This study reveals a novel regulatory mechanism that links the versatile receptor kinase FERONIA with plant C/N responses.
Keywords: Arabidopsis thaliana, ATL6, carbon/nitrogen response, E3 ubiquitin ligase, FERONIA, phosphorylation, RALF1, 14-3-3 protein
Abstract
The ratio between carbon (C) and nitrogen (N) utilization must be precisely coordinated to enable plant growth. Although numerous physiological studies have examined carbon/nitrogen (C/N) ratios, the mechanisms of sensing the C/N balance and C/N signaling remain elusive. Here, we report that a mutation of FERONIA (FER), a receptor kinase that plays versatile roles in plant cell growth and stress responses, caused hypersensitivity to a high C/N ratio in Arabidopsis. In contrast, FER-overexpressing plants displayed more resistant phenotypes. FER can interact with and phosphorylate ATL6, an E3 ubiquitin ligase that has been shown to regulate plant C/N responses. FER-mediated ATL6 phosphorylation enhanced the interaction between ATL6 and its previously identified target 14-3-3 proteins, thus decreasing 14-3-3 protein levels, leading to an increased insensitivity to high C/N ratios. Further analyses showed that the rapid alkalinization factor peptide (RALF1), which is a ligand of FER, also influenced the stability of 14-3-3 proteins via a FER–ATL6-mediated pathway. These findings reveal a novel regulatory mechanism that links the RALF1/FER–ATL6 pathway to whole-plant C/N responses and growth.
Introduction
Carbon (C) and nitrogen (N) are essential nutrients for plants to maintain routine and fundamental cellular activities. Various carbohydrates provide not only the energy but also the C skeletons for the biosynthesis of myriad compounds including amino acids (Coruzzi and Bush, 2001; Coruzzi and Zhou, 2001). N is essential in many aspects of plant growth and metabolism, such as amino acid metabolism and energy homeostasis (Novoa and Loomis, 1981; Frink et al., 1999). In addition to the importance of C and N individually, the precise coordination of cellular C and N (referred to as the C/N ratio or response) is critically important for the growth, development, biomass accumulation, and metabolism of plants (Vidal and Gutiérrez, 2008; Eveland and Jackson, 2012; Gutiérrez, 2012). Genome-wide studies show that a series of pathways involved in plant growth and development, including glycolysis/gluconeogenesis, protein metabolism, protein targeting, and activity, are controlled by the coordination of C and N status (Palenchar et al., 2004; Gutiérrez et al., 2007). Although the physiological impacts of the C/N ratio have been studied in considerable depth, the molecular mechanisms underlying the perception and regulation of this fundamentally important ratio in plant cellular metabolism are not well understood.
In the past two decades, researchers have identified several regulatory proteins that directly or indirectly mediate C/N responses, including the nitrate transporter NTR2.1, the glutamate receptor GLR1.1, the methyltransferase OSU1, the phosphatase type 2C protein ABI1, the glutamine synthetase, and the soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor SPY21 (Hsieh et al., 1998; Kang and Turano, 2003; Little et al., 2005; Gao et al., 2008; Maekawa et al., 2014; Bao et al., 2015; Lu et al., 2015). The wide variety of protein types represented in this list underscores the complexity of the sophisticated regulatory network that controls the plant C/N ratio. Although progress has been made in characterizing C/N-related regulatory proteins, it is likely that other novel mediators and/or pathways remain to be characterized.
The ATL (Arabidopsis Tóxicos en Levadura) family encodes plant-specific E3 ubiquitin ligase proteins, all of which contain a variable RING-H2 domain and a hydrophobic region located at their N-terminus. The Arabidopsis genome harbors thirteen 14-3-3 isoforms, and each isoform shows different binding affinity for its client proteins (Bachmann et al., 1996; Rosenquist et al., 2000). ATL31 has, to date, been demonstrated to associate with isoforms of 14-3-3s, 14-3-3λ, and 14-3-3χ in a phosphorylation-dependent manner; these associations target the 14-3-3 proteins for ubiquitination (Sato et al., 2009; Yasuda et al., 2014). It is known that 14-3-3 proteins regulate diverse biological processes by binding phosphorylated target proteins in eukaryotes (Ferl et al., 1999; Cotelle and Mackintosh, 2001; Aitken, 2006; Chevalier et al., 2009). In plants, 14-3-3 proteins have been shown to associate with a great many enzymes involved in primary C and N metabolism and to modulate their catalytic activities and their proteolytic degradation (Huber et al., 2002). These enzymes include sucrose-phosphate synthase, glutamine synthetase, nitrate reductase, and trehalose-phosphate synthase, among others (Moorhead et al., 1996; Cotelle et al., 2000; Riedel et al., 2001; Kulma et al., 2004). Diaz et al. (2011) performed parallel analyses of metabolites and enzyme activities with 14-3-3-overexpressing transgenic and knockout plants, and found that the modifications of 14-3-3 proteins were associated with the drastic changes in plant C/N responses. Transgenic plants overexpressing 14-3-3χ exhibited increased sensitivity to a high C/N ratio (Sato et al., 2011). The hypersensitivity phenotype of atl31 mutant plants to a high C/N ratio may result from the excessive accumulation of 14-3-3 proteins. A kinase target to ATL31 (KTA) was identified using a proteomics approach, but remains to be biochemically characterized (Yasuda et al., 2014). A subsequent study showed that CBL-interacting protein kinases, CIPK7, CIPK12, and CIPK14, negatively regulated C/N responses by phosphorylating ATL31 (Yasuda et al., 2017).
ATL6 is the closest homolog of ATL31 in Arabidopsis (65% identity at the amino acid level). Previous studies demonstrated that ATL6 and ATL31 have similar functions in C/N response and in defense responses (Sato et al., 2011; Maekawa et al., 2012). Similar to ATL31, ATL6 was also shown to interact with 14-3-3λ and 14-3-3χ (Sato et al., 2011; Yasuda et al., 2014). Further studies demonstrated that multiple 14-3-3 proteins are substrates of ATL6 (Sato et al., 2011; Yasuda et al., 2014). We propose that ATL6 probably targets 14-3-3 proteins for ubiquitination in a phosphorylation-dependent manner. However, the key kinase that functions upstream of the ATL6-mediated signaling pathway is still unknown.
FERONIA (FER) is an important receptor-like kinase that is known to play critical roles in fertilization, pathogen resistance, seed development, vegetative growth, root hair elongation, hormone responses, and starch accumulation (Huck et al., 2003; Escobar-Restrepo et al., 2007; Guo et al., 2009; Deslauriers and Larsen, 2010; Duan et al., 2010; Kessler et al., 2010; Keinath et al., 2010; Huang et al., 2013; Yu et al., 2012, 2014; Yang et al., 2015; Li et al., 2016; Liao et al., 2016; Stegmann et al., 2017; Feng et al., 2018). It is expressed broadly, and FER loss-of-function mutants have highly pleiotropic phenotypes affecting cell growth, suggesting that FER acts at the center of multiple signaling pathways. Here, we discovered that FER plays a regulatory role in plant response to C/N ratios. We used genetic and physiological approaches to demonstrate that FER is an essential component that modulates the phosphorylation of ATL6, a process which regulates the stability of 14-3-3 proteins in response to altered C/N ratios.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana was maintained with a 16 h light/8 h dark photoperiod at 22 °C in a growth chamber (Percival, Perry, IA, USA). The fer-4, fer-5, and srn mutants were previously described (Rotman et al., 2003; Escobar-Restrepo et al., 2007; Duan et al., 2010). Wild-type (WT) Columbia (Col-0) was used as control for fer-4 and fer-5. C24 was used as control for the srn mutant. The T-DNA insertion line SALK_083652 (atl6) was obtained from the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org). For seedlings grown on plates, seeds were surface-sterilized with 75% ethanol for 2 min and 10% NaClO (v/v) for 15–20 min. After washing at least three times with sterile water, seeds were stratified at 4 °C for 2 d in the dark and were then grown on Murashige and Skoog (MS) medium supplemented with 0.8% sucrose and 0.75% agar.
Plant transformation
To generate transgenic Arabidopsis overexpressing ATL6, ATL6T240A/T276A/S278A (hereafter designated ATL63A), and RALF1 (rapid alkalinization factor 1) constructs, ATL6, mutated forms of ATL6, and RALF1 coding sequence tagged with Myc that were driven by the Actin2 promoter were individually cloned into a pDT1 vector (He et al., 2016). The constructed plasmids were introduced into Agrobacterium tumefaciens strain GV3101 (pMP90) and transformed into both the Col-0 and fer-4 mutant backgrounds using the Agrobacterium-mediated floral dip method (Clough and Bent, 1998). The homozygous lines were obtained by self-pollination. At the T3 generation, two independent homozygous lines with high expression levels for each gene were chosen for further experiments. The target gene expression levels in transgenic lines were determined by semi-quantitative real-time PCR (RT-PCR) or quantitative RT-PCR (qRT-PCR). The Actin2 gene was used as an internal control. FER-overexpressing transgenic Arabidopsis was generated by Du et al. (2016). Primer pairs used for cloning are listed in Supplementary Table S1 at JXB online.
Carbon/nitrogen response assays
A modified MS medium containing various concentrations of sucrose and nitrogen was used in this assay, as described previously by Yasuda et al. (2014). The medium contained a constant sucrose concentration (200 mM) but had varying concentrations of nitrogen (3 mM and 1 mM): 200C/3N (200 mM sucrose, 1 mM KNO3, 1 mM NH4NO3) and 200C/1N (200 mM sucrose, 0.33 mM KNO3, 0.33 mM NH4NO3). After 2 d of vernalization, plates were transferred to a growth chamber for germination and growth. The green cotyledon rates were determined 8 d after germination from data recorded from three independent experiments, each of which had at least three replicates per group. Each treatment included at least 30 seedlings.
Gene expression analysis
For qRT-PCR analysis, 8-day-old seedlings grown on MS medium were treated with different C/N ratios for 2 d (see Fig. 1 for ratios). Materials were harvested and then immediately ground to a fine powder in liquid nitrogen. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and treated with RNase-free DNase I (Promega, Madison, WI, USA) to remove potential DNA contamination. A 2 µg aliquot of total RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (Takara, Japan). The reaction was performed using an ABI 7900HT instrument (Applied Biosystems, USA) with Power SYBR Green PCR master mix (Applied Biosystems). The data were analyzed by the comparative Ct method as described by Livak and Schmittgen (2001). The Actin2 gene was used as the internal control. Three biological replicates and three technical replicates were performed for each sample. For semi-quantitative RT-PCR, cDNA was prepared as described above. PCR was performed in a total volume of 20 μl with 2 μl of the reverse transcription reactions, 0.2 μM gene-specific primers, and 1 U of rTaq DNA polymerase (Takara). A total of 25–27 cycles were performed. The primer pairs used in these analyses are listed in Supplementary Table S2.
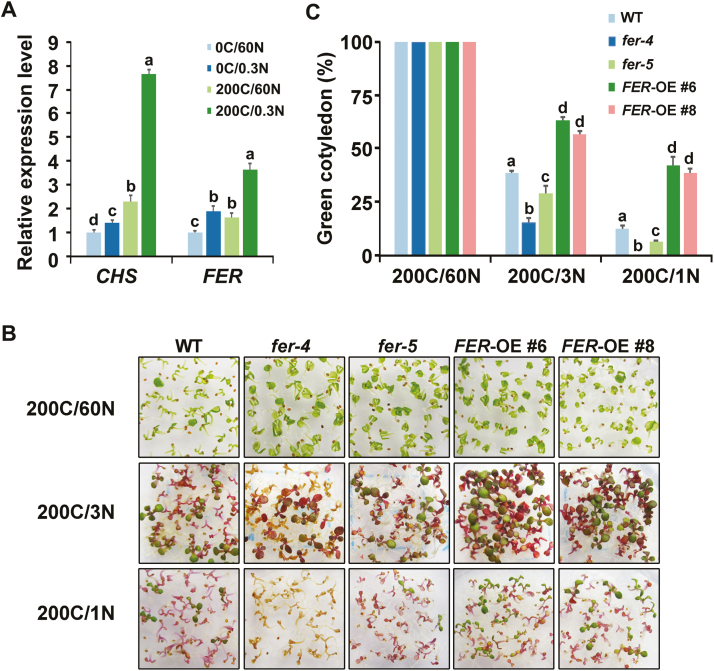
Fig. 1.
FER is involved in plant C/N response. (A) Expression level of FER under different C/N ratios. qRT-PCR was performed using total RNA isolated from 8-day-old WT seedlings (Col-0) treated with different C/N ratios for 2 d. Actin2 was used as an internal control. Expression data relative to 0C/60N were normalized to those of Actin2. Values are means ±SD from three independent experiments. (B) Phenotypic analysis of WT, fer-4, fer-5, and two FER-OE lines grown on medium containing a constant level of sucrose (200 mM) but with varying concentrations of nitrogen (3 mM and 1 mM). Photographs were taken 8 d after germination, and the number of plants with green cotyledons were scored. #6 and #8 represent transgenic line 6 and line 8. (C) Green cotyledons of WT, fer-4, fer-5, and two FER-OE lines grown under different C/N conditions as described in (B). Data are means ±SD from three independent experiments analyzing at least 30 seedlings each. Different letters above the bars indicate significant differences in each condition assessed by one-way ANOVA. C, carbon (sucrose); N, nitrogen. (This figure is available in color at JXB online.)
Yeast two-hybrid (Y2H) assays
Assays for protein–protein interactions by Y2H were performed using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech Laboratories Inc., CA, USA) according to the manufacturer’s manual. The full-length coding regions of ATL6 and ATL31 were cloned into pGADT7 individually. BD-FER-KD (FER-KD, FER kinase domain, amino acids 469–896) was reported previously (Du et al., 2016). The full-length coding region of 14-3-3χ (AT4G09000) was ligated into pGBKT7. Primers used for cloning are listed in Supplementary Table S1. The substitution forms of AD-ATL6173–398 were synthesized (Thr240, Thr276, and Ser278 were substituted with Ala to block phosphorylation, either individually or in combination) by Sangon Biotech (Shanghai); all mutation sites were confirmed by DNA sequencing. The desired pairs of pGADT7 and pGBKT7 vectors were co-transformed into yeast strain AH109 using LiAc-mediated transformation. Protein–protein interactions were identified by growth assays on SD/-Trp/-Leu/-His medium supplemented with 10 mM 3-aminotriazole (3-AT) or by β-galactosidase. Empty vectors were co-transformed as negative controls. The β-galactosidase assay was performed with a Yeast β-Galactosidase Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s manual. Briefly, colonies were resuspended in Y-PER reagent, and the OD660 values of homogeneous solutions were determined. Then, β-galactosidase assay buffer was added to tubes and placed at 37 °C until a yellow color was observed. The total reaction time was recorded at the end of the experiment. After centrifugation, absorbance of the supernatant at 420 nm was measured.
Bimolecular fluorescence complementation (BiFC) assays
For BiFC assays, the coding sequences of ATL6 and ATL31 were amplified with specific primers (Supplementary Table S1) and cloned into the pE3449 plasmid. The pE3308-FER-nVenus construct was that of Chen et al. (2016). To prepare Arabidopsis protoplast cells, 4-week-old Arabidopsis rosette leaves were slit using a razor blade. Leaf strips were incubated in an enzyme solution (1.5% cellulase R10, 0.4% macerozyme R10, 20 mM KCl, 10 mM CaCl2, 0.1% BSA, 20 mM MES, pH 5.7) for 3 h in the dark with gentle rotation (40–50 rpm). Protoplast transfection was performed with a standard polyethylene glycol transformation method. The protoplasts were treated with FM4-64 (2 µM) for 5 min before imaging. After incubation in the dark for 12–18 h at 23 °C, fluorescence was observed using a TCS STED CW confocal laser microscope (LEICA, Wetzlar, Germany).
Co-immunoprecipitation (Co-IP) assay
For Co-IP assay using Flag agarose, the full-length coding sequences of FER and ATL6 were cloned into the pCAMBIA1301-Flag and pDT1 backbones, respectively, to generate the FER-Flag and ATL6-Myc expression vectors. Both constructs were then transiently co-expressed in Nicotiana benthamiana. The co-infiltrated parts of N. benthamiana leaves were harvested 48 h post-infiltration. Total proteins were extracted with NEB-T buffer (20 mM HEPES, pH 7.5, 40 mM KCl, 5 mM MgCl2, 0.1% Triton X-100) containing a protease inhibitor (Roche, Basel, Switzerland). After centrifuging at 13 000 g for 20 min, the supernatant was incubated with ANTI-FLAG® M1 Agarose Affinity Gel (Sigma) for 3–4 h at 4 °C with gentle shaking, and precipitates were washed six times with NEB-T buffer. Immunoprecipitates were boiled in 1× SDS–PAGE loading buffer for 5 min before immunoblot analysis. Anti-Myc and anti-Flag antibodies were used for detecting ATL6 and FER, respectively.
For the Co-IP assays using anti-c-Myc magnetic beads (Thermo Fisher Scientific), total protein extracts of WT ATL6-Myc and ATL63A-Myc transgenic plants were prepared from 8-day-old seedlings as described above. Protein extracts were incubated with pre-washed beads for 2 h at room temperature with gentle mixing. The beads were washed three times with washing buffer (125 mM Tris, 0.75 M NaCl, 0.25% Tween-20 detergent), and boiled in 1× SDS–PAGE loading buffer. Proteins were separated by 10% SDS–PAGE for immunoblotting assays using anti-14-3-3 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-ACTIN antibodies (CMCTAG, Milwaukee, WI, USA).
In vitro phosphorylation assay and LC-MS/MS analysis
The abscisic acid (ABA)-induced phosphorylation co-expression system developed for studying the FER-mediated phosphorylation reaction was precisely described by Li et al. (2018). Since ATL6 is a transmembrane protein, we expressed ATL6 in Escherichia coli without a transmembrane domain. In brief, plasmids pACYC-PYL1-FER, pACYC-PYL1-FERK565R, and pRSF-ABI1-ATL688–398 were constructed. pRSF-ABI1-ATL688–398 together with pACYC-PYL1-FER (or pACYC-PYL1-FERK565R) were transformed into BL21 E. coli cells. The transformed E. coli strains were then inoculated into LB medium (containing kanamycin and chloromycetin) at 37 °C. Isopropyl-β-d-thiogalactoside (IPTG; 0.5 mM) was added to induce the expression of proteins when the OD600 was 0.6. After 2 h of induction, 50 µM ABA was added to release the phosphorylation activity of FER for 10 min. Anti-His antibody was used to detect the phosphorylation band shift via an immunoblotting assay. The bands of interest were isolated, and gel strips were subjected to alkylation/tryptic digestion followed by LC-MS/MS as described by Du et al. (2016). MS was performed using an EASY-nano LC system (Proxeon Biosystems) coupled with an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Xcalibur v.2.1 and Proteome Discoverer v.1.3 beta (Thermo Fisher Scientific) were employed to analyze raw data against the Arabidopsis database; this resource can be accessed via UniProt/Swiss-Prot and UniProt/TrEMBL.
Analysis of the amount of 14-3-3 proteins
To detect the accumulation of 14-3-3 proteins in transgenic Arabidopsis overexpressing different forms of ATL6, seedlings were grown on medium containing 200C/60N or 200C/3N for 8 d. For the detection of 14-3-3 proteins in fer4 and FER transgenic plants, 8-day-old seedlings were transferred to medium containing 200C/1N for 4 d. Total proteins were extracted by NEB-T buffer as described above. A 10% SDS–PAGE gel was used to separate proteins. Anti-14-3-3 and anti-ACTIN antibodies were used for immunoblotting analysis.
RALF1 and MG132 treatment
RALF1 treatment was performed as described by Li et al. (2018) with minor modifications. Eight-day-old seedlings grown on MS were transferred to liquid MS medium containing 1 μM RALF1 for 0.5 h. Medium with an equal volume of buffer was used as a mock control. For MG132 coupled with RALF1 treatment, seedlings of the WT were first treated with 50 μM MG132 (Selleckchem). After 1 h incubation, seedlings were transferred to solution with or without 1 μM RALF1 for 0.5 h. Harvested seedlings were ground in liquid nitrogen for further protein extraction and immunoblotting analysis using anti-Myc and anti-14-3-3 antibodies. For determining whether the degradation of 14-3-3 proteins was dependent on the ubiquitin–proteasome system (UPS), 7-day-old seedlings grown on MS medium were transferred to medium containing 200C/3N with or without 20 μM MG132 for 3 d. Total protein extraction and western blotting were performed as described above.
Statistical analysis
All data were tested based on one-way ANOVA using SPSS 13.0 (SPSS Inc., USA).
Results
FER is involved in plant C/N responses
Extensive studies have demonstrated that FER has pleiotropic roles in a variety of cellular processes (Escobar-Restrepo et al., 2007; Duan et al., 2010; Chen et al., 2016; Stegmann et al., 2017; Feng et al., 2018, Li et al., 2018), but there are few studies concerning its potential role in nutrition or metabolism. Previous works illustrated that the fer mutant exhibits sucrose-conditional ectopic starch accumulation and a cellulose deficiency (Yang et al., 2015; Yeats et al., 2016). These observations led us to propose that FER may function in nutrition or energy metabolism. We initially found that FER exhibited higher expression levels under a high C/N ratio than under control growth conditions (Fig. 1A); similar expression trends were reported for CHALCONE SYNTHASE (CHS), a known C/N response marker gene involved in anthocyanin biosynthesis (Martin et al., 2002). High C/N represents a severe form of C/N stress during the post-germination stage, and is known to prevent the transition from heterotrophic to photoautotrophic growth (Sato et al., 2009). Previous work established a method to study C/N responses with sugar supplementation but varied nitrogen media during the early post-germination stage in Arabidopsis (Sato et al., 2009). With this method, seedlings exposed to high C/N conditions exhibit strong purple pigmentation as well as a lower proportion of green cotyledons and decreased survival rates. Specifically, the percentage of green cotyledons is used as an indicator to evaluate the sensitivity of plants to high C/N.
To examine the role of FER in plant C/N responses, we performed a series of diagnostic phenotypic assays with fer-4 and fer-5 mutants, and FER overexpression (FER-OE) plants, together with their corresponding WT controls. Duan et al. (2010) showed that fer-4 is a null mutant with its T-DNA insertion in the extracellular domain-coding region of the FER locus and that fer-5 expresses truncated transcripts with its T-DNA insertion positioned at the end of its kinase domain. Whereas no full-length or truncated transcripts of FER are detected in the fer-4 mutant, fer-5 expresses truncated transcripts of its extracellular domain-coding region. Since a 200C/60N ratio is close to the typical C/N ratio of normal MS medium, we designated this as the control condition for our study. All the examined seedlings developed green cotyledons at the 200C/60N ratio (Fig. 1B, C). As N concentrations in media were decreased, growth of WT seedlings was arrested, and the percentage of green cotyledons gradually reduced. Compared with the WT, fer-4 exhibited increased hypersensitivity, with seedlings showing significant lethality under the 200C/1N condition. The sensitivity of fer-5 to high C/N was between that of the WT and the fer-4 mutant. Two different FER-OE transgenic lines (FER-OE #6 and #8) showed reduced sensitivity to a high C/N ratio, exhibiting a higher percentage of green cotyledons and reduced growth arrest. These results suggest that the expression level of FER may be a determinant of plant response to high C/N ratios. siréne (srn), whose mutation is allelic with fer-4 in the C24 background (Rotman et al., 2003; Huck et al., 2003; Escobar-Restrepo et al., 2007), was also included in this study. srn seedlings showed hypersensitivity to high C/N ratios similar to fer-4 (Supplementary Fig. S1). Collectively, these results establish that FER functions in C/N responses in Arabidopsis.
FER physically interacts with ATL6
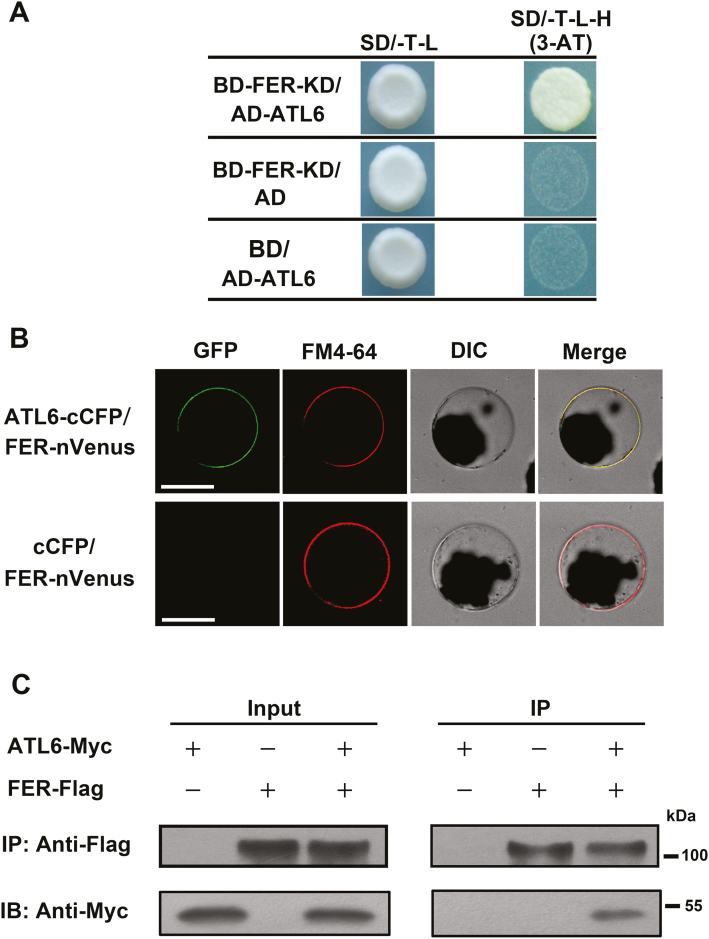
A Y2H assay was performed using the FER-KD (amino acids 469–896) as a bait to screen new interactors involved in C/N responses. ATL6, an E3 ubiquitin ligase known to regulate the C/N response in Arabidopsis (Supplementary Fig. S2; Sato et al., 2009; Maekawa et al., 2012), was thus identified as a potential interactor. An independent Y2H assay confirmed the interaction between FER-KD and full-length ATL6 in yeast cells (Fig. 2A). To confirm the physical interaction in vivo, BiFC combined with FM4-64 staining (to label the plasma membrane) were conducted in Arabidopsis protoplasts. We generated a fusion protein comprising ATL6 and the C-terminal fragment of the cyan fluorescent protein (cCFP), as well as a fusion protein comprising FER and the N-terminal fragment of Venus (nVenus). The combination of the cCFP and nVenus fragments induced an observable fluorescence signal (Du et al., 2016). In Arabidopsis mesophyll protoplasts co-expressing ATL6–cCFP and FER–nVenus, we observed a cell surface-localized fluorescence signal that overlapped with the FM4-64 signal, indicating a physical interaction. However, no fluorescence was observed when FER–nVenus was co-expressed with an empty cCFP control construct (Fig. 2B). However, the closest homolog of ATL6 in Arabidopsis, ATL31, did not show any interaction with the FER-KD in the Y2H or BiFC assays (Supplementary Fig. S3). Co-IP analysis was also conducted to confirm the in vivo interaction between FER and ATL6. Specifically, Flag-tagged FER (FER-Flag; Du et al., 2016) was co-expressed with Myc-tagged ATL6 (ATL6-Myc) in N. benthamiana leaves via agroinfiltration. As shown in Fig. 2C, ATL6-Myc was co-immunoprecipitated with FER-Flag, indicating that FER physically associates with ATL6 in vivo. When FER-Flag or ATL6-Myc was expressed alone, ATL6-Myc was not immunoprecipitated with the anti-Flag agarose beads. Taken together, these results demonstrate that FER physically interacts with ATL6, findings that led us to explore further the mechanism of this interaction.
Fig. 2.
FER physically interacts with ATL6. (A) Y2H assays show the interaction between FER and ATL6. Synthetic dropout medium/-Trp/-Leu/-His (SD/-T-L-H) supplemented with 10 mM 3-AT was used for evaluating protein interaction. (B) BiFC assays indicating the interaction between FER and ATL6 in Arabidopsis protoplast cells. Before imaging, the protoplasts were treated with FM4-64 (2 µM) for 5 min. The experiments were repeated three times with similar results. Scale bar=25 µm. (C) FER co-immunoprecipitated with ATL6. FER-Flag was co-transformed with ATL6-Myc in N. benthamiana leaves via agroinfiltration. At 48 h post-infiltration, protein extracts were immunoprecipitated with anti-Flag beads and the immunoprecipitates were assessed by western blotting using anti-Flag and anti-Myc antibodies. (This figure is available in color at JXB online.)
FER phosphorylates ATL6 and promotes its interaction with 14-3-3 proteins
In previous studies, ATL31 has been reported to regulate plant C/N responses by regulating the stability of 14-3-3 proteins in a phosphorylation-dependent manner (Sato et al., 2009, 2011; Maekawa et al., 2012; Yasuda et al., 2014). A
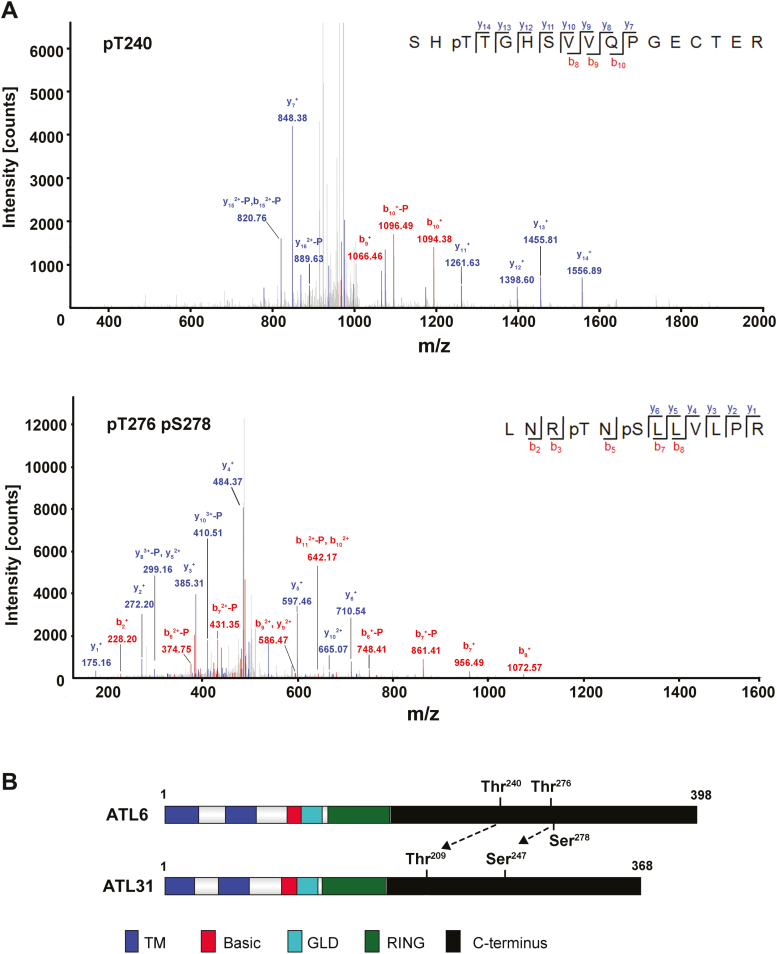
phosphorylation-dependent role for ATL6 in C/N responses is suggested by ATL6’s highest identity to ATL31 among ATL family proteins. Recall that the putative upstream protein kinase which functions in the ATL6-mediated signaling pathway remains elusive. Considering that FER has kinase activity (Chen et al., 2016; Du et al., 2016; Li et al., 2018), and given our finding that it physically interacts with ATL6, we hypothesized that FER may target ATL6 for phosphorylation. We experimentally investigated FER-mediated phosphorylation reactions using a modified E. coli co-expression system developed by our research group (Li et al., 2018). We co-expressed PYL1, ABI1, FER-KD, and ATL688–398 (PYL1/ABI1/FER-KD/ATL6) in a single E. coli strain. In parallel, a mutant version of FER-KD (FER-KDK565R), in which its kinase activity was abolished, was also co-expressed with the same proteins (PYL1/ABI1/FER-KDK565R/ATL688–398) as a negative control. The co-expression system combined with LC-MS/MS analysis was performed to identify the FER phosphorylation residues on ATL6. Three phosphorylated residues on the C-terminal region of ATL6 could be reproducibly detected (Fig. 3A); we did not detect these residues in the PYL1/ABI1/FER-KDK565R/ATL688–398 system. Among these phosphorylation sites, Thr240 and Ser278 were found to be putative 14-3-3 protein-binding sites when motif analysis of ATL6 was performed using Scansite 3 (Supplementary Fig. S4A). Alignment of the amino acids between ATL6 and ATL31 showed that the phosphorylation sites on ATL6 were conserved with those on ATL31 (Supplementary Fig. S4B): Thr240 and Ser278 of ATL6 are equivalent to Thr209 and Ser247 of ATL31, respectively (Fig. 3B), suggesting that these phosphorylation sites are conserved and essential in plant responses to varied C/N ratios as regulated by different upstream kinases.
Fig. 3.
FER phosphorylates ATL6 in vitro. (A) MS/MS spectra show the phosphorylation of Thr240 (upper panel), Thr276, and Ser278 (lower panel). (B) Summary of in vitro phosphorylation sites identified in ATL6 by FER through LC-MS/MS analyses. Dotted arrows represent conserved phosphorylation sites between ATL6 and ATL31. TM, transmembrane-like hydrophobic region; Basic, region rich in basic amino acids; GLD, highly conserved region including Gly–Leu–Asp residues; RING, a RING-H2 type zinc finger. (This figure is available in color at JXB online.)
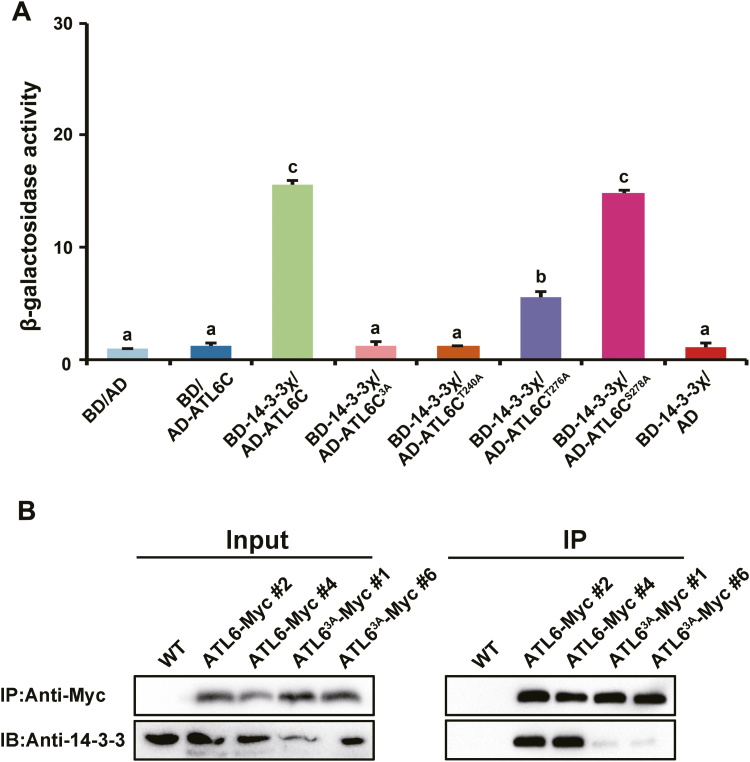
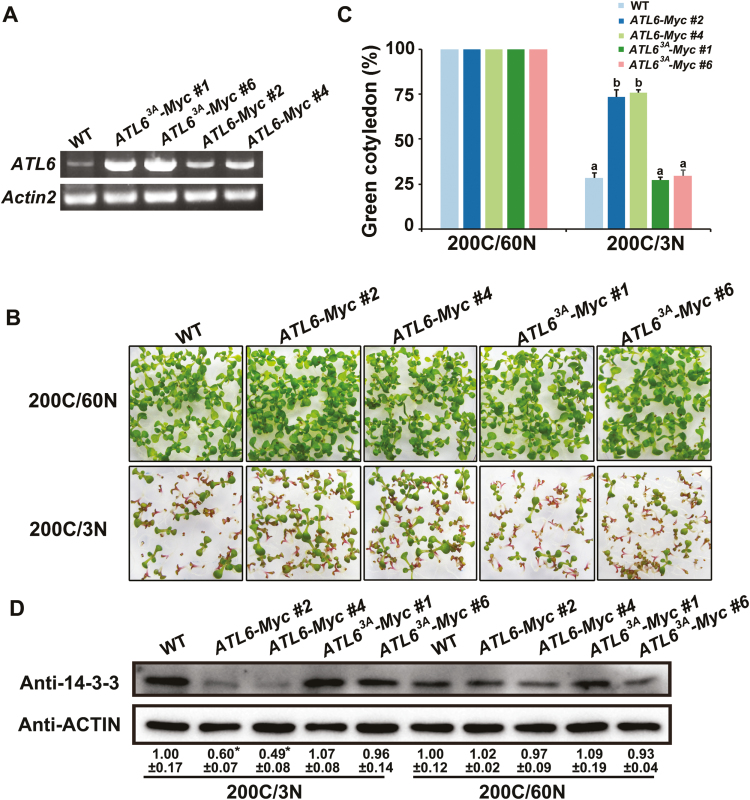
To explore whether the Thr240, Thr276, and Ser278 residues on ATL6 are required for binding to 14-3-3 proteins, we substituted Thr240, Thr276, and Ser278 with Ala to blocks phosphorylation (either individually or in combination), and tested their 14-3-3 binding capacity using β-galactosidase assays in a Y2H system. Guided by previously reported findings (Sato et al., 2011), we selected 14-3-3χ as a representative isoform to study the role of phosphorylation sites on ATL6 in the interaction between 14-3-3 proteins and ATL6. As the C-terminal region of ATL6 (amino acids 173–398) was shown to interact with 14-3-3χ in a Y2H assay (Sato et al., 2011), AD-ATL6173–398 (hereafter referred to as AD-ATL6C) was constructed and used for the β-galactosidase assay. The triple mutation in ATL6C3A abolished its interaction with 14-3-3χ, displaying low β-galactosidase activity (no difference from negative controls). As indicated in Fig. 4A, ATL6CT240A had a reduced interaction with 14-3-3χ compared with the interaction between ATL6C and 14-3-3χ, findings similar to that obtained for ATL6C3A and the negative controls, thereby suggesting that Thr240 is apparently a key phosphorylation site for 14-3-3χ binding. ATL6CT276A exhibited a less substantial reduction for its interaction as compared with the native form of ATL6C. ATL6CS278A did not show any obvious effects on the interaction. To support these findings with evidence for Arabidopsis, we generated transgenic plants overexpressing WT and mutant ATL6 tagged with Myc, ATL6-Myc, and ATL63A-Myc (phosphor-inactive mutant) in the Col-0 background. Multiple lines of each construct were obtained, and two lines with relatively higher protein expression levels for each construct were chosen for further Co-IP (Fig. 5A). As shown in Fig. 4B, Co-IP results further confirmed that mutation of the phosphorylation sites on ATL6 reduced the interaction of ATL6 and 14-3-3 proteins, consistent with our results obtained in the β-galactosidase activity assay. Taken together, these results indicate that FER-mediated phosphorylation on ATL6 is essential for its interaction with 14-3-3 proteins.
Fig. 4.
Phosphorylation of ATL6 by FER promotes its interaction with 14-3-3 proteins. (A) β-Galactosidase assay in the Y2H system. 14-3-3χ and different forms of ATL6 were cloned into pBGKT7 (BD) and pGADT7 (AD) vectors, respectively. Experiments were repeated three times with similar results. 3A, Thr240, Thr276, and Ser278 were substituted to Ala to block phosphorylation. Different letters above the bars indicate significant differences in each condition assessed by one-way ANOVA. (B) Co-IP assays. Crude proteins were extracted and subjected to immunoprecipitation using anti-Myc beads followed by immunoblot analysis with anti-14-3-3 and anti-Myc antibodies. Transgenic Arabidopsis overexpressing different forms of ATL6 were used in this experiment. # represents lines. (This figure is available in color at JXB online.)
Fig. 5.
FER-mediated phosphorylation of ATL6 enhances its function in response to a high C/N ratio. (A) RT-PCR analysis of the expression level of ATL6 in WT and transgenic Arabidopsis plants expressing ATL63A-Myc or ATL6-Myc. Total RNA was extracted from 8-day-old seedlings, and Actin2 was used as an internal control. (B) Phenotypes of WT, ATL6-Myc, or ATL63A-Myc transgenic plants grown on medium containing 200C/3N for 8 d. (C) The percentage of seedlings with green cotyledons in (B). Data are means ±SD from three independent experiments analyzing at least 30 seedlings each. Different letters above the bars indicate significant differences assessed by one-way ANOVA. (D) Comparison of the amount of 14-3-3 proteins in WT and transgenic Arabidopsis plants expressing ATL63A-Myc or ATL6-Myc under 200C/60N and 200C/3N conditions. Crude proteins were extracted from 8-day-old seedlings grown on medium as indicated above. Western blotting was performed using anti-14-3-3. Anti-ACTIN was used as a loading control. The numbers below the blot indicate the relative signal intensity compared with the WT. Data shown are means. Error bars indicate the SD (n=3). An asterisk indicates a significant difference compared with the WT (P<0.05). (This figure is available in color at JXB online.)
FER-mediated phosphorylation of ATL6 is required for C/N responses
To decipher the biological significance of FER-mediated ATL6 phosphorylation in regulating C/N responses, transgenic plants overexpressing ATL6-Myc and ATL63A-Myc were employed in C/N response assays (Fig. 5A). All seedlings exhibited expanded green cotyledons under the 200C/60N condition. When grown with a 200C/3N ratio, WT seedlings showed a strong growth arrest phenotype, displaying a strong purple pigmentation; only 28.6% of the cotyledons maintained greening. In contrast, transgenic plants overexpressing ATL6-Myc were less sensitive to high C/N ratios compared with the WT, with ~74% of seedlings exhibiting expanded green cotyledons. However, the percentage of green cotyledons in ATL63A-Myc transgenic plants decreased to a similar level to that observed in the WT (Fig. 5B, C), indicating that the phosphorylation of 14-3-3 protein-binding sites on ATL6 plays an important physiological role in plant C/N responses. We also overexpressed ATL63A-Myc or ATL6-Myc in the fer-4 background. ATL63A-Myc transgenic Arabidopsis plants did not rescue the hypersensitive phenotype of fer-4 to a high C/N ratio. However, the overexpression of ATL6-Myc slightly rescued the hypersensitive phenotype of fer-4, displaying a higher percentage of green cotyledons than fer-4 (Supplementary Fig. S5). This result suggested that FER-mediated ATL6 phosphorylation plays an essential role in plant C/N responses. Overexpression of ATL6-Myc only partially rescued the C/N response defects of fer-4, either because the ATL6 protein cannot be phosphorylated due to the lack of functional FER protein in the fer-4 mutant plants or owing to other unknown factor(s).
A previous study reported that ATL31 regulates the stability of 14-3-3 proteins in C/N responses (Yasuda et al., 2014). To explore whether the phosphorylation of 14-3-3 protein-binding sites on ATL6 regulates 14-3-3 protein stability during plant C/N responses, we examined the accumulation of 14-3-3 proteins in WT, ATL6-Myc, and ATL63A-Myc transgenic plants. The WT accumulated higher 14-3-3 protein levels under a high C/N ratio compared with the normal condition, findings in line with previously reported results (Sato et al., 2011). In contrast, accumulation of 14-3-3 proteins was reduced in ATL6-Myc transgenic plants under a high C/N ratio (Fig. 5D). Interestingly, this reduction was rescued in ATL63A-Myc transgenic plants; which had 14-3-3 levels similar to the WT (Fig. 5D). As reported in a previous study, ATL6 targeted 14-3-3 proteins for degradation via the UPS in the in vitro protein ubiquitination assay (Sato et al., 2011). To confirm this observation in plants, MG132, which effectively blocks the proteolytic activity of the 26S proteasome, was employed to treat WT, ATL6-Myc, and ATL63A-Myc transgenic plants under a high C/N ratio. ATL6-Myc transgenic plants accumulated fewer 14-3-3 proteins than WT and ATL63A-Myc transgenic plants. However, the reduction was rescued when plants were treated with MG132. However, 14-3-3 levels in ATL63A-Myc transgenic plants were similar to those in the WT in the presence or absence of MG132 (Supplementary Fig. S6), suggesting that ATL6-mediated degradation of 14-3-3 proteins is dependent on the UPS in Arabidopsis. Collectively, these observations suggest that ATL6 regulates C/N responses in a phosphorylation-dependent manner by mediating the stability of 14-3-3 proteins.
The FER–ATL6 interaction mediates RALF1’s modulation of 14-3-3 protein levels during C/N responses
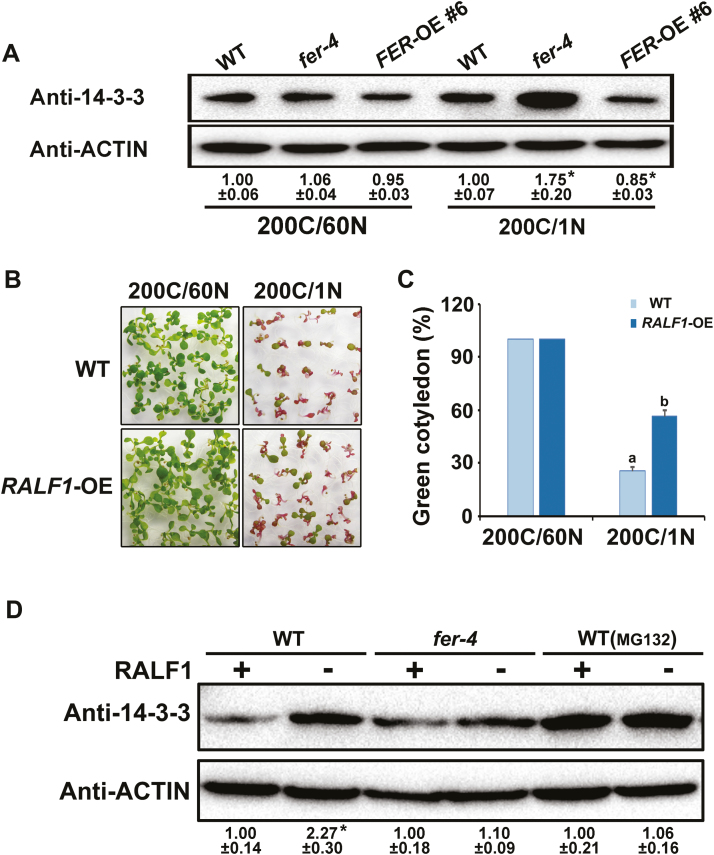
It is known that ATL31 interacts with 14-3-3 proteins, and the strong accumulation of 14-3-3 proteins in atl31 is associated with C/N sensitivity phenotypes (Sato et al., 2011). The hypersensitivity phenotype of fer-4 to a high C/N ratio, considered alongside our discovery of the FER–ATL6 interaction, led us to examine the 14-3-3 protein levels in fer-4 mutants. fer-4 plants accumulated higher 14-3-3 protein levels than WT plants under a high C/N ratio. In contrast, FER-OE transgenic plants accumulated reduced 14-3-3 protein levels compared with the WT (Fig. 6A). This result is consistent with the hypothesis that FER regulates C/N responses by mediating the stability of 14-3-3 proteins.
Fig. 6.
RALF1 peptide participates in C/N response by affecting the stability of 14-3-3 proteins. (A) Comparison of the amounts of 14-3-3 proteins in WT, fer-4, and FER-OE seedlings under 200C/60N and 200C/1N conditions. The numbers below the blot indicate the relative signal intensity compared with the WT. Data shown are means. Error bars indicate the SD (n=3). An asterisk indicates a significant difference compared with the WT (P<0.05). (B) Phenotypes of WT and RALF1-OE transgenic plants grown on medium containing 200C/1N for 8 d. (C) The percentage of seedlings with green cotyledons in (B). The bar represents the mean ±SD from three independent experiments examining at least 30 seedlings each. Different letters above the bars indicate significant differences assessed by one-way ANOVA. (D) RALF1 peptide affected the stability of 14-3-3 proteins in an FER-dependent manner. Eight-day-old seedlings were treated with 0 μM or 1 μM RALF1 peptide for 0.5 h. Total proteins were extracted, and western blotting was performed using anti-14-3-3 as described above. The ACTIN protein was used as a loading control. C, carbon (sucrose); N, nitrogen. (This figure is available in color at JXB online.)
RALF1 is a secreted peptide that is known to directly bind to and activate FER (Haruta et al., 2014). To investigate whether RALF1 participates in plant C/N responses, we examined the sensitivity of RALF1-OE transgenic plants to a high C/N ratio. The percentage of green cotyledons in RALF1-OE transgenic plants was higher than in the WT (Fig. 6B, C), suggesting that RALF1 is indeed involved in plant C/N responses. We also conducted experiments wherein recombinant RALF1 peptide was added to seedling growth media and we found that such treatment caused obviously reduced accumulation of 14-3-3 proteins. Importantly, no such reduction was observed for samples grown in media which contained both the RALF1 peptide and the proteasome inhibitor MG132, thereby demonstrating the functional contribution of proteasomal degradation to this RALF1-mediated regulation of 14-3-3 protein levels (Fig. 6D). It was also very interesting to note that the 14-3-3 protein accumulation level in the fer-4 mutant did not differ significantly upon treatment with the RALF1 peptide, revealing that RALF1 mediates the stability of 14-3-3 proteins in a FER-dependent manner. This result is consistent with the hypothesis that the activation of FER by RALF1 increases phosphorylation of ATL6, a biochemical event that we have also shown to alter the stability of 14-3-3 proteins.
Discussion
Extensive studies have revealed that the C/N balance is important not only for fundamental research but also for applied research in plants. A better understanding of the mechanisms of C/N balance will be instructive for crop engineering, such as improvement of crop yield and biofuel production. However, the sensing and signaling mechanisms underlying C/N balance have long remained elusive. These mechanisms started to be clarified when ATL31 and ATL6 were identified to play regulatory roles in C/N responses in several biological processes, such as during the post-germination stage and in defense response (Sato et al., 2009, 2011; Maekawa et al., 2012). In the present study, we present data showing that FER, a key hub of cell signaling networks mediating hormone, stress, and immune responses, also regulates plant C/N responses by modulating the stability of 14-3-3 proteins via an ATL6-mediated signaling pathway.
ATL31 and ATL6 are close homologs in Arabidopsis. Previous studies reported that these proteins have similar functions in plant C/N responses (Sato et al., 2009; Maekawa et al., 2012). ATL31/6 target 14-3-3 proteins for ubiquitination and promote 14-3-3 protein degradation by the 26S proteasome in C/N responses (Sato et al., 2011). ATL31 binds to 14-3-3 proteins depending on its phosphorylation, and CIPK7/12/14 are the protein kinases that function upstream of ATL31 (Yasuda et al., 2014, 2017). We found that FER interacts with (Fig. 2) and phosphorylates ATL6 (Fig. 3). However, we failed to observe any interaction between FER and ATL31 in Y2H and BiFC assays (Supplementary Fig. S3), suggesting that ATL31 and ATL6 are perhaps phosphorylated by different protein kinases (such as other FER family-related kinase members) and may be involved in different kinase-mediated signaling pathways. Despite ATL6 and ATL31 being phosphorylated by different kinases, their phosphorylation sites are conserved to some extent. Our data showed that three residues (Thr240, Thr276, and Ser278) on ATL6 were phosphorylated by FER. Thr240 and Ser278 were equivalent to Thr209 and Ser247 on ATL31 (Fig. 3B). Consistent with the conclusion that Thr209 of ATL31 plays a central role in binding 14-3-3 proteins (Maekawa et al., 2014), we also found that Thr240 of ATL6 is a key site for the interaction of ATL6 and 14-3-3 proteins (Fig. 4A). However, ATL6S278A did not show an obvious effect on this interaction, which is different from the previously reported finding that Ser247 of ATL31 affects the interaction between ATL31 and 14-3-3 proteins (Maekawa et al., 2014). Additionally, our study identified a novel phosphorylation site of ATL6, Thr276. Mutating Thr276 to Ala reduced the interaction of ATL6 and 14-3-3 (Fig. 4A), indicating that Thr276 is an ATL6-specific phosphorylation site that plays a role in this protein’s interaction with 14-3-3 proteins. Mutation of the phosphorylation sites of ATL6 reduced the interaction with 14-3-3χ in yeast as well as in plants. In a previous study, ATL31 was also found to interact with 14-3-3s in yeast cells (Sato et al., 2011; Yasuda et al., 2014), and the authors of those studies suggested that a kinase for ATL31 is broadly conserved in eukaryotes. As FER is a plant-specific receptor-like kinase, yeast do not harbor FER homologs. We speculate that the native form of ATL6 has low affinity for 14-3-3 proteins, and the mutated form has even lower affinity in yeast cells. In addition to this speculation, another possibility is that an unknown mechanism may exist in yeast that mediates the interaction between 14-3-3s and ATL6 or ATL31. Based on all presently available information, we suggest that the two signaling pathways mediated by ATL6 and ATL31 appear to converge to target 14-3-3 proteins for degradation in plant C/N responses. Thus, the identification of more kinases that can regulate phosphorylation of ATL6/31 will probably deepen our understanding of the molecular mechanisms responsible for plant C/N responses.
It is well established that FER functions as a versatile mediator that can differentially coordinate cell growth in distinct cell types and in response to various environmental factors (Duan et al., 2010; Mao et al., 2015; Li et al., 2018). Cell growth and proliferation are energy-consuming processes, suggesting that FER may participate in energy regulation. Several studies have provided clues that FER may be involved in energy management. For instance, FER was shown to regulate starch accumulation by interacting with GAPC1/2, enzymes that are critical for glycolysis (Yang et al., 2015). Yeats et al. (2016) showed that mutation of FER facilitates preferential utilization of carbon for synthesizing starch rather than cellulose under enhanced intracellular sucrose conditions. We here confirmed that FER plays a regulatory role in C/N responses (Fig. 1) by recruiting an ATL6-mediated signaling pathway, demonstrating a novel link for this protein to nutrient metabolism. As FER is a critical node of hormone and stress responses (Chen et al., 2016; Li et al., 2016), it would be interesting to determine whether different hormones or stress cues regulate C/N responses through the FER–ATL6 interaction. These observations together lead to the proposition that FER is a node linking cell growth, energy, and nutrient metabolism. The target of rapamycin (TOR) protein is known to function as a central integrator of nutrient, energy, and stress signaling networks (Wullschleger et al., 2006; Dobrenel et al., 2013; Yuan et al., 2013). TOR also affects carbon and nitrogen metabolism in Arabidopsis (Caldana et al., 2013). A recent study reported that TOR influences biomass accumulation and cell cycle progression by altering C/N balance in synchronized Chlamydomonas reinhardtii cells (Jüppner et al., 2018). Future work should test whether FER regulates energy metabolism through crosstalk with some other classical energy metabolism pathways, such as pathways mediated by SNF1-related kinase 1 (SnRK1) or TOR.
FER serves as a receptor for the RALF1 peptide (Haruta et al., 2014), which causes the rapid alkalinization of plant cell walls and inhibits root cell growth (Pearce et al., 2001). Several studies have reported that the RALF1–FER pathway is involved in many biological processes, such as cell growth, responses to pathogen invasion, abiotic responses, and stomatal movement (Haruta et al., 2014; Thynne et al., 2017; Stegmann et al., 2017; Li et al., 2018; Yu et al., 2018). Our work shows that RALF1 also participates in the regulation of plant C/N responses (Fig. 6). RALF1-OE transgenic plants displayed less sensitivity to a high C/N ratio than WT plants (Fig. 6B, C). 14-3-3 proteins were previously reported to regulate primary C and N metabolism by directly interacting with essential enzymes (Comparot et al., 2003; Shin et al., 2011). Transgenic plants overexpressing 14-3-3 showed increased sensitivity to high C/N ratios (Sato et al., 2011). The WT accumulated less 14-3-3 proteins when treated with RALF1 peptide, while the amount of 14-3-3 proteins in fer-4 was comparable before and after treatment (Fig. 6D), indicating that RALF1 regulates the stability of 14-3-3 proteins in a FER-dependent manner. It will be interesting to examine whether other members of the RALF family, such as RALF23 and RALF33, which are known to play a role in plant immunity (Stegmann et al., 2017; Guo et al., 2018), are also involved in C/N responses via the FER-mediated signaling pathway.
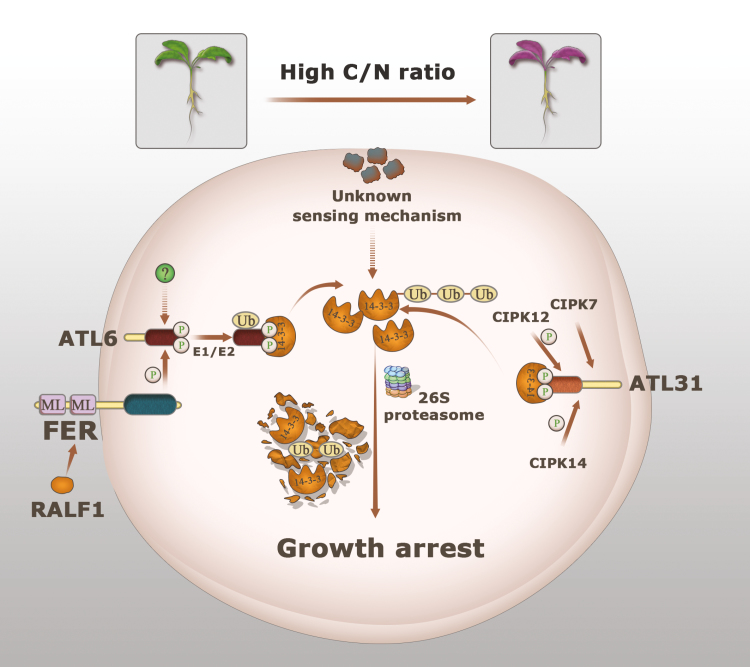
Based on our findings and previous reports in the literature, we propose a working model for how FER regulates C/N responses in Arabidopsis (Fig. 7). When plants experience a high C/N ratio, the altered C/N signal is sensed by an unknown nutrient-sensing mechanism, which causes excess accumulation of 14-3-3 proteins, eventually leading to growth arrest. ATL6 and ATL31 are two regulators that positively control plant C/N responses by mediating the stability of 14-3-3 proteins in a phosphorylation-dependent manner. CIPK7/12/14 are the key kinases which function upstream of the ATL31-mediated signal pathway. The protein kinase FER interacts with and phosphorylates ATL6, providing the binding sites for 14-3-3 proteins, which are eventually degraded via the UPS. Moreover, we propose that the RALF1 peptide is also involved in C/N responses via this FER–ATL6-mediated signal pathway.
Fig. 7.
A proposed working model of the FER-mediated C/N response. 14-3-3 proteins are accumulated in excess when plants experience a high C/N ratio, which leads to growth arrest. For the ATL6-mediated C/N response, receptor-like protein kinase FER interacts with and phosphorylates ATL6, which provides binding sites for 14-3-3 protein, and eventually targets it for degradation by the UPS. In addition to FER, ATL6 may be phosphorylated by other protein kinases in the C/N response. Moreover, we propose that RALF1 peptide may also be involved in the C/N response via the FER–ATL6-mediated signal pathway. Dashed lines with arrowheads represent possible suggested connections for which there is no direct experimental evidence yet. (This figure is available in color at JXB online.)
Supplementary data
Table S1. Primers used for vector construction.
Table S2. Primers used for qRT-PCR and semi-quantitative RT-PCR.
Fig. S1. Sensitivity of C24 and the srn mutant to high C/N ratios.
Fig. S2. ATL6 positively regulates high C/N response.
Fig. S3. Interaction between FER and ATL31 in the Y2H and BiFC asssys.
Fig. S4. 14-3-3 protein-binding sites on ATL6 and ATL31 were conserved.
Fig. S5. Phenotypes of fer-4, ATL6-Myc, or ATL63A-Myc transgenic plants in the fer-4 background grown under high C/N condition.
Fig. S6. ATL6-mediated degradation of 14-3-3 proteins was dependent on the ubiquitin–proteasome system in Arabidopsis.
Acknowledgements
We are grateful to Dr Li Jianglin (Hunan University) for LC/MS analysis, and the Arabidopsis Biological Resource Center for providing the atl6 mutant. This work was supported by grants from the National Natural Science Foundation of China (NSFC-31371244, 31571444, 31871396, and 31201012), China Postdoctoral Science Foundation (grant no. 2015M580634), and The Young Elite Scientist Sponsorship Program by CAST (grant no. YESS20160001). The authors declare no conflict of interest.
References
- Aitken A. 2006. 14-3-3 proteins: a historic overview. Seminars in Cancer Biology 16, 162–172. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. 1996. 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Letters 398, 26–30. [DOI] [PubMed] [Google Scholar]
- Bao A, Zhao Z, Ding G, Shi L, Xu F, Cai H. 2015. The stable level of glutamine synthetase 2 plays an important role in rice growth and in carbon–nitrogen metabolic balance. International Journal of Molecular Sciences 16, 12713–12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. 2013. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. The Plant Journal 73, 897–909. [DOI] [PubMed] [Google Scholar]
- Chen J, Yu F, Liu Y, et al. 2016. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, E5519–E5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier D, Morris ER, Walker JC. 2009. 14-3-3 and FHA domains mediate phosphoprotein interactions. Annual Review of Plant Biology 60, 67–91. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Comparot S, Lingiah G, Martin T. 2003. Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. Journal of Experimental Botany 54, 595–604. [DOI] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR. 2001. Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiology 125, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L. 2001. Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects’. Current Opinion in Plant Biology 4, 247–253. [DOI] [PubMed] [Google Scholar]
- Cotelle V, Mackintosh C. 2001. Do 14-3-3s regulate ‘resource allocation in crops’? Annals of Applied Biology 138, 1–7. [Google Scholar]
- Cotelle V, Meek SE, Provan F, Milne FC, Morrice N, MacKintosh C. 2000. 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. The EMBO Journal 19, 2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslauriers SD, Larsen PB. 2010. FERONIA is a key modulator of brassinosteroid and ethylene responsiveness in Arabidopsis hypocotyls. Molecular Plant 3, 626–640. [DOI] [PubMed] [Google Scholar]
- Diaz C, Kusano M, Sulpice R, Araki M, Redestig H, Saito K, Stitt M, Shin R. 2011. Determining novel functions of Arabidopsis 14-3-3 proteins in central metabolic processes. BMC Systems Biology 5, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Marchive C, Azzopardi M, Clément G, Moreau M, Sormani R, Robaglia C, Meyer C. 2013. Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Frontiers in Plant Science 4, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du CQ, Li XS, Chen J, et al. 2016. Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, E8326–E8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM. 2010. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proceedings of the National Academy of Sciences, USA 107, 17821–17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, Grossniklaus U. 2007. The FERONIA receptor-like kinase mediates male–female interactions during pollen tube reception. Science 317, 656–660. [DOI] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP. 2012. Sugars, signalling, and plant development. Journal of Experimental Botany 63, 3367–3377. [DOI] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, et al. 2018. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Current Biology 28, 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferl RJ, Chung HJ, Sehnke PC. 1999. The 14-3-3 proteins: cellular regulators of plant metabolism. Trends in Plant Science 4, 463. [DOI] [PubMed] [Google Scholar]
- Frink CR, Waggoner PE, Ausubel JH. 1999. Nitrogen fertilizer: retrospect and prospect. Proceedings of the National Academy of Sciences, USA 96, 1175–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Xin Z, Zheng ZL. 2008. The OSU1/QUA2/TSD2-encoded putative methyltransferase is a critical modulator of carbon and nitrogen nutrient response in Arabidopsis. PLoS One 3, 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y. 2009. Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 106, 7648–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Nolan TM, Song G, Liu S, Xie Z, Chen J, Schnable PS, Walley JW, Yin Y. 2018. FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Current Biology 28, 3316–3324. [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. 2007. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biology 8, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA. 2012. Systems biology for enhanced plant nitrogen nutrition. Science 336, 1673–1675. [DOI] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. 2014. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Liu B, Wang X, Bian M, He R, Yan J, Zhong M, Zhao X, Liu X. 2016. Construction and validation of a dual-transgene vector system for stable transformation in plants. Journal of Genetics and Genomics 43, 199–207. [DOI] [PubMed] [Google Scholar]
- Hsieh MH, Lam HM, van de Loo FJ, Coruzzi G. 1998. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proceedings of the National Academy of Sciences, USA 95, 13965–13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GQ, Li E, Ge FR, Li S, Wang Q, Zhang CQ, Zhang Y. 2013. Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytologist 200, 1089–1101. [DOI] [PubMed] [Google Scholar]
- Huber SC, MacKintosh C, Kaiser WM. 2002. Metabolic enzymes as targets for 14-3-3 proteins. Plant Molecular Biology 50, 1053–1063. [DOI] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U. 2003. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130, 2149–2159. [DOI] [PubMed] [Google Scholar]
- Jüppner J, Mubeen U, Leisse A, Caldana C, Wiszniewski A, Steinhauser D, Giavalisco P. 2018. The target of rapamycin kinase affects biomass accumulation and cell cycle progression by altering carbon/nitrogen balance in synchronized Chlamydomonas reinhardtii cells. The Plant Journal 93, 355–376. [DOI] [PubMed] [Google Scholar]
- Kang J, Turano FJ. 2003. The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 100, 6872–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R. 2010. PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. Journal of Biological Chemistry 285, 39140–39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U. 2010. Conserved molecular components for pollen tube reception and fungal invasion. Science 330, 968–971. [DOI] [PubMed] [Google Scholar]
- Kulma A, Villadsen D, Campbell DG, Meek SEM, Harthill JE, Nielsen TH, MacKintosh C. 2004. Phosphorylation and 14-3-3 binding of Arabidopsis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. The Plant Journal 37, 654–667. [DOI] [PubMed] [Google Scholar]
- Li C, Liu X, Qiang X, et al. 2018. EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biology 16, e2006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wu HM, Cheung AY. 2016. FERONIA and her pals: functions and mechanisms. Plant Physiology 171, 2379–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Tang R, Zhang X, Luan S, Yu F. 2017. FERONIA receptor kinase at the crossroads of hormone signaling and stress responses. Plant & Cell Physiology 58, 1143–1150. [DOI] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. 2005. The putative high affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proceedings of the National Academy of Sciences, USA 102, 13693–13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu Y, Sasaki Y, Li X, Mori IC, Matsuura T, Hirayama T, Sato T, Yamaguchi J. 2015. ABI1 regulates carbon/nitrogen-nutrient signal transduction independent of ABA biosynthesis and canonical ABA signalling pathways in Arabidopsis. Journal of Experimental Botany 66, 4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Inada N, Yasuda S, Fukao Y, Fujiwara M, Sato T, Yamaguchi J. 2014. The carbon/nitrogen regulator ARABIDOPSIS TOXICOS EN LEVADURA31 controls papilla formation in response to powdery mildew fungi penetration by interacting with SYNTAXIN OF PLANTS121 in Arabidopsis. Plant Physiology 164, 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Sato T, Asada Y, Yasuda S, Yoshida M, Chiba Y, Yamaguchi J. 2012. The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Molecular Biology 79, 217–227. [DOI] [PubMed] [Google Scholar]
- Mao D, Yu F, Li J, et al. 2015. FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresse s S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant, Cell & Environment 38, 2566–2574. [DOI] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA. 2002. Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiology 128, 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Morrice N, Scarabel M, Aitken A, MacKintosh C. 1996. Phosphorylated nitrate reductase from spinach leaves is inhibited by 14-3-3 proteins and activated by fusicoccin. Current Biology 6, 1104–1113. [DOI] [PubMed] [Google Scholar]
- Novoa R, Loomis RS. 1981. Nitrogen and plant production. Plant and Soil 58, 177–204. [Google Scholar]
- Palenchar PM, Kouranov A, Lejay LV, Coruzzi GM. 2004. Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biology 5, R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr. 2001. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proceedings of the National Academy of Sciences, USA 98, 12843–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel J, Tischner R, Mäck G. 2001. The chloroplastic glutamine synthetase (GS-2) of tobacco is phosphorylated and associated with 14-3-3 proteins inside the chloroplast. Planta 213, 396–401. [DOI] [PubMed] [Google Scholar]
- Rosenquist M, Sehnke P, Ferl RJ, Sommarin M, Larsson C. 2000. Evolution of the 14-3-3 protein family: does the large number of isoforms in multicellular organisms reflect functional specificity? Journal of Molecular Evolution 51, 446–458. [DOI] [PubMed] [Google Scholar]
- Rotman N, Rozier F, Boavida L, Dumas C, Berger F, Faure JE. 2003. Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Current Biology 13, 432–436. [DOI] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, et al. 2009. CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. The Plant Journal 60, 852–864. [DOI] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, Domeki Y, Sueyoshi K, Fujiwara M, Fukao Y, Goto DB, Yamaguchi J. 2011. Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. The Plant Journal 68, 137–146. [DOI] [PubMed] [Google Scholar]
- Shin R, Jez JM, Basra A, Zhang B, Schachtman DP. 2011. 14-3-3 proteins fine-tune plant nutrient metabolism. FEBS Letters 585, 143–147. [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C. 2017. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. [DOI] [PubMed] [Google Scholar]
- Thynne E, Saur IML, Simbaqueba J, et al. 2017. Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Molecular Plant Pathology 18, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA. 2008. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Current Opinion in Plant Biology 11, 521–529. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124, 471–484. [DOI] [PubMed] [Google Scholar]
- Yang T, Wang L, Li C, Liu Y, Zhu S, Qi Y, Liu X, Lin Q, Luan S, Yu F. 2015. Receptor protein kinase FERONIA controls leaf starch accumulation by interacting with glyceraldehyde-3-phosphate dehydrogenase. Biochemical and Biophysical Research Communications 465, 77–82. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Aoyama S, Hasegawa Y, Sato T, Yamaguchi J. 2017. Arabidopsis CBL-interacting protein kinases regulate carbon/nitrogen-nutrient response by phosphorylating ubiquitin ligase ATL31. Molecular Plant 10, 605–618. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Sato T, Maekawa S, Aoyama S, Fukao Y, Yamaguchi J. 2014. Phosphorylation of Arabidopsis ubiquitin ligase ATL31 is critical for plant carbon/nitrogen nutrient balance response and controls the stability of 14-3-3 proteins. Journal of Biological Chemistry 289, 15179–15193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Sorek H, Wemmer DE, Somerville CR. 2016. Cellulose deficiency is enhanced on hyper accumulation of sucrose by a H+-coupled sucrose symporter. Plant Physiology 171, 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Li J, Huang Y, Liu L, Li D, Chen L, Luan S. 2014. FERONIA receptor kinase controls seed size in Arabidopsis thaliana. Molecular Plant 7, 920–922. [DOI] [PubMed] [Google Scholar]
- Yu F, Qian L, Nibau C, et al. 2012. FERONIA receptor kinase pathway suppresses abscisic acid signaling in Arabidopsis by activating ABI2 phosphatase. Proceedings of the National Academy of Sciences, USA 109, 14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chakravorty D, Assmann SM. 2018. The G protein β-subunit, AGB1, interacts with FERONIA in RALF1-regulated stomatal movement. Plant Physiology 176, 2426–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan HX, Xiong Y, Guan KL. 2013. Nutrient sensing, metabolism, and cell growth control. Molecular Cell 49, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.