promoting CmWIP1 gene via induction of H3K27me3 (Latrasse et al., 2017). In naturally occurring gynoecious lines of C. melo, the transition from male to female flowers results from a transposon insertion proximal to the CmWIP1 promoter, which induces DNA methylation and silencing of the gene (Martin et al., 2009). In maize, maintenance of the monoecious pattern of sex determination is achieved by epigenetic restriction of SILKLESS1 (SK1), a uridine diphosphate glycosyltransferase (UGT), from the apical inflorescence: SK1 is required for female flower development, and constitutive expression of SK1 in transgenic maize results in complete feminization (Parkinson et al., 2007; Hayward et al., 2016). In Diospyros lotus, a dioecious plant with heterogametic males (XY), a Y-specific sex-determinant, OGI, encodes a small RNA that suppresses the MeGI gene that encodes a feminizing homeodomain transcription factor (Akagi et al., 2014). Interestingly, our data and those obtained in other studies on a variety of dioecious species suggest that mutations leading to evolution of dioecy have not directly affected (genetically or epigenetically) floral homeotic transcription factors; rather, these transcriptional regulators seem to act as downstream effectors of sex-determining gene(s) for sex-organ specification.
Conclusions
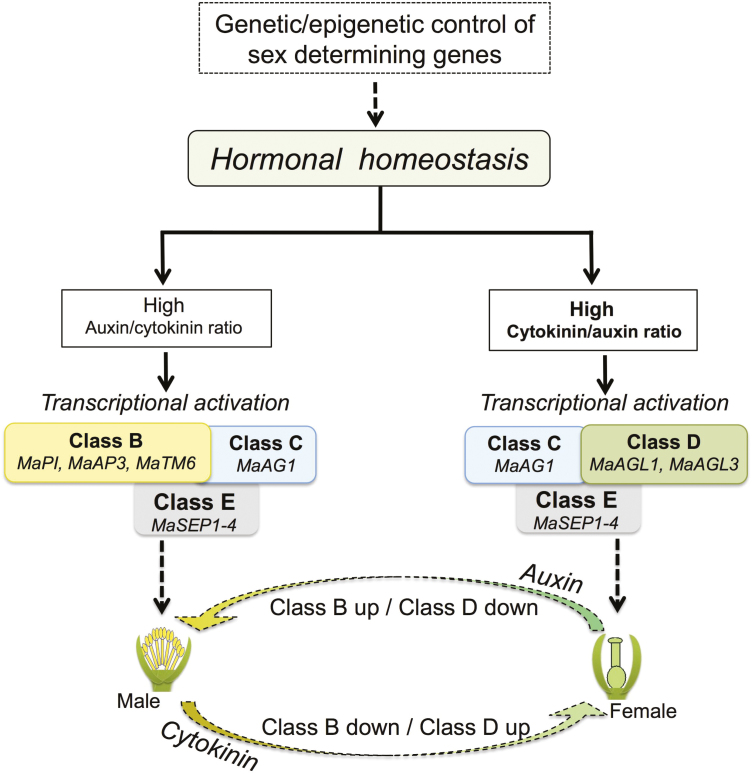
In conclusion, we propose a comprehensive model of sex determination in dioecious M. annua (Fig. 8). According to this model, sex conversion in M. annua does not primarily involve epigenetic regulation of floral homeotic genes. Instead, sex identity in this species seems to be controlled genetically/epigenetically upstream in the regulatory pathway by a gender-specific regulator(s) that affects, at least partly, hormonal homeostasis. Thus, high cytokinin induces transcriptional activation of female identity genes and the production of female flowers, while high auxin induces transcriptional activation of male identity genes and production of male inflorescences. This is supported by recent analysis of the DNA sequences of the sex-determining region in M. annua that has failed to show any floral homeotic genes or other strong candidate genes for sex determination (Veltsos et al., 2018). It appears that keeping the functioning of the floral homeotic genes, i.e. the effector proteins that specify sex organs in dioecious plants, enables sex conversion (Fig. 8) and this might have an adaptive value, particularly in cases where the two sexes exhibit differential tolerance to stresses (Orlofsky et al., 2016). The sexual bi-potency of dioecious plants may also explain their capacity for multiple cycles of transitions from dioecy to monoecy during the evolution of plants.
Fig. 8.
A proposed model of sex determination in the dioecious plant Mercurialis annua. Differentiation of unisexual flowers is controlled genetically/epigenetically by as yet unknown sex-determining genes that affect hormonal homeostasis. A high cytokinin/auxin ratio activates the transcription of effector genes such as female-identity class D genes (MaAGL1 and MaAGL3). The class D proteins together with class C and class E proteins promote female flower development. Alternatively, a high auxin/cytokinin ratio is presumed to lead to transcriptional activation of male-identity class B genes (MaPI, MaAP3, and MaTM6). The class B proteins together with class C (MaAG1) and class E (MaSEP1–4) promote male flower development. Exogenous application of cytokinin feminizes males by inducing down-regulation of class B genes and up-regulation of class D genes. Exogenous auxin masculinizes females (Delaigue et al., 1984), probably via up-regulation of class B genes concomitantly with down-regulation of class D genes. (This figure is available in colour at JXB online.)
Supplementary data
Supplementary data can be found at JXB online.
Fig. S1. Phylogenetic analysis of class B genes from M. annua, Arabidopsis, and various taxonomic groups.
Fig. S2. Phylogenetic analysis of AG-like genes from M. annua, Arabidopsis, and various taxonomic groups.
Fig. S3. Phylogenetic analysis of SUPERMAN-like genes from M. annua, Arabidopsis, and various taxonomic groups.
Table S1. List of primers used in this study.
Table S2. List of proteins identified in proteomic analysis (repeat I).
Table S3. List of proteins identified in proteomic analysis (repeat II).
Table S4. Proteins exclusively present in female flower buds that appeared following BAP treatment.
Table S5. Proteins exclusively present in male flower buds that had lower expression following BAP treatment.
Table S6. Proteins normally absent in male and female flower buds were up-regulated following BAP treatment.
Table S7. Proteins present in male and female flower buds that had higher expression following BAP treatment.
Acknowledgements
We acknowledge the support of the Albert Katz International School for Desert Studies for a graduate scholarship to JK. We also thank the Blaustein Center for Scientific Cooperation (BCSC) and the PBC Program of Israeli Council for Higher Education for postdoctoral fellowships to NSY. This project was supported by the Frances and Elias Margolin Trust and by ICA in Israel, as well as by The Hans-Fischer-Gesellschaft, Munich and BAYHOST funding provided by the Bayerisches Staatsministerium für Bildung und Kultur, Wissenschaft und Kunst.
References
- Ackerman CM, Yu Q, Kim S, Paull RE, Moore PH, Ming R. 2008. B-class MADS-box genes in trioecious papaya: two paleoAP3 paralogs, CpTM6-1 and CpTM6-2, and a PI ortholog CpPI. Planta 227, 741–753. [DOI] [PubMed] [Google Scholar]
- Agarwal P, Dabi M, Agarwal PK. 2014. Molecular cloning and characterization of a group II WRKY transcription factor from Jatropha curcas, an important biofuel crop. DNA and Cell Biology 33, 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth C, Crossley S, Buchanan-Wollaston V, Thangavelu M, Parker J. 1995. Male and female flowers of the dioecious plant sorrel show different patterns of MADS box gene expression. The Plant Cell 7, 1583–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi T, Henry IM, Tao R, Comai L. 2014. Plant genetics. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 346, 646–650. [DOI] [PubMed] [Google Scholar]
- Aquino VH, Figueiredo LT. 2004. Linear amplification followed by single primer polymerase chain reaction to amplify unknown DNA fragments: complete nucleotide sequence of Oropouche virus M RNA segment. Journal of Virological Methods 115, 51–57. [DOI] [PubMed] [Google Scholar]
- Bahar Halpern K, Vana T, Walker MD. 2014. Paradoxical role of DNA methylation in activation of FoxA2 gene expression during endoderm development. The Journal of Biological Chemistry 289, 23882–23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Camps C, et al. . 2015. A cucurbit androecy gene reveals how unisexual flowers develop and dioecy emerges. Science 350, 688–691. [DOI] [PubMed] [Google Scholar]
- Boualem A, Troadec C, Kovalski I, Sari MA, Perl-Treves R, Bendahmane A. 2009. A conserved ethylene biosynthesis enzyme leads to andromonoecy in two Cucumis species. PLoS ONE 4, e6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM. 1992. SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599–615. [DOI] [PubMed] [Google Scholar]
- Bräutigam K, Soolanayakanahally R, Champigny M, Mansfield S, Douglas C, Campbell MM, Cronk Q. 2017. Sexual epigenetics: gender-specific methylation of a gene in the sex determining region of Populus balsamifera. Scientific Reports 7, 45388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 1978. A model for the evolution of dioecy and gynodioecy. The American Naturalist 112, 975–997. [Google Scholar]
- Charlesworth D. 2002. Plant sex determination and sex chromosomes. Heredity 88, 94–101. [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Cossard GG, Toups MA, Pannell JR. 2019. Sexual dimorphism and rapid turnover in gene expression in pre-reproductive seedlings of a dioecious herb. Annals of Botany 123, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1877. The different forms of flowers on plants of the same species. London: John Murray. [Google Scholar]
- Delaigue M, Poulain T, Durand B. 1984. Phytohormone control of translatable RNA populations in sexual organogenesis of the dioecious plant Mercurialis annua L. (2n = 16). Plant Molecular Biology 3, 419–429. [DOI] [PubMed] [Google Scholar]
- Di Stilio VS, Kramer EM, Baum DA. 2005. Floral MADS box genes and homeotic gender dimorphism in Thalictrum dioicum (Ranunculaceae) – a new model for the study of dioecy. The Plant Journal 41, 755–766. [DOI] [PubMed] [Google Scholar]
- Durand B, Durand R. 1991. Sex determination and reproductive organ differentiation in Mercurialis. Plant Science 80, 49–65. [Google Scholar]
- Eckardt NA. 2006. Functional divergence of AP3 genes in the MAD world of flower development. The Plant Cell 18, 1779–1781. [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L. 2003. MADS-box protein complexes control carpel and ovule development in Arabidopsis. The Plant Cell 15, 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu QT, Niu LJ, Zhang QF, Pan BZ, He HY, Xu ZF. 2014. Benzyladenine treatment promotes floral feminization and fruiting in a promising oilseed crop Plukenetia volubilis. Industrial Crops and Products 59, 295–298. [Google Scholar]
- Gibney ER, Nolan CM. 2010. Epigenetics and gene expression. Heredity 105, 4–13. [DOI] [PubMed] [Google Scholar]
- Golan-Goldhirsh A, Jones R, Rowland LJ. 2001. AFLP markers for sex determination in an Ilex species. Acta Horticulturae 546, 591–595. [Google Scholar]
- Golenberg EM, West NW. 2013. Hormonal interactions and gene regulation can link monoecy and environmental plasticity to the evolution of dioecy in plants. American Journal of Botany 100, 1022–1037. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. 2009. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proceedings of the National Academy of Sciences, USA 106, 16529–16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. 1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes & Development 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R. 2008. Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR Jr, Quatrano RS. 1990. A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250, 267–271. [DOI] [PubMed] [Google Scholar]
- Guo X, Hu Z, Yin W, Yu X, Zhu Z, Zhang J, Chen G. 2016. The tomato floral homeotic protein FBP1-like gene, SlGLO1, plays key roles in petal and stamen development. Scientific Reports 6, 20454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DJ, Bellman SK, Sacher RM. 1976. Gibberellic acid-controlled sex expression of corn tassels. Crop Science 16, 371–374. [Google Scholar]
- Hardenack S, Ye D, Saedler H, Grant S. 1994. Comparison of MADS box gene expression in developing male and female flowers of the dioecious plant white campion. The Plant Cell 6, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Zhou J, Xu C, et al. . 2017. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nature Communications 8, 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SH, Krien MJ, O’Connell MJ. 2002. Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases. Genome Biology 3, reviews3003.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward AP, Moreno MA, Howard TP 3rd, et al. . 2016. Control of sexuality by the sk1-encoded UDP-glycosyltransferase of maize. Science Advances 2, e1600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergeth SP, Schneider R. 2015. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Reports 16, 1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herridge RP, Day RC, Macknight RC. 2014. The role of the MCM2-7 helicase complex during Arabidopsis seed development. Plant Molecular Biology 86, 69–84. [DOI] [PubMed] [Google Scholar]
- Ishihara S, Varma R, Schwartz RH. 2010. A new fractionation assay, based on the size of formaldehyde-crosslinked, mildly sheared chromatin, delineates the chromatin structure at promoter regions. Nucleic Acids Research 38, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janousek B, Siroký J, Vyskot B. 1996. Epigenetic control of sexual phenotype in a dioecious plant, Melandrium album. Molecular & General Genetics 250, 483–490. [DOI] [PubMed] [Google Scholar]
- Kapolas G, Beris D, Katsareli E, Livanos P, Zografidis A, Roussis A, Milioni D, Haralampidis K. 2016. APRF1 promotes flowering under long days in Arabidopsis thaliana. Plant Science 253, 141–153. [DOI] [PubMed] [Google Scholar]
- Kazama Y, Fujiwara MT, Koizumi A, Nishihara K, Nishiyama R, Kifune E, Abe T, Kawano S. 2009. A SUPERMAN-like gene is exclusively expressed in female flowers of the dioecious plant Silene latifolia. Plant & Cell Physiology 50, 1127–1141. [DOI] [PubMed] [Google Scholar]
- Khadka DK, Nejidat A, Tal M, Golan-Goldhirsh A. 2002. DNA markers for sex: molecular evidence for gender dimorphism in dioecious Mercurialis annua L. Molecular Breeding 9, 251–257. [Google Scholar]
- Khadka DK, Nejidat A, Tal M, Golan-Goldhirsh A. 2005. Molecular characterization of a gender-linked DNA marker and a related gene in Mercurialis annua L. Planta 222, 1063–1070. [DOI] [PubMed] [Google Scholar]
- Kotomura N, Harada N, Ishihara S. 2015. The proportion of chromatin graded between closed and open states determines the level of transcripts derived from distinct promoters in the CYP19 gene. PLoS ONE 10, e0128282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM, Dorit RL, Irish VF. 1998. Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149, 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. 2005. Molecular mechanisms of flower development: an armchair guide. Nature Reviews Genetics 6, 688–698. [DOI] [PubMed] [Google Scholar]
- Latrasse D, Rodriguez-Granados NY, Veluchamy A, et al. . 2017. The quest for epigenetic regulation underlying unisexual flower development in Cucumis melo. Epigenetics & Chromatin 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G. 1996. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Lindsay DL, Sawhney VK, Bonham-Smith PC. 2006. Cytokinin-induced changes in CLAVATA1 and WUSCHEL expression temporally coincide with altered floral development in Arabidopsis. Plant Science 170, 1111–1117. [Google Scholar]
- Lisch D. 2009. Epigenetic regulation of transposable elements in plants. Annual Review of Plant Biology 60, 43–66. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, Morin H, Pitrat M, Dogimont C, Bendahmane A. 2009. A transposon-induced epigenetic change leads to sex determination in melon. Nature 461, 1135–1138. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nature Protocols 8, 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami K, Sakamoto A, Iwabuchi M. 1994. The HBP-1 family of wheat basic/leucine zipper proteins interacts with overlapping cis-acting hexamer motifs of plant histone genes. The Journal of Biological Chemistry 269, 9974–9985. [PubMed] [Google Scholar]
- Mitchell CH, Diggle PK. 2005. The evolution of unisexual flowers: morphological and functional convergence results from diverse developmental transitions. American Journal of Botany 92, 1068–1076. [DOI] [PubMed] [Google Scholar]
- Murmu J, Bush MJ, DeLong C, Li S, Xu M, Khan M, Malcolmson C, Fobert PR, Zachgo S, Hepworth SR. 2010. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiology 154, 1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau C, Di Stilio VS, Wu HM, Cheung AY. 2011. Arabidopsis and Tobacco SUPERMAN regulate hormone signalling and mediate cell proliferation and differentiation. Journal of Experimental Botany 62, 949–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth CE, Schmitz RJ. 2017. Putting DNA methylation in context: from genomes to gene expression in plants. Biochimica et Biophysica Acta 1860, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen MI, Osborne AR, Yang H, Rastogi S, Chellappan S, Cheng JQ, Boss JM, Blanck G. 2005. Activation of a methylated promoter mediated by a sequence-specific DNA-binding protein, RFX. The Journal of Biological Chemistry 280, 38914–38922. [DOI] [PubMed] [Google Scholar]
- Nitiss JL. 2009. DNA topoisomerase II and its growing repertoire of biological functions. Nature Reviews Cancer 9, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeso JR. 2002. The costs of reproduction in plants. New Phytologist 155, 321–348. [DOI] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Wuest SE, Rae L, et al. . 2013. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. The Plant Cell 25, 2482–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlofsky ME, Kozhoridze G, Lyubenova L, et al. . 2016. Sexual dimorphism in the response of Mercurialis annua to stress. Metabolites 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan BZ, Xu ZF. 2011. Benzyladenine treatment significantly increases the seed yield of the biofuel plant Jatropha curcas. Journal of Plant Growth Regulation 30, 166–174. [Google Scholar]
- Parkinson SE, Gross SM, Hollick JB. 2007. Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Developmental Biology 308, 462–473. [DOI] [PubMed] [Google Scholar]
- Paxson-Sowders DM, Dodrill CH, Owen HA, Makaroff CA. 2001. DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiology 127, 1739–1749. [PMC free article] [PubMed] [Google Scholar]
- Pfent C, Pobursky KJ, Sather DN, Golenberg EM. 2005. Characterization of SpAPETALA3 and SpPISTILLATA, B class floral identity genes in Spinacia oleracea, and their relationship to sexual dimorphism. Development Genes and Evolution 215, 132–142. [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. 2003. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. [DOI] [PubMed] [Google Scholar]
- Poupin MJ, Federici F, Medina C, Matus JT, Timmermann T, Arce-Johnson P. 2007. Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene 404, 10–24. [DOI] [PubMed] [Google Scholar]
- Prunet N, Yang W, Das P, Meyerowitz EM, Jack TP. 2017. SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proceedings of the National Academy of Sciences, USA 114, 7166–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puterova J, Kubat Z, Kejnovsky E, Jesionek W, Cizkova J, Vyskot B, Hobza R. 2018. The slowdown of Y chromosome expansion in dioecious Silene latifolia due to DNA loss and male-specific silencing of retrotransposons. BMC Genomics 19, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS, Ricklefs RE. 1995. Dioecy and its correlates in the flowering plants. American Journal of Botany 82, 596–606. [Google Scholar]
- Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C. 2010. CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes. Proceedings of the National Academy of Sciences, USA 107, 20311–20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JR, Pannell JR. 2015. Sex determination in dioecious Mercurialis annua and its close diploid and polyploid relatives. Heredity 114, 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather DN, Jovanovic M, Golenberg EM. 2010. Functional analysis of B and C class floral organ genes in spinach demonstrates their role in sexual dimorphism. BMC Plant Biology 10, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena PK, Fowke LC, King J. 1985. An efficient procedure for isolation of nuclei from plant protoplasts. Protoplasma 128, 184–189. [Google Scholar]
- Schwartz U, Németh A, Diermeier S, Exler JH, Hansch S, Maldonado R, Heizinger L, Merkl R, Längst G. 2019. Characterizing the nuclease accessibility of DNA in human cells to map higher order structures of chromatin. Nucleic Acids Research 47, 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Chanderbali AS, Kim S, Buzgo M, Soltis PS. 2007. The ABC model and its applicability to basal angiosperms. Annals of Botany 100, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Ma K, Ci D, Tian X, Zhang Z, Zhang D. 2013. The SUPERMAN gene family in Populus: nucleotide diversity and gene expression in a dioecious plant. Plant Cell Reports 32, 1277–1288. [DOI] [PubMed] [Google Scholar]
- Stewart D, Graciet E, Wellmer F. 2016. Molecular and regulatory mechanisms controlling floral organ development. The FEBS Journal 283, 1823–1830. [DOI] [PubMed] [Google Scholar]
- Sundaram S, Kertbundit S, Shakirov EV, Iyer LM, Jurícek M, Hall TC. 2013. Gene networks and chromatin and transcriptional regulation of the phaseolin promoter in Arabidopsis. The Plant Cell 25, 2601–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, Vega-Montoto L, Rudnick PA, et al. . 2010. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. Journal of Proteome Research 9, 761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan QK, Irish VF. 2006. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiology 140, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G, Melzer R. 2007. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Annals of Botany 100, 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theißen G, Melzer R, Rümpler F. 2016. MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. [DOI] [PubMed] [Google Scholar]
- Theißen G, Saedler H. 2001. Plant biology. Floral quartets. Nature 409, 469–471. [DOI] [PubMed] [Google Scholar]
- Trebitsh T, Staub JE, O’Neill SD. 1997. Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiology 113, 987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tröbner W, Ramirez L, Motte P, Hue I, Huijser P, Lönnig WE, Saedler H, Sommer H, Schwarz-Sommer Z. 1992. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. The EMBO Journal 11, 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltsos P, Cossard G, Beaudoing E, Beydon G, Savova BD, Roux C, González-Martínez CS, Pannell RJ. 2018. Size and content of the sex-determining region of the Y chromosome in dioecious Mercurialis annua, a plant with homomorphic sex chromosomes. Genes 9, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltsos P, Ridout KE, Toups MA, et al. . 2019. Early sex-chromosome evolution in the diploid dioecious plant Mercurialis annua. Genetics 212, 815–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genetics 39, 457–466. [DOI] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Story GW, Sunderland PA, Jiang Q, Bray CM. 2002. Disruption of the Arabidopsis AtKu80 gene demonstrates an essential role for AtKu80 protein in efficient repair of DNA double-strand breaks in vivo. The Plant Journal 31, 517–528. [DOI] [PubMed] [Google Scholar]
- West NW, Golenberg EM. 2018. Gender-specific expression of GIBBERELLIC ACID INSENSITIVE is critical for unisexual organ initiation in dioecious Spinacia oleracea. New Phytologist 217, 1322–1334. [DOI] [PubMed] [Google Scholar]
- Winter KU, Becker A, Münster T, Kim JT, Saedler H, Theißen G. 1999. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proceedings of the National Academy of Sciences, USA 96, 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Maoileidigh DSO, Rae L, et al. . 2012. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proceedings of the National Academy of Sciences, USA 109, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav NS, Khadka J, Domb K, Zemach A, Grafi G. 2018. CMT3 and SUVH4/KYP silence the exonic Evelknievel retroelement to allow for reconstitution of CMT1 mRNA. Epigenetics & Chromatin 11, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav NS, Shukla PS, Jha A, Agarwal PK, Jha B. 2012. The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biology 12, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubov B, Barazani O, Golan-Goldhirsh A. 2005. Combination of SCAR primers and touchdown-PCR for sex identification in Pistacia vera L. Scientia Horticulturae 103, 473–478. [Google Scholar]
- Yun JY, Weigel D, Lee I. 2002. Ectopic expression of SUPERMAN suppresses development of petals and stamens. Plant & Cell Physiology 43, 52–57. [DOI] [PubMed] [Google Scholar]
- Zhang T, Zhao Y, Juntheikki I, et al. . 2017. Dissecting functions of SEPALLATA‐like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytologist 216, 939–954. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, et al. . 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126, 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhao J, Morozova N, Williams L, Libs L, Avivi Y, Grafi G. 2001. Two phases of chromatin decondensation during dedifferentiation of plant cells: distinction between competence for cell fate switch and a commitment for S phase. The Journal of Biological Chemistry 276, 22772–22778. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature Genetics 39, 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.