Oxidation of proline in response to dark-induced senescence in Arabidopsis leaves allows this amino acid to be used as an alternative respiratory substrate and contributes to the production of glutamate and energy that can be used for nutrient remobilization.
Keywords: Arabidopsis thaliana, dark-induced leaf senescence, mitochondria, primary metabolism, proline dehydrogenase, proline metabolism
Abstract
Leaf senescence is a form of developmentally programmed cell death that allows the remobilization of nutrients and cellular materials from leaves to sink tissues and organs. Among the catabolic reactions that occur upon senescence, little is known about the role of proline catabolism. In this study, the involvement in dark-induced senescence of proline dehydrogenases (ProDHs), which catalyse the first and rate-limiting step of proline oxidation in mitochondria, was investigated using prodh single- and double-mutants with the help of biochemical, proteomic, and metabolomic approaches. The presence of ProDH2 in mitochondria was confirmed by mass spectrometry and immunogold labelling in dark-induced leaves of Arabidopsis. The prodh1 prodh2 mutant exhibited enhanced levels of most tricarboxylic acid cycle intermediates and free amino acids, demonstrating a role of ProDH in mitochondrial metabolism. We also found evidence of the involvement and the importance of ProDH in respiration, with proline as an alternative substrate, and in remobilization of proline during senescence to generate glutamate and energy that can then be exported to sink tissues and organs.
Introduction
Leaf senescence is the final stage of leaf development and is critical for converting cellular materials into exportable nutrients that can be transported to sink tissues and organs (Lim et al., 2007). Senescence starts with programmed dismantling of chloroplasts, which leads to a general coordinated remodelling of cellular metabolism. In order to fuel the energetic demand of cells after the loss of the photosynthetically active chloroplasts, mitochondria persist long after chloroplast degradation. Mitochondria are involved in oxidative energy metabolism and release energy to allow the maintenance of primary metabolism prior to cell death. Mitochondria numbers have been shown to decrease by 30% during developmental leaf senescence, although the integrity of the remaining mitochondria is preserved (Keech et al., 2007; Chrobok et al., 2016). It has also been postulated that mitochondria acquire additional functions, such as selective catabolism of lipids and amino acids, that compensate the loss of energy supplied by chloroplasts and maintain cell viability until the completion of nutrient remobilization from the cell (Keech et al., 2007).
Glutamate, which is released from several mitochondrial catabolic pathways, is proposed to play a crucial role in nitrogen remobilization during leaf senescence (Chrobok et al., 2016). It is one of the main amino acids present in phloem sap in senescing Arabidopsis (Masclaux-Daubresse et al., 2010) and in poplar (Couturier et al., 2010). Glutamate may originate from proline catabolism and from conversion through arginase activity of arginine to ornithine, which in turn enters in the proline catabolic pathway (Funck et al., 2008). Taken together, although little is known about its role at this developmental stage, these findings suggest that proline catabolism could play a role in recycling proline and arginine during leaf senescence. Proline accumulation has been described in excised rice leaves during dark-induced senescence (Wang et al., 1982; Mondal et al., 1985), although a decrease in proline content has been reported during developmental leaf senescence in Arabidopsis (Chrobok et al., 2016).
Energy deprivation triggers dramatic reprogramming of transcription, which supports metabolic adjustment. Proline metabolism and its regulation have been the subject of numerous studies in plants (Szabados and Savouré, 2010; Sharma et al., 2011; Servet et al., 2012). Anabolism and catabolism are each composed of two steps and take place in different cellular compartments. In response to environmental stresses such as drought and salinity, proline is synthesized from glutamate in the cytosol and most probably in chloroplasts via the activities of bifunctional pyrroline-5-carboxylate synthetase (P5CS) and P5C reductase (P5CR) (Székely et al., 2008). Upon recovery from stress, proline is rapidly oxidized to glutamate in mitochondria via sequential action of proline dehydrogenase (ProDH) and P5C dehydrogenase (P5CDH). In Arabidopsis, proline oxidation occurs via two non-redundant ProDH isoforms (Funck et al., 2010). ProDH1 and ProDH2 share high sequence homology, with 75% identical amino acids (Servet et al., 2012). ProDH1 expression has been shown to be repressed during water stress but is induced by rehydration (Kiyosue et al., 1996; Verbruggen et al., 1996). In addition to its developmental regulation, ProDH1 expression appears to be triggered by hypo-osmolarity and exogenously applied proline (Verbruggen et al., 1996; Nakashima et al., 1998). Expression of ProDH1 in response to proline or hypo-osmolarity is controlled by a proline-responsive element (PRE) cis-acting element via direct binding of the S1-bZIP transcriptional activator to its promoter (Satoh et al., 2002, 2004; Weltmeier et al., 2006). In addition bZIP1, bZIP2, and bZIP53 have been shown to regulate ProDH1 transcript levels and proline content during dark-induced starvation (Dietrich et al., 2011; Pedrotti et al., 2018). Transcriptional analysis of ProDH1 indicates abundant expression in flowers, particularly in pollen and stigma (Funck et al., 2010). In contrast with ProDH1, low expression is generally observed for ProDH2 with the exception of vascular tissues and the abscission zones of floral organs and senescent leaves, where high expression levels have been recorded (Funck et al., 2010; Faës et al., 2015).
ProDH1 subcellular localization has unambiguously been shown to be mitochondrial (Mani et al., 2002; Schertl and Braun, 2014; Cabassa-Hourton et al., 2016). In response to proline treatment, ProDH1 has been shown to be part of low molecular mass complexes in mitochondria and to play an essential role in mitochondrial proline oxidation, which may provide reducing power, energy, and nitrogen sources for growth (Cabassa-Hourton et al., 2016). The function of ProDH2 was addressed later because its gene had for a long time been considered to be a pseudo gene. The use of ProDH2-GFP fusion proteins suggests either mitochondrial or plastid localization (Van Aken et al., 2009; Funck et al., 2010). Arabidopsis ProDH2 has been shown to complement a yeast ∆put1 mutant and, when overexpressed in a GFP-tagged form, to rescue an Arabidopsis prodh1 mutant using proline as sole source of nitrogen (Funck et al., 2010), demonstrating that ProDH2 as well as ProDH1 can mediate proline oxidation. However, in Arabidopsis supplied with an excess of proline and other nitrogen sources, ProDH2 is neither able to overcome the proline sensitivity of the prodh1 mutant (Funck et al., 2010; Cabassa-Hourton et al., 2016) nor to restore the lack of ProDH1 in mitochondrial proline oxidation (Cabassa-Hourton et al., 2016). Therefore the two ProDHs appear to have distinct and non-redundant physiological functions in proline oxidation, at least in Arabidopsis (Funck et al., 2010; Rizzi et al., 2017; Cabassa-Hourton et al., 2016).
All of these data suggest that remobilization of proline and its oxidized derivatives could participate in the senescence process; however, the role of ProDHs remains to be demonstrated. Therefore, in this study we investigated the contribution of ProDHs during dark-induced leaf senescence in Arabidopsis. Our analyses of the prodh1 prodh2 double-mutant in comparison to the wild-type revealed an important function of ProDHs in proline oxidation and its contribution to respiration during senescence. Distinct metabolome patterns demonstrated a link between the tricarboxylic acid cycle and ProDH, demonstrating a key role of proline oxidation for providing energy and glutamate during senescence.
Materials and methods
Plant growth and senescence treatment
All the Arabidopsis thaliana lines tested were in the Columbia-0 (Col-0) background. The prodh1-4 (SALK_119334) and prodh2-2 (GABI_328G05) insertion mutants were obtained from the Salk Institute, La Jolla, CA, USA (Alonso and Stepanova, 2003) and from the Center for Biotechnology, Universität Bielefeld, Germany (Kleinboelting et al., 2012), respectively, and genetically characterized (Cabassa-Hourton et al., 2016). A double-mutant prodh1-4 prodh2-2 (hereafter called prodh1 prodh2) was obtained by crossing prodh1-4 and prodh2-2 as described by Cabassa-Hourton et al. (2016). Seedlings were sown and grown in soil under a 16/8-h light/dark cycle at 80–100 µmol photons m−2 s−1 at 21 °C for 4 weeks.
Senescence experiments were carried out on excised leaves that were left to age in the dark. Leaves were collected from 4-week-old plants and placed on wet Whatman® paper in Petri dishes. The dishes were then wrapped in aluminum foil and kept in the growth chamber until further analysis. For mitochondrial respiration, ProDH activity, transcript and metabolomic analyses, leaves 7, 8, and 9 from the base of the plant were harvested from 4-week-old plants and were processed as indicated above to trigger dark-induced senescence.
Immunocytochemical studies
For electron microscopy, 2×5-mm pieces of leaves were cut and placed in a fixation solution containing 3% (v/v) formaldehyde and 0.5% (v/v) glutaraldehyde in 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4). The samples were then subjected to a low vacuum for 20 min, and washed twice in PBS and once in distilled water for 20 min each. A dehydration procedure was then performed with 25% and 50% ethanol successively for 20 min each under gentle shaking. The samples were then stored in 70% ethanol overnight at 4 °C. Dehydration was achieved by successive incubation in 80% and 90% ethanol for 1 h each. Embedding was started in a mix of 25% London Resin White (LR) in 90% ethanol for 1 h under gentle shaking, followed by incubation in 50% LR in 90% ethanol overnight, 75% LR in 90% ethanol for 2 h, and finally in 100% LR for 3 h. The embedded samples were then placed in gelatine capsules at 55 °C for 48 h. Ultra-thin sections were then cut with a diamond knife to a thickness of 70 nm. Sections were collected on 200 mesh Formwar-coated nickel grids.
For immunological detection of ProDH, the grids were firstly incubated in goat serum 5% (v/v) in T1 buffer [0.05 M Tris-HCl, 2% (w/v) NaCl, 0.1% (w/v) BSA grade V, and 0.05% (v/v) Tween-20, pH 7.4] for 1 h. Antibodies raised against the ProDH1 protein were diluted 50-fold in T1 buffer and applied to the grids overnight at 4 °C. The grids were then washed five times with T1 buffer and three times with T2 buffer [0.02 M Tris-HCl, 2.5% (w/v) NaCl, 0.1% (w/v) BSA grade V, and 0.05% Tween-20 (v/v), pH 8] for 5 min each. The grids were incubated with a 10-nm colloidal gold–goat anti-rabbit IgG whole molecule complex in T2 buffer for 2 h at room temperature and were then washed five times with T2 and five times with ultra-pure water for 5 min each. The grids were finally contrasted with 5% uranyl acetate in water for 5 min, washed three times with ultra-pure water and then with lead citrate tri-hydrate in water for 30 s, and washed 10 times with ultra-pure water. Observations were made using a Zeiss 912 TEM with Omega filter.
RNA extraction and droplet digital PCR analysis
RNAs were extracted from three leaves at the same developmental stage (~100 mg of plant tissue) that were ground in liquid nitrogen using a mixer mill (MM301; Retsch, Haan, Germany) according to Ben Rejeb et al. (2015). RNA quality and concentration were determined using a NP80 Nanophotometer® (Implen). A mixture of cDNAs corresponding to four independent biological experiments was prepared for each time point and used for droplet digital PCR (ddPCR). ddPCR was conducted and data were analysed using a QX200 ddPCR system (Bio-Rad) according to the manufacturer’s protocol. The 20-μl reaction was conducted using 2× ddPCR supermix, and the cDNA sample was at a concentration of 20 ng µl−1. The sample mix and 70 μl of droplet generation oil were added into their corresponding wells of the DG8TM cartridge in order to be emulsioned into 20 000 oil encapsulated nanodroplets. Data analysis was performed using the Quantasoft™ software (Bio-Rad) to calculate the number of copies per cDNA. The absolute expression quantitation of the ProDH genes was normalized using adenine phosphoribosyltransferase 1 (APT1, At1g27450) and the ubiquinol-Cyt c reductase iron–sulfur subunit (AT5G, At5g13440) as housekeeping genes (Ben Rejeb et al., 2015). All the primers used for ddPCR are given in Supplementary Table S1 at JXB online.
Crude mitochondria extracts, and measurement of respiration and ProDH activity
Mitochondria were isolated at 4 °C from 3 g of fresh leaves (numbers 7, 8, and 9 from the base of the plant) of 4-week-old Arabidopsis Col-0 lines and the double-mutant prodh1 prodh2 after either 0 d or 5 d of dark-induced leaf senescence. The harvested leaves were ground in a cold mortar with 30 ml of grinding buffer, and crude mitochondria were obtained by filtration followed by several differential centrifugation steps as described by Cabassa-Hourton et al. (2016). The mitochondrial protein concentration was determined according to Lowry et al. (1951) using BSA as a standard.
Oxygen consumption was measured on 120 µg of crude mitochondria using a Clark-type oxygen electrode (Hansatech) in 1 ml of air-saturated electrode medium (0.3 M sucrose, 5 mM KH2PO4, 10 mM TES buffer pH 7.4, 10 mM KCl, 2 mM MgCl2, and 0.1 % fatty-acid free BSA from Sigma-Aldrich) at 25 °C. The respiratory chain was first activated with 50 µM of ADP whichever substrate was being used. Respiration was measured from three entry points independently. A mix of 10 mM malate, 1 mM glutamate, and 5 mM pyruvate was used as substrates for investigating respiration from complex I, 10 mM succinate for respiration from complex II, and 6 mM L-proline for respiration from proline. From these entry points, electron flux through the cytochrome pathway (CP) was measured as the rate of oxygen uptake sensitive to cyanide and was then inhibited with 1.5 mM of KCN. We refer to it here as CP capacity. For complex II and proline, the capacity for electron flux through the alternative oxidase (AOX) pathway was obtained as the rate of oxygen uptake in the presence of 1.5 mM KCN. The AOX pathway was activated by a mix of 5 mM pyruvate and 1 mM dithiotreitol (DTT) and then inhibited with 0.1 mM n-propyl gallate. We refer to it here as AOX capacity.
ProDH activity was measured using the 2,6-dichlorophenolindophenol (DCIP-)based assay as described by Cabassa-Hourton et al. (2016).
Measurement of proline and chlorophyll contents
Measurement of the free proline content in 50–70 mg samples of fresh rosette leaf tissues was carried out using the procedure described by Ben Rejeb et al. (2015), which was adapted from the method developed by Bates et al. (1973). The absorbance of the proline–ninhydrin complex in toluene was measured at 520 nm using a NP80 Nanophotometer® (Implen). A standard curve of known L-proline quantities allowed the calculation of the proline content in the samples.
For measurement of chlorophyll contents, 100 mg of frozen leaves were ground to powder in liquid nitrogen and resuspended in 1 ml of 92% acetone. Samples were centrifuged for 10 min at 10 000 g. The supernatants were collected and the volume adjusted to 2 ml with 80% acetone. The absorbance was read at 663 nm and 645 nm to determine chlorophyll concentrations using the equations of Arnon (1949).
Protein extraction and western blotting
Leaves (100–150 mg) were ground in a fine powder in liquid nitrogen. The frozen powder was then suspended into 2 volumes of extraction buffer containing 8 M urea, 5% (w/v) SDS, 40 mM Tris-HCl pH 6.8, 0.1 mM EDTA, and 0.4 mg ml–1 bromophenol blue, supplemented with 1× protease inhibitor cocktail (Merck). Samples were shaken three times for 5 min with 5 min intervals on ice. The samples were then centrifuged at 18 000 g for 10 min at 10 °C and the supernatants (30 µl) were loaded and separated onto a 8% polyacrylamide gel. The proteins were then transferred onto nitrocellulose membranes (Amersham Hybond-ECL® from GE Healthcare) and were immunoblotted using purified antibodies directed against either Arabidopsis recombinant ProDH1, ProDH2, P5CDH, or P5CS proteins. Antibody binding was detected using a secondary antibody conjugated with a horseradish peroxidase and the ECL prime system from GE Healthcare using the G:BOX® system (Syngene).
Identification of ProDH2 by MS analysis
Preparation of samples and identification of proteins by MS was performed as described by Fromm et al. (2016). Briefly, 50 µg of proteins from crude mitochondria isolated from leaves of prodh1 seedlings were loaded onto polyacrylamide gels (stacking gel 10% acrylamide, separating gel 14% acrylamide). Gel runs were stopped when the bromophenol blue dye front reached the end of the stacking gel, and were subsequently stained with Coomassie Brilliant Blue G250. Gel pieces containing the proteins that concentrated at the stacking gel–separation gel interface were subjected to in-gel digestion followed by peptide extraction. MS analyses were performed using a Dionex Ultimate 3000 (ThermoFisher Scientific) equipped with a 50-cm reverse-phase analytical column (C18, ID 75 µm, particle diameter 3 µm, 100Å pore size) in line to a 2-cm reverse-phase pre-column (C18, ID 100 µm, particle diameter 5 µm, 100Å pore size). Peptides were eluted using a 1-h non-linear acetonitrile gradient and submitted to tandem MS employing a top-10 duty cycle. MS/MS data were subsequently evaluated using the Mascot (Matrix Science, London, UK) search engine querying against an in-house Arabidopsis (TAIR10) protein database.
Metabolic profiling using GC-MS
Sample preparation, analysis, and data processing were performed as previously described by Fiehn (2006). Ground, frozen samples (100 mg) from harvested leaves (7, 8, and 9 from the base of the plant) exposed to 5 d of either dark or light conditions were resuspended in 1 ml of frozen (−20 °C) water:acetonitrile:isopropanol (2:3:3) containing ribitol at 4 μg ml−1 and extracted by shaking for 10 min at 4 °C. Insoluble material was removed by centrifugation. Three blank tubes were subjected to the same steps as the samples. A quality control was made by pooling an equal volume from each light/dark condition. A sample of 30 μl was collected from each tube and dried overnight at 35 °C in a vacuum centrifuge.
Derivatization was performed according to Clément et al. (2018), 4 h after the end of which the whole sample series was first injected in splitless mode and then in split mode (1/30). Four different standard mixes were injected at the beginning and the end of the analysis as well as three independent derivatizations of the quality control (beginning, middle, and end) for monitoring the derivatization stability. Samples were randomized. The instrument was an Agilent 7890A gas chromatograph coupled to an Agilent 5977B mass spectrometer. The column was a Rxi-5SilMS from Restek (30 m with 10-m integraguard column). The liner (Restek # 20994) was changed before analysis. The oven temperature ramp was 70 °C for 7 min then 10 °C min−1 to 330 °C for 5 min (run length 38 min). The helium constant flow was 0.7 ml min−1. The following temperatures were used: injector, 250 °C; transfer line, 290 °C; source, 250 °C; and quadripole, 150 °C. Five scans per second were acquired spanning a 50–600 Da range. The instrument was tuned with perfluorotributylamine (PFTBA) with the 69 m/z and 219 m/z of equal intensities. The split mode conditions were 70 °C for 2 min, then 30 °C min–1 to 330 °C for 5 min. The helium constant flow was 1 ml min−1.
The data were processed as follows. The raw Agilent data files were converted into NetCDF format and analysed with AMDIS (http://chemdata.nist.gov/mass-spc/amdis/). A home retention indices/mass spectra library built from the NIST, Golm (http://gmd.mpimp-golm.mpg.de/), and Fiehn databases and standard compounds was used for metabolite identification. Peak areas were also determined with the Targetlynx software (Waters) after conversion of the NetCDF files into masslynx format. AMDIS, Target Lynx in splitless, and split 30 mode data were compiled in one single Excel file for comparisons. After subtraction of the blank mean, peak areas were normalized to ribitol and expressed on a fresh weight basis. Only metabolites showing repeatable and significant differences (based on t-tests) with respect to light and dark conditions are presented in heat maps. A complete list of all the metabolites that were detected can be found in Supplementary Data Set S1.
Statistical analysis
Two-way ANOVAs were performed for the qPCR analysis. The effects of senescence on chlorophyll and proline contents in wild-type and mutants, including time effects, as well as across days was assessed by ANOVA. Tukey tests were performed for comparisons of ProDH activities.
Results
Knockout Arabidopsis prodh mutants accumulate proline during dark-induced senescence
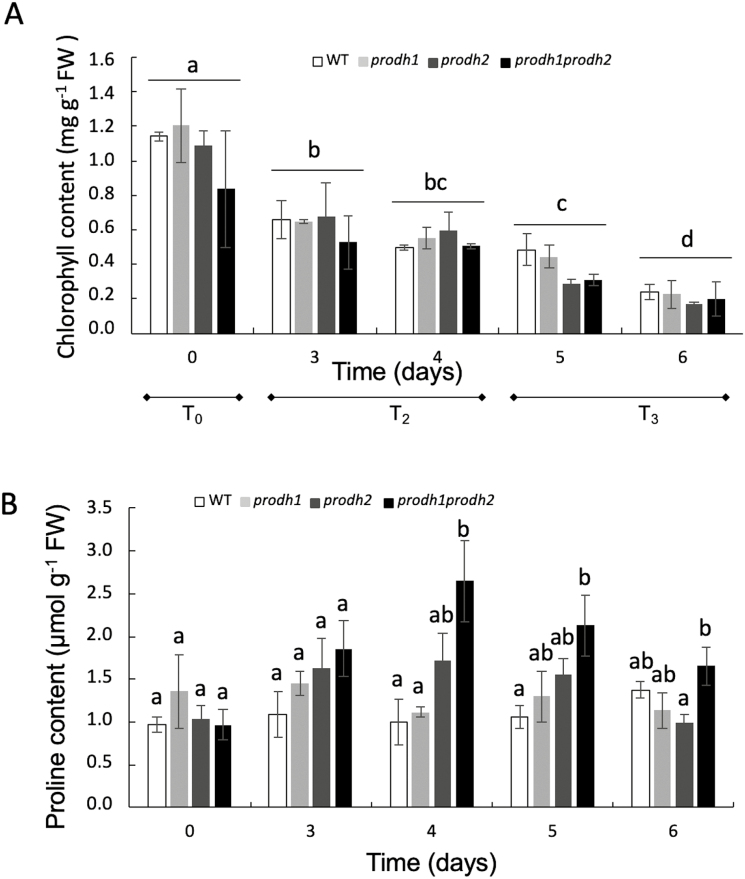
Dark-induced senescence (DIS) was triggered by cutting leaves of 4-week-old Arabidopsis Col-0 (wild-type, WT), prodh1, prodh2, and prodh1 prodh2 plants at the same developmental stage and placing them in the dark for 6 d. The progression of DIS was followed by measuring chlorophyll degradation over time. As shown in Fig. 1A, chlorophyll contents progressively decreased during DIS in all genotypes to less than 0.2 mg chlorophyll g−1 FW at Day 6 (contents could not be determined at 7 d due to degradation of the leaf tissue). The kinetics of chlorophyll degradation were similar to the WT in the both prodh single- and double- mutants. Chrobok et al. (2016) determined five time periods of senescence from T0, which corresponds to leaf expansion, to T4, which corresponds to the mature leaf at the late stage of senescence. Based on this reference, the chlorophyll contents indicated that Day 3 and Day 4 in our study were equivalent to Chrobok’s T2 period, and Day 5 and Day 6 were equivalent to the T3 period. We also monitored the proline content, and no significant variations were observed in the WT and the single-mutants up to Day 6 of DIS (Fig. 1B). However, significant increases in proline accumulation were measured in the prodh1 prodh2 double-mutant compared to the WT at Day 4 and Day 5, suggesting a putative role of ProDH in the oxidation of proline during senescence.
Fig. 1.
Chlorophyll contents and proline accumulation during dark-induced leaf senescence in Arabidopsis. (A) Total chlorophyll content was measured in a time-course experiment over 6 d of dark-induced senescence in Col-0 (wild-type, WT), prodh1, prodh2, and the prodh1 prodh2 double-mutant. Data are means (±SE) of four independent rosettes from two independent experiments. The time periods T0, T2, and T3 are delineated by the progression of senescence in leaves according to Chrobok et al. (2016). Variations within each time-point were not statistically different (ANOVA, P>0.05). Different letters indicate significant differences in overall chlorophyll content between time-points (ANOVA, P<0.05). (B) Proline content was measured in the same time-course experiments as shown in (A). Data are means (±SE) of four independent experiments. Different letters indicate significant differences between means on individual days (ANOVA, P<0.05).
Expression of ProDH during DIS
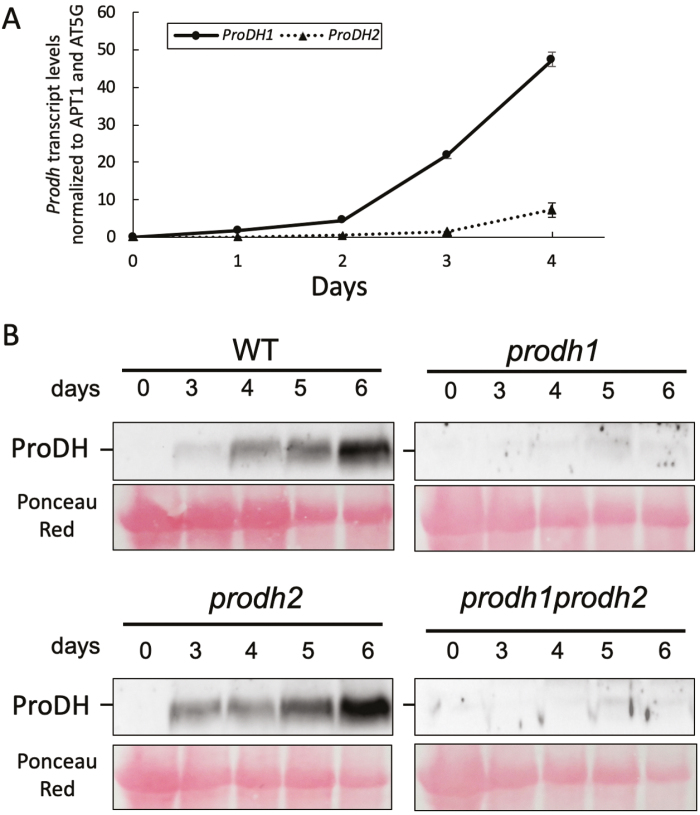
ProDH expression was investigated in dark-induced senescent leaves in a time-course experiment. Leaves from 4-week-old Arabidopsis WT and prodh1 prodh2 plants were collected during senescence. Transcript analyses using the highly sensitive droplet digital PCR (ddPCR) method revealed very low transcript levels of both ProDH1 and ProDH2 in the WT at Day 0 (Fig. 2A), although the ProDH1 transcript level was 5-fold higher than ProDH2. During the 4 d of the senescence experiment, a consistent increase of ProDH1 transcripts (575-fold induction between 0–4 d), and to a lesser extent ProDH2, occurred in the WT (at Day 5, ProDH transcript levels could not be determined due to high degradation of RNAs). At Day 4, the ProDH1 transcript level was 6.5-fold higher than that of ProDH2. These results prompted us to monitor ProDH protein accumulation in a time-course experiment using an antibody directed against ProDH1 recombinant protein (Cabassa-Hourton et al., 2016). In the WT total protein extracts, no ProDH signal was detected at Day 0 (Fig. 2B). A faint signal appeared at Day 3, increased at Day 4 and Day 5, and reached a maximum at the end of the senescence period. A similar progression of ProDH accumulation was observed in the prodh2 mutant, but no ProDH signal could be detected in either the prodh1 or prodh1 prodh2 mutants. These results suggested that at least the ProDH1 isoform accumulated during DIS. ProDH2 could not be detected in the total protein extracts but an increase of ProDH2 transcripts was observed. As the antibody directed against ProDH1 could detect recombinant ProDH2 but with a lower sensitivity than recombinant ProDH1 (Cabassa-Hourton et al., 2016), the lack of detection of ProDH2 in the total protein extracts could have been due to the fact that ProDH2 is a very low abundant enzyme, as postulated by Cabassa-Hourton et al. (2016).
Fig. 2.
ProDH expression during dark-induced leaf senescence in Arabidopsis. (A) ProDH1 and ProDH2 expression in the Col-0 wild-type (WT) was quantified by droplet digital PCR after 0–4 d of dark-induced senescence. Data are means (±SE) of four biological replicates. (B) Western blots of total proteins extracted from 100 mg fresh leaves from Col-0 (WT), prodh1, prodh2, and the prodh1 prodh2 double-mutant in a time-course experiment over 6 d of dark-induced senescence. Proteins were separated by SDS/PAGE and blots were probed with an anti-ProDH1 antibody. (This figure is available in colour at JXB online.)
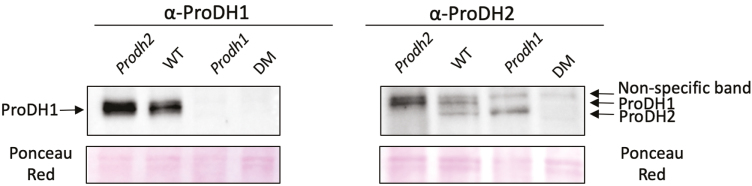
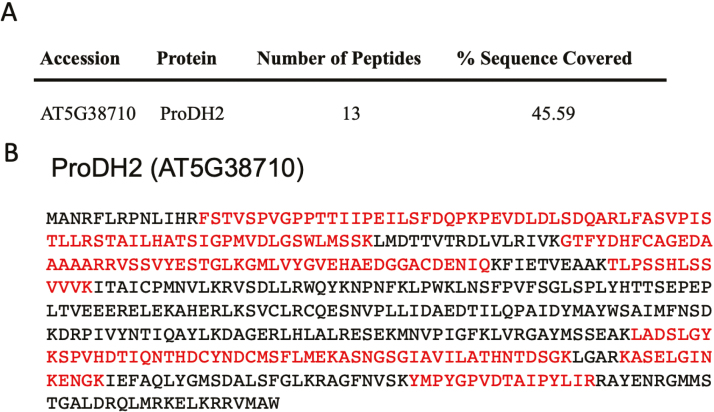
ProDH2 was therefore investigated by western blotting using a new antibody directed against ProDH2 recombinant protein. Leaves from 4-week-old plants were sampled after 5 d of DIS and crude mitochondria were isolated (Cabassa-Hourton et al., 2016). Western blots showed different signal patterns with antibodies directed against either ProDH1 or ProDH2 (Fig. 3). The anti-ProDH1 antibody showed a signal at an apparent molecular mass of ~60 kDa that was not found in the prodh1 and prodh1 prodh2 mutants. In contrast, the use of the new antibody directed against ProDH2 recombinant protein resulted in the observation of both ProDH isoforms (Fig. 3). ProDH2 was only detected in the WT and prodh1 mutant with the anti-ProDH2 antibody. However, this antibody also detected ProDH1 in the WT and prodh2 mutant but to a lesser extent than with the antibody raised against ProDH1. A non-specific signal was observed at a higher apparent molecular mass for all the genotypes. In order to further confirm the nature of the signal detected by the anti-ProDH2 antibody, a MS analysis was performed using crude mitochondria isolated from dark-induced senescent leaves of the prodh1 mutant. This resulted in the identification of 13 unique peptides exactly matching the ProDH2 protein, which covered 45% of the protein sequence with a MASCOT protein score (sum of the peptide score) of 633.16 (Fig. 4). These results confirmed the presence of the ProDH2 isoform in crude mitochondria of leaves subjected to DIS.
Fig. 3.
ProDH expression in Arabidopsis leaf mitochondria after 5 d of dark-induced senescence. Antibodies raised against recombinant ProDH1 and ProDH2 isoforms were used for western blots of proteins from crude mitochondria extracted from Col-0 (wild-type, WT), prodh1, prodh2 and the prodh1 prodh2 double-mutant (DM). The blots were loaded with 5 µg (ProDH1) and 10 µg (ProDH2) of crude mitochondrial extracts. (This figure is available in colour at JXB online.)
Fig. 4.
Identification of the ProDH2 isoform in Arabidopsis mitochondria by mass spectrometry using crude mitochondrial extracts from leaves of prodh1 plants after 5 d of dark-induced senescence. (A) Summary of MS results. (B) The ProDH2 protein sequence with the identified peptides marked in red.
The mitochondrial localization of both ProDHs was confirmed by preparation of ultra-thin leaf sections that were immunogold-labelled with the anti-ProDH1 antibody and observed using TEM. The gold labelling showed a mitochondrial localization of ProDH in the WT at 5 d of DIS (Fig. 5). No signal was detected in the chloroplasts. Additional experiments were performed with the prodh single- and double-mutants in order to assess the identity of the isoform. Signals were detected in mitochondria of the prodh1 (Fig. 5B) and prodh2 (Fig. 5C) mutants, which indicated the presence of both ProDH isoforms in mitochondria during DIS. No signal was detected in the prodh1 prodh2 double-mutant (Fig. 5D). Taken together, these results demonstrated that both the ProDHs were expressed and accumulated in mitochondria during DIS.
Fig. 5.
Immunolocalization using antibodies directed against ProDH during dark-induced senescence in leaves of Arabidopsis. The electron micrographs show sections of leaves after 5 d of dark-induced senescence for (A) Col-0 (wild-type, WT), (B) prodh1, (C) prodh2, and (D) the prodh1 prodh2 double-mutant, and show typical labelling within the mitochondria. Arrows indicate the presence of ProDH. No signal was detected in prodh1 prodh2. m, mitochondrion; cp, chloroplast.
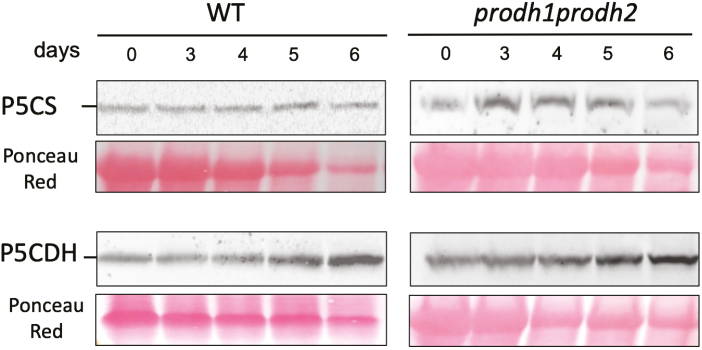
P5CDH but not P5CS accumulates during DIS
We also investigated the effect of dark-induced leaf senescence on P5CS and P5CDH, markers of proline biosynthesis and proline catabolism, respectively, by western blotting using antibodies raised against P5CS1 and P5CDH recombinant enzymes (Fig. 6). In contrast to ProDH, P5CS and P5CDH were already present at Day 0. The level of P5CS protein was not really affected during the time-course experiment in the WT and the prodh1 prodh2 double-mutant. On the other hand, the P5CDH content slightly increased during DIS. These results showed that DIS enhanced proline catabolism but not proline biosynthesis.
Fig. 6.
Analysis of the proline metabolism enzymes P5CS and P5CDH in Arabidopsis leaves over 5 d of dark-induced senescence. Western blots of total proteins extracts taken from Col-0 (wild-type, WT) and the prodh1 prodh2 double-mutant. The blots were obtained using antibodies raised against P5CS and P5CDH recombinant enzymes. (This figure is available in colour at JXB online.)
DIS triggers ProDH activity and proline respiration
The enzymatic activity of ProDH was investigated in crude mitochondria extracts during DIS. After 5 d of DIS, ProDH activity increased by 5-fold in the WT (Fig. 7), but no activity could be measured in the prodh1 prodh2 double-mutant whatever the growth conditions, as has previously been reported by Cabassa-Hourton et al. (2016). Taken together, these results indicated that ProDH was functional during DIS.
Fig. 7.
ProDH activity in Arabidopsis leaves in response to dark-induced senescence. Activity was measured in crude mitochondria extracts from leaves 7, 8, and 9 from the base of the plant that were sampled at 0 d and 5 d from Col-0 (wild-type, WT) and the prodh1 prodh2 double-mutant. Data are means (±SE) of at least for independent biological experiments. Different letters indicate significant differences between means as determined by ANOVA followed by Tukey's test (P<0.05).
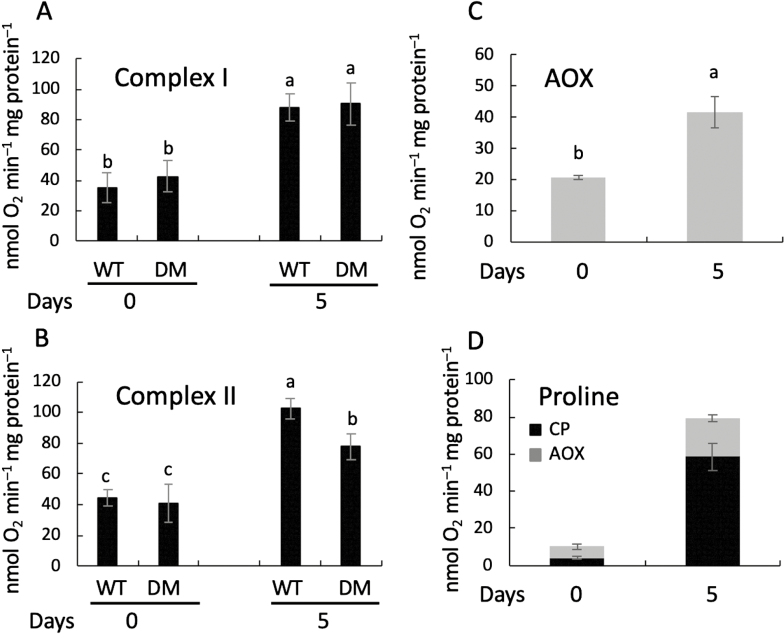
Oxygen consumption by crude mitochondria extracts was investigated in the WT and the prodh1 prodh2 double-mutant. Complex I capacity through the cytochrome pathway increased by 2.5-fold after 5 d of DIS in both the WT and the double-mutant (Fig. 8A). Complex II capacity increased in the same range for the WT but to a lower extent for the double-mutant (Fig. 8B). The potential contribution of the AOX pathway was determined by completely inhibiting the CP with KCN and by activating the AOX pathway with DTT in the presence of excess of pyruvate. The capacity for electron flux through the AOX pathway increased by 2-fold in the WT during DIS when succinate was used as the substrate (Fig. 8C). In the WT, proline respiration increased through both the CP and AOX pathway by 14- and 3-fold, respectively, after 5 d of DIS (Fig. 8D). Interestingly, the potential contribution of the AOX pathway upon proline respiration was higher than the CP at Day 0 while the opposite was observed after 5 d of DIS, when 65% of the electron flux passed through the CP. No proline respiration was measured in the double-mutant (data no shown), as was previously observed by Cabassa-Hourton et al. (2016).
Fig. 8.
Oxygen consumption in mitochondria isolated from Arabidopsis leaves from Col-0 (wild-type, WT) and the prodh1 prodh2 double-mutant (DM) in response to dark-induced senescence. Oxygen consumption was determined using a Clark-type electrode in crude mitochondria extracts from leaves 7, 8, and 9 from the base of the plant sampled at 0 d and 5 d of dark-induced senescence. The capacity of the cytochrome pathway (CP) was measured from (A) complex I, (B) complex II, and (D) proline. The capacity of the alternative oxidase pathway (AOX) was measured in WT mitochondria from (C) complex II and (D) proline. Data are means (±SE) of at least four biological replicates. Different letters indicate significant differences between means as determined using ANOVA (P<0.05).
Taken together, these results indicated that there was a higher proline oxidation rate during DIS, making it a potential respiratory substrate, together with a shift of the major electron pathway for proline respiration from AOX to cytochrome.
Metabolic changes in detached leaves subjected to DIS
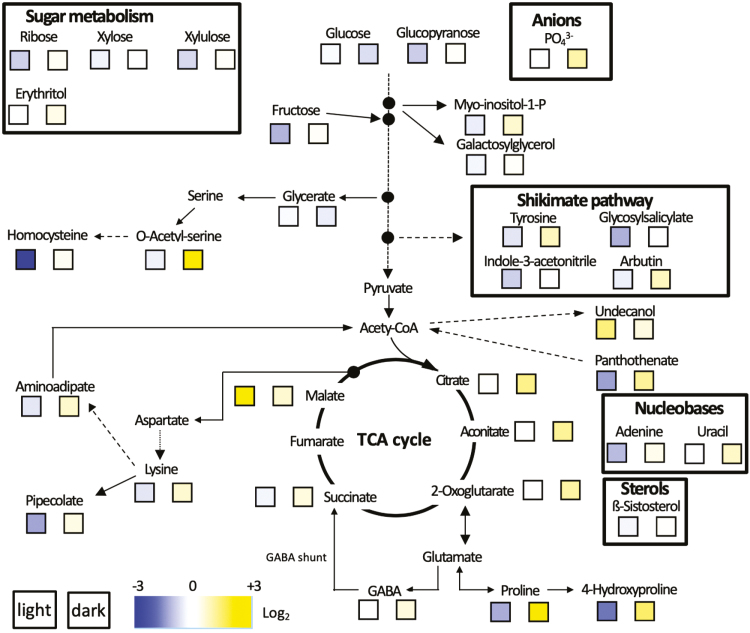
To gain more insights into ProDH function during senescence, a metabolic profiling analysis was carried out in detached leaves of the WT and prodh1 prodh2 double-mutant exposed for 5 d to either light or dark conditions. A GC-MS untargeted approach identified 63 metabolites, of which 32 were unknown, that were differentially accumulated in the double-mutant compared with the WT (Fig. 9). Clearly opposite behavior was generally observed mainly for differentially accumulated metabolites of the primary metabolism in the two genotypes exposed to light or dark conditions. Under light conditions, the identified metabolites were generally accumulated to a higher level in the WT than in the double-mutant, which was in contrast to DIS where the opposite response was observed. In the TCA cycle, levels of citrate, aconitate, 2-oxoglutarate, succinate, and malate were significantly higher in prodh1 prodh2 than in the WT under DIS. This was also the case with some amino acids such as proline, hydroxyproline, lysine, acetyl-serine, tyrosine, GABA, aminoadipate, and homocysteine, with nucleobases such as adenine and uracil, and with sugars such as ribose, xylose, xylulose, erythritol, and fructose. Interestingly, proline and its derivatives such as hydroxyproline and GABA followed the same trend. Only glycerate and glucose were present at lower levels in the prodh1 prodh2 double-mutant compared to the WT in dark conditions. In contrast, the glutamate level was not significantly changed between light and dark conditions in the WT and the double-mutant.
Fig. 9.
Primary metabolism pathways of metabolites measured in detached leaves of Arabidopsis under light or dark conditions compared between Col-0 (wild-type) and the prodh1 prodh2 double-mutant. Heat-maps are shown for changes in metabolites in pooled samples of leaves 7–9 from the base of the plant after 5 d under light (left square) or dark (right square) conditions: the blue-to-yellow scale shows the log2 ratio of the fold-change between the double-mutant and the wild-type. Data represents the mean values of four biological replicates for each time-point.
Discussion
The remobilization of metabolites during stress and senescence plays an important role in plant adaptation to the environment. Here, we hypothesized a requirement for ProDHs for alternative respiration and for the remobilization of proline during senescence, and this was demonstrated by using a combination of genetic, biochemical, proteomic, and immunolocalization approaches.
Increased transcript abundance of both ProDH1 and ProDH2 during DIS
We found that the transcript levels of both ProDH1 and ProDH2 steadily increased over 4 d in detached leaves incubated in the dark (Fig. 2), although much higher levels of ProDH1 than ProDH2 were observed (6.5-fold higher at 4 d). In the literature, data concerning ProDH expression during senescence are very mixed depending on the way it is induced. In developmental leaf senescence (DLS), Chrobok et al. (2016) observed an equal increase of both ProDH1 and ProDH2 transcript abundance in Col-0 leaves. van der Graaff et al. (2006) also observed an increase of ProDH1 transcript levels in DLS but to a lesser extent than ProDH2. Previous studies have also described accumulation of ProDH2 transcripts during senescence in Arabidopsis (Buchanan-Wollaston et al., 2005; Funck et al., 2010; Allu et al., 2014; Chrobok et al., 2016) and in Brassica napus (Faës et al., 2015). In our experiments, leaf senescence was induced by placing detached leaves in the dark. In these conditions, much higher amounts of ProDH1 transcripts than of ProDH2 were measured, as was also observed by van der Graaff et al. (2006). More generally, ProDH transcript levels have been found to be higher in dark-induced senescence in detached leaves, in individually darkened leaves or darkened whole plants than during DLS (van der Graaff et al., 2006; Yu et al., 2016).
Both ProDH isoforms are located in the mitochondria
We found an increase in abundance of both ProDH transcripts but mRNA and protein levels are not always strictly correlated, as has been demonstrated for ProDH1 in response to water- and salt-stress conditions (Parre et al., 2007). Indeed, the protein concentration in a cell is determined by several factors such as transcription rate, transcript stability, and/or protein degradation. Van Aken et al. (2009) showed that variations in transcript levels of genes that putatively encode mitochondrial proteins are generally greater than those of proteins under stress-related conditions. Moreover, microarray analyses have shown that the half-life times of ProDH1 and ProDH2 transcripts are different, being estimated at 6.3 h and 2.2 h, respectively (Narsai et al., 2007). It was there essential to examine whether the observed changes in ProDH1 and ProDH2 transcript levels were reflected in the amounts of their corresponding protein. Accumulation of ProDH1 was clearly observed during DIS in total protein extracts (Fig. 2) and in crude mitochondrial fractions (Fig. 3), while ProDH2 was only found in crude mitochondria but not in total protein extracts (data not shown) when using antibodies raised against recombinant ProDH2. The absence of any signal in the total protein extracts when using antibodies raised against ProDH2 may probably be explained by a co-migration of ProDH2 with Rubisco, with the abundance of the latter masking ProDH2; however, the presence of ProDH2 in crude mitochondria was also confirmed by MS (Fig. 4). To our knowledge, this is the first time that ProDH2 has been identified using a proteomic approach (Klodmann et al., 2011; Rao et al., 2017).
ProDH1 has previously been shown to be clearly localized in mitochondria (Funck et al., 2010; Cabassa-Hourton et al., 2016) while dual targeting to chloroplasts and mitochondria has been proposed for ProDH2 as a result of GFP fusion experiments (Van Aken et al., 2009). Our immuno-microscopy results clearly showed that ProDH2 was only detected in mitochondria during DIS (Fig. 5).
ProDHs contribute to fuelling respiration and the TCA cycle during senescence
We found a higher respiration capacity in complex I and II after 5 d of DIS, whatever the electron pathway (Fig. 8). In senescence induced by covering individual Arabidopsis leaves with aluminum foil, Keech et al. (2007) also reported higher levels of respiration at 6 d of senescence. An increase in respiration has also been shown in leaves of Populus tremuloides and Quercus rubra during autumnal senescence (Collier and Thibodeau, 1995). Interestingly, our work showed for the first time that senescence stimulated a striking increase in proline respiration (Fig. 8D). Cabassa-Hourton et al. (2016) demonstrated that treatment of Arabidopsis seedlings with proline triggers ProDH1 accumulation and activity. Concomitantly, proline respiration increases but to a significantly lesser extent than during senescence: it was two-fold higher in DIS than in proline-treated seedlings. To our knowledge, our data represents one of the highest rates of proline respiration that has been measured. Electron fluxes generated from proline respiration in proline-treated seedlings (Cabassa-Hourton et al., 2016), as well as in leaves before senescence (Day 0, this study), are mainly directed through the AOX pathway and do not efficiently contribute to ATP synthesis. However, upon DIS in our study, the electron flux from proline oxidation was preferentially directed through the CP, allowing a better coupling to ATP synthesis. Taken together, these observations suggest the importance of proline as an alternative respiratory substrate during senescence. Proline oxidation may contribute to the generation of ATP that is necessary for the maintenance of leaf viability while the remobilization of nutrients is carried out. It has previously been reported that the total oxidation of one proline molecule can generate reductants that can fuel the mitochondrial electron transfer chain and support the formation of 30 ATP molecules (Atkinson, 1977). DIS leads to intensive metabolite recycling through protein degradation and the generation of amino acids that are used as alternative substrates for respiration. More generally, energy-limited conditions require alternative pathways of respiration to provide the necessary respiratory function (Barros et al., 2017). Interestingly, proline has also been shown to be the main energy substrate to fuel flight in some insects (Gäde and Auerswald, 2002).
In addition to its role in respiratory electron flux, ProDH is more widely involved in mitochondrial metabolism. Our untargeted metabolomic approach based on GC-MS provided a more comprehensive view of the role of ProDH in primary metabolic pathways during senescence (Fig. 9). Interestingly, this analysis identified increased levels of citrate, aconitate, 2-oxoglutarate, succinate, malate, proline, and hydroxyproline in the prodh1 prodh2 double-mutant in detached leaves subjected to DIS when compared to the WT. These metabolites are linked to the TCA cycle and associated pathways. The increase of TCA intermediates in the double-mutant may indicate a lower availability of alternative substrates such as proline, which could no longer be oxidized to supply the carbon backbone in the form of glutamate or 2-oxoglutarate to sustain the TCA cycle. An increase in TCA intermediates has previously been observed in plants lacking D-2-hydroxyglutarate dehydrogenase or isovaleryl-CoA dehydrogenase, demonstrating the participation of lysine and branched amino acids as alternative substrates for respiration during senescence (Araújo et al., 2010; Engqvist et al., 2011). More generally, a reduced flow of carbon into the TCA cycle seems to lead to an increase of intermediates, probably due to a reduced overall activity of the TCA cycle (Araujo et al., 2010; Yu et al., 2012; Huang et al., 2013; Pires et al., 2016).
Metabolites from sugar metabolism and the shikimate pathway together with homocysteine, a sulfur-containing amino acid, also accumulated more in the double-mutant than in the WT during DIS (Fig. 9). Interestingly, Law et al. (2018) have shown that genes encoding proteins involved in primary energy production (respiration, fermentation, and ß-oxidation), amino acids, lipid or nucleotide catabolism, sulfur metabolism, and the shikimate pathway are overexpressed during senescence of individually darkened leaves. ProDH and proline may thus be crucial actors of this metabolic switch under senescence.
Proline generated by DIS can be metabolized by ProDH enzymes to produce P5C. This can also be synthesized by ornithine aminotransferase, the mRNA levels of which are also up-regulated during senescence (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006; Chrobok et al., 2016). We also found that the P5CDH protein, which allows the conversion of P5C to glutamate, was up regulated in DIS (Fig. 6). Previous results for DLS have shown either no response or up-regulation (van der Graaff et al., 2006). No induction of P5CS was observed under our conditions, which is consistent with previous results reported for leaves subjected to DIS (van der Graaff et al., 2006). Thus, DIS triggers proline catabolism rather than proline biosynthesis and allows the generation of glutamate, which is the metabolic precursor of glutamine and asparagine that are involved in nitrogen remobilization during senescence (Finnemann and Schjoerring, 2000; Lin and Wu, 2004; Masclaux-Daubresse et al., 2010). Proline content has been shown to decrease during developmental senescence in wild-type Arabidopsis and in tobacco (Masclaux et al., 2000; Chrobok et al., 2016), which indicates an important role of ProDH in proline oxidation to release glutamate. Moreover, it has been shown that proline regulates the expression of Glutamine Synthase 1 (GS1) and Glutamate Dehydrogenase (GDH), which encode two enzymes involved in nitrogen remobilization during senescence (Masclaux-Daubresse et al. (2005), thus supporting an important role of this amino acid as a signal molecule during senescence.
Proline could originate from the degradation of cellular and cell wall proteins, which are notably rich in proline and hydroxy (Ihsan et al., 2017), as has also been described for the mammalian extracellular matrix (Pandhare et al., 2009). Degradation of cell wall proteins could therefore lead to an increase of free proline concentration in the cell that would act as an activator of ProDH transcription and as a substrate for both ProDH isoforms, contributing to nitrogen remobilization through glutamine generation from glutamate and its subsequent transport to sinks. In conclusion, our results indicate a dual role of ProDH under DIS. Proline oxidation allows it to be used as an alternative respiratory substrate and contributes to the production of glutamate and energy that can subsequently participate in the remobilization of nutrients from senescent tissues to developing plant organs.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of primers used for ddPCR.
Data Set S1. Raw data and statistical analysis of metabolite profiles in the leaf experiments.
Acknowledgements
We are indebted to Dr Dietmar Funck for kindly providing an antibody directed against P5CDH. We thank the electron microscopy team of the IBPS Imaging Facility, Sorbonne University for their help with the experiments. This study received financial support from Sorbonne University.
Glossary
Abbreviations
- AOX
alternative oxidase
- CP
cytochrome pathway
- DIS
dark-induced senescence
- DLS
developmental leaf senescence
- P5C
pyrroline-5-carboxylate
- P5CDH
P5C dehydrogenase
- P5CR
pyrroline-5-carboxylate reductase
- P5CS
pyrroline-5-carboxylate synthetase
- ProDH
proline dehydrogenase
- TCA
tricarboxylic acid
References
- Allu AD, Soja AM, Wu A, Szymanski J, Balazadeh S. 2014. Salt stress and senescence: identification of cross-talk regulatory components. Journal of Experimental Botany 65, 3993–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN. 2003. T-DNA mutagenesis in Arabidopsis. Methods in Molecular Biology 236, 177–188. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, et al. . 2010. Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. The Plant Cell 22, 1549–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DI. 1949. Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiology 24, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson DE. 1977. Cellular energy metabolism and its regulation. New York: Academic Press. [Google Scholar]
- Barros JAS, Cavalcanti JHF, Medeiros DB, Nunes-Nesi A, Avin-Wittenberg T, Fernie AR, Araújo WL. 2017. Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana. Plant Physiology 175, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID. 1973. Rapid determination of free proline for water-stress studies. Plant and Soil 39, 205–207. [Google Scholar]
- Ben Rejeb K, Lefebvre-De Vos D, Le Disquet I, Leprince AS, Bordenave M, Maldiney R, Jdey A, Abdelly C, Savouré A. 2015. Hydrogen peroxide produced by NADPH oxidases increases proline accumulation during salt or mannitol stress in Arabidopsis thaliana. The New Phytologist 208, 1138–1148. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, et al. . 2005. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. The Plant Journal 42, 567–585. [DOI] [PubMed] [Google Scholar]
- Cabassa-Hourton C, Schertl P, Bordenave-Jacquemin M, et al. . 2016. Proteomic and functional analysis of proline dehydrogenase 1 link proline catabolism to mitochondrial electron transport in Arabidopsis thaliana. The Biochemical Journal 473, 2623–2634. [DOI] [PubMed] [Google Scholar]
- Chrobok D, Law SR, Brouwer B, et al. . 2016. Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant Physiology 172, 2132–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément G, Moison M, Soulay F, Reisdorf-Cren M, Masclaux-Daubresse C. 2018. Metabolomics of laminae and midvein during leaf senescence and source-sink metabolite management in Brassica napus L. leaves. Journal of Experimental Botany 69, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier DE, Thibodeau BA. 1995. Changes in respiration and chemical content during autumnal senescence of Populus tremuloides and Quercus rubra leaves. Tree Physiology 15, 759–764. [DOI] [PubMed] [Google Scholar]
- Couturier J, Doidy J, Guinet F, Wipf D, Blaudez D, Chalot M. 2010. Glutamine, arginine and the amino acid transporter Pt-CAT11 play important roles during senescence in poplar. Annals of Botany 105, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K, Weltmeier F, Ehlert A, Weiste C, Stahl M, Harter K, Dröge-Laser W. 2011. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. The Plant Cell 23, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist MK, Kuhn A, Wienstroer J, Weber K, Jansen EE, Jakobs C, Weber AP, Maurino VG. 2011. Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. The Journal of Biological Chemistry 286, 11382–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faës P, Deleu C, Aïnouche A, et al. . 2015. Molecular evolution and transcriptional regulation of the oilseed rape proline dehydrogenase genes suggest distinct roles of proline catabolism during development. Planta 241, 403–419. [DOI] [PubMed] [Google Scholar]
- Fiehn O. 2006. Metabolite profiling in Arabidopsis. Methods in Molecular Biology 323, 439–447. [DOI] [PubMed] [Google Scholar]
- Finnemann J, Schjoerring JK. 2000. Post-translational regulation of cytosolic glutamine synthetase by reversible phosphorylation and 14-3-3 protein interaction. The Plant Journal 24, 171–181. [DOI] [PubMed] [Google Scholar]
- Fromm S, Senkler J, Eubel H, Peterhänsel C, Braun HP. 2016. Life without complex I: proteome analyses of an Arabidopsis mutant lacking the mitochondrial NADH dehydrogenase complex. Journal of Experimental Botany 67, 3079–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funck D, Eckard S, Müller G. 2010. Non-redundant functions of two proline dehydrogenase isoforms in Arabidopsis. BMC Plant Biology 10, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funck D, Stadelhofer B, Koch W. 2008. Ornithine-delta-aminotransferase is essential for arginine catabolism but not for proline biosynthesis. BMC Plant Biology 8, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gäde G, Auerswald L. 2002. Beetles’ choice—proline for energy output: control by AKHs. Comparative Biochemistry and Physiology Part B: Biochemistry & Molecular Biology 132, 117–129. [DOI] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Ströher E, Fenske R, Millar AH. 2013. Succinate dehydrogenase assembly factor 2 is needed for assembly and activity of mitochondrial complex II and for normal root elongation in Arabidopsis. The Plant Journal 73, 429–441. [DOI] [PubMed] [Google Scholar]
- Ihsan MZ, Ahmad SJ, Shah ZH, Rehman HM, Aslam Z, Ahuja I, Bones AM, Ahmad JN. 2017. Gene mining for proline based signaling proteins in cell wall of Arabidopsis thaliana. Frontiers in Plant Science 8, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech O, Pesquet E, Ahad A, Askne A, Nordvall D, Vodnala SM, Tuominen H, Hurry V, Dizengremel P, Gardeström P. 2007. The different fates of mitochondria and chloroplasts during dark-induced senescence in Arabidopsis leaves. Plant, Cell & Environment 30, 1523–1534. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. 1996. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. The Plant Cell 8, 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. 2012. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Research 40, D1211–D1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP. 2011. Defining the protein complex proteome of plant mitochondria. Plant Physiology 157, 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SR, Chrobok D, Juvany M, et al. . 2018. Darkened leaves use different metabolic strategies for senescence and survival. Plant Physiology 177, 132–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136. [DOI] [PubMed] [Google Scholar]
- Lin J-F, Wu S-H. 2004. Molecular events in senescing Arabidopsis leaves. The Plant Journal 39, 612–628. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. The Journal of Biological Chemistry 193, 265–275. [PubMed] [Google Scholar]
- Mani S, Van De Cotte B, Van Montagu M, Verbruggen N. 2002. Altered levels of proline dehydrogenase cause hypersensitivity to proline and its analogs in Arabidopsis. Plant Physiology 128, 73–83. [PMC free article] [PubMed] [Google Scholar]
- Masclaux C, Valadier MH, Brugière N, Morot-Gaudry JF, Hirel B. 2000. Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211, 510–518. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Carrayol E, Valadier MH. 2005. The two nitrogen mobilisation- and senescence-associated GS1 and GDH genes are controlled by C and N metabolites. Planta 221, 580–588. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. 2010. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Annals of Botany 105, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal WA, Dey BB, Choudhuri MA. 1985. Proline accumulation as a reliable indicator of monocarpic senescence in rice cultivars. Experientia 41, 346–348. [Google Scholar]
- Nakashima K, Satoh R, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. 1998. A gene encoding proline dehydrogenase is not only induced by proline and hypoosmolarity, but is also developmentally regulated in the reproductive organs of Arabidopsis. Plant Physiology 118, 1233–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Millar AH, O’Toole N, Small I, Whelan J. 2007. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. The Plant Cell 19, 3418–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandhare J, Donald SP, Cooper SK, Phang JM. 2009. Regulation and function of proline oxidase under nutrient stress. Journal of Cellular Biochemistry 107, 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parre E, Ghars MA, Leprince AS, Thiery L, Lefebvre D, Bordenave M, Richard L, Mazars C, Abdelly C, Savouré A. 2007. Calcium signaling via phospholipase C is essential for proline accumulation upon ionic but not nonionic hyperosmotic stresses in Arabidopsis. Plant Physiology 144, 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti L, Weiste C, Nägele T, et al. . 2018. Snf1-RELATED KINASE1-controlled C/S1-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. The Plant Cell 30, 495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires MV, Pereira Júnior AA, Medeiros DB, et al. . 2016. The influence of alternative pathways of respiration that utilize branched-chain amino acids following water shortage in Arabidopsis. Plant, Cell & Environment 39, 1304–1319. [DOI] [PubMed] [Google Scholar]
- Rao RS, Salvato F, Thal B, Eubel H, Thelen JJ, Møller IM. 2017. The proteome of higher plant mitochondria. Mitochondrion 33, 22–37. [DOI] [PubMed] [Google Scholar]
- Rizzi YS, Cecchini NM, Fabro G, Alvarez ME. 2017. Differential control and function of Arabidopsis ProDH1 and ProDH2 genes on infection with biotrophic and necrotrophic pathogens. Molecular Plant Pathology 18, 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh R, Fujita Y, Nakashima K, Shinozaki K, Yamaguchi-Shinozaki K. 2004. A novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant & Cell Physiology 45, 309–317. [DOI] [PubMed] [Google Scholar]
- Satoh R, Nakashima K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2002. ACTCAT, a novel cis-acting element for proline- and hypoosmolarity-responsive expression of the ProDH gene encoding proline dehydrogenase in Arabidopsis. Plant Physiology 130, 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertl P, Braun HP. 2014. Respiratory electron transfer pathways in plant mitochondria. Frontiers in Plant Science 5, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servet C, Ghelis T, Richard L, Zilberstein A, Savouré A. 2012. Proline dehydrogenase: a key enzyme in controlling cellular homeostasis. Frontiers in Bioscience 17, 607–620. [DOI] [PubMed] [Google Scholar]
- Sharma S, Villamor JG, Verslues PE. 2011. Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiology 157, 292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Savouré A. 2010. Proline: a multifunctional amino acid. Trends in Plant Science 15, 89–97. [DOI] [PubMed] [Google Scholar]
- Székely G, Abraham E, Cseplo A, et al. . 2008. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. The Plant Journal 53, 11–28. [DOI] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J. 2009. Defining the mitochondrial stress response in Arabidopsis thaliana. Molecular Plant 2, 1310–1324. [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge UI, Kunze R. 2006. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiology 141, 776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hua XJ, May M, Van Montagu M. 1996. Environmental and developmental signals modulate proline homeostasis: evidence for a negative transcriptional regulator. Proceedings of the National Academy of Sciences, USA 93, 8787–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Cheng SH, Kao CH. 1982. Senescence of rice leaves. VII. Proline accumulation in senescing excised leaves. Plant Physiology 69, 1348–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Alonso R, Harter K, Vicente-Carbajosa J, Dröge-Laser W. 2006. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. The EMBO Journal 25, 3133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Du X, Zhang F, Zhang F, Hu Y, Liu S, Jiang X, Wang G, Liu D. 2012. A mutation in the E2 subunit of the mitochondrial pyruvate dehydrogenase complex in Arabidopsis reduces plant organ size and enhances the accumulation of amino acids and intermediate products of the TCA cycle. Planta 236, 387–399. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang Y, Di C, et al. . 2016. JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. Journal of Experimental Botany 67, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.