Abstract
The capsular polysaccharide (CPS) structure of Campylobacter jejuni contributes to its robust fitness. Many strains contain heptose moieties in their CPS units. The precursor heptose is GDP-d-glycero-α-d-manno-heptose, from which modifications to the stereochemistry at C3, C4, C5, and C6, as well as additions of methyl and phosphoramidate groups lend to the hypervariability of the C. jejuni CPS structures. Synthesis of GDP-d-glycero-α-d-manno-heptose has been described previously, but using enzymes from Aneurinibacillus thermoaerophilus DSM 10155. Here we describe the complete synthesis of GDP-d-glycero-α-d-manno-heptose using enzymes from C. jejuni NTCC 11168: Cj1152, Cj1423, Cj1424, and Cj1425. Our results yield kinetic parameters for these enzymes and outline a successful strategy for milligram-gram scale synthesis of GDP-d-glycero-α-d-manno-heptose. This achievement is critical for the characterization of other carbohydrate tailoring enzymes, which are expected to utilize GDP-d-glycero-α-d-manno-heptose for the biosynthesis of more complex carbohydrates in the CPS of C. jejuni.
Graphical Abstract
Introduction
Campylobacter jejuni is a Gram-negative pathogenic bacterium commonly found in chickens and is the leading cause of bacterial gastrointestinal infections in the United States (1, 2). The enhanced pathogenicity of C. jejuni is due to its high adaptability, growing antibiotic resistance, and ability to evade the host immune response (2). These factors have made C. jejuni a specific target for the development of novel antimicrobial agents. Like many Gram-negative bacterial species, C. jejuni displays several carbohydrate-based structures on the surface of the cell, including lipooligosaccharides (LOS) and capsular polysaccharides (CPS). These polysaccharides are critical for the integrity and maintenance of the bacterial cell wall and the evasion of the host immune response upon infection (3, 4). Disrupting the synthesis of these carbohydrate structures drastically lowers the pathogenicity of C. jejuni and thus the enzymes responsible for the synthesis of these carbohydrates are potential therapeutic targets (5). Heptose residues are common components of both the LOS and CPS of many C. jejuni strains (3, 4).
The LOS structure contains three main components: the outer core comprised of short oligosaccharides, the inner core, and the lipid A anchor into the cellular membrane. Unlike the lipopolysaccharide (LPS) structures found in most bacteria, the LOS of C. jejuni lacks the O-antigen component normally found in the outer core region. The oligosaccharides of the outer core are highly variable among different C. jejuni strains with at least 12 different structures identified (6, 7). Typically, these structures contain sialic acids, which mimic the oligosaccharide structures found on gangliosides. This feature may help with the evasion of the host immune response and lead to the triggering of autoimmune mechanisms, which may result in Guillain-Barre syndrome (6, 8). Unlike the high variability of the outer core, the inner core region of the LOS is highly conserved among characterized serotypes and has been shown to be comprised of a single 3-deoxy-d-manno-octulosonic acid (Kdo) residue, which is attached to the lipid A anchor and two heptose carbohydrates (8, 9). The heptose residues have been shown to be l-glycero-d-manno-heptose, which is the same carbohydrate found in other Gram-negative bacteria, including Escherichia coli and Salmonella (9).
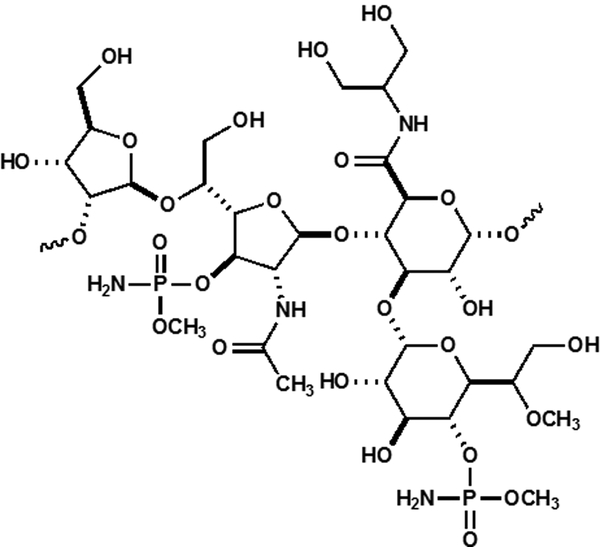
The capsular polysaccharide (CPS) is composed of repeating units of monosaccharides joined by glycosidic linkages. In C. jejuni, the CPS is critical for colonization and pathogenicity (10). The CPS is hypervariable among C. jejuni strains in which at least 47 different serotypes have been identified (2). The repeating CPS structure of C. jejuni NCTC 11168 is shown in Scheme 1. This CPS unit contains several unique features including a 2-amino-2-deoxyglycerol or ethanolamine derivative of glucuronic acid and two instances of O-methyl phosphoramidate modification. The CPS contains a single heptose moiety, identified as d-glycero-l-gluco-heptose (11, 12).
Scheme 1:
Structure of the CPS unit of C. jejuni NCTC 11168.
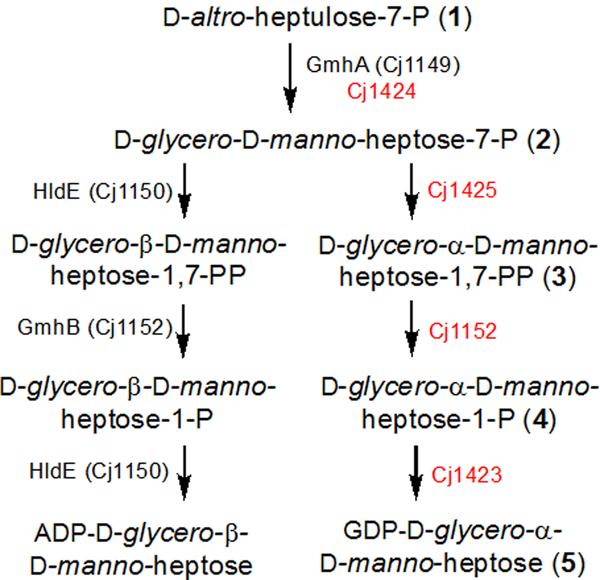
The CPS and LOS polysaccharides of C. jejuni contain heptose residues of different configurations, but the starting material, in all cases, is predicted to be subsequently transformed to d-glycero-d-manno-heptose by separate biosynthetic pathways (Scheme 2) (9, 13). The starting material is predicted to be d-altro-heptulose-7-P (more commonly known as d-sedoheptulose-7-P). The LOS pathway utilizes the β-anomer of ADP-d-glycero-d-manno-heptose, whereas the CPS pathway utilizes the α-anomer of GDP-d-glycero-d-manno-heptose. The enzymes used to synthesize ADP-d-glycero-β-d-manno-heptose are predicted to be Cj1149, Cj1150, and Cj1152. In the context of the LOS biosynthetic pathway, Cj1149 (GmhA) and Cj1152 (GmhB) have been experimentally investigated by knockout mutation analysis (5). The catalytic activity of Cj1150 (HldE) has been inferred based on the genomic context and sequence identity to previously characterized enzymes (9, 14).
Scheme 2.
Proposed synthesis of two heptose molecules found in the LOS and CPS.
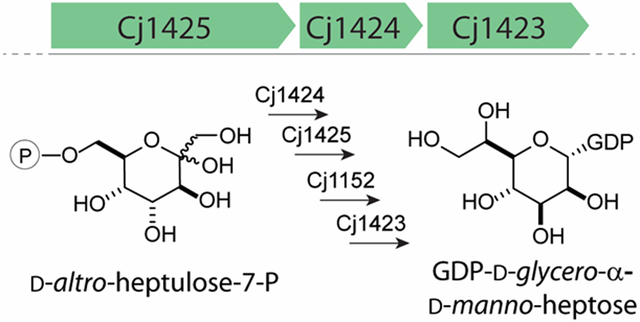
For the CPS pathway, several publications describe the biosynthesis of GDP-d-glycero-α-d-manno-heptose (15–18). However, these investigations have all used enzymes from Aneurinibacillus thermoaerophilus DSM 10155, where d-glycero-d-manno-heptose is found as a modification to an S-layer glycoprotein (13). The protein sequences of Cj1423, Cj1424, and Cj1425 share 48%, 70%, and 55% sequence identity with their A. thermoaerophilus homologues, HddC (Uniprot: Q9AGY6), GhmA (Uniprot: Q9AGY7), and HddA (Uniprot: Q9AGY8), respectively. The genomic context of the C. jejuni NCTC 11168 enzymes and A. thermoaerophilus homologues differ as well. C. jejuni NCTC 11168 lacks a nearby phosphatase enzyme and contains a second heptose isomerase enzyme, GhmA (Cj1149), in its genome (5). Interestingly, Cj1149 and Cj1424 only share 46% sequence identity, but were predicted to catalyze the same reaction on the same substrate. Given these differences, we decided to experimentally characterize the catalytic functions of Cj1423, Cj1424, Cj1152, and Cj1425. Moreover, the product of the Cj1423 through Cj1425 catalyzed reaction pathway is anticipated to be GDP-d-glycero-α-d-manno-heptose, which appears to be the starting material for all of the known heptoses used in the CPS of C. jejuni (19). Therefore, it is essential that significant amounts of this product can be produced such that the biosynthetic pathways for the other heptoses in C. jejuni can be elucidated. Here, we outline our bioinformatic analysis of the enzymes responsible for the biosynthesis of the heptose moieties found in the CPS of C. jejuni, establish that the starting material of this pathway is d-altro-heptulose-7-P, and present a preparative method for enzymatic synthesis of GDP-d-glycero-α-d-manno-heptose.
Materials and Methods
Materials and Equipment.
All materials used in this study were obtained from Sigma-Aldrich, Carbosynth, or GE Healthcare Bio-Sciences, unless stated otherwise. E. coli strains XL1 Blue, Rosetta (DE3), and BL21-Gold (DE3) were obtained from New England Biolabs. NMR spectra were collected on a Bruker Avance III 400 MHz system equipped with a broadband probe and sample changer. Mass spectrometry samples were collected on an MDS-Sciex 4000 Qtrap system or a Thermo Scientific Q Exactive Focus system run in the negative ion mode. UV spectra were collected on a SpectraMax340 UV-visible plate reader using 96-well NucC plates. d-Mannoheptulose and l-galactoheptulose were obtained from Emmanuel Zissis via the laboratory of W. W. Cleland (University of Wisconsin-Madison). l-Galactoheptulose-7-P was synthesized as previously described (20).
Bioinformatic Analysis of Heptose Formation in C. jejuni.
The protein FASTA sequences for 484 completely sequenced C. jejuni strains were obtained from the NCBI assembly database (21). These genomes were compiled into a custom C. jejuni sequence database using the command-line applications provided by NCBI (22). The protein sequences for Cj1423, Cj1424, Cj1425, and Cj1152 were used as BLAST search queries against the custom database containing all protein FASTA sequences for the 484 strains of C. jejuni with only the top result returned. Sequence identity comparisons for all four genes were used to draw conclusions about the molecular structure of the CPS for individual C. jejuni strains.
Sequence Similarity Networks and Genome Neighborhood Analysis.
Sequence similarity networks (SSN) were generated by directly submitting FASTA sequences to the EFI-EST webtool (23). Genome neighborhoods were generated by submitting the UniProt codes for KpsF and KpsC of the various strains to the EFI-GNT webtool (24). If UniProt codes for KpsF or KpsC for the strain of interest were unavailable (e.g. HS3), the genome neighborhood was constructed manually using the available sequence information gathered from the NCBI database. All network layouts were created and visualized using Cytoscape 3.4 (25).
Cloning of cj1423, cj1424, cj1425, and cj1152.
The DNA for cj1423 (Uniprot ID: Q0P8J1) was chemically synthesized and codon optimized by Genscript (Piscataway, NJ). Initial expression tests with the non-codon optimized gene showed little to no expression in BL21-Gold (DE3) or Rosetta (DE3) cell lines. From the chemically synthesized DNA fragment, cj1423 was amplified using the primer pair:
5’- ATATAGGATCCATGCAGGCGATCATTC-3’
5’- GCATGCTCGAGTTAGTTGTTAATCAGAAAGTGATAGTAATC -3’
The gene for cj1424 (Uniprot ID: Q0P8J0) was amplified using the primer pair:
5’- AGCGCGGATCCATGGAGAATTTAAATTCCTATATAAAAGGACAT-3’
5’- TATGCCTCGAGTTATTGCTTGCAAGAAAAACCTTTACCA-3’
The gene for cj1425 (Uniprot ID: Q0P8I9) was amplified using the primer pair:
5’- AGCGCGGATCCATGGAGAATTTAAATTCCTATATAAAAGGACAT -3’
5’- TATGCCTCGAGTTAAATTCTCCATGATTTAACCCCCTC -3’
The gene for cj1152 (Uniprot ID: Q6TG07) was amplified using the primer pair:
5’-GCGCGGGATCCATGTTTATTTTCTTTTTAAAAAAGCTATATTATAATG-3’
5’-GCTAGCTCGAGTTAAATATCCTTTTCCTTAAAAAAGTTC-3’
All genes were amplified using the genomic DNA of C jejuni NCTC 11168 (ATCC 700819D-5) as the template. For all primers, restriction sites for BamHI and XhoI (underlined) were introduced into the forward and reserve primers, respectively. These restriction sites allowed for the addition of an N-terminal 6x-His-tag in the pET30a+ expression vector for all four enzymes. Procedures for gene amplification, restriction digest, ligation, and plasmid isolation were followed as previously reported (26). Plasmids were fully sequenced to confirm correctly cloned genes in the appropriate vector.
Cloning of gmhB and tktA.
Several different growth and expression conditions were attempted for Cj1152, but only a small amount of protein was able to be purified. As a result, GmhB from E. coli was used as a substitute for Cj1152 in large-scale reactions. The gene for GmhB (Uniprot ID: P63228) was amplified from the genomic DNA of E. coli K12 with the primer pair:
5’- ATTATGGATCCATGGCGAAGAGCGTACCC -3’
5’- GCTAGAAGCTTTCATTGTGCCGGTTTTTGCTG -3’
Restriction sites for BamHI and HindIII (underlined) were introduced into the forward and reserve primers, respectively.
The gene for transketolase A, (Uniprot ID: P27302) was amplified from the genomic DNA of E. coli K12 with the primer pair:
5’- ATGCTGGATCCATGTCCTCACGTAAAGAGCTT -3’
5’- ATTATGCGGCCGCTTACAGCAGTTCTTTTGCTTT -3’
Restriction sites for BamHI and NotI (underlined) were introduced into the forward and reserve primers, respectively. For both constructs, these restriction sites allowed for the addition of an N-terminal 6x-His-tag in the pET30a+ expression vector. There were no changes to gene amplification, restriction digest, ligation, or plasmid isolation as outlined previously (26). Plasmids were sequenced to confirm the correctly cloned genes in the vector.
Expression and Purification of Enzymes.
Recombinant plasmids containing the cj1423, cj1424, cj1425, gmhB, or tktA genes were used to transform E. coli BL21 (DE3) cells using the method of heat-shock (27). Conditions for expression and purification were followed as previously published with the following minor adjustments (26), except for Cj1152. For the expression of all proteins studied, except Cj1152, cells were grown in lysogeny broth (LB) medium at 37 °C until OD600 = 0.6 followed by the addition of 1.0 mM isopropyl β-thiogalactoside (IPTG) to induce protein expression for 4–5 h at 37 °C. Growth and expression of the genes for Cj1423 and Cj1425 proceeded with no other additions to the growth medium. During growth of cells containing the plasmid for Cj1424, 1.0 mM zinc acetate was added, and during the expression of gmhB and tktA, 1.0 mM MgCl2 was included in the medium. All enzymes were purified as outlined previously (26). Briefly, cells were resuspended in 50 mL of 50 mM HEPES/K+, pH 8.5 with 250 mM KCl and 10 mM imidazole and then lysed by sonication at 4 °C. Cell debris was removed by centrifugation at 12,000 g for 20 min followed by passing the cell lysate through a 0.45 μm filter attached to a syringe. This solution was loaded onto a 5-mL HisTrap HP (GE Healthcare) nickel affinity column and eluted from the column using 50 mM HEPES/K+, pH 8.5, with 250 mM KCL and 500 mM imidazole over a gradient of 30 column volumes. Fractions containing the purest samples based on SDS-PAGE gel electrophoresis were pooled and buffer exchanged by dialysis into 50 mM HEPES/K+, pH 8.5 with 250 mM KCl overnight at 4 °C. Protein was concentrated using Vivaspin 20 centrifugal concentrators (GE Healthcare) and the final concentration of enzyme determined by absorbance at 280 nm. The extinction coefficients were estimated based on the protein sequence including the His-tag and linkers (28). The extinction coefficients used were as follows: Cj1423 (E280 = 18,000 M−1 cm−1); Cj1424 (E280 = 9,315 M−1 cm−1); Cj1425 (E280 = 47,455 M−1 cm−1); GmhB (E280 = 24,075 M−1 cm−1); and TktA (E280 = 94,770 M−1 cm−1). Yields of purified Cj1423 were 50–60 mg, 150–200 mg for Cj1424, 100–150 mg for Cj1425, 50–75 mg of GmhB, and 50–100 mg of TktA from 2.0 L of culture.
For Cj1152, Rosetta (DE3) E. coli cells were transformed with plasmid containing the gene for Cj1152 by electroporation and plated on LB media containing 1% agar and 50 μg/mL kanamycin. A 20-mL starter culture supplemented with 50 μg/mL kanamycin and 25 μg/mL chloramphenicol was inoculated with a single colony and grown overnight at 37 °C. This starter culture was used to inoculate 2× 1 L of Terrific Broth (TB) containing 1.0 mM MgCl2, 1.0 mM zinc acetate, 50 μg/mL kanamycin and 25 μg/mL chloramphenicol and allowed to grow at 37 °C until OD600 = 0.4 was reached. IPTG was added to a final concentration 100 μM and protein was expressed for 16 h at 18 °C. Cell were harvested by centrifugation at 7,000 g for 10 min and the cell pellet (~30 g) was stored at −80 °C until purification. Protein purification was followed as outlined above. The final concentration of enzyme was determined by absorbance at 280 nm. The extinction coefficient for Cj1152 was E280 = 12,295 M−1 cm−1 and includes the His-tag and linker sequence. Final yield of purified for Cj1152 was 12 mg per 1.0 L of cell culture.
Enzymatic Synthesis of d-altro-heptulose-7-P (1).
d-Altro-heptulose-7-P (d-sedoheptulose-7-P) was synthesized following a procedure outlined previously (18, 29). In general, 60 mg (0.22 mmol) of d-ribose-5-P was mixed with a 1.5 molar excess of hydroxypyruvate (0.33 mmol) in 4.5 mL of 50 mM HEPES/K+ buffer, pH 7.4. To this solution, 0.5 mM thiamine pyrophosphate (TPP) and 1.0 mM MgCl2 were added. Transketolase A (10 μM) was added and the reaction monitored by 31P NMR spectroscopy for 2–4 h. After the reaction was complete, TktA was removed by centrifugation filtration and d-altro-heptulose-7-P was purified by anion-exchange chromatography as described previously (20, 26). The net yield of d-altro-heptulose-7-P was approximately 60 mg or 80%. The structure of d-altro-heptulose-7-P was confirmed by 13C NMR spectroscopy and mass spectrometry.
Determination of Kinetic Constants for Cj1424 and Cj1425.
For Cj1424 and Cj1425, the kinetic constants for product formation were determined by monitoring the oxidation of NADH to NAD+ at 340 nm using a coupled assay for kinase activity as described previously at 30 °C (20). For Cj1424, 0.5–2 μM of enzyme was used in the presence of a 10-fold excess of Cj1425. For Cj14525, 1–2 μM of enzyme was used in the presence of a 10-fold excess of Cj1424. Enzyme was added to the wells containing d-altro-heptulose-7-P (0–4000 μM) and other assay components (pyruvate kinase, lactate dehydrogenase, ATP, PEP, and MgCl2). The equilibrium constant for the conversion of d-altro-heptulose-7-P to d-glycero-α-d-manno-heptose-7-P catalyzed by Cj1424 was determined to be 0.22 by 31P NMR and 1H NMR spectroscopy. The equilibrium constant was used to correct the initial substrate concentrations for d-glycero-α-d-manno-heptose-7-P with Cj1425. The values of kcat and kcat/Km were determined by fitting the initial velocity data to eq. 1 using GraFit 5, where v is the initial velocity of the reaction, E is the enzyme concentration, kcat is the turnover number, and Km is the Michaelis constant.
| (1) |
Synthesis of d-Glycero-α-d-manno-heptose-1,7-bisphosphate.
d-ribose-5-P (0.24 mmol) was mixed with 0.48 mmol hydroxypruvate (2-fold excess) with 2 mM TPP and 10 mM MgCl2 in 50 mM HEPES/K+ buffer, pH 7.4. TktA (3 μM) was added to the solution and allowed to incubate for 1 h. After 1 h, 0.24 mmol of PEP and ATP (1.0 mM final concentration) was added to the d-altro-heptulose-7-P solution with 30 μM Cj1424 and Cj1425 and 10 U of pyruvate kinase. After 24 h, d-glycero-d-manno-heptose-1,7-bisphosphate (3) was purified by anion exchange chromatography as described previously and quantified by 31P NMR using an internal standard (20).
Determination of Kinetic Constants for Cj1152.
Kinetic constants for Cj1152 were determined using a malachite green phosphate detection assay. A 0.0012% (w/v) malachite green solution and an 11% Tween 20 solution were prepared as described previously (30). A 4.2 % (w/v) of ammonium molybdate solution was prepared as described previously (31). Fresh color reagent solution was prepared by mixing 1 mL of malachite green solution, 0.45 mL of ammonium molybdate solution, and 0.05 mL of Tween 20. Cj1152 (1 μM) was mixed with various concentrations of 3 (20–1000 μM) in 50 mM HEPES/K+ buffer, pH 7.4 with 1 mM MgCl2 added to a total volume of 200 μL. At 30 or 60 s time intervals, 20 or 50 μL aliquots were removed and added to 96-well plate containing 200 or 170 μL of 50 mM HEPES/K+ buffer, pH 7.4. The lower concentration range of substrate (20–200 μM) used long time points (60 s) and larger aliquots (50 μL) of reaction. To each well, 30 μL of fresh color reagent solution was added to give a total volume of 250 μL. A total of eight time points were collected for up to 4–8 min. The color reagent was allowed to develop for 20 min before reading A650 with a UV-vis spectrophotometer. Absorbance data were converted to concentration of phosphate using a standard curve generated using a commercial phosphate standard solution (Sigma Aldrich). Observed rates of product formation were obtained from the slope of a linear fit of the amount of product versus time. The resulting observed rates were plotted versus substrate concentration and fit to eq. 1 yielding kinetic constants for kcat and kcat/Km.
Synthesis of d-Glycero-α-d-manno-heptose-1-P (4).
d-altro-heptulose-7-P (0.050 mmol) was mixed with 0.055 mmol ATP and 0.055 mmol MgCl2 (10% excess over d-altro-heptulose-7-P) in 50 mM HEPES/K+ buffer, pH 7.4. To this solution, 20 μM of Cj1424 and Cj1425 and 6 μM GmhB were added. After 1 h, the reaction was complete, yielding d-glycero-α-d-manno-heptose-1-P. Enzymes where removed by centrifugal filtration and d-glycero-α-d-manno-heptose-1-P was purified by anion exchange chromatography as described previously (20).
Determination of Kinetic Constants for Cj1423.
Kinetic constant for Cj1423 were determined by 31P NMR spectroscopy and HPLC anion exchange chromatography. For 31P NMR experiments, Cj1423 (200 nM) was mixed with 6.0 mM GTP, 6 mM MgCl2, and 5.0 mM d-glycero-α-d-manno-heptose-1-P into 50 mM HEPES/K+ buffer, pH 7.4. One unit of yeast pyrophosphatase (Sigma Aldrich) was included, allowing the reaction to proceed to completion. NMR spectra were collected every 2.5 min for 1 h. The initial data were fit to a linear equation with the slope equal to kcat. For the HPLC method, a previously development methodology was followed (32). Briefly, Cj1423 (10 nM) was mixed with various concentrations of d-glycero-α-d-manno-heptose-1-P (0.1–8.0 mM) in 50 mM HEPES/K+, pH 7.4 containing 1.0 mM GTP, 2.0 mM MgCl2 and 1.0 U of pyrophosphatase. Every 12 min, a 100 μL aliquot was removed and injected into a BioRad FPLC system fitted with a 1.0 mL Resource Q Column (GE Healthcare). The column was washed with 10 mM triethanolamine, pH 8.0, and the products were eluted with a linear gradient of 10 mM triethanolamine, pH 8.0 and 2.0 M KCl. The products were monitored by the change in absorbance at 255 nm and quantified by relative peak area integration. Each reaction was conducted at 25 °C with a minimum of five time points for each concentration collected. The amount of product was plotted versus time and fit to a linear equation with the resulting slope equal to the rate of product formation. Subsequently, the observed rate of product formation was plotted versus the substrate concentration and fitted to eq. 1 to yield the steady-state kinetic constants for kcat and kcat/Km.
Preparative Scale Synthesis of GDP-d-glycero-α-d-manno-heptose (5).
The synthesis of GDP-d-glycero-α-d-manno-heptose was conducted by addition of Cj1423, Cj1424, Cj1425, GmhB, and TktA to a single reaction vessel. For this, 330 mg (0.8 mmol) of d-ribose-5-P and 254 mg (1.6 mmol) hydroxypyruvate were mixed with 50 mM HEPES/K+ buffer, pH 7.4 containing 2.0 mM TPP, and 100 mM MgCL2. ATP (650 mg, 0.6 mmol) and 332 mg (0.63 mmol) GTP were dissolved in 500 mM HEPES/K+ buffer, pH 7.4 and added to the above mixture to a final volume of 12 mL. The enzymes TktA, Cj1424, Cj1425, GmhB, and Cj1423 were added to be a concentration of 20 μM each. One unit of pyrophosphatase was included. The reaction was followed by 31P NMR spectroscopy. After 24 h, the reaction was approximately 80% complete and no additional product formation was observed over the next 24 h. After 48 h, 20 U of recombinant shrimp alkaline phosphatase (rSAP, New England BioLabs) was added and the reaction monitored over a period of 5 h. Test reactions shows that rSAP will dephosphorylate the remaining d-ribose-5-P, ATP, ADP, and GTP with no measurable loss of GDP-d-glycero-α-d-manno-heptose in 72 h. With only free phosphate and GDP-d-glycero-α-d-manno-heptose visible by 31P NMR spectroscopy, all of the enzymes were removed by centrifugal filtration using a 6-mL Vivaspin 10 kDa filter. GDP-d-glycero-α-d-manno-heptose was purified by DEAE anion exchange chromatography using the same method as outlined previously (20, 26). Fractions were monitored by UV-Vis spectroscopy by measuring the absorbance at 255 nm. The final yield of GDP-d-glycero-α-d-manno-heptose was 192 mg (~50% net yield).
RESULTS
Bioinformatic Analysis of Heptose Containing Polysaccharides.
The structurally characterized oligosaccharides of the CPS contained within the sequenced strains of C. jejuni exhibit an array of chemical modifications. To better understand the sequence diversity among the enzymes responsible for the synthesis of d-glycero-d-manno-heptose, a bioinformatic analysis was conducted. The complete FASTA protein sequences for 484 strains of C. jejuni were retrieved from the NCBI and compiled into a local custom database. The genomes were annotated as either “complete” or “scaffold” assembly level by NCBI. The sequences for Cj1423, Cj1424, Cj1425, and Cj1152 from C. jejuni NCTC 11168 were used as BLAST queries against the local database. The BLAST results and sequence similarity network construction show that 373 out of 484 strains of C. jejuni have protein sequences that share 88% sequence identity or higher to all four genes within their genomes (Figure S1). There are seven additional strains that have genes that share at least a 66% sequence identity to all four genes. A total of 482 out of the 484 strains have sequences that share greater than 83% sequence identity with Cj1152 and 57% sequence identity to Cj1424. This result is mostly likely due to the isomerase (GmhA) and phosphatase (GmhB) requirement for the synthesis of the heptose moiety found in the inner core region of the LOS structure. It is important to note that not all C. jejuni strains have a heptose moiety in their CPS structure. A total of 99 of the 484 strains do not contain homologs with greater than 34% sequence identity to either Cj1425 or Cj1423. The strains that are known to lack a heptose moiety in their CPS (e.g. HS1/HS44, HS6/7, and HS19) are found among the 99 strains without homologs to either Cj1425 or Cj1423. Based on the high sequence conservation of each of the four genes among the 373 C. jejuni strains, the starting material for the synthesis of the ultimate heptose moiety in the CPS of C. jejuni will most likely be the same GDP-heptose carbohydrate from which a variety of possible modifications can be synthesized.
Catalytic Activity of Cj124 and Cj1425.
The gene for Cj1424 was cloned, expressed, and the enzyme purified to homogeneity. The protein was tested for catalytic activity with d-altro-heptulose and d-altro-heptulose-7-P (1) by monitoring the changes in the 1H and 31P NMR spectra. d-Altro-heptulose-7-P was synthesized using transketolase A (TktA), d-ribose-5-P, hydroxypyruvate (HPA), thiamine pyrophosphate (TPP), and MgCl2 (Figure 1a). The experiments containing d-altro-heptulose with Cj1424 showed no new resonances for the formation of products after 24 h (data not shown). However, in the presence of d-altro-heptulose-7-P, a new resonance at 4.78 ppm (Figure 1b) appears, corresponding to the formation of d-glycero-d-manno-heptose-7-P (2). The equilibrium constant for the isomerization of d-altro-heptulose-7-P to d-glycero-d-manno-heptose-7-P was determined to be 0.22. This value agrees with the previously estimated equilibrium constant for this reaction using GmhA from A. thermoaerophilus (13). Due to the unfavorable equilibrium constant, direct measurement of the kinetic constants for this reaction were not attempted. However, the kinetic constants for the reaction catalyzed by Cj1424 were obtained using a coupled assay in the presence of excess Cj1425 (vida infra), which enabled the reaction to be followed spectrophotometrically by monitoring the formation of ADP with pyruvate kinase/lactate dehydrogenase in the presence of PEP (Table 1). l-Galacto-heptulose, l-galacto-heptulose-7-P (20), and d-manno-heptulose were also tested as initial substrates for the Cj1424/Cj1425 catalyzed reaction sequence, but no activity was observed.
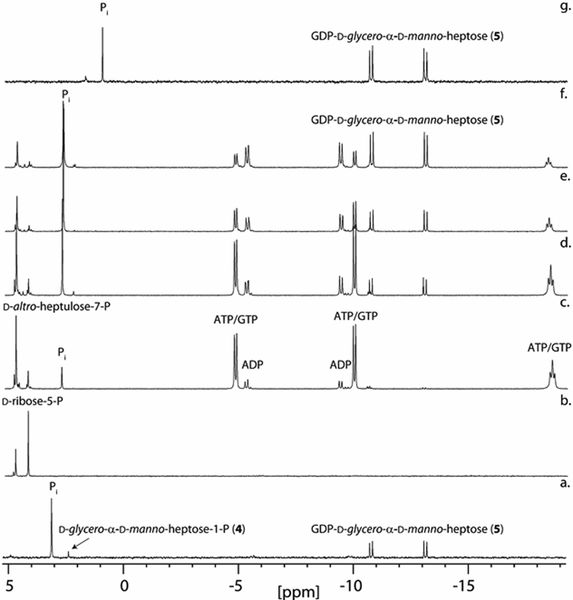
Figure 1.
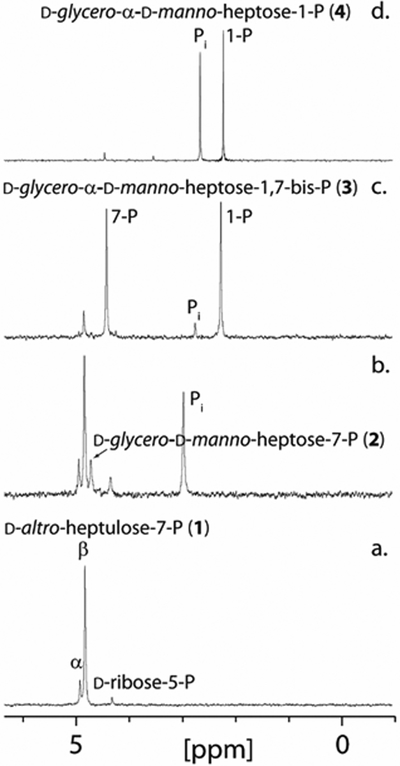
31P NMR spectra for the synthesis of d-glycero-α-d-manno-heptose-1-P (4) starting from d-altro-heptulose-7-P (1). (a) 31P NMR spectrum of d-altro-heptulose-7-P (1). (b) d-glycero-d-manno-heptose-7-P (2) formed from the addition of Cj1424 to d-altro-heptulose-7-P. (c) d-glycero-α-d-manno-heptose-1,7-bisphosphate (3) formed after the addition of Cj1425 and MgATP to d-glycero-d-manno-heptose-7-P. (d) d-glycero-α-d-manno-heptose-1-P (4) formed after the addition of Cj1152 to d-glycero-α-d-manno-heptose-1,7-bisphosphate. Additional details are provided in the text.
Table 1.
Steady-state kinetic parameters for Cj1424, Cj1425 and Cj1423a
| Enzyme | Substrate | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| Cj1424b | d-altro-heptulose-7-P (1) | 0.14 ± 0.01 | 320 ± 25 | (4.4 ± 0.4) × 102 |
| Cj1425b | d-glycero-d-manno-heptose-7-P (2)c | 0.48 ± 0.01 | 103 ± 9 | (4.7 ± 0.4) × 103 |
| Cj1152d | d-glycero-α-d-manno-heptose-1,7-bisphosphate (3) | 0.12 ± 0.01 | 186 ± 25 | (6.5 ± 1.0) × 102 |
| GmhBe | d-glycero-α-d-manno-heptose-1,7-bisphosphate (3) | 4.6 ± 0.1 | 67 ± 1 | (7.0 ± 0.1) × 104 |
| GmhBe | d-glycero-β-d-manno-heptose-1,7-bisphosphate | 36.0 ± 0.2 | 5.0 ± 0.1 | (7.2 ± 0.2) × 106 |
| Cj1423f | d-glycero-α-d-manno-heptose-1-P (4) | 1.35 ± 0.04 | 495 ± 65 | (2.7 ± 0.4) × 103 |
Errors were calculated from the standard deviation of the fitting results.
Reactions were monitored by UV coupled assay at pH 7.4 and 30 °C in the presence of NADH (300 μM), MgCl2 (5 mM), ATP (2 mM), PEP (1.0 mM), 1 U of lactate hydrogenase and pyruvate kinase.
The substrate for Cj1425, d-glycero-d-manno-heptose-7-P (2), was generated in situ using a 10-fold excess of Cj1424 and d-altro-heptulose-7-P (1). Kinetic constants were determined from the calculated concentrations of (2) using the equilibrium constant of 0.22 for the Cj1424 catalyzed reaction.
Reaction was followed by discontinuous UV assay using malachite green free phosphate detection at pH 7.5 at 25°C.
Reactions were monitored at pH 7.5 and 25 °C as previously reported (17).
Reaction was monitored by HPLC at pH 7.4 and 25 °C with GTP (1.0 mM), MgCl2 (2.0 mM) and pyrophosphatase (1 U).
The gene for Cj1425 was cloned and expressed, and the protein purified to homogeneity. In the presence of the Cj1424 isomerase, d-altro-heptulose-7-P (1) and MgATP, Cj1425 catalyzes the phosphorylation of d-glycero-d-manno-heptose-7-P (2) to yield d-glycero-α-d-manno-heptose-1,7-bisphosphate (3). A new 31P NMR resonance for the phosphate at C1 is observed at 2.18 ppm (Figure 1c). The kinetic constants for Cj1425 were obtained using a coupled enzyme assay in the presence of excess Cj1424 and d-altro-heptulose-7-P (Table 1). For this experiment, the steady state concentration of the substrate (d-glycero-d-manno-heptose-7-P, 2) was defined by the thermodynamic equilibrium constant observed for the isomerization of d-altro-heptulose-7-P using an excess of Cj1424 (Keq = 0.22).
Catalytic Activity of Cj1152.
Initial attempts to express Cj1152 showed a significant loss of soluble protein compared to Cj1424 and Cj1423. Multiple conditions were screened, including variations of cell lines, IPTG concentrations, temperature, and media, with best conditions for Cj1152 expression determined to be in Rosetta (DE3) E. coli cells grown in Terrific Broth (TB) with 0.1 mM IPTG for 16 h at 18 °C. In the presence of MgCl2 (1.0 mM), Cj1152 catalyzes the dephosphorylation of d-glycero-d-manno-heptose-1,7-bisphosphate (3) to yield d-glycero-d-manno-heptose-1-P (4) as shown in Figure 1d. The formation of Pi is observed at 2.71 ppm with retention of the resonance for the phosphoryl group attached to C1. Kinetic constants for Cj1152 were obtained by quantifying free phosphate formation over time using the malachite green assay. The kinetic constants are summarized in Table 1.
Cj1152 shares 32% sequence identity with GmhB from E. coli, which has been shown to function in the same LOS biosynthetic pathway as Cj1152 (17). Previously, GmhB was shown to catalyze the dephosphorylation of both the α- and β-forms of d-glycero-d-manno-heptose-1,7-bisphosphate (17). Kinetic constants for this enzyme for both d-glycero-α-d-manno-heptose-1,7-bisphosphate and d-glycero-β-d-manno-heptose-1,7-bisphosphate have been reproduced in Table 1. Due to the small amount of soluble Cj1152 produced and slow rate of dephosphorylation compared to GmhB, GmhB was cloned and purified and used as a substitute for Cj1152 in all other reactions requiring the production of d-glycero-α-d-manno-heptose-1-P (4).
Catalytic Activity of Cj1423.
Similar to Cj1152, the expression of the gene for the production of Cj1423 was problematic in both E. coli (DE3) BL21 and E. coli Rosetta cell lines. Under all conditions tested, no expression of native Cj1423 protein was observed. Similar problems were described previously for the Cj1423 homolog, GmhD from A. thermoaerophilus (13). As a result, the gene for Cj1423 was codon optimized (GeneScript), chemically synthesized, and cloned into a pET30a expression vector. This construct yields significant improvement in the expression of Cj1423, and ~30 mg of Cj1423 was able to be purified from 1 L of media. In the presence of d-glycero-α-d-manno-heptose-1-P (4) and MgGTP, Cj1423 catalyzes the formation of GDP-d-glycero-α-d-manno-heptose (5). The equilibrium constant is approximately 0.24. In the presence of pyrophosphatase, the reaction proceeds to completion (Figure 2a). Cj1423 was shown to utilize ATP, UTP, and CTP as substitutes for GTP in the presence of pyrophosphatase, but the rates are significantly slower (Figures S2a–d for GTP, ATP, UTP, and CTP, respectively). Cj1423 will also combine α-d-mannose-1-P with GTP to form GDP-α-d-mannose (Figure S2e). Kinetic constants for Cj1423 were determined using two different experiments. The formation of product was followed by 31P NMR spectroscopy and a plot of the concentration of GDP-d-glycero-α-d-manno-heptose as a function of time gave an apparent kcat of 3.0 ± 0.3 s−1. However, this assumes that 5.0 mM d-glycero-α-d-manno-heptose-1-P (4) and 5.0 mM MgGTP were saturating. The reaction was repeated and product formation was determined by anion exchange chromatography. The concentration of d-glycero-α-d-manno-heptose-1-P (4) was varied from 0.1 to 8.0 mM at a fixed concentration of 1.0 mM GTP (1). The resulting values of the kinetic constants are summarized in Table 1.
Figure 2.
31P NMR spectra for the time course of the preparative-scale biosynthesis of GDP-d-glycero-α-d-manno-heptose from d-ribose-5-P. (a) GDP-d-glycero-α-d-manno-heptose (5) after the addition of Cj1423, GTP, and pyrophosphatase to d-glycero-α-d-manno-heptose-1-P. (b) Spectrum of d-ribose-5-P with d-altro-heptulose-7-P added for reference. (c) 1 h after addition of all enzymes (TktA, Cj1423, Cj1424, Cj1425, GmhB, and pyrophosphatase) and co-factors as described in the text. (d) 6 h. (e) 24 h. (f) 48 h. (g) GDP-d-glycero-α-d-manno-heptose after rSAP degradation of remaining phosphate compounds (ATP/ADP, GDP, d-altro-heptulose-7-P) and anion-exchange column purification. The shift in the phosphate resonance is due to the change in pH after ammonium bicarbonate has been removed by acidification and then neutralized (final pH ~6).
Preparative Scale Synthesis of GDP-d-glycero-α-d-manno-heptose.
The apparent widespread utilization of GDP-d-glycero-α-d-manno-heptose as the entry point for the biosynthesis of heptoses of various stereochemical arrangements makes it an attractive molecule to obtain in significant quantities. Two experimental trials were utilized to determine the effectiveness of synthesizing this intermediate using the combined catalytic activities of Cj1423, Cj1424, and Cj1425. In the first example, all three enzymes were mixed, along with pyrophosphatase and GmhB from E. coli and the required cofactors and co-substrates (ATP, GTP, and MgCl2) in a single reaction vessel. The reaction was initiated by the addition of d-altro-heptulose-7-P, and GDP-d-glycero-α-d-manno-heptose (5) was efficiently formed (Figures S3a and S3b). Moreover, the reaction could be initiated directly from d-ribose-5-phosphate and hydroxypyruvate in the presence of TktA and TPP to yield GDP-d-glycero-α-d-manno-heptose. The final yield of GDP-d-glycero-α-d-manno-heptose from d-ribose-5-P and hydroxypyruvate in a single reaction vessel was 60% after 72 h (Figure 2b–f). GDP-d-glycero-α-d-manno-heptose was purified by DEAE anion exchange and separated from the remaining compounds after incubation with recombinant shrimp alkaline phosphatase (rSAP) to degrade all remaining phosphate containing compounds. rSAP does not measurably degrade GDP-d-glycero-α-d-manno-heptose over 72 h. The 1H and 13C NMR spectra for GDP-d-glycero-α-d-manno-heptose are shown in Figure S4.
Discussion
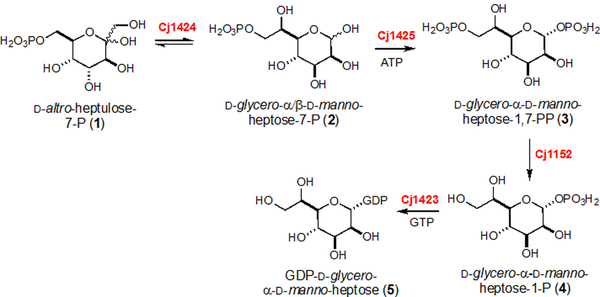
Heptoses are key components of many extracellular structures including the LOS and CPS units of C. jejuni. The requirement for two d-altro-heptulose-7-P isomerases and the apparent absence of a nearby phosphatase raised a question about d-altro-heptulose-7-P as the predicted starting material for the Cj1423/Cj1424/Cj1425 cluster of enzymes in C. jejuni NCTC 1168. A bioinformatic analysis of 484 strains of C. jejuni revealed that approximately 77% of these strains contain the appropriate enzymes for the biosynthesis of a heptose. Moreover, this analysis showed that all of these enzymes are highly similar (>88%) to one another (Figure S1) suggesting that the starting material, the intermediates, and ultimate product are the same (Scheme 3).
Scheme 3:
Synthesis of GDP-d-glycero-α-d-manno-heptose (5) in C. jejuni NCTC 11168
Assays with Cj1424 show that d-altro-heptulose-7-P (1) is a substrate, while no activity was observed with d-altro-heptulose. Catalytic activity of Cj1424 with d-altro-heptulose-7-P, but not d-altro-heptulose, confirms the requirement for a phosphatase to remove the phosphate group from C7. The equilibrium constant for the isomerization of d-altro-heptulose-7-P to d-glycero-d-manno-heptose-7-P (2) is 0.22, which agrees with previously reported results using homologs of Cj1424 (13). Although the measured specificity constant (kcat/Km) and turnover number (kcat) for Cj1424 are not large, these values are similar to the kinetic constants for the E. coli homologue, GmhA, which were previously reported to be 0.44 s−1 and 500 M−1 s−1 for kcat and kcat/Km, respectively (33).
Cj1425 was shown to phosphorylate d-glycero-d-manno-heptose-7-P (2) resulting in a single anomer of d-glycero-d-manno-heptose-1,7-bisphosphate. The configuration at C1 was previously shown to be of the α-configuration (9, 13, 17). This is different from the LOS heptose synthesis pathway where the dual-function enzyme HldE (Cj1150) catalyzes the formation of d-glycero-β-d-manno-heptose-1,7-bisphosphate. Kinetic constants for Cj1425 are slightly higher than Cj1424 with a turnover number of 0.48 s−1 and a specificity constant of 4200 M−1 s−1.
A previous report demonstrated that a knockout of Cj1152 affected the production of both the LOS and CPS in C. jejuni (5). Based on this observation, it was concluded that Cj1152 is required to dephosphorylate the α- and β-anomers of d-glycero-d-manno-1,7-bisphosphate in both biosynthetic pathways. A previous bioinformatic study suggested that bacterial species with two heptose synthesis pathways, as found in C. jejuni, is relatively rare, and these species commonly utilize two phosphatase enzymes, one for each pathway (17). To experimentally demonstrate the ability of Cj1152 to dephosphorylate d-glycero-α-d-manno-1,7-bisphosphate (3), the enzyme was purified and characterized. A 31P NMR experiment showed that Cj1152 catalyzes the dephosphorylation of compound 3 to yield d-glycero-α-d-manno-heptose-1-P (4). The rate of turnover (kcat) of Cj1152 is about 40-fold slower than the value reported for the E. coli GmhB phosphatase with 3. However, the preferred substrate for GmhB is d-glycero-β-d-manno-1,7-bisphosphate, which is about 100-fold better than 3 (comparing values of kcat/Km). Homologues of these phosphatases tend to show evolved anomer specificity based on their function in the cell (i.e. degradation of α- or β-anomers d-glycero-d-manno-1,7-bisphosphate) (17).
The native gene for Cj1423 could not be expressed in E. coli cell lines and needed to be codon-optimized for protein expression. Cj1423 was shown to combine GTP and d-glycero-α-d-manno-heptose-1-P (4) yielding GDP-d-glycero-α-d-manno-heptose (5). Cj1423 will slowly accept ATP and UTP as a substitute for GTP. It will also accept CTP, but the reaction is very slow with only a small amount of product observed after 72 h of incubation (Figure S2d). The turnover number for Cj1423 was shown to be the fastest of the three enzymes with a kcat = 3.0 s−1 and 1.4 s−1 determined by 31P NMR and HPLC, respectively.
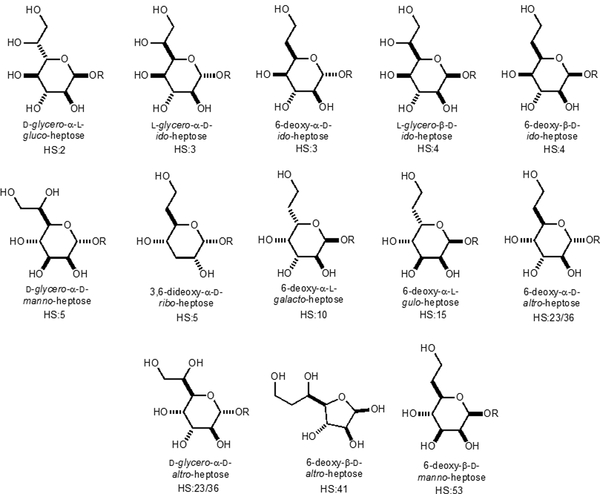
Significant quantities of GDP-d-glycero-α-d-manno-heptose (5) can be generated from this pathway in a single reaction vessel starting from d-altro-heptulose-7-P (1) or d-ribose-5-P and hydroxypyruvate upon the addition TktA and relevant cofactors. Since GDP-d-glycero-α-d-manno-heptose is the expected starting point for all of the other heptose moieties in the CPS of C. jejuni, the ability to efficiently generate a significant amount of GDP-d-glycero-α-d-manno-heptose is valuable for the analysis of the other enzymes involved in heptose biosynthesis for the assembly of the CPS in C. jejuni. As of May 2019, the CPS structures of 12 C. jejuni serotypes (Figures S5–S16) have been reported (11). From these 12 structures, 9 contain heptose residues with 13 different heptose variations identified (Scheme 4). Methylation and phosphoramidate modifications enable more variations to these heptose structures (not shown).
Scheme 4:
Observed variations in the heptose residues of the CPS from C. jejuni.
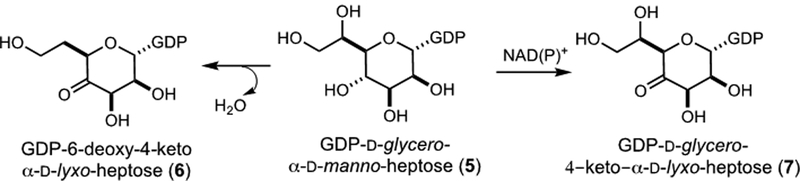
All of the CPS gene clusters for these serotypes contain enzymes with high sequence identity (>88%) for the four enzymes described above (Cj1423, Cj1424, Cj1152, and Cj1425) (Figures S5–S16) and thus GDP-d-glycero-α-d-manno-heptose (5) is the most likely starting point in the biosynthesis of each of these different heptose molecules (13, 15, 19, 34). Heptoses that are dehydrated at C6 appear to be synthesized through a GDP-6-deoxy-4-keto-d-lyxo-heptose (6) intermediate (15, 34) whereas the other heptoses are likely to be synthesized from a GDP-d-glycero-4-keto-d-lyxo-heptose intermediate (7) (19). For C. jejuni strain 81–176, it has been shown that DhahA (Cjj1426) catalyzes the formation of GDP-6-deoxy-4-keto-d-lyxo-heptose (6) via the oxidation, dehydration and reduction of GDP-d-glycero-α-d-manno-heptose (15, 34). Unfortunately, the enzyme that catalyzes the putative formation of GDP-d-glycero-4-keto-d-lyxo-heptose (7) has not been identified (19). These two transformations are summarized in Scheme 5.
Scheme 5:
Metabolic fate of GDP-d-glycero-α-d-manno-heptose (5).
In this report, we have shown that the four enzymes from C. jejuni NCTC 11168, Cj1423, Cj1424, Cj1425, Cj1152 are responsible for the initial synthesis of GDP-d-glycero-α-d-manno-heptose (5). The pathway utilizes d-altro-heptose-7-P (1) and requires the action of a dual-function phosphatase, Cj1152, found in the LOS gene cluster. A bioinformatic analysis of sequence identities show that there is high sequence conservation among these four genes in all C. jejuni strains that have heptose residues in their CPS structures. These results suggest that the starting material, intermediates, and final product of the pathway are the same for all C. jejuni strains. We have also shown the ability to generate significant quantities of the final product, GDP-d-glycero-α-d-manno-heptose (5), in a single reaction vessel. This result is critical for mechanistic characterization of the subsequent enzymes of the various heptose biosynthetic pathways and the enzymes responsible for the modifications to the heptose-containing polysaccharides (e.g. changes in stereochemistry and addition of methyl and phosphoramidate groups).
Supplementary Material
Funding
This work was supported in parts by grants from the Robert A. Welch Foundation (A-840) and the National Institutes of Health (GM122825).
Footnotes
Supporting Information
Sequence similarity network containing enzymes for Cj1423, Cj1424, Cj1425, and Cj1152 from 373 C. jejuni strains. 31P NMR spectra of CJ1423 with GTP, ATP, UTP, CTP, and α-d-mannose-1-P. 31P NMR spectra of full reaction starting with d-altro-heptulose-7-P to yield GDP-d-glycero-α-d-manno-heptose. Structures and gene clusters for the 12 previously determined CPS structures for various serotypes.
The authors declare no competing financial interest.
REFERENCES
- (1).García-Sánchez L, Melero B, and Rovira J (2018) Campylobacter in the Food Chain, In Advances in Food and Nutrition Research (Rodríguez-Lázaro D, Ed.), pp 215–252, Academic Press. [DOI] [PubMed] [Google Scholar]
- (2).Burnham PM, and Hendrixson DR (2018) Campylobacter jejuni: collective components promoting a successful enteric lifestyle, Nat. Rev. Microbiol, 16, 551–565. [DOI] [PubMed] [Google Scholar]
- (3).Gaudet RG, and Gray-Owen SD (2016) Heptose sounds the alarm: innate sensing of a bacterial sugar stimulates immunity, PLoS Pathog., 12, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kosma P (2008) Occurrence, synthesis and biosynthesis of bacterial heptoses, Curr. Org. Chem, 12, 1021–1039. [Google Scholar]
- (5).Karlyshev AV, Champion OL, Churcher C, Brisson JR, Jarrell HC, Gilbert M, Brochu D, St Michael F, Li J, Wakarchuk WW, Goodhead I, Sanders M, Stevens K, White B, Parkhill J, Wren BW, and Szymanski CM (2005) Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses, Mol. Microbiol, 55, 90–103. [DOI] [PubMed] [Google Scholar]
- (6).Godschalk PC, Kuijf ML, Li J, St Michael F, Ang CW, Jacobs BC, Karwaski MF, Brochu D, Moterassed A, Endtz HP, van Belkum A, and Gilbert M (2007) Structural characterization of Campylobacter jejuni lipooligosaccharide outer cores associated with Guillain-Barre and Miller Fisher syndromes, Infect. Immun, 75, 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Gilbert M, Karwaski MF, Bernatchez S, Young NM, Taboada E, Michniewicz J, Cunningham AM, and Wakarchuk WW (2002) The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. biosynthesis of sialylated ganglioside mimics in the core oligosaccharide, J. Biol. Chem, 277, 327–337. [DOI] [PubMed] [Google Scholar]
- (8).Kanipes MI, Tan X, Akelaitis A, Li J, Rockabrand D, Guerry P, and Monteiro MA (2008) Genetic analysis of lipooligosaccharide core biosynthesis in Campylobacter jejuni 81–176, J. Bacteriol, 190, 1568–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kneidinger B, Marolda C, Graninger M, Zamyatina A, McArthur F, Kosma P, Valvano MA, and Messner P (2002) Biosynthesis pathway of ADP-L-glycero-β-d-manno-heptose in Escherichia coli, J. Bacteriol, 184, 363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wong A, Lange D, Houle S, Arbatsky NP, Valvano MA, Knirel YA, Dozois CM, and Creuzenet C (2015) Role of capsular modified heptose in the virulence of Campylobacter jejuni, Mol. Microbiol, 96, 1136–1158. [DOI] [PubMed] [Google Scholar]
- (11).Monteiro MA, Baqar S, Hall ER, Chen Y-H, Porter CK, Bentzel DE, Applebee L, and Guerry P (2009) Capsule polysaccharide conjugate vaccine against diarrheal disease caused by Campylobacter jejuni, Infect. Immun, 77, 1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Michael FS, Szymanski CM, Li J, Chan KH, Khieu NH, Larocque S, Wakarchuk WW, Brisson J-R, and Monteiro MA (2002) The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168, Eur. J. Biochem, 269, 5119–5136. [DOI] [PubMed] [Google Scholar]
- (13).Kneidinger B, Graninger M, Puchberger M, Kosma P, and Messner P (2001) Biosynthesis of nucleotide-activated d-glycero-d-manno-heptose, J. Biol. Chem, 276, 20935–20944. [DOI] [PubMed] [Google Scholar]
- (14).McArthur F, Andersson CE, Loutet S, Mowbray SL, and Valvano MA (2005) Functional analysis of the glycero-manno-heptose 7-phosphate kinase domain from the bifunctional HldE protein, which is involved in ADP-L-glycero-d-manno-heptose biosynthesis, J. Bacteriol, 187, 5292–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).McCallum M, Shaw GS, and Creuzenet C (2011) Characterization of the dehydratase WcbK and the reductase WcaG involved in GDP-6-deoxy-d-manno-heptose biosynthesis in Campylobacter jejuni, Biochem. J, 439, 235–248. [DOI] [PubMed] [Google Scholar]
- (16).Butty FD, Aucoin M, Morrison L, Ho N, Shaw G, and Creuzenet C (2009) Elucidating the formation of 6-deoxyheptose: biochemical characterization of the GDP-d-glycero-d-manno-heptose C6 dehydratase, DmhA, and its associated C4 reductase, DmhB, Biochemistry, 48, 7764–7775. [DOI] [PubMed] [Google Scholar]
- (17).Wang L, Huang H, Nguyen HH, Allen KN, Mariano PS, and Dunaway-Mariano D (2010) Divergence of biochemical function in the HAD superfamily: d-glycero-d-manno-heptose-1,7-bisphosphate phosphatase (GmhB), Biochemistry, 49, 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Huang H, Pandya C, Liu C, Al-Obaidi NF, Wang M, Zheng L, Toews Keating S, Aono M, Love JD, Evans B, Seidel RD, Hillerich BS, Garforth SJ, Almo SC, Mariano PS, Dunaway-Mariano D, Allen KN, and Farelli JD (2015) Panoramic view of a superfamily of phosphatases through substrate profiling, Proc. Natl. Acad. Sci. U.S.A, 112, E1974–E1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).McCallum M, Shaw GS, and Creuzenet C (2013) Comparison of predicted epimerases and reductases of the Campylobacter jejuni D-altro- and l-gluco-heptose synthesis pathways, J. Biol. Chem, 288, 19569–19580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Huddleston JP, and Raushel FM (2019) Functional characterization of YdjH, a sugar kinase of unknown specificity in Escherichia coli K12, Biochemistry, 58, 3354–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kitts PA, Church DM, Thibaud-Nissen F, Choi J, Hem V, Sapojnikov V, Smith RG, Tatusova T, Xiang C, Zherikov A, DiCuccio M, Murphy TD, Pruitt KD, and Kimchi A (2016) Assembly: a resource for assembled genomes at NCBI, Nucleic Acids Res, 44, D73–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009) BLAST+: architecture and applications, BMC Bioinform, 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, and Whalen KL (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks, Biochim. Biophys. Acta, Proteins Proteomics, 1854, 1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gerlt JA (2017) Genomic enzymology: web tools for leveraging protein family sequence-function space and genome context to discover novel functions, Biochemistry, 56, 4293–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome. Res, 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Huddleston JP, Thoden JB, Dopkins BJ, Narindoshvili T, Fose BJ, Holden HM, and Raushel FM (2019) Structural and functional characterization of YdjI, an aldolase of unknown specificity in Escherichia coli K12, Biochemistry, 58, 3340–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Inoue H, Nojima H, and Okayama H (1990) High efficiency transformation of Escherichia coli with plasmids, Gene, 96, 23–28. [DOI] [PubMed] [Google Scholar]
- (28).Gasteiger E, Hoogland C, Gattiker A, Duvaud S. e., Wilkins MR, Appel RD, and Bairoch A (2005) Protein identification and analysis tools on the ExPASy server, In The Proteomics Protocols Handbook (Walker JM, Ed.), pp 571–607, Humana Press, Totowa, NJ. [Google Scholar]
- (29).Charmantray F, Hélaine V, Legeret B, and Hecquet L (2009) Preparative scale enzymatic synthesis of d-sedoheptulose-7-phosphate from β-hydroxypyruvate and d-ribose-5-phosphate, J. Mol. Catal. B Enzym, 57, 6–9. [Google Scholar]
- (30).Baykov AA, Evtushenko OA, and Avaeva SM (1988) A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay, Anal. Biochem, 171, 266–270. [DOI] [PubMed] [Google Scholar]
- (31).Lanzetta PA, Alvarez LJ, Reinach PS, and Candia OA (1979) An improved assay for nanomole amounts of inorganic phosphate, Anal. Biochem, 100, 95–97. [DOI] [PubMed] [Google Scholar]
- (32).Taylor ZW, Brown HA, Holden HM, and Raushel FM (2017) Biosynthesis of nucleoside diphosphoramidates in Campylobacter jejuni, Biochemistry, 56, 6079–6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Taylor PL, Blakely KM, de Leon GP, Walker JR, McArthur F, Evdokimova E, Zhang K, Valvano MA, Wright GD, and Junop MS (2008) Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants, J. Biol. Chem, 283, 2835–2845. [DOI] [PubMed] [Google Scholar]
- (34).McCallum M, Shaw SD, Shaw GS, and Creuzenet C (2012) Complete 6-Deoxy-d-altro-heptose biosynthesis pathway from Campylobacter jejuni, J. Biol. Chem, 287, 29776–29788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.