Abstract
Recognition of foreign and dysregulated antigens by the cellular innate and adaptive immune systems is in large part dependent on the cell surface display of peptide/MHC (pMHC) complexes. The formation of such complexes requires the generation of antigenic peptides, proper folding of MHC molecules, loading of peptides onto MHC molecules, glycosylation, and transport to the plasma membrane. This complex series of biosynthetic, biochemical, and cell biological reactions is known as “antigen processing and presentation”. Here, we summarize recent work, focused on the structural and functional characterization of the key MHC-I-dedicated chaperones, tapasin, and TAPBPR. The mechanisms reflect the ability of conformationally flexible molecules to adapt to their ligands, and are comparable to similar processes that are exploited in peptide antigen loading in the MHC-II pathway.
Keywords: Antigen presenting cell (APC), major histocompatibility complex (MHC), peptide-loading complex (PLC), tapasin, TAP-binding protein related (TAPBPR), X-ray crystallography, nuclear magnetic resonance spectroscopy (NMR)
Introduction
Antigen processing and presentation, the cellular steps by which fragments of proteins are generated, loaded into major histocompatibility complex (MHC)-encoded class I and class II molecules, trafficked to the cell surface, and displayed for recognition by CD8+ and CD4+ T lymphocytes via their clonally expressed T cell receptors (TCR) is a central aspect of the adaptive immune response (Germain and Margulies 1993; York and Rock 1996; Barber et al. 2001; Blum et al. 2013). The peptide/MHC (pMHC) interaction with TCR is critical to the development, selection, survival, tolerance, and activation of T cells. Although the process of antigen presentation has been studied in molecular and cellular detail for several decades, recent data, drawing from structural and modeling studies, including X-ray crystallography, cryo-electron microscopic (cryo-EM) images, nuclear magnetic resonance (NMR) spectroscopy, and computational molecular dynamics simulations have expanded our understanding of molecular details that contribute to the loading of peptide antigens onto MHC-I and MHC-II molecules. This review focuses on several molecules that facilitate peptide loading in the MHC-I pathway, specifically the MHC-I chaperones, tapasin (also known as TAP-binding protein, TAPBP) and TAPBPR (TAP-binding protein related). MHC-II molecules are loaded by distinct molecular and cellular mechanisms that have some similarities to the MHC-I pathway. Our focus here will be on MHC-I, and we will describe some similarities and differences to MHC-II. Although the key chaperones involved in stabilizing nascent MHC-I structures and in facilitating peptide loading were identified over two decades ago, it is only relatively recently that the subtleties of their molecular and structural interactions have been revealed—through X-ray crystallography, multidimensional NMR, cryo EM, and molecular dynamics simulations. A common theme of the molecular mechanisms is that the chaperones are dynamic molecules, containing both highly structured and mobile regions that allow them to stabilize otherwise metastable conformations of the highly polymorphic MHC molecules, to facilitate the loading of high affinity antigenic peptides.
Overview of the MHC-I antigen presentation pathway
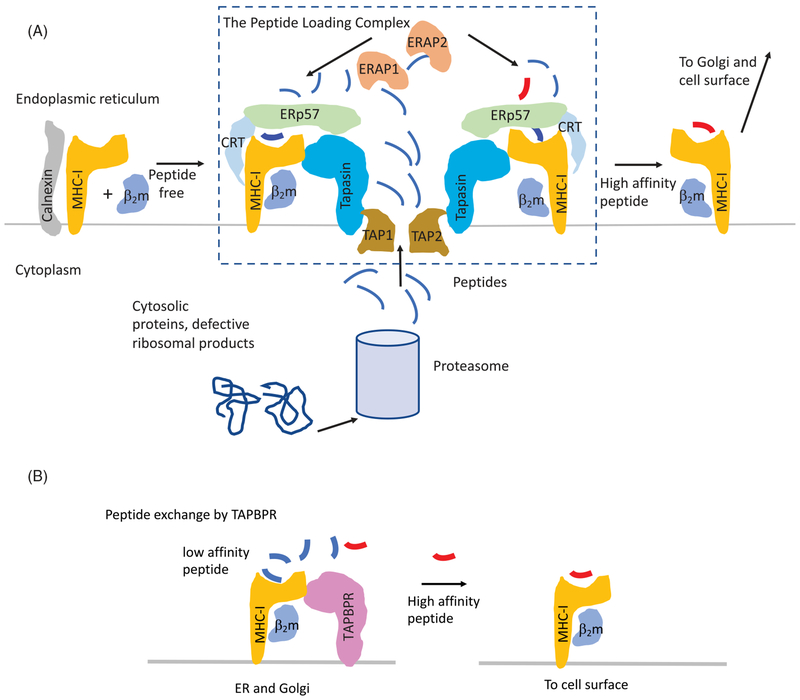
In the ER newly synthesized, partially folded MHC-I heavy chains, chaperoned by the carbohydrate-binding protein calnexin, assemble with the MHC-I light chain β2m. Peptides destined for presentation by MHC-I originate from proteasome-mediated degradation of endogenous proteins (Huang et al. 2011; Blum et al. 2013) or from defective protein translation (Wei and Yewdell 2018) (Figure 1). The resulting peptides are transported from the cytosol into the ER by the TAP1/TAP2 heterodimer, a member of the ATP-binding cassette family of transporters. The TAP transporter preferentially transports peptides of 9–16 amino acids in length (van Endert et al. 1994). Peptide-receptive MHC-I/β2m complexes are incorporated into a multiprotein assembly termed the peptide-loading complex (PLC) (Hulpke and Tampe 2013). The PLC includes TAP, the MHC-I/β2m heterodimer, the peptide editor tapasin [also known as TAP-binding protein (TAPBP)], the protein disulfide isomerase ERp57 which is disulfide-linked via its active site cysteine to tapasin, and the carbohydrate-binding chaperone calreticulin. The PLC thus juxtaposes a peptide-receptive, scaffolded form of the MHC-I with the abundant and diverse supply of peptides in the ER to facilitate loading of peptides of correct size and sequence for the particular MHC-I allelomorph (Figure 1(A)). Longer peptides are trimmed by the ER resident amino peptidases ERAP1 and ERAP2 (only ERAP, the equivalent to human ERAP1 is found in the mouse) to the 8–10 amino acid length necessary for optimal MHC-I binding (Hammer et al. 2006). Although ERAP proteins may trim MHC-I-bound precursor peptides, the efficiency is lower than for the corresponding free precursors (Chen et al. 2016). X-ray structures of ERAP1 suggest that the active site may not readily accommodate an MHC-I bound precursor peptide (Gandhi et al. 2011; Kochan et al. 2011; Nguyen et al. 2011). Following loading with a high affinity peptide, the now stable MHC-I molecule is released from the PLC for export to the Golgi complex, modifications of its carbohydrate moiety, and transport to the cell surface. Suboptimally loaded, and therefore unstable, MHC-I molecules released from the PLC may be salvaged by re-glucosylation by UDP-glucose:glycoprotein glucosyl transferase (UGGT1) in the cis-Golgi (Ritter et al. 2005), allowing rebinding to calreticulin and reincorporation of MHC-I into the PLC for further rounds of peptide editing. In addition to tapasin-mediated peptide editing that occurs in the PLC, a second peptide editor that has been recently characterized, designated TAPBPR, functions independently of the PLC (Figure 1(B)) (Hermann et al. 2013, 2015; Morozov et al. 2016). The gene encoding TAPBPR was originally identified by homology search of the human genome with tapasin, and the encoded protein was shown to be about 22% identical to tapasin in amino acid sequence (Teng et al. 2002; Boyle et al. 2013; Hermann et al. 2013, 2015). TAPBPR differs from tapasin in several respects: (1) the TAPBPR gene maps to human chromosome 12 rather than to chromosome 6 where the extended HLA locus and tapasin are located (Teng et al. 2002); (2) TAPBPR is not a biochemical component of the PLC; (3) unlike tapasin, TAPBPR lacks a discernible ER retention motif (Jackson et al. 1990; Schoenhals et al. 1999); and (4) TAPBPR continues to be associated with MHC-I through the medial Golgi, while MHC-I is released from the PLC in the ER as it binds higher affinity peptide. The contribution of TAPBPR to the shaping of the functional peptide repertoire remains unclear. Its role appears to be subtler than that of tapasin, as a cell line lacking TAPBPR showed little loss of surface MHC-I expression, a hallmark of tapasin deficiency, although the repertoire of bound peptides was altered (Neerincx and Boyle 2017).
Figure 1.
Schematic representation of peptide-loading steps in the classical (PLC-dependent) and PLC-independent pathways. (A) PLC-dependent pathway. MHC-I heavy chains, folding cotranslationally in the ER, are chaperoned first by calnexin, as the light chain, β2m, coassembles. The MHC-I/β2m heterodimer then joins the peptide-loading complex (PLC) where calnexin is replaced by calreticulin (CRT) that bridges the MHC-I heavy chain to ERp57. CRT additionally interacts with the chaperone tapasin, which is also known as TAP-binding protein (TAPBP) due to its physical association with the TAP transporter. Peptides, generated in the cytoplasm by proteasomal degradation of defective translation, are actively transported via the TAP1/2 heterodimer where they assemble with the MHC-I heavy chain, are trimmed by ERAP1 or ERAP2. High affinity peptides lead to dissociation of the pMHC–I/β2m complex from the rest of the PLC. This complex then proceeds through the Golgi to the cell surface. Steps in glycosylation are not shown. (B) PLC-independent pathway. Low affinity pMHC–I/β2m complexes are bound by TAPBPR, either in the ER or cis-Golgi. Exposure to high affinity peptide leads to dissociation of TAPBPR from the pMHC–I/β2m complex (see colour version of this figure at www.tandfonline.com/ibmg).
Thus, a coordinated series of protein:protein interactions, some broadly required for protein folding and some unique to the requirements of peptide loading onto MHC-I, are employed to generate a stable pMHC-I for display at the cell surface. The following discussion focuses on the dynamics of the interaction between MHC-I and the two chaperones, tapasin and TAPBPR, based on recent X-ray crystallographic structures, cryo-EM reconstructions, NMR analyses, and molecular dynamics simulations.
Dynamics of tapasin-mediated peptide exchange
Tapasin was first identified biochemically as a 48-kD glycoprotein that co-immunoprecipitated with MHC-I in cell lysates (Ortmann et al. 1994). The encoding cDNA was subsequently cloned (Ortmann et al. 1997) and tapasin was revealed to be a transmembrane protein belonging to the immunoglobulin superfamily and containing an ER retention signal in its cytoplasmic domain. In mutant cell lines deficient in tapasin, MHC-I/TAP association is lost but can be restored by transfection of the tapasin gene (Sadasivan et al. 1996). These experiments established tapasin as a component of the PLC in the ER, functioning as a bridge between the source of incoming peptides, the TAP transporter, and the target of peptide loading, the MHC-I molecule.
The role of tapasin in peptide loading is illustrated by the greatly reduced levels of cell surface MHC-I in the tapasin-deficient human lymphoblastoid line 721.220 and in tapasin-deficient mice as a result of a failure to load with high affinity peptides. The defect in 721.220 cells can be corrected by expressing a recombinant soluble tapasin that interacts with MHC-I but not with TAP, indicating that tapasin association with MHC-I is sufficient for peptide loading (Lehner et al.1998). The effect of tapasin on MHC-I surface expression is allele dependent (Peh et al. 1998; Grandea et al. 2000; Barber et al. 2001; Williams et al. 2002; Garbi et al. 2006; Rizvi et al. 2014), strikingly illustrated by HLA-B alleles B*44:02 and B*44:05, which although differing by just a single amino acid at position 116 in the F pocket of the peptide binding groove (Asp116 in B*44:02 and Tyr116 in B*44:05), show distinct differences in tapasin requirement: B*44:02 is highly tapasin dependent for assembly whereas B*44:05 is tapasin independent (Macdonald et al. 2003).
Although the crystal structure of a tapasin/ERp57 complex has been reported (Dong et al. 2009), there is as yet no crystallographic data on a tapasin/MHC-I complex to explain how tapasin stabilizes MHC-I in a peptide-deficient form and catalyzes peptide exchange. Instead, molecular dynamic (MD) simulations have been employed to discern differences between tapasin-dependent and tapasin-independent MHC-I alleles (Sieker et al. 2007, 2008; Fisette et al. 2017). X-ray crystal structures of HLA-B*44:02 and HLA-B*44:05 bound to the same peptide reveal no major differences around the F pocket (aside from the Asp116 to Tyr 116 polymorphism) to easily explain the tapasin dependence (Macdonald et al. 2003; Zernich et al. 2004). However MD simulations of the α1/α2 domains of peptide-free versions of the two alleles revealed significant differences in the dynamics of the F pocket. When devoid of peptide, the tapasin-dependent HLA-B*44:02 allelomorph displayed a more open F-pocket that revealed a broader range of molecular motions than the tapasin-independent molecule which resembled the peptide filled state even in the absence of peptide. Opening of the F-pocket was associated with dynamics of the C-terminus of the α1 helix and of the α2–1 helical segment which distorted the peptide binding groove near the F pocket. This behavior leads to the speculation that tapasin stabilizes this “open” state of HLA-B*44:02 until release by the binding of high affinity peptide. MD simulations modeling the tapasin/MHC-I interaction indicated that tapasin binding widens the groove near the α2–1 helix promoting the release of low affinity peptides (Fleischmann et al. 2015). This interpretation is supported further by MD simulations of HLA-B*44:02 loaded with peptides truncated at the N-or C-terminus (Fisette et al. 2017), consistent with the concept that low affinity peptides bound to tapasin/MHC-I complexes cause a widening of the peptide-binding groove as a result of lack of pocket interactions, particularly by the C terminus of the peptide at the F pocket.
Examination of the interaction of tapasin and MHC-I has been studied by pull-down assays in cell lysates (Wearsch and Cresswell 2007, 2013). Attempts to use purified soluble tapasin and MHC-I ER lumenal domains to examine mechanistic details of peptide exchange have been stymied by the apparent weak affinity of the interaction (Chen and Bouvier 2007), necessitating the use of jun/fos leucine zippers to tether tapasin and MHC-I. Using this approach the effect of tapasin/jun on the kinetics of peptide association and dissociation from HLA-B*08:01/fos was analyzed by fluorescence anisotropy (Chen and Bouvier 2007). Tapasin increased the rates of dissociation of a variety of peptides by factors of 17–104 although there was no correlation between intrinsic dissociation half-lives of the peptides and the dissociation rate enhancement by tapasin. An increased association rate of high affinity peptides to peptide-deficient HLA-B*08:01 was also seen in the presence of tapasin. Interestingly, tapasin increased the total amount of loaded HLA-B*08:01 consistent with a stabilization of a peptide-receptive form of HLA-B*08:01 (Chen and Bouvier 2007). Using peptides modified at either their N- or C-termini the authors found that sensitivity to tapasin-mediated dissociation is influenced by peptide interactions with MHC-I at both the A and F pockets and likely throughout the peptide binding groove—as suggested by NMR and X-ray data on the related chaperone TAPBPR discussed below.
Dynamics of TAPBPR-mediated peptide exchange
The tapasin homolog, TAPBPR, shares sequence identity with tapasin and, like tapasin, is a membrane protein whose lumenal face contains an N-terminal domain followed by IgV and IgC domains (Teng et al. 2002). TAPBPR was subsequently shown to be broadly expressed, IFN-γ inducible, and to associate with MHC-I/β2m complexes but not with components of the PLC (Boyle et al. 2013). Recombinant TAPBPR promotes peptide exchange onto a variety of MHC-I allelomorphs without the requirement for jun/fos linkers to stabilize the TAPBPR/MHC-I complex (Hermann et al. 2015; Morozov et al. 2016) suggestive of a higher intrinsic affinity of the TAPBPR/MHC-I interaction compared with tapasin. Indeed, direct binding measurements by sedimentation velocity-analytical ultracentrifugation indicated a Kd of 0.18 μM for TAPBPR interaction with peptide deficient HLA-A2 an affinity 60-fold greater than that measured for peptide-complexed HLA-A2 (Morozov et al. 2016). Direct interaction of tapasin with peptide-deficient HLA-A2 by the same method was not detectable. TAPBPR dissociates from peptide-free HLA-A2 on exposure to high affinity peptides (Morozov et al. 2016), and similar results have been obtained for TAPBPR in several different MHC-I systems, confirming the role of TAPBPR as a bona fide peptide editor (Hermann et al. 2015; Jiang et al. 2017; McShan et al. 2018). Because of the sequence and functional similarities of TAPBPR and tapasin, as well as their ability to mutually compete for MHC-I interaction (Hermann et al. 2013), we expect that both molecules interact with the same general footprint on MHC-I, and destabilize the peptide binding groove similarly. Nevertheless, the apparent differences in affinity, cellular site of action, and the requirement of tapasin for additional components of the PLC, suggest that molecular details may prove distinct.
The high affinity of the interaction between TAPBPR and peptide-deficient MHC-I permitted the crystallization of TAPBPR/MHC-I complexes, revealing molecular details of the chaperone-stabilized peptide-receptive state of MHC-I (Jiang et al. 2017; Thomas and Tampe 2017). Two different strategies were used to generate peptide-deficient MHC-I for crystallization with TAPBPR. One approach was to generate an H2-Dd molecule folded with a truncated, disulfide-linked 5-mer peptide (Jiang et al.2017). This idea was provoked by molecular dynamics simulations of the tapasin/MHC-I interaction (Sieker et al. 2008; Fleischmann et al. 2015) and mutagenesis data on the TAPBPR/MHC-I interaction (Hermann et al. 2013) which implicated the region around the F-pocket for the TAPBPR/MHC-I interaction. The binding groove was further stabilized by the addition of a Gly-Leu dipeptide to the otherwise empty F-pocket (Saini et al. 2015). This strategy leaves the F-pocket of the MHC-I free to undergo conformational dynamics to promote TAPBPR binding. Indeed, the binding affinity of TAPBPR to H2-Dd complexed to the truncated peptide, is 9nM as measured by surface plasmon resonance (Jiang et al. 2017). A second strategy employed the mouse MHC-I molecule H2-Db folded with a UV-sensitive peptide. Exposure to UV-light released the bound peptide and in the presence of TAPBPR a stable complex with the chaperone was formed (Thomas and Tampe 2017).
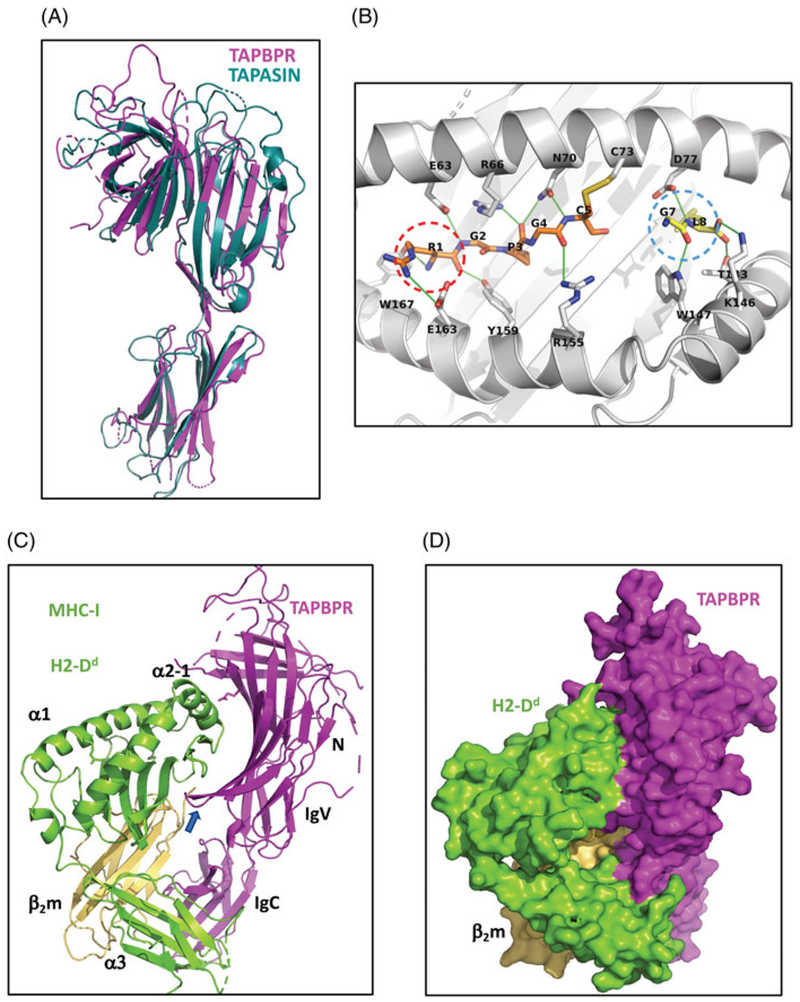
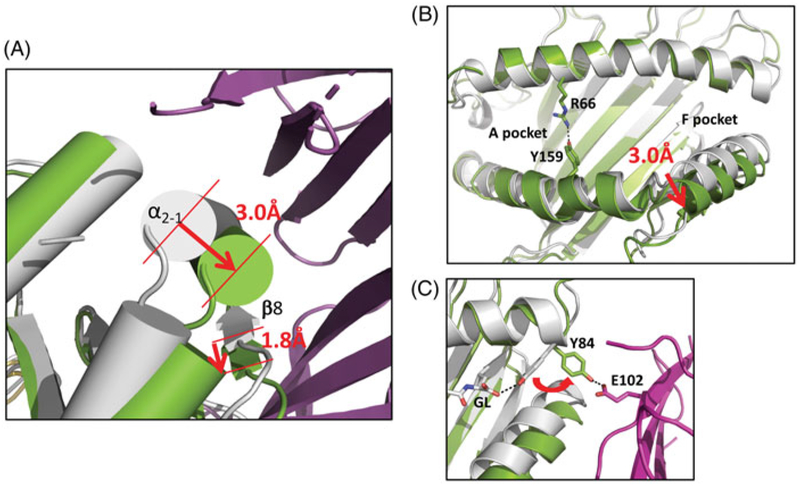
As the two structures are broadly similar, with the exception of one structural detail described below, the following description is derived primarily from the structure of the TAPBPR/H2-Dd complex (Jiang et al. 2017) (Figure 2). Prediction of structural similarity of TAPBPR and tapasin based on amino acid sequence homology, mutational analysis (Van Hateren et al. 2010; Hermann et al. 2013), and comparison of small angle X-ray scattering (SAXS) structures (Morozov et al. 2016) was confirmed as the TAPBPR structure was solved by molecular replacement using tapasin as a model (Figure 2(A)). The peptide receptive form of MHC-I as stabilized by TAPBPR, and compared with the H2-Dd/5-mer complex (Figure 2(B)), reveals dynamic changes in the peptide binding groove that accompany the transition from the peptide-free to the peptide-bound state (Figure 3). The N- and IgV domains of TAPBPR form a basket that cradles the H2-Dd α2–1 helix and extend a loop that contacts a site beneath the peptide-binding platform that senses peptide occupancy (Figure 2(C)). In the peptide-free state, the groove is widened at the F-pocket due to a 3.0 Å outward displacement of the α2–1 helix that is stabilized by interaction with TAPBPR (Figure 3(A)). A downward displacement of the β-strands that form the floor of this pocket lends further distortion to the F-pocket (Figure 3(A)). This appears to be in part due to incursions of a loop of TAPBPR encompassing residues 211–213 that interact with the floor of the binding groove (Figure 2(C)). Further, the side chain of the conserved MHC-I residue Tyr84, which usually coordinates both the C-terminus of the bound peptide and Lys146 of the α2–1 helix, is rearranged to interact with TAPBPR (Figure 3(C)). Regions of the peptide binding groove that are not in direct contact with TAPBPR are also affected as shown by the side chain of Arg66 in the α1 helix which in the TAPBPR bound state extends across the groove to Tyr159 impeding peptide access (Figure 3(B)). An unexpected finding of the crystallographic structures is that the interaction of TAPBPR with MHC-I is also dependent on the contact of the membrane proximal IgC domain of TAPBPR with an interface of the membrane proximal α3-domain of the MHC and the β2m-subunit (Figure 2(C,D)). As a result of these multiple structural changes, the peptide is no longer bound as evidenced by a lack of electron density throughout the peptide binding groove (Figure 3(B)).
Figure 2.
X-ray structures of H2-Dd/5-mer and TAPBPR/H2-Dd complexes. (A) Ribbon diagram of superposition of TAPBPR (magenta) (PDB ID: 5WER) with tapasin (blue-green) (PDB ID:3F8U). (B) The covalently bound 5-mer (RGPGC) in complex with H2-DdT73C/β2m with GL dipeptide visualized in the F pocket (PDB ID: 5WES). The locations of pocket A and F are indicated by dashed red and light blue circles, respectively. (C) Ribbon diagram of TAPBPR/H2-Dd/β2m complex (PDB ID: 5WER). Arrow indicates loop that senses peptide occupancy. (D) Surface representation of TAPBPR/H2-Dd/β2m. For (A) TAPBPR is magenta, tapasin is blue-green; (B) peptide is orange, GL dipeptide is yellow, H2-Dd is gray; (C,D), TAPBPR is magenta, H2-Dd is green, and β2m is yellow (see colour version of this figure at www.tandfonline.com/ibmg).
Figure 3.
Conformational changes of MHC-I accompany interaction with TAPBPR. (A) α2–1 helix of H2-Dd/5-mer (gray) moves 3.0 Å on interaction with TAPBPR, and platform strand, β8, is displaced by 1.8 Å. (Green indicates H2-Dd in complex with TAPBPR (purple) (B) TAPBPR complexed H2-Dd shows rearrangement of binding groove with closing of A and B pockets and opening of F pocket. (C) TAPBPR interaction rearranges orientation of side chain of Y84 of H2-Dd, a conserved residue that normally coordinates with carboxyl terminus of peptide, forms hydrogen bond to E102 of TAPBPR (see colour version of this figure at www.tandfonline.com/ibmg).
In the TAPBPR/H2-Db complex (Thomas and Tampe 2017), but not in the TAPBPR/H2-Dd complex (Jiang et al. 2017), an extended loop encompassing residues 22–36 in TAPBPR was modeled as an α-helix that inserts into the F-pocket region and was hypothesized to function in catalytic exchange of peptide. Despite careful crystallographic refinement guided by omit maps, there was no evidence for this loop in the TAPBPR/H2-Dd complex (Jiang et al. 2017). Large crystallographic B-factors for the atoms of the α-helix in the TAPBPR/H2-Db model are consistent with weak electron density in this region, indicative of a loop of great mobility and uncertain structure. Nevertheless a functional role for the TAPBPR 22–36 loop has been adduced by Ilca et al. (2018) who found that mutant TAPBPR lacking this loop was unable to facilitate peptide exchange onto HLA-A*68:02. A recent crytallographic study addressed the role of the 22–32 loop of tapasin (homologous to the 22–36 loop of TAPBPR) (Hafstrand et al. 2019). Here, the authors took advantage of the ability of the Gly-Leu dipeptide to stabilize some MHC-I molecules either lacking peptide or complexed with truncated peptides. They determined the crystal structure of a complex of the MHC-I molecule H2-Db with a 7-mer peptide truncated at the C-terminus and containing a second peptide representative of the tapasin-derived loop and concluded that steric effects of the tapasin (and by analogy the TAPBPR) loop residues on the F pocket may contribute to the selection of particular antigenic peptides by some MHC-I alleles. Further experimental studies are clearly needed to clarify mechanistic details at the amino acid and atomic levels.
A detailed study on TAPBPR modulation of the solution dynamics of H2-Dd has recently been reported (McShan et al. 2018). Employing multimensional NMR first to experimentally analyze the dynamics of unliganded H2-Dd in solution, the authors identified several sites that underwent conformational exchange in the μs-ms timescale, including the β-sheet floor of the peptide binding groove, the heavy chain interface with β2m, the α2–1 helix, and regions of the α3-domain. Strikingly, these sites of increased flexibility are implicated in TAPBPR interactions as noted in the crystal structures, thus implying a key role for protein dynamics in chaperone interactions with MHC-I. Peptide-deficient, partially loaded, or fully loaded H2-Dd in complex with TAPBPR showed distinct changes in resonances of amino acids lining the peptide binding groove consistent with progressive dampening of groove dynamics as pockets A through F are occupied. On testing peptides of varying lengths and affinities for their ability to promote TAPBPR dissociation from H2 to Dd, the authors found that TAPBPR senses peptide/MHC-I interactions along the entire length of the groove and not only at the TAPBPR/MHC-I interface, implying an allosterically driven release of the peptide in the presence of TAPBPR (McShan et al. 2018).
Although the structures of the TAPBPR/MHC-I complex illustrate how TAPBPR stabilizes a peptide-receptive form of MHC-I, they do not readily explain the apparent MHC-I allelomorph preference of TAPBPR (Morozov et al. 2016). Nor do these structures explain the influence of a single amino acid difference between HLA-B*44:02 and HLA-B*44:05 on their differential requirements for tapasin (Macdonald et al.2003). Sequence comparisons of interface residues do not offer an obvious explanation for these differences. Instead dynamic features of the interacting surfaces must be invoked, both of the chaperones and MHC-I, as influencing the likelihood of a stable interaction. Conformational dynamics of MHC-I molecules have been examined both by computational and experimental methods. Fodor et al. (2018) applied a short, steered MD simulation to each of 20 crystal structures of human MHC-I molecules to obtain an ensemble of structures revealing conformational flexibility of the bound peptide with implications for TCR recognition and vaccine design. Using NMR methods, MHC-I molecules were shown to undergo conformational dynamics at sites of interaction with β2m (Beerbaum et al. 2013), the T cell receptor (Hawse et al. 2014), CD8αβ (McShan et al. 2018), and TAPBPR (McShan et al. 2018). In this context, it is noteworthy that the structure of TAPBPR contained several surface loops that could not be modeled due to lack of electron density (Jiang et al. 2017), including in regions interfacing MHC-I, indicating that TAPBPR also displays conformational heterogeneity. Similarly, the X-ray structure of tapasin reveals loop regions of poor electron density, likely indicative of plasticity (Dong et al. 2009). It is probable that these intrinsically disordered regions of TAPBPR, as well as similar regions in tapasin, are dynamic and flexible to enable broad recognition of MHC-I allelomorphs as a necessary requirement for their chaperone function. Such plasticity is a general feature of MHC molecules and their recognition by T cell receptors (Natarajan et al. 2018). The experimental challenge is to determine how sequence dependent differences in conformational dynamics of different MHC-I allelomorphs correlate with their ability to interact with the chaperones. The answer may reveal insights into not only chaperone-MHC-I interactions but more broadly into other protein-protein interactions which ultimately underlie all of biology.
Complementing the recent X-ray structures of MHC-I/TAPBPR, a cryo-EM structure of the entire PLC has been determined (Blees et al. 2017). The purified complex consists of two editing modules, each containing tapasin, calreticulin, ERp57, and MHC-I, that are pseudo-symmetrically oriented around the TAP heterodimer. Calreticulin’s lectin domain interacts with the MHC-I while its tail extends towards ERp57, permitting tapasin to grab the MHC-I, in an orientation similar to that visualized for TAPBPR/MHC-I. Several different PLC structures seen in the cryo-EM reconstructions suggest how peptide-loaded MHC-I may be released. Indeed, this graphic description of the PLC is competely consistent with earlier biochemical studies characterizing the associations of the components (Cresswell 2019).
Comparison of peptide exchange facilitated by MHC-I and MHC-II chaperones
In contrast to the loading of peptides onto MHC-I in the ER by the PLC-dependent tapasin or in the ER or cis-Golgi by TAPBPR, peptide loading onto MHC-II occurs in acidic endosomal or lysosomal compartments. Transport and initial chaperone function are provided by the CD74 (invariant chain, Ii) molecule that provides transport signals and protects the peptide binding site with its class II-associated invariant chain peptide (CLIP) (Landsverk et al. 2011; Blum et al. 2013; Cresswell and Roche 2014). CLIP is removed and higher affinity peptides are selected by the interaction of HLA-DM (in the mouse, H2-DM), which binds the MHC-II molecule near the P1 pocket that accommodates the major amino terminal p1 anchor side chain (Pos et al. 2012). HLA-DM, an MHC-II-like molecule, binds MHC-II, leads to distortion of the MHC-II peptide binding groove, and is dissociated by binding of a high affinity peptide. Thus, its activity resembles that of tapasin in the PLC, and of TAPBPR, but functions at the opposite end of the peptide binding site. The peptide editing function of HLA-DM is modulated by another MHC-II-like molecule, HLA-DO (H2-DO in the mouse), which appears to bind competitively for the MHC-II binding site of HLA-DM (Guce et al. 2013; Mellins and Stern 2014). Alternative noncompetitive mechanistic models for HLA-DO function have been proposed (Poluektov, Kim, Hartman, et al. 2013; Poluektov, Kim, Sadegh-Nasseri 2013). Although HLA-DM functions analogously to tapasin and TAPBPR, no modulators that mimic HLA-DO have been yet identified for the MHC-I system. Recent evidence suggests that MHC-II molecules (either HLA-DR or -DM) may interact with internalized antigen-bound B cell receptors, raising the possibility of a B cell receptor-mediated MHC-II peptide-loading complex similar to the PLC that serves MHC-I (Macmillan et al. 2014; Barroso et al. 2015).
Conclusion
The immunological necessity of displaying self or pathogen-derived fragments of peptides for recognition by T cells has driven the evolution of the complex and dynamic molecular systems for the generation of peptide antigens, their transport, loading onto MHC molecules, and their display at the antigen presenting cell surface. In recent years, the combined application of computational, functional, genetic, and structural studies has provided insight into the dynamic actions of molecules of the peptide-loading pathways. In particular, structural changes observed as MHC molecules interact with their chaperones and are released from them by binding of high affinity peptides offer understanding of these key steps to the stimulation of both CD8 (MHC-I restricted) and CD4 (MHC-II restricted) T lymphocytes. Such knowledge promises to contribute to our ability to design better immunogens as components of vaccines for infections and tumors.
Acknowledgements
We thank the members of our laboratory for their help, and accept responsibility for any errors or omissions.
Funding
This work was supported by the intramural research program of the NIAID, NIH.
Footnotes
This work was authored as part of the Contributor’s official duties as an Employee of the United States Government and is therefore a work of the United States Government. In accordance with 17 U.S.C. 105, no copyright protection is available for such works under U.S. Law.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Barber LD, Howarth M, Bowness P, Elliott T. 2001. The quantity of naturally processed peptides stably bound by HLA-A*0201 is significantly reduced in the absence of tapasin. Tissue Antig. 58:363–368. [DOI] [PubMed] [Google Scholar]
- Barroso M, Tucker H, Drake L, Nichol K, Drake JR. 2015. Antigen-B cell receptor complexes associate with intracellular major histocompatibility complex (MHC) class II molecules. J Biol Chem. 290:27101–27112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerbaum M, Ballaschk M, Erdmann N, Schnick C, Diehl A, Uchanska-Ziegler B, Ziegler A, Schmieder P. 2013. NMR spectroscopy reveals unexpected structural variation at the protein-protein interface in MHC class I molecules. J Biomol NMR. 57:167–178. [DOI] [PubMed] [Google Scholar]
- Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, Tampe R. 2017. Structure of the human MHC-I peptide-loading complex. Nature. 551: 525–528. [DOI] [PubMed] [Google Scholar]
- Blum JS, Wearsch PA, Cresswell P. 2013. Pathways of antigen processing. Annu Rev Immunol. 31:443–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle LH, Hermann C, Boname JM, Porter KM, Patel PA, Burr ML, Duncan LM, Harbour ME, Rhodes DA, Skjodt K, et al. 2013. Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc Natl Acad Sci USA. 110:3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li L, Weimershaus M, Evnouchidou I, van Endert P, Bouvier M. 2016. ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Sci Rep. 6:28902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Bouvier M. 2007. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 26:1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P 2019. A personal retrospective on the mechanisms of antigen processing. Immunogenetics. 71:141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P, Roche PA. 2014. Invariant chain-MHC class II complexes: always odd and never invariant. Immunol Cell Biol. 92:471–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Wearsch PA, Peaper DR, Cresswell P, Reinisch KM. 2009. Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity. 30:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisette O, Wingbermuhle S, Schafer LV. 2017. Partial dissociation of truncated peptides influences the structural dynamics of the MHCI binding groove. Front Immunol. 8:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann G, Fisette O, Thomas C, Wieneke R, Tumulka F, Schneeweiss C, Springer S, Schafer LV, Tampe R. 2015. Mechanistic basis for epitope proofreading in the peptideloading complex. J Immunol. 195:4503–4513. [DOI] [PubMed] [Google Scholar]
- Fodor J, Riley BT, Borg NA, Buckle AM. 2018. Previously hidden dynamics at the TCR-peptide-MHC interface revealed. J Immunol. 200:4134–4145. [DOI] [PubMed] [Google Scholar]
- Gandhi A, Lakshminarasimhan D, Sun Y, Guo HC. 2011. Structural insights into the molecular ruler mechanism of the endoplasmic reticulum aminopeptidase ERAP1. Sci Rep. 1:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbi N, Tanaka S, Momburg F, Hammerling GJ. 2006. Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat Immunol. 7:93–102. [DOI] [PubMed] [Google Scholar]
- Germain RN, Margulies DH. 1993. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 11:403–450. [DOI] [PubMed] [Google Scholar]
- Grandea AG, Golovina TN, Hamilton SE, Sriram V, Spies T, Brutkiewicz RR, Harty JT, Eisenlohr LC, Van Kaer L. 2000. Impaired assembly yet normal trafficking of MHC class I molecules in Tapasin mutant mice. Immunity. 13:213–222. [DOI] [PubMed] [Google Scholar]
- Guce AI, Mortimer SE, Yoon T, Painter CA, Jiang W, Mellins ED, Stern LJ. 2013. HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nat Struct Mol Biol. 20:90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafstrand I, Sayitoglu EC, Apavaloaei A, Josey BJ, Sun R, Han X, Pellegrino S, Ozkazanc D, Potens R, Janssen L, et al. 2019. Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin. Proc Natl Acad Sci USA. 116:5055–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer GE, Gonzalez F, Champsaur M, Cado D, Shastri N. 2006. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat Immunol. 7:103–112. [DOI] [PubMed] [Google Scholar]
- Hawse WF, De S, Greenwood AI, Nicholson LK, Zajicek J, Kovrigin EL, Kranz DM, Garcia KC, Baker BM. 2014. TCR scanning of peptide/MHC through complementary matching of receptor and ligand molecular flexibility. J Immunol. 192:2885–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann C, Strittmatter LM, Deane JE, Boyle LH. 2013. The binding of TAPBPR and tapasin to MHC class I is mutually exclusive. J Immunol. 191:5743–5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann C, van Hateren A, Trautwein N, Neerincx A, Duriez PJ, Stevanovic S, Trowsdale J, Deane JE, Elliott T, Boyle LH. 2015. TAPBPR alters MHC class I peptide presentation by functioning as a peptide exchange catalyst. Elife. 4:pii: e09617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Marvin JM, Tatsis N, Eisenlohr LC. 2011. Cutting edge: selective role of ubiquitin in MHC class I antigen presentation. J Immunol. 186:1904–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpke S, Tampe R. 2013. The MHC I loading complex: a multitasking machinery in adaptive immunity. Trends Biochem Sci. 38:412–420. [DOI] [PubMed] [Google Scholar]
- Ilca FT, Neerincx A, Hermann C, Marcu A, Stevanovic S, Deane JE, Boyle LH. 2018. TAPBPR mediates peptide dissociation from MHC class I using a leucine lever. Elife. 7: pii:e40126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 9:3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Natarajan K, Boyd LF, Morozov GI, Mage MG, Margulies DH. 2017. Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation. Science. 358:1064–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan G, Krojer T, Harvey D, Fischer R, Chen L, Vollmar M, von Delft F, Kavanagh KL, Brown MA, Bowness P, et al. 2011. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci USA. 108: 7745–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk OJ, Barois N, Gregers TF, Bakke O. 2011. Invariant chain increases the half-life of MHC II by delaying endosomal maturation. Immunol Cell Biol. 89:619–629. [DOI] [PubMed] [Google Scholar]
- Lehner PJ, Surman MJ, Cresswell P. 1998. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line .220. Immunity. 8:221–231. [DOI] [PubMed] [Google Scholar]
- Macdonald WA, Purcell AW, Mifsud NA, Ely LK, Williams DS, Chang L, Gorman JJ, Clements CS, Kjer-Nielsen L, Koelle DM, et al. 2003. A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J Exp Med. 198:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macmillan H, Strohman MJ, Ayyangar S, Jiang W, Rajasekaran N, Spura A, Hessell AJ, Madec AM, Mellins ED. 2014. The MHC class II cofactor HLA-DM interacts with Ig in B cells. J Immunol. 193:2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShan AC, Natarajan K, Kumirov VK, Flores-Solis D, Jiang J, Badstubner M, Toor JS, Bagshaw CR, Kovrigin EL, Margulies DH, et al. 2018. Peptide exchange on MHC-I by TAPBPR is driven by a negative allostery release cycle. Nat Chem Biol. 14:811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellins ED, Stern LJ. 2014. HLA-DM and HLA-DO, key regulators of MHC-II processing and presentation. Curr Opin Immunol. 26:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov GI, Zhao H, Mage MG, Boyd LF, Jiang J, Dolan MA, Venna R, Norcross MA, McMurtrey CP, Hildebrand W, et al. 2016. Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc Natl Acad Sci USA. 113:E1006–E1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Jiang J, May NA, Mage MG, Boyd LF, McShan AC, Sgourakis NG, Bax A, Margulies DH. 2018. The role of molecular flexibility in antigen presentation and T cell receptor-mediated signaling. Front Immunol. 9:1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neerincx A, Boyle LH. 2017. Properties of the tapasin homologue TAPBPR. Curr Opin Immunol. 46:97–102. [DOI] [PubMed] [Google Scholar]
- Nguyen TT, Chang SC, Evnouchidou I, York IA, Zikos C, Rock KL, Goldberg AL, Stratikos E, Stern LJ. 2011. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol. 18:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann B, Androlewicz MJ, Cresswell P. 1994. MHC class I/beta 2-microglobulin complexes associate with TAP transporters before peptide binding. Nature. 368:864–867. [DOI] [PubMed] [Google Scholar]
- Ortmann B, Copeman J, Lehner PJ, Sadasivan B, Herberg JA, Grandea AG, Riddell SR, Tampe R, Spies T, Trowsdale J, et al. 1997. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 277:1306–1309. [DOI] [PubMed] [Google Scholar]
- Peh CA, Burrows SR, Barnden M, Khanna R, Cresswell P, Moss DJ, McCluskey J. 1998. HLA-B27-restricted antigen presen-tation in the absence of tapasin reveals polymorphism in mechanisms of HLA class I peptide loading. Immunity. 8: 531–542. [DOI] [PubMed] [Google Scholar]
- Poluektov YO, Kim A, Hartman IZ, Sadegh-Nasseri S. 2013. HLA-DO as the optimizer of epitope selection for MHC class II antigen presentation. PLoS One. 8:e71228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluektov YO, Kim A, Sadegh-Nasseri S. 2013. HLA-DO and its role in MHC class II antigen presentation. Front Immunol. 4:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pos W, Sethi DK, Call MJ, Schulze MS, Anders AK, Pyrdol J, Wucherpfennig KW. 2012. Crystal structure of the HLA-DM-HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell. 151:1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter C, Quirin K, Kowarik M, Helenius A. 2005. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 24:1730–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi SM, Salam N, Geng J, Qi Y, Bream JH, Duggal P, Hussain SK, Martinson J, Wolinsky SM, Carrington M, et al. 2014. Distinct assembly profiles of HLA-B molecules. J Immunol. 192:4967–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. 1996. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 5:103–114. [DOI] [PubMed] [Google Scholar]
- Saini SK, Schuster H, Ramnarayan VR, Rammensee HG, Stevanovic S, Springer S. 2015. Dipeptides catalyze rapid peptide exchange on MHC class I molecules. Proc Natl Acad Sci USA. 112:202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenhals GJ, Krishna RM, Grandea AG 3rd, Spies T, Peterson PA, Yang Y, Fruh K. 1999. Retention of empty MHC class I molecules by tapasin is essential to reconstitute antigen presentation in invertebrate cells. EMBO J. 18:743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieker F, Springer S, Zacharias M. 2007. Comparative molecular dynamics analysis of tapasin-dependent and -independent MHC class I alleles. Protein Sci. 16:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieker F, Straatsma TP, Springer S, Zacharias M. 2008. Differential tapasin dependence of MHC class I molecules correlates with conformational changes upon peptide dissociation: a molecular dynamics simulation study. Mol Immunol. 45:3714–3722. [DOI] [PubMed] [Google Scholar]
- Teng MS, Stephens R, Du Pasquier L, Freeman T, Lindquist JA, Trowsdale J. 2002. A human TAPBP (TAPASIN)-related gene, TAPBP-R. Eur J Immunol. 32:1059–1068. [DOI] [PubMed] [Google Scholar]
- Thomas C, Tampe R. 2017. Structure of the TAPBPR-MHC I complex defines the mechanism of peptide loading and editing. Science. 358:1060–1064. [DOI] [PubMed] [Google Scholar]
- van Endert PM, Tampe R, Meyer TH, Tisch R, Bach JF, McDevitt HO. 1994. A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity. 1:491–500. [DOI] [PubMed] [Google Scholar]
- Van Hateren A, James E, Bailey A, Phillips A, Dalchau N, Elliott T. 2010. The cell biology of major histocompatibility complex class I assembly: towards a molecular understanding. Tissue Antig. 76:259–275. [DOI] [PubMed] [Google Scholar]
- Wearsch PA, Cresswell P. 2007. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol. 8:873–881. [DOI] [PubMed] [Google Scholar]
- Wearsch PA, Cresswell P. 2013. In vitro reconstitution of the MHC class I peptide-loading complex. Methods Mol Biol. 960:67–79. [DOI] [PubMed] [Google Scholar]
- Wei J, Yewdell JW. 2018. Immunoribosomes: where’s there’s fire, there’s fire. Mol Immunol. 10.1016/j.molimm.2017.12.026 [DOI] [PubMed] [Google Scholar]
- Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. 2002. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 16:509–520. [DOI] [PubMed] [Google Scholar]
- York IA, Rock KL. 1996. Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 14:369–396. [DOI] [PubMed] [Google Scholar]
- Zernich D, Purcell AW, Macdonald WA, Kjer-Nielsen L, Ely LK, Laham N, Crockford T, Mifsud NA, Bharadwaj M, Chang L, et al. 2004. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J Exp Med. 200:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]