Abstract
Infections by pathogens that contain DNA trigger the production of type-I interferons and inflammatory cytokines through the DNA-sensing enzyme cyclic GMP–AMP synthase, which produces 2′3′-cyclic GMP–AMP (cGAMP) that binds to and activates stimulator of interferon genes (STING; also known as TMEM173, MITA, ERIS and MPYS)1–8. STING is an endoplasmic-reticulum membrane protein that contains four transmembrane helices followed by a cytoplasmic ligand-binding domain9–13. The ligand-binding domain of STING forms a dimer, which undergoes a conformational change upon binding to cGAMP9,14. However, it remains unclear how this conformational change leads to STING activation. Here we present cryo-electron microscopy structures of full-length STING from human and chicken in the inactive dimeric state (about 80 kDa in size), as well as cGAMP-bound chicken STING in both the dimeric and tetrameric states. The structures show that the transmembrane and cytoplasmic regions interact to form an integrated, domain-swapped dimeric assembly. Closure of the ligand-binding domain, induced by cGAMP, leads to a 180° rotation of the ligand-binding domain relative to the transmembrane domain. This rotation is coupled to a conformational change in a loop on the side of the ligand-binding-domain dimer, which leads to the formation of the STING tetramer and higher-order oligomers through side-by-side packing. This model of STING oligomerization and activation is supported by our structure-based mutational analyses.
The crystal structures of the ligand-binding domain (LBD) of STING in its apo and cGAMP-bound forms reveal that STING forms a butterfly-shaped dimer, and that binding to cGAMP induces an inward rotation of both monomers towards the ligand-binding pocket as well as the formation of a four-stranded β-sheet ‘lid’ that seals the pocket9,14–16. How this conformational change leads to activation of STING remains unclear. STING activation requires ligand-induced oligomerization and translocation from the endoplasmic reticulum through the endoplasmic reticulum–Golgi intermediate compartment and Golgi-to-post-Golgi vesicles17,18. The importance of STING translocation in signalling implies that the transmembrane domain of STING has a role in regulating its activity. Our understanding of the transmembrane domain of STING is poor, owing to the lack of a structure for full-length STING. The structure and function of several other parts in STING—including the N-terminal cytoplasmic segment and the linkers between the transmembrane helices—are likewise unknown. To address these questions, we determined the structures of full-length STING from chicken (Gallus gallus) and human (Homo sapiens) in the inactive dimeric state, as well as cGAMP-bound chicken STING in both the dimeric and tetrameric states.
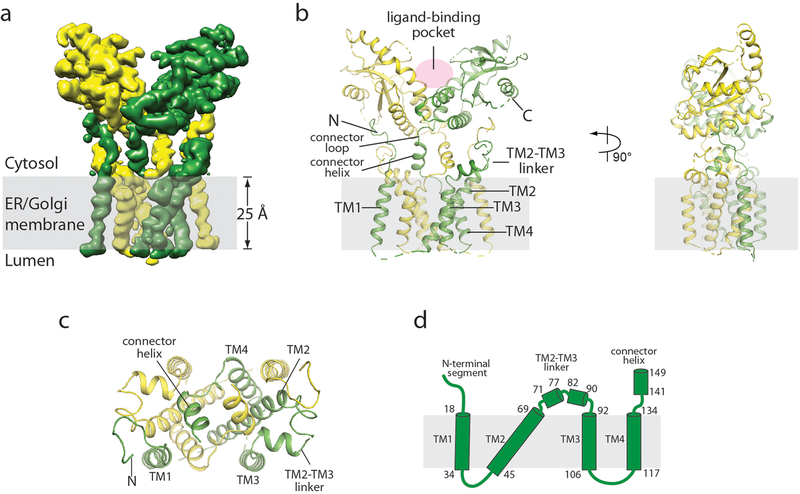
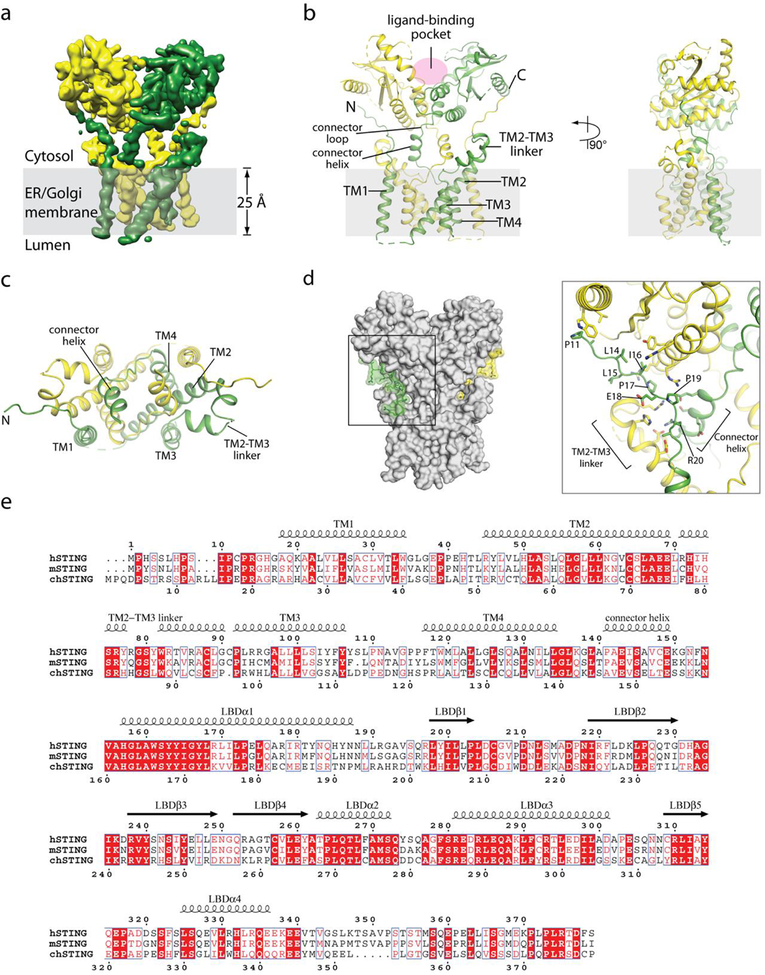
The small size of the STING dimer (about 80 kDa) makes it one of the most challenging membrane proteins for cryo-electron microscopy (cryo-EM) analyses19. To enhance the contrast for better image alignment, we collected the data using a Volta phase plate20 and obtained the final reconstructions of human and chicken STING in the apo state at overall resolutions of 4.1 Å and 4.0 Å, respectively (Extended Data Figs. 1, 2). The structures of human (Fig. 1) and chicken (Extended Data Fig. 3a–d) STING are very similar to each other. Residue numbers in the following descriptions will refer to human STING unless otherwise stated. The LBD of full-length STING adopts an inactive conformation that is similar to that of the LBD of STING that lacks the N terminus10–13,16,21. The C-terminal tail (residue 337 to the C terminus)—which contains the phosphorylation site for TANK-binding kinase 1 (TBK1)22 that is essential for signalling—is not visible in the cryo-EM maps, owing to structural flexibility. The four transmembrane helices adopt a domain-swapped architecture in the STING dimer, in which transmembrane helix 1 (TM1) from one subunit packs with transmembrane helices 2, 3 and 4 (TM2, TM3 and TM4) from the other (Fig. 1a, b). The eight transmembrane helices in the STING dimer organize into two layers: TM2 and TM4 from both subunits form the central layer, which is surrounded by TM1 and TM3 at the periphery (Fig. 1c). TM2 (residues 45–69), which is the longest transmembrane helix in STING, is highly tilted relative to the membrane plane (Fig. 1d).
Fig. 1 |. Structure of full-length human STING in the apo state.
a, Side view of the cryo-EM 3D reconstruction. The two subunits in the dimer are coloured in yellow and green. b, Cartoon representation of the structure in two orthogonal side views. c, Cartoon representation of the transmembrane domain dimer, viewed from the cytosolic side. d, Topological diagram of the transmembrane domain.
The structure of full-length STING reveals several features that were not included in previous studies. For instance, the linker between TM4 and the first helix in the LBD (LBDα1, residues 157–187) contains a two-turn amphiphilic helix (residues 141–149; hereafter referred to as the connector helix) (Fig. 1b, Extended Data Fig. 3b). Two copies of the connector helix in the dimer form a small, right-handed coiled-coil through the hydrophobic face, and sit on the concave cytoplasmic face of the dimeric transmembrane domain. A connector loop (residues 150–156) links the connector helix to LBDα1. The two connector LBDα1 elements in the dimer form a right-handed crossover that makes close intermolecular interactions at the junction.
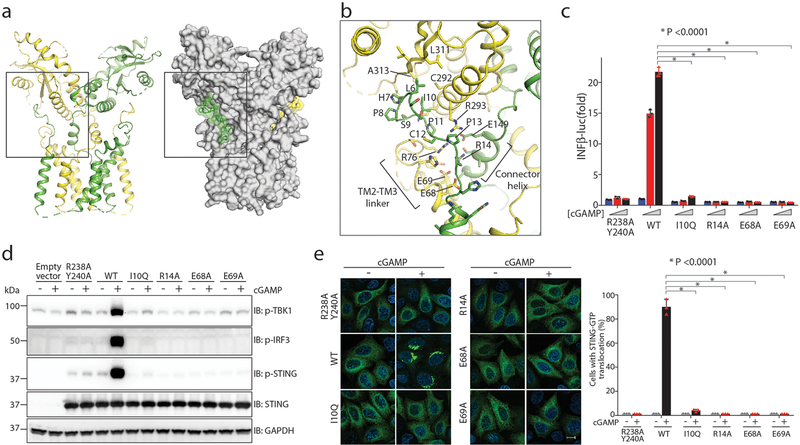
The linker between TM2 and TM3 (residues 70–91) forms two short helices (Figs. 1d, 2a, b). The C-terminal end of the second helix in this linker packs closely with the N-terminal end of the connector helix. The TM2–TM3 linker, the connector helix and the LBD together form a surface groove between the cytoplasmic and transmembrane domains. The N-terminal segment that precedes TM1 (residues 4–15) sits snugly in this groove (Fig. 2a, b). In detail, Leu6, His7, Ile10, Pro11 and Pro13 in the N-terminal segment form hydrophobic interactions with the membrane-proximal surface of the LBD, which includes Cys292 and Arg293 in LBDα3 and Leu311 and Ala313 in strand 5 of the LBD (LBDβ5). Arg14 in the N-terminal segment sticks into a deep pocket in the surface groove, and forms a charge–charge interaction network together with conserved charged residues Glu68, Glu69, Arg76, Glu149 and Arg293 in the groove (Fig. 2b). This inter-domain interface is also present in chicken STING (Extended Data Fig. 3d), which suggests that it is a conserved structural feature that is important for the stability and function of the protein.
Fig. 2 |. Interaction between the N-terminal segment and the body of human STING.
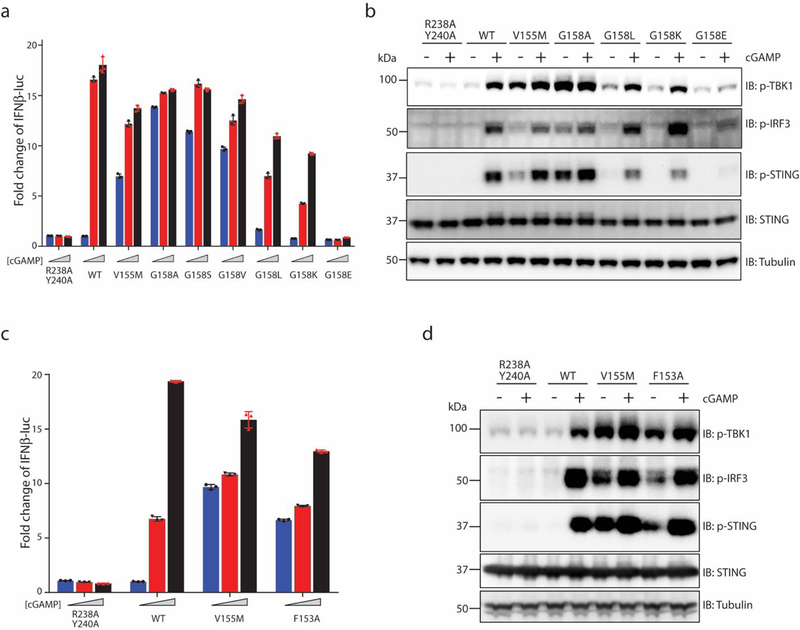
a, Overview of STING N-terminal and body interaction in cartoon and surface representations. b, Detailed view of the interface. The view is expanded from the region in the box in a. c, Effects of STING mutation on the inter-domain interface in cGAMP-induced expression of the interferon-β 1 (IFNB1) gene. HEK293T cells that stably express IFNβ–luciferase reporter were transfected with expression vectors that encode wild-type (WT) or mutants of human STING. Cells were stimulated with three different concentrations of cGAMP (0, 0.3 and 1.4 μM). The R238A/Y240A mutant, which cannot bind cGAMP, was used as a negative control. Data are mean ± s.d. and representative of three biological replicates. d, Effects of STING mutations on the phosphorylation of STING, TBK1 and IRF3. HEK293T cells that express wild-type or mutant human STING were stimulated with cGAMP (1 μM) for 3 h and subjected to immunoblot analyses with indicated proteins. Data are representative of three biological replicates. e, Effects of STING mutations on the translocation and oligomerization of STING in cells. cGAS-deficient HeLa cells were transfected with expression vectors encoding wild-type or mutant STING–GFP. Representative confocal images with or without cGAMP stimulation are shown. Scale bar, 10 μm. The graph on the right is calculated from at least 150 cells. Data are mean ± s.d. and representative of three biological replicates.
Guided by the structure, we introduced substitutions at several residues in human STING (I10Q, R14A, E68A and E69A) to disrupt the inter-domain interface that is formed by the N-terminal segment, and then examined the effects of these substitutions on STING signalling. The results show that, in HEK293T cells, the substitutions abolished cGAMP-induced expression of an interferon-β (IFNβ) reporter (Fig. 2c). Similarly, these substitutions in STING abolished cGAMP-induced phosphorylation of TBK1, STING and the downstream transcription factor interferon regulatory factor 3 (IRF3) (Fig. 2d). cGAMP triggers the translocation of STING from the endoplasmic reticulum to post-Golgi compartments in which it forms puncta-like structures that are indicative of oligomers3. The translocation and oligomerization were abrogated by the substitutions (Fig. 2e). These results validate the importance of interactions mediated by the N-terminal segment in the structure and function of STING.
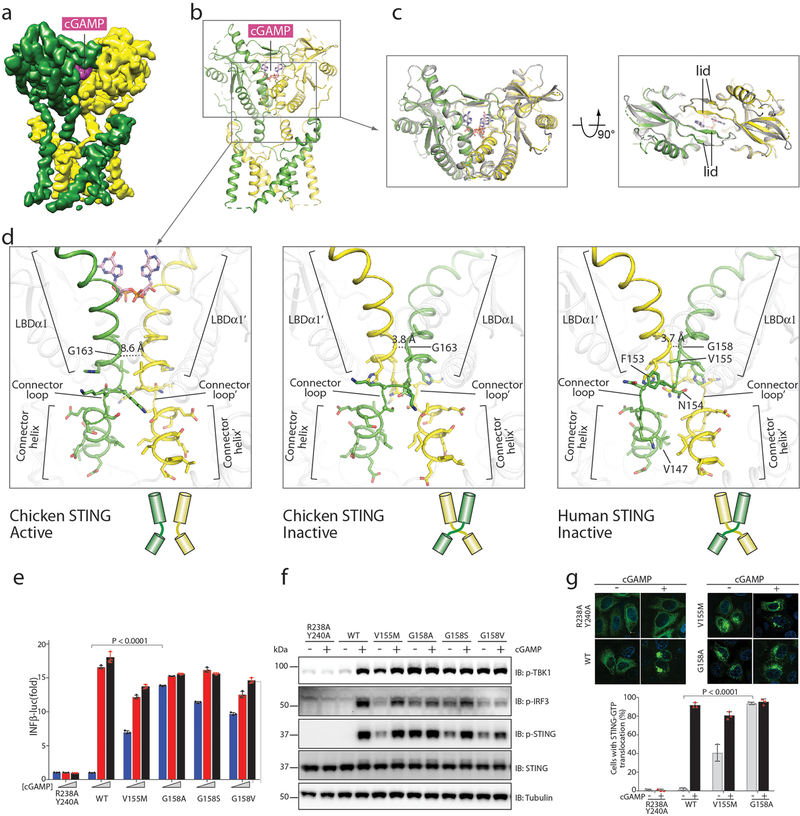
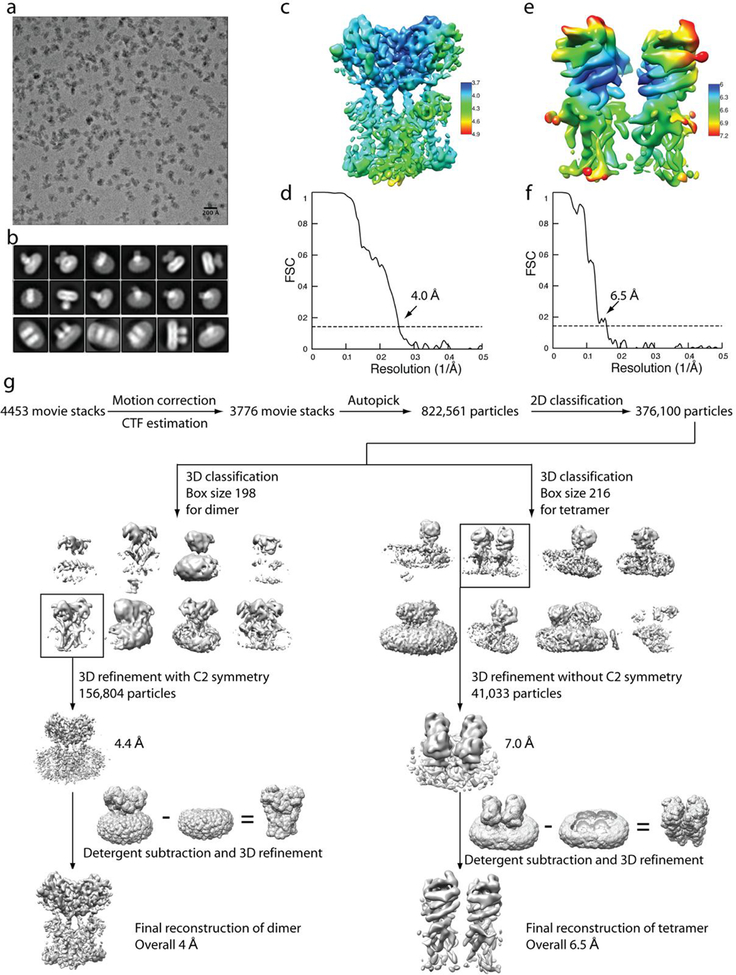
Consistent with previous studies that show that DNA-activated cyclic GMP–AMP synthase (cGAS) induces STING oligomerization23,24, cryo-EM micrographs of cGAMP-bound chicken STING contained both the dimer and higher-order oligomers, at a ratio of approximately 4:1 (Extended Data Fig. 4). Higher-order oligomers were composed of multiple (predominantly two) dimers arranged in a linear manner. We separated the dimer and tetramer by 3D classification and refined each sub-class separately. The reconstruction of dimer cGAMP-bound chicken STING reached a resolution of 4.0 Å. Similar to the structure of the soluble form of the cGAMP-bound LBD dimer9,14–16, the LBD dimer in full-length STING adopts a closed conformation that tightly embraces cGAMP in the ligand-binding pocket (Fig. 3a–c).
Fig. 3 |. Structure of full-length chicken STING bound to cGAMP.
a, Side view of the 3D reconstruction. b, Cartoon representation of the structure. c, Comparison of the cGAMP-bound LBD in full-length STING with that of STING that lacks the transmembrane domain (PDB code 4KSY, shown in grey). d, The 180° rotation of the LBD relative to the transmembrane when converting from the inactive to the active state of STING. The connector and LBDα1, which are at the centre of the rotation, are highlighted. e, Effects of mutations in the connector and LBDα1 on IFNB1 gene expression, stimulated by three different concentrations of cGAMP (0, 0.3 and 1.4 μM). Data are mean ± s.d. f, Effects of STING mutations on phosphorylation of STING, TBK1 and IRF3. One representative set of results from three independent experiments is shown. g, Effects of STING mutations on STING translocation and oligomerization. Representative confocal images of STING–GFP with or without cGAMP stimulation are shown. Scale bar, 10 μm. Data are mean ± s.d. The analyses in e, f and g were conducted as in Fig. 2c, d and e, respectively. Data in e–g are representative of three biological replicates.
In addition to the closure of the ligand-binding pocket, a notable structural rearrangement of cGAMP-bound chicken STING, as compared with STING in the apo state, is a 180° rotation of the LBD relative to the transmembrane domain (Fig. 3b, d, Supplementary Video 1). As a result, the connector-LBDα1 elements of the two subunits in the dimer no longer form the right-handed crossover (as observed in the apo structures). Instead, these elements are arranged in a parallel fashion, and the connector loop undergoes a sharp kink to link the connector helix and LBDα1 located on the same side (Fig. 3d). The rotation must be clockwise (as viewed from the top of the LBD towards the membrane), as it is required to unwind the right-handed crossover (Supplementary Video 1). In the apo structures of both human and chicken STING, the two LBDα1 helices in the dimer pack very closely at the N-terminal end, and converge at a conserved glycine residue (Gly158 and Gly163 in human and chicken STING, respectively) in the interface (Fig. 3d). The inter-Cα distance between this pair of glycine residues in the dimer is less than 4 Å, which cannot accommodate any other type of residue with a larger side chain. In the cGAMP-bound STING structure, the direct binding of cGAMP to the middle section of LBDα1 pushes the two LBDα1 helices in the dimer away from each other, which increases the distance between the two glycine residues to 8.6 Å. This movement of LBDα1 would lead to clashes in the connector loop if the crossover conformation was maintained, which indicates that the unwinding of the crossover by the 180° rotation of the LBD is driven by conformational changes induced by cGAMP.
Activating mutations in STING (also known as TMEM173) have been linked to the human autoinflammatory disease, STING-associated vasculopathy with onset in infancy. Several of these mutants—including V147L, N154S and V155M—are located at the connector helix loop25,26. On the basis of our structural analyses, we suggest that these mutants probably promote the 180° rotation of the LBD and thus cause STING activation. Consistent with previous studies25–27, our functional assays confirmed that the V155M mutant of human STING led to cGAMP-independent signalling (Fig. 3e–g). To further test the importance of the 180° rotation in STING activation, we substituted Gly158 in human STING with various residues. The functional assays showed that the G158A mutant is highly active in the absence of cGAMP, and that the G158S and G158V mutants also exhibit cGAMP-independent activity—albeit at lower levels (Fig. 3e–g). These results are consistent with the idea that residues of larger side chains at this position disfavour the tight packing at the connector–LBDα1 junction, and promote the 180° rotation. By contrast, replacing Gly158 with leucine, lysine or glutamate led to diminished signalling (Extended Data Fig. 5), which may be due to STING destabilization caused by the incompatibility of these bulky or polar residues with both the inactive and active states of the protein.
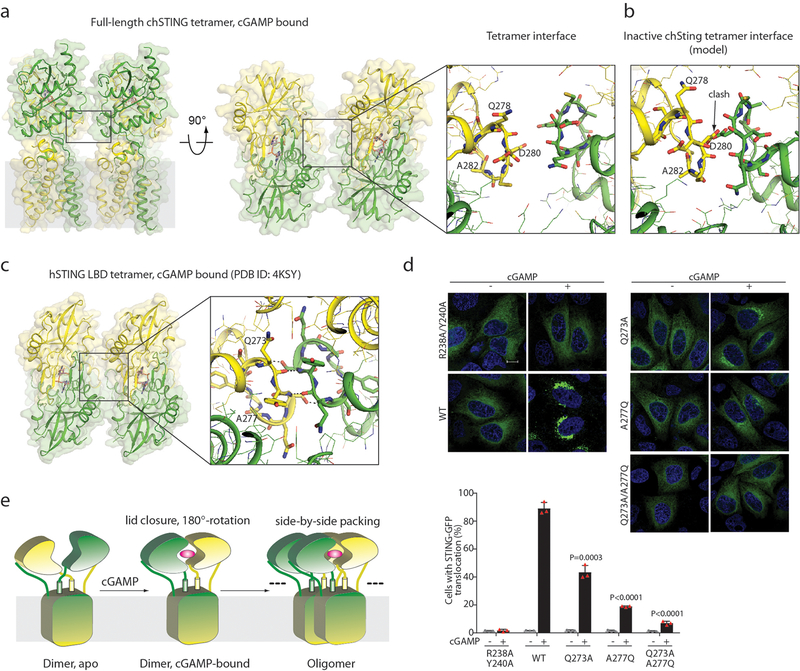
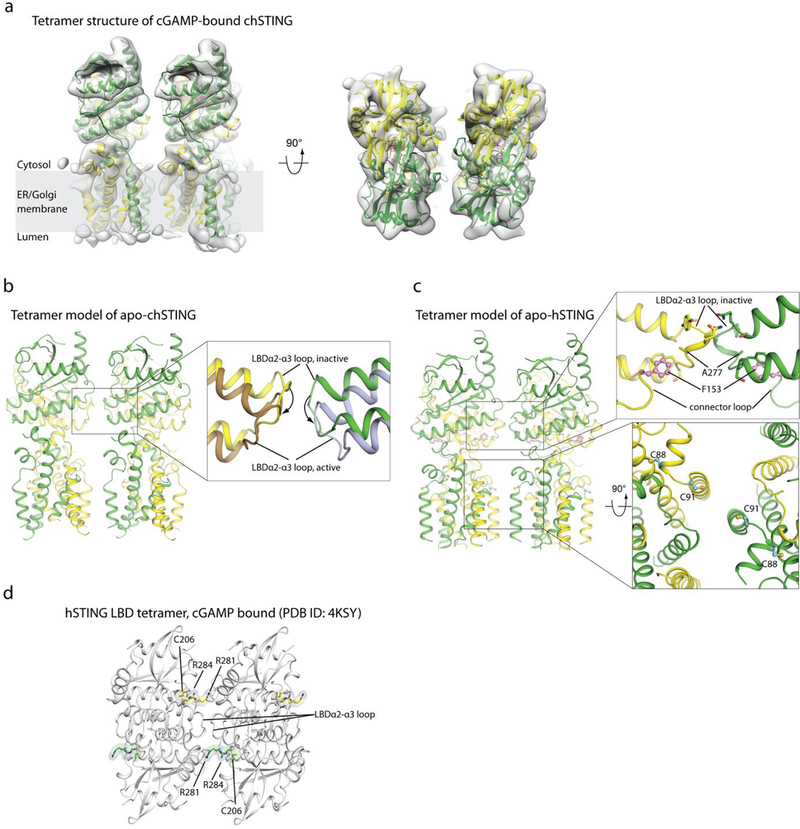
The cryo-EM map of the cGAMP-bound chicken STING tetramer had a resolution of 6.5 Å, which only allowed us to generate a tetramer model by rigid-body docking of two active dimers (Fig. 4, Extended Data Fig. 6). The two dimers interact in an open-ended, side-by-side fashion, which suggests that larger STING oligomers could be formed by a linear array of dimers. Indeed, larger oligomers were seen occasionally in the micrographs (Extended Data Fig. 4a). One of the TM3 helices from the first STING dimer in the tetramer is placed close to its counterpart from the second dimer, suggesting that TM3 may be involved in mediating the tetramerization (Fig. 4). Another structural element in the tetramer interface is the loop (residues 278–285 in chicken STING, which corresponds to residues 273–280 in human STING) that connects LBDα2 and LBDα3 (Fig. 4a). The details of the tetramer interface were, however, not well-resolved. We constructed a tetramer model of chicken STING in the apo state by superimposing the transmembrane region of apo STING onto that of the active tetramer, because the transmembrane region adopts similar conformations in both states (Extended Data Fig. 6b). The two copies of the LBDα2–LBDα3 loop clash with one another in this tetramer model of apo chicken STING (Fig. 4b). The binding of cGAMP induces downward tilting of LBDα2 and LBDα3 (Extended Data Fig. 6b), which resolves the clashes and favours the formation of the tetramer interface. These analyses suggest that the LBDα2–LBDα3 loop has a dual role in both the autoinhibition and activation of STING.
Fig. 4 |. Structure of STING tetramer.
a, Tetramer structure of cGAMP-bound chicken STING in the side and top views. The right panel shows an expanded view of the LBDα2–LBDα3 loop at the tetramer interface. b, Clashes at the LBDα2–LBDα3 loop in the tetramer model of inactive chicken STING, as constructed in Extended Data Fig. 6b. c, Tetramer of the human STING LBD bound to cGAMP. Two symmetry-related human STING LBD dimers in the structure (PDB code 4KSY) form the tetramer, which is similar to the chicken STING tetramer. d, Effects of mutations in the STING tetramer interface on cGAMP-induced formation of STING cellular puncta. Representative confocal images with or without cGAMP stimulation are shown. Scale bar, 10 μm. The experiments and analyses were conducted as in Fig. 2e. Data are mean ± s.d. and representative of three biological replicates. e, Cartoon model of cGAMP-induced conformational changes and oligomerization of STING.
A close inspection of the packing interactions in the crystal structures of the cGAMP-bound human STING LBD in the RCSB Protein Data Bank (PDB) database revealed a side-by-side packing mode that is similar to the mode seen in the chicken STING tetramer; this packing mode is present in all but one of the human STING LBD structures in the cGAMP-bound conformation9,14,28 (Fig. 4c, Extended Data Table 1). Similar packing interactions are also found in cGAMP-bound structures of mouse and anemone STING (PDB codes 4LOJ and 5CFQ, respectively)14,15. By contrast, none of the human STING LBD structures in the inactive conformation contains this type of packing. These observations suggest that this side-by-side packing underlies the oligomerization and activation of human STING. The tetramer interface is mediated largely by the LBDα2–LBDα3 loop (residues 273–280), and contains both backbone-mediated hydrogen bonding and side-chain packing interactions (Fig. 4c). The LBDα2–LBDα3 loop is buttressed by Phe153 in the connector loop, which suggests that there is a coupling between the 180° rotation around the connector loop and the conformational change in the LBDα2–LBDα3 loop (Fig. 3d, Extended Data Fig. 6c). An F153A mutation caused partial constitutive activation of STING (Extended Data Fig. 5d), consistent with the idea that a reduced size of residue at this position increases the flexibility of both the connector and LBDα2–LBDα3 loops, and relieves the autoinhibition on STING oligomerization. A recent study has shown that alterations of Cys206, Arg281 and Arg284—all of which are located near LBDα2–LBDα3 loop (Extended Data Fig. 6d)—in patients with STING-associated vasculopathy with onset in infancy causes cGAMP-independent activation of STING27, suggesting that these mutations relieve the autoinhibition of STING oligomerization. By contrast, single or double mutants for two residues in the tetramer interface (Q273A, A277Q and Q273A/A277Q) abolish cGAMP-induced STING translocation and puncta formation in cells (Fig. 4c, d). In the accompanying paper29, we show that mutations that affect these residues also result in disruption of the oligomerization of STING, as well as the phosphorylation of TBK1 and STING, in response to cGAMP stimulation. These observations together support a model in which cGAMP induces STING activation by promoting the side-by-side oligomerization (Fig. 4e).
Our structural analyses of full-length STING in three different states provide insights into how cGAMP triggers STING oligomerization and activation. The unanticipated cGAMP-induced 180° rotation of the LBD relative to the transmembrane highlights the important regulatory role of the transmembrane–cytoplasmic connector, which explains the gain-of-function that occurs with some disease-causing mutations of STING. The oligomerization of STING is probably enhanced in cells in which it can reach higher local concentrations and is confined to two-dimensional membranes. Recent studies have shown that palmitoylation of two cysteine residues, Cys88 and Cys91, in human STING is important for its activation30,31. Based on our structures, Cys88 is buried inside the TM2–TM3 linker whereas Cys91 is exposed and therefore more likely to be palmitoylated (Extended Data Fig. 6c). Cys91 is located close to TM3, which suggests that its palmitoylation may enhance STING activation by contributing to the tetramer interface. STING signalling requires phosphorylation of its C-terminal tail by TBK1, which occurs only after STING is activated22,24. The C-terminal tail is invisible in all of the available structures of STING. The accompanying paper29 on the STING–TBK1 complex reveals the details of the interactions between the STING C-terminal tail and TBK1, and provides a mechanism by which the oligomerization of STING regulates TBK1 activation and phosphorylation of the STING C-terminal tail.
METHODS
No statistical methods were used to predetermine sample size. The experiments were not randomized and investigators were not blinded to allocation during experiments and outcome assessment.
Protein expression and purification
The coding sequences for human STING residues 1–379 or 1–343 fused to a cleavage site for the human rhinovirus 3C protease and T6SS secreted immunity protein 3 (Tsi3) from Pseudomonas aeruginosa at the C terminus in tandem were inserted into the pEZT-BM vector32. The vectors were used to generate recombinant baculovirus for proteins expression in HEK293 GnTI− cells (ATCC) with the BacMam system33. These cells and other cells used in the study were assumed to be authentic as from the commercial sources and therefore not independently authenticated. Cells were routinely checked to ensure no mycoplasma contamination by using methods such as DAPI staining and the e-Myco Mycoplasma PCR Detection Kit (Bulldog Bio). Cells were grown in FreeStyle medium (Thermo Fisher Scientific), supplemented with 5 mM sodium butyrate 12 h after transduction. Cells were collected after a 60-h culture. Cells re-suspended in buffer A (20 mM Tris pH8.0, 150 mM NaCl and 1 mM PMSF) were disrupted by French press and centrifuged for 10 min at 5,000g to remove debris. Membrane pellet was obtained by centrifugation at 100,000g for 1 h. Membrane proteins were extracted by 1% n-dodecyl-β-d-Maltoside (DDM) and 0.2% cholesteryl hemisuccinate tris salt (CHS) in buffer A. Insoluble fraction was removed by another round of centrifugation. The first purification step was based on the high-affinity interaction between Tsi3 and the T6SS effector protein Tse334. Detergent-solubilized STING was captured by Tse3-conjugated Sepharose 4B resin (GE Healthcare) equilibrated in buffer B (20 mM Tris pH8.0, 150 mM NaCl, 1mM CaCl2, 0.02% DDM and 0.004% CHS). After extensive washing with buffer B to remove unbound proteins, human STING was cleaved from the Tsi3 affinity tag by treatment with the 3C protease on resin at 4 °C for 8 h. The eluted protein was further purified on a Superdex S200 increase 10/300 column (GE healthcare) in buffer C (20 mM Tris pH8.0, 150 mM NaCl, 0.02% DDM, 0.004% CHS and 1 mM DTT). Peak fractions were collected, concentrated and kept at −80 °C before use.
The coding region of chicken STING with a C-terminal Flag-tag was cloned into CSII-EF-DEST-IRES-blasticidin lentivirus vector, which was used to infect HEK293 GnTI− cells grown on 10 × 20-mm dishes. Cells were further grown on 145 × 20-mm dishes with 8 μg/ml blasticidin for 10 days to select for cells stably expressing chicken STING–Flag. Selected cells were cultured in FreeStyle 293 medium with 3% fetal calf serum at 37 °C under 8% CO2 in an incubator shaker (Eppendorf, 130 r.p.m). Cells were collected when density reached 2 × 106–3 × 106 cells/ml, and resuspended in buffer A. The same procedures as described above were used for cell disruption and membrane preparation. Membrane proteins were solubilized in buffer A supplemented with 1.5% DDM/CHS (10:1, Anatrace). The protein was purified with anti-Flag M2 affinity resin (Sigma) in a buffer containing 25 mM Tris pH 7.5, 300 mM NaCl and 0.1% DDM/CHS (10:1). The chicken STING protein was eluted with an elution buffer containing 25mM Tris pH 7.5, 200 mM NaCl, 0.1% DDM/CHS (10:1) and 250 μg/ml Flag peptide (Sigma). The protein was further purified through a Superdex 200 Increase 10/300 column. Peak fractions were pooled, concentrated and kept at −80 °C before use.
Cryo-EM data collection
Purified human and chicken STING of either the apo or cGAMP-bound form at 4.5 mg/ml was applied to a glow-discharged Quantifoil R1.2/1.3 300-mesh gold holey carbon grid (Quantifoil, Micro Tools), blotted under 100% humidity at 4 °C and plunged into liquid ethane using a Mark IV Vitrobot (FEI). Micrographs were acquired on a Titan Krios microscope (FEI) with a K2 Summit direct electron detector (Gatan) in the super-resolution counting mode, operated at 300 kV using the EPU software (FEI). The slit width of the GIF-Quantum energy filter was set to 20 eV. A Volta phase plate was used to enhance low-resolution features20. Micrographs were dose-fractioned into 20 frames with a total exposure time of 10 s at the dose rate of 2 e− per pixel per second.
Image processing and 3D reconstruction
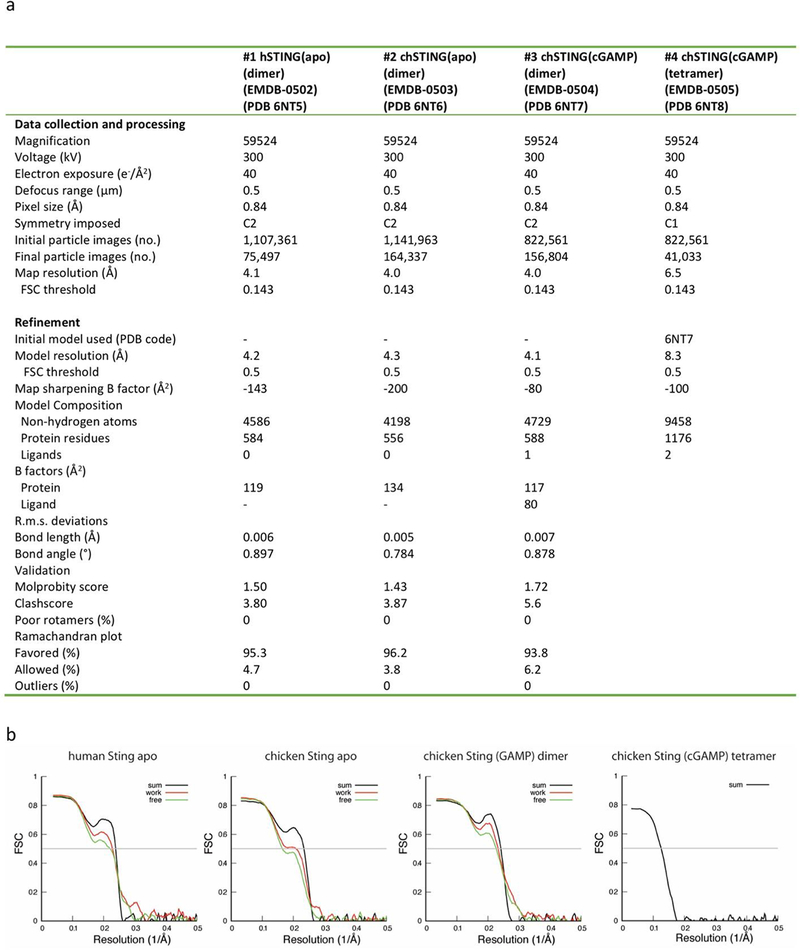
Movie frames of chicken STING micrographs were motion-corrected and binned twofold, resulting in the pixel size of 0.84 Å, and dose-weighted using the Motioncorr2 program35 (Extended Data Fig. 1). CTF corrections were performed using the GCTF program36. The rest of the image processing steps were carried out using Relion 2.037. A few micrographs from the apo STING dataset were used for manual picking of ~1,000 particles. These particles were subjected to 2D classification. Class averages representing projections of the STING dimer in different orientations were used as templates for automated particle picking from the full datasets. A total of 1,141,963 particles were picked from 3,617 micrographs. Particles were extracted and binned 3 times (leading to 2.52 Å per pixel), and subjected to 2D classification. Particles in good 2D classes were chosen (283,027 in total) for 3D classification using an initial model generated from a subset of the particles. Three of the 3D classes showing clear secondary structural features were selected. A total of 164,337 particles from these 3D classes was re-extracted to the original pixel size of 0.84 Å. Using these particles, 3D refinement with C2 symmetry imposed resulted in a reconstruction at 4.4 Å resolution, which allowed initial model building. To further improve the resolution, we performed a refinement with density subtraction of the detergent belt38, which yielded a map at 4.0 Å resolution.
Templates from the apo STING data were used for particle picking from 3,776 micrographs of cGAMP-bound chicken STING (Extended Data Fig. 4). A total of 822,561 particles, containing both the active dimer and tetramer, were picked. Subsequent 2D and 3D classification steps separated particles of the dimer (156,804) and tetramer (41,033). A map of the apo STING dimer was low-pass-filtered and used as the reference for the cGAMP-bound dimer. The subsequent 3D refinement with C2 symmetry imposed and focused refinement with density subtraction were the same as those used for the apo STING dimer, which led to a final reconstruction at 3.96 Å resolution. It is worth mentioning that 3D refinement with C1 symmetry results in a map very similar to the C2 map, which suggests that the binding of asymmetrical cGAMP does not break the C2 symmetry of STING. The reconstruction for the tetramer was performed using a similar procedure. The initial reference map of the cGAMP-bound tetramer was generated from a subset of particles. The two dimers in the tetramer are related by approximate two-fold symmetry, with the rotation between them being 165° instead of 180°. Refinement with the detergent belt subtracted and without symmetry imposed led to a reconstruction at a resolution of 6.5 Å.
The same image processing procedure was used to obtain the 3D reconstruction of human STING in the apo state (Extended Data Fig. 2), which yielded a map at a resolution of 4.1 Å. Local resolution was calculated in Relion. Resolution was estimated by applying a soft mask around the protein density with the Fourier shell correlation (FSC) 0.143 criterion.
Model building, refinement and validation
Density maps of the STING dimers were of sufficient quality for de novo model building in Coot39 (Extended Data Fig. 7), facilitated by previous crystal structures of the STING LBD, sequence alignment of STING from different species and secondary structure predictions. The density of the transmembrane region is relatively poor. Model building of this region was initiated by determining the identities of the transmembrane helices. TM2 was readily recognized because it is much longer than the other three transmembrane helices on the basis of secondary structure predictions. The identity of TM1 was determined by its connection with the well-ordered N-terminal segment. Similarly, TM3 and TM4 were determined by their well-resolved connections with TM2 and the connector helix, respectively. The TM1–TM2 and TM3–TM4 loops are disordered, which results in ambiguity in the connections among these helices in the dimeric structure. The disordered TM1–TM2 loop in chicken STING contains 5 residues, which can bridge the gap of ~17 Å in the current domain-swapped model. It cannot do so in the alternative model, in which the gap is ~33 Å. The density of TM3 is poor, and therefore not fully modelled. On the basis of both our structures and secondary structure predictions, the TM3–TM4 loop in both chicken and human STING is ~10 residues in length. In the current model, the C-terminal end of TM3 is in close proximity to the N terminus of TM4. By contrast, the linear distance between them is ~30 Å in the alternative connection. For the soluble TM3–TM4 loop to bridge the gap, it must loop above the lipid bilayer, which would further increase the distance. The alternative connection is therefore highly improbable. In addition, in one isoform of mouse STING (Gene Bank number AMD16372), the TM3–TM4 loop contains only three residues, which would make it impossible for the loop to assume the alternative connection. The assignment of the sequence register in the transmembrane region was facilitated by conserved polar residues in the transmembrane helices. For example, human STING Arg45 (which corresponds to chicken STING Arg51) and human STING Glu68 and Glu69 (which correspond to chicken STING Glu74 and Glu75) at the N and C termini, respectively, of TM2 served as anchor residues. These residues restrained the modelling of the entire TM2 helix, because any register shift in the helix would either lead to burying these polar residues in the membrane or lead to clashes with other residues in the protein. Similarly, human STING Gln128 (which corresponds to chicken STING Gln133) helped with the modelling of TM4. In addition, tight packing among the transmembrane helices helped to position the human STING Gly57 (which corresponds to chicken STING Gly63) in TM2, as a register shift would place a larger residue at this position and lead to clashes.
Real space refinement of the models was carried out using Phenix, with secondary structure restraints and non-crystallographic symmetry restraints40. No de novo model building was performed for the chicken STING tetramer, owing to the low resolution of the map. Two copies of cGAMP-bound chicken STING dimers were rigid-body-docked into the density to generate the tetramer model. Model geometries were assessed with MolProbity as a part of the Phenix validation tools41 (Extended Data Fig. 8a). To test for overfitting, the coordinates were randomly shifted by 0.2 Å using Phenix. The displaced models were refined in Phenix against one of the half maps calculated from half of the dataset in Relion (half-map 1). FSC curves were calculated for the models in relation to half-map 1 and compared with that for the summed map. The good agreement between the two curves indicates no overfit (Extended Data Fig. 8b). Figures were rendered in PyMOL (The PyMOL Molecular Graphics System, v.2.0, Schrödinger), Coot or Chimera39,42.
Constructs for cell-based assays
For transient transfection, Flag-tagged STING constructs were cloned into the p-CMV-Flag-N2 vector. For the generation of stable cell lines, STING–Flag and its mutants were cloned into the gateway cloning vector CSII-EF-DEST-IRES-blasticidin, which is a derivative of a set of lentiviral expression vectors. STING–Flag and mutants were also cloned into pCDH-CMV-MCS-IRES-Puro vectors to generate lentivirus expression constructs. GFP-tagged STING constructs (STING–GFP) were cloned into p-CMV-GFP-N2 and CSII-EF-DEST-IRES-hygromycin or blasticidin vectors.
Antibodies
Rabbit antibodies against STING, pSTING (S366), pTBK1 (S172), pIRF3 (S396) and GAPDH were purchased from Cell Signaling Technology. Anti-Flag antibody (M2), anti-tubulin antibody and M2 affinity gel were purchased from Sigma.
Cell culture, transfection and reagent treatment
HEK293T and HeLa cells were from ATCC. HeLa cells deficient in cGAS were generated by CRISPR–Cas9. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (Thermo Fisher Scientific), penicillin and streptomycin (100 IU/ml and 100 μg/ml, respectively, Gibco by Life Technologies) and 1× nonessential amino acids (Gibco by Life Technologies). Endogenous STING expression is undetectable in these cells. Cell lines stably expressing various forms of STING through lentivirus transduction were selected with 8 μg/ml blasticidin (Thermo Fisher Scientific), 100 μg/ml hygromycin (Alexis Biochemicals) or 2 μg/ml puromycin (Thermo Fisher Scientific), according to the corresponding selection markers of the constructs.
Luciferase reporter assay
HEK293T cells stably expressing the gene for the interferon-β 1–luciferase–puromycin reporter were transfected with the human STING wild type, or mutant constructs. Cells were stimulated with indicated concentrations of cGAMP in the presence of 10 μg/ml digitonin (Sigma) 10 h after transfection. After an additional 14-h incubation in medium, luciferase activity was determined following the standard protocol (Promega, E1501).
Western blot analyses of phosphorylation of STING, TBK1 and IRF3
HEK293T cells were transfected with the human STING wild type or mutant constructs, and stimulated with cGAMP. Phosphorylation of STING, TBK1 and IRF3 was detected by immunoblotting. Alternatively, STING-deficient HeLa cells were infected with lentiviruses containing the human STING constructs in the pCDH-CMV-MCS-IRES-Puro vector and the cells stably expressing the human STING wild type or mutants were selected with puromycin. The cells were stimulated with cGAMP and analysed as above.
Immunofluorescence microscopy
HeLa cells deficient in cGAS were cultured on cover glasses or four-well glass-bottom chambers (Laboratory-Tek). Cells were transfected with human STING expression vectors in the p-CMV-GFP-N2 vector. Twenty-four hours after transfection, cells were stimulated with cGAMP (1 μM) for 1 h. Cells were then fixed with 4% paraformaldehyde (Electron Microscope Sciences) for 15 min. Confocal images were acquired on a Nikon A1R Confocal microscope using a 60× (NA 1.45) objective.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The cryo-EM maps generated in this study have been deposited in the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-0502 (human STING, apo dimer), EMD-0503 (chicken STING, apo dimer), EMD-0504 (chicken STING, cGAMP-bound dimer) and EMD-0505 (chicken STING, cGAMP-bound tetramer). The atomic coordinates have been deposited in the PDB under the accession numbers 6NT5 (human STING, apo dimer), 6NT6 (chicken STING, apo dimer), 6NT7 (chicken STING, cGAMP bound dimer) and 6NT8 (chicken STING, cGAMP bound tetramer).
Extended Data
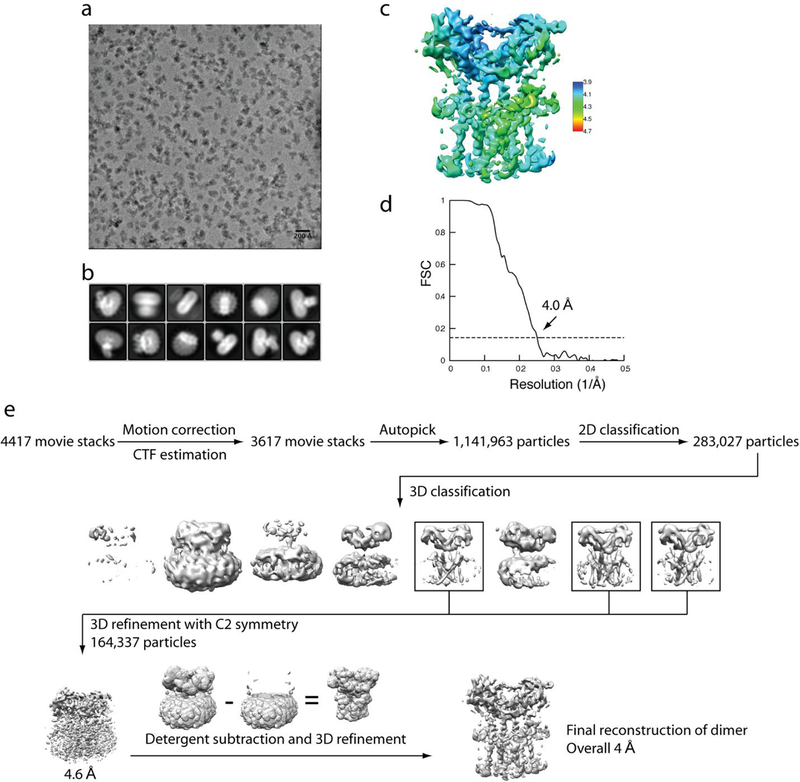
Extended Data Fig. 1 |. Flow chart of cryo-EM image processing for chicken STING in the apo state.
a, Representative micrograph. b, Representative 2D classes. c, Final reconstruction with colours based on local resolution. d, Gold-standard FSC curve of the final 3D reconstruction. e, Image processing procedure.
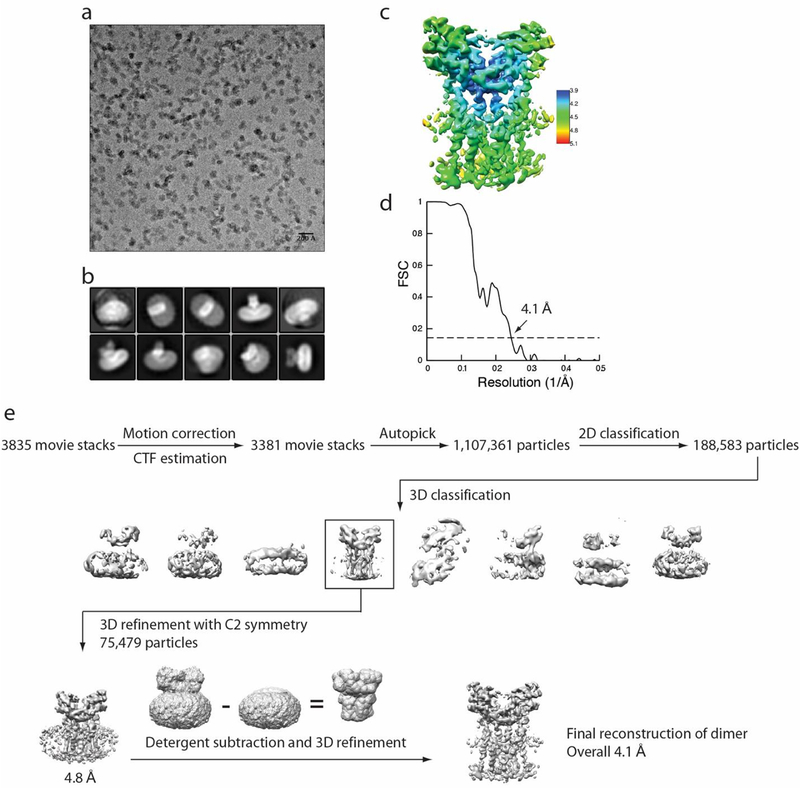
Extended Data Fig. 2 |. Flow chart of cryo-EM image processing for human STING in the apo state.
a, Representative micrograph. b, Representative 2D classes. c, Final reconstruction with colours based on local resolution. d, Gold-standard FSC curve of the final 3D reconstruction. e, Image processing procedure.
Extended Data Fig. 3 |. Structure of full-length chicken STING in the apo state.
a, Side view of the cryo-EM 3D reconstruction. The two subunits in the dimer are coloured in yellow and green. b, Cartoon representation of the structure in two orthogonal side views. c, Cartoon representation of the transmembrane domain dimer in the top view, from the cytosolic side. d, Interactions between the N-terminal segment and the body of chicken STING. e, Sequence alignment of STING from human, mouse and chicken (denoted by h-, m- and ch- prefixes, respectively). Secondary structure assignments are based on the structures. Residue numbers of human and chicken STING are shown above and below the aligned sequences, respectively.
Extended Data Fig. 4 |. Flow chart of cryo-EM image processing for cGAMP-bound chicken STING.
a, Representative micrograph. b, Representative 2D classes of the dimer (two top panels) and tetramer (bottom panel). c, e, Final reconstructions of the dimer and tetramer, respectively, coloured on the basis of local resolution. d, f, Gold-standard FSC curves of the final 3D reconstructions of the dimer and tetramer. g, Image processing procedure.
Extended Data Fig. 5 |. Additional mutational analyses of the transmembrane–LBD connector of human STING.
a, c, Effects of mutations in the connector and LBDα1 on cGAMP-stimulated IFNβ expression. Data are mean ± s.d. and representative of three biological replicates. b, d, Effects of STING mutations on phosphorylation of STING, TBK1 and IRF3. The result is representative of two independent experiments. The analyses for gene expression in a, c and phosphorylation in b, d were carried out in the same manner as in Fig. 2c and 2d, respectively.
Extended Data Fig. 6 |. Tetramer of full-length STING.
a, Rigid docking of the atomic model of the cGAMP-bound full-length chicken STING dimer (green and yellow) to the 3D reconstruction of the tetramer (grey). b, Left, tetrameric model of chicken STING in the apo state. The model is generated by superposition of two chicken STING inactive dimers on the active tetramer, on the basis of the transmembrane domain. Right, expanded view of the LBDα2–LBDα3 loop at the tetramer interface. The same loop in the active tetramer structure is shown for comparison; arrows indicate conformational differences between the two states. c, Model of full-length human STING tetramer. The model is constructed by superimposing two inactive human STING dimers on the active chicken STING tetramer, on the basis of the transmembrane domain. Right, top panel, packing between Phe153 in the connector loop and the LBDα2–LBDα3 loop. Right, bottom panel, Cys88 and Cys91—which have been shown to be palmitoylated—are highlighted. d, Cys206, Arg281 and Arg284—alterations of which cause constitutive activation of human STING—are located near the LBDα2–LBDα3 loop that forms the tetramer interface. These residues are highlighted in the tetramer model of the human STING LBD bound to cGAMP (based on PDB code 4KSY, as shown in Fig. 4c).
Extended Data Fig. 7 |. Sample density maps.
a–c, Sample density maps for various parts of chicken (a) and human (b) STING in the apo state, and cGAMP-bound chicken STING (c).
Extended Data Fig. 8 |. Data collection and model statistics.
a, Data collection and model refinement statistics. b, FSC curves between the maps and models.
Extended Data Table 1 |.
Crystal-packing analyses of crystal structures of the human STING LBD in the ligand-bound, lid-closed conformation
| PDB ID | Ligand | Side-by-side packing | Lid | Space group | Crystallization condition |
|---|---|---|---|---|---|
| 4F5D | c-di-GMP | Yes | closed | P41 | 0.025 M MgSO4, 0.0 5M Tris-HCl pH 8.5, 1.8M (NH4)2SO4 |
| 4KSY | 2’,3’-cGAMP | Yes | closed | C2 | 27% PEG 3,350, 0.1 M Bis-Tris, pH 6.8, 0.2 M NaCl |
| 5BQX | 3’,2’-cGAMP | Yes | closed | P41212 | 0.2 M ammonium acetate, 0.1 M trisodium citrate, pH 5.8, 25% PEG4000 |
| 4LOH | 2’,3’-cGAMP | Yes | Closed | P1 | 0.01 M NiCl2, 0.1 M Tris, 20% PEG2000, pH 8.5 |
| 4LOI | 2’,2’-cGAMP | Yes | Closed | P1 | 1.6 M NaH2PO4, 0.4 M Na2HPO4, 0.1 M phosphate-citrate, pH 4.2 |
| 4QXP | DMXAA | Yes | Closed | P21 | 0.2 M Li2SO4, 20% PEG 3350, 0.015 mM CYMAL-7, 0.1 M Tris, pH 8.1 |
| 4QXQ | DMXAA | Yes | Closed | P6 | 0.2 M Ca(Ac)2, 15% PEG3000, 0.01 M L-Proline, 0.1 M NaAc, pH 5.1 |
| 4QXR | DMXAA | Yes | Closed | P6 | 0.2 M Ca(Ac)2, 15% PEG3000, 0.1 M LiCl, 0.1 M NaAc, pH 5.1 |
| 4QXO | DMXAA | No | Closed | P6122 | 2 M LiCl, 10% PEG6000, 0.01 M CaCl2, 0.1 M Tris, pH 7.8 |
All of these structures (except one) contain the side-by-side packing interaction that mediates the formation of the tetramer and higher-order oligomers.
Supplementary Material
Acknowledgements
Cryo-EM data were collected at the University of Texas Southwestern Medical Center (UTSW) Cryo-Electron Microscopy Facility, which is funded by the Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Support Award RP170644. We thank D. Nicastro for facility access and data acquisition. This work is supported in part by the Howard Hughes Medical Institute (Z.J.C.), grants from the National Institutes of Health (GM088197 and R35GM130289 to X.Z.), grants from the Welch foundation (I-1389 to Z.J.C.; I-1702 to X.Z.; I-1944 to X.-c.B.), grants from CPRIT (RP150498 to Z.J.C.; RP160082 to X.-c.B.). X.-c.B. and X.Z. are Virginia Murchison Linthicum Scholars in Medical Research at UTSW. Z.J.C. is an investigator of Howard Hughes Medical Institute.
Footnotes
Competing interests: The authors declare no competing interests.
Reviewer information Nature thanks Andrea Ablasser, Philip Kranzusch and Osamu Nureki for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online content Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at
References
- 1.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J et al. Cyclic GMP–AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai X, Chiu YH & Chen ZJ The cGAS–cGAMP–STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 54, 289–296 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa H & Barber GN STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong B et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Burdette DL et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin L et al. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol 28, 5014–5026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun W et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl Acad. Sci. USA 106, 8653–8658 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X et al. Cyclic GMP–AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang G et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol 19, 725–727 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Huang YH, Liu XY, Du XX, Jiang ZF & Su XD The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol 19, 728–730 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Ouyang S et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu C, Yi G, Watts T, Kao CC & Li P Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol 19, 722–724 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao P et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kranzusch PJ et al. Ancient origin of cGAS–STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H et al. Rat and human STINGs profile similarly towards anticancer/antiviral compounds. Sci. Rep 5, 18035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q, Sun L & Chen ZJ Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol 17, 1142–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Li T & Chen ZJ The cGAS–cGAMP–STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med 215, 1287–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai XC, McMullan G & Scheres SH How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci 40, 49–57 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Danev R & Baumeister W Cryo-EM single particle analysis with the Volta phase plate. eLife 5, e13046 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Q et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Li Z et al. PPM1A regulates antiviral signaling by antagonizing TBK1-mediated STING phosphorylation and aggregation. PLoS Pathog 11, e1004783 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka Y & Chen ZJ STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal 5, ra20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med 371, 507–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeremiah N et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest 124, 5516–5520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melki I et al. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J. Allergy Clin. Immunol 140, 543–552.e545 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Shi H, Wu J, Chen ZJ & Chen C Molecular basis for the specific recognition of the metazoan cyclic GMP–AMP by the innate immune adaptor protein STING. Proc. Natl Acad. Sci. USA 112, 8947–8952 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukai K et al. Activation of STING requires palmitoylation at the Golgi. Nat. Commun 7, 11932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haag SM et al. Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Morales-Perez CL, Noviello CM & Hibbs RE Manipulation of subunit stoichiometry in heteromeric membrane proteins. Structure 24, 797–805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dukkipati A, Park HH, Waghray D, Fischer S & Garcia KC BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr. Purif 62, 160–170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu D et al. Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism. Mol. Microbiol 92, 1092–1112 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang K Gctf: real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai XC, Rajendra E, Yang G, Shi Y & Scheres SH Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps generated in this study have been deposited in the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-0502 (human STING, apo dimer), EMD-0503 (chicken STING, apo dimer), EMD-0504 (chicken STING, cGAMP-bound dimer) and EMD-0505 (chicken STING, cGAMP-bound tetramer). The atomic coordinates have been deposited in the PDB under the accession numbers 6NT5 (human STING, apo dimer), 6NT6 (chicken STING, apo dimer), 6NT7 (chicken STING, cGAMP bound dimer) and 6NT8 (chicken STING, cGAMP bound tetramer).