GABA has beneficial effects on salinity stress tolerance in Arabidopsis linked to increased activity of H+-ATPase, reduced ROS-induced K+ efflux from root epidermis, and increased SOS1 and NHX1 transcript levels in plant roots.
Keywords: Arabidopsis, H+-ATPase, hydrogen peroxide, potassium retention, reactive oxygen species, sodium sequestration
Abstract
The non-protein amino acid γ-aminobutyric acid (GABA) rapidly accumulates in plant tissues in response to salinity. However, the physiological rationale for this elevation remains elusive. This study compared electrophysiological and whole-plant responses of salt-treated Arabidopsis mutants pop2-5 and gad1,2, which have different abilities to accumulate GABA. The pop2-5 mutant, which was able to overaccumulate GABA in its roots, showed a salt-tolerant phenotype. On the contrary, the gad1,2 mutant, lacking the ability to convert glutamate to GABA, showed oversensitivity to salinity. The greater salinity tolerance of the pop2-5 line was explained by: (i) the role of GABA in stress-induced activation of H+-ATPase, thus leading to better membrane potential maintenance and reduced stress-induced K+ leak from roots; (ii) reduced rates of net Na+ uptake; (iii) higher expression of SOS1 and NHX1 genes in the leaves, which contributed to reducing Na+ concentration in the cytoplasm by excluding Na+ to apoplast and sequestering Na+ in the vacuoles; (iv) a lower rate of H2O2 production and reduced reactive oxygen species-inducible K+ efflux from root epidermis; and (v) better K+ retention in the shoot associated with the lower expression level of GORK channels in plant leaves.
Introduction
Soil salinization affects ~10% of the land surface (950 million ha) and costs agriculture over $27 bn per annum in lost opportunities; this problem is expected to be exacerbated by the current climate changes. As most crop species are sensitive to salt, breeding for salt tolerance remains one of the current priorities of plant breeders. To illustrate this point, the number of published papers having keywords ‘salinity stress’ and ‘breeding’ has increased from just four to five per year in early 1990 to 35–40 per year in mid-2000 and exceeded a 150 papers threshold in 2017 (Web of Science data). This reflects the alarming trend in developing soil salinity, which takes about 3 ha of arable land out of production every minute (Shabala et al., 2014). However, progress in crop breeding for salinity tolerance has been disappointingly slow, and all attempts to improve crop tolerance by targeting salt exclusion traits have not resulted in creating salt-tolerant cultivars for farmers’ fields (Shabala, 2013; Ismail and Horie, 2017). This implies that traits other than Na+ exclusion should be explored in more details. It has also become apparent that it is the mode of regulation of various ion transporters and their upstream signaling, rather than the molecular identity of transporters per se, that is critical to confer salinity stress tolerance.
γ-Aminobutyric acid (GABA) is a four-carbon non-proteinogenic amino acid that was first isolated from potato tubers over half a century ago (Steward et al., 1949). In animals, GABA acts as an inhibitory neurotransmitter and also as a signaling molecule; this role has been extensively studied over the last six decades. In plants, the evidence has been mounting since the 1990s that GABA may also act as a signaling molecule (Ramesh et al., 2017), besides being a defensive compound against insect herbivory (Scholz et al., 2015). GABA is metabolized via a short pathway known as the GABA shunt, in which it is mainly synthesized by the irreversible reaction of the cytosolic enzyme glutamate decarboxylase (GAD; EC 4.1.l.15) using glutamate as a substrate (Fait et al., 2008). GABA catabolism occurs in the mitochondrial matrix of multicellular organisms by the action of GABA transaminase (GABA-T; EC 2.6.1.19) to produce succinic semi-aldehyde (Shelp et al., 2012). The GABA-T encoding gene (At3g22200) is present as a single copy in the Arabidopsis genome and was initially termed pollen–pistil incompatibility 2 (POP2; Palanivelu et al., 2003).
Variability and a large, rapid increase in GABA concentration in plant tissue (0.02 to 4 µmol g−1 FW) is one of the main pieces of evidence that GABA acts as a signal in plants (Ramesh et al., 2017). A multitude of abiotic stresses can drive GABA accumulation in plants, including salinity, oxidative stress, hypoxia, extreme temperature, drought, and waterlogging (Ramesh et al., 2015). Of these, the important role of GABA in plant adaptive responses to hypoxia and flooding has received major attention (Miyashita and Good, 2008). Salt-induced GABA accumulation has been also observed broadly in a the number of plant species, including alfalfa, Arabidopsis, barley, tobacco, and soybean (Widodo et al., 2009; Renault et al., 2010; Zhang et al., 2011). The Arabidopsis seedlings produced approximately 20-fold higher GABA under 150 mM NaCl (Renault et al., 2010). Central carbon metabolism and cell-wall modification were changed in an Arabidopsis GABA-T knockout mutant (pop2) under salt stress (Renault et al., 2013). Public data from AtGenExpress (http://atpbsmd.yokohama-cu.ac.jp) show that POP2 transcription in Arabidopsis shoot is up-regulated by 24 h of salt stress, and expression of GAD in cells of root stele, root cortex, and root epidermis is down-regulated by 140 mM NaCl at 8 and 48 h. However, it remains unclear whether these changes in GABA levels are essential for plants to deal with salt load, or whether they are merely ‘physiological noise’, e.g. a by-product of some adaptive responses. To the best of our knowledge, no causal link between salt stress-induced GABA accumulation and activity of key cellular transporters mediating plant ionic homeostasis and metabolism under saline conditions has been reported in the literature.
In Arabidopsis, the pop2 mutant is unable to produce a functional GABA-T enzyme (Van Cauwenberghe et al., 2002), which leads to the inhibition of GABA degradation, and, as a consequence, causes more GABA accumulation. On the contrary, disruption of the GAD gene prevented the accumulation of GABA, because GAD1 and GAD2 play a major role in GABA synthesis in plants (Bouché et al., 2004; Scholz et al., 2015). In this study, we used Arabidopsis gad1,2 double mutant and pop2-5 mutant lines to investigate the role of GABA in plant adaptive responses to salinity and its control over the activity of key plasma membrane transporters mediating Na+ and K+ homeostasis in plant roots and the modes of their operation. Our work provides a mechanistic explanation for the role of GABA operating upstream of K+ and Na+ transporters thus mediating salt-related adaptive responses, and also provides valuable information for improving crop salt tolerance by genetic engineering.
Materials and methods
Plant materials and growth conditions
Arabidopsis Columbia-0 (Col-0) was used as wild-type (WT). Seeds of gad1,2 and pop2-5 mutants were kindly provided by Frank Ludewig (University of Erlangen-Nuremberg, Germany). Seeds were surface-sterilized by 20% commercial bleach (1% (v/v) NaClO) for 10 min and thoroughly washed with sterilized distilled water. Seeds were then sown in Petri dishes containing 1% (w/v) phytogel, half-strength Murashige and Skoog (MS) medium, and 0.5% (w/v) sucrose at pH 5.7–5.8, sealed with 3 M micro-pore tape (3M Health Care, St Paul, MN, USA). After 2 d of stratification at 4 °C, Petri dishes were placed in a growth chamber and positioned vertically to allow root growth along the surface of the medium. The conditions in the growth chamber were 16 h/8 h light/dark cycles, with 100 μmol m−2 s−1 photon flux density during the light period, and temperature at 22 °C. Unless specified, all chemicals were of analytical grade from Sigma-Aldrich (Castle Hill, NSW, Australia). The relative expression of GAD1–GAD5 and POP2 genes in plant roots was measured to demonstrate that mutant lines used are true knockout (see Supplementary Fig. S1 at JXB online). In some experiments, two additional mutant lines—pop2 (CS415022, T-DNA insertion, https://www.arabidopsis.org/servlets/TairObject?type=germplasm&id=6530372490; GABA overaccumulating) and gad1 (SALK_017810, T-DNA insertion, https://www.arabidopsis.org/servlet/TairObject?id=230900&type=stock; with reduced GABA production ability)—were used to validate most critical conclusions made. Their homozygosis was shown (Supplementary Fig. S2). These seeds were kindly provided by Alberto Macho (Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences, China).
Whole-plant physiological assessment
Two types of phenotyping experiments were conducted, for plants grown on either soil or agar medium. In the latter case, surface-sterilized seeds were sown in Petri dishes in 1/2 strength MS medium containing appropriate salt concentration (0, 50, 100, and 150 mM NaCl). After 2 d of vernalization at 4 °C, Petri dishes were placed in a growth chamber as described above. For root length measurements, Petri dishes were oriented vertically, and plants were grown for 1 week. To determine the shoot fresh weight, Petri dishes were placed horizontally, and plants were allowed to grow for 3 weeks. For pot phenotyping experiments, seeds were sown in 0.2-liter pots filled with peat moss, perlite, vermiculite, and coarse sand (2:1:1:1, v/v), and grown for 4 weeks at 21 °C using a 12/12 h light (photosynthetically active radiation 100 μmol m−2 s−1)/dark regime. Seedlings were then treated with either 50 or 100 mM NaCl for another 2 weeks added with irrigation on a daily basis. The fresh and dry weight of each plant was then measured. Leaf chlorophyll content was measured using a SPAD meter (SPAD-502, Minolta, Japan). The maximum photochemical efficiency of PSII (chlorophyll fluorescence Fv/Fm ratio) was measured using an OS-30p chlorophyll fluorometer (Opti-Sciences, USA). All non-distractive measurements were taken on rosette leaves from at least five biological replicates for each treatment in a single experiment.
Leaf sap Na+ and K+ content measurement
The Arabidopsis leaves were harvested and quickly frozen in Eppendorf tubes. To measure leaf sap K+ and Na+ content, the leaves were thawed and the sap extracted by hand-squeezing the leaf samples as described elsewhere (Cuin et al., 2009). A volume of 50 μl of collected sap was diluted to 5 ml using distilled water and used for the determination of K+ and Na+ concentration (in mM) using a flame photometer (Corning 410C, Halstead, UK). Five replicates for each treatment were assessed.
GABA content determination
GABA extraction and quantification was carried out as described previously (Scholz et al., 2017). Briefly, fresh tissue from roots was harvested, frozen in liquid nitrogen and weighed for the determination of GABA content per gram fresh weight. After maceration, GABA was extracted twice with a total volume of 2 ml methanol, and supernatants were combined, dried, and re-suspended in 500 μl methanol. This extract was diluted 1:20 (v/v) with water containing the internal standard (algal amino acid mix 13C, 15N (Isotec, Miamisburg, OH, USA), at a mix concentration of 10 μg ml−1). GABA content was analysed by LC-MS/MS according to Scholz et al. (2017) using an API 5000 tandem mass spectrometer (Applied Biosystems, Darmstadt, Germany) operated in positive ionization mode with GABA-specific multiple reaction monitoring to monitor analyte parent ion → product ion—GABA: m/z 104.1→87.1; DP 51, CE 17.
Viability staining
Viability of root cells was assessed 2 h after the salt treatment using fluorescein diacetate (FDA)-propidium iodide (PI) double staining method as described in Koyama et al. (1995). FDA (cat. no. F7378; Sigma-Aldrich, St Louis, MO, USA) is permeant through the intact plasma membrane and shows a green color under a fluorescence microscope in viable cells after hydrolysis by the internal esterases (Rotman and Papermaster, 1966). On the other hand, PI (cat. no. P4864; Sigma-Aldrich) is not permeant to live cells, and is commonly used to detect dead cells in a population, showing a red color upon PI–nuclear DNA conjugate formation (Riccardi and Nicoletti, 2006). Accordingly, roots were stained with a freshly prepared 5 g ml−1 FDA for 3 min followed by 3 g ml−1 PI treatment for 10 min. The double-stained roots were observed under a fluorescence microscope (Leica MZ12; Leica Microsystems, Wetzlar, Germany) illuminated by an ultra-high-pressure mercury lamp (Leica HBO Hg 100 W; Leica Microsystems) and fitted with a Leica I3-wavelength filter cube (Leica Microsystems). The excitation and emission wavelengths were 488/505–530 and 543/585 nm for FDA and PI, respectively. Photographs were taken by a Leica (DFC295; Leica Microsystems) camera fitted on the microscope using image acquisition and processing software LAS V3.8 (Leica Microsystems). During the image acquisition, all the automatic exposure features of the LAS V3.8 were disabled, and exposure time (2.6 s), gain (×2.1), saturation (2.10), and gamma (0.87) were set to constant values as indicated in parentheses for the entire period of the experiment.
Quantification of root viability by TTC method
The 2,3,5-triphenyltetrazolium chloride (TTC) method was used to determine root viability as described elsewhere (Clemensson-Lindell, 1994). This method is based on the existence of a positive correlation between the activity of dehydrogenase in root and the root viability. The dehydrogenase in plant root can reduce TTC to triphenylmethylhydrazone (TTF), which can then be detected by spectrophotometry. Roots (0.2 g) were placed in 10 ml of 0.4% (w/v) TTC in 0.06 mM Na2HPO4–KH2PO4 solution and vacuum-infiltrated for 15 min. After that, the incubation stood at 37 °C for 10 h. The samples were then extracted in 95% (v/v) ethanol followed by incubation in a water bath at 90 °C for 15 min. The absorbance was recorded at 520 nm.
H2O2 production
Two methods were used to compare salinity-stress-induced H2O2 production in plant roots. The first method utilized H2O2-sensitive fluorescent probe cell-permeant 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA or DCF, Thermo Fisher Scientific, Waltham, MA, USA). The indicator was dissolved in dimethyl sulfoxide (Sigma-Aldrich) to a stock concentration of 1 mM. Then, 20 μM DCF was added to a measuring buffer (10 mM KCl, 5 mM Ca2+-MES, pH 6.1). After 2, 6, 12, and 24 h of NaCl treatment, Arabidopsis seedling roots were incubated in the dye-containing measuring buffers for 30 min in the dark. The stained roots were washed in distilled water for 3 min to remove residual dye before measuring fluorescence intensity in root cells. The fluorescence images of H2O2 were collected using excitation at 488 nm and emission at 517–527 nm. Then, fluorescence images were analysed with ImageJ software (NIH, USA) based on integrated density. For each treatment, at least eight biological replicates (individual roots) were measured.
The second (quantitative) method followed the procedure of Hossain et al. (2010); 0.5 g fresh roots were ground in 5 ml of 50 mM sodium phosphate buffer (pH 7.8), then centrifuged at ~11 900 g for 15 min at 4 °C. The yellow color was developed after reaction of 3 ml supernatant with 1 ml 0.1% TiCl4 containing 20% H2SO4 for 10 min at room temperature. The absorbance was then recorded at 410 nm.
Non-invasive ion flux measurements
Net H+, K+ and Na+ fluxes were measured using the non-invasive ion-selective microelectrode ion flux estimation (MIFE) technique (University of Tasmania, Hobart, Australia) as described previously (Shabala and Shabala, 2002; Shabala et al., 2006). Net ion fluxes were calculated using MIFEFLUX software for cylindrical diffusion geometry (Shabala et al., 2006). The principles of MIFE ion flux measurements and all the details of microelectrode fabrication and calibration are available in our previous publications (Shabala et al., 2006). Liquid ionic exchangers used in this work are the commercially available ionophore cocktails.
Prior to the MIFE measurement, 7-day-old Arabidopsis seedlings were immobilized in a 5 ml Perspex measuring chamber (10.5 × 0.8 × 2.0 cm), and 3 ml of basic salt medium (BSM; 0.5 mM KCl + 0.1 mM CaCl2, pH 5.6) was added into the chamber. After 40 min, the net ion fluxes were measured by microelectrodes from either elongation (0.4 mm from the root tip) or mature (5 mm from the root tip) root zone. Steady-state fluxes were measured for 5 min, and then the treatment was applied, followed by another 40 min of measurements. At least 15 replicates were used for each treatment.
Membrane potential measurements
Conventional KCl-filled Ag–AgCl microelectrodes were used for membrane potential measurements in the elongation and mature root zones (Bose et al., 2013). The roots of an intact 7-day-old seedling were immobilized in BSM for 40 min. Resting membrane potential was recorded for 1 min before adding NaCl to the BSM solution. The resulting change in transient membrane potential was continuously monitored for up to 25 min. Membrane potential values of 10 individual seedlings were averaged for each treatment.
RT-qPCR analysis
Total RNA was isolated from root tissues using Trizol extraction reagent (Thermo Fisher Scientific) and the RNA purity was verified by the ratio (>1.9) of 260/280 nm absorbance. DNA-free total RNA (5 g) from different treatments was used for first-strand cDNA synthesis in a 20 μl reaction volume (RevertAid First Strand cDNA Synthesis Kit, Thermo Fisher Scientific) according to the manufacturer’s instructions. Real-time quantitative PCR reactions were performed using a Mastercycler® ep realplex real-time PCR system (ABI7500, Thermo Fisher Scientific) with Bestar® Sybr Green qPCR mastermix (DBI Bioscience Inc., Ludwigshafen, Germany) in a 20 μl reaction volume according to the user manual. All primers (see Supplementary Table S1) were synthesized by Eurofins Ltd Company (Melbourne, Australia). The relative expression level was presented relative to that of the corresponding control sample at the indicated time, after normalization to actin transcript levels. All experiments were repeated three times.
Statistical analysis
Values presented are means ± either standard deviation (SD) or standard error (SE). Data were subjected to analysis of variance (ANOVA), and mean values were compared by Duncan’s and Tukey’s multiple range test (P<0.05). All statistical analysis was performed using SPSS 19.0 for Windows.
Results
Plants’ ability to produce GABA alters their sensitivity to saline stress
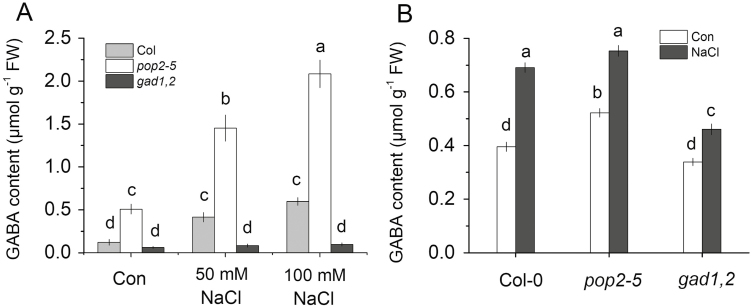
In Arabidopsis, the gene POP2 encodes GABA-T, which degrades GABA. The pop2 mutant is unable to produce a functional GABA-T enzyme. As a result, the GABA content in roots of the pop2-5 line was significantly higher (over 3-fold) than that in wild-type Columbia (Col) (Fig. 1A). GAD catalyses the conversion of glutamate to GABA; only a small difference (not significant at P<0.05) in GABA content was found between WT and gad1,2 plants under control conditions. However, when the seedlings were subjected to NaCl stress, the GABA content in both Col and pop2-5 roots was significantly increased (about 4-fold at the highest 100 mM NaCl treatment) while it remained largely unchanged in gad1,2 roots. Regardless of the salt concentration used, the GABA content was several-fold higher in pop2-5 compared with WT (Fig. 1A). This NaCl-induced increase in GABA concentration in roots was rapid and detected within 15 min of stress onset (Fig. 1B).
Fig. 1.
GABA content in the roots of Arabidopsis Col-0, pop2-5, and gad1,2 seedlings. (A) Arabidopsis seeds were sown in half-strength MS medium with 2% w/v phytogel infused with different salt concentrations in Petri dishes and grown for 7 d. (B) Plants were first grown in Petri dishes for 1 week in half-strength MS medium with 2% w/v phytogel and then transferred into quarter-strength Hoagland nutrient solution for 3 weeks. Seedlings were then treated with or without 100 mM NaCl for 15 min. Data are mean ±SD (n=3). Data labelled with different lowercase letters are significantly different at P<0.05 level.
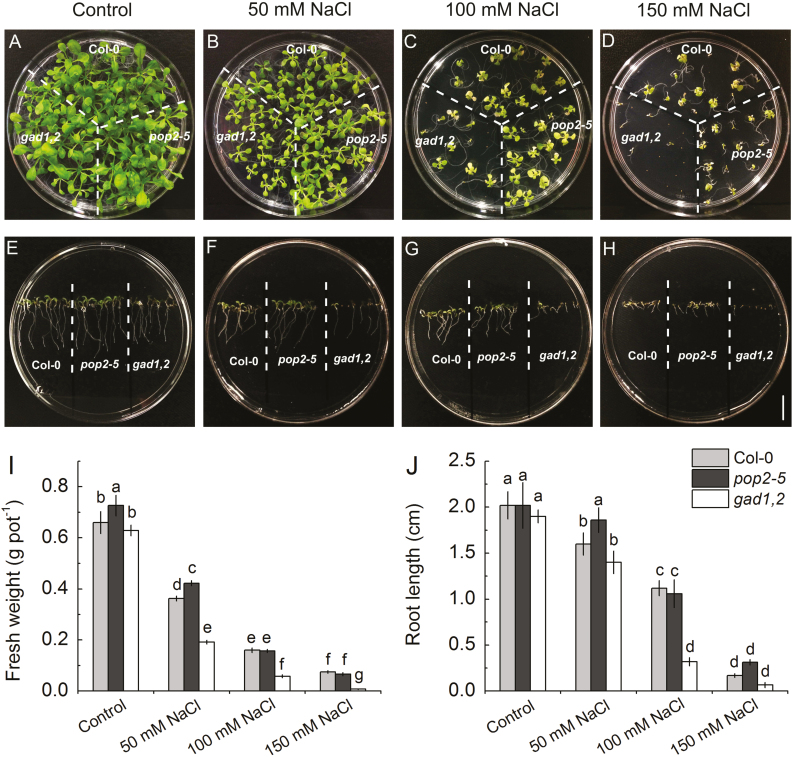
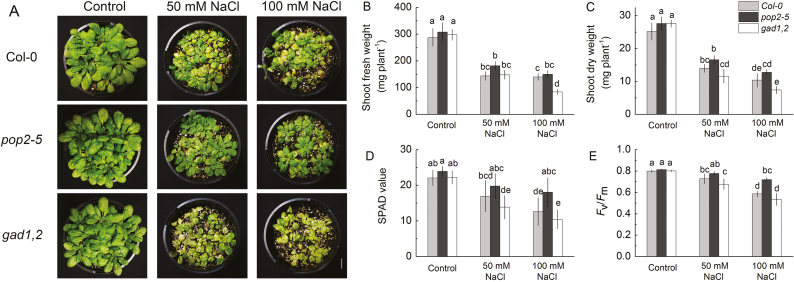
We then tested the sensitivity of various growth parameters to NaCl. A Petri dish experiments showed that under saline conditions the germination ability of gad1,2 was reduced distinctly (Fig. 2A–D). Without NaCl, the fresh weight and root length were nearly the same in Col, pop2-5, and gad1,2 seedlings. With the increase of NaCl concentration, both these characteristics were reduced (Fig. 2E–J). This salinity-induced inhibition of plant growth was most pronounced in gad1,2 plants lacking an ability to respond to salt stress by increasing GABA levels. Similar trends were revealed for plants grown in soil (Fig. 3), where plants were exposed to salinity for the longer period, and plants were actively transpiring thus bringing the salt to the shoot. No difference was found in the shoot fresh and dry weight, chlorophyll content (SPAD value) and Fv/Fm among Col, pop2-5, and gad1,2 under control condition. However, under salt treatment, pop2-5 showed a salt-tolerant phenotype, having higher shoot fresh and dry weight, SPAD value and Fv/Fm compared with Col, whereas the gad1,2 mutant showed a tendency to higher sensitivity (Fig. 3).
Fig. 2.
Effects of NaCl stress on the morphology (A–H), fresh weight (I), and root length (J) of Arabidopsis Col, pop2-5, and gad1,2 plants. (A–D) Plants were grown in Petri dishes for 3 weeks in half-strength MS medium with 2% w/v phytogel infused with different salt concentrations. The seedlings were photographed and the fresh weight was measured (I). (E–H) Root length of Col, pop2-5, and gad1,2 Arabidopsis plants grown in vertically oriented Petri dishes for 1 week. Data are mean ±SD (n=5). Data labelled with different lowercase letters are significantly different at P<0.05 level. (This figure is available in color at JXB online.)
Fig. 3.
Effects of salinity on morphology (A), shoot fresh weight (B), shoot dry weight (C), chlorophyll content (SPAD value; D), and chlorophyll fluorescence (Fv/Fm; E) of Arabidopsis Col, pop2-5, and gad1,2 plants grown at various NaCl levels. All seedlings were grown in pots for 4 weeks under control conditions, followed by 2 weeks of salinity treatment. Data are mean ±SE (n=6). Data labelled with different lowercase letters are significantly different at P<0.05 level. (This figure is available in color at JXB online.)
The ability of GABA-accumulating plants to develop a salt-tolerant phenotype when grown under saline conditions was further confirmed in experiments using two additional mutant lines (pop2 and gad1; Supplementary Fig. S3). Also, supplying exogenous GABA was able to rescue salt-sensitive gad1,2 phenotype (see Supplementary Fig. S4).
Ionic relations in Col, pop2-5, and gad1,2 mutants under salt stress
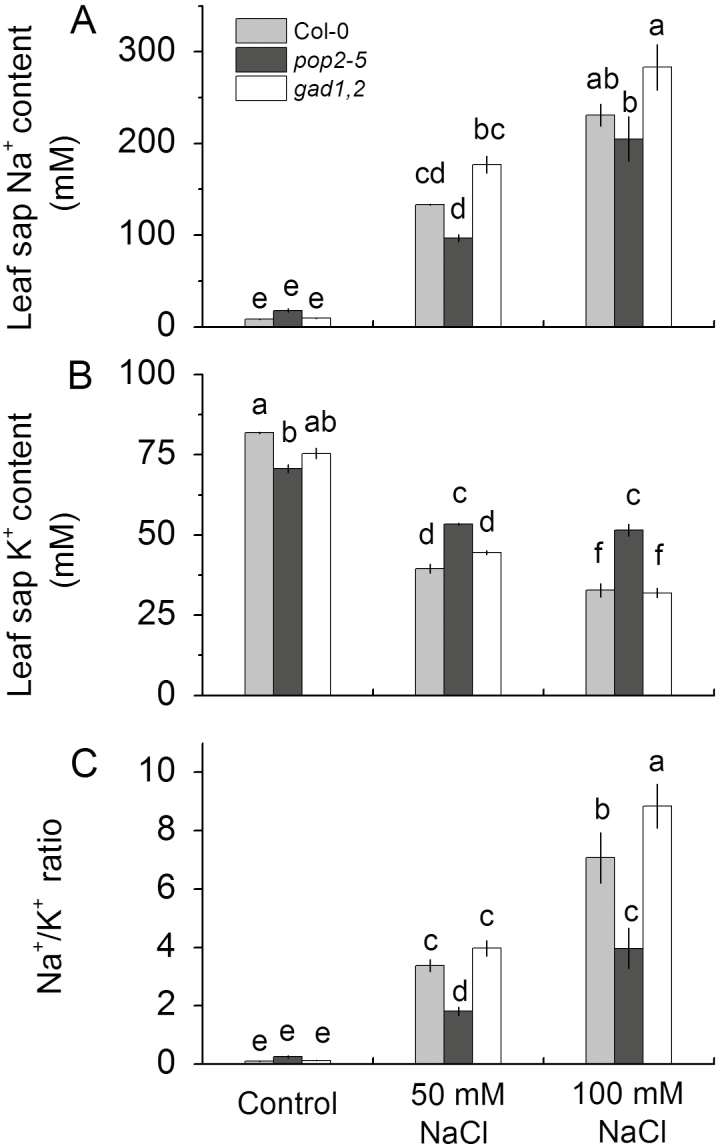
Because the different growth responses to NaCl treatment reported in Figs 2 and 3 may result from difference in their ability to prevent accumulation of Na+ in the shoot, we tested this hypothesis. As shown in Fig. 4, the leaf sap Na+ concentration was not different between Col and mutant plants under control condition, while under 50 and 100 mM NaCl treatments, the Na+ content was significantly higher in gad1,2 than that in Col and reduced in pop2-5. On the contrary, when subjected to salt stress, pop2-5 accumulated considerably more K+ in leaf than did gad1,2 and Col, though there was no difference between Col and gad1,2 (Fig. 4). These difference in Na+ and K+ content resulted in a substantially lower Na+/K+ ratio in pop2-5 under salt stress, compared with both WT and gad1,2 (Fig. 4C).
Fig. 4.
The leaf sap Na+ (A) and K+ (B) content and shoot Na+/K+ ratio (C) of Arabidopsis Col, pop2-5, and gad1,2 plants grown at various NaCl levels for 2 weeks. Salinity stress was applied to 4-week-old pot-grown plants. Data are mean ±SE (n=6). Data labelled with different lowercase letters are significantly different at P<0.05 level.
Elevated GABA levels improve root cell viability under conditions of oxidative and saline stress
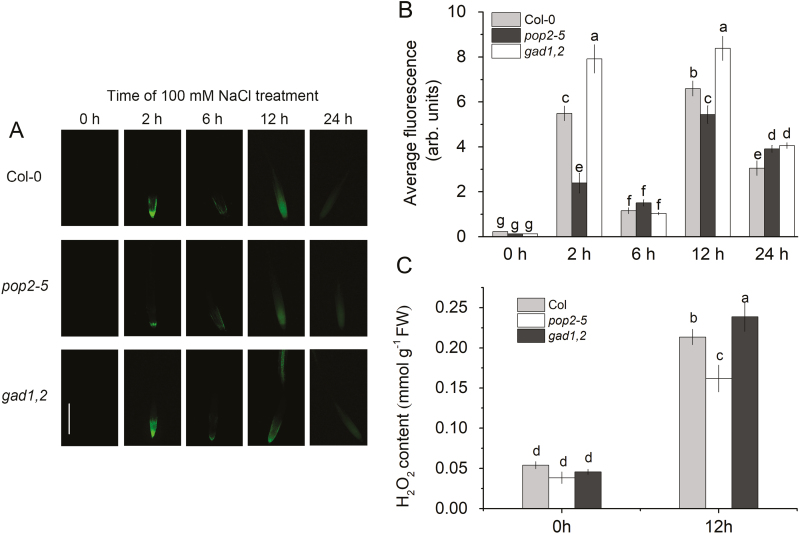
Salinity stress results in a rapid accumulation of reactive oxygen species in plant roots, in a time-dependent and tissue-specific manner (Miller et al., 2010; Nguyen et al., 2017). Accordingly, we have quantified effects of salinity on H2O2 production in Arabidopsis roots at different times after stress exposure using two different methods. In vivo detection using fluorescent dye imaging revealed a biphasic response in H2O2 production, with two peaks observed at 2 and 12 h (Fig. 5A, B). Here, the H2O2 production was highest in gad1,2 and lowest in pop2-1, suggesting a possible causal link between the amount of accumulated GABA and NaCl-induced reactive oxygen species (ROS) production in plant roots. These results were fully consistent with in vitro quantification of amount of accumulated H2O2 (Fig. 5C). Here, root H2O2 content increased about 4-fold (from ~0.05 to ~0.2 mmol g−1 FW) within 12 h of salinity treatment, with pop2-5 plants showing significantly lower H2O2 levels compared with two other lines.
Fig. 5.
Detection of H2O2 production in the roots in vivo (A, B) and in vitro (C) of Col-0, pop2-5, and gad1,2 seedlings. (A, B) Four-day-old seedlings were treated with 100 mM NaCl for different time (0–24 h). Root samples were stained with 2′,7′-dichlorofluorescein diacetate for imaging under a fluorescence microscope. (A) Representative images. (B) Mean fluorescence intensity from elongation zone. (C) In vitro quantification of root H2O2 content at two time points by TiCl4 method. Plants were first grown in Petri dishes for 1 week in half-strength MS medium with 2% w/v phytogel and then transferred into quarter-strength Hoagland nutrient solution for 3 weeks. Seedlings were then treated with or without 100 mM NaCl for 12 h. Data are mean ±SE (n=6). Data labelled with different lowercase letters are significantly different at P<0.05 level.
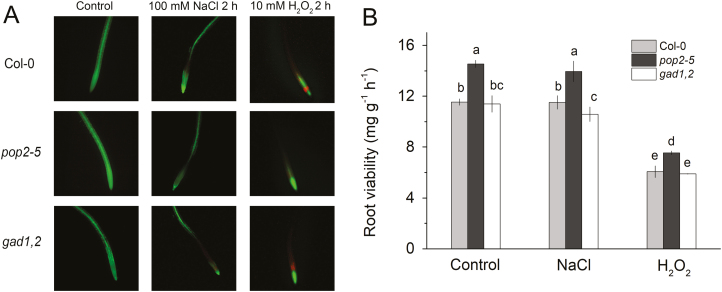
Both salinity and oxidative stress affect root cell viability (Jayakannan et al., 2015). Accordingly, 4-day-old Arabidopsis seedlings were exposed to either 100 mM NaCl or 10 mM H2O2 treatments for 2 h, and then double stained with a fluorescein diacetate–propidium iodide dyes (Fig. 6A). Under the fluorescence microscope, viable cells fluoresced bright green, whereas dead or damaged cells fluoresced bright red. No dead or damaged cells were found in the root tips in either line without NaCl treatment. Salinity exposure for 2 h resulted in a loss of cell viability in the root apex (less green signal), with gad1,2 roots being affected most. The bright red signal appeared when the roots were treated with H2O2 for 2 h; this signal was strongest in WT and gad1,2 roots, and was absent in pop2-5 seedlings (Fig. 6A). The root viability was also measured by the TTC method. As shown in Fig. 6B, under all conditions pop2-5 plants possessed the highest root viability. Two hours of NaCl treatment induced a decrease in a root viability only in gad1,2, with no changes observed in Col and pop2-5. While H2O2 inhibited root viability in all three lines; still the pop2-5 line performed best of the three lines.
Fig. 6.
Viability staining (A) and quantification (B) of Arabidopsis Col-0, pop2-5, and gad1,2 roots. (A) Four-day-old plants were treated with either 100 mM NaCl or 10 mM H2O2 for 2 h and then double stained with fluorescein diacetate–propidium iodide for imaging under a fluorescence microscope. One (of six) typical images is shown for each treatment. (B) Root viability quantified by TTC method. Arabidopsis seedlings were cultivated in quarter-strength Hoagland solution for 4 weeks and then roots were treated with 100 mM NaCl or 10 mM H2O2 for 2 h. Data are mean ±SD (n=3). Data labelled with different lowercase letters are significantly different at P<0.05 level.
Higher GABA content results in the optimal Na+/K+ ratio in the root apex under saline stress conditions
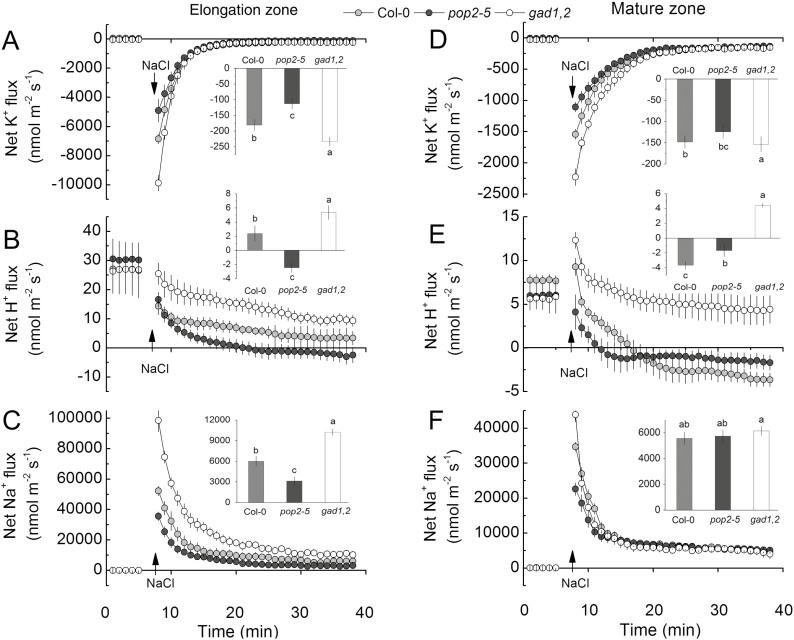
The non-invasive microelectrode MIFE technique was further used to measure kinetics of NaCl-induced fluxes of K+, H+, and Na+ in the roots of Arabidopsis GABA mutants. Addition of 100 mM NaCl to the bath induced a massive efflux of K+, both in the elongation (Fig. 7A) and mature (Fig. 7B) root zones. Responses from the elongation zone were 4-fold stronger, indicating their higher sensitive to salinity. The peak K+ efflux followed the trend pop2-5<WT<gad1,2 (Fig. 7A) indicating a possible causal link with the level of endogenous GABA accumulated in roots. The difference in steady-state K+ flux was significant (at P<0.05) in the elongation but not the mature root zone, 30 min after stress onset (see insets in Fig. 7A, B). Salinity treatment also shifted H+ flux towards net efflux (in both zones; Fig. 7C, D). This shift was smallest in the gad1,2 mutant. As a result, 30 min after salinity exposure, net H+ efflux was measured in the pop2-5 mutant in both zones while gad1,2 roots showed net H+ uptake (Fig. 7C, D). Responses in WT Col plants were in between. NaCl addition also caused a significant transient increase in the net Na+ influx in all genotypes in both elongation and mature zones. The peak value of Na+ influx in gad1,2 was nearly 2-fold higher than that in pop2-5, with Col plants in between (Fig. 7E, F). The steady state Na+ uptake was statistically different in plant roots after 30 min of exposure (pop2-5<WT<gad1,2) both in the elongation and in the mature zones (Fig. 7E, F). The above observations obtained for pop2-5 and gad1,2 lines were further confirmed in experiments using two additional mutant lines (pop2 and gad1). As shown in Supplementary Fig. S5, for NaCl-induced K+ loss from the elongation zone, pop2<WT<gad1, while the opposite trend was observed for net Na+ uptake (see Supplementary Fig. S5). Taken together, these results indicate that the differential ability of Arabidopsis plants to produce GABA has strongly affected plant ionic relations in the elongation root zone, with plants producing more GABA being able to maintain a more optimal Na+/K+ ratio in the root apex.
Fig. 7.
Transient K+, H+ and Na+ fluxes measured from the elongation zone (A–C) and mature (D–F) root zone of 4-day-old Col, pop2-5, and gad1,2 seedlings in response to 100 mM NaCl treatment. The insets show steady fluxes after 30 min exposure to 100 mM NaCl. Data are mean ±SE (n=6). The sign convention is ‘efflux negative’. Data labelled with different lowercase letters are significantly different at P<0.05 level.
H2O2-induced changes in net K+ >flux
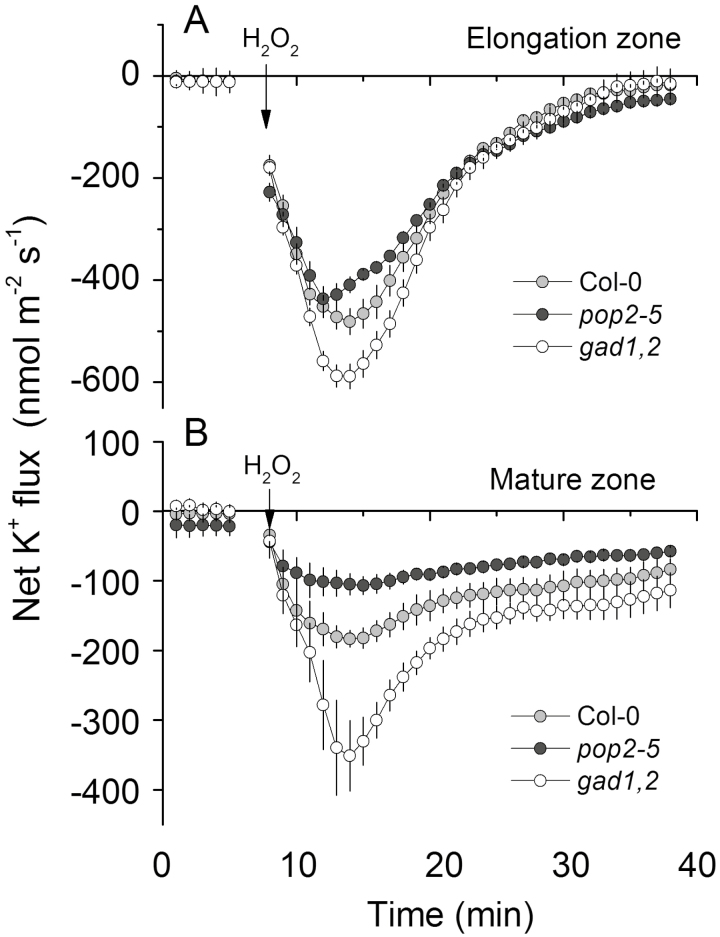
As shown in Fig. 6, the three genotypes respond differently to H2O2 treatment. Thus, we measured H2O2-induced changes in K+ flux. Similar to previous reports (Wang et al., 2018), application of 10 mM H2O2 caused K+ efflux from both the elongation and the mature zone of the root in all three Arabidopsis genotypes (Fig. 8). This H2O2-induced K+ efflux was increased gradually over time, reaching peak values after 5–7 min of stress treatment. The K+ efflux then gradually decreased but remained negative in the next 20 min. Compared with the response in the elongation zone, cells in the mature zone were less sensitive to H2O2 and exhibited a lower K+ efflux level. Similarly, in both zones, the magnitude of K+ efflux was the lowest in pop2-5 and the highest in gad1,2, and they followed the sequence gad1,2>WT>pop2-5.
Fig. 8.
Transient K+ fluxes measured from the elongation (A) and mature (B) root zones of the 4-day-old Col, pop2-5, and gad1,2 seedlings in response to acute 10 mM H2O2 treatment. Data are mean ±SE (n=6). The sign convention is ‘efflux negative’.
GABA-overproducing pop2-5 plants maintain more negative membrane potential values in root epidermis when exposed to salinity
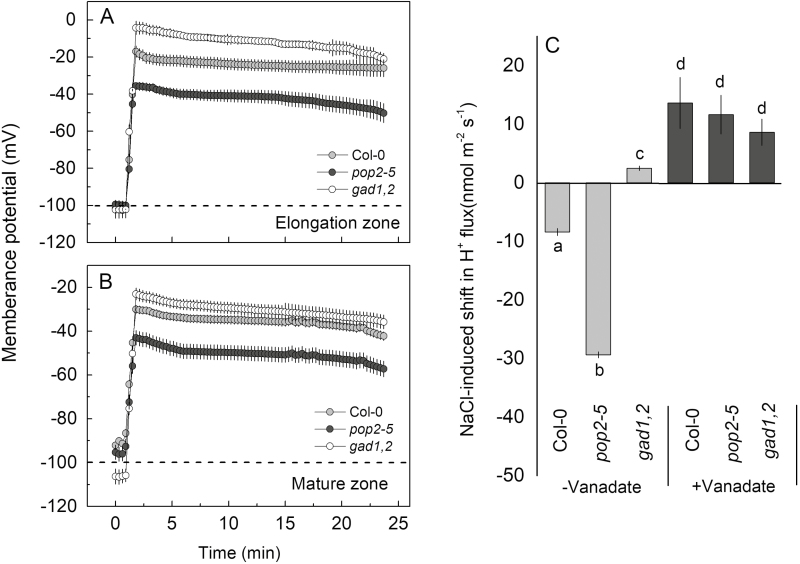
Next the effect of NaCl on membrane potential in the different lines was analysed. The resting membrane potential in the elongation and mature zones in the three genotypes was nearly the same, approximately −100 mV, with no clear differences noted. NaCl addition induced a quick depolarization in a pronounced genotype-specific manner. The maximum depolarization was observed in gad1,2, especially in the elongation zone, followed by Col and then by pop2-5. The latter showed a minimum depolarization in both zones (Fig. 9A, B).
Fig. 9.
(A, B) Salinity-induced changes in the plasma membrane potential in roots of 4-day-old Col-0, pop2-5, and gad1,2 seedlings. Membrane potential was measured in the elongation (A) and mature (B) root zones of plants exposed to 100 mM NaCl. (C) Effect of the H+-ATPase inhibitor sodium orthovanadate (Van) on the shift in net H+ fluxes induced by NaCl measured from mature root zone. Light grey bars: changes in net H+ fluxes from roots without sodium orthovanadate treatment; dark grey bars: changes in net H+ flux values from roots with sodium orthovanadate pre-treatment. Data are mean ±SE (n=5).
In plant systems, membrane potential is maintained primarily by operation of the plasma membrane-based H+-ATPase. In both WT and pop2-5 mutant, the measured NaCl-induced shift towards H+ efflux was completely suppressed by vanadate (Fig. 9C) suggesting that GABA operates upstream of H+-ATPase.
Stress-induced transcriptional changes
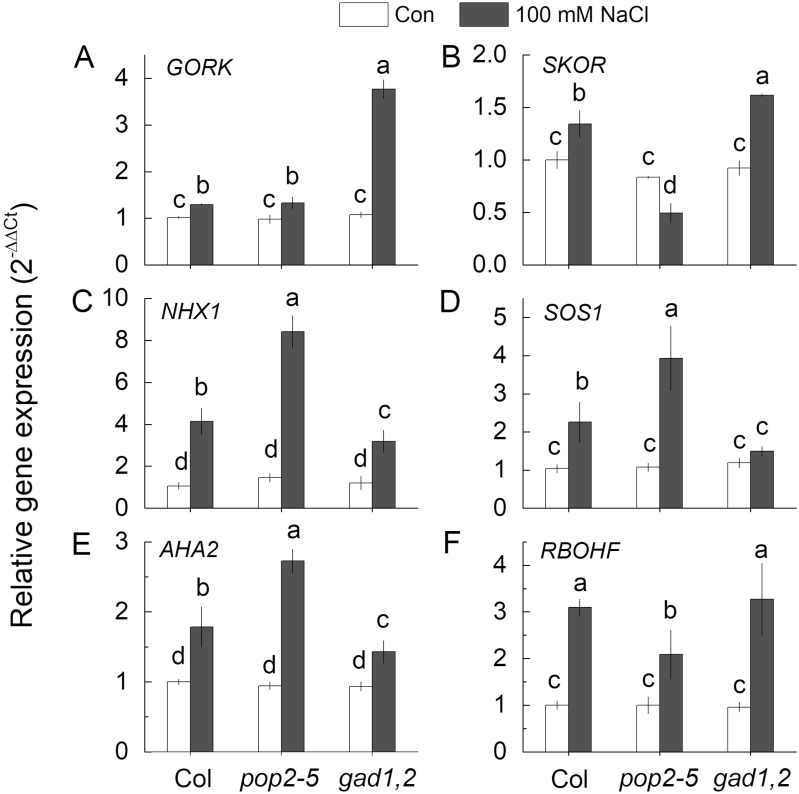
The relative expressions of the key genes mediating cellular K+ and Na+ homeostasis were compared between the three genotypes. This included genes encoding: outward-rectifying K+ efflux channels (GORK and SKOR); Na+/H+ exchangers (SOS1 and NHX1); H+-ATPase (AHA2); and ROS production (RBOHF). Under control condition, the expression of these genes in Col, pop2-5, and gad1,2 did not show any significant difference (at P<0.05), while NaCl treatment up-regulated their transcription levels by different magnitudes (Fig. 10). The expression of GORK was significantly up-regulated by NaCl treatment in all three types, but it was nearly 3-fold higher in gad1,2 than in Col and pop2-5 (Fig. 10A). After salt treatment, the transcription of SKOR was increased particularly in Col and gad1,2, but was decreased in pop2-5 (Fig. 10B). The expression levels of NHX1, SOS1, and AHA2 showed the same trend, with pop2-5 plants showing the highest increase followed by Col upon salinity exposure, while gad1,2 showed only a modest increase (Fig. 10C–E). The RBOHF transcription level was up-regulated by NaCl treatment in all types, with pop2-5 being least responsive (Fig. 10F).
Fig. 10.
Relative gene expression of GORK, SKOR, NHX1, SOS1, AHA2, and RBOHF in the roots of 4-week-old Col-0, pop2-5, and gad1,2 seedlings under control and saline conditions. Plants were first grown in Petri dishes for 1 week in half-strength MS medium with 2% w/v phytogel and then transferred into quarter-strength Hoagland nutrient solution for 3 weeks. Seedlings were then treated with 100 mM NaCl for 12 h under standard condition (day/night cycle, 16/8 h; light intensity 100–150 mmol m−2 s−1; temperature day/night, 25/20 °C). Data are mean values of three independent experiments with each containing three replicates. Data labelled with different lowercase letters are significantly different at P<0.05 level.
Discussion
Higher GABA accumulation results in salt-tolerant phenotype
As expected, the pop2-5 mutant accumulated more GABA in the roots (more than 2-fold under control and 3–4-fold upon salt stress than Col), and the GAD-deficient mutant gad1,2 accumulated much less GABA, which even could not be induced by NaCl (Fig. 1A). As a result, the difference in the GABA content between these two mutants was about 20-fold in plants exposed to the highest salinity level used (Fig. 1A). This difference in the endogenous GABA content was reflected in differential sensitivity to salinity stress, with pop2-5 having a salt-tolerant and gad1,2 a salt-sensitive phenotype (Figs 2–4). These findings are in contrast to the results from Renault et al. (2010), in which pop2-1 was oversensitive to salt tress. The reason for this discrepancy is not clear. One of the obvious differences is that those authors used Arabidopsis Ler ecoptype while our work was in the Columbia background. Also, the GABA content in their pop2-1 mutant roots exposed to salinity treatment was more than 40 μmol g−1 DW (Renault et al., 2010), while in our experiments the reported numbers were 2-fold lower (2 μmol g−1 FW, or 20 μmol g−1 DW, under 100 mM NaCl; Fig. 1A). Therefore, one possible explanation for this discrepancy may be that GABA may efficiently operate only in a certain concentration ‘window’, similar to that reported for other growth regulators such as polyamines (Pandolfi et al., 2010). If the endogenous GABA concentration exceeds a certain threshold, deleterious or pleiotropic effects may be observed. Also, in their work Renault et al. (2010) used only one knock-out GABA-overproducing line, while our study used the gad1,2 mutant with a lost ability for GABA biosynthesis, which showed increased sensitivity to NaCl.
Moreover, our results are also in a good agreement with reports that the gad1,2 mutant is more sensitive to drought stress compared with WT (Mekonnen et al., 2016) and that GABA priming improved osmotic stress tolerance in Piper nigrum (Vijayakumari and Puthur, 2016) and creeping bentgrass (Agrostis stolonifera; Li et al., 2017). Given the fact that salinity is often called a ‘physiological drought’, these and our data suggest that the work by Renault et al. (2010) is an exception from the general trend, and that higher GABA levels are beneficial for salinity stress tolerance in plants.
GABA improves salinity stress tolerance by conferring optimal Na+/K+ ratio in plant tissues
Maintaining cellular ion homeostasis is an important adaptive trait of glycophytes under salt stress (Munns and Tester, 2008; Yang and Guo, 2018), with higher intracellular K+/Na+ ratios being associated with higher salinity stress tolerance (Yue et al., 2012). When Arabidopsis seedlings were subjected to NaCl stress, the highest Na+ and lowest K+ contents were detected in the leaves of gad1,2; the opposite trend was found for pop2-5 seedlings. As a result, the leaf sap Na+/K+ ratio was gad1,2,>WT>pop2-5 (Fig. 4), correlating with the amount of GABA accumulated in plant tissues (Fig. 1).
The lower Na+ accumulation in the shoots may be due to several factors including (i) lower net Na+ uptake by roots; (ii) reduced rate of Na+ transport to the shoot; and (iii) stronger Na+ retrieval from the shoot (Munns and Tester 2008; Jayakannan et al., 2015). To provide insights into this process, the transient net Na+ flux was measured from plant roots upon salinity exposure. The magnitude of net Na+ uptake in gad1,2 was more than 2-fold that in pop2-5 (Fig. 7E, F), explaining the difference in tissue Na+ content.
When the Na+ is absorbed and transported into shoots, the capability of excluding Na+ from cytoplasm to apoplast and partitioning vacuolar Na+ into the vacuole becomes a primary adaptive mechanism for reducing cytoplasmic ion toxicity in plants grown under high salt concentration (Hasegawa et al., 2000). The plasma membrane Na+/H+ antiporter SOS1 is a key determinant of Na+ transport from the cytoplasm to the apoplast (Shi et al., 2000). The SOS1 operation is driven by the proton gradient created by the plasma membrane H+-ATPase encoded by the AHA genes. The vacuolar Na+ compartmentalization is mediated by the vacuolar Na+/H+ exchanger (NHX1) (Brini and Masmoudi, 2012), and a constitutive overexpression of AtNHX1 and its orthologs in Arabidopsis led to increased plant salinity tolerance (Apse et al., 1999). Here, we observed that under salt stress, the expressions of SOS1, NHX1 and AHA2 in roots were all up-regulated in the pop2-5 mutant with elevated GABA content (Fig. 10). At the same time, gad1,2 showed little increase in the transcription levels of NHX1 and AHA2, and SOS1 transcript levels barely changed (Fig. 10C–E). These results are consistent with the scenario that the salt-tolerant pop2-5 mutant had an enhanced ability to reduce accumulation of cytotoxic Na+ through two concurrent mechanisms. One is a superior Na+ exclusion to the apoplast by SOS1, fueled by the higher plasma membrane H+-ATPase activity. Supporting this notion is our observation of smaller net Na+ uptake in plant roots and more negative H+ flux values (indicating a higher rate of H+-ATPase operation) in the GABA-overproducing pop2-5 mutant. The second mechanism may include better Na+ compartmentation into the vacuole (conferred by a higher level of NHX transcripts). All these features were lost (less functional) in the salt-oversensitive gad1,2 mutant.
The mechanisms by which GABA levels control expression and activity of SOS1 and NHX1 genes remains to be determined. It was shown previously that GABA acts as a signaling molecule during drought stress by binding to GABA receptors (ALMT proteins) and also modulates the activity of H+-ATPase via 14-3-3 proteins, thereby regulating stomatal movement (Mekonnen et al., 2016). Given the fact that SOS1 is relieved from auto-inhibition upon phosphorylation of the auto-inhibitory domain by the SOS2–SOS3 kinase complex (Quintero et al., 2011), the phosphorylation model of GABA regulation of SOS1 and NHX1 operation is plausible.
Another way of achieving a more optimal cytosolic Na+/K+ ratio is to increase tissue K+ content. In this context, cytosolic K+ retention under salt stress is now firmly established as a key trait associated with salinity stress tolerance (Shabala and Cuin, 2008; Wu et al., 2015; Shabala et al., 2016). Under saline conditions, the massive entry of the positively charged Na+ ions cause dramatic depolarization of the plasma membrane (Fig. 9A, B). Salt-tolerant genotypes typically show intrinsically more negative membrane potential values compared with their salt-sensitive counterparts (Chen et al., 2007a; Smethurst et al., 2008). Consistent with this, the root membrane potential of pop2-5 was depolarized by about 60 mV, while this depolarization was nearly 100 mV in gad1,2, in the elongation zone. Also obvious was the difference in membrane potential depolarization between pop2-5 and gad1,2 in the mature zone (Fig. 9A, B).
In higher plants, the plasma membrane H+-ATPase is considered to be a major contributor to the maintenance of the membrane potential (Palmgren and Nissen, 2011). Here we showed that NaCl treatment shifted H+ fluxes towards net H+ efflux in both zones. This shift was much stronger in pop2-5 then in gad1,2 (Fig. 7C, E), and was prevented by vanadate, a known inhibitor of the H+-ATPase (Fig. 9C), indicating the involvement of the H+-ATPase as a downstream target of GABA signaling. The substantial NaCl-induced depolarization reported here (Fig. 9A, B) resulted in an immediate K+ leak via depolarization-activated outward rectifying K+ GORK (AT5G37500) channels, as shown in experiments with knock-out gork lines (Shabala and Cuin, 2008). The reported difference in membrane potential values between gad1,2 and pop2-5 lines (about 30 mM less negative in salt-sensitive gad1,2; Fig. 9A, B) may explain a 2-fold higher K+ efflux in this genotype. A stronger depolarization in the elongation zone may also explain a higher extent of K+ efflux in this tissue. The process was further exacerbated by NaCl-induced upregulation of GORK transcript levels (Fig. 10A). This upregulation was only 30% in the salt-tolerant pop2-5 but nearly 4-fold in the salt-sensitive gad1,2.
GABA accumulation mitigates salt stress through ROS pathway
Differential GABA accumulation in plant roots also affected roots’ sensitivity to ROS, as indicated by both viability staining (Fig. 6) and the extent of sensitivity of K+ transport systems to H2O2 (Fig. 8). The role of ROS in plant signaling and adaptation to salinity stress (as well as other stresses) is not straightforward. Low ROS concentrations can function as a signal that activate salt-stress responses, while high ROS concentrations damage proteins, lipids, DNA, and carbohydrates (Miller et al., 2010). Amongst other sources, apoplastic ROS production by NADPH oxidase is considered to be a major source of salt-stress-induced increase in ROS levels in plant roots (Kaye et al., 2011; Bose et al., 2014; Ben Rejeb et al., 2015). In plants, NADPH oxidase is encoded by Rboh/Nox genes (Marino et al., 2012; Morales et al., 2016). Amongst 10 Nox genes in Arabidopsis, RbohD and RbohF are strongly up-regulated under salt stress (Ma et al., 2012). Moreover, a RbohF mutant showed strong Na+ hypersensitivity in its shoots (Jiang et al., 2012). In our experiments, the amount of H2O2 in the roots of salt-tolerant pop2-5 seedlings was minimal, while salt-sensitive gad1,2 produced much more H2O2 than did pop2-5 at 2 and 12 h after salt treatment (Fig. 5). Consistent with this, the expression of RBOHF in the shoot was also significantly higher in gad1,2 compared with Col and pop2-5 under salt stress (Fig. 10F), indicating that loss of GABA accumulation under salt stress resulted in more ROS production in both roots and shoots. This is in good agreement with reports by Shi et al. (2010) who showed that GABA treatment resulted in a lower NaCl-induced H2O2 production in the legume species Caragana intermedia.
Our previous experiments showed that higher H2O2-induced K+ efflux in cereals, such as barley and wheat, correlated with their lower salinity tolerance (Chen et al., 2007b; Maksimovic et al., 2013; Wang et al., 2018). Here we show that salt-tolerant pop2-5 showed lowest K+ leak when treated with H2O2, while the highest K+ efflux was observed in the roots of the salt-sensitive gad1,2 line (Fig. 8). Apoplastic H2O2 is known to interact with transition metals (Cu2+ and Fe2+) in the cell wall, leading to formation of hydroxyl radicals (Demidchik, 2014). The latter were shown to be able directly activate GORK channels (Demidchik et al., 2010), prompting massive K+ efflux from Arabidopsis roots. In this context, stronger induction of net K+ efflux by exogenous application of H2O2 in the salt-sensitive gad1,2 line may be explained in several possible ways. First, these plants may possess intrinsically lower levels of endogenous antioxidants preventing hydroxyl radical formation. Second, the relative availability of Fe or Cu in the apoplast, or their reducing power, may differ between lines. Finally, there is the possibility of desensitization of GORK channels to ∙OH−. Future studies are required to differentiate between these possibilities.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Relative gene expressions of GAD1–GAD5 and POP2 in the roots of 4-week-old Col-0, pop 2-5, and gad1,2 seedlings.
Fig. S2. The homozygosis of gad1 and pop2.
Fig. S3. Phenotypes of pop2, gad1, and Col-0 Arabidopsis seedlings treated with and without 100 mM NaCl.
Fig. S4. Exogenous GABA application rescues salt-sensitive gad1,2 phenotype.
Fig. S5. Transient Na+, K+, and H+ fluxes measured from the elongation zone and mature root zone of 4-day-old Col-0, pop2, and gad1 seedlings in response to 100 mM NaCl treatment.
Table S1. The sequences of primers for qPCR.
Acknowledgements
This work was supported by National Natural Science Foundation of China (NSFC, No.31772360) to JC; Australian Research Council grant to SS; and funding from Foshan University to SS and MY. The co-first author QW was a recipient of China Scholarship Council (CSC). We thank M. Reichelt for support in GABA analysis.
References
- Apse MP, Aharon GS, Snedden WA, Blumwald E. 1999. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 85, 1256–1258. [DOI] [PubMed] [Google Scholar]
- Ben Rejeb K, Lefebvre-De Vos D, Le Disquet I, Leprince AS, Bordenave M, Maldiney R, Jdey A, Abdelly C, Savouré A. 2015. Hydrogen peroxide produced by NADPH oxidases increases proline accumulation during salt or mannitol stress in Arabidopsis thaliana. New Phytologist 208, 1138–1148. [DOI] [PubMed] [Google Scholar]
- Bose J, Shabala L, Pottosin I, Zeng F, Velarde-Buendía AM, Massart A, Poschenrieder C, Hariadi Y, Shabala S. 2014. Kinetics of xylem loading, membrane potential maintenance, and sensitivity of K+-permeable channels to reactive oxygen species: physiological traits that differentiate salinity tolerance between pea and barley. Plant, Cell & Environment 37, 589–600. [DOI] [PubMed] [Google Scholar]
- Bose J, Xie Y, Shen W, Shabala S. 2013. Haem oxygenase modifies salinity tolerance in Arabidopsis by controlling K⁺ retention via regulation of the plasma membrane H⁺-ATPase and by altering SOS1 transcript levels in roots. Journal of Experimental Botany 64, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Fait A, Zik M, Fromm H. 2004. The root-specific glutamate decarboxylase (GAD1) is essential for sustaining GABA levels in Arabidopsis. Plant Molecular Biology 55, 315–325. [DOI] [PubMed] [Google Scholar]
- Brini F, Masmoudi K. 2012. Ion transporters and abiotic stress tolerance in plants. ISRN Molecular Biology 3, 927436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Cuin TA, Zhou M, Twomey A, Naidu BP, Shabala S. 2007a Compatible solute accumulation and stress-mitigating effects in barley genotypes contrasting in their salt tolerance. Journal of Experimental Botany 58, 4245–4255. [DOI] [PubMed] [Google Scholar]
- Chen Z, Pottosin II, Cuin TA, et al. 2007. b Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiology 145, 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemensson-Lindell A. 1994. Triphenyltetrazolium chloride as an indicator of fine-root vitality and environmental stress in coniferous forest stands: Applications and limitations. Plant and Soil 159, 297–300. [Google Scholar]
- Cuin TA, Tian Y, Betts SA, Chalmandrier R, Shabala S. 2009. Ionic relations and osmotic adjustment in durum and bread wheat under saline conditions. Functional Plant Biology 36, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Demidchik V. 2014. Mechanisms and physiological roles of K+ efflux from root cells. Journal of Plant Physiology 171, 696–707. [DOI] [PubMed] [Google Scholar]
- Demidchik V, Cuin TA, Svistunenko D, Smith SJ, Miller AJ, Shabala S, Sokolik A, Yurin V. 2010. Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. Journal of Cell Science 123, 1468–1479. [DOI] [PubMed] [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR. 2008. Highway or byway: the metabolic role of the GABA shunt in plants. Trends in Plant Science 13, 14–19. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. 2000. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology 51, 463–499. [DOI] [PubMed] [Google Scholar]
- Hossain MA, Hasanuzzaman M, Fujita M. 2010. Up-regulation of antioxidant and glyoxalase systems by exogenous glycinebetaine and proline in mung bean confer tolerance to cadmium stress. Physiology and Molecular Biology of Plants 16, 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Horie T. 2017. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annual Review of Plant Biology 68, 405–434. [DOI] [PubMed] [Google Scholar]
- Jayakannan M, Bose J, Babourina O, Shabala S, Massart A, Poschenrieder C, Rengel Z. 2015. The NPR1-dependent salicylic acid signalling pathway is pivotal for enhanced salt and oxidative stress tolerance in Arabidopsis. Journal of Experimental Botany 66, 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Belfield EJ, Mithani A, Visscher A, Ragoussis J, Mott R, Smith JA, Harberd NP. 2012. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. The EMBO Journal 31, 4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye Y, Golani Y, Singer Y, Leshem Y, Cohen G, Ercetin M, Gillaspy G, Levine A. 2011. Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiology 157, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Toda T, Yokota S, Zuraida D, Hara T. 1995. Effects of aluminium and pH on root growth and cell viability in Arabidopsis thaliana strain Landsberg in hydroponic culture. Plant and Cell Physiology 36, 201–205. [Google Scholar]
- Li Z, Yu J, Peng Y, Huang B. 2017. Metabolic pathways regulated by abscisic acid, salicylic acid and γ-aminobutyric acid in association with improved drought tolerance in creeping bentgrass (Agrostis stolonifera). Physiologia Plantarum 159, 42–58. [DOI] [PubMed] [Google Scholar]
- Ma L, Zhang H, Sun L, Jiao Y, Zhang G, Miao C, Hao F. 2012. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na⁺/K⁺ homeostasis in Arabidopsis under salt stress. Journal of Experimental Botany 63, 305–317. [DOI] [PubMed] [Google Scholar]
- Maksimovic JD, Zhang JY, Zeng FR, Zivanovic BD, Shabala L, Zhou MX, Shabala S. 2013. Linking oxidative and salinity stress tolerance in barley: Can root antioxidant enzyme activity be used as a measure of stress tolerance? Plant and Soil 365, 141–155. [Google Scholar]
- Marino D, Dunand C, Puppo A, Pauly N. 2012. A burst of plant NADPH oxidases. Trends in Plant Science 17, 9–15. [DOI] [PubMed] [Google Scholar]
- Mekonnen DW, Flügge UI, Ludewig F. 2016. Gamma-aminobutyric acid depletion affects stomata closure and drought tolerance of Arabidopsis thaliana. Plant Science 245, 25–34. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. 2010. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell & Environment 33, 453–467. [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Good AG. 2008. Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant & Cell Physiology 49, 92–102. [DOI] [PubMed] [Google Scholar]
- Morales J, Kadota Y, Zipfel C, Molina A, Torres MA. 2016. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. Journal of Experimental Botany 67, 1663–1676. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Nguyen HM, Sako K, Matsui A, Suzuki Y, Mostofa MG, Ha CV, Tanaka M, Tran LP, Habu Y, Seki M. 2017. Ethanol enhances high-salinity stress tolerance by detoxifying reactive oxygen species in Arabidopsis thaliana and rice. Frontiers in Plant Science 8, 1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Preuss D. 2003. Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114, 47–59. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Nissen P. 2011. P-type ATPases. Annual Review of Biophysics 40, 243–266. [DOI] [PubMed] [Google Scholar]
- Pandolfi C, Pottosin I, Cuin T, Mancuso S, Shabala S. 2010. Specificity of polyamine effects on NaCl-induced ion flux kinetics and salt stress amelioration in plants. Plant & Cell Physiology 51, 422–434. [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Martinez-Atienza J, Villalta I, Jiang XY, Kim WY, Ali Z, Fujii H, Mendoza I, Yun DJ, Zhu JK, Pardo JM. 2011. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proceedings of the National Academy of Sciences, USA 108, 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh SA, Tyerman SD, Gilliham M, Xu B. 2017. γ-Aminobutyric acid (GABA) signalling in plants. Cellular and Molecular Life Sciences 74, 1577–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh SA, Tyerman SD, Xu B, et al. 2015. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nature Communications 6, 7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault H, El Amrani A, Berger A, Mouille G, Soubigou-Taconnat L, Bouchereau A, Deleu C. 2013. γ-Aminobutyric acid transaminase deficiency impairs central carbon metabolism and leads to cell wall defects during salt stress in Arabidopsis roots. Plant, Cell & Environment 36, 1009–1018. [DOI] [PubMed] [Google Scholar]
- Renault H, Roussel V, El Amrani A, Arzel M, Renault D, Bouchereau A, Deleu C. 2010. The Arabidopsis pop2-1 mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biology 10, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi C, Nicoletti I. 2006. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nature Protocols 1, 1458–1461. [DOI] [PubMed] [Google Scholar]
- Rotman B, Papermaster BW. 1966. Membrane properties of living mammalian cells as studied by enzymatic hydrolysis of fluoregenic esters. Proceedings of the National Academy of Sciences, USA 55, 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz SS, Malabarba J, Reichelt M, Heyer M, Ludewig F, Mithöfer A. 2017. Evidence for GABA-induced systemic GABA accumulation in Arabidopsis upon wounding. Frontiers in Plant Science 8, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz SS, Reichelt M, Mekonnen DW, Ludewig F, Mithöfer A. 2015. Insect herbivory-elicited GABA accumulation in plants is a wound-induced, direct, systemic, and jasmonate-independent defense response. Frontiers in Plant Science 6, 1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala L, Ross T, McMeekin T, Shabala S. 2006. Non-invasive microelectrode ion flux measurements to study adaptive responses of microorganisms to the environment. FEMS Microbiology Reviews 30, 472–486. [DOI] [PubMed] [Google Scholar]
- Shabala S. 2013. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Annals of Botany 112, 1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Bose J, Fuglsang AT, Pottosin I. 2016. On a quest for stress tolerance genes: membrane transporters in sensing and adapting to hostile soils. Journal of Experimental Botany 67, 1015–1031. [DOI] [PubMed] [Google Scholar]
- Shabala S, Bose J, Hedrich R. 2014. Salt bladders: do they matter? Trends in Plant Science 19, 687–691. [DOI] [PubMed] [Google Scholar]
- Shabala S, Cuin TA. 2008. Potassium transport and plant salt tolerance. Physiologia Plantarum 133, 651–669. [DOI] [PubMed] [Google Scholar]
- Shabala S, Shabala L. 2002. Kinetics of net H+, Ca2+, K+, Na+, NH4+, and Cl − fluxes associated with post-chilling recovery of plasma membrane transporters in Zea mays leaf and root tissues. Physiologia Plantarum 114, 47–56. [DOI] [PubMed] [Google Scholar]
- Shelp BJ, Mullen RT, Waller JC. 2012. Compartmentation of GABA metabolism raises intriguing questions. Trends in Plant Science 17, 57–59. [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK. 2000. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences, USA 97, 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SQ, Shi Z, Jiang ZP, Qi LW, Sun XM, Li CX, Liu JF, Xiao WF, Zhang SG. 2010. Effects of exogenous GABA on gene expression of Caragana intermedia roots under NaCl stress: regulatory roles for H2O2 and ethylene production. Plant, Cell & Environment 33, 149–162. [DOI] [PubMed] [Google Scholar]
- Smethurst CF, Rix K, Garnett T, Auricht G, Bayart A, Lane P, Wilson SJ, Shabala S. 2008. Multiple traits associated with salt tolerance in lucerne: revealing the underlying cellular mechanisms. Functional Plant Biology 35, 640–650. [DOI] [PubMed] [Google Scholar]
- Steward FC, Thompson JF, Dent CE. 1949. γ-Aminobutyric acid: a constituent of the potato tuber? Science 110, 439–440. [Google Scholar]
- Van Cauwenberghe OR, Makhmoudov A, McLean MD, Clark SM, Shelp BJ. 2002. Plant pyruvate-dependent gamma aminobutyrate transaminase: identification of an Arabidopsis cDNA and its expression in Escherichia coli. Canadian Journal of Botany 80, 933–941. [Google Scholar]
- Vijayakumari K, Puthur JT. 2016. γ-Aminobutyric acid (GABA) priming enhances the osmotic stress tolerance in Piper nigrum Linn. plants subjected to PEG-induced stress. Plant Growth Regulation 78, 57–67. [Google Scholar]
- Wang H, Shabala L, Zhou M, Shabala S. 2018, Hydrogen peroxide-induced root Ca2+ and K+ fluxes correlate with salt tolerance in cereals: towards the cell-based phenotyping. International Journal of Molecular Sciences 19, 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widodo, Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U. 2009. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. Journal of Experimental Botany 60, 4089–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhu M, Shabala L, Zhou M, Shabala S. 2015. K+ retention in leaf mesophyll, an overlooked component of salinity tolerance mechanism: a case study for barley. Journal of Integrative Plant Biology 57, 171–185. [DOI] [PubMed] [Google Scholar]
- Yang Y, Guo Y. 2018. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytologist 217, 523–539. [DOI] [PubMed] [Google Scholar]
- Yue Y, Zhang M, Zhang J, Duan L, Li Z. 2012. SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. Journal of Plant Physiology 169, 255–261. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Y, Du Y, Chen S, Tang H. 2011. Dynamic metabonomic responses of tobacco (Nicotiana tabacum) plants to salt stress. Journal of Proteome Research 10, 1904–1914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.