Abstract
G-quadruplex (G4) is a non-canonical nucleic acid structure that arises from the stacking of planar G-tetrads, stabilized by monovalent cations. G4 forming sequences exist throughout the genome and G4 structures are shown to be involved in many processes including DNA replication and gene expression. The single molecule total internal reflection fluorescence (TIRF) microscopy has been employed to study G4 structure formation and protein binding interactions. Here, we describe methods by which we tested the folding and unfolding of G-quadruplexes structure and studied the dynamics of its interaction with POT1 protein. The methods presented here can be applied to study other putative G4 sequences and potential binding partners.
Keywords: G-quadruplex, G4, Single-molecule, FRET, telomere, POT1
1. Introduction
G-quadruplex (G4) is a non-canonical nucleic acid structure that can form from a guanine-rich single strand. The core structure of G4 is comprised of stacks of guanine(G)-tetrad planes formed by Hoogsteen hydrogen bonds, stabilized by monovalent cations, especially potassium (1). Putative G4 forming sequences are unevenly distributed throughout the human genome (2, 3). G4 structures have been reported to be involved in DNA replication, gene regulation, genome instability and human diseases (4–7). One well-characterized G-quadruplex is the human telomeric 3’ overhang of 50–200 nucleotide region containing a repeat sequence (TTAGGG)n. Four repeats of this sequence can fold into a G-quadruplex structure in the presence of monovalent cations, such as sodium or potassium (8, 9). Additional repeat sequences can associate with the G4, but extra repeats can destabilize G4 in terms of thermostability and enthalpy (10). The G4 structure can also influence the accessibility of the telomeric DNA to proteins such as telomerase and helicase (11). However, determining how the telomeric overhang affects G-quadruplex formation and the accessibility of protein binding is difficult to monitor by traditional biochemical methods, especially when the process is dynamic. Single molecule methods can offer an advantage of providing structural dynamics at the molecular level.
Total internal reflection fluorescence (TIRF) microscopy is a popular single molecule detection method which yields reduced background noise and enables collecting data from hundreds of molecules in one measurement (12, 13). In our system, DNA samples are labeled with two dyes, Cy3 and Cy5, in the FRET-sensitive distance. FRET is a distance-dependent energy transfer process used to monitor the interaction between the two dyes, donor and acceptor which report on structural dynamics of labeled molecules. For example, when two dyes are labeled on one molecule, such as in two positions within the same DNA, the FRET change induced by protein binding would report on how the protein changes the conformation of the DNA strand within the labeled position.
We have previously reported single molecule fluorescence study on the G4 folding of telomeric overhang and the interaction with POT1 binding (14, 15). POT1 is a component of the telomere binding protein complex termed shelterin, which specifically associates with ten nucleotide of telomeric repeat sequence (TTAGGGTTAG). POT1 protects telomeric overhang by preventing DNA repair machinery from associating with non-damaged telomere DNA (16). The binding of POT1 on telomeric G4 can unfold the structure. Such binding and unfolding of the target G4 can be probed and quantified by smFRET. Here, we describe detailed protocols of smFRET studies on telomeric G-quadruplexes and demonstrate how to interpret protein-DNA interactions observed with smFRET.
2. Materials
2.1. Total internal reflection fluorescence (TIRF) microscope
2.2. DNA constructs
GQ strand (G2): 5’-TGGCGACGGCAGCGAGGCTTAGGGTTAGGG/3’Cy3/
GQ strand (G3): 5’-TGGCGACGGCAGCGAGGCTTAGGGTTAGGGTTAGGG/3’Cy3/
GQ strand (G4): 5’-TGGCGACGGCAGCGAGGCTTAGGGTTAGGGTTAGGGTTAGGG/3’Cy3/
GQ strand (G5): 5’-TGGCGACGGCAGCGAGGCTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG/3’Cy3/
GQ strand (G6): 5’-TGGCGACGGCAGCGAGGCTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG/3’Cy3/
GQ strand (G7): 5’-TGGCGACGGCAGCGAGGCTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG/3’Cy3/
Biotin primer: /5’Cy5/GCCTCGCTGCCGTCGCCA/3’Biotin/
Annealing buffer: 20 mM Tris-HCl pH 7.5, 50 mM NaCl
Heating block
2.3. Sample chamber preparation
Rectangular cover slips (24 × 40 mm No. 1½).
Quartz microscope slides, 1″ × 3″, 1 mm thick.
Epoxy, 5 min epoxy (Devcon).
Double-sided tape.
Neutravidin, ImmunoPure NeutrAvidin protein.
2.4. Imaging buffer
glucose oxidase
glucose
6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic (Trolox).
catalase
KCl
2.5. Flowing system
Tubing, WEICO ETT-28 (inner diameter = 0.015 ″, outer wall = 0.016 ″)
Needle: BD (26 gauge, 3/8 ″)
1-mL syringe
10-uL, 20–200-uL pipette tips
2.6. Protein
POT1 protein. The purification protocol was previously published in (17).
3. Methods
3.1. DNA annealing
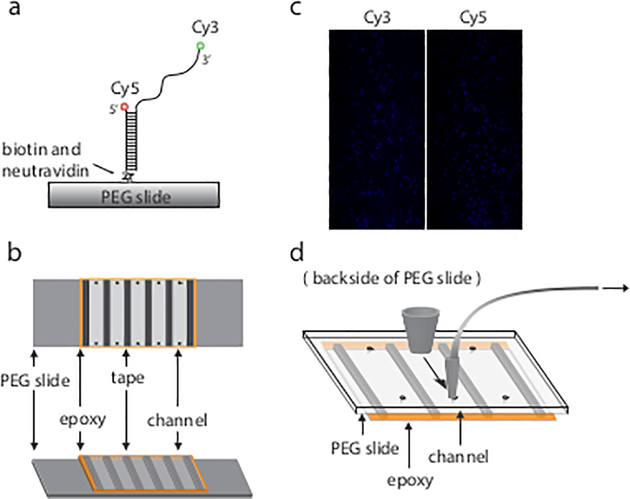
Single molecule FRET constructs were prepared by annealing two single strands which were labeled with either Cy3 or Cy5 dye. One end of the Cy5-labeled strand was also biotinylated to immobilize DNA on the slide surface. (Fig. 1a)
Fig. 1.
Examples of DNA construct slide and image. (a) Single molecule FRET construct are immobilized on PEG slide through biotin and neutravidin interaction. (b) Assembled channels. Each channel is separated by double-side tape and sealed by epoxy. (c) Image captured by camera. Each dot indicates one molecule. (d) Additional pipette tips are used to build a flowing system.
DNA sequences were purchased from a vendor and labeled with either Cy3 or Cy5 dyes. The DNA oligos were dissolved with T50 (50 mM Tris-HCl pH 7.5) to a final concentration of 100 μM. The stock DNA samples were stored at −20°C. (see Note 1)
The GQ strands were annealed with the biotinylated, Cy5-containing primer at a molar ratio of 1:1.5 in annealing buffer. (see Note 2)
The mixtures were incubated in a pre-heated 95°C heat block for 5 min and slowly cooled to room temperature. (see Note 3 and 4)
The annealed constructs were stored at −20°C. (see Note 5)
3.2. Slide preparation and sample immobilization
To perform single molecule experiments, the quartz microscope slide and coverslip was cleaned and chemically coated with biotin-PEG to create a passivated surface. This prevents non-specific adsorption of sample to surface (see Note 6). A practical procedure was written in previous literature (14).
The PEG-coated surface of both the slide and coverslip must face each other. The coverslip was taped to the slide by one layer (about 100μm, see Note 7) of double-side tape to create a passivated channel. For multiple channels, each channel was separated by one layer of tape (see Note 8).
Overhanging extra tape was cut to fit the slide, and all borders of the coverslip/slide surface were sealed by epoxy (Fig. 1b).
To immobilize the DNA substrate, the PEG-coated slide was incubated with neutravidin solution (0.05 mg/mL, see Note 9) for 1 min, and then rinsed with 100 μL of T50 buffer. (see Note 10)
DNA samples were diluted to 25 pM and added to the channel. After a 5 minute incubation, the channel was rinsed with 100 μL of T50 (see Note 11).
3.3. Imaging
In general, single molecule FRET measurements can provide real-time traces of FRET efficiency, which informs the dynamics of FRET states. In addition, the trace data can be used to generate a FRET histogram, which indicates the overall FRET distribution under certain equilibrium states. Both smFRET traces and histograms are able to answer different questions. Here is a procedure to test for the formation of the G-quadruplex structure and to obtain smFRET data.
A prism-type TIRF microscope was used in this protocol. A detailed protocol of TIRF setup can be found previously published in (12).
After DNA immobilization and prior to any sample excitation, the image buffer was mixed with the oxygen scavenging system (1 mg/mL glucose oxidase, 0.8% v/v glucose, ~10 mM Trolox, and 0.03 mg/mL catalase) to reduce photobleaching events and added into the channel. (see Note 12)
The donor dye (Cy3) was excited by a 532 nm solid-state laser, and emitted fluorescence (both of Cy3 and Cy5) was collected with an EMCCD camera by using a custom C++ program.
Data was collected over time as a movie. Depending on the purpose of the experiment, the movie can be short (20 to 40 frames in 100 ms time interval, see Note 13) or long (lasting several minutes). Collecting 15 to 20 short movies, each in different imaging fields, was usually required to generate one FRET histogram. (see Note 14)
The recorded movie data was processed by IDL software to identify each individual fluorescent spot and correlate donor and acceptor channels, described in detail in (12) (Fig. 1c).
The output file was sorted by signal spots (molecules) and the intensity was plotted as a function of time to generate a single molecule signal trace.
To generate a histogram, FRET was first calculated with this equation: (see Note 15). The average FRET efficiencies were then calculated from the initial 10 to 20 frames of each short movie. This FRET efficiency data was plotted as a histogram by a homemade Matlab code.
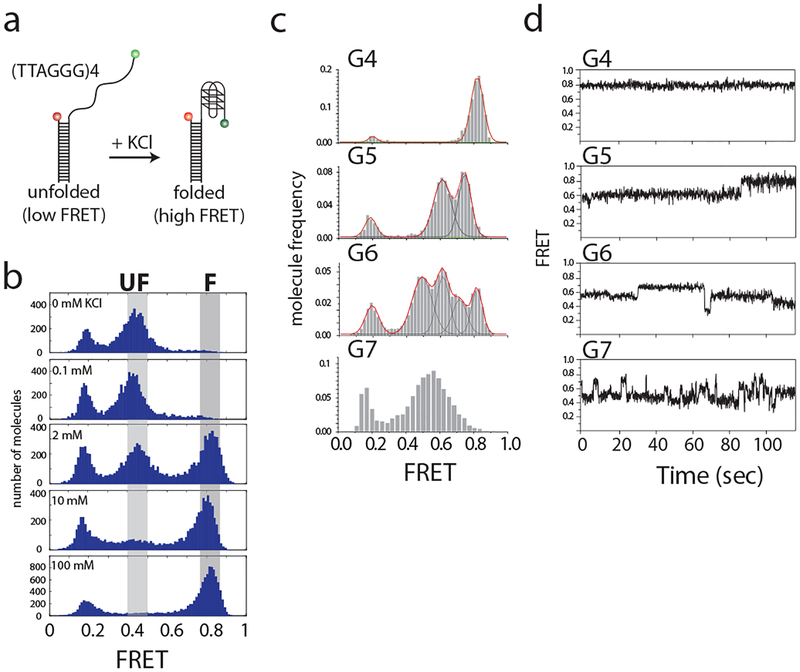
Since G4 is stabilized by a potassium ion, the formation of G4 can be confirmed by titrating increasing potassium concentrations into the system. The image buffer was prepared with 0, 0.1, 2, 10 or 100 mM potassium. FRET histograms were generated for each concentration by repeating steps 1 to 6. (Fig. 2a and 2b)
In order to observe the dynamics of FRET efficiency, a 3-minute long movie was recorded (see Note 16). Traces generated from long movies can be used to confirm if FRET efficiency over time is stable or fluctuating (Fig. 2c and 2d).
FRET histograms of other DNA constructs with longer overhangs (G5, G6, or G7) can also be studied in a fully-folding buffer condition (100 mM KCl) by repeating the above steps.
Fig. 2.
FRET histograms show G-quadruplex folding. (a) Scheme of single molecule FRET constructs. Formation of G-quadruplex can be induced by addition of potassium ions. (b) FRET histograms showing G-quadruplex folding after titration of K+. Increasing K+ concentration enhances the folded population. Three major peaks observed here are at FRET efficiencies of 0.8, 0.5 and 0.2, representing folded, unfolded and donor-only constructs. (c) FRET histograms of constructs with increasing 3’ overhang lengths. Red line indicates overall shape of fitting; black line indicates the Gaussian distribution fit to each peak. (d) Time trace examples of each overhang construct. This figure was reproduced from published work (Helen Hwang, 2014) with permission from Elsevier. (14)
3.4. Protein binding
Unlike when imaging DNA only, introduction of proteins into this DNA system produces an additional challenge: protein binding to DNA may be a pseudo- irreversible process. Therefore, binding events could only be studied directly following the protein being flowed into the channel. Once the reaction reaches an equilibrium state, protein binding can no longer be observed. Therefore, a flow system and real-time imaging was implemented to capture the initial binding event. Here is the procedure to construct a flowing system and use it to test POT1 protein binding to telomeric G-quadruplex sequences.
To adapt the slide for a flowing system, a short segment of a p10 tip and a thin tubule were used to connect one side of the channel to a syringe. The syringe could be controlled manually or by a syringe pump. A wide portion of a p200 tip was adhered to the other side of the channel by epoxy to create a buffer well (Fig. 1d) (see Note 17).
POT1 protein was mixed with image buffer for a final concentration of 100 nM and loaded into the buffer well.
A long movie recording was initiated. Then, the buffer was drawn through by pulling the syringe manually or controlling with a syringe pump (see Note 18 and 19). This way, the movie will record both pre-flow and post-flow states for later comparison.
POT1 protein was incubated for 5–10 min (including the time to complete step 3, ie to record a long movie) to reach an equilibrium state. Then, 15–20 short movies were taken to generate a FRET histogram of equilibrium protein binding. FRET histograms can also be collected at several time points following the addition of protein, and the changes in FRET patterns would then indicate a change in equilibrium state due to protein binding.
This method (steps 3–4) was used to also study other DNA constructs (G2, G3, G5, G6 and G7).
3.5. Data Analysis and Interpretation
All codes are part of a homemade smFRET tool package, which is available to download at the Center for the Physics of Living Cells (http://cplc.illinois.edu/software/, Biophysics Department, University of Illinois at Urbana-Champaign.)
Analysis of the FRET histograms were used to define whether the G quadruplex was folded or unfolded. In our system, KCl titration revealed 2 identifiable states: the unfolded structure has low FRET efficiency, but the donor and acceptor fluorophores come into close proximity upon folding to exhibit a high FRET efficiency (Fig. 2a and 2b).
Since evidence of the overhang length affecting G-quadruplex stability was published previously (10), here we also test 5, 6 and 7 repeats of TTAGGG (G5, G6 and G7) in the presence of 100 mM KCl (Fig. 2c). The FRET histograms show that while G4 has a single sharp peak, other constructs have multiple peaks or a broad peak, suggesting that 3’ overhangs exceeding the length of G4 can result in multiple states. Histograms were fitted by Gaussian distribution with multiple states in order to distinguish FRET states.
The multiple states of FRET efficiency observed in the histograms maybe be due to one of two situations: either more than two FRET species exist at same time or the dynamics of FRET transition is too fast to be resolved in this time interval. To resolve this, long-movie single molecule FRET traces were used to determine FRET states present (Fig. 2d). Analysis of FRET efficiency shows that G4 has flat FRET trace over time, indicating a lack of dynamics within this structure. G5 and G6 show that a FRET transition occurs within one trace and results in multiple FRET peaks. G7 shows that the FRET signal fluctuates in one trace, resulting in a broad FRET histogram (see Note 13).
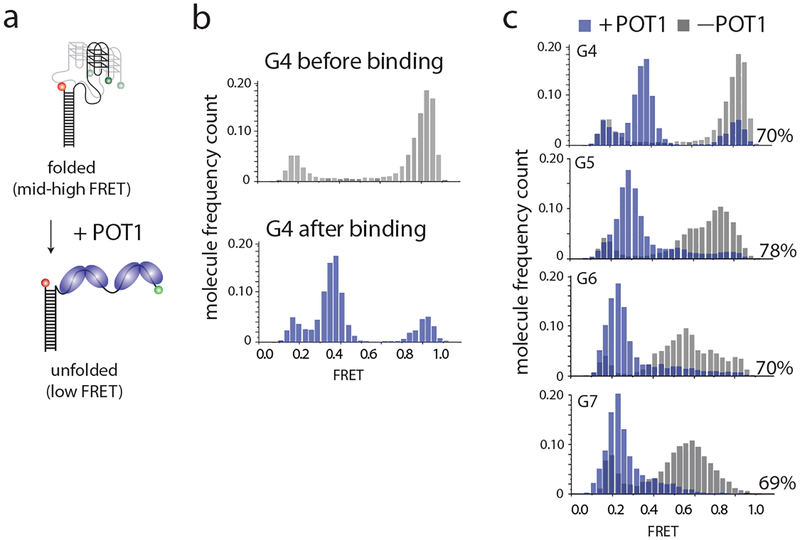
POT1 binding can unfold G4 structures, visualized by a decrease in FRET efficiency. To determine the ratio of POT1 binding, peaks within each histogram were fit by a Gaussian distribution. The bound and unbound states were defined by comparing the post–flow to pre-flow FRET histogram. The fraction in the bound state was determined by calculating the area under the curve. A shift in the peak would indicate a difference in the binding ratio due to an increase in the distance between donor and acceptor fluorophores from protein binding (Fig. 3a and 3b).
Further, the effect of overhang length on POT1 binding can be examined. Figure 3c shows histograms of the FRET efficiency observed with G4, G5, G6 and G7 constructs before and after POT1 binding. After fitting each peak to a Gaussian distribution and determining the area under the bound peak, the binding ratios are 70%, 78%, 70% and 69%, respectively, which are not significantly different among the constructs. It suggests that although overhang length affects G4 folding and stability, POT1 binding is independent of GQ folding and overhang length.
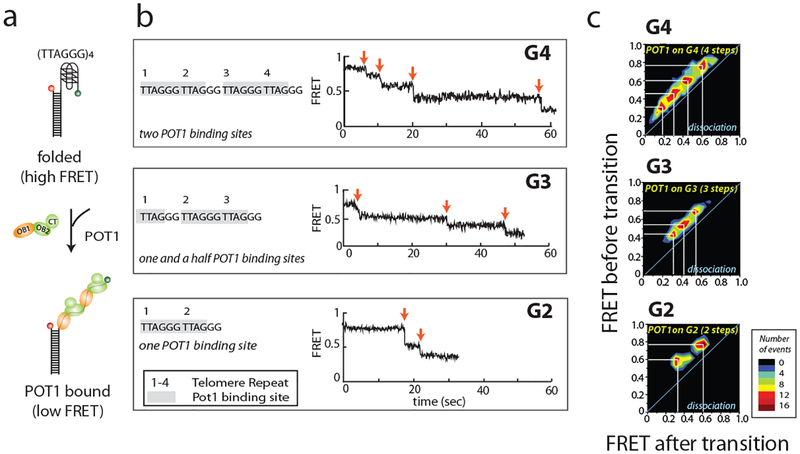
POT1 binding to the DNA sequence TTAGGGTTAG reduces FRET by unfolding the G-quadruplex structure. POT1 contains two subunits OB1 and OB2 which can bind to DNA. (Fig. 4a) To determine the stoichiometric effects of POT1 binding, the number of POT1 binding sites was reduced from two in G4 to 1.5 and one in G3 and G2, respectively. Here, the real-time binding process can be observed in the long movie (Fig. 4b). There are four transitions that occur upon POT1 binding to the G4 construct. Because the G4 sequence contains two POT1 binding sites, and two subunits (OB1 and OB2) bind to each binding site, we interpret these four transitions as four steps of binding. This was further confirmed by testing G2 and G3, because these constructs have 1 and 1.5 POT1 binding sites and the number of transition steps become 2 and 3, respectively.
The transitions of FRET within all the traces can be plotted as transition density plots (TDP) (Fig. 4c). TDP is a 2D accumulative cluster map. The x and y axis refer to the FRET value before and after transition. The colormap indicates the density of the transition events. If the TDP is symmetric along diagonal line, it means the transition is reversible. Here, the TDP contains four clusters from upper-right to lower-left and no symmetric plot, indicating that there are four irreversible transitions from high FRET to low FRET. This suggests POT1 binding to G4 occurs in 4 steps and is irreversible. TDP is built by using the software HaMMy, which is based on hidden Markov modeling (available for download at http://ha.med.jhmi.edu/resources/#1464200861600-0fad9996-bfd4) (16)
Fig. 3.
FRET histograms show POT1 binding. (a) Scheme of POT1 binding on DNA. POT1 binding unfolds the G4 structure and reduces FRET efficiency. (b, c) FRET patterns change after POT1 binding. Grey depicts FRET efficiency distributions prior to POT1 binding, and blue shows the distributions after POT1 binding. The binding ratio, calculated from the area beneath each curve, is displayed to the bottom right of each histogram. This figure was reproduced from published work (Helen Hwang, 2014) with permission from Elsevier. (14)
Fig. 4.
Dissecting POT1 binding site from smFRET time trace. (a) Scheme of POT1 binding. POT1 consists of three domains, OB1, OB2 and CT. Both OB1 and OB2 bind to telomere sequence TTAGGGTTAG. (b) Representative smFRET time trace of each construct. Time zero is the time the protein was flowed into the channel. Each red arrow indicates a FRET transition, which is correlated to the number of binding sites. (c) Transition density plots (TDP) of each construct. The x axis and y axis are the FRET state after and before transition. This figure was reproduced from published work (Helen Hwang, 2012) with permission from Elsevier. (15)
4. Notes
We recommend purchasing DNA with HPLC purification to improve DNA sample purity. If the DNA will be labeled in the lab, depending on the labeling method, a proper buffer system will require for dissolving DNA instead of Tris buffer.
Excess biotin primer is used to maximize the density of fluorescent G4-content DNA. Here, G4-content DNA is labeled with Cy3 dye, which is the source of fluorescence when it is excited by 532 nm laser.
Depending on the experiment, the annealing buffer may be adjusted to contain different salt ions and concentrations. For example, G4 is known to be stabilized by potassium ions, so when studying G4 folding, one should avoid potassium ions while annealing the DNA.
The cooling process is critical for G4 formation. Fast cooling may cause kinetically stable products, such as a hairpin or double helix. Slower cooling will better stabilize G4 formation.
Repeated freezing and thawing of DNA samples will induce G4 conformational changes as well as fluorophore degradation; therefore, we recommend using small, single-use aliquots of DNA samples.
A clean and passivated surface becomes hydrophobic, and so it can be differentiated from a non-treated surface by testing the behavior of a drop of water on the surface. Also, the surface should not be physically touched to protect the passivated surface.
The tape must be sufficiently wide (~ 2 mm) to prevent channel-channel leakage, but narrow enough to allow the solution to flow through the length of the channel.
Bubbles inside the tape will cause leakage and contaminate the neighbor channels. In order to prevent bubbles, the tape should be placed flatly and carefully pressed from one side. Once the tape is placed on the slide, it should not be removed to prevent damage to the passivated surface.
The dilution factor of neutravidin depends on slide quality. The lower the density of biotin-PEG, the higher the concentration of neutravidin should be. In our case, the slide contains 3% biotin-PEG, and a 50-fold dilution is sufficient to immobilize 1000 molecules in one image.
The solution can be added by directly pipetting into the channel and cleaning extra buffer with a kimwipe. Another option is to use a flow setup system, which is described later in the Protein Binding section.
Generally, 25 pM to 100 pM DNA is low enough to yield 300 to 400 molecules in an imaging field of view of 25 × 75 μm2. Higher than this density may cause signal overlap because of Abbe’s law. However, many factors impact immobilization, and so starting with a low concentration and increasing until 300–400 molecules/image is reached is recommended.
10 mM trolox was prepared by dissolving 25 mg in 10 mL ddH2O with 10M NaOH (about 10 μL, adjusting to pH 8). After inverting up and down to mix the powder, the solution was wrapped in aluminum foil and rotated in the cold room overnight until it is fully dissolved. The next day, the solution was filtered using a 0.22-μm membrane and aliquotted (1 mL per tube). The trolox solution can be stored for 2–3 weeks at 4°C and longer in −80°C.
Taking short movies (containing only 20 to 40 frames) will prevent photobleaching. However, the time interval that these frames are taken over can vary. For example, if the FRET dynamics are faster than 100 ms, there will be one broad peak observed at 100 ms scale. Therefore, a short time interval is able to capture and distinguish different FRET states.
The number of short movies taken depends on molecule density. Reliable FRET histograms must be built from a sufficient number of effective molecules (approximately 5000 to 6000 molecules). If DNA labeling efficiency is low, the ratio of donor- or acceptor-only molecules becomes higher, so more movies must be taken to obtain a representative FRET histogram. To ensure that areas are only imaged once (to reduce photobleaching), we recommend snaking the microscope stage across and down the length of the channel.
Signal leakage comes from the instrument setup, usually the dichroic mirror. A perfect dichroic mirror would be able to completely separate light at a certain wavelength; however, in actuality part of the signal leaks through the mirror (ie Cy3 emission into the red channel and Cy5 emission into the green channel). Because our system uses Cy3 and Cy5 intensities to calculate FRET efficiency, this leakage causes incorrect calculation of FRET efficiency. The calibration was described in the reference (12).
In our oxygen scavenging system conditions, about 50% of both dyes photobleached after 3 min (1801 frames in 100 ms resolution). Another way to prevent fast photobleaching is to decrease the laser power. However, lower laser power will cause lower fluorescence emission. In this case, lengthening frame resolution can increase average signal intensity to overcome the decreased fluorescence. Ensuring that the lasers are properly focused will help limit the laser power needed.
To prevent buffer leakage by atmospheric pressure, the flowing system must be isolated from outside airflow. Therefore, the gap between the tubing and tip should be epoxied and tested for air leakage by flowing water through before attaching the flowing system to the slide - the volume of water pulled through should enter the syringe immediately. Any leakage will cause bubbles in the tubing and slow down the flow speed.
Do not pull all of the buffer into the channel, because the balance of flow pressure will draw more solution than the expected volume. If 50 μL of solution is loaded into the well, then the flowing volume should be less than 40μL. Before attaching the p10 tip onto the slide channel, a small amount of T50 buffer can be pulled through the tubing first to fill the volume of the tubing itself.
The pulling action may perturb the sample stage or microscope, causing imaging drift. Be careful while pulling the syringe.
5. Acknowledgement
Most of the data was taken and analyzed by our alumni, Helen Hwang. We thank the members of the Sua Myong and Taekjip Ha laboratory for their input.
6. References
- 1.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res 34(19):5402–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maizels N, Gray LT (2013) The G4 genome. Plos Genet 9(4):e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers VS, Marsico G, Boutell JM, Antonio MD, Smith GP, Balasubramanian S (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol 33(8):877–881 [DOI] [PubMed] [Google Scholar]
- 4.Valton AL, Prioleau MN (2016) G-quadruplexes in DNA replication: a problem or a necessity? Trends Genet 32(11):697–706 [DOI] [PubMed] [Google Scholar]
- 5.Rhodes D, Lipps H (2015) G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res 43(18):8627–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M (2014) A polymerase theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun 5:3216. [DOI] [PubMed] [Google Scholar]
- 7.Maizels N (2015) G4-associated human diseases. EMBO Rep. 16(8):910–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghuraman MK, Cech TR (1990) Effect of monovalent cation-induced telomeric DNA structure on the binding of Oxytricha telomeric protein. Nucleic Acids Res. 18(15):4543–4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardin C, Henderson E, Watson T, Prosser JK (1991) Monovalent cation induced structural transitions in telomeric DNAs: G-DNA folding intermediates. Biochemistry 30(18):4460–4472 [DOI] [PubMed] [Google Scholar]
- 10.Viglasky V, Bauer L, Tluckova K, Javorsky P (2010) Evaluation of human telomeric G-quadruplexes: the influence of overhanging sequences on quadruplex stability and folding. Journal of Nucleic Acids 2010:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Liu J, Chen Z et al. (2011) G-quadruplex formation at the 3’ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 39(14):6229–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joo C, Ha T (2008) Single molecule FRET with total internal reflection microsopy In: Selvin P, Ha T (eds) Single molecule techniques: A laboratory manual, Cold Spring Harbor Laboraroty Press, Cold Spring Harbor, New York: ISBN 978-087969775-4, 507 pp [Google Scholar]
- 13.Roy R, Hohng S, Ha T (2008) A practical guide to single-molecule FRET. Nature Methods, 5(6):507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang H, Kreig A, Calvert J et al. (2014) Telomeric overhang length determines structural dynamics and accessibility to telomerase and ALT-associated proteins. Structure 22(6):842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang H, Buncher N, Opresko PL, Myong S (2012) POT1-TPP1 regulates telomeric overhang structural dynamics. Structure 20(11):1872–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448(7157):1068–1071 [DOI] [PubMed] [Google Scholar]
- 17.Sowd G, Lei M, Opresko PL (2008) Mechanism and substrate specificity of telomeric protein POT1 stimulation of the Werner syndrome helicase. Nucleic Acids Res 36(13):4242–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKinney SA, Joo C, Ha T (2006) Analysis of single-molecule FRET trajectories using hidden Markkov modeling. Biophys J 91:1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]