Abstract
Due to the efficacy of tropomyosin receptor kinase (TRK) inhibitor therapy and the recent Food and Drug Administration approval of larotrectinib, it is now clinically important to accurately and efficiently identify patients with neurotrophic TRK (NTRK) fusion-driven cancer. These oncogenic fusions occur when the kinase domain of NTRK1, NTRK2 or NTRK3 fuse with any of a number of N-terminal partners. NTRK fusions are characteristic of a few rare types of cancer, such as secretory carcinoma of the breast or salivary gland and infantile fibrosarcoma, but they are also infrequently seen in some common cancers, such as melanoma, glioma and carcinomas of the thyroid, lung and colon. There are multiple methods for identifying NTRK fusions, including pan-TRK immunohistochemistry, fluorescence in situ hybridisation and sequencing methods, and the advantages and drawbacks of each are reviewed here. While testing algorithms will obviously depend on availability of various testing modalities and economic considerations for each individual laboratory, we propose triaging specimens based on histology and other molecular findings to most efficiently identify tumours harbouring these treatable oncogenic fusions.
Keywords: NTRK fusions, ancillary testing, next-generation sequencing, tyrosine kinase inhibitor
Key messages
NTRK fusions are seen in a few rare cancer types and occur infrequently in some common cancers. Accurate identification of NTRK fusion-driven cancer is clinically important and may be achieved using multiple methods. We propose triaging specimens for NTRK fusion testing based on histology and other molecular findings to most efficiently identify patients with these treatable oncogenic fusions.
Oncogenic neurotrophic tropomyosin receptor kinase fusions
The neurotrophic tropomyosin receptor kinases are a family of transmembrane tyrosine kinases that are important players in neural development. The three members of the family, TRKA (NTRK1), TRKB (NTRK2) and TRKC (NTRK3), are encoded by the NTRK1, NTRK2 and NTRK3 genes, respectively, and each consists of an extracellular ligand-binding domain, a transmembrane region and an intracellular kinase domain [1]. Normally, physiological activation of the receptor through ligand binding activates the kinase domain, leading to receptor homodimerisation, phosphorylation and activation of downstream signalling pathways [2]. Although highly homologous, each receptor has a preferred ligand: TRKA has the highest affinity for neurotrophin nerve growth factor, TRKB has the highest affinity for brain-derived neurotrophic factor and neurotrophin-4 and TRKC has the highest affinity for neurotrophin-3 [1−5]. A number of splice variants have been characterised, particularly involving NTRK1, and these variants have been observed both in normal tissues and in human cancers such as neuroblastoma and acute myeloid leukaemia where it is thought that they may play a role in tumourigenesis [1, 2]. While some studies have identified somatic point mutations or amplification in the NTRK genes, such alterations have so far not been shown to be a driver of oncogenesis [2].
Constitutive activation of the tropomyosin receptor kinase (TRK) receptors and subsequent downstream pathways can occur through chromosomal inversions, deletions or translocations that result in an in-frame fusion of the C-terminal tyrosine kinase domain of any of the NTRK genes with an N-terminal fusion partner. A multitude of 5′ fusion partners have been described, and in virtually all cases, the fusion eliminates the ligand binding site, resulting in ligand-independent dimerisation and phosphorylation [2]. The first TRK fusion protein was originally described in a colorectal adenocarcinoma cell line, but even at the time of discovery, it was recognised that involvement of this particular oncogene in such a fusion was an uncommon event in colon cancer [6]. It was later discovered that infantile fibrosarcoma was characterised by an ETV6-NTRK3 fusion involving a translocation of chromosomes 12 and 15 [7, 8], and this same fusion was subsequently also reported in secretory carcinoma of the breast and salivary gland, which now defines these subsets of carcinomas [9, 10]. NTRK fusions have also been reported in a subset of carcinomas of the thyroid, especially in patients with a history of exposure to radiation [11], and they are also rarely found in many other tumours, including carcinomas of the lung [12] and colon [13], gliomas [14, 15], other sarcomas [16], inflammatory myofibroblastic tumours [17] and melanocytic tumours [18, 19].
In recent years, clinical trials have shifted away from site-of-origin and histological subtype-specific designs and more towards basket trials, which are designed to test therapies targeted towards specific molecular mechanisms [20], and trials targeting NTRK fusions have been particularly successful. In one such recent trial, larotrectinib showed remarkable and durable efficacy against locally advanced and metastatic solid tumours harbouring an NTRK fusion [21]. Entrectinib, active against NTRK fusions as well as fusions involving ROS1 and ALK, has also shown great efficacy in recent clinical trials [22, 23]. The success of larotrectinib has resulted in its subsequent fast-track approval by the Food and Drug Administration (FDA),* and therefore standard of care will now require accurate identification of patients who could benefit from this practice-changing therapy.
Methods for detection
Immunohistochemistry
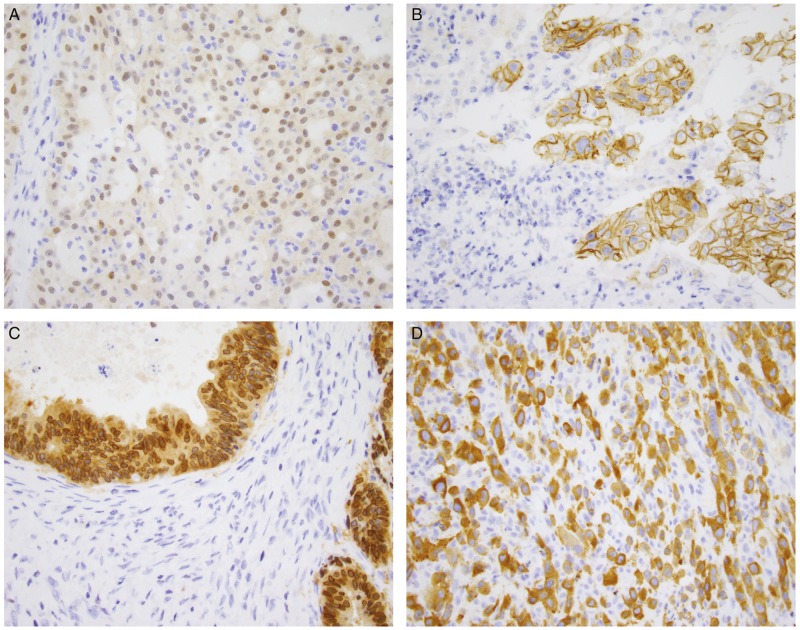
Immunohistochemistry to examine protein expression has several advantages. It is commonly used in clinical labs and is therefore relatively straightforward to implement and validate. It also has the benefits of being inexpensive, requiring only a single unstained slide and having a rapid turnaround time. The clone that is most often used and well-studied is clone EPR17341 (Abcam and Roche/Ventana), which reacts with a conserved proprietary peptide from the C-terminus of TRKA, TRKB and TRKC, and is therefore reactive with any of the oncogenic NTRK fusions. Positive staining has been defined as staining above background in at least 1% of tumour cells [24]. Initial studies have shown sensitivity ranging from 75% to 96.7% and specificity ranging from 92% to 100% [24−27]. However, the staining intensity has been shown to be variable, and staining pattern correlates with fusion partner (Figure 1) [25]. The fusion partner can direct the fusion protein to localise to other cellular compartments, in contrast to the membrane-associated expression of native TRK. One caveat is that recent studies have shown reduced sensitivity for NTRK3 fusions [24]. In our experience, e.g. we have found that sensitivity for NTRK1 and NTRK2 fusions was 96% and 100%, respectively, while sensitivity for NTRK3 fusions was 79% [28]. In addition, immunohistochemistry seems to have variable specificity according to tumour type. While the antibody appears to have 100% specificity in carcinomas of the colon, lung, thyroid, pancreas and biliary tract, decreased specificity is seen in breast and salivary gland carcinomas, as cytoplasmic staining can occasionally be seen. Specificity is lower in sarcomas, particularly those with neural or smooth muscle differentiation as wild-type TRK protein is physiologically expressed in neural and smooth muscle tissue [27, 28].
Figure 1.
Patterns of immunohistochemical staining in NTRK fusion-positive tumours. (A) Secretory carcinoma of the salivary gland with an ETV6-NTRK3 fusion shows weak to moderate nuclear and cytoplasmic staining. (B) Intrahepatic cholangiocarcinoma with a PLEKHA6-NTRK1 fusion shows prominent membranous staining. (C) Gallbladder adenocarcinoma with an LMNA-NTRK1 fusion shows strong cytoplasmic and perinuclear staining. (D) Metastatic thyroid carcinoma to soft tissue with a TPM3-NTRK1 fusion shows strong cytoplasmic and membranous staining.
Fluorescence in situ hybridisation
Fluorescence in situ hybridisation (FISH) can detect large structural variants at the DNA level and is often used in the clinical laboratory to detect oncogenic fusions in solid tumours. A commercial break-apart probe is available for the ETV6 gene (Abbott, Chicago, IL) where separation of a green signal at the centromeric 3′ end of ETV6 and an orange signal at the 5′ end of ETV6 indicates a structural variant involving the gene. In cases that are histologically suggestive of ETV6-NTRK3 fusions, such as infantile fibrosarcoma, congenital mesoblastic nephroma or secretory carcinoma of the salivary gland or breast, such testing can be useful to confirm the translocation [29]. Since fusions in other cancers can involve any of the NTRK genes and any of a number of partners through either balanced or unbalanced translocation or large deletions, only examining the ETV6 gene would miss many oncogenic NTRK fusions. To this end, break-apart probes for the three NTRK genes have been used to identify fusions and are commercially available from multiple sources [16, 30–32]. Theoretically, break-apart probes have adequate sensitivity and specificity for chromosomal abnormalities, but there are practical technical considerations in the interpretation of break-apart FISH assays. In one study, short inversions and intrachromosomal translocations involving ALK resulted in a short split length using a break-apart probe. These short split lengths were difficult to distinguish from those seen in some normal cells, and therefore can result in false-negative results [33]. These findings would have particular relevance for NTRK1 fusions, a majority of which are intrachromosomal events involving chromosome 1 [34]. For example, LMNA-NTRK1 fusions are formed through an intrachromosomal deletion, which can result in a false-negative FISH due to insufficient splitting of the signals. In addition, while a positive result with a break-apart probe shows the presence of a structural variant involving the probed gene, whether the abnormality results in a functional transcribed fusion cannot be determined. Advantages of FISH include that the amount of material required is only a few unstained slides—usually one unstained slide per probe examined—and the turnaround time is usually only a few days.
Reverse transcriptase polymerase chain reaction
Reverse transcriptase polymerase chain reaction (RT-PCR) can be used to detect the presence of transcribed RNA, and it can be used either qualitatively or quantitatively to detect the presence of a single oncogenic fusion for which both fusion partners are known. For NTRK fusions, however, because of the number of different fusion partners and breakpoints involved, the utility of RT-PCR for individual fusion transcripts is limited and has been used in the past mainly to detect canonical ETV6-NTRK3 fusions. Even though this method may prove difficult to obtain direct evidence of a fusion, RT-PCR could be used to examine differences in expression of the 5′ versus the 3′ ends of a gene, as this has been shown to be associated with the presence of a translocation [35, 36]. For NTRK, which is not expressed in most normal tissues, the 3′ kinase domain would be transcribed at a much higher level than the 5′ extracellular domain in tumour tissue that harbours an NTRK fusion. Such an assay could thereby provide indirect evidence of an NTRK fusion.
DNA-based next-generation sequencing
In DNA-based next-generation sequencing (NGS), tumour DNA is extracted from formalin-fixed paraffin-embedded (FFPE) tissue and then sequenced to investigate whether specific alterations are present in the tumour. Although there are a number of different library preparation and sequencing solutions available, there are two main approaches to isolating the genes of interest for sequencing. Amplicon-based methods use PCR primers to amplify the areas of interest, and while this method is suitable for detecting point mutations and small indels in a small panel of genes, the detection of gene fusions, which usually involve intronic breakpoints, is limited. Targeted hybridisation capture-based NGS assays on the other hand use capture probes that hybridise to the areas of interest in the genome. This non-biased approach enables deep sequencing of exons of key cancer-related genes for the detection of single point mutations, indels and copy number variations. In addition, the introns of specific genes known to be involved in functional gene fusions can be tiled with probes (baits) to detect these rearrangements [25, 37]. It should be noted, however, that some introns, such as those in NTRK3, are extremely long (spanning 193 KB) and would not be feasible to cover. If covered, those introns would constitute a large percentage of the panel size resulting in coverage reduction of other exonic regions and the overall assay sensitivity. Furthermore, some of these intronic regions cannot be effectively captured even if desired because they contain repetitive elements which cannot be tiled with unique capture probes and therefore cannot yield reads that can be reliably mapped back to that intron (poor mapping quality). Therefore, sensitivity of DNA-based NGS suffers if fusion breakpoints involve long intronic regions that cannot be covered by hybridisation-capture probes. For example, MSK-IMPACT, the DNA-based NGS assay used at Memorial Sloan Kettering Cancer Center, interrogates introns 3 and 7 through 12 in NTRK1, intron 15 in NTRK2 and introns 4 and 5 in ETV6, the most common NTRK3 fusion partner. However, because of the aforementioned issues involving coverage of the NTRK3 introns, fusions involving NTRK3 other than ETV6, are not covered by the assay [25, 28, 37]. These considerations are not limited to MSK-IMPACT, as other widely used cancer gene panels such as FoundationOne CDx also only assess these same intronic regions [38].
One drawback to DNA-based NGS is that when novel structural variants are detected, it can be difficult to determine whether such an event results in a functional expressed fusion. In these cases, ancillary testing with an orthogonal method, such as RNA-based NGS can be carried out. Other drawbacks include turnaround time, which is significantly longer than immunohistochemistry or FISH, and that more material is required for testing. On the other hand, a major advantage of DNA-based NGS testing is that many genomic events can be interrogated, allowing for simultaneous direct assessment of point mutations, indels, copy number variants and tumour mutation burden in addition to DNA-level gene fusions. Information gained from using this method includes MAPK driver status, which can be used to triage any follow-up testing (see Testing algorithm considerations section). Finally, this method is also effective for monitoring patients with NTRK fusions for development of resistance mutations. Recent studies have observed p.G667C and p.G595R mutations in NTRK1 and p.G696A mutations in NTRK3 that confer resistance to TRK inhibitor therapy [21, 39]. Using DNA-based NGS to monitor for tumour evolution is therefore useful in patients with NTRK fusion-positive cancers treated with TRK inhibitor therapy.
RNA-based NGS
RNA-based sequencing presents several advantages over DNA. The introns are spliced out in the RNA, which removes the technical limitations of intronic coverage. In addition, detection of RNA-level fusions provides direct evidence that they are functionally transcribed, and analysis of the spliced sequence can determine whether the protein would be translated and in-frame. Fusion transcripts can also be detected with high confidence in the RNA of low tumour purity samples because gene fusions are often highly expressed in the tissue.
Detection of the fusion transcript by RNA-based NGS can be carried out using a few different enrichment methods. In all technologies, the RNA library is first converted to cDNA through reverse transcription. Then, for amplicon-based panels, PCR is used to amplify the sequences of interest. In assays that use standard multiplex PCR, both the driver gene and the fusion partner must be known and the two gene-specific PCR primers must be present in the assay for amplification to occur. In one study that used such a multiplexed amplicon approach using a panel of primer pairs for 169 gene fusions between 19 target genes and 94 fusion partners, sensitivity for fusion detection was determined to be 86% [40, 41]. However, when incorporating the read count information to determine 5′/3′ ratios, as described above for RT-PCR, sensitivity was increased to 100% [40].
A more sensitive and specific targeted amplicon-based method for fusion gene detection is anchored multiplex PCR that is commercially available through ArcherDX. In this technology, an Illumina sequencing adaptor is ligated to both ends of the cDNA. In the PCR steps, a gene-specific primer hybridises to the gene of interest, while a universal primer hybridises to the ligated adaptor sequence. Since fusion breakpoints are usually within introns, the gene-specific primers are often complementary to regions at the ends of exons such that the PCR products span exon boundaries. By using this method of sequencing, only one fusion partner needs to be targeted, and therefore novel fusion partners can be characterised [42, 43]. In our laboratory at Memorial Sloan Kettering, we have shown that this technology is able to sensitively and specifically identify fusion transcripts and alternative transcripts resulting from splicing alterations [43, 44].
Capture-based approaches for either targeted or whole transcriptome sequencing assays can also be used. After reverse transcription to convert extracted RNA to cDNA, hybridisation capture is carried out using capture probes in a method similar to that used in DNA-based sequencing. With this method, only one fusion partner needs to be known. The clinically validated assay, OSU-SpARKFuse, which uses such an approach, demonstrated 93.3% sensitivity and 100% specificity for fusion detection [45].
One of the main drawbacks to working with RNA is its lability. RNA can be extracted from FFPE tissue, but because it is susceptible to fragmentation and degradation, especially in older material, adequate quality control is required. Methods to interrogate RNA quality can be analysis of RNA fragment size distribution and examining amplification of a housekeeping gene in a quantitative PCR-based assay [46]. For assessing quality of the sequencing assay, one can determine the ratio of RNA reads to DNA reads and examine sequencing coverage and depth for the RNA reads.
Finally, it should be noted that some commercially available platforms are able to simultaneously assess both RNA and DNA. The DNA and RNA libraries are prepared separately, but then can be combined for analysis in a single sequencing run. The Oncomine Comprehensive Assay by ThermoFisher, which uses amplicon-based technology, covers 161 cancer-related genes and can detect fusions involving all three NTRK genes. Per ThermoFisher’s technical specifications, the assay can be carried out with as few as three FFPE slides or as little as 10 ng of DNA or RNA. The TruSight Oncology 500 assay by Illumina uses hybridisation-capture technology to interrogate DNA-level alterations involving 523 cancer-related genes, and it also sequences RNA-transcripts to detect fusions involving any of 55 genes, including all three NTRK genes.
Testing algorithm considerations
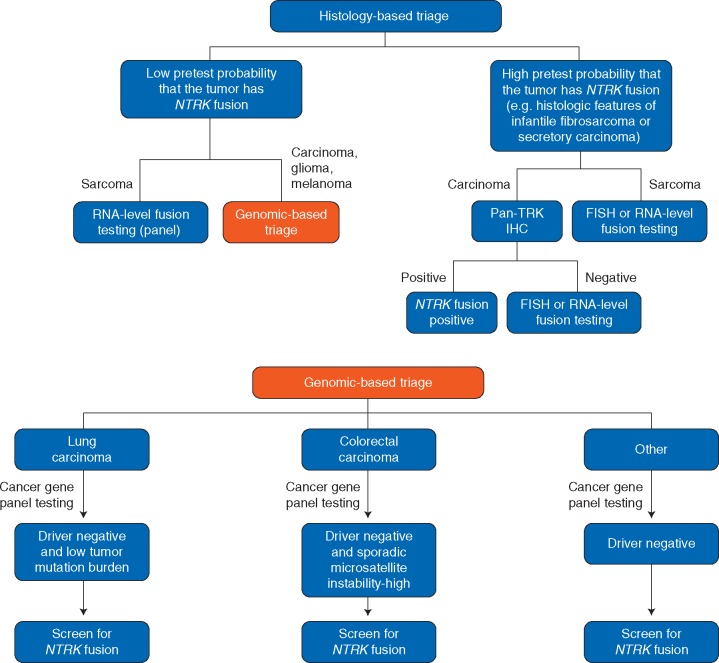
While the importance of identifying patients that could benefit from targeted therapy cannot be understated, feasibility and economic considerations should also be taken into account when creating testing algorithms and guidelines. Triaging specimens should be based not only on tumour type and its pre-test probability for NTRK fusions, but also on the availability of clinically validated methodologies along with their positive and negative predictive values (Figure 2). Overall, a comprehensive diagnostic algorithm that is appropriate for all clinical situations and laboratories is difficult to propose and depends on all of these factors. Here, we discuss a few considerations based on the observation that a few rare cancer types commonly harbour NTRK fusions whereas common cancer types rarely harbour NTRK fusions. However, in absolute numbers, the latter group contributes the majority of patients with NTRK fusions.
Figure 2.
Diagnostic algorithm for NTRK testing. Histology-based triaging should first be carried out to separate the rare cancer subtypes that commonly have NTRK fusions from those that have a low pre-test probability of NTRK fusions. In the tumours that often have oncogenic NTRK fusions, confirmatory methods can be used. In secretory carcinomas, pan-TRK immunohistochemistry can be used as an initial screen, but if negative, additional testing with FISH or RNA-level fusion testing should be used. In sarcomas, immunohistochemistry should be eschewed due to its lower specificity. It is also worth noting that comprehensive fusion testing (for all major sarcoma fusions) is increasingly being carried out as a first-line test in sarcomas rather than waiting for results from a DNA-based triage as one would in carcinoma. We therefore recommend inclusion of NTRK primers in comprehensive sarcoma fusion test panels. In cancers with a low pre-test probability of NTRK fusion, such as most carcinomas, gliomas and melanomas, molecular testing such as DNA-based cancer gene panels is often carried out, and driver status can therefore be used to narrow down the tumours that should undergo further screening for oncogenic fusions, as NTRK fusions are typically mutually exclusive with other common mitogenic driver alterations that activate MAPK signalling. The resulting ‘driver-negative’ cases are therefore likely enriched for NTRK fusions and these can be screened for by IHC or an RNA-based fusion panel assay. For lung and colorectal cancer, we highlight how to further enrich for NTRK fusions in settings where broad, routine screening is not possible.
Histology-based triaging
The rare cancer subtypes that commonly harbour NTRK fusions include secretory carcinomas of the breast and salivary glands, infantile fibrosarcoma, congenital mesoblastic nephroma and paediatric papillary thyroid cancer. For these tumours, a histology-based testing algorithm is preferred and confirmation of the NTRK fusion should be carried out using a preferred method that has high specificity, such as FISH or DNA-based NGS. If negative by a single method, additional testing should be carried out.
Mass screening-based detection
The second group includes many cancer types with a low probability (<1%) of harbouring NTRK fusions. These low probability cancers include lung cancer, breast cancer, colorectal cancer, pancreatic cancer, cholangiocarcinomas, adult papillary thyroid cancer (2%), melanomas, gliomas and sarcomas (gastrointestinal stromal tumour, uterine and soft tissue). Mass screening approaches include comprehensive molecular evaluation using broad NGS that includes assessment for NTRK fusions as part of broad panels, as described above, or routine screening by immunohistochemistry, with the caveats and limitations stated above. Mass screening-based detection is not algorithmic and involves no triaging; it refers to testing all patients either by immunohistochemistry or by comprehensive DNA- and RNA-based panels. Therefore, it is intrinsically inefficient if one is solely screening for an alteration with very low prevalence, such as NTRK fusions.
Genomic-based triaging
In many cases in the group of cancer types with a low probability (<1%) of harbouring NTRK fusions, one can triage tumours that are more likely to harbour NTRK fusions based on their genomic profiles. NTRK fusions are typically mutually exclusive with KRAS, NRAS, BRAF, MAP2K1, EGFR, ALK, RET, ROS1, KIT, PDGFRA and other common mitogenic ‘driver’ alterations that activate MAPK signalling. Thus, excluding cases in which a known mitogenic driver has been identified (‘driver-negative’) can significantly narrow down the number of cases to be systematically screened for NTRK fusions. For example, in a recent study, we observed a high yield of RNA sequencing for targetable kinase fusions, including NTRK fusions, in driver-negative lung adenocarcinomas that also showed low tumour mutation burden [44]. In another similar study, Cocco et al. examined a cohort of 2314 colorectal adenocarcinomas with MSK-IMPACT, identifying 21 cases with kinase fusions, including 8 with NTRK fusions. Of the 21 cases with kinase fusions, a majority (57%, 12/21) were present in cases that had microsatellite instability. Of a cohort of 24 colorectal carcinoma cases that exhibited microsatellite instability due to MLH1 promoter methylation and that did not have driver mutations in RAS or BRAF, 10 harboured kinase fusions, including 6 that had NTRK fusions. Overall, the findings from this study identify a subset of colorectal carcinomas for which there should be a high suspicion of kinase fusions [47]. Therefore, it may be most efficient to focus ancillary testing on colonic adenocarcinomas that have microsatellite instability and that lack other conventional driver mutations. For lung and colorectal cancer, such approaches highlight how to further enrich for NTRK fusions in settings where broad, routine screening is not possible. We do not propose restricting further testing to these subsets of driver-negative lung or colorectal cases but mainly wish to emphasise which types of cases should be of highest priority for further screening.
Discussion
Conclusions
Oncogenic NTRK gene fusions occur in many different tumour types. While present in a majority of certain rare tumours, they are also rarely present in many common cancers. With the recent FDA approval of NTRK targeted therapy and the marked and durable responses these agents produce in patients with NTRK fusion-positive cancers, identification of tumours harbouring these fusions has become essential. Our approach echoes the recently published recommendations of the ESMO Translational Research and Precision Medicine Working Group [48], but we place more emphasis on genomic-based triage. Further downstream in the cancer care timeline, as we more routinely detect NTRK fusions that make patients eligible for TRK inhibitors, we will also have to consider how our molecular diagnostic tests detect mechanisms of acquired resistance in these patients, both on-target second site mutations in the NTRK kinase domain [49, 50] and alterations that activate bypass signalling pathways [51].
Acknowledgements
Medical writing support, including assisting authors with the development of the outline was provided by Penny Butcher, PhD and editorial support, including figure preparation, formatting and proofreading was provided by Annabel Ola, MSc, both of Scion, UK, supported by Bayer according to Good Publication Practice guidelines. Bayer was involved in data checking of information provided in the manuscript. However, ultimate responsibility for opinions, conclusions and data interpretation lies with the authors.
Funding
This paper was published as part of a supplement financially supported by Bayer AG and Loxo Oncology, Inc., a wholly owned subsidiary of Eli Lilly and Company.
Disclosure
JFH has received honoraria from WebMD, Axiom Healthcare Strategies and Cor2Ed, and research funding from Loxo Oncology, Bayer and Boehringer Ingelheim. ML has received research funding and advisory board compensation from Loxo Oncology and advisory board compensation from Bayer. All remaining authors have declared no conflicts of interest.
Footnotes
Note added in proof: The European Medicines Agency granted marketing authorisation for larotrectinib on 23 September 2019 as monotherapy for the treatment of adult and paediatric patients with solid tumours that display a neurotrophic tyrosine receptor kinase (NTRK) gene fusion, and who have disease that is locally advanced, metastatic or where surgical resection is likely to result in severe morbidity, and who have no satisfactory treatment options.
References
- 1. Vaishnavi A, Le AT, Doebele RC.. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov 2015; 5(1): 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cocco E, Scaltriti M, Drilon A.. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018; 15(12): 731–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davies AM, Horton A, Burton LE. et al. Neurotrophin-4/5 is a mammalian-specific survival factor for distinct populations of sensory neurons. J Neurosci 1993; 13(11): 4961–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soppet D, Escandon E, Maragos J. et al. The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the trkB tyrosine kinase receptor. Cell 1991; 65(5): 895–903. [DOI] [PubMed] [Google Scholar]
- 5. Kaplan DR, Martin-Zanca D, Parada LF.. Tyrosine phosphorylation and tyrosine kinase activity of the trk proto-oncogene product induced by NGF. Nature 1991; 350(6314): 158–160. [DOI] [PubMed] [Google Scholar]
- 6. Martin-Zanca D, Hughes SH, Barbacid M.. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature 1986; 319(6056): 743–748. [DOI] [PubMed] [Google Scholar]
- 7. Knezevich SR, McFadden DE, Tao W. et al. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet 1998; 18(2): 184–187. [DOI] [PubMed] [Google Scholar]
- 8. Bourgeois JM, Knezevich SR, Mathers JA. et al. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol 2000; 24(7): 937–946. [DOI] [PubMed] [Google Scholar]
- 9. Vasudev P, Onuma K.. Secretory breast carcinoma: unique, triple-negative carcinoma with a favorable prognosis and characteristic molecular expression. Arch Pathol Lab Med 2011; 135(12): 1606–1610. [DOI] [PubMed] [Google Scholar]
- 10. Skalova A, Vanecek T, Sima R. et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol 2010; 34(5): 599–608. [DOI] [PubMed] [Google Scholar]
- 11. Ricarte-Filho JC, Li S, Garcia-Rendueles ME. et al. Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J Clin Invest 2013; 123(11): 4935–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farago AF, Taylor MS, Doebele RC. et al. Clinicopathologic features of non-small-cell lung cancer harboring an NTRK gene fusion. JCO Precis Oncol 2018; doi: 10.1200/PO.18.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pietrantonio F, Di Nicolantonio F, Schrock AB. et al. ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J Natl Cancer Inst 2017; 109(12): doi:10.1093/jnci/djx089. [DOI] [PubMed] [Google Scholar]
- 14. Xu T, Wang H, Huang X. et al. Gene fusion in malignant glioma: an emerging target for next-generation personalized treatment. Transl Oncol 2018; 11(3): 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferguson SD, Zhou SH, Huse JT. et al. Targetable gene fusions associate with the IDH wild-type astrocytic lineage in adult gliomas. J Neuropathol Exp Neurol 2018; 77(6): 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chiang S, Cotzia P, Hyman DM. et al. NTRK fusions define a novel uterine sarcoma subtype with features of fibrosarcoma. Am J Surg Pathol 2018; 42(6): 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alassiri AH, Ali RH, Shen Y. et al. ETV6-NTRK3 is expressed in a subset of ALK-negative inflammatory myofibroblastic tumors. Am J Surg Pathol 2016; 40(8): 1051–1061. [DOI] [PubMed] [Google Scholar]
- 18. Wang L, Busam KJ, Benayed R. et al. Identification of NTRK3 fusions in childhood melanocytic neoplasms. J Mol Diagn 2017; 19(3): 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lezcano C, Shoushtari AN, Ariyan C. et al. Primary and metastatic melanoma with NTRK fusions. Am J Surg Pathol 2018; 42(8): 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cunanan KM, Gonen M, Shen R. et al. Basket trials in oncology: a trade-off between complexity and efficiency. J Clin Oncol 2017; 35(3): 271–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drilon A, Laetsch TW, Kummar S. et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 2018; 378(8): 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farago AF, Le LP, Zheng Z. et al. Durable clinical response to entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol 2015; 10(12): 1670–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Drilon A, Siena S, Ou SI. et al. Safety and antitumor activity of the multitargeted pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov 2017; 7(4): 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gatalica Z, Xiu J, Swensen J. et al. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol 2019; 32(1): 147–153. [DOI] [PubMed] [Google Scholar]
- 25. Hechtman JF, Benayed R, Hyman DM. et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol 2017; 41(11): 1547–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rudzinski ER, Lockwood CM, Stohr BA. et al. Pan-Trk immunohistochemistry identifies NTRK rearrangements in pediatric mesenchymal tumors. Am J Surg Pathol 2018; 42(7): 927–935. [DOI] [PubMed] [Google Scholar]
- 27. Hung YP, Fletcher CDM, Hornick JL.. Evaluation of pan-TRK immunohistochemistry in infantile fibrosarcoma, lipofibromatosis-like neural tumour and histological mimics. Histopathology 2018; 73(4): 634–644. [DOI] [PubMed] [Google Scholar]
- 28. Solomon JP, Linkov I, Rosado A. et al. NTRK fusion detection across multiple assays and 33, 997 cases: diagnostic implications and pitfalls. Mod Pathol 2019; doi: 10.1038/s41379-019-0324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Connor A, Perez-Ordonez B, Shago M. et al. Mammary analog secretory carcinoma of salivary gland origin with the ETV6 gene rearrangement by FISH: expanded morphologic and immunohistochemical spectrum of a recently described entity. Am J Surg Pathol 2012; 36(1): 27–34. [DOI] [PubMed] [Google Scholar]
- 30. Doebele RC, Davis LE, Vaishnavi A. et al. An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer Discov 2015; 5(10): 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Milione M, Ardini E, Christiansen J. et al. Identification and characterization of a novel SCYL3-NTRK1 rearrangement in a colorectal cancer patient. Oncotarget 2017; 8(33): 55353–55360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skalova A, Vanecek T, Simpson RH. et al. Mammary analogue secretory carcinoma of salivary glands: molecular analysis of 25 ETV6 gene rearranged tumors with lack of detection of classical ETV6-NTRK3 fusion transcript by standard RT-PCR: report of 4 cases harboring ETV6-X gene fusion. Am J Surg Pathol 2016; 40(1): 3–13. [DOI] [PubMed] [Google Scholar]
- 33. Martin V, Bernasconi B, Merlo E. et al. ALK testing in lung adenocarcinoma: technical aspects to improve FISH evaluation in daily practice. J Thorac Oncol 2015; 10(4): 595–602. [DOI] [PubMed] [Google Scholar]
- 34. Hsiao SJ, Zehir A, Sireci AN. et al. Detection of tumor NTRK gene fusions to identify patients who may benefit from TRK inhibitor therapy. J Mol Diagn 2019; 21(4): 553–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L, Motoi T, Khanin R. et al. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosom Cancer 2012; 51(2): 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suehara Y, Arcila M, Wang L. et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res 2012; 18(24): 6599–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zehir A, Benayed R, Shah RH. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10, 000 patients. Nat Med 2017; 23(6): 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.U.S. Food and Drug Administration. Summary of safety and effectiveness data for FoundationOne CDx. https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019B.pdf (26 June 2019, date last accessed).
- 39. Russo M, Misale S, Wei G. et al. Acquired resistance to the TRK inhibitor entrectinib in colorectal cancer. Cancer Discov 2016; 6(1): 36–44. [DOI] [PubMed] [Google Scholar]
- 40. Beadling C, Wald AI, Warrick A. et al. A multiplexed amplicon approach for detecting gene fusions by next-generation sequencing. J Mol Diagn 2016; 18(2): 165–175. [DOI] [PubMed] [Google Scholar]
- 41. Lange AM, Lo HW.. Inhibiting TRK proteins in clinical cancer therapy. Cancers (Basel) 2018; 10(4): 105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zheng Z, Liebers M, Zhelyazkova B. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014; 20(12): 1479–1484. [DOI] [PubMed] [Google Scholar]
- 43. Zhu G, Benayed R, Ho C. et al. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol 2019; 32(5): 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benayed R, Offin M, Mullaney K. et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res 2019; 25(15): 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reeser JW, Martin D, Miya J. et al. Validation of a targeted RNA sequencing assay for kinase fusion detection in solid tumors. J Mol Diagn 2017; 19(5): 682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murphy DA, Ely HA, Shoemaker R. et al. Detecting gene rearrangements in patient populations through a 2-step diagnostic test comprised of rapid IHC enrichment followed by sensitive next-generation sequencing. Appl Immunohistochem Mol Morphol 2017; 25(7): 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cocco E, Benhamida J, Middha S. et al. Colorectal carcinomas containing hypermethylated MLH1 promoter and wild type BRAF/KRAS are enriched for targetable kinase fusions. Cancer Res 2019; 79(6): 1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marchio C, Scaltriti M, Ladanyi M. et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann Oncol 2019; doi: 10.1093/annonc/mdz204. [DOI] [PubMed] [Google Scholar]
- 49. Drilon A, Nagasubramanian R, Blake JF. et al. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov 2017; 7(9): 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drilon A, Li G, Dogan S. et al. What hides behind the MASC: clinical response and acquired resistance to entrectinib after ETV6-NTRK3 identification in a mammary analogue secretory carcinoma (MASC). Ann Oncol 2016; 27(5): 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cocco E, Schram AM, Kulick A. et al. Resistance to TRK inhibition mediated by convergent MAP kinase pathway activation. Nat Med 2019; 25(9): 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]