Abstract
Most eukaryotic messenger RNA precursors must undergo 3′-end cleavage and polyadenylation for maturation. We and others recently reported the structure of the AAUAAA polyadenylation signal (PAS) in complex with the protein factors CPSF-30, WDR33, and CPSF-160, revealing the molecular mechanism for this recognition. Here we have characterized in detail the interactions between the PAS RNA and the protein factors using fluorescence polarization experiments. Our studies show that AAUAAA is recognized with ∼3 nM affinity by the CPSF-160–WDR33–CPSF-30 ternary complex. Variations in the RNA sequence can greatly reduce the affinity. Similarly, mutations of CPSF-30 residues that have van der Waals interactions with the bases of AAUAAA also lead to substantial reductions in affinity. Finally, our studies confirm that both CPSF-30 and WDR33 are required for high-affinity binding of the PAS RNA, while these two proteins alone and their binary complexes with CPSF-160 have much lower affinity for the RNA.
Keywords: RNA processing, RNA recognition, protein structure
INTRODUCTION
Most eukaryotic messenger RNA precursors (pre-mRNAs) must undergo extensive processing to become functional mRNAs, which includes 5′-end capping, splicing, and 3′-end cleavage and polyadenylation (Proudfoot 2011; Yang and Doublié 2011; Curinha et al. 2014; Xiang et al. 2014; Shi and Manley 2015). The recognition of a polyadenylation signal (PAS) is a crucial step for 3′-end processing, which helps to define the position of cleavage in the pre-mRNA as the PAS is often located 10–30 nt upstream of the cleavage site. The PAS is a hexanucleotide, and the most common motif is AAUAAA (∼55% frequency) for mammalian pre-mRNAs, followed by the AUUAAA motif (∼16% frequency) (Proudfoot and Brownlee 1976; Beaudoing et al. 2000; Tian et al. 2005). Many other motifs can also support 3′-end processing, but are much rarer (<4% frequency). AAUAAA, AUUAAA, and 10 other single nucleotide variants account for ∼92% of PAS in human and mouse pre-mRNAs (Tian et al. 2005).
Many proteins are involved in pre-mRNA 3′-end processing (Zhao et al. 1999; Mandel et al. 2008; Shi et al. 2009), and several subcomplexes of this 3′-end processing machinery have been identified, including the cleavage and polyadenylation specificity factor (CPSF) and the cleavage stimulation factor (CstF). The 73-kDa subunit of CPSF (CPSF-73) is the endoribonuclease for the cleavage reaction (Mandel et al. 2006), and two other CPSF subunits, WDR33 (Shi et al. 2009) and CPSF-30, are required for recognizing the PAS (Chan et al. 2014; Schönemann et al. 2014). CPSF-30, containing five zinc fingers and a zinc knuckle, also interacts with Fip1 (Kaufmann et al. 2004), another subunit of CPSF, which helps to recruit the poly(A) polymerase to the 3′-end processing machinery. CstF recognizes a G/U-rich sequence motif downstream from the cleavage site, and it also has a role in alternative polyadenylation (Elkon et al. 2013; Gruber et al. 2014; Tian and Manley 2017).
We and others recently reported the structures of a quaternary complex of human CPSF-160, CPSF-30, WDR33, and an AAUAAA PAS RNA (Fig. 1A,B; Clerici et al. 2018; Sun et al. 2018), the structure of a ternary complex of the yeast protein homologs (Cft1, Yth1, and Pfs2, without RNA) (Casañal et al. 2017), as well as the structure of a binary complex of human CPSF-160 and WDR33 (Clerici et al. 2017). The structures of the quaternary complexes revealed extensive and specific interactions between the AAUAAA PAS and WDR33 and CPSF-30 (Fig. 1C), while CPSF-160 serves a crucial scaffold role in the complex. In addition, there is a Hoogsteen base pair between U3 and A6 of the PAS. A few aspects of the interactions between the ternary complex and PAS RNA have been studied by fluorescence polarization assays (Clerici et al. 2017), including Kd values for RNAs containing AAUAAA and AAGAAA PAS, the contributions of different CPSF-30 zinc fingers to RNA binding, and the effects of two mutations in WDR33 (K46A/R47A and R49A/K50A) on RNA binding. We had also characterized the interactions among the protein factors and the RNA by gel shift assays (Sun et al. 2018).
FIGURE 1.
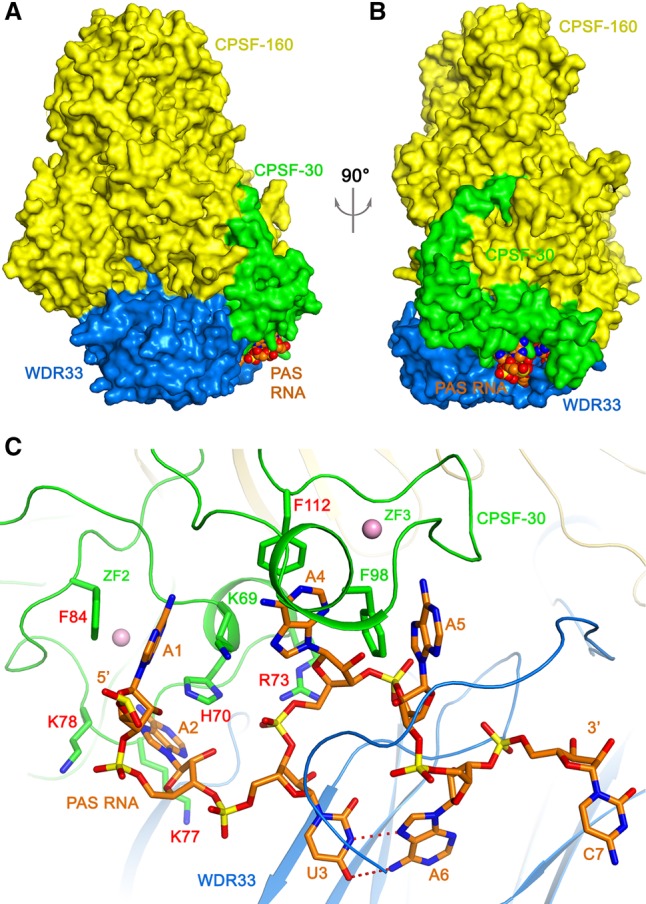
Overall structure of the human CPSF-160–WDR33–CPSF-30–PAS RNA quaternary complex. (A) Structure of the quaternary complex. CPSF-160 (yellow), WDR33 (blue), and CPSF-30 (green) are shown as molecular surfaces. The PAS RNA is shown as a sphere model (orange). (B) Structure of the quaternary complex, viewed after a 90° rotation of around the vertical axis. (C) Recognition of the AAUAAA PAS (orange) by CPSF-30 zinc fingers ZF2-ZF3 (green) and WDR33 (blue). Hydrogen bonds in the U3–A6 Hoogsteen base pair are indicated with dashed lines in red. Side chains of CPSF-30 that contact the RNA bases are shown as stick models, and those selected for mutagenesis studies are labeled in red. Zinc atoms are shown as spheres in pink. Produced with PyMOL (www.pymol.org).
We report here detailed characterizations of the interactions between CPSF-160, WDR33, CPSF-30, and various PAS RNAs. We show that the ternary complex has high affinity for the AAUAAA PAS RNA, with Kd of ∼3 nM, and the AUUAAA PAS RNA has a Kd of ∼17 nM. In comparison, other sequence motifs that can also support 3′-end processing, as well as changes to the U3–A6 Hoogsteen base pair, lead to a substantial reduction in the binding affinity. In addition, mutations of CPSF-30 residues that are in contact with the RNA bases can also give rise to reductions in binding affinity. The CPSF-160–WDR33 and CPSF-160–CPSF-30 binary complexes have much lower affinity for the RNA, confirming that both WDR33 and CPSF-30 are required for PAS recognition. We also show that CPSF-160 is required for high-affinity recognition of the PAS RNA, as WDR33 and CPSF-30 alone have much lower affinity.
RESULTS
Variation of the AAUAAA RNA length
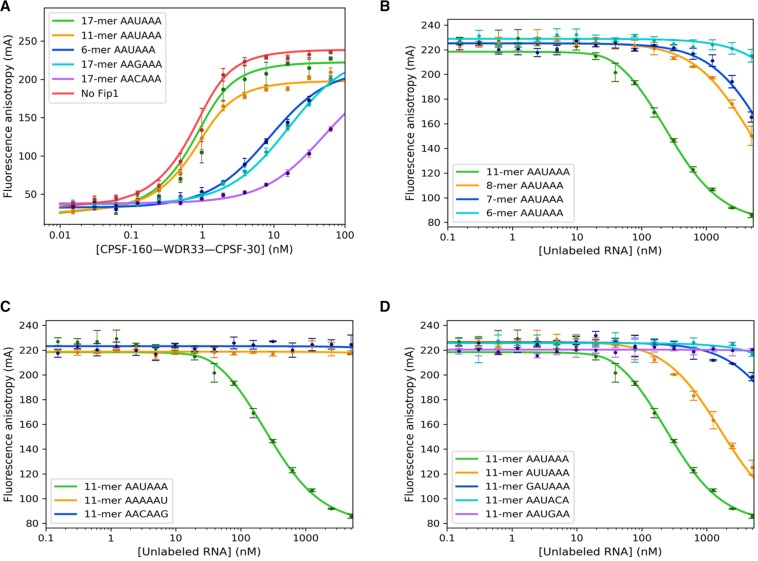
The structures of the quaternary complexes show that WDR33 and CPSF-30 primarily recognize the PAS hexanucleotide itself (Fig. 1C). Although a 17-mer RNA was used for the structural study, only the PAS was found to be well ordered (Sun et al. 2018). The nucleotide directly following the PAS was weakly ordered, and the other nucleotides were disordered. To assess this structural observation, we used RNA oligos of various lengths, 17-mer (FAM-AACCUCCAAUAAACAAC), 11-mer (FAM-CAAUAAACAAC) and 6-mer (AAUAAA-FAM), and carried out fluorescence polarization binding assays. All these oligo RNAs carry a FAM fluorescent label at the 5′ or 3′ end, allowing direct observation of the fluorescence polarization signal.
The experimental data confirmed that the 17-mer and 11-mer RNAs have nearly the same binding affinity to the CPSF-160–WDR33–CPSF-30 ternary complex, with Kd values of 0.28 ± 0.07 nM and 0.32 ± 0.04 nM (P value of 0.34, and therefore the difference is not significant), respectively (Table 1; Fig. 2A), consistent with that reported in an earlier study (Clerici et al. 2017). Fip1 (residues 159–200) was included in these experiments to help stabilize full-length CPSF-30. Without Fip1, the Kd value for the 17-mer RNA decreased slightly to 0.25 ± 0.05 nM (P value 0.24) (Table 1; Fig. 2A), suggesting that Fip1 has essentially no effect on binding this 17-mer RNA. Earlier studies have shown that this segment of Fip1 is involved in binding CPSF-30, while a carboxy-terminal Arg-rich region of Fip1 (missing in the protein studied here) interacts with U-rich RNA (Kaufmann et al. 2004).
TABLE 1.
Summary of observed dissociation constants
FIGURE 2.
Effects of variations in the RNA on PAS recognition. (A) Fluorescence polarization binding assays of the CPSF-160–WDR33–CPSF-30 ternary complex with labeled 17-mer, 11-mer, and 6-mer AAUAAA PAS RNAs, as well as labeled 17-mer RNAs with AAGAAA and AACAAA as the PAS (nucleotides distinct from AAUAAA are indicated by underline). The curves represent fitting to the binding data. (B) Competition fluorescence polarization binding assays of the CPSF-160–WDR33–CPSF-30 ternary complex with unlabeled 11-mer, 8-mer, 7-mer, and 6-mer AAUAAA PAS RNAs. Labeled 17-mer AAUAAA PAS RNA was used as the reporter. (C) Competition fluorescence polarization binding assays of the CPSF-160–WDR33–CPSF-30 ternary complex with unlabeled 11-mer RNAs containing variations of the U3–A6 Hoogsteen base pair, AAAAAU, and AACAAG. (D) Competition fluorescence polarization binding assays of the CPSF-160–WDR33–CPSF-30 ternary complex with unlabeled 11-mer RNAs containing variations of the A1, A2, A4, and A5 bases. Fip1 (159–200) is included in all the assays. Error bars are ±1 standard deviations from triplicate experiments.
In comparison, the 6-mer oligo had a higher Kd value of 8.05 ± 0.45 nM (P value <0.001). This could be due to interference by the FAM label at its 3′ end and/or contribution from nucleotides outside the PAS. For example, the phosphate group of the nucleotide directly following the PAS has some interactions with WDR33 (Fig. 1C). To assess these different scenarios, we used unlabeled AAUAAA 6-mer oligo, AAUAAAC 7-mer oligo, and CAAUAAAC 8-mer oligo and carried out competition fluorescence polarization binding assays against the FAM-labeled 17-mer oligo (Fig. 2B). We included the unlabeled 11-mer CAAUAAACAAC oligo as a control. The Kd value determined from this competition assay for the AAUAAA 11-mer oligo was 3.10 ± 0.17 nM (Table 1), 10-fold higher than the FAM-labeled 11-mer oligo. This indicates that the FAM label enhances the binding of the 11-mer oligo.
The Kd value determined from the competition assay for the AAUAAA 8-mer oligo was 57 ± 18 nM (Table 1), 18-fold higher than the 11-mer oligo in the same assay (P value <0.001), and that for the 7-mer oligo was 35-fold higher (Table 1). The 6-mer oligo showed some binding only at above 2000 nM concentration (Fig. 2B), in contrast to the good affinity for the FAM-labeled 6-mer oligo (Fig. 2A). The FAM label in this oligo is located at the 3′ end, which introduces a phosphate group at that position, while the unlabeled 6-mer oligo has only a hydroxyl group at the 3′ end. Therefore, the higher affinity of the FAM-labeled 6-mer oligo is likely due to contributions from both the 3′ phosphate group and the body of the FAM. Overall, these data indicate that nucleotides outside the AAUAAA hexamer, especially the phosphate group of the 7th nt, also contribute substantially to the binding.
Variation of the U3–A6 Hoogsteen base pair
A U3–A6 Hoogsteen base pair was observed in the PAS when bound to the CPSF-160–WDR33–CPSF-30 ternary complex (Fig. 1C; Clerici et al. 2018; Sun et al. 2018). The binding mode of the PAS suggests that other Hoogsteen base pairs, such as C3–G6, could be accommodated, while a wobble U3–G6 base pair would not fit. To assess whether the alternative Hoogsteen base pair can support binding to the ternary complex, we determined the binding affinity of unlabeled 11-mer RNA with AACAAG (variations from the AAUAAA PAS are indicated by underlines) as the equivalent of the AAUAAA PAS by competition fluorescence polarization assays. The experimental data showed no binding of the oligo even at 5000 nM concentration (Table 1; Fig. 2C), indicating that the alternative Hoogsteen base pair could not be accommodated in the binding site. The 11-mer RNA with AAAAAU as the PAS showed no binding either.
We also characterized the binding of FAM-labeled 17-mer RNAs with AAGAAA and AACAAA as the PAS, to test the effect of breaking the U3–A6 Hoogsteen base pair. These RNAs did show binding to the ternary complex, with Kd values of 15.5 ± 1.3 and 50.3 ± 9.2 nM, respectively (Table 1; Fig. 2A), roughly 55- and 180-fold higher than the 17-mer AAUAAA RNA. The Kd value for the AAGAAA oligo is consistent with that reported in an earlier study (Clerici et al. 2017).
Variation of other positions of the AAUAAA PAS
Besides the U3–A6 Hoogsteen base pair, the structures show that A1 and A4 of the AAUAAA PAS are specifically recognized by CPSF-30, with hydrogen-bonds to the N1 and N6 atoms of the adenine base (Clerici et al. 2018; Sun et al. 2018). A2 and A5 establish good hydrogen-bonding interactions only from their N1 atoms. To assess the binding affinity of other PAS hexamers that can also support 3′-end processing, we selected from those hexamers identified in mammalian pre-mRNAs (Beaudoing et al. 2000; Tian et al. 2005), changing each of these four positions at a time. The unlabeled 11-mer RNAs that we studied included GAUAAA (∼1% frequency, first position), AUUAAA (∼16%, second position), AAUGAA (∼1%, fourth position), and AAUACA (∼2%, fifth position) as the PAS.
The experimental data showed that these variant PAS RNAs have much lower affinity for the ternary complex, except for AUUAAA, which is the second most frequently observed PAS (Fig. 2D). The Kd value for AUUAAA 11-mer oligo is 17.2 ± 2.3 nM (Table 1), about sixfold higher than the corresponding 11-mer AAUAAA oligo (P value <0.001). The GAUAAA 11-mer oligo showed some binding above 2000 nM, while the AAUGAA and AAUACA 11-mer oligos showed only minor binding at 5000 nM concentration (Fig. 2D). AAGAAA is another PAS hexamer with low frequency (∼3%), and it had Kd of 15.5 ± 1.3 nM (55-fold higher, although the RNA carried a FAM label) (Fig. 2A).
Mutations of CPSF-30
Besides hydrogen-bonding interactions between the adenine bases and the backbone of CPSF-30, there are also extensive van der Waals interactions. Specifically, the A1, A2, A4, and A5 bases are each involved in π-stacking interaction with an aromatic side chain of CPSF-30: A1 with Phe84, A2 with His70, A4 with Phe112, and A5 with Phe98 (Fig. 1C). In addition, A2 is flanked on the other face by two Lys side chains, Lys77 and Lys78, and Arg73 has ionic interactions with the phosphate connecting nucleotides U3 and A4.
To test the importance of these interactions for PAS RNA binding, we produced the H70A, R73A, K77A/K78A, F84A, and F112A mutants of CPSF-30. Each mutant and the wild-type CPSF-30 was coexpressed with Fip1, which produced sharp peaks on a gel filtration column (Supplemental Fig. S1), suggesting the complex is well-folded and monodisperse. The purified complex was mixed with the CPSF-160–WDR33 binary complex (which also produced a sharp peak on a gel filtration column, Supplemental Fig. S1), with CPSF-30 at fivefold molar excess, which we showed was sufficient to achieve maximal binding to the RNA (see next). The affinity of these mixtures for the FAM-labeled 17-mer RNA was then determined (Fig. 3). The K77A/K78A mutant had roughly twofold higher Kd (0.34 ± 0.05 nM, Table 1) compared to wild-type CPSF-30 (P value of 0.19), which had a Kd of 0.19 ± 0.03 nM in this assay. On the other hand, the F84A and R73A mutations had larger effects on the binding, with eight- and 17-fold higher Kd values (P value <0.001). Finally, the H70A and F112A mutants showed the largest effects, with 90- and 140-fold higher Kd values (P values <0.001).
FIGURE 3.
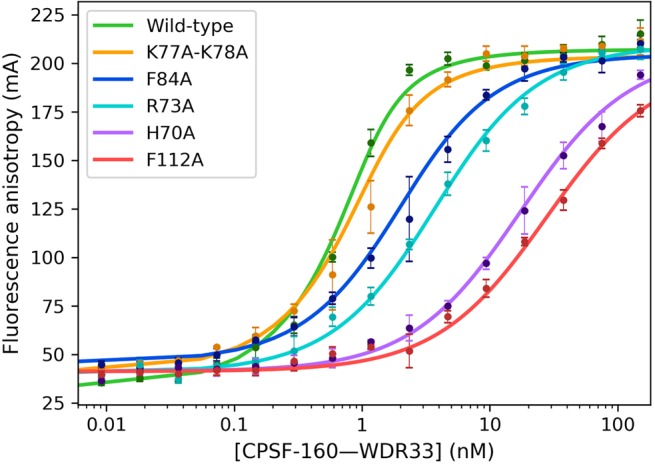
Effects of mutations in CPSF-30 on PAS recognition. Fluorescence polarization binding assays between the labeled 17-mer AAUAAA PAS RNA and mixtures of the CPSF-160–WDR33 binary complex and CPSF-30 wild-type and mutants (coexpressed with Fip1) at fivefold molar ratio. Error bars are ±1 standard deviations from triplicate experiments.
Requirement of both WDR33 and CPSF-30 for RNA binding
The AAUAAA RNA is bound at the interface between WDR33 and CPSF-30 (Fig. 1C), and our earlier electrophoretic mobility shift assays showed that CPSF-30 alone, or the CPSF-160–WDR33 and CPSF-160–CPSF-30 binary complexes could not bind the RNA (Sun et al. 2018). To characterize these interactions more quantitatively, we mixed the CPSF-30–Fip1 complex at increasing molar ratios (0-, 0.5-, 1-, 5-, 10-, and 20-fold) relative to the CPSF-160–WDR33 binary complex, and observed a clear enhancement of the apparent affinity of the mixture for the RNA when CPSF-30 concentration was increased (Fig. 4A). Above fivefold molar ratio of CPSF-30-Fip1 relative to CPSF-160–WDR33, maximal RNA binding was obtained, with essentially the same Kd values (Table 1). In fact, even an equal molar amount of CPSF-30–Fip1 was sufficient to achieve nearly maximal binding, with threefold higher Kd (P value 0.17), indicating a high affinity of CPSF-30 for CPSF-160–WDR33. On the other hand, in the absence of CPSF-30, no binding was observed even at 10 nM concentration of the CPSF-160–WDR33 binary complex. Similarly, no RNA binding was observed for the mixture of CPSF-160 and CPSF-30 (with Fip1) below 10 nM, and for CPSF-30–Fip1 below 200 nM concentration (Fig. 4B), consistent with both WDR33 and CPSF-30 being required for RNA binding. The segment of Fip1 included in the complex with CPSF-30 (residues 159–200) showed no binding to this RNA (Fig. 4B).
FIGURE 4.
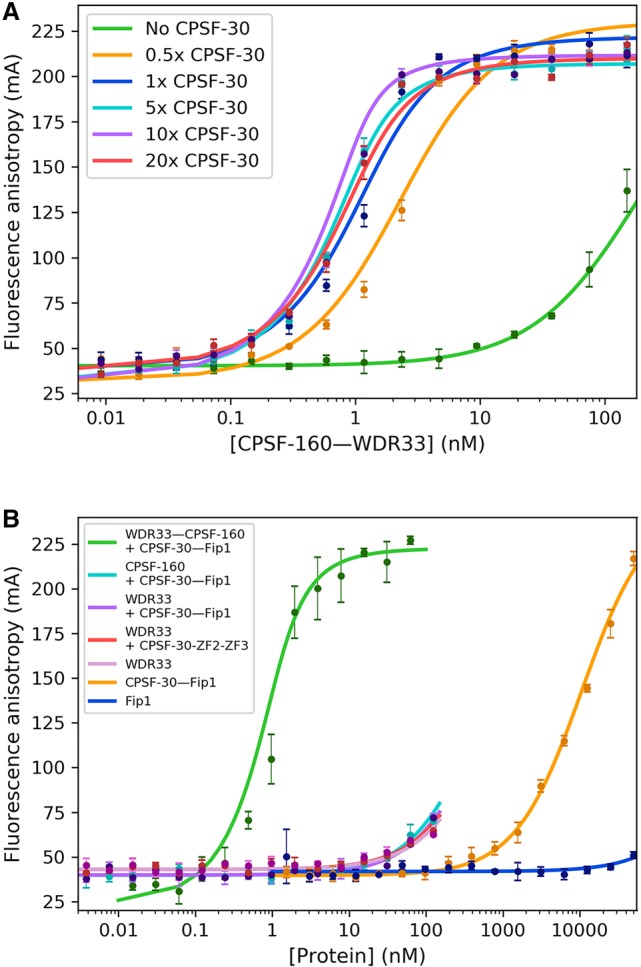
Characterization of the roles of CPSF-160, WDR33, and CPSF-30 in RNA binding. (A) Both CPSF-30 and WDR33 are required for PAS RNA binding. Fluorescence polarization binding assays between the labeled 17-mer AAUAAA PAS RNA and mixtures of the CPSF-160–WDR33 binary complex and CPSF-30 (full-length, coexpressed with Fip1) at increasing molar ratios. (B) WDR33 (containing residues 1–425 as a MBP fusion protein) and CPSF-30 (full-length, coexpressed with Fip1) cannot achieve high-affinity RNA binding in the absence of CPSF-160. WDR33 and CPSF-30 alone also have low affinity for the RNA. Error bars are ±1 standard deviations from triplicate experiments.
CPSF-160 is a scaffold in the complex, recruiting both WDR33 and CPSF-30 and positioning them correctly for PAS RNA binding. On the other hand, the RNA directly contacts both WDR33 and CPSF-30 (Fig. 1A), raising the question whether WDR33 and CPSF-30 can form a complex with the PAS RNA in the absence of CPSF-160. We carried out fluorescence polarization binding assays using purified WDR33 alone (residues 1–425, as an MBP fusion protein), CPSF-30 (full-length) in complex with Fip1, and their mixture. The affinity of the proteins for the RNA was very low, with binding observed only above 10 nM concentration, and we did not observe any enhanced binding of the RNA for the mixture of WDR33 and CPSF-30 (Fig. 4B). We also carried out these experiments with ZF2–ZF3 of CPSF-30 and obtained essentially the same results (Fig. 4B). Therefore, WDR33 and CPSF-30 cannot synergistically give rise to high-affinity binding of the PAS RNA in the absence of CPSF-160.
DISCUSSION
Overall, our studies have provided detailed knowledge on PAS recognition by the CPSF-160–WDR33–CPSF-30 ternary complex. They extend beyond the earlier fluorescence polarization experiments (Clerici et al. 2017), by characterizing the binding affinity of additional PAS hexamers, the importance of the Hoogsteen base pair, the effect of RNA length, the effects of mutating CPSF-30 residues in the binding site. These studies also extend beyond our gel shift experiments (Sun et al. 2018) on these interactions, by examining additional complexes and by providing quantitative measures of the binding affinity.
Our results confirm that both hydrogen-bonding and van der Waals interactions are important for the recognition of the A1, A2, A4, and A5 bases. Changing the identity of these bases generally has strong deleterious effects on the binding affinity, with the exception of the AUUAAA sequence, consistent with it also being frequently observed for 3′-end processing. On the other hand, loss of π-stacking interactions with these bases is also detrimental for the recognition. The structure suggests that the U2 base of AUUAAA could maintain the hydrogen-bond with the main-chain amide of Lys78 in CPSF-30 with a small conformational change, as well as the π-stacking with His70 (Fig. 1C).
Our studies demonstrate the importance of the U3–A6 Hoogsteen base pair for high-affinity binding to the ternary complex. The AAAAAU hexamer, swapping the positions of the U3 and A6 nucleotides, cannot maintain a Hoogsteen base pair as the AAUAAA hexamer (Supplemental Fig. S2), explaining the lack of binding for this RNA. On the other hand, the AACAAG hexamer appears to fit nicely into the binding site (Supplemental Fig. S2), with the guanine base flanked on either side by Phe43 and Phe153 of WDR33 and picking up a hydrogen-bond between its 2-amino group and the main-chain carbonyl of Thr115 in WDR33. The exact reason why this hexamer cannot bind with high affinity is not clear, although it is consistent with the fact that it is not frequently observed for 3′-end processing (Tian et al. 2005). At the same time, the AAGAAA and AACAAA hexamers are able to bind the ternary complex, albeit with substantially reduced affinity, indicating that a base pair here may not be absolutely required for binding.
The less frequently observed PAS motifs studied here appear to have a much lower affinity for the ternary complex. The AAUAAA hexamer is most often associated with the last PAS in human and mouse pre-mRNAs, while the less frequently observed hexamers are associated with upstream PAS of pre-mRNAs with two or more processing sites (Tian et al. 2005). Especially, the AAGAAA hexamer is often found as the PAS in an upstream exon. This suggests that these less frequently observed PAS hexamers have a more prominent role in alternative polyadenylation. A possible mechanism is that other protein factors (such as CstF) as well as recognition of auxiliary sequence motifs may have a larger contribution to the binding of the pre-mRNA, thereby facilitating processing at these sites.
MATERIALS AND METHODS
Protein expression and purification
A human CPSF-160–WDR33 (residues 1–425) binary complex was expressed in baculovirus-infected Hi5 insect cells as described earlier (Sun et al. 2018). WDR33 carried an amino-terminal His-tag followed by MBP and a TEV protease cleavage site. Full-length human CPSF-30 was cloned into ampicillin-resistant pET15_NESG, with amino-terminal His and SUMO tags. Human Fip1 (residues 159–200) was cloned into the pET28a vector (Novagen), also with amino-terminal His and SUMO tags. The proteins were expressed in E. coli BL21(DE3) cells. Coexpression with the Fip1 fragment helped to prevent aggregation of CPSF-30. The components were mixed together for the binding assays, allowing the variation of the molar ratios of CPSF-30 relative to CPSF-160–WDR33.
A CPSF-160–WDR33–CPSF-30 ternary complex was expressed in Hi5 insect cells with the Multibac expression system (Sari et al. 2016) (Geneva Biotech). WDR33 (residues 1–425) carried an amino-terminal His-tag, followed by MBP and a TEV protease cleavage site. CPSF-160 and CPSF-30 were untagged.
The CPSF-160–WDR33–CPSF-30 and the CPSF-160–WDR33 complexes were purified following the same protocol. The insect cells were lysed by sonication in a buffer containing 50 mM Tris (pH 8.0), 500 mM NaCl, 30 mM imidazole, 10 mM β-mercaptoethanol, and one SIGMAFAST protease inhibitor cocktail tablet. The lysate was mixed with 2 mL Ni-NTA beads (Qiagen), washed with 15 mL buffer containing 2 M NaCl to remove bound nucleic acids, then the protein was eluted with 5 mL buffer containing 250 mM imidazole. One milligram TEV protease was added, and the sample was incubated overnight at 4°C. It was then run over a Superdex200 16/60 column (GE Healthcare), using a buffer containing 20 mM (Tris 8.0), 350 mM NaCl, and 10 mM DTT.
For Fip1, the cells were lysed by sonication in a buffer containing 50 mM Tris (pH 8.0), 200 mM NaCl, 30 mM imidazole, 10 mM β-mercaptoethanol, and 2 mM PMSF. The lysate was incubated with 5 mL Ni-NTA, washed with 30 mL buffer, then Fip1 was eluted with 10 mL buffer containing 250 mM imidazole and 100 mM NaCl. To remove the His-SUMO tag, 100 µg UlpI protease was added, and the sample was incubated at 4°C overnight. It was then purified further using a 5 mL Fastflow MonoQ column followed by a Superdex200 16/60 column using a buffer containing 20 mM Tris (pH 8.0), 100 mM NaCl, and 10 mM DTT.
The cells coexpressing Fip1 and CPSF-30 were lysed by sonication in a buffer containing 50 mM Tris (pH 7.5), 500 mM NaCl, 30 mM imidazole, 10 mM β-mercaptoethanol, 100 µM ZnSO4, and 2 mM PMSF. The lysate was incubated with 5 mL Ni-NTA, washed with 20 mL buffer containing 2 M NaCl, then eluted with 10 mL buffer containing 250 mM imidazole, 100 µM ZnSO4, and 500 mM NaCl. To remove the His-SUMO tag, 100 µg UlpI protease was added, and the sample was incubated at 4°C overnight. It was then incubated again with 5 mL Ni-NTA to remove the His-tagged SUMO, and the flow-through was run over a Superdex 200 16/60 column using a buffer containing 20 mM Tris (pH 7.5), 500 mM NaCl, 100 µM ZnSO4, and 10 mM DTT. The excess Fip1 was removed by this gel filtration step.
Fluorescence polarization binding assays
The assays were performed at room temperature using a Neo2S plate reader (Biotek). The buffer for all the assays contained 20 mM Tris (pH 8.0), 150 mM NaCl, 10 mM DTT, 0.01% (v/v) NP-40, and 100 nM BSA. Oligonucleotides that included a polyadenylation site and the surrounding bases from the SV40 virus and a 6-carboxyfluorescein (6-FAM) label at the 5′ or 3′ end were used in direct binding assays. Oligonucleotides without a FAM label were used in competition binding assays. A 17-mer oligonucleotide with a 5′-end FAM label was used as the probe at a concentration of 1 nM in these experiments, and the protein was at 25 nM. For assays that involved titration with CPSF-30, various molar ratios of CPSF-30 (in complex with Fip1) were added to CPSF-160–WDR33. For other binding assays, 1 µM Fip1 was also included. All the mixtures were incubated on ice for 1 h and then transferred to 384-well plates at room temperature. Oligonucleotides were purchased from IDT. All titration experiments were carried out in triplicate.
Both the direct and competition binding experiments were fit to analytical equations (Wang 1995; Lundblad et al. 1996) using the optimize package from SciPy version 1.1.0 (Oliphant 2007). The fit to direct binding data took into account the depletion of the free probe during the titration. Statistical significance was tested by performing an F-test using both the global fit and individual fits of pairs of curves, with the stats package from SciPy version 1.1.0.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This research is supported by National Institutes of Health (NIH) grant R35GM118093 (to L.T.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.070870.119.
REFERENCES
- Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res 10: 1001–1010. 10.1101/gr.10.7.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casañal A, Kumar A, Hill CH, Easter AD, Emsley P, Degliesposti G, Gordiyenko Y, Santhanam B, Wolf J, Wiederhold K, et al. 2017. Architecture of eukaryotic mRNA 3'-end processing machinery. Science 358: 1056–1059. 10.1126/science.aao6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR III, Ule J, Manley JL, Shi Y. 2014. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev 28: 2370–2380. 10.1101/gad.250993.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Faini M, Aebersold R, Jinek M. 2017. Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex. Elife 6: e33111 10.7554/eLife.33111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Faini M, Muckenfuss LM, Aebersold R, Jinek M. 2018. Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. Nat Struct Mol Biol 25: 135–138. 10.1038/s41594-017-0020-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curinha A, Oliveira Braz S, Pereira-Castro I, Cruz A, Moreira A. 2014. Implications of polyadenylation in health and disease. Nucleus 5: 508–519. 10.4161/nucl.36360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506. 10.1038/nrg3482 [DOI] [PubMed] [Google Scholar]
- Gruber AR, Martin G, Keller W, Zavolan M. 2014. Means to an end: mechanism of alternative polyadenylation of messenger RNA precursors. Wiley Interdiscip Rev RNA 5: 183–196. 10.1002/wrna.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626. 10.1038/sj.emboj.7600070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad JR, Laurance M, Goodman RH. 1996. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol 10: 607–612. 10.1210/mend.10.6.8776720 [DOI] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956. 10.1038/nature05363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122. 10.1007/s00018-007-7474-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant TE. 2007. Python for scientific computing. Comput Sci Eng 9: 10–20. 10.1109/MCSE.2007.58 [DOI] [Google Scholar]
- Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782. 10.1101/gad.17268411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Brownlee GG. 1976. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature 263: 211–214. 10.1038/263211a0 [DOI] [PubMed] [Google Scholar]
- Sari D, Gupta K, Thimiri Govinda Raj DB, Aubert A, Drncová P, Garzoni F, Fitzgerald D, Berger I. 2016. The MultiBac baculovirus/insect cell expression vector system for producing complex protein biologics. Adv Exp Med Biol 896: 199–215. 10.1007/978-3-319-27216-0_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönemann L, Kuhn U, Martin G, Schafer P, Gruber AR, Keller W, Zavolan M, Wahle E. 2014. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev 28: 2381–2393. 10.1101/gad.250985.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Manley JL. 2015. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev 29: 889–897. 10.1101/gad.261974.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376. 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhang Y, Hamilton K, Manley JL, Shi Y, Walz T, Tong L. 2018. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc Natl Acad Sci 115: E1419–E1428. 10.1073/pnas.1718723115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. 10.1038/nrm.2016.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acid Res 33: 201–212. 10.1093/nar/gki158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZX. 1995. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett 360: 111–114. 10.1016/0014-5793(95)00062-E [DOI] [PubMed] [Google Scholar]
- Xiang K, Tong L, Manley JL. 2014. Delineating the structural blueprint of the pre-mRNA 3′ end processing machinery. Mol Cell Biol 34: 1894–1910. 10.1128/MCB.00084-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Doublié S. 2011. Structural biology of poly(A) site definition. Wiley Interdiscip Rev RNA 2: 732–747. 10.1002/wrna.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore CL. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.