The rbcS multigene family evolved through complex duplication events leading to species-specific gene copies. Selection and coevolution with rbcL constrained rbcS evolution thereby limiting the divergence of each gene copy.
Keywords: Coevolution, duplication, gene copies, homology modelling, molecular evolution, multigene family, photosynthesis, positive selection, rbcS, RuBisCO
Abstract
Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is considered to be the main enzyme determining the rate of photosynthesis. The small subunit of the protein, encoded by the rbcS gene, has been shown to influence the catalytic efficiency, CO2 specificity, assembly, activity, and stability of RuBisCO. However, the evolution of the rbcS gene remains poorly studied. We inferred the phylogenetic tree of the rbcS gene in angiosperms using the nucleotide sequences and found that it is composed of two lineages that may have existed before the divergence of land plants. Although almost all species sampled carry at least one copy of lineage 1, genes of lineage 2 were lost in most angiosperm species. We found the specific residues that have undergone positive selection during the evolution of the rbcS gene. We detected intensive coevolution between each rbcS gene copy and the rbcL gene encoding the large subunit of RuBisCO. We tested the role played by each rbcS gene copy on the stability of the RuBisCO protein through homology modelling. Our results showed that this evolutionary constraint could limit the level of divergence seen in the rbcS gene, which leads to the similarity among the rbcS gene copies of lineage 1 within species.
Introduction
Gene duplication is one of the main mechanisms creating novel features at the molecular level during evolution (Flagel and Wendel, 2009). The functional role played by duplicated genes has been discussed in detail (Hughes, 1994; Lynch and Force, 2000) and the mechanisms at work in this process are now relatively well understood (Hughes, 1994; Studer et al., 2008; Innan and Kondrashov, 2010; Roulin et al., 2012; Rensing, 2014). At the molecular level, it was initially proposed that relaxation of the selective constraints on one of the gene copies following gene duplication allows an accumulation of mutations that can permit the evolution of novel or sub-gene function or lead to a total loss of function (Ohta, 1988; Wagner, 1998; Moore and Purugganan, 2005). However, the advantages brought by gene duplication could not only stem from the effects of mutations but also from the protection against deleterious mutations or the mechanisms of dosage effect (Papp et al., 2003; Kafri et al., 2008; Cheeseman et al., 2016).
The creation of new gene copies by duplication is further affected by species divergence and the evolutionary history of the resulting gene family. Members of most gene families are therefore connected by a complex history of duplication and speciation events that have produced paralogous and orthologous gene copies. The identification of the proper sets of orthologous genes is challenging (Altenhoff et al., 2011). Correct identification of relationships of gene copies is further complicated by the presence of gene conversion that may alter the origin of similarities between homologous regions (Mansai and Innan, 2010; Song et al., 2012). The members of a multigene family can further be modified by crossing over and/or recombination (Ohta, 1977, 1979, 1983; Nei and Rooney, 2005; Mano and Innan, 2008; Dumont and Eichler, 2013). Each gene copy might further differ not only by the evolutionary process but also by the function, cellular localization of encoded protein, stability, and/or expression levels (Hudsona et al., 1992; Ku et al., 1996; Clark et al., 2001; Petter et al., 2008; Niimura, 2009). The different gene copies can, therefore, play a core role in organizing the novel or modified functions that are often required during adaptive evolution (Ohta, 1991; Nei et al., 1997; Niimura, 2009; McGlothlin et al., 2016).
An example of this adaptive evolution is the evolution of photosynthesis. Atmospheric CO2 drastically decreased in the Oligocene (Pearson et al., 2009; Edwards et al., 2010; Beerling and Royer, 2011) and some plant species adapted to the depleted CO2 concentration by evolving a mechanism, called C4 photosynthesis, to concentrate CO2 by modifying the biochemical cascade and the cellular structures (Sage, 2004). C4 plants have diverged from C3 plants through the acquisition of novel enzymes. Most of the C4-specific enzymes are encoded by multigene families and the co-option of genes pre-existing in the ancestral C3 plants plays an important role during the transition from C3 to C4 type (Monson, 2003; Christin et al., 2013; Bianconi et al., 2018). The first enzyme of the Calvin–Benson cycle and the one that fixes CO2 into sugar is ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (Hatch and Slack, 1968; Kanai and Edwards, 1999). RuBisCO has slower catalytic efficiency than other photosynthetic enzymes because of its affinities to both O2 and CO2 (Rawsthorne, 1992). The fixation of O2 results in a loss of energy and CO2, so-called photorespiration (Kubien et al., 2008; Peterhansel et al., 2010). In C4 plants, the CO2-concentrating mechanism (CCM) enabled RuBisCO to be surrounded by highly concentrated CO2, which led to the fixation of substitutions along the protein sequence that increased the catalytic efficiency of RuBisCo and decreased the affinity of CO2, compared with C3 plants (Badger and Andrews, 1987; von Caemmerer and Quick, 2000; Sage and Coleman, 2001).
Evidence for the adaptive evolution of RuBisCO has come from the study of the evolution of the chloroplast rbcL gene encoding RBCL, the large subunit of RuBisCO. Positive selection for rbcL has been detected in independent C4 lineages (Kapralov and Filatov, 2007; Christin et al., 2008; Piot et al., 2018). In Flaveria, the signal of positive selection of the rbcL gene is almost 20 times stronger than that detected for rbcS, the gene encoding the small subunit (Kapralov et al., 2011). The RBCL subunit is considered to determine the catalytic properties of RuBisCO, because it contains the catalytic site of the enzyme (Andersson, 2008). However, RBCS has been reported to have an influence on the catalytic efficiency, CO2 specificity, activity, quantity, assembly, and stability of RuBisCO (Andrews and Ballment, 1983; Furbank et al., 2000; Spreitzer, 2003; Genkov and Spreitzer, 2009; Genkov et al., 2010; Bracher et al., 2011). Studer et al. (2014) have suggested that some positively selected codons encoding amino acid residues that are located at the interface between RBCL and RBCS may affect the stability and the catalytic properties of RuBisCO. All these studies suggest that the interaction between RBCS and RBCL, and the rbcS gene itself, may play important roles in the evolution of RuBisCO.
A better understanding of the evolutionary history of rbcS is thus essential to obtain a deeper insight into the evolution of RuBisCO. We extracted the nucleotide sequences of the rbcS gene from available full genomes of angiosperms and reconstructed the phylogenetic relationships of the rbcS gene copies. We then tested for the presence of positive selection acting on the rbcS gene across the evolution of angiosperms. Positive selection of rbcS has already been tested within some genera but has never been measured on a wider range of plants. Therefore, we aimed to elucidate the differences between gene copies of rbcS in higher plants and to infer their respective evolutionary histories. Firstly, we hypothesized that each rbcS copy may have a different interaction with rbcL, and we tested this hypothesis by inferring the coevolution between rbcS and rbcL. Secondly, we hypothesized that RBCS encoded by different rbcS gene copies may have a different degree of influence on the stability of RuBisCO. We tested this by modelling a RuBisCO structure with eight RBCS units encoded by a unique rbcS copy. We did the same for each rbcS copy and compared the stability between models. Our study provides new insights into the evolutionary mechanism of the rbcS multigene family and sheds light on its influence on RuBisCO evolution.
Materials and methods
Phylogenetic tree of rbcS among angiosperms
We downloaded the annotated rbcS gene of all angiosperms available in Phytozome v12 (https://phytozome.jgi.doe.gov/pz/portal.html). We aligned the sequences obtained using MAFFT (Katoh and Standley, 2013) and removed unreliable sequences that were poorly aligned using GUIDANCE2 with default settings (http://guidance.tau.ac.il/ver2/;Sela et al., 2015). We then converted these amino acid alignments back into codon alignment using PAL2NAL (http://www.bork.embl.de/pal2nal/#RunP2N) to obtain the final nucleotide alignment of 171 rbcS gene copies for 43 angiosperm species. The TN93 model of substitution was identified as the best model using Jmodeltest v2.1.4 (Darriba et al., 2012). We reconstructed the phylogenetic tree with PhyML v3.0 (Guindon and Gascuel, 2003) using the BEST algorithm for tree rearrangement while estimating all parameters of the TN93 model and the branch lengths. Branch support values were estimated based on 1,000 bootstrap replicates.
Gene conversion
We tested for the signatures of recombination and gene conversion in the rbcS gene copies using the Recombination Detection Program v4.56 software (RDP4; Martin et al., 2015). We used Chimaera, 3seq, GENECONV, MaxChi, and SiScan with their default parameters. The nucleotide alignment created for the phylogenetic reconstruction was used as an input for the gene conversion analyses.
Selection
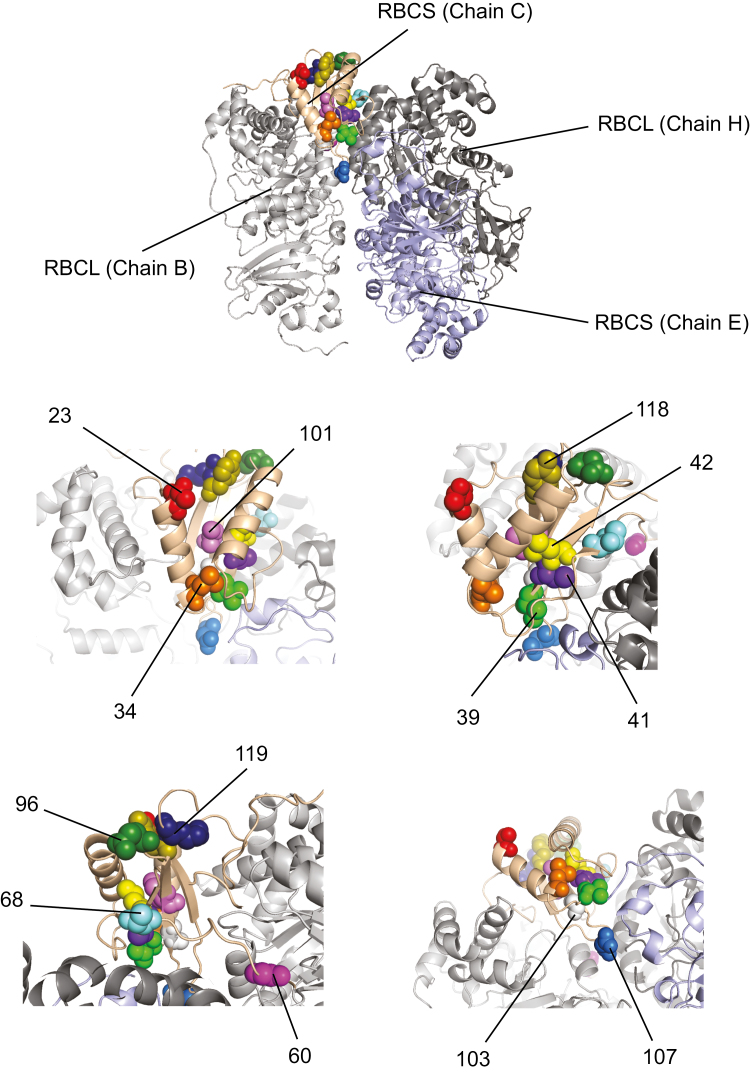
Positive selection analysis in rbcS was performed using the mixed effects model of evolution (MEME) implemented in HyPhy v2.2.6 (Pond et al., 2005). We used the MG94 codons substitution base model (Muse and Gaut, 1994) and we corrected for multiple testing using a false discovery rate (FDR; Benjamini and Hochberg, 1995) with a threshold of 0.1. We selected the MEME model because it is more suitable than the branch-site mode (Zhang et al., 2005) for estimating site-specific probabilities (Lu and Guindon, 2014). Positions under positive selection were plotted on the known protein structure of Spinacia oleracea (Chains B, C, E and H of 1RCX of the Protein Data Bank; Taylor and Andersson, 1997) using the software PyMol v1.3 (The PyMOL Molecular Graphics System; Schrödinger, LLC).
Coevolution between rbcS and rbcL
We downloaded the complete genome of the rbcL sequences from NCBI (NCBI Resource Coordinators, 2018) and used the filtered alignment from Guidance (see above) to ensure that the pattern of substitutions is not simply due to alignment errors but does indeed represent genuine evolutionary signals. We retained only the 30 species for which both rbcS and rbcL sequences were available. The resulting alignments were 721 and 1,461 bp long for rbcS and rbcL, respectively.
Coevolution analysis of rbcS and rbcL was performed using the maximum likelihood implementation of model Coev (Dib et al., 2014, 2015). For each pair of sites, we compared the likelihood of a dependent and an independent model of substitution using the Akaike information criterion (AIC). The difference in AIC (dAIC) between the two models varies depending on the tree structure and characteristics of the alignment. To estimate the expected distribution of dAIC under no coevolution given the rbcS gene tree, we obtained a null distribution of dAIC by simulating sequences of 2,000 bp on the rbcS tree under the independent substitution model. We estimated the support for the Coev model for the ca. 4 million pairs of sites based on this simulated dataset and determined the dAIC value representing the 95% percentile of this distribution (for details, see Dib et al., 2014). We then combined the dAIC values with an s/d ratio (ratio between the parameters s, representing the rate of change away from the coevolution profile, and d, representing the rate of change towards the coevolution profile) lower than 0.1 to identify sites under coevolution (see Dib et al., 2014 for details). Since rbcL has a single gene copy per species and rbcS shows a variable copy number between species, we duplicated the rbcL sequences for each species to match the rbcS copy number. The rbcS and rbcL alignments were concatenated into a single matrix and conserved positions with a percentage of identity higher than 95% were removed because they do not provide enough information to estimate coevolution (Dib et al., 2014, 2015). The final concatenated alignment of rbcL–rbcS contained 590 nucleotide positions (345 bp of rbcL and 245 bp of rbcS), which led to a total of 84,525 tests of coevolution for pairs of sites. In every pair, one of the sites belonged to rbcL, while the other belonged to rbcS. There were 7,828 pairs of sites that passed the dAIC and s/d ratio thresholds defined above and that were considered as coevolving (Supplementary Fig. S1 at JXB online). Among those, the ones with the strongest signals (dAIC more than 35 and s/d ratio less than 0.1; Supplementary Fig. S1) were selected and the R package qgraph (Epskamp et al., 2012) was used to visualize them. We also plotted the pairs of sites selected on the known protein structure of S. oleracea (Supplementary Fig. S2; Chain B, C, E and H of 1RCX of Protein Data Bank: Taylor and Andersson, 1997) using PyMol v1.3 (Schrödinger, 2015).
Protein stability of RuBisCO structure
The RuBisCO quaternary structure is a hexadecamer composed of eight subunits of RBCL and eight subunits of RBCS. Since RBCL is encoded by a single gene, the eight RBCL subunits are always the same for a given species. On the other hand, the exact combination of the eight RBCS subunits is unknown. We assumed here that, for a given RuBisCO protein, the eight RBCS subunits are encoded by the same copy of rbcS. This assumption made the modelling of protein stability feasible by limiting the number of combinations and allowed us to study differences between gene copies.
We performed homology modelling and estimated the Gibbs free energy, estimated as the difference of thermodynamic stability between the folded and unfolded states of a protein, to compare the stability of the whole RuBisCO structures. When the Gibbs free energy is below 0, the folded state is preferred over the unfolded state and protein models with a smaller value of Gibbs free energy can be considered to be more stable. To model the RuBisCO stability in angiosperms, the RBCS and RBCL amino acid sequences of several species of Brassicaceae and Poaceae were downloaded from UniProt (UniProt Consortium, 2015). We selected these two clades because they are well defined in the rbcS phylogenetic tree and are representative of the evolution of rbcS (see Results). To create RBCS encoded by a single gene copy, we duplicated eight times the rbcS sequence in each pair protein structure file. However, when different gene copies of the same species differed only by synonymous substitutions or when amino acids differed in a region outside the crystallized structure, only one complex was tested for these gene copies since amino acid sequences were identical (e.g. Setaria italica copies 4 and 5). Homology modelling was performed using Modeller v9.17 (Eswar et al., 2008). The RuBisCO structure of Oryza sativa (1WDD of Protein Data Bank; Matsumura et al., 2012) was used as a template. The homology modelling was run 100 times for each structural complex of rbcL–rbcS and the best model (the one with the lowest DOPE score) was selected for further analyses. These models were then repaired with FoldX v4.0 (Schymkowitz et al., 2005) using the RepairPDB function. The repair step is mandatory for removing potential bad contacts (i.e. Van der Waals clashes) in the structures, which may cause instability of modelled protein. Also using FoldX v4.0, we predicted the differences of Gibbs free energy between maximum likelihood model and null model (ΔG) of each estimated structure using the ‘Stability’ function, with default parameters. Three-dimensional structures were visualized with PyMol v1.3 (Schrödinger, 2015). Estimated ΔG values for the Brassicaceae and Poaceae were visualized on their respective rbcS gene trees using the function phenogram of the R package phytools (Revell, 2012).
Results
Phylogenetic tree of rbcS among angiosperms
We identified two rbcS lineages (rbcS lineages 1 and 2) that could represent a deep duplication event that occurred before the divergence of eudicots and monocots. One gene lineage includes genes that cluster together with a known expressed gene in photosynthetic organs of rice (OsRbcS2; Morita et al., 2014); we refer to this gene lineage as rbcS lineage 1 (Fig. 1). The second gene lineage includes gene copies expressed in non-photosynthetic organs such as OsRbcS1 in rice (Morita et al., 2014); we refer to this lineage as rbcS lineage 2 (Fig. 2).
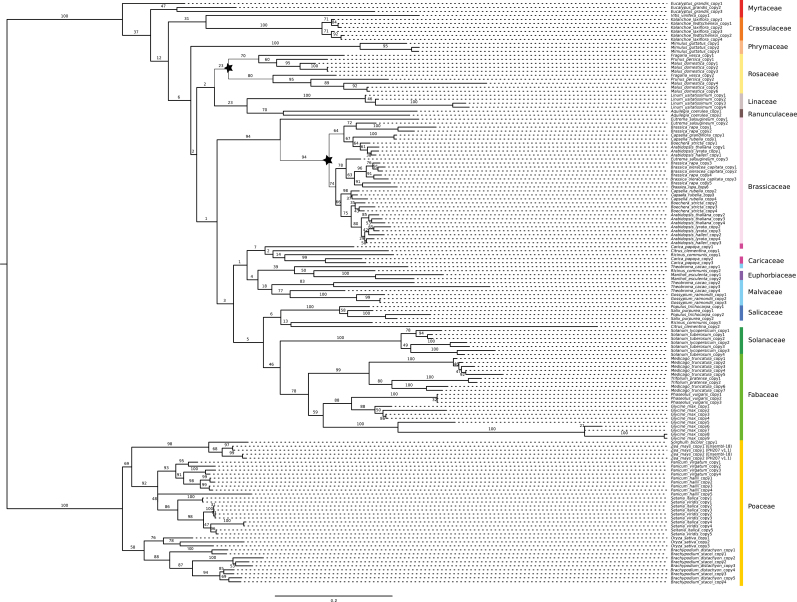
Fig. 1.
Maximum likelihood tree of rbcS lineage 1 in angiosperms. The phylogenetic tree was reconstructed in PhyML v3.0 (Guindon and Gascuel, 2003) using a TN93 model. Each gene copy of a given species is identified by the species name and distinguished by a number (see Supplementary Table S1 for correspondence of gene copy name used in this figure and gene ID in Phytozome database). Names of plant families are indicated on the right next to the species names except for Citrus clementina and Ricinus communis. Because the gene copies of these two species were spread all over the tree and did not cluster within a family, their family names are not indicated in this tree. Branch support was estimated using 1000 bootstraps replicates. Values above or below the branches represent the bootstrap support for each branch (%). The scale bar is shown below the phylogeny. Stars indicate the duplication events within a family.
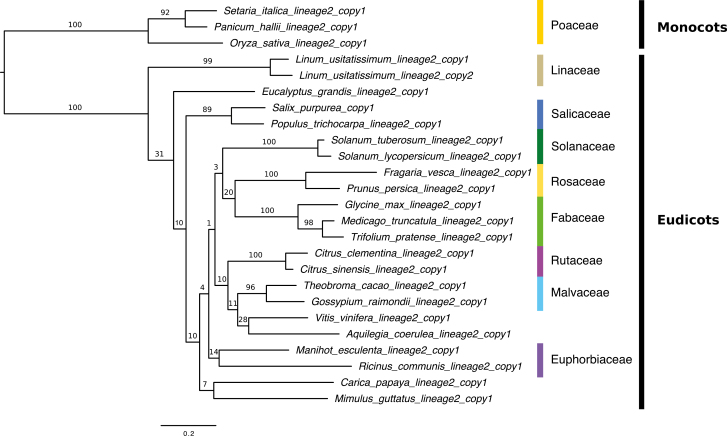
Fig. 2.
Maximum likelihood of rbcS lineage 2 in angiosperms. The phylogenetic tree of the rbcS gene copies in angiosperms was reconstructed in PhyML v3.0 (Guindon and Gascuel, 2003) using a TN93 model. Names of plant families are indicated on the right next to the species names when the gene copies of the same family cluster together. Branch support was estimated using 1000 bootstrap replicates. Values above the branches represent the bootstrap support for each branch (%). The scale bar is shown below the phylogeny.
The phylogenetic tree of rbcS lineage 1, including 146 sequences available for 42 species, is shown in Fig. 1 (see Supplementary Table S1 for correspondence of gene copy name in Fig. 1 and gene ID in the Phytozome v12 database). Each plant family is well defined with subtending branches well supported (bootstrap support >78%; Fig. 1), except for the three families Caricaceae, Malvaceae, and Rosaceae. The relationships obtained within each family or subfamily are further well supported. Globally, the topology of the gene tree follows the expected species tree of angiosperms (e.g. clear division between monocots and eudicots; see Magallón et al., 2015) but the relationships between several plants families in eudicots (i.e. Linaceae, Malvaceae, Phrymaceae, Rosaceae, Salicaceae, and Solanaceae) were not supported by high bootstrap values (Fig. 1). The low support obtained could be due to short branch lengths and the peculiar evolutionary history of the rbcS gene (see below). The rbcS gene tree estimated by PhyML shows a particular topology with the gene copies of the same species clustering together with high bootstrap support (Fig. 1). The phylogenetic analyses showed also deeper duplication events in several plant families (e.g. Brassicaceae and Rosaceae; shown by stars in Fig. 1) and there are a few exceptions, such as Citrus clementina and Ricinus communis, which have gene copies widely spread across the tree. The branch lengths leading to the Vitis vinifera_copy1, Malus domestica_copy4, and Citrus clementina_copy2 of rbcS lineage 1 are longer (Fig. 1), suggesting the accumulation of more substitutions in these specific gene copies. We observed a deletion of 192 bp in the sequence of Vitis vinifera_copy1, an insertion of 57 bp and a deletion of 72 bp in Malus domestica_copy4, and deletion of 285 bp in Citrus clementina_copy2. These large insertions/deletions could be signs of the loss (pseudogenization) or change of function of these gene copies, but a deeper investigation of these sequences should be carried out to fully understand these patterns.
The phylogenetic tree of rbcS lineage 2, including 25 sequences available for 24 species, is shown in Fig. 2. As observed in the tree of rbcS lineage 1, there is a clear division between monocots and eudicots. Although the number of gene copies is limited, we observed the rbcS gene copies of each family (Euphorbiaceae, Fabaceae, Linaceae, Malvaceae, Rosaceae, Rutaceae, Salicaceae, and Solanaceae) cluster together.
In the end, we excluded gene copies of rbcS lineage 2 from further analyses because (i) the copies of lineage 2 are more divergent than those of lineage 1, and the sequences of lineage 2 cannot be aligned reliably, and thus cannot be tested for positive selection and coevolution, and (ii) our focus was on the molecular evolution of the gene copies involved in photosynthesis.
Minimum numbers of gene copies per species are shown in Supplementary Table S2. We show this table to demonstrate that the estimated number of rbcS gene copies varies between species, but given uncertainty in the quality of genome assemblies, it indicates a minimum number of rbcS gene copies per species. It is also possible that there are additional gene copies that were not available in the current version of genome assemblies.
Gene conversion and positive selection
We did not detect any significant signal of gene conversion (P>0.05). We tested rbcS sequences for signs of positive selection using the MEME model of HyPhy (Pond et al., 2005). A strong signal of positive selection was detected in 13 sites (Table 1; Fig. 3). The episodes of positive selection were not associated with specific branches or duplication events.
Table 1.
Codon position of the aminoacid sequences of RBCS under positive selection
| Amino acid residues in RuBisCO structure of Spinacia oleracea (1RCX of Protein Data Bank) | p-value | q-value |
|---|---|---|
| 23 | 1.46E-03 | 2.66E-02 |
| 34 | 1.31E-04 | 5.39E-03 |
| 39 | 3.44E-04 | 8.88E-03 |
| 41 | 5.60E-03 | 8.27E-02 |
| 42 | 1.84E-04 | 6.32E-03 |
| 60 | 4.67E-03 | 7.62E-02 |
| 68 | 2.59E-03 | 4.46E-02 |
| 96 | 6.93E-03 | 9.34E-02 |
| 101 | 1.07E-05 | 1.10E-03 |
| 103 | 9.33E-06 | 1.10E-03 |
| 107 | 7.87E-03 | 9.87E-02 |
| 118 | 4.89E-04 | 1.01E-02 |
| 119 | 7.96E-03 | 9.87E-02 |
Fig. 3.
RBCS residues under positive selection. Thirteen positions of rbcS showed strong signals of positive selection (see also Table 1). We plotted corresponding amino acid residues to RuBisCO structure of Spinacia oleracea (1RCX of Protein Data Bank; Taylor and Andersson, 1997). The light pink cartoon ribbons indicate RBCS chain C of 1RCX. The light grey, light blue, and dark grey cartoon ribbons indicate RBCL chains B, E, and H of 1RCX, respectively. The positions of RBCS under positive selection are shown as spheres in different colours. The upper panel shows the overview of the positions under positive selection. The other four panels show the zoom view of each sphere.
Coevolution between rbcS and rbcL
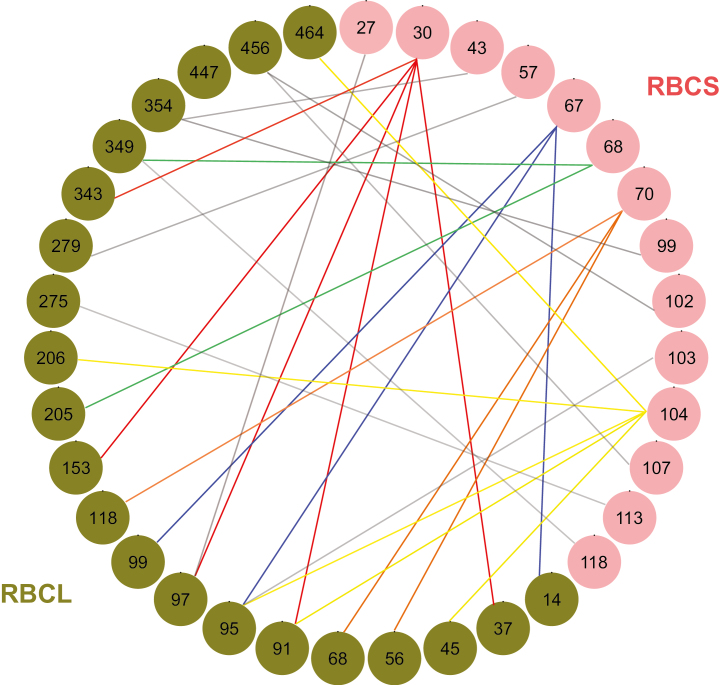
We tested a total of 84,525 pairs of sites to detect coevolution between rbcS and rbcL. Signs of coevolution were, as expected, pervasive between these two genes and 26,338 pairs had a dAIC value between the null and alternative model higher than the threshold of 9.893, which represented the 95% percentile of the distribution of dAIC obtained by simulating the evolution of the independent model along the rbcS gene tree (Supplementary Fig. S1A). Among these 26,338 pairs, we further looked at the strength of the signal by considering the ratio of the parameters s and d, which indicates a strong signal if its value is close to zero (Dib et al., 2014, 2015). The distribution of s/d ratios is shown in Supplementary Fig. S1B and we identified 7,828 profiles with an s/d ratio less than 0.1. The 28 pairs with the strongest signal of coevolution (dAIC more than 35 and s/d ratio less than 0.1) are listed in Table 2. Among these 28 pairs, we found five positions along the rbcS sequence, which encode residues 30, 67, 68, 70, and 104 of RBCS of 1RCX (Table 2; Fig. 4), that were each coevolving with multiple positions of rbcL encoding RBCL. Similarly, six positions of rbcL encoding residues 91, 95, 97, 349, 354, and 456 of RBCL of 1RCX (Table 2; Fig. 4) were found to be coevolving with multiple positions of rbcS encoding RBCS.
Table 2.
Coevolving sites between rbcS and rbcL and corresponding amino acid residues plotted to known RuBisCO structure of spinach
| Nucleotide position of rbcS in our alignment | Corresponding amino acid residue of RBCS of 1RCX | Nucleotide position of rbcL in our alignment | Corresponding amino acid residue of RBCL of 1RCX | s/d ratio | dAIC |
|---|---|---|---|---|---|
| 183 | — | 1362 | 447 | 9.11E-02 | 44.5468 |
| 320 | 27 | 311 | 97 | 5.49E-02 | 36.26688 |
| 328 | 30 | 132 | 37 | 6.30E-02 | 35.73412 |
| 328 | 30 | 292 | 91 | 4.40E-02 | 37.72452 |
| 328 | 30 | 480 | 153 | 6.36E-02 | 35.06962 |
| 328 | 30 | 1050 | 343 | 5.01E-02 | 35.14996 |
| 369 | 43 | 1081 | 354 | 2.49E-02 | 41.5562 |
| 409 | 57 | 857 | 279 | 6.00E-10 | 36.22426 |
| 441 | 67 | 61 | 14 | 5.42E-10 | 38.19676 |
| 441 | 67 | 304 | 95 | 4.47E-02 | 49.84318 |
| 441 | 67 | 316 | 99 | 8.87E-02 | 35.08008 |
| 441 | 67 | 317 | 99 | 8.87E-02 | 35.08008 |
| 444 | 68 | 636 | 205 | 8.63E-02 | 35.08624 |
| 444 | 68 | 1068 | 349 | 7.73E-02 | 35.55796 |
| 500 | 70 | 189 | 56 | 5.31E-02 | 35.22768 |
| 500 | 70 | 225 | 68 | 5.79E-10 | 42.60568 |
| 500 | 70 | 375 | 118 | 5.93E-10 | 39.87838 |
| 589 | 99 | 1081 | 354 | 5.21E-10 | 38.31622 |
| 596 | 102 | 1389 | 456 | 1.95E-02 | 38.76078 |
| 601 | 103 | 304 | 95 | 8.21E-02 | 40.1506 |
| 602 | 104 | 292 | 91 | 3.47E-02 | 35.81118 |
| 604 | 104 | 156 | 45 | 4.91E-02 | 38.94026 |
| 604 | 104 | 304 | 95 | 7.99E-02 | 38.06524 |
| 604 | 104 | 639 | 206 | 7.46E-02 | 40.35832 |
| 604 | 104 | 1412 | 464 | 4.02E-02 | 36.44858 |
| 611 | 107 | 1389 | 456 | 3.18E-10 | 38.82068 |
| 634 | 113 | 846 | 275 | 3.24E-02 | 35.55106 |
| 652 | 118 | 1068 | 349 | 4.16E-02 | 35.65656 |
Fig. 4.
Coevolving residues between RBCS and RBCL. Coevolution between paired combinations of rbcS sites and rbcL sites was estimated by a maximum-likelihood implementation of Coev and dependent model using nucleotide sequences. The differences in AIC between pairs of models (dAIC) were calculated. AIC of the null model (9.893) was used as a threshold. We then filtered further with the s/d ratio threshold (0.1) according to previous studies (Dib et al., 2014, 2015). The 28 coevolving profiles with the strongest signals (dAIC >35 and s/d ratio <0.1) were selected (see Table 2). We identified the corresponding amino acid residues and plotted these sites using the qgraph function of R (Epskamp et al., 2012). The residues of RBCS and RBCL are shown by pink and green filled circles, respectively and the numbers in the circles indicate the residues of known RuBIsCO structure (1RCX of Protein Data Bank; Taylor and Andersson, 1997). A coevolving profile pair is connected with a line. The coevolution profiles including the same RBCS residues are shown as lines of the same colour connected with multiple RBCL residues (e.g. lines in red, blue, yellow, orange, and green). Grey lines show the other coevolving profiles including the residues of RBCS that coevolve with a single RBCL among listed 28 profiles.
Among the 28 pairs of coevolving sites, one pair occurred in the region encoding the transit-peptide (nucleotide position 183 of our alignment; Table 2). In RBCS, the 11 residues (residues 27, 30, 43, 68, 70, 99, 102, 103, 104, 113, and 118) among the 14 residues with the strongest signal of coevolution (dAIC>35; Table 2) form the elements of the secondary structure (see Knight et al., 1990). Among those residues, the three residues 33, 99, and 115 are in the core of the small subunit. In RBCL, the 12 residues among 21 with the strongest signal of coevolution listed in Table 2 also form the elements of the secondary structure (residues 447 and 456 in the C-terminal domain; 37, 56, 97, 99, 118, 275, 279, 343, 349, and 354 in the N-terminal domain; see Knight et al., 1990) and residues 45, 118, 205, 275, and 279 of RBCL are at the interface between two RBCLs (see the bold residues in Supplementary Fig. S2; see Knight et al., 1990). Furthermore, residues 107 and 118 of RBCS (reference sequence 1RCX) were detected to be evolving under positive selection and are coevolving with RBCL.
Protein stability of RuBisCO structure
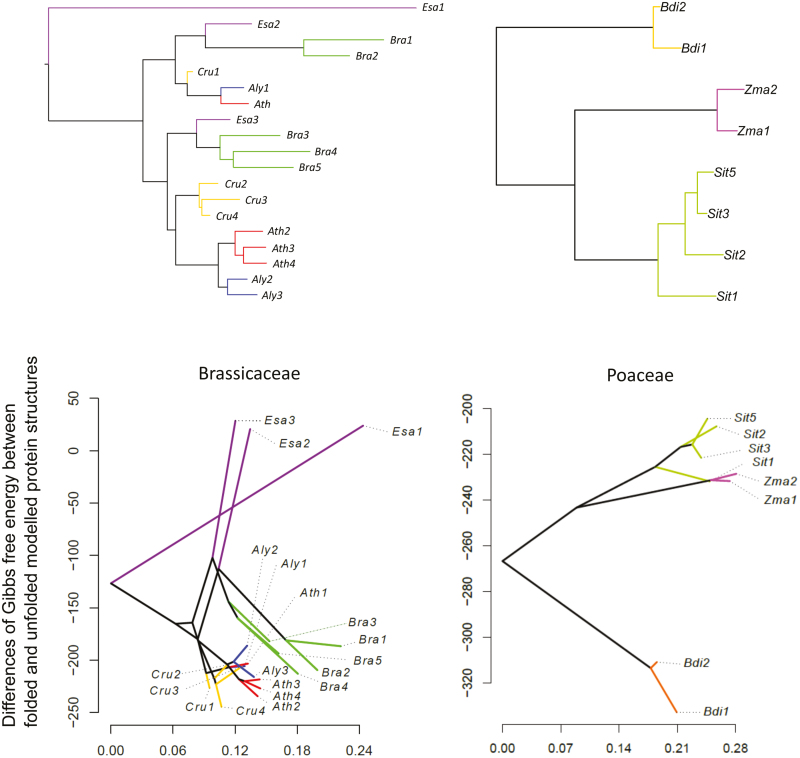
Our phylogenetic analyses indicated that at least two plant families (Rosaceae and Brassicaceae; Fig. 1) had old duplication events during their evolutionary history. In contrast, the Poaceae family did not show any signs of old duplication event within the family (Fig. 1). The large sequence divergence between gene copies in Brassicaceae could lead to a variable stability of the heterodimers formed with the single RBCL protein when different rbcS gene copies are involved. We therefore compared the characteristics of each gene copy from both the Brassicaceae and Poaceae by estimating the Gibbs free energy of the RuBisCO structure.
In Poaceae, the Gibbs free energy values estimated were similar for gene copies of the same species (Fig. 5; Table 3). There was also a clear distinction between the values for the Pooideae, represented by Brachypodium distachyon, and for representatives of the PACMAD clade (Zea mays and Setaria italica). Oryza sativa was not included in our analysis because (i) the translated amino acid sequences of LOC_Os12g17600 and LOC_Os12g19470 are identical, and (ii) the translated amino acid sequence of the other copy, LOC_Os12g19381, had the insertion of one nucleotide that causes a frame shift. In Brassicaceae, we expected differences of Gibbs free energy values between gene copies because their duplication is relatively old, having taken place during the early steps of diversification of the family. However, the estimated Gibbs free energy values showed a clear clustering by species (Fig. 5; Table 3). This shows that stabilities for the RuBisCO complex within the species are consistent, despite different evolutionary histories of gene copies within the same species.
Fig. 5.
Stability of modelled RuBisCO structure. The phylogenetic trees of rbcS in Brassicaceae and Poaceae are shown in the upper panels. RuBisCO protein structures with RBCS encoded by each rbcS were estimated by homology modeling using Modeller (Eswar et al., 2008). The structure was repaired by the RepairPDB function of FoldX v4 (Schymkowitz et al., 2005). The stability of the whole RuBisCO was estimated using the ‘Stability’ function of FoldX4. Then, the result of protein stability was taken as a trait and phylogenetic relationships were given as input trees. We then drew a phenogram using the phytools package (Revell, 2012) in R (lower panels). Sit5 is shown as representative of Sit4/Sit5 because of synonymous substitutions.
Table 3.
Delta Gibbs free energy of modelled RuBisCO structure
| Species names | Name of each gene copy | Differences of Gibbs free energy between maximum likelihood model and null model | |
|---|---|---|---|
| Brassicaceae | Arabidopsis lyrata | Aly1 | −205.782 |
| Aly2 | −186.133 | ||
| Aly3 | −216.206 | ||
| Arabidopsis thaliana | Ath1 | −203.385 | |
| Ath2 | −234.677 | ||
| Ath3 | −218.445 | ||
| Ath4 | −227.182 | ||
| Brassica rapa | Bra1 | −186.759 | |
| Bra2 | −209.734 | ||
| Bra3 | −182.142 | ||
| Bra4 | −213.151 | ||
| Bra5 | −193.802 | ||
| Capsella rubella | Cru1 | −226.842 | |
| Cru2 | −204.699 | ||
| Cru3 | −206.047 | ||
| Cru4 | −244.759 | ||
| Eutrema solsugineum | Esa1 | 23.9451 | |
| Esa2 | 20.741 | ||
| Esa3 | 28.6711 | ||
| Poaceae | Brachypodium distachyon | Bdi1 | −332.978 |
| Bdi2 | −310.927 | ||
| Setaria italica | Sit1 | −231.832 | |
| Sit2 | −207.691 | ||
| Sit3 | −221.541 | ||
| Sit4/Sit5 | −204.342 | ||
| Zea mays | Zma1 | −231.694 | |
| Zma2 | −228.502 |
Discussion
In this study, we investigated the evolution of the small subunit of the RuBisCO protein in 43 species of angiosperms. We characterized the differences between each rbcS gene copy by testing coevolution between rbcS and rbcL and the influence of each copy on the stability of the enzyme.
We reconstructed the phylogenetic relationships of the rbcS gene copies, and this showed a pattern in which gene copies of the same species were more closely related to each other than those of different species. We did not detect a significant signal of gene conversion but found extensive coevolution between the RBCS and RBCL subunits. The presence of coevolution between these two genes that encode tightly linked proteins was expected. Besides that, we detected that the same nucleotide positions of each rbcS copy coevolve with the exact same positions of rbcL. This suggests that the coevolution between rbcS and rbcL did not involve specific rbcS gene copies, but represented rather a pervasive process throughout the evolution of these genes. We finally identified several sites that are evolving under positive selection in rbcS and showed through homology modelling that the incorporation of any of the rbcS sequence for a given species does not affect significantly the stability of the RuBisCO protein.
Number of rbcS gene copies per species
The number of rbcS gene copies per species used in this study is shown in Supplementary Table S2. The number of rbcS gene copies has been estimated with different methods in several studies (Galili et al., 1991; Ogihara et al., 1994; Sasanuma, 2001; Thomas-Hall et al., 2007; Kapralov et al., 2011; Miller, 2014). For example, the number of rbcS copies in wheat has been estimated to range from 21 gene copies (Southern hybridization analysis; Galili et al., 1991) to 100 gene copies (slot-blot analysis; Ogihara et al., 1994). Absolute detection of the number of gene copies is remarkably complex (Cantsilieris et al., 2013). The estimation of the gene copy number variation became more reliable thanks to the released genomic data. However, the reported number of gene copies changes when using newly released genomes with an improved method of assembly. Also, we should note that the precision of our method is limited by the genome assembly quality, and could be substantially degraded for various reasons, e.g. tandem copies of the gene are missing from the assembly (Panchy et al., 2016).
Phylogenetic reconstruction of the rbcS gene family
The topology of the rbcS gene tree within each angiosperm family mostly follows the topology of the expected species tree. In most species of angiosperms, gene copies of the same species were more closely related than those of different species. This pattern has already been reported within some species of the same genus such as Solanum and Flaveria (Pichersky and Cashmore, 1986; Kapralov et al., 2011). Our analysis is, however, the first to show that this pattern is not restricted to specific genera and is present across all angiosperms using nucleotide sequences. We also found family-specific duplication events in Brassicaceae and Rosaceae (Fig. 1).
In general, the evolution of multigene families is affected by a number of processes that involve either divergent, concerted, or birth-and-death evolution (Nei and Rooney, 2005). Nei and Rooney (2005) defined divergent evolution as a mechanism by which gene copies of the common ancestral species are retained after speciation in descendant species while diverging through the accumulation of substitutions. However, we observed copy number variation between species and also non-expressed copies of the rbcS gene, which makes divergent evolution unlikely to be the main process behind the rbcS evolution.
Gene copies of the same species were more similar than gene copies of different species (Fig. 1). Such similarity between gene copies within species is often the result of frequent gene conversions between gene copies during concerted evolution. Sugita and colleagues have suggested that the high similarity of the rbcS copies of Solanum lycopersicum is likely to be explained by gene conversion (Sugita et al., 1987). We tested for gene conversion using RDP4 (Martin et al., 2015) and CHAP2 (Song et al., 2012). However, we could not detect any significant signal of gene conversion across angiosperms. This result is congruent with the results of Miller (2014) who did not find clear evidence of gene conversion between rbcS gene copies of Solanaceae species. Additionally, we observed that the gene copies of the same species are separated by long branches, such as those found in Linum usitatissimum or Mimulus guttatus (Fig. 1). These genes are unlikely to be affected by concerted evolution because in such cases gene copies would be less genetically distant due to frequent gene conversions and crossing-over.
Our results suggest, therefore, that rbcS evolved following a birth-and-death process (Nei and Rooney, 2005). The observed pattern of the rbcS tree may have occurred by frequent recent duplications followed by pseudogenization and/or gene loss.
Retention rate of duplicates and two lineages of rbcS
The topology of the rbcS tree suggests that gene copies that may have originated from the ancient duplication events have been lost (except the event that led to the emergence of rbcS lineages 1 and 2, and ones before the divergence of Brassicaceae and Rosaceae), while gene copies that may have originated from recent events have been retained. The two lineages of rbcS have already been identified using amino acid sequences of angiosperms, gymnosperms, pteridophytes, and bryophytes (Pottier et al., 2018). Our phylogenetic tree based on the nucleotide sequences showed that the two rbcS lineages (rbcS lineage 1 and rbcS lineage 2) have originated from a duplication event before the divergence of monocots and eudicots. rbcS lineage 1 (shown in Fig. 1) includes gene copies that are expressed in photosynthetic organs (Cheng et al., 1998; Yoon et al., 2001). rbcS lineage 2 (shown in Fig. 2) includes gene copies that are expressed in non-photosynthetic organs such as OsRbcS1 (Morita et al., 2016). All sampled species of angiosperms carry gene copies of rbcS lineage 1 except Citrus sinensis, but only a few carry copies of rbcS lineage 2 (see Supplementary Table S2). The gene copies of lineage 1, given their ubiquitous presence in almost all the species sampled and their expressions associated with photosynthetic organs, are probably the functionally important gene copies involved in photosynthesis. However, the rbcS gene copies of lineage 2 have been kept in a few species of angiosperms since the divergence of monocots and eudicots. Also, higher numbers of rbcS copies of lineage 2 were found in seedless plants (bryophytes and pteridophytes) than in angiosperms, and they were shown to be more similar to the copies of lineage 2 than to those of lineage 1 in angiosperms (Pottier et al., 2018). Although the exact function of the rbcS copies from lineage 2 is still unclear, a study found that the incorporation of RBCS encoded by the OsRbcS1 gene, which belongs to lineage 2 of rice, into RuBisCO increased the catalytic turnover rate of this enzyme (Morita et al., 2014). Furthermore, the RuBisCO of Chlamydomonas reinhardtii showed higher carboxylation rate and higher affinity to CO2 if the rbcS gene copy of lineage 2 from Nicotiana tabacum was inserted rather than the rbcS gene copy of lineage 1 (Laterre et al., 2017). There is clearly the need to study in more detail the exact roles played by gene copies of rbcS lineage 2 in angiosperms, in particular, to understand if these copies are functional in all the species carrying this lineage. If this is the case, we will still have to investigate how gene copies of rbcS lineage 2 may contribute to the improvement of catalytic properties of RuBisCO given they do not exist in all the species. Pottier et al. (2018) have suggested that the rbcS lineage 2 may have been the predominant copies when RuBisCO was surrounded by a lower concentration of O2. Investigating the environmental habitat of the species carrying the gene copies of lineage 2 may help to understand the reason why they are carried only by a few species in angiosperms.
Positive selection and coevolution analyses
Another goal was to estimate the selective pressure acting on rbcS and uncover the coevolution between rbcS and rbcL encoding the subunits of the RuBisCO protein by estimating the coevolution between pairs of sites from these two genes. We detected positive selection in 13 positions along the rbcS sequence (Table 1), which indicates that the evolution of the rbcS gene is affected by episodic events of positive selection. The adaptation of the RuBisCO protein, which has been previously attributed mainly to the evolution of rbcL (Kapralov and Filatov, 2007; Christin, et al., 2008), could thus also be mediated by changes occurring within the gene encoding the small subunit. We further detected extensive signals of coevolution between the two subunits, which reinforces our understanding of the tight interaction between the two subunits. Our result from the coevolution analysis is consistent with the analyses based on the correlation of codon usage bias between two genes (Pei et al., 2013). We can go a step further with our analyses by identifying the residues that are potentially interacting in the 3D structure formed by RBCS and RBCL.
Our study shows that there are extensive signals of coevolution between the residues of the two subunits. Some of the residues (residues 14, 56, and 95) that we identified as coevolving were also previously reported as coevolving intra-RBCL in gymnosperms species (Sen et al., 2011). These residues of RBCL coevolve with specific residues of RBCS, and then, again, these residues of RBCS coevolve with multiple residues of RBCL (e.g. residue 14 of RBCL is the coevolving site intra-RBCL, but it also coevolves with residue 67 of RBCS; then, residue 67 of RBCS coevolves also with residues 95 and 99 of RBCL; see Fig. 4). The coevolution between some specific residues of RBCS and RBCL may have been driven by the tight interaction between the two subunits.
We found that the majority of coevolving RBCL positions are located in the N-terminal domain (10 residues among 12; Supplementary Fig. S2). Knight et al. (1990) observed that the residues involved in the interaction of subunits are mostly found in the N-terminal domain. It is suggested that residues 30 and 70 of RBCS coevolving with multiple RBCL residues in the N-terminal domain might be key sites for the interaction of the subunits. Similarly, the residues of RBCS coevolving with residues 45, 118, 205, 275, and 279 of RBCL at the interface between two RBCL subunits could be potentially involved in the assembly of RuBisCO structures. Furthermore, Knight et al. (1990) have observed that residues in the C-terminal domain are often involved in the catalysis and binding of substrates. The catalytic sites of RuBisCO are found in the RBCL subunit, and the coevolving residues of RBCS (e.g. residue 102) that mostly interact with residues of RBCL in the C-terminus (e.g. residue 456) are probably not directly involved in the activity of the RuBisCO. However, these RBCS residues may coordinately react to the functional changes on the large subunits and thus be involved in the maintenance of the 3D structure.
One of the coevolving residues of RBCL (residue 449 of 1RCX; dAIC value=23.46404 and s/d ratio=7.43E-10; with a significant signal of coevolution but not included in the 28 residues with the strongest signals) is part of a codon that is highly conserved between higher plants and the algae Chlamydomonas (Marín-Navarro and Moreno, 2006). The change of this amino acid from a cysteine to a serine has been shown to drastically increase the degradation of RuBisCO in Chlamydomonas (Marín-Navarro and Moreno, 2006). Residue 16 of RBCS (1RCX), coevolving with residue 449 of RBCL, further coevolves with another residue of RBCL (residue 40 of 1RCX; dAIC=15.4967 and s/d ratio=9.44E-10; with a significant signal of coevolution but not included in the 28 residues with the strongest signals), which was also described as important for the degradation of the RuBisCO (Kokubun et al., 2002). Our results could indicate that residue 16 in the small subunit may also be involved in the protection against degradation of RuBisCO.
In our study, we show positive selection acting on the rbcS gene, and positively selected rbcS sites that are coevolving with rbcL. The residues 107 and 118 are both under positive selection and coevolving with rbcL. These results may suggest that the substitution of an amino acid of RBCL may coordinately lead to the substitution of an amino acid of RBCS, and vice versa. Chakrabarti and Panchenko (2010) have suggested that functionally important sites undergo coevolution. Some of the positively selected sites or coevolving sites are on the interface of RBCS and RBCL. We suppose that the evolutionary processes of RBCS and RBCL are profoundly influenced by each other. These reported positively selected positions of rbcS and coevolving positions of rbcS with rbcL may be important sites for the structure and the function of RBCS and these results may help to elucidate the function of RBCS.
Protein stability of RuBisCO structure
Another goal was to understand the differences of stability between different gene copies. The composition of the RBCS subunits within the RuBisCO complex in vivo is not known. Structural stability is an important feature in an enzyme, which tends to evolve in a narrow range of stability. RuBisCO is no exception and it was observed that some amino acid substitutions under positive selection can slightly shift the stability during adaptation, in order to improve the catalytic efficiency while keeping the global fold intact (Studer et al., 2014). We were thus interested to see if the differences in the multiple copies of rbcS could significantly impact the stability of the RuBisCO complex.
Our protein stability modelling suggests that gene copies of rbcS lineage 1 of the same species may have similar functions in spite of their different evolutionary histories. Sasanuma (2001) investigated the fate of newly duplicated rbcS genes in Triticum spp. and found evidence of homogenization and pseudogenized genes, but no evidence of gaining new functions was detected. Therefore, multiple gene copies may exist for robustness (Wagner, 2005; Plata and Vitkup, 2014) to maintain the important function of RuBisCO.
Like Sasanuma’s, our results suggest that _rbcS_ is robust to gene dosage effect. As RuBisCO is necessary for plants to survive, the robustness of the rbcS gene can assist plant adaptation to drastic environmental change or prevent lack of RBCS when some of the copies are lost. Further investigation is required if we are to understand rbcS evolution in more detail. The evolutionary history of rbcS is complex to track but we suppose that studying rbcS will allow for a deeper understanding of the multigene family.
Concluding remarks
Investigating the mechanisms that have shaped the evolution of the RuBisCO complex is important for understanding the function of this key enzyme in photosynthesis. This is usually done by looking at the chloroplast gene rbcL, but this approach only provides half of the picture and it is important to consider the evolution of the small subunit encoded by the nuclear gene family rbcS. Although rbcS has a more complex evolutionary history than rbcL, involving the appearance of multiple gene copies, there are strong connections between the two subunits, as detected in the coevolution analysis of rbcS and rbcL. Some coevolving or positively selected positions are at the interface of RBCS and RBCL. A striking example is positions 107 and 118 of RBCS, which are both under positive selection and coevolving with rbcL. These results suggest substantial interactions between the subunits. However, the coevolution is not occurring between a specific rbcS gene copy and rbcL. Further, the differences of the evolutionary history of each of the gene copies do not lead to differences in the stability of the RuBisCO. We thus propose: (i) that rbcS gene copies are created under neutral evolutionary processes, or (ii) that different copies are kept by the selective pressure that allows plants to cope with different environmental conditions or to be expressed differently in each organ. We need to further investigate the mechanism and the rate of gain and loss of rbcS. Transcriptome data of rbcS in different organs and different conditions (e.g. temperature, aridity) may help us to understand if these copies are playing a role in maintaining stoichiometry.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. dAIC and s/d ratio distributions of frequency of coevolving profiles by Coev model.
Fig. S2. The residues of RBCS and RBCL under coevolution plotted on known RuBisCO structure of spinach.
Table S1. Correspondence of gene copy names in Fig. 1 and Gene ID in Phytozome database.
Table S2. Minimum number of rbcS gene copies per species in angiosperms.
Acknowledgements
We sincerely thank Dr. Romain A. Studer for his help in setting up the experiment and with the advice and invaluable help that he provided for the analyses of homology modelling. We would like to thank Victor Rossier for his help during data collection and two anonymous reviewers who helped us improve the manuscript. The bioinformatic analyses were performed at the Vital-IT facilities of the Swiss Institute of Bioinformatics. This project was funded by Swiss National Science Foundation (grant number 31003A_138282) to NS. GAB was supported by the LABEX “TULIP” managed by Agence Nationale de la Recherche (ANR-10-LABX-0041) and LABEX “CEBA” (ANR-10-LABX-25-01).
References
- Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. 2011. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Research 39, D289–D294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson I. 2008. Catalysis and regulation in Rubisco. Journal of Experimental Botany 59, 1555–1568. [DOI] [PubMed] [Google Scholar]
- Andrews TJ, Ballment B. 1983. The function of the small subunits of ribulose bisphosphate carboxylase-oxygenase. The Journal of Biological Chemistry 258, 7514–7518. [PubMed] [Google Scholar]
- Badger MR, Andrews TJ. 1987. CO-evolution of Rubisco and CO2 concentrating mechanisms. Progress in Photosynthesis Research 9, 601–609. [Google Scholar]
- Beerling DJ, Royer DL. 2011. Convergent Cenozoic CO2 history. Nature Geoscience 4, 418–420. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B 57, 289–300. [Google Scholar]
- Bianconi ME, Dunning LT, Moreno-Villena JJ, Osborne CP, Christin PA. 2018. Gene duplication and dosage effects during the early emergence of C4 photosynthesis in the grass genus Alloteropsis. Journal of Experimental Botany 69, 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracher A, Starling-Windhof A, Hartl FU, Hayer-Hartl M. 2011. Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco. Nature Structural & Molecular Biology 18, 875–880. [DOI] [PubMed] [Google Scholar]
- Cantsilieris S, Baird PN, White SJ. 2013. Molecular methods for genotyping complex copy number polymorphisms. Genomics 101, 86–93. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Panchenko AR. 2010. Structural and functional roles of coevolved sites in proteins. PLoS One 5, e8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IH, Miller B, Tan JC, et al. 2016. Population structure shapes copy number variation in malaria parasites. Molecular Biology and Evolution 33, 603–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Moore B, Seemann JR. 1998. Effects of short- and long-term elevated CO2 on the expression of ribulose-1,5-bisphosphate carboxylase/oxygenase genes and carbohydrate accumulation in leaves of Arabidopsis thaliana (L.) Heynh. Plant Physiology 116, 715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Boxall SF, Gregory R, Edwards EJ, Hartwell J, Osborne CP. 2013. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biology and Evolution 5, 2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Muasya AM, Roalson EH, Russier F, Besnard G. 2008. Evolutionary switch and genetic convergence on rbcL following the evolution of C4 photosynthesis. Molecular Biology and Evolution 25, 2361–2368. [DOI] [PubMed] [Google Scholar]
- Clark GB, Sessions A, Eastburn DJ, Roux SJ. 2001. Differential expression of members of the annexin multigene family in Arabidopsis. Plant Physiology 126, 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib L, Meyer X, Artimo P, Ioannidis V, Stockinger H, Salamin N. 2015. Coev-web: a web platform designed to simulate and evaluate coevolving positions along a phylogenetic tree. BMC Bioinformatics 16, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib L, Silvestro D, Salamin N. 2014. Evolutionary footprint of coevolving positions in genes. Bioinformatics 30, 1241–1249. [DOI] [PubMed] [Google Scholar]
- Dumont BL, Eichler EE. 2013. Signals of historical interlocus gene conversion in human segmental duplications. PLoS One 8, e75949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Osborne CP, Strömberg CA, et al. ; C4Grasses Consortium 2010. The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328, 587–591. [DOI] [PubMed] [Google Scholar]
- Epskamp S, Cramer A, Waldorp L, Schmittmann V, Borsboom D. 2012. qgraph: network visualizations of relationships in psychometric data. Journal of Statistical Software 48, 1–18. [Google Scholar]
- Eswar N, Eramian D, Webb B, Shen MY, Sali A. 2008. Protein structure modeling with MODELLER. Methods in Molecular Biology 426, 145–159. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. 2009. Gene duplication and evolutionary novelty in plants. New Phytologist 183, 557–564. [DOI] [PubMed] [Google Scholar]
- Furbank RT, Hatch MD, Jenkins CLD. 2000. C4 photosynthesis: mechanism and regulation. In: Leegood RC, Sharkey TD, von Caemmerer S, eds. Photosynthesis: physiology and metabolism. Dordrecht: Kluwer Academic Publishers, 435–457. [Google Scholar]
- Galili S, Galili G, Feldman M. 1991. Chromosomal location of genes for Rubisco small subunit and Rubisco-binding protein in common wheat. Theoretical and Applied Genetics 81, 98–104. [DOI] [PubMed] [Google Scholar]
- Genkov T, Meyer M, Griffiths H, Spreitzer RJ. 2010. Functional hybrid rubisco enzymes with plant small subunits and algal large subunits: engineered rbcS cDNA for expression in Chlamydomonas. The Journal of Biological Chemistry 285, 19833–19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genkov T, Spreitzer RJ. 2009. Highly conserved small subunit residues influence rubisco large subunit catalysis. The Journal of Biological Chemistry 284, 30105–30112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Hatch MD, Slack CR. 1968. A new enzyme for the interconversion of pyruvate and phosphopyruvate and its role in the C4 dicarboxylic acid pathway of photosynthesis. The Biochemical Journal 106, 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudsona GS, Dengler RE, Hattersleya PW, Dengler NG. 1992. Cell-specific expression of rubisco small subunit and rubisco activase genes in C3 and C4 species of Atriplex. Australian Journal of Plant Physiology 19, 89–96. [Google Scholar]
- Hughes AL. 1994. The evolution of functionally novel proteins after gene duplication. Proceedings of the Royal Society B: Biological Sciences 256, 119–124. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nature Reviews. Genetics 11, 97–108. [DOI] [PubMed] [Google Scholar]
- Kafri R, Dahan O, Levy J, Pilpel Y. 2008. Preferential protection of protein interaction network hubs in yeast: evolved functionality of genetic redundancy. Proceedings of the National Academy of Sciences, USA 105, 1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai R, Edwards GE. 1999. The biochemistry of C4 photosynthesis. In: Sage RF, Monson RK, eds. C4 plant biology. San Diego: Academic Press, 49–87. [Google Scholar]
- Kapralov MV, Filatov DA. 2007. Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evolutionary Biology 7, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov MV, Kubien DS, Andersson I, Filatov DA. 2011. Changes in Rubisco kinetics during the evolution of C4 photosynthesis in Flaveria (Asteraceae) are associated with positive selection on genes encoding the enzyme. Molecular Biology and Evolution 28, 1491–1503. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight S, Andersson I, Brändén CI. 1990. Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 Å resolution. Subunit interactions and active site. Journal of Molecular Biology 215, 113–160. [DOI] [PubMed] [Google Scholar]
- Kokubun N, Ishida H, Makino A, Mae T. 2002. The degradation of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase into the 44-kDa fragment in the lysates of chloroplasts incubated in darkness. Plant & Cell Physiology 43, 1390–1395. [DOI] [PubMed] [Google Scholar]
- Ku MS, Kano-Murakami Y, Matsuoka M. 1996. Evolution and expression of C4 photosynthesis genes. Plant Physiology 111, 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubien DS, Whitney SM, Moore PV, Jesson LK. 2008. The biochemistry of Rubisco in Flaveria. Journal of Experimental Botany 59, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Laterre R, Pottier M, Remacle C, Boutry M. 2017. Photosynthetic trichomes contain a specific rubisco with a modified pH-dependent activity. Plant Physiology 173, 2110–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Guindon S. 2014. Performance of standard and stochastic branch-site models for detecting positive selection among coding sequences. Molecular Biology and Evolution 31, 484–495. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207, 437–453. [DOI] [PubMed] [Google Scholar]
- Mano S, Innan H. 2008. The evolutionary rate of duplicated genes under concerted evolution. Genetics 180, 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansai SP, Innan H. 2010. The power of the methods for detecting interlocus gene conversion. Genetics 184, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Navarro J, Moreno J. 2006. Cysteines 449 and 459 modulate the reduction-oxidation conformational changes of ribulose 1.5-bisphosphate carboxylase/oxygenase and the translocation of the enzyme to membranes during stress. Plant, Cell & Environment 29, 898–908. [DOI] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evolution 1, vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura H, Mizohata E, Ishida H, Kogami A, Ueno T, Makino A, Inoue T, Yokota A, Mae T, Kai Y. 2012. Crystal structure of rice Rubisco and implications for activation induced by positive effectors NADPH and 6-phosphogluconate. Journal of Molecular Biology 422, 75–86. [DOI] [PubMed] [Google Scholar]
- McGlothlin JW, Kobiela ME, Feldman CR, et al. 2016. Historical contingency in a multigene family facilitates adaptive evolution of toxin resistance. Current Biology 26, 1616–1621. [DOI] [PubMed] [Google Scholar]
- Miller R. 2014. Evolution of the rbcS gene family in Solanaceae: concerted evolution and gain and loss of introns, with a description of new statistical guidelines for determining the number of unique gene copies. PhD thesis, University of Washington. [Google Scholar]
- Monson RK. 2003. Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. International Journal of Plant Sciences 164, S43–S54. [Google Scholar]
- Moore RC, Purugganan MD. 2005. The evolutionary dynamics of plant duplicate genes. Current Opinion in Plant Biology 8, 122–128. [DOI] [PubMed] [Google Scholar]
- Morita K, Hatanaka T, Misoo S, Fukayama H. 2014. Unusual small subunit that is not expressed in photosynthetic cells alters the catalytic properties of Rubisco in rice. Plant Physiology 164, 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Hatanaka T, Misoo S, Fukayama H. 2016. Identification and expression analysis of non-photosynthetic Rubisco small subunit, OsRbcS1-like genes in plants. Plant Gene 8, 26–31. [Google Scholar]
- Muse SV, Gaut BS. 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Molecular Biology and Evolution 11, 715–724. [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators 2018. Database resources of National Center for Biotechnology Information. Nucleic Acids Research 46, D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gu X, Sitnikova T. 1997. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proceedings of National Academy of Sciences, USA 94, 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP. 2005. Concerted and birth-and-death evolution of multigene families. Annual Review of Genetics 39, 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y. 2009. Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents. Human Genomics 4, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y, Shimizu H, Hasegawa K, Tsujimoto H, Sasakuma T. 1994. Chromosome assignment of four photosynthesis-related genes and their variability in wheat species. Theoretical and Applied Genetics, 88, 383–394. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1977. Genetic variation in multigene families. Nature, 267, 515–517. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1979. An extension of a model for the evolution of multigene families by unequal crossing over. Genetics 91, 591–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 1983. On the evolution of multigene families. Theoretical Population Biology 23, 216–240. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1988. Time for acquiring a new gene by duplication. Proceedings of the National Academy of Sciences, USA 85, 3509–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 1991. Multigene families and the evolution of complexity. Journal of Molecular Evolution 33, 34–41. [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH. 2016. Evolution of gene duplication in plants. Plant Physiology 171, 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424, 194–197. [DOI] [PubMed] [Google Scholar]
- Pearson PN, Foster GL, Wade BS. 2009. Atmospheric carbon dioxide through the Eocene-Oligocene climate transition. Nature 461, 1110–1113. [DOI] [PubMed] [Google Scholar]
- Pei ZY, Mu GL, Pan J, Zhang DM. 2013. Codon usage and coevolution of the large and small subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase. Journal of Systematics and Evolution 51, 511–521. [Google Scholar]
- Peterhansel C, Horst I, Niessen M, Blume C, Kebeish R, Kürkcüoglu S, Kreuzaler F. 2010. Photorespiration. The Arabidopsis Book 8, e0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter M, Bonow I, Klinkert MQ. 2008. Diverse expression patterns of subgroups of the rif multigene family during Plasmodium falciparum gametocytogenesis. PLoS One 3, e3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Cashmore AR. 1986. Evidence for selection as a mechanism in the concerted evolution of Lycopersicon esculentum (tomato) genes encoding the small subunit of ribulose-1, 5-bisphosphate carboxylase/oxygenase. Proceedings of National Academy of Sciences, USA 83, 3880–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piot A, Hackel J, Christin PA, Besnard G. 2018. One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta 247, 255–266. [DOI] [PubMed] [Google Scholar]
- Plata G, Vitkup D. 2014. Genetic robustness and functional evolution of gene duplicates. Nucleic Acids Research 42, 2405–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. [DOI] [PubMed] [Google Scholar]
- Pottier M, Gilis D, Boutry M. 2018. The hidden face of Rubisco. Trends in Plant Science 23, 382–392. [DOI] [PubMed] [Google Scholar]
- Rawsthorne S. 1992. C3–C4 intermediate photosynthesis: linking physiology to gene expression. The Plant Journal 2, 267–274. [Google Scholar]
- Rensing SA. 2014. Gene duplication as a driver of plant morphogenetic evolution. Current Opinion in Plant Biology 17, 43–48. [DOI] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3, 217–223. [Google Scholar]
- Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA. 2012. The fate of duplicated genes in a polyploid plant genome. The Plant Journal 73, 143–153. [DOI] [PubMed] [Google Scholar]
- Sage RF. 2004. The evolution of C4 photosynthesis. New Phytologist 161, 341–371. [DOI] [PubMed] [Google Scholar]
- Sage RF, Coleman JR. 2001. Effects of low atmospheric CO2 on plants: more than a thing of the past. Trends in Plant Science 6, 18–24. [DOI] [PubMed] [Google Scholar]
- Sasanuma T. 2001. Characterization of the rbcS multigene family in wheat: subfamily classification, determination of chromosomal location and evolutionary analysis. Molecular Genetics and Genomics 265, 161–171. [DOI] [PubMed] [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. 2005. The FoldX web server: an online force field. Nucleic Acids Research 33, W382–W388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Research 43, W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen L, Fares MA, Liang B, Gao L, Wang B, Wang T, Su YJ. 2011. Molecular evolution of rbcL in three gymnosperm families: identifying adaptive and coevolutionary patterns. Biology Direct 6, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Riemer C, Dickins B, et al. 2012. Revealing mammalian evolutionary relationships by comparative analysis of gene clusters. Genome Biology and Evolution 4, 586–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ. 2003. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Archives of Biochemistry and Biophysics 414, 141–149. [DOI] [PubMed] [Google Scholar]
- Studer RA, Christin PA, Williams MA, Orengo CA. 2014. Stability-activity tradeoffs constrain the adaptive evolution of RubisCO. Proceedings of the National Academy of Sciences, USA 111, 2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Penel S, Duret L, Robinson-Rechavi M. 2008. Pervasive positive selection on duplicated and nonduplicated vertebrate protein coding genes. Genome Research 18, 1393–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Manzara T, Pichersky E, Cashmore A, Gruissem W. 1987. Genomic organization, sequence analysis and expression of all five genes encoding the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase from tomato. Molecular & General Genetics 209, 247–256. [DOI] [PubMed] [Google Scholar]
- Taylor TC, Andersson I. 1997. The structure of the complex between rubisco and its natural substrate ribulose 1,5-bisphosphate. Journal of Molecular Biology 265, 432–444. [DOI] [PubMed] [Google Scholar]
- Thomas-Hall S, Campbell PR, Carlens K, Kawanishi E, Swennen R, Sági L, Schenk PM. 2007. Phylogenetic and molecular analysis of the ribulose-1,5-bisphosphate carboxylase small subunit gene family in banana. Journal of Experimental Botany 58, 2685–2697. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium 2015. UniProt: a hub for protein information. Nucleic Acids Research 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer S, Quick PW. 2000. Rubisco: physiology in vivo. In: Leegood RC, Sharkey TD, von Caemmerer S, eds. Photosynthesis: physiology and metabolism. Dordrecht, The Netherlands: Kluwer Academic Publishers, 85–113. [Google Scholar]
- Wagner A. 1998. The fate of duplicated genes: loss or new function? BioEssays 20, 785–788. [DOI] [PubMed] [Google Scholar]
- Wagner A. 2005. Robustness, evolvability, and neutrality. FEBS Letters 579, 1772–1778. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Lee Y, Kim T, Ahn JS, Jung Y, Kim B, Lee S. 2001. High resolution resonance enhanced two photon ionization spectroscopy of RbCs in a cold molecular beam. The Journal of Chemical Physics 114, 8926–8931. [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Molecular Biology and Evolution 22, 2472–2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.