Abstract
Background
The introduction of industrially produced antibiotics was a milestone in the history of medicine. Now, almost a century later, the adverse consequences of these highly effective drugs have become evident in the form of antibiotic-resistant infections, which are on the rise around the world. The search for solutions to this problem has involved both the introduction of newer types of antibiotics and, increasingly, the development of alternative strategies to prevent infections due to multidrug-resistant bacteria. In this article, we review the pathophysiological connection between the use of antibiotics and the occurrence of such infections. We also discuss some alternative strategies that are currently under development.
Methods
This review is based on pertinent articles that appeared from January 2000 to April 2019 and were retrieved by a selective search in the PubMed database employing the search term “(microbiota OR microbiome) AND infection.” Further suggestions by our author team regarding relevant literature were considered as well.
Results
The spectrum of preventive strategies encompasses measures for the protection of the intestinal microbiota (antimicrobial stewardship, neutralization of antibiotic residues in the bowel, use of phages and species-specific antibiotics) as well as measures for its reconstitution (prebiotics, probiotics, and fecal microbiota transfer).
Conclusion
In view of the major problem that multidrug-resistant bacteria pose for the world’s population and the resources now being spent on the search for a solution, derived both from public funding and from the pharmaceutical industry, we hope to see new, clinically useful approaches being developed and implemented in the near future.
The human body is home to a multitude of microorganisms—bacteria, viruses, protozoa, fungi, and archae—with which it has a symbiotic relationship. As a whole, these microorganisms are referred to as microbiota and their collective genome as microbiome. Bacteria play a particularly important role within the microbiota due to their comparatively large total biomass. Every body surface and internal body surface has its own individual bacteriome (= share of bacterial genetic information in the microbiome). While around 1200 species that colonize humans have been identified to date, there are almost 200 bacterial strains in each individual that can be assigned to 160 different species. The majority of these are found in the colon.
Thanks to continuous developments in high-throughput molecular biological analytics, which make rapid sequencing of bacterial genomes possible, new focus has been put on the role of bacterial ecosystems in maintaining human homeostasis. Based on the rapidly increasing insights in this area, several relevant axes of interaction have been described in recent years, such as the gut–brain, gut–lung, and gut–liver axes (1– 4).
With regard to the role of microbiota in the prevention of infection, the gut–immune axis and the phenomenon of colonization resistance, i.e., the ability to protect the host from the invasion of pathogens into the organism, warrant emphasis (5, 6). These effects play an important role particularly in the prevention of infection by multidrug-resistant bacteria. These bacteria are characterized by acquired resistance to a number of classes of antibiotics. Examples include methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, extended spectrum betalactamases, and carbapenemase-producing Enterobacteriaceae.
Many details still need to be clarified in the search for the definition of a healthy microbiota; however, numerous studies already indicate that high biodiversity in the microbiota is essentially correlated with health. Disease states, on the other hand, are often associated with a depletion or shift in the composition of the microbiota towards dominance by potential pathogens, that is to say, dysbiosis. This observation also applies to the field of infection medicine (7– 10).
Colonization, domination, and infection
In 1945, the Nobel Prize for Medicine was awarded to Alexander Fleming, Ernst Boris Chain, and Howard Walter Florey for their discovery and development of penicillin. The possibility to use antibiotics therapeutically represents a milestone in the history of human medicine. However, not even 100 years later, relevant side effects of antibiotic exposure are becoming ever more apparent.
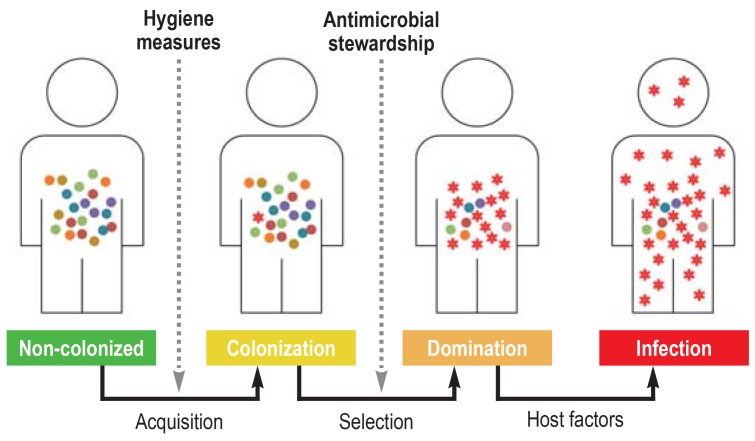
Our commensal microbiota, in particular our gut microbiota, is a reservoir for bacteria that can become the origin of endogenous infections. As a result of contact with multidrug-resistant bacteria, e.g., through the food chain, our environment, or a hospital stay, colonization can occur, with these pathogens initially settling in the gut in extremely low numbers. It is not until an individual is exposed to systemic antibiotics or other substances with an antibacterial effect that an ecological niche emerges, which is filled with multidrug-resistant bacteria undergoing clonal expansion under the selection pressure of the antibiotics. Sustained selection pressure can lead to the dominance of these pathogens within the gut microbiota. If, in addition, host factors emerge that promote a barrier dysfunction in the intestinal mucosa, such as chemotherapy or surgery, bacterial translocation and subsequent blood stream infection may occur (figure 1). Up until only a few years ago, the chief concern was that overly low-dose or overly short treatments could amplify resistant clones of the treated pathogen. However, the view held today is that, if clinical efficacy has been proven, short antibiotic treatment prevents resistance development, since it minimizes the risk of selecting a resistant bacterial population outside the targeted treatment area.
Figure 1.
Phases of infection pathogenesis: Under the influence of antibiotic exposure, multidrug-resistant bacteria that were initially present at low density in the gut—the main resistance reservoir— proliferate in a clonal manner. The dominance of multidrug-resistant bacteria created in this way predisposes to the development of invasive infections with the same pathogens.
A number of studies support this model of pathogenesis. For example, the administration of oral antibiotics results in a significant depletion of intestinal microbiota that can persist for weeks to months (11, 12). Reduced colonization resistance during this phase promotes the colonization of multidrug-resistant bacteria in the resulting ecological niches (11). The results from a US cohort of patients that received stem cell transplantation were among the pioneering findings made over the last decade. Patients post stem cell transplant often receive above averagely intensive broad-spectrum antibiotic treatment. On the basis of microbiome analysis, it was shown that exposure of this kind promotes the dominance—defined as a relative percentage of the bacterial composition on the genus level of over 30%—of Enterococcus, Streptococcus, and Proteobacteria. Proteobacteria include many of the most relevant nosocomial gram-negative pathogens, for example, Enterobacteriaceae and Pseudomonas spp.. Moreover, dominance by particular bacteria in the gut was significantly associated with subsequent bloodstream infection with these bacteria (13).
A similar pattern is also seen in other organs. Although our lungs were long considered to be sterile, it is assumed today that there is a pulmonary microbiota (14). In the case of structural lung impairment, e.g., due to fibrosis or bronchiectasia, an atypical microbiota dominated by multidrug-resistant strains can become established. Similar processes are seen in the setting of mechanical ventilation. Here, the pulmonary microbiota undergoes gradual rarefication. As in the gut, the occurrence of ventilator-associated pneumonia is also associated with domination by single pathogens (14, 15).
Gut–immune axis
Dominance by potentially pathogenic bacteria and subsequent translocation can help to explain in particular the occurrence of nosocomial bacterial infections. However, the evidence is growing for other effects of dysbiosis on the immune system, especially T-cell regulation (figure 2). For example, it was shown in independent mouse models that a reduction in gut microbiota diversity significantly increases susceptibility to viral diseases, depending on T cell–based immunity (16, 17).
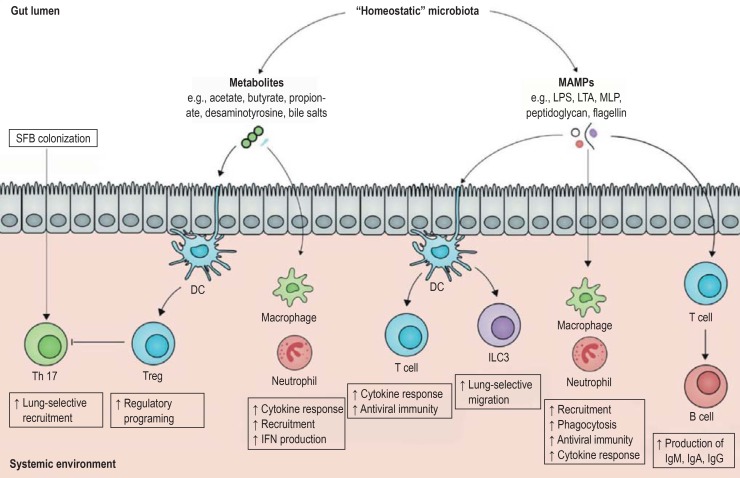
Figure 2.
The gut–immune axis (modified from [6]): metabolites synthesized by the human gut microbiota as well as bacterial surface markers play a crucial role in the regulation of the immune system via direct contact with immune system cells in the gut. DC, dendritic cell; Ig, immunoglobulin; ILC3, type-3 innate lymphoid cell; IFN, interferon; LPS, lipopolysaccharide; LTA, lipoteichoic acid; MAMP, microbe associated molecular pattern; MLP, murein lipoprotein; Treg, regulatory T cells; SFB, segmented filamentous bacteria; Th, T helper
The question arises as to whether these pathomechanisms are also detectable in humans. Initial findings come from the above-mentioned cohort of allogeneic stem cell transplantation recipients. Here, the role of butyrate-producing bacteria in particular was investigated. Butyrate is a short-chain fatty acid present as a fermentation product of anaerobic commensal bacteria in the gut and plays an important role in the regulation of the host immune response. It was shown that, in the absence of butyrate-producing bacteria, there was an increased probability of developing viral respiratory complications. Although that particular study was not able to demonstrate a causal link between microbiota constellation and immune status, it does provide a further indication of the potential relevance of the gut–immune axis for the prevention of infectious diseases (18). Further research in humans will be required in order to elucidate the precise mechanisms and relevance of these effects.
Approaches to protecting the gut microbiota
The pathomechanisms described above suggest that maintaining a microbiota that is as diverse as possible could be a valuable tool in the prevention of infections. Microbiota-based strategies that are already available or are in development are presented below.
Antimicrobial stewardship
Around 25% of all inpatients in Germany receive antibiotics, 70% of which are administered with a therapeutic goal, 30% as prophylaxis. Approximately a third to half of antibiotic prescriptions in the inpatient setting are issued with too little attention to indication and/or treatment duration (19, 20). An analysis of prescription practice in the German primary care setting showed that the treatment of infectious respiratory diseases did not comply with the treatment guidelines in around three out of four cases and that unnecessary or non–guideline-adherent antibiotics were often prescribed (21). More than half of perioperative prophylaxis in Germany is continued for over more than 24 h (22).
Antimicrobial stewardship (AMS) programs are designed to improve the quality of prescribing practices in terms of choice of antibiotic, dose, route of administration, and treatment duration. Restrictive (e.g., special prescription regulations, automated stop-order systems) as well as persuasive or qualifying strategies (e.g., guidelines, training, antibiotic visits, consultation services) are used to this end by interdisciplinary teams.
According to data from cohort studies, randomized studies, and meta-analyses, a correspondingly rational use of antibiotics reduces not only overall mortality (23), but also the rate of sepsis (24) as well as the rate of infection with Clostriduim difficile (25) and multidrug-resistant gram-negative bacteria (e1). A randomized controlled study, in which use of the biomarker procalcitonin to guide the duration of antibiotic treatment was compared to a control group, made a groundbreaking contribution in terms of the effects of reduced antibiotic exposure. On day 28 following the initiation of antibiotic therapy, significantly fewer patients had died in the intervention group (149/761 [20%] compared to 196/785 [25%]) despite shorter antibiotic exposure (95% confidence interval [CI]: [1.2; 9.5], p = 0.0122) (23). It seems reasonable to assume that at least part of this effect is mediated by maintaining a diverse microbiota. The question of whether and to what extent AMS programs are actually able to have a positive effect on the intestinal microbiota needs to be answered in current and future studies.
Neutralizing antibiotic residues in the gut
In addition to AMS, drug options to protect the intestinal microbiota are being developed. One promising option is based on the inactivation of non-absorbed antibiotic residues in the colon, which significantly contribute to the selection of multidrug-resistant pathogens in the gut reservoir. Two substances are currently undergoing phase-II clinical trials:
The oral granulate DAV132 is co-administered with oral or intravenous antibiotics. The core of the granules is made up of neutralizing activated charcoal. The granules have a protective coating that dissolves at pH values from 7. Thus, the substance is released only once it reaches the colon and, according to evidence to date, the serum levels of the respective antibiotics are not affected. The administration of DAV132 to healthy subjects was able to maintain the concentration and diversity of the intestinal microbiota during the use of moxifloxacin (e2).
The orally administered beta-lactamase ribaxamase follows a similar principle. It protects the gastrointestinal microbiota from residues of beta-lactam antibiotics in the intestinal lumen that are excreted, for example, via the biliary route (e.g., ceftriaxone). Since ribaxamase is only activated from a pH value of over 5.5, its action exclusively affects the excreted portion of the antibiotic (e3).
Phase-III trials are currently at the planning stage for both compounds.
Targeted antibiotics and phages
Although in the past it appeared beneficial in particular to promote the development of broad-spectrum antibiotics, findings from microbiome research also highlight the potential of antibiotics with a very narrow spectrum. In situations in which the triggering pathogen requiring treatment has already been unequivocally identified, the species-specific use of antibiotics could minimize the collateral damage to the microbiota and, thus, the selection pressure. The use of tailored antibiotics in the sense of targeted decolonization would also be conceivable. There are only a handful of such substances to date, one example being the agent fidaxomicin, which is approved for the treatment of Clostridium difficile infection. Even though this drug is not species-specific, a randomized study showed that it is able to maintain a more diverse microbiota thanks to its comparatively narrow spectrum of action. This results in significantly fewer recurrences of Clostridium difficile infection. A randomized controlled trial reported that 124/177 patients (70%) in the fidaxomicin group and 106/179 patients (59%) in the vancomycin group responded to treatment and also experienced no recurrence 30 days after the end of treatment (95% CI): [1.0; 20.7], p = 0.030) (e4). Moreover, fewer colonization events with vancomycin-resistant enterococci are observed under fidaxcomicin treatment (e5).
A very similar principle is applied in the use of phages for the targeted treatment of infections. However, the number of published clinical studies is limited, and the findings do not permit a solid assessment of this method as yet (table 1). Dose determination, optimizing the stability of phages as they pass through the intestine, as well as their effect on other bacteria other than those to be eradicated currently still represent considerable scientific challenges (e6).
Table 1. Clinical trials on the use of bacteriophages to treat bacterial infections.
| Indication | Study type | Result | Reference |
| Chronic venous leg ulcers | Monocentric, placebo-controlled phase-I study |
No evidence of relevant side effects |
Rhoads et al., 2009 (e14) |
| Chronic otitis due to multidrug-resistant Pseudomonas aeruginosa | Monocentric, randomized, double-blind, placebo-controlled phase-I/-II study |
– No evidence of relevant side effects – Significant symptom reduction in the treatment arm |
Wright et al., 2009 (e15) |
| Burn wounds colonized with Staphylococcus aureus and Pseudomonas aeruginosa |
Monocentric, placebo-controlled phase-I study |
No evidence of relevant side effects | Rose et al., 2014 (e16) |
|
Escherichia coli–associated diarrhea in childhood |
Monocentric, placebo-controlled phase-II study |
– No evidence of relevant side effects – No significant symptom reduction in the treatment arm |
Sarker et al., 2016 (e17) |
| Chronic rhinosinusitis due to Staphylococcus aureus |
Open-label phase-I study | – No evidence of relevant side effects – Microbial and clinical response in 22.2 % of treated patients (2/9) |
Ooi et al., 2019 (e18) |
Modulation and reconstitution of the microbiota
In addition to the prevention of infections and protection of the microbiota, numerous approaches aimed at modulating the microbiota are currently under development with the goal of increasing species diversity and thereby improving resistance to colonization with multidrug-resistant pathogens.
Fecal microbiota transfer
Today, fecal microbiota transfer (FMT) is recognized worldwide as a clinically highly effective treatment of recurrent Clostridium difficile infection. It is a process by which the microbiota from a healthy donor is transferred to the colon of a patient, either endoscopically, via rectal enema, or by oral administration of capsule preparations.
Building on the considerable success of this treatment in the area of Clostridium difficile infection, FMT’s potential for other indications is currently being explored. The treatment results in a significant reduction in the resistance genes in the stool of patients receiving FMT (e7, e8). This suggests that it could be an approach that has the potential to eliminate multidrug-resistant bacteria from the gut microbiota. Some clinical data on this are already available. One study that investigated the efficacy of an industrially produced microbiota preparation on recurrent Clostridium difficile infection documented clearance of colonization with vancomycin-resistant enterococci as a secondary effect (e9). Thus, the question arises as to whether a similar effect can be demonstrated for decolonization with multidrug-resistant gram-negative bacteria. Since colonization with these bacteria does not necessarily occur as a result of niche formation in the gut, it is uncertain whether microbiota transfer could be effective here. Although there are now numerous case reports and uncontrolled studies, as well as a randomized study, on this question (table 2), it cannot be definitively answered as yet.
Table 2. Evidence on the use of fecal microbiota transfer to eradicate gram-negative pathogens.
| Pathogen | Status | Study type | Eradication rate | Reference |
| ESBL-producing E. coli | Colonization and infection | Case report | 1/1 | Singh et al., 2014 (e19) |
| OXA-48 carbapenemase–producing K. pneumoniae | Colonization | Case report | 1/1 | Lagier et al., 2015 (e20) |
| OXA-48 carbapenemase-producing K. pneumoniae, OXA-48 carbapenemase–producing E. coli, NDM-1 carbapenemase– producing K. pneumoniae | Colonization | Uncontrolled multicenter study | 2/6 | Davido et al., 2017 (e21) |
| NDM-1 carbapenemase–producing and ESBL-producing K. pneumoniae, ESBL- and OXA-48-producing E. coli, MBL carbapenemase–producing P. aeruginosa, carbapenemase-producing E. cloacae, MBL-producing Acinetobacter ursingii, multidrug-resistant Stenotrophomonas maltophilia | Colonization | Uncontrolled monocenter study | 1. FMT: 13/20 2. FMT: 2/4 3. FMT: 0/1 Total: 15/20 |
Bilinski et al., 2017 (e22) |
| VIM carbapenemase–producing P. aeruginosa, ESBL-producing E. coli |
Colonization (E. coli) and infection (P. aeruginosa) |
Case report | 1/1 (P. aeruginosa), but persistent ESBL-producing E. coli | Stalenhoef et al., 2017 (e23) |
| ESBL-producing E. coli | Colonization and infection | Case report | 1/1 | Lahtinen et al., 2017 (e24) |
| ESBL-producing E. coli, ESBL-producing K. pneumoniae | Colonization and infection | Uncontrolled monocenter study | 1. FMT: 3/15 2. FMT: 3/7 Total: 6/15 |
Singh et al., 2018 (e25) |
| NDM- and OXA-48 carbapenemase–producing K. pneumoniae, OXA-48 carbapenemase–producing E. coli | Colonization | Uncontrolled monocenter study | 4/8 | Dinh et al., 2018 (e26) |
| Carbapenemase-producing Acinetobacter spp. | Colonization and infection | Single-arm monocenter study with historically matched control group | 1. FMT: 4/10 2. FMT: 4/5 Total FMT: 8/10 Total controls: 2/20 |
Saïdani et al., 2019 (e27) |
| Carbapenemase- and ESBL-producing Enterobacteriaceae | Colonization | Randomized multicenter open-label study | Non-absorbable antibiotics followed by FMT: 9/22 (41%) vs. 5/17 (29%); OR for decolonization: 1.7 (95% CI [0.4; 6.4]) |
Huttner et al., 2019 (e28) |
ESBL, extended spectrum beta-lactamase; E. coli, Escherichia coli; E. cloacae, Enterobacter cloacae; FMT, fecal microbiota transfer; CI, confidence interval;
K. pneumoniae, Klebsiella ?pneumoniae; MBL, metallo-beta-lactamase; NDM, New Delhi metallo-beta-lactamase; OR, odds ratio; OXA, oxacillinase; P. aeruginosa, Pseudomonas aeruginosa;
vs, versus; VIM, Verona ?integron-encoded metallo-beta-lactamase
Pre- and probiotics
Prebiotics are non-digestible food components that have a favorable effect on the health of the host by selectively promoting the proliferation and/or activity of one or more species of bacteria in the colon. Probiotics, on the other hand, are products that contain live microorganisms and, when consumed orally in sufficient quantities, have a health-promoting effect on the host organism.
The potential of these products becomes clear if one assumes that certain microbiota signatures are able to improve resistance to colonization with multidrug-resistant bacteria. Attempts are currently underway to identify defined bacterial consortiums and/or prebiotics that promote the proliferation of precisely these bacteria. In contrast to FMT, these preparations could be produced under controlled, and thus safer, conditions. Products of this kind are already being developed. Noteworthy examples in the prebiotics class include the human milk oligosaccharides (HMO), which are an important component of breast milk. Synthetically produced HMO help restore the balance between Firmicutes and Bacteroidetes in healthy subjects following antibiotic exposure (e10). Furthermore, recruitment to a randomized study on the clinical efficacy of a synthetic oligosaccharide in the decolonization of patients colonized with multi-resistant bacteria is just about to be initiated in the US.
Little can be said as yet about the clinical efficacy of prebiotics or bacterial consortia. However, early results confirm the observations made in the area of FMT, i.e., the effect varies according to the multidrug-resistant pathogen. Whereas two randomized studies reported success in the decolonization of patients with vancomycin-resistant enterococci using Lactobacillus rhamnosus GG (e11, e12), the combination of Lactobacillus bulgaricus and Lactobacillus rhamnosus in a suspension with fructo-oligosacharides had no effect on the colonization rate in the gram-negative range (e13).
Conclusion
The human microbiota plays an important role in maintaining colonization resistance to multidrug-resistant pathogens. Therefore, the protection and restoration of its diversity represent important goals in translational research. While AMS is already receiving growing worldwide recognition as an implementable approach, other strategies are still in preclinical or clinical development. These include products capable of neutralizing antibiotic residues, targeted antibiotics, phages, pre- and probiotics, as well as FMT. In view of the major problem that multidrug-resistant bacteria pose for the world’s population and the resources now being spent on the search for a solution, derived both from public funding and from the pharmaceutical industry, we hope to see new, clinically useful approaches being developed and implemented in the near future.
Key Messages.
The intestinal microbiota is essential for the maintenance of colonization resistance.
Antibiotic exposure promotes colonization with multidrug-resistant bacteria, their proliferation, and ultimately infection caused by them.
Antimicrobial stewardship (AMS) is the most effective tool available to us at present for the prevention of infection with multidrug-resistant bacteria.
A number of innovative microbiota-based strategies for the prevention of infection with multidrug-resistant bacteria are currently in development.
Acknowledgments
Translated from the original German by Christine Rye.
Footnotes
Conflict of interest statement Yascha Khodamoradi received lecture fees and reimbursement for travel expenses from Merck/MSD.
Prof. Maria Vehreschild received lecture fees and travel expenses support from 3M, Astellas Pharma, Basilea, Gilead Sciences, Merck/MSD, Organobalance, and Pfizer. She received research funding from 3M, Astellas Pharma, DaVolterra, Evonik, Gilead Sciences, MaaT Pharma, Merck/MSD, Morphochem, Organobalance, and Seres Therapeutics. She has provided expert appraisals for Alb-Fils Kliniken GmbH, Ardeypharm, Astellas Pharma, Berlin Chemie, DaVolterra, Ferring, MaaT Pharma, and Merck/MSD.
Prof. Jörg Janne Vehreschild received lecture fees and travel expenses support from Merck/MSD, Gilead, Pfizer, Astellas Pharma, Basilea, and Back Bay Strategies. He received research funding from Merck/MSD, Gilead, Pfizer, Astellas Pharma, and Basilea.
Dr. Kessel states that she has no conflicts of interest.
References
- 1.Agusti A, Garcia-Pardo MP, Lopez-Almela I, et al. Interplay between the gut-brain axis, obesity and cognitive function. Front Neurosci. 2018;12 155. doi: 10.3389/fnins.2018.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibbo S, Ianiro G, Dore MP, Simonelli C, Newton EE, Cammarota G. Gut microbiota as a driver of inflammation in nonalcoholic fatty liver disease. Mediators Inflamm. 2018;2018 doi: 10.1155/2018/9321643. 9321643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tweedle JL, Deepe GS Jr. TNFalpha antagonism reveals a gut/lung axis that amplifies regulatory T cells in a pulmonary fungal infection. Infect Immun. 2018;86 doi: 10.1128/IAI.00109-18. pii: e00109-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. 2016;22:713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 5.Caballero S, Kim S, Carter RA, et al. Cooperating commensals restore colonization resistance to vancomycin-resistant enterococcus faecium. Cell Host Microbe. 2017;21:592–602. doi: 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haak BW, Prescott HC, Wiersinga WJ. Therapeutic potential of the gut microbiota in the prevention and treatment of sepsis. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taur Y, Jenq RR, Perales MA, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karkman A, Lehtimaki J, Ruokolainen L. The ecology of human microbiota: dynamics and diversity in health and disease. Ann N Y Acad Sci. 2017;1399:78–92. doi: 10.1111/nyas.13326. [DOI] [PubMed] [Google Scholar]
- 9.Milani C, Duranti S, Bottacini F, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017;81 doi: 10.1128/MMBR.00036-17. pii: e00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 11.Isaac S, Scher JU, Djukovic A, et al. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J Antimicrob Chemother. 2017;72:128–136. doi: 10.1093/jac/dkw383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haak BW, Lankelma JM, Hugenholtz F, Belzer C, de Vos WM, Wiersinga WJ. Long-term impact of oral vancomycin, ciprofloxacin and metronidazole on the gut microbiota in healthy humans. J Antimicrob Chemother. 2019;74:782–786. doi: 10.1093/jac/dky471. [DOI] [PubMed] [Google Scholar]
- 13.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zakharkina T, Martin-Loeches I, Matamoros S, et al. The dynamics of the pulmonary microbiome during mechanical ventilation in the intensive care unit and the association with occurrence of pneumonia. Thorax. 2017;72:803–810. doi: 10.1136/thoraxjnl-2016-209158. [DOI] [PubMed] [Google Scholar]
- 15.Kelly BJ, Imai I, Bittinger K, et al. Composition and dynamics of the respiratory tract microbiome in intubated patients. Microbiome. 2016;4 doi: 10.1186/s40168-016-0151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosshart SP, Vassallo BG, Angeletti D, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171:1015–1028. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thackray LB, Handley SA, Gorman MJ, et al. Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Rep. 2018;22:3440–3453. doi: 10.1016/j.celrep.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haak BW, Littmann ER, Chaubard JL, et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood. 2018;131:2978–2986. doi: 10.1182/blood-2018-01-828996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davey P, Marwick CA, Scott CL, et al. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst Rev. 2017;2 doi: 10.1002/14651858.CD003543.pub4. CD003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willemsen I, van den Broek R, Bijsterveldt T, et al. A standardized protocol for perioperative antibiotic prophylaxis is associated with improvement of timing and reduction of costs. J Hosp Infect. 2007;67:156–160. doi: 10.1016/j.jhin.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Kraus EM, Pelzl S, Szecsenyi J, Laux G. Antibiotic prescribing for acute lower respiratory tract infections (LRTI)—guideline adherence in the German primary care setting: an analysis of routine data. PLoS One. 2017;12 doi: 10.1371/journal.pone.0174584. e0174584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plachouras D, Karki T, Hansen S, et al. Antimicrobial use in European acute care hospitals: results from the second point prevalence survey (PPS) of healthcare-associated infections and antimicrobial use, 2016 to 2017. Euro Surveill. 2018 doi: 10.2807/1560-7917.ES.23.46.1800393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Jong E, van Oers JA, Beishuizen A, et al. Efficacy and safety of procalcitonin guidance in reducing the duration of antibiotic treatment in critically ill patients: a randomised, controlled, open-label trial. Lancet Infect Dis. 2016;16:819–827. doi: 10.1016/S1473-3099(16)00053-0. [DOI] [PubMed] [Google Scholar]
- 24.Baggs J, Jernigan JA, Halpin AL, Epstein L, Hatfield KM, McDonald LC. Risk of subsequent sepsis within 90 days after a hospital stay by type of antibiotic exposure. Clin Infect Dis. 2018;66:1004–1012. doi: 10.1093/cid/cix947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barlow G, Patterson J, Stultz J, Pakyz AL. Associations between antimicrobial stewardship program elements and clostridium difficile infection performance. Am J Infect Control. 2017;45:1399–1401. doi: 10.1016/j.ajic.2017.06.022. [DOI] [PubMed] [Google Scholar]
- E1.Baur D, Gladstone BP, Burkert F, et al. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis. 2017;17:990–1001. doi: 10.1016/S1473-3099(17)30325-0. [DOI] [PubMed] [Google Scholar]
- E2.de Gunzburg J, Ghozlane A, Ducher A, et al. Protection of the human gut microbiome from antibiotics. J Infect Dis. 2018;217:628–636. doi: 10.1093/infdis/jix604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E3.Kokai-Kun JF, Roberts T, Coughlin O, et al. Use of ribaxamase (SYN-004), a beta-lactamase, to prevent clostridium difficile infection in beta-lactam-treated patients: a double-blind, phase 2b, randomised placebo-controlled trial. Lancet Infect Dis; 01919. :487–496. doi: 10.1016/S1473-3099(18)30731-X. [DOI] [PubMed] [Google Scholar]
- E4.Guery B, Menichetti F, Anttila VJ, et al. Extended-pulsed fidaxomicin versus vancomycin for clostridium difficile infection in patients 60 years and older (EXTEND): a randomised, controlled, open-label, phase 3b/4 trial. Lancet Infect Dis. 2018;18:296–307. doi: 10.1016/S1473-3099(17)30751-X. [DOI] [PubMed] [Google Scholar]
- E5.Nerandzic MM, Mullane K, Miller MA, Babakhani F, Donskey CJ. Reduced acquisition and overgrowth of vancomycin-resistant enterococci and candida species in patients treated with fidaxomicin versus vancomycin for clostridium difficile infection. Clin Infect Dis. 2012;55(Suppl 2):S121–S126. doi: 10.1093/cid/cis440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E6.Lin DM, Koskella B, Lin HC. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther. 2017;8:162–173. doi: 10.4292/wjgpt.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E7.Jouhten H, Mattila E, Arkkila P, Satokari R. Reduction of antibiotic resistance genes in intestinal microbiota of patients with recurrent clostridium difficile infection after fecal microbiota transplantation. Clin Infect Dis. 2016;63:710–711. doi: 10.1093/cid/ciw390. [DOI] [PubMed] [Google Scholar]
- E8.Leung V, Vincent C, Edens TJ, Miller M, Manges AR. Antimicrobial resistance gene acquisition and depletion following fecal microbiota transplantation for recurrent clostridium difficile infection. Clin Infect Dis. 2018;66:456–457. doi: 10.1093/cid/cix821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E9.Dubberke ER, Mullane KM, Gerding DN, et al. Clearance of vancomycin-resistant enterococcus concomitant with administration of a microbiota-based drug targeted at recurrent clostridium difficile infection. Open Forum Infect Dis. 2016;3 doi: 10.1093/ofid/ofw133. ofw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E10.Elison E, Vigsnaes LK, Rindom Krogsgaard L, et al. Oral supplementation of healthy adults with 2‘-O-fucosyllactose and lacto-N-neotetraose is well tolerated and shifts the intestinal microbiota. Br J Nutr. 2016;116:1356–1368. doi: 10.1017/S0007114516003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E11.Manley KJ, Fraenkel MB, Mayall BC, Power DA. Probiotic treatment of vancomycin-resistant enterococci: a randomised controlled trial. Med J Aust. 2007;186:454–457. doi: 10.5694/j.1326-5377.2007.tb00995.x. [DOI] [PubMed] [Google Scholar]
- E12.Szachta P, Ignys I, Cichy W. An evaluation of the ability of the probiotic strain lactobacillus rhamnosus GG to eliminate the gastrointestinal carrier state of vancomycin-resistant enterococci in colonized children. J Clin Gastroenterol. 2011;45:872–877. doi: 10.1097/MCG.0b013e318227439f. [DOI] [PubMed] [Google Scholar]
- E13.Salomao MC, Heluany-Filho MA, Menegueti MG, Kraker ME, Martinez R, Bellissimo-Rodrigues F. A randomized clinical trial on the effectiveness of a symbiotic product to decolonize patients harboring multidrug-resistant gram-negative bacilli. Rev Soc Bras Med Trop. 2016;49:559–566. doi: 10.1590/0037-8682-0233-2016. [DOI] [PubMed] [Google Scholar]
- E14.Rhoads DD, Wolcott RD, Kuskowski MA, Wolcott BM, Ward LS, Sulakvelidze A. bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial. J Wound Care. 2009;18:237–238. doi: 10.12968/jowc.2009.18.6.42801. 240-3. [DOI] [PubMed] [Google Scholar]
- E15.Wright A, Hawkins CH, Anggard EE, Harper DR. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol. 2009;34:349–357. doi: 10.1111/j.1749-4486.2009.01973.x. [DOI] [PubMed] [Google Scholar]
- E16.Rose T, Verbeken G, Vos DD, et al. Experimental phage therapy of burn wound infection: difficult first steps. Int J Burns Trauma. 2014;4:66–73. [PMC free article] [PubMed] [Google Scholar]
- E17.Sarker SA, Sultana S, Reuteler G, et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine. 2016;4:124–137. doi: 10.1016/j.ebiom.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E18.Ooi ML, Drilling AJ, Morales S, et al. Safety and tolerability of bacteriophage therapy for chronic rhinosinusitis due to staphylococcus aureus. JAMA Otolaryngol Head Neck Surg. 2019 doi: 10.1001/jamaoto.2019.1191. doi: 10.1001/jamaoto.2019.1191 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- E19.Singh R, van Nood E, Nieuwdorp M, et al. Donor feces infusion for eradication of extended spectrum beta-lactamase producing Escherichia coli in a patient with end stage renal disease. Clin Microbiol Infect. 2014;20:O977–O978. doi: 10.1111/1469-0691.12683. [DOI] [PubMed] [Google Scholar]
- E20.Lagier JC, Million M, Fournier PE, Brouqui P, Raoult D. Faecal microbiota transplantation for stool decolonization of OXA-48 carbapenemase-producing klebsiella pneumoniae. J Hosp Infect. 2015;90:173–174. doi: 10.1016/j.jhin.2015.02.013. [DOI] [PubMed] [Google Scholar]
- E21.Davido B, Batista R, Michelon H, et al. Is faecal microbiota transplantation an option to eradicate highly drug-resistant enteric bacteria carriage? J Hosp Infect. 2017;95:433–437. doi: 10.1016/j.jhin.2017.02.001. [DOI] [PubMed] [Google Scholar]
- E22.Bilinski J, Grzesiowski P, Sorensen N, et al. Fecal microbiota transplantation in patients with blood disorders inhibits gut colonization with antibiotic-resistant bacteria: results of a prospective, single-center study. Clin Infect Dis. 2017;65:364–370. doi: 10.1093/cid/cix252. [DOI] [PubMed] [Google Scholar]
- E23.Stalenhoef JE, Terveer EM, Knetsch CW, et al. Fecal microbiota transfer for multidrug-resistant gram-negatives: a clinical success combined with microbiological failure. Open Forum Infect Dis. 2017;4 doi: 10.1093/ofid/ofx047. ofx047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E24.Lahtinen P, Mattila E, Anttila VJ, et al. Faecal microbiota transplantation in patients with clostridium difficile and significant comorbidities as well as in patients with new indications: a case series. World J Gastroenterol. 2017;23:7174–7184. doi: 10.3748/wjg.v23.i39.7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E25.Singh R, de Groot PF, Geerlings SE, et al. Fecal microbiota transplantation against intestinal colonization by extended spectrum beta-lactamase producing enterobacteriaceae: a proof of principle study. BMC Res Notes. 2018;11 doi: 10.1186/s13104-018-3293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E26.Dinh A, Fessi H, Duran C, et al. Clearance of carbapenem-resistant enterobacteriaceae vs vancomycin-resistant enterococci carriage after faecal microbiota transplant: a prospective comparative study. J Hosp Infect. 2018;99:481–486. doi: 10.1016/j.jhin.2018.02.018. [DOI] [PubMed] [Google Scholar]
- E27.Saïdani N, Lagier JC, Cassir N, et al. Faecal microbiota transplantation shortens the colonisation period and allows re-entry of patients carrying carbapenamase-producing bacteria into medical care facilities. Int J Antimicrob Agents. 2019;53:355–361. doi: 10.1016/j.ijantimicag.2018.11.014. [DOI] [PubMed] [Google Scholar]
- E28.Huttner BD, de Lastours V, Wassenberg M, et al. A 5-day course of oral antibiotics followed by faecal transplantation to eradicate carriage of multidrug-resistant Enterobacteriaceae: a randomized clinical trial. Clin Microbiol Infect. 2019;25:830–838. doi: 10.1016/j.cmi.2018.12.009. [DOI] [PubMed] [Google Scholar]