Abstract
The cytoskeletal interacting protein Septin 9 (SEPT9), a member of the septin gene family, has been proposed to have oncogenic functions. It is a known hot spot of retroviral tagging insertion and a fusion partner of both de novo and therapy-induced mixed lineage leukemia (MLL). Of all septins, SEPT9 holds the strongest link to cancer, especially breast cancer. Murine models of breast cancer frequently exhibit Sept9 amplification in the form of double minute chromosomes, and about 20% of human breast cancer display genomic amplification and protein over expression at the SEPT9 locus. Yet, a clear mechanism by which SEPT9 elicits tumor-promoting functions is lacking.
To obtain unbiased insights on molecular signatures of SEPT9 upregulation in breast tumors, we overexpressed several of its isoforms in breast cancer cell lines. Global transcriptomic profiling supports a role of SEPT9 in invasion. Functional studies reveal that SEPT9 upregulation is sufficient to increase degradation of the extracellular matrix, while SEPT9 downregulation inhibits this process. The degradation pattern is peripheral and associated with focal adhesions (FA), where it is coupled with increased expression of matrix metalloproteinases. SEPT9 overexpression induces MMP upregulation in human tumors and in culture models and promotes MMP3 secretion to the media at FAs. Downregulation of SEPT9 or chemical inhibition of septin filament assembly impairs recruitment of MMP3 to FAs. Our results indicate that SEPT9 promotes upregulation and both trafficking and secretion of MMPs near FAs, thus enhancing migration and invasion of breast cancer cells.
Keywords: Septin 9, invasion, migration, breast cancer, focal adhesions, oncogene, metalloproteinases, MMP3
Introduction
Septin 9 (SEPT9) is a GTP-binding protein that assembles into hetero-oligomeric complexes in the form of filaments and rings1. At the genomic level, it is the most complex member of the septin family of genes, coding for up to 18 isoform variants, and is, to date, the septin with the strongest association to cancer. SEPT9 was first linked to tumorigenesis in 1999 by the discovery of therapy induced chromosomal translocations between the SEPT9 locus and the mixed lineage leukemia (MLL) gene in an acute myeloid leukemia (AML) patient2. Septins are the protein family most frequently involved in rearrangements with MLL3. MLL-SEPT9 fusions are highly heterogeneous in terms of both patients’ age (4 months–72 years), and clinical presentation (myelodysplastic syndromes as well as acute myeloid cases), suggesting a broad implication in the tumorigenic process3. SEPT9 transcripts are generated by splicing or coded by alternative promoters4, 5. SEPT9 isoforms include a central common core (exons 4–11) and differing 5’ and 3’ ends3. While mechanistically the contribution of MLL partners to the tumorigenic process remains largely unknown, it is well established that fusion genes have oncogenic functions. This suggests that SEPT9 acts as a bona fide oncogene.
In solid tumors, SEPT9 was originally proposed as a tumor suppressor gene as it maps to a region of loss of heterozygosity in some breast and ovarian tumors6. However, several reports now suggest that this is not the case. SEPT9 was identified as a hot spot of viral integration in retroviral insertion mutagenesis experiments7, and it is amplified in murine models of breast cancer in the form of double minute chromosomes8, both of which are molecular properties of strong oncogenes9. Genomic amplification of SEPT9 leads to its overexpression in human and murine breast adenocarcinomas and in a variety of breast cancer cell lines8. The likely mechanism of SEPT9 tumorigenic activity is via upregulation of its transcripts, given that mutations of this gene are rare in cancer10. This is supported by mining The Cancer Genome Atlas11. Upregulation of the transcript variant SEPT9_v1 has been reported in human cell lines as well as in matched tumor and peritumoral breast cancer-tissue specimens5, 12, 13. Expression of the SEPT9_v3 isoform is regulated by DNA methylation upstream of the SEPT9_v3 transcription start site14. Overexpression of SEPT9 is detected more commonly in high-grade carcinomas, which are also associated with a worse clinical prognosis5.
Septins play pivotal roles in cytoskeleton dynamics; accordingly, they are linked to a broad range of cellular functions, many of which have the potential to foster tumorigenic phenotypes. SEPT9_v1 promotes tumor growth by stabilizing HIF-1α15, while SEPT9_v4 has been shown to increase resistance to microtubules-interacting drugs16. SEPT9_v1 is upregulated in breast cancer and is associated with oncogenic potential via its interaction with c-Jun12, 17.
One proposed mechanism for SEPT9 contribution to tumorigenesis is to promote tumor metastasis via enhancing migration18. We previously performed migration assays using MCF7 cells expressing GFP-tagged SEPT9 isoforms and showed that SEPT9_v1, SEPT9_v3, SEPT9_v4, and SEPT9_v5 have increased numbers of migratory cells, suggesting that SEPT9 contributes to the migratory phenotype. Analysis of morphogenesis of renal cysts using MDCK cells grown in 3-D revealed that SEPT9 overexpression doubled the number of cellular extensions and their length, suggesting a more aggressive tumor phenotype due to SEPT9 upregulation19.
While it is recognized that SEPT9 contributes to tumorigenesis, a clear mechanism of action by which SEPT9 elicits its tumor-promoting functions is lacking. To begin addressing this significant gap of knowledge, we overexpressed three SEPT9 isoforms deregulated in breast primary tumors into the hormone responsive luminal A subtype MCF7 cell line, therefore mimicking the occurrence of the most common breast cancer subtype (luminal A, estrogen and progesterone responsive), and performed global transcriptomic analysis20. Our results uncovered a significant association of SEPT9 overexpression with differential expression (DE) of genes expressed in vesicle membranes and in the lumen of the endoplasmic reticulum (ER). Functional analysis of SEPT9-overexpressing breast tumor cells revealed increased degradation of the ECM. SEPT9 overexpression promoted MMP upregulation and a significant increase of matrix metalloproteinases secreted to the culture media. In human breast cancer patients in whom SEPT9 expression is upregulated, MMP levels are also significantly increased. ECM degradation pattern co-localized with vinculin and overlapped with FAs at the cell membrane, suggesting that one of the oncogenic functions of SEPT9 is to promote degradation of the ECM by mediating the transport of protease-containing vesicles from the ER to the tumor microenvironment.
Results
Upregulation of SEPT9 isoforms SEPT9_v1, SEPT9_v2, and SEPT9_v3 in MCF7 cells activates different but complementary oncogenic pathways.
To directly test the consequences of overexpression of the SEPT9 isoforms SEPT9_v1, SEPT9_v2, and SEPT9_v3 we performed global transcriptomic profiling using our previously characterized MCF7 clones as a model5. Fluorescence Activated Cell Sorting (FACS) was used to remove GFP negative cells in order to achieve homogeneity of chimeric GFP-SEPT9 protein expression across the pool of transfected lines (>98% GFP-SEPT9 positive cells) (Supplementary Figure 1A). To identify SEPT9 isoform-specific gene expression changes occurring as a consequence of SEPT9 isoform overexpression, we performed global transcriptomic analysis using RNA-sequencing on MCF7_SEPT9_v1, MCF7_SEPT9_v2, and MCF7_SEPT9_v3 and compared the transcriptional profile to MCF7 control cells (MFC7_c) (Supplementary Table 1 and Supplementary Figure 1B–D).
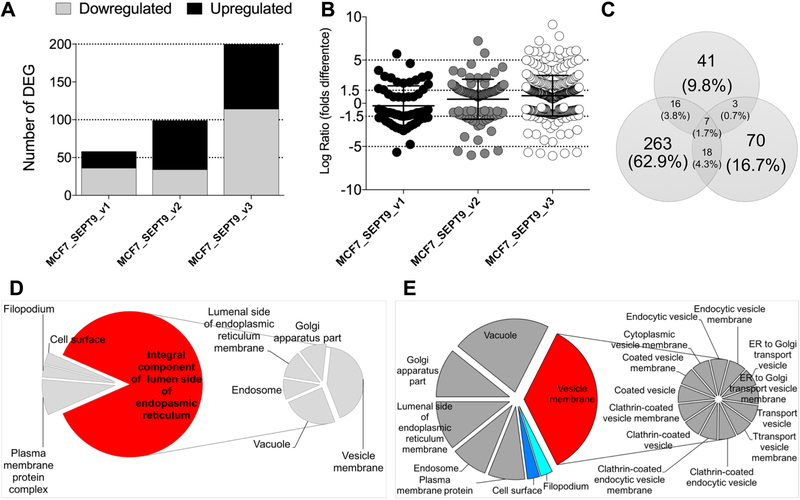
Relative to MCF7_c (FDR < 0.05) we found that, globally, 459 normalized genes were differentially expressed (DE): 184 genes were downregulated and 275 were upregulated. Because we anticipated the differential gene expression between the SEPT9 isoforms to be relatively small, we refrained from setting an arbitrary cut-off for fold change, allowing us to detect all possible statistically significant changes in expression. Of the 459 genes that met the FDR<0.05 criteria, 58 were DE between MCF7_SEPT9_v1 and MCF7_c (36 downregulated and 22 upregulated)(Figure 1A), 99 were DE between MCF7_SEPT9_v2 and MFC7_c (34 downregulated and 65 upregulated), and 302 were DE between MCF7_SEPT9_v3 and MCF7_c (114 downregulated and 188 upregulated). The fold difference in log normalized expression varied between 6.072 and +9.11(Figure 1B), with 204 genes DE by 1.5 folds or more. Surprisingly, we found minimal overlap between the DE genes identified as result of overexpression of different isoforms supporting previous reports that SEPT9 isoforms may provide complementary but unique functions4, 5, 14, 21–24. Seven genes were DE in all MCF7 clones, of which two were upregulated (Figure 1C and Table 1). Among those, the TCR Gamma Alternate Reading Frame Protein (TARP) which encodes for an alternative T-cell receptor protein, is a novel breast and prostate tumor-associated antigen25. TARP is localized to the outer mitochondrial membrane26 and its overexpression increase the growth rate of epithelial cells and induce caveolins expression27. RNA (UPK1AS1), a novel uncharacterized noncoding RNA, is also expressed in the mammary epithelium28 and in the mammary gland29.
Figure 1: MCF7 clones overexpressing isoforms MCF7_SEPT9_v1, MCF7_SEPT9_v2, and MCF7_SEPT9_v3 differentially express isoform-specific genes.
A. Plot of number of DE genes in MCF7-SEPT9-overexpressing isoforms relative to MCF7 control cells. Y-axis: number of DE genes; grey bars depict genes downregulated relative to MCF7_c; black bars depict genes upregulated relative to MCF7_c. B. Fold change of DE genes relative to MCF7_c. Each dot corresponds to a single gene. Values on the Y-axis are expressed as log ratio of normalized fold change of differential expression when compared to MCF7_c. C. Venn diagram depicting the overlap of DE genes between the three SEPT9 isoforms analyzed. D-E. Pie charts depicting the cellular component significantly enriched in DE genes. D. MCF7_SEPT9_v1. E. MCF7_SEPT9_v3. Cellular components are indicated as percentage over the whole components identified for each specific MCF7 overexpressing clone.
Table 1:
Common DE genes in MCF7_SEPT9 clones
| Gene | MCF7_SEPT9_v1 | MCF7_SEPT9_v2 | MCF7_SEPT9_v3 |
|---|---|---|---|
| AMPH | 1.710 | −2.502 | 3.554 |
| SEC14L4 | −4.751 | −4.262 | −3.601 |
| SHISA2 | −0.973 | −1.295 | −0.822 |
| SULF1 | −1.246 | 1.386 | −1.067 |
| SLC17A9 | −4.262 | 2.205 | −1.313 |
| TARP | 1.616 | 1.528 | 3.461 |
| UPK1A-AS1 | 3.809 | 3.554 | 4.135 |
Values are expressed as folds of DE relative to MCF7_c
Two genes were downregulated in all three MCF7_SEPT9 isoform clones: SEC14-Like Lipid Binding 4 (SEC14L4), which encodes a member of Sec14-like phosphatidylinositol transfer proteins that functions as one of the key regulators of phosphoinositide signaling of trafficking through the trans-Golgi network30; and Shisa Family Member 2 (SHISA2), which encodes an endoplasmic reticulum (ER) protein that cell-autonomously inhibits FGF and Wnt signaling by preventing the maturation and the cell-surface expression of their receptors31. Remarkably, two genes, Amphiphysin (AMPH) and Solute Carrier Family 17 Member 9 (SLC17A9), both associated with vesicle mediated transport, were both upregulated in MCF7_SEPT9_v1 and MCF7_SEPT9_v3 but were downregulated in MCF7_SEPT9_v2 cells. On the contrary, Sulfatase 1 (SULF1), an extracellular heparan sulfate endosulfatase enzyme that is secreted through the Golgi, was downregulated in MCF7_SEPT9_v1 and MCF7_SEPT9_v3 but was upregulated in MCF7_SEPT9_v2 cells. When considered globally, all DE genes common to the three SEPT9 isoforms were significantly enriched in genes expressed in the lipid bilayer of membranes (GO:0016020), suggesting that membrane dynamics is an important consequence of SEPT9 over expression in epithelial breast cancer cells.
Of the DE genes that were found common in two SEPT9 isoforms (Supplementary Figure 2A and Supplementary Table 2), three were common to MCF7_SEPT9_v1 and MCF7_SEPT9_v2: Cadherin 4 (CDH4), Caveolin 1 (CAV1) and ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 (ATP8A1), suggesting again a pivotal role of membrane bound proteins. Eighteen genes were common to MCF7_SEPT9_v2 and MCF7_SEPT9_v3, enriched in the cellular component GO term vesicle lumen (GO:0031983, p=1.61e-02), Aldolase C, fructose-bisphosphate (ALDOC), Leucine-rich alpha-2-glycoprotein 1 (LRG1), and vascular endothelial growth factor C (VEGFC)(Supplementary Figure 2B), as well as positive regulation of cell migration (GO:0030335). Significant enrichment of the 18-gene set was also found in GO terms associated with regulation of secretion. When overlaid with genes of the phospholipid signaling pathway (Supplementary Figure 2B blue lines) we identified significant enrichment (p<0.05) within genes common to MCF7_SEPT9_v2 and MCF7_ SEPT9_v3.
A total of 16 genes common to MCF7_SEPT9_v1 and MCF7_SEPT9_v3 were found to be involved in the Protein Kinase A (PKA) signaling pathway (Supplementary Figure 2C).
Because of the limited overlap of DE genes between the SEPT9 isoforms, we proceeded to analyze individually the transcriptome of each MCF7 clone overexpressing the different isoforms.
At the level of single genes, the top 10 up- and downregulated MCF7_SEPT9_v1 genes were associated with functions important for antigen binding and processing (Supplementary Table 3). These genes are conventionally expressed in the cellular compartment of the Golgi apparatus. The top 10 up- and downregulated genes identified in MCF7_SEPT9_v2 and MCF7_SEPT9_v3 cells were not significantly associated, as a group, to specific GO terms.
Globally, Gene Set Enrichment Analysis (GSA) of all significantly DE genes relative to MCF7_c cells revealed that MCF7_SEPT9_v1-overexpressing cells were mainly expressed in the endoplasmic reticulum and the Golgi apparatus (Figure 1D), and were enriched for antigen processing and response to cytokine stimulus pathways (FDR= 6.02E-06), as well as tissue remodeling and regulation of cell to cell adhesion (Supplementary Figure 3A and Supplementary Table 4). We identified a significant association of DE genes in response to SEPT9_v1 upregulation with hallmarks of protein secretion and apical surface markers (Supplementary Figure 3A blue and red lines). Interestingly, we found a significant enrichment (FDR =5.78E-03) with regulation of pinocytosis (CAV1 and the AXL Receptor Tyrosine Kinase AXL1 gene). AXL1 is a signal transducer regulating communication of epithelial cells with the extracellular matrix and it has been involved in several functions including promoting cell invasion32.
MCF7_SEPT9_v2 cells were enriched for cell growth functions (FDR= 1.79E-03) and response to growth factor stimulus (FDR= 6.77E-03) (Supplementary Figure 3B and Supplementary Table 4). MCF7_SEPT9_v3 cells were mainly associated with vesicles (Figure 1E) and mostly enriched in pathways controlling excretion (FDR= 2.9E-02) and lipid localization and transport (FDR=3.3E-03). Overlay of the Biocarta TGFβ pathway gene members with enriched GO terms as result of SEPT9_v3 overexpression revealed an extensive overlap between these two groups (Supplementary Figure 3C and Supplementary Table 4). To verify the biological finding that SEPT9_v1, SEPT9_v2 and SEPT9_v3 may promote isoform specific differential gene expression, we randomly selected six genes (Versican -VCAN, T-Box Transcription Factor TBX15, Sulfatase 1 -SULF1, Glypican 6 -GPC6, Keratin 20 -KRT20 and Fibroblast Growth Factor 21 -FGF21). These targets, which included high and low expressing genes as well as transcripts upregulated or downregulated in one or more isoform specific MCF7 lines, showed similar trend of expression in MCF7 cells as determined by quantitative PCR across 3 independent lines, each expressing one SEPT9 isoform (Supplementary Figure 4).
When considered as a whole, the transcriptomic profiling analysis of MCF7 cells overexpressing SEPT9_v1, SEPT9_v2, and SEPT9_v3 isoforms points to altered cell communication between cells and the microenvironment via vesicle formation and membrane dynamics.
SEPT9 overexpression promotes invasion and migration
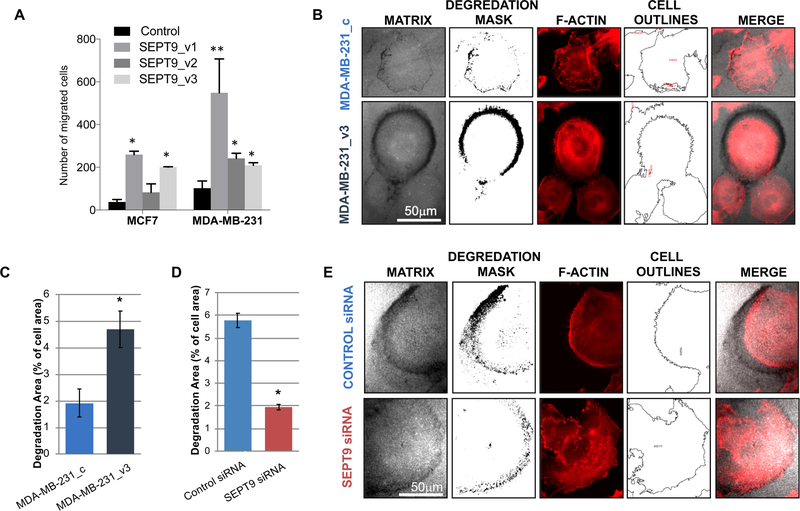
Because the RNA-sequencing results strongly supports a role for SEPT9 in the dynamics of vesicles and their interaction with the extracellular matrix, we reasoned that one of the oncogenic functions linked to its overexpression in breast cancer cells is to promote invasion. We therefore proceeded to functionally test this hypothesis in vitro. Among the features that contribute to cell invasiveness are cell motility and their ability to degrade the surrounding extracellular matrix (ECM). We previously showed that SEPT9 expression in MCF7 cells contributes to the migratory phenotype of these cells (Figure 2A and14). To verify that this effect was not restricted to MCF7 cells, we generated stable MDA-MB-231 clones, expressing GFP-tagged SEPT9 isoforms SEPT9_v1, SEPT9_v2, or SEPT9_v3. The pro-migratory and invasive potential of each of the MDA-MB-231 clones was tested using the transwell migration assay with collagen-coated inserts14. The ability of the cells to migrate/invade in the transwell assay relies on their motility as well as their ability to degrade the matrix through which they migrate. Expression of all three SEPT9 isoforms showed, as in MCF7 cells, a significant increase in the number of migratory/invading cells (p<0.05), suggesting that the pro-migratory and invasive effect of SEPT9 in mammary epithelial cells is likely a general phenomenon (Figure 2A.
Figure 2: SEPT9 promotes fluorescent matrix degradation by MDA-MB-231 cells.
A. Transwell migration assay comparing MCF7 and MDA-MB-231 clones, each expressing a different GFP-SEPT9 isoform fusion construct. Cells were added to transwell inserts and allowed to migrate for 10 hours at 37°C, after which migrated cells were counted. Plotting reflects the average number of migrated cells from two biological replicate assays conducted for each isoform. Statistical differences were calculated using ANOVA, *p<0.05 **p<0.02. B. Degradation patterns of MDA-MB-231_c (control) and MDA-MB-231_SEPT9_v3 cells. Images of matrix channel are shown together with the degradation mask and the F-actin staining (red) together with the outlines of cell areas. Bar = 50μm. C. Bar plot depicting the percentage of gelatin degradation area (representative 4×5 panel use for analysis is shown in Supplementary Figure 4A). Bars represent the mean of the degradation area as % of total cell area and error bars represent the standard error of the mean (SEM). *, p < 0.033. Data was confirmed at least by three independent experiments. D-E. Degradation patterns of MDA-MB-231 transfected with SEPT9 siRNA or scramble (control) siRNA. Images of matrix channel are shown with the degradation mask and the F-actin staining (red) together with the outlines of cell areas (E). Bar = 50μm Bar plot depicting the percentage of gelatin degradation area (representative 4×5 panel use for analysis is shown in Supplementary Figure 4A). Bars represent the mean of the degradation area as % of total cell area and error bars represent the standard error of the mean (SEM).
*, p < 0.001. Data was confirmed at least by three independent experiments.
SEPT9 overexpression increases cellular degradation of fluorescent matrix
Among the genes enriched in SEPT9-overexpressing MCF7 cells, several were involved in cytoskeletal organization, cell-cell adhesion, and secretion, all of which can contribute to metastasis by promoting ECM degradation. We subsequently wanted to test whether the expression of the SEPT9 isoforms in mammary epithelial cells affects the cells’ ability to digest the ECM. We performed matrix degradation assays, using 2-dimensional fluorescent analysis using MDA-MB-231 clones because of their high degrading phenotype33, 34. Initially we tested the three different GFP-SEPT9 variants to determine to what extent they differed in their matrix degradation activity. MDA-MB-231_SEPT9_v3 was the most degrading SEPT9 isoform. This agrees with our RNA-Seq analysis indicating that SEPT9_v3 overexpressing cells are enriched for the expression of genes controlling cell-cell adhesion and excretion when compared with WT cells. Therefore, we proceeded to perform a more detailed analysis comparing matrix degradation in MDA-MB-231_SEPT9_v3 relative to MDA-MB-231_c cells. In order to estimate the degradation potential of the cells, we quantified gelatin degradation by calculating the total degradation area for each panel of images as percentage of total cells’ area (Supplementary Figure 5A). This approach provides a normalized and highly reproducible way to compare control cells to SEPT9 overexpressing cells. We found that SEPT9_v3 overexpression significantly increases the gelatin degradation activity of MDA-MB-231 cells of about 2.5 folds (4.69±0.69% vs. 1.92±0.53%, respectively; P<0.033, Figure 2B–C). These results suggest that, in addition to contributing to migration, SEPT9_v3 overexpression in MB-MDA-231 cells also promotes matrix degradation under these experimental conditions.
SEPT9 knockdown decreases cellular degradation of fluorescent matrix
To confirm that the effect of gelatin degradation was indeed a function of SEPT9 activity in MDA-MB-231 cells, we tested the consequence of SEPT9 downregulation on gelatin degradation. To this end, MDA-MB-231_c cells were transfected with siRNA targeting SEPT9 or with a scramble siRNA (control). Cells were analyzed 48 hours post-SEPT9 silencing to ensure decreased levels of SEPT9 mRNA as well as SEPT9 protein (82% mRNA reduction relative to scramble siRNA treated cells - Supplementary Figure 6A; and 75% protein expression decrease -Supplementary Figure 6B–C). The SEPT9 silenced and control cells were next tested for their ability to degrade fluorescent gelatin. The results revealed that reduction of ~80% of SEPT9 mRNA levels relative to controls leads to significantly lower degradation activity compared with that of cells treated with control siRNA (1.94±0.15% vs. 5.77±0.39%, respectively; P<0.001; Figure 2D–E and Supplementary Figure 5B). These results support the hypothesis that SEPT9 is required for extracellular matrix degradation in breast cancer cells.
SEPT9 overexpression promotes secretion of proteases into the cell media
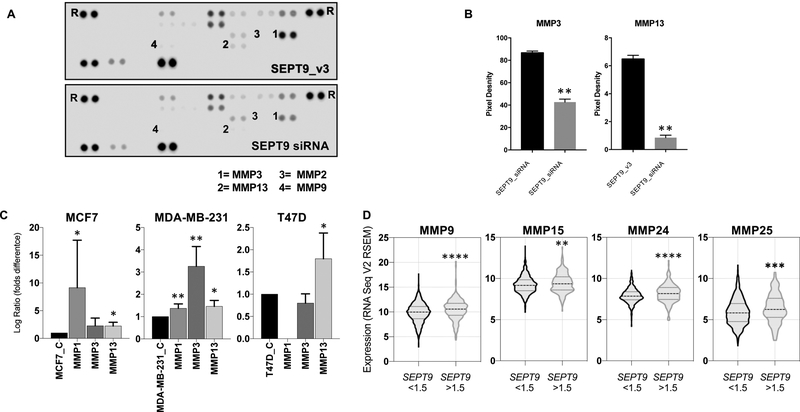
As suggested by our global transcriptomic results, SEPT9 may promote ECM degradation through enhanced secretion of matrix metalloproteinases (MMPs) to the media. In fact, MCF7 cells overexpressing SEPT9_v3 are enriched in excretion pathways, and significantly over express genes associated with MMPs activity (Supplementary Figure 7). Consequently, to examine if SEPT9_v3 overexpression affects the levels of proteases secreted to the media we used a protease array to analyzed the cell culture medium for the presence of 34 different proteases (Supplementary Figure 6E–F). To maximize the potential difference in secreted proteases, we compared the conditioned medium of MDA-MB-231 cells transfected with the SEPT9 siRNA to the conditioned medium of MDA-MB-231 cells expressing SEPT9_v3 (Supplementary Figure 6B–D). We found that the levels of two secreted proteases, Matrix Metalloproteinase MMP3 and MMP13, were significantly decreased in the culture medium of MDA-MB-231 cells transfected with SEPT9 siRNA (~40%) (Figure 3A–B). These results indicate that SEPT9 promotes secretion of MMPs to the media. Interestingly, there was no change in the secreted MMP2 and MMP9 levels, the metalloproteinases implicated in invadopodia-mediated matrix degradation35, 36.
Figure 3: SEPT9 overexpression induces MMP upregulation and promotes secretion of proteases into the cell media.
A. Representative images of protease arrays incubated with cell culture supernatants from MDA-MB-231_SEPT9_v3 cells (Top) and MDA-MB-231 cells transfected with SEPT9 siRNA (Bottom). B. Quantification of MMP3 and MMP13 signals that were most suppressed in conditioned media from MDA-MB-231 transfected with SEPT9 siRNA (** p<0.005) is shown in A. Error bars indicate SEM. Abbreviations: MMP3 - matrix metalloproteinase-3 (Stromelysin-1); MMP13 - matrix metalloproteinase-13 (Collagenase 3). C. Bar graphs depicting the quantification of MMP1, MMP3 and MMP13 in MCF7, MDA-MB-231 and T47D cells over expressing GFP-SEPT9_v3 relative their parental control lines (* p<0.005; ** p<0.001) are shown. Error bars indicate SEM. D. Violin plots depicting the expression levels of MMP9, MMP15, MMP24, and MMP25 in TCGA breast cancer provisional cases expressing SEPT9 mRNA 1.5 folds higher than the mean (n=210) or 1.5 folds lower than the mean (n=883).
(**** p<0.0001; *** p<0.0003; ** p<0.0035).
To better understand the correlation between SEPT9 and MMPs, we wanted to establish if SEPT9 overexpression directly promoted MMPs expression and also evaluate the generality of this process by analyzing multiple breast cancer cell lines. We designed primers for the quantitative measurement of mRNA coded by an array of MMPs: MMP3 and MMP13 were selected based on the protein array results (Figure 3A–B), and MMP1, MMP2, and MMP9 are widely studied matrix-degrading enzymes enriched at invadopodia35, 37–42. The levels of these five degrading enzymes were quantified by qRT-PCR in MCF7, MDA-MB-231, and T47D either overexpressing SEPT9_v3 or not overexpressing SEPT9_v3. MMP13 expression was significantly upregulated as a consequence of SEPT9_v3 overexpression in all three cell lines tested (Figure 3C). MMP1 was significantly upregulated in both MCF7 and MDA-MB-231, the latter of which also upregulated MMP3. The mRNA of MMP2 and MMP9, in concordance with the protease array results (Figure 3C), was expressed at low levels in MDA-MB-231 as well as the other cell lines tested and therefore we could not detect significant changes (data not shown).
Because SEPT9 is amplified in ~20% of breast cancer cases43 and because MMPs are key regulators of the mammary tumor microenvironment that mediate ECM degradation and remodeling44, we mined The Cancer Genome Atlas (TCGA) to study a possible correlation between SEPT9 and MMPs in human sporadic breast cancer. Of the n=1093 cases analyzed as part of the provisional breast cancer dataset, ~19% (n=210) express SEPT9 at 1.5 folds higher than the median levels across all cases. In these samples, from the global transcriptomic profiling, MMP levels were detected across all 210 cases for 8 out of the 23 known MMPs (MMP1, MMP9, MMP11, MMP14, MMP15, MMP19, MMP24, MMP25). We found that 4 out of these 8 MMPs (MMP9, MMP15, MMP24 and MP25) were significantly upregulated in those patients that also overexpressed SEPT9 (Figure 3D), while no significant expression changes were detected in the other MMPs.
Thus, SEPT9 overexpression directly promoted MMPs upregulation in cultured cells, and human tumors with high SEPT9 expression also significantly upregulating the expression of several MMPs.
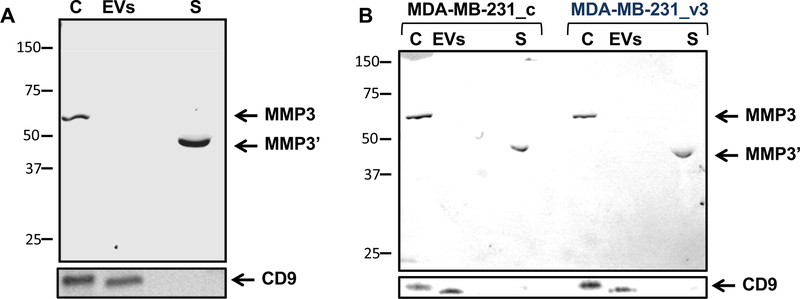
Involvement of MMP secretion with extracellular vesicles (EVs) activity is an emerging field45, 46; we next wanted to test whether SEPT9 affects EV secretion. We isolated EVs from conditioned medium of MDA-MB-231 overexpressing SEPT9_v3 and MDA-MB-231 transfected with control RNAi or SEPT9 RNAi. The isolated EVs were analyzed by dynamic light scattering47, as well as laser scattering microscopy48, 49. Both analyses failed to detect significant differences in secreted EV numbers or size (Supplemental Table 5). This implied that SEPT9 affects either the EV cargo, or the MMP’s secretion pathway by directly enhancing exocytosis into the medium and/or activation of MMP activity during secretion. To distinguish among these mechanisms, we separated the EVs from the MDA-MB-231 culture media to obtain EV-free media. To ensure EV depletion we tested for the presence of CD9, a marker for EVs50, which was present in whole cell lysate and in the EV fraction but not in the conditioned medium (Figure 4A). MMP3 expression was detected in the whole cells extract and in the supernatant fraction but not in the EV-free conditioned medium. This supports the hypothesis that MMP3 is secreted directly to the medium and not via EVs and SEPT9 affects this activation and/or secretion. Moreover, while the protein size of MMP3 expressed in the cells was the expected 57kDa size, MDA-MB-231 cells expressing SEPT9_v3 contained a truncated form. MMP3, like other MMPs, contains a pro-peptide of ~80 amino acids51, which is cleaved upon secretion resulting in a smaller but active MMP3 form (45 kDa, MMP3’). When comparing the supernatant fraction of MDA-MB-231_c and MDA-MB-231_SEPT9_v3 cells, the latter secreted higher levels of MMP3’ (4.78+/− 0.07 folds increase) (Figure 4B), strengthening the notion that SEPT9 promotes secretion of protease activated MMPs to the media.
Figure 4: SEPT9 overexpression promotes secretion of the active form of MMP3 into the cell media.
A. WB analysis of Cells (C), EV fraction (EVs), and supernatant fraction (S) of conditioned medium from MDA-MB-231 cells. MMP3 is secreted from the cells directly to the medium but not in EVs. Size of MMP3 is ~54 kDa and size of secreted MMP3’ is ~45kDa. The CD9 was used as a marker for EVs. B. WB analysis of Cells (C), EVs fraction (EVs), and supernatant fraction (S) of conditioned medium from MDA-MB-231_c and MDA-MB-231_v3 cells. The CD9 signal in the secreted EV fraction was used as a reference for the quantification of the levels of the secreted MMP3’.
SEPT9 is associated with Focal Adhesions degradation sites and MMP3
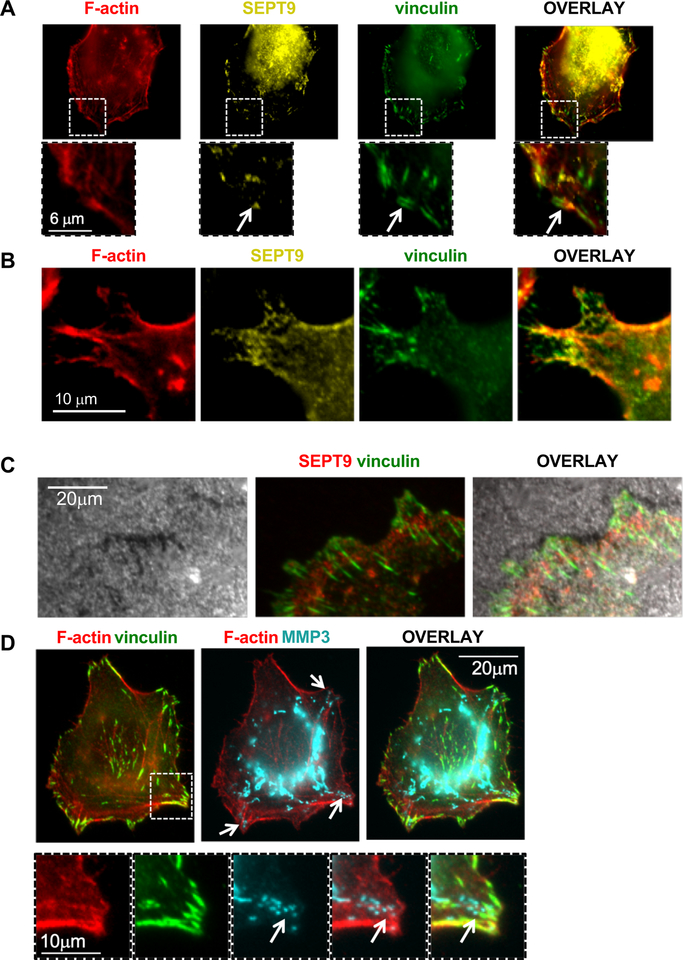
We next sought to further characterize the relation between SEPT9 activity and extracellular matrix degradation in MDA-MB-231 cells. The degradation pattern exhibited by MDA-MB-231 is peripheral (Figure 2B) and resembles the pattern of focal adhesion (FA) ECM degradation52. To determine whether FAs drive the observed degradation pattern and whether SEPT9 is associated with these structures, we stained MDA-MB-231 cells grown on gelatin with pan-SEPT9 and the FA marker vinculin. As expected, SEPT9 assembled into filaments that partially colocalize with F-actin. However, this localization was uneven along the F-actin filaments, as previously described for SEPT2 and other septins53 (Figure 5A–B). We also found that SEPT9 localized adjacent to FAs, which appeared to extend from the SEPT9 and F-actin filaments (Figure 5A, white arrow). These findings are in accordance with previous studies showing that SEPT9 directly cross-links prepolymerized actin filaments into bundles and promotes maturation of nascent FAs19. Moreover, the vinculin staining patterns were very similar to the gelatin degradation patterns (Figure 5C), implying that the ECM degradation observed under these conditions is associated with FAs.
Figure 5: SEPT9 localizes to Focal Adhesions, gelatin degradation sites, and MMP3.
A-B. Representative images depicting F-actin (red), SEPT9 (yellow), and vinculin (green) staining of MDA-MB-231 cells. SEPT9 filaments partly colocalize with F-actin filaments (arrow points at FAs extending from SEPT9 filaments). Lower panels in A show enlarged views of the white-boxed regions. C. Representative images depicting matrix (grey), SEPT9 (red), and vinculin (green) staining. Matrix degradation pattern colocalizes with vinculin. MDA-MB-231 cells were seeded on Alexa Fluor 405–labeled gelatin. D. Representative images depicting filamentous MMP3 signals associated with F-actin filaments emerging from the cell center towards focal adhesions (vinculin staining, green). Arrows point at MMP3 foci localized at sites of FAs. Lower panels show enlarged views of the white-boxed region. Bar=10μm. Cells were seeded on non-fluorescent gelatin.
In addition to the interaction with F-actin and the association with FAs, SEPT9 has been shown to interact with microtubules8, 19, 54–56. Moreover, studies indicate that septins participate in Golgi-to-plasma membrane vesicle transport57, 58 suggesting that SEPT9 may be involved in trafficking of MMPs from the Golgi to mature FAs. Hence, we examined the association between SEPT9, FAs, F-actin, and MMP3. First, MDA-MB-231 cells were seeded on gelatin and stained for F-actin, vinculin, and MMP3. Interestingly, the MMP3 signal appeared as a tubular staining, which in some instances localized along F-actin filaments. The MMP3 tubular staining extended from the cell center towards the cell edge, and specifically towards FA, where MMP3 is expressed in foci (Figure 5D, arrows).
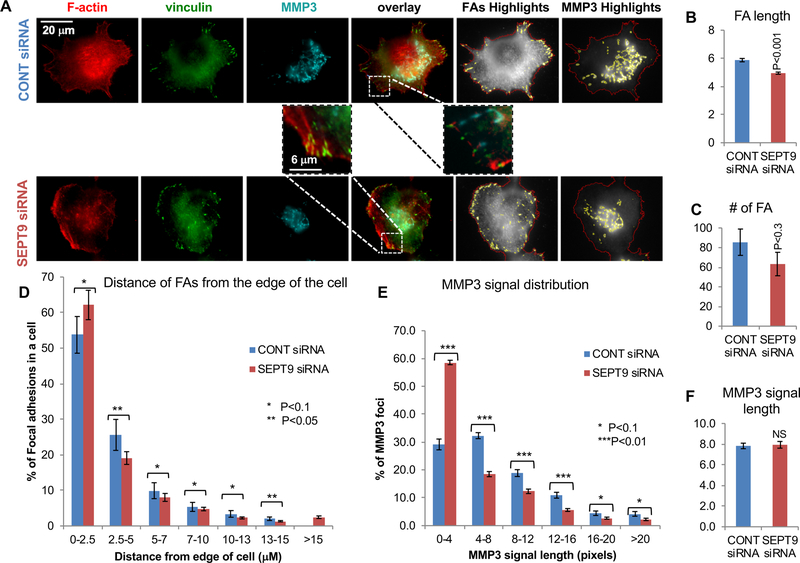
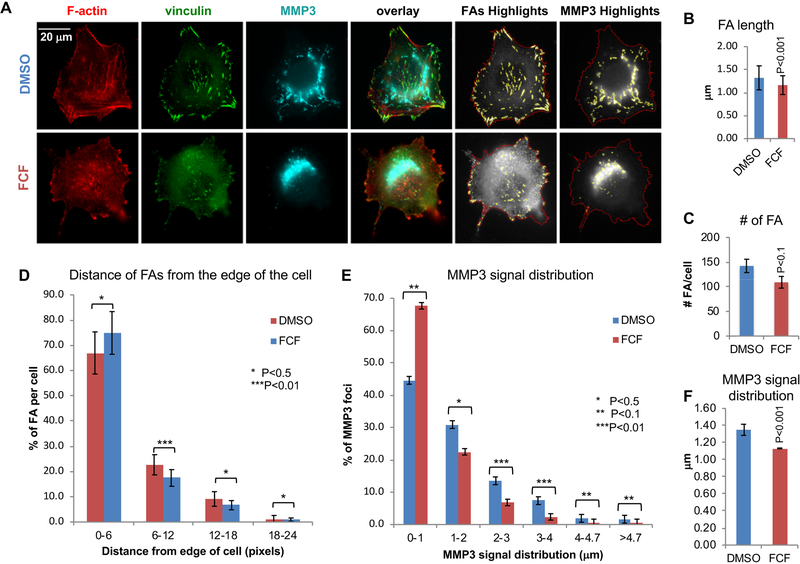
Next, we asked whether SEPT9 depletion affects either FAs, as previously demonstrated19, or the tubular morphology of the MMP3 staining. To address this, MDA-MB-231 cells transfected with siRNA targeting SEPT9 or with a scramble siRNA (control), were seeded on gelatin before staining with F-actin, vinculin, and MMP3 and analyzed for FA morphology and number. Notably, SEPT9 depletion decreased the number of mature FAs and had some effect on the MMP3 signal (Figure 6; Supplementary Figure 8). Image analysis revealed that cells transfected with SEPT9 siRNA had significantly shorter FAs (Figure 6B; SEPT9, 1.1±0.26 μm; CONT, 1.3±0.2 μm; p<0.02) than control cells. Moreover, SEPT9 depletion significantly decreased the number of FAs larger than 472 nm (2 pixels)(Figure 6C; SEPT9, 48±6.2; CONT, 80±9.7; p<0.01), while increasing the percentage of FAs localizing distal to the cell edge relative to control cells (Figure 6D). These results demonstrate that SEPT9 depletion affects the maturation of FAs, which is characterized by both elongated FAs and FA position closer to the cell center19. However, the effect of SEPT9 depletion on MMP3 was not as striking (Supplementary Figure 9). While the size distribution of the MMP3 signals was affected slightly (Figure 6E; SEPT9, 59% of the signals were shorter than 944 nm and 41% longer; CONT, 29 % of the signals were shorter than 944 nm and 71% longer, P<0.01), the average signal distribution of the MMP3 signals was similar between control and SEPT9 siRNA cells (Figure 6F). This could be partly due to cell-to-cell variability of SEPT9 silencing using siRNA, as well as limitations in the ability of SEPT9 siRNA to equally target all the 18 known SEPT9 isoforms that assemble the septin complex resulting in residual filament assembly12, 59. To test whether a more severe phenotype on MMP3 secretion could be observed using a septin inhibitor, we exposed MDA-MB-231 cells to forchlorfenuron (FCF). FCF impairs septin dynamics and mammalian septin organization, and it has been shown to inhibit exocytosis of secreted proteins in various cells55, 60. If our hypothesis is correct, stabilization of septin filaments which impairs their dynamics should severely affect the maturation of FAs and destroy the tubular pattern of MMP3 expression. MDA-MB-231 cells were seeded on gelatin and incubated with 50 μM FCF or DMSO control followed by staining for F-actin, vinculin, and MMP3. As predicted (Figure 7A), FCF impaired the formation of mature FAs as well as the tubular expression of MMP3. FCF-treated cells had statistically significant smaller and fewer central FAs than DMSO-treated cells, indicating that FCF inhibits the maturation of the FAs (Supplementary Figure 10). Likewise, the tubular appearance of MMP3 was impaired by FCF and most of the FCF-treated cells showed a main centrally located MMP3 signal with only few distal foci (Supplementary Figure 11). Quantification of the images revealed that DMSO-treated control MDA-MB-231 cells had significantly longer FAs (Figure 7B; DMSO, 1.3±0.1 μm; FCF, 1.1±0 μm, P<0.001) than FCF-treated cells. Additionally, FCF treatment, like SEPT9 depletion, significantly decreased the number of FAs larger than 472 nm (2 pixels) (Fig. 7C; DMSO, 142±13.5; FCF, 109±11.8, P<0.1), while increasing the percentage of distal FAs near the cell edge relative to control cells (Fig. 7D). Like SEPT9 depletion, FCF altered the size distribution of the MMP3 signals, as we predicted (Figure 7E; FCF, 68% of the signals were shorter than 944 nm −4 pixels- and 32% were longer; DMSO, 45% of the signals were shorter than 944 nm and 56% were longer, P<0.01). In addition, FCF significantly shortened the average length of the MMP3 signals (Figure 7F; DMSO, 1.3±0.07 μm; FCF, 1.1±0 μm, P<0.001).
Figure 6: SEPT9 promotes FA stabilization and MMP3 trafficking.
A. Representative images of MDA-MB-231 cells transfected with SEPT9 siRNA or scramble (control) siRNA grown on gelatin; F-actin (red), vinculin (green), and MMP3 (cyan). Also shown are highlighted images depicting FA highlights (FAs in yellow and cell outlines in red) and MMP3 highlights (MMP3 in yellow and cell outlines in red). Bar=20 μm. B-C. Bar graphs depicting the quantification of FA server analysis for FAs in MDA-MB-231 cells transfected with control siRNA (n= 18 cells, 1480 FAs) or SEPT9 siRNA (n= 30 cells, 1940 FAs). See corresponding FA highlights in supplementary Figure 7. B. Bar graph indicating the FA average length (μM). C. Bar graph indicating FA number per cell. D. Histogram of the distribution of the FAs according to their distance from the cell edge (μM). E-F. Bar graphs indicating the quantification of MMP3 signals in MDA-MB-231 cells transfected with control siRNA (n= 20 cells, 1922 MMP3 signals) or SEPT9 siRNA (n= 30 cells, 1669 FAs). See corresponding MMP3 highlights in supplementary Figure 8. E. Histogram indicating the distribution of the MMP3 signals according to their length (μm). F. Graph indicates the MMP3 signal average length (μm). We detected no significant difference between transfected with control or SEPT9 siRNA. Error bars indicate SEM.
*, p<0.1; **, p < 0.05; ***, p < 0.01.
Figure 7: Inhibition of Septins by FCF results in impairment of FAs and MMP3 trafficking.
A. Representative images of MDA-MB-231 control cells (DMSO) or FCF treated; F-actin (red), vinculin (green), and MMP3 (cyan). Also shown are FA highlights (FAs in yellow and cell outlines in red) and MMP3 highlights (MMP3 in yellow and cell outlines in red). Bar = 20μm. B–C. Bar graphs indicating the quantification of FAs in MDA-MB-231 cells (n= 15 cells) treated with DMSO (n= 2523 FAs) and FCF (n=1307 FAs). See corresponding FA highlights in supplementary Figure 9. B. Bar graph indicating FA average length (μm). C. Bar graph indicating the number of FAs per cell. D. Histogram indicating the distribution of the FAs according to their distance from the edge of the cell (μm). E-F. Bar graphs indicating the quantification of MMP3 signals in MDA-MB-231 cells (n= 15 cells) treated with DMSO (n=1510 MMP3 signals) and FCF (n=853 MMP3 signals). See corresponding MMP3 highlights in supplementary Figure 10. E. Histogram indicating the distribution of MMP3 signals according to their length (μm). F. Bar graph indicating the MMP3 signal average length (μm). Error bars indicate SEM.
*, P<0.5; **, P < 0.1; ***, P < 0.01.
Collectively, these results support the hypothesis that SEPT9 promotes invasiveness by increasing the maturation of FAs and facilitating MMP3 trafficking and secretion at FAs.
Discussion
The main goal of this work was to study the consequences of SEPT9 overexpression in breast cancer cells. Given the strong association between SEPT9 amplification and overexpression in breast cancer5, 8, 14, 61 and the previously reported SEPT9 functions in enhancing migration14, 19, we reasoned that this protein would function as a bona fide oncogene, granting cells tumor-promoting characteristics. The transcriptomic profiling of MCF7 cells overexpressing three of the major SEPT9 isoforms found in mammary epithelial cells clearly indicate that SEPT9 promotes invasion. Altogether, the enrichment analysis on gene sets differentially expressed in MCF7-overexpressing SEPT9 isoforms points to regulation of cell motility, cell-cell adhesion, lipid metabolism, vesicle transport and secretion.
Some of these properties have been previously linked to septins, while others were a novel discovery of our study. Epithelial-Mesenchymal Transition (EMT) requires epithelial cells to gain migratory and invasive potential. It has been previously shown that septins promote motility of renal epithelial cells19. Here we demonstrate that the enhanced motility is also maintained in mammary epithelial cells. We previously demonstrated, using a transwell migration assay, the pro-migratory effect of SEPT9 expression on the low-migratory MCF7 cells. Here we show that even highly motile and invasive MDA-MB-231 cells further increase this phenotype when SEPT9 is overexpressed. Thus, SEPT9 strongly contributes to migration. This conclusion is consistent with previous in vivo studies showing that SEPT9_v1 stabilizes HIF-1α15 and that the HIF-1a signaling pathway is directly associated with invasive tumor cell migration in breast tumors62.
The greatest effect on migration was achieved by expression of SEPT9_v1, both in MCF7 and MDA-MB-231 cells. This further supports the growing concept that SEPT9 isoforms contribute differently to tumorigenesis. It was recently proposed that SEPT9_v2 acts as an inhibitor of migration in MCF7 cells and that this specific isoform is downregulated in primary breast tumors13. The weak tumorigenic potential of SEPT9_v2 is supported by our transcriptomic analysis of MCF7 cells overexpressing this isoform when compared to SEPT9_v1 or SEPT9_v3. In fact, we found DE genes as consequence of SEPT9_v2 overexpression to be the least enriched in molecular, cellular, and biological functions compared to the other two isoforms tested.
The major outcome of the transcriptomic profiling analysis was the enrichment of tissue remodeling and cell adhesion pathways along with the novel findings linking SEPT9 overexpression in mammary epithelial cells to excretion. Notably, genes differentially expressed as consequence of SEPT9 overexpression were significantly associated with cellular component ontologies related to the plasma membrane, especially the lumen side of the endoplasmic reticulum and membrane vesicles. This is in concordance with recent findings based on proteomic analysis63. Septins can bind to membranes, specifically to phosphoinositides (PIs)64, where they provide membrane stability and serve as diffusion barriers for membrane proteins65, 66. While some mechanisms linking SEPT9 functions to phospholipids and vesicle transports in mammalian cells are emerging67, 68, most of our knowledge remains restricted to yeast biology. However, it is important to note that the phosphoinosities are essential regulatory scaffolds linking invadopodium precursor formation to the proximal tip of FAs69, which is consistent with the results of our study linking SEPT9 to the emergence of proteolysis at FAs.
One of the features of invasion is the ability of cells to degrade their surrounding matrix. Here we provide evidence that SEPT9_v3 overexpression in MDA-MB-231 cells increases their ability to degrade gelatin, whereas global knockdown of SEPT9 in the same cells decreases their degradation potential. These results suggest that increased levels of SEPT9 in mammary epithelial cells promotes degradation of the extracellular matrix, providing a functional link between SEPT9 amplification and overexpression and its oncogenic properties. The most profound effect on degradation was demonstrated by expression of SEPT9_v3 which, when expressed in MCF7 cells, was accountable for enrichment in genes involved in secretion. The different effect of the SEPT9 isoform variants on migration vs. degradation implies that SEPT9 isoforms may be accountable for complementary but unique functions.
Degradation activity of the ECM is a key feature of invadopodia, which is achieved by the secretion of proteases that degrade the basement membrane surrounding the tumor70. The characteristic pattern of gelatin degradation by cancer cells is a centrally situated and dot-shaped degradation pattern that localizes with invadopodia69–72. However, the degradation pattern exhibited by MDA-MB-231 cells in our study was mostly peripheral and localized at the basal membrane that bridges the actin cytoskeleton to the ECM, and the pattern resembled ECM degradation by FAs. This type of proteolytic degradation mechanism has been previously described both in fibroblasts and in epithelial cells73 and shown to be associated with invadopodium precursor assembly at the proximal tips of FAs in breast cancer cells69. In fact, we show here that in MDA-MB-231 cells SEPT9 localizes in adjacent to FAs and that the degradation patterns formed by these cells are similar to the FA staining patterns. FAs are integrin-containing, multi-protein structures that form mechanical connections between intracellular actin bundles and the ECM. The dynamic assembly and disassembly of FAs plays a central role in cell migration. In order to allow movement, FAs have to release and disassemble at the ventral side of the cell body. It was suggested that FA disassembly is facilitated by FA-targeted, local exocytosis leading to ECM degradation, causing disassembly of integrin-matrix connections. Microtubules anchored near FAs serve as tracks for transport of EVs and deliver MT1-MMP to the cell surface membrane, which can activate MMPs and degrade the extracellular matrix around FAs, resulting in integrin detachment, loss of tension, and FA turnover52, 74. Dolat and colleagues previously showed that septin filaments expressed at the leading edge of motile renal epithelia cells promote cell motility by reinforcing the organization of the lamellar stress fiber network and the stability of nascent FAs. Furthermore, this septin function was shown to rely on the ability of SEPT9 to directly cross-link preassembled F-actin filaments, promoting the maturation of nascent FAs19. The direct binding of SEPT9 to F-actin was demonstrated by electron microscopy and it was suggested that SEPT9 could maintain the integrity of growing and contracting actin filaments75. Septins are also implicated in microtubule-dependent transport, which involves the regulation of microtubule motor interactions with cargo. Intriguingly, it has been shown that SEPT9 interacts with a microtubule motor (KIF17) and it was proposed that SEPT9 may act as a regulator of cargo-motor binding by either affecting the loading of cargo to KIF17 or triggering the release of cargo from KIF1776. It is possible that in addition to promoting FA stabilization, SEPT9 functions as a mediator of MMP trafficking, activation, and secretion, thereby promoting ECM degradation.
Our transcriptomic analysis of MCF7 cells overexpressing SEPT9_v3 uncovered a significant enrichment in genes involved in MMP secretion. Here, we functionally show that SEPT9 expression directly results in MMPs upregulation in several breast cancer cell lines, and that SEPT9 upregulation promotes secretion of MMP3 and MMP13 to the cell media in MDA-MB-231. We also provide evidence that the link between SEPT9 upregulation and MMPs activity is maintained in human breast cancer. Secreted MMPs play a crucial role in cellular invasion through modulation of cell–cell and cell–ECM interactions77, 78. Most secreted MMPs require activation post-secretion by cell surface MMPs such as MT1-MMP, which is known to mediate ECM degradation at FAs73; some of these secreted MMPs, such as MMP2 and MMP9, which are activated by MT1-MMP, are involved in direct degradation of the ECM. However, other MMPs can bind to their respective membrane receptors after their activation and initiate signal transduction pathways that influence the cell’s migration and invasion. MMP3 (stromelysin-1) is a known inducer of epithelial–mesenchymal transition (EMT) that has been shown to promote mammary carcinogenesis79–81. It is also involved in the activation process of MMP9, consequently promoting breast tumor progression82, 83. Interestingly, MMP3 is also required for branching morphogenesis in the mammary gland84–86 and has been shown to be markedly upregulated during pregnancy and involution of the mammary gland87, 88. MMP13 (Collagenase 3) is known to be efficiently activated by MT1-MMP89. Its expression is associated with advanced local invasion in human squamous cell carcinomas of the larynx90, and it was suggested to play a potentially significant role in breast cancer invasion and metastasis91. It is intriguing to speculate that SEPT9 promotes cell invasion at least partly by enhancing the levels of the secreted MMP3 and MMP13. Because SEPT9 acts in concert with other members of the septin family92, and septins other than SEPT9 have been found expressed in FA19, it is likely that SEPT9 acts in concert with other septin filaments that, as a whole, provide a structural network and/or docking sites for MMPs and other proteolytic enzymes at FAs, contributing to extracellular matrix degradation. Indeed, FCF is not SEPT9 specific, but stabilizes higher-order assembly of the septin family as a whole. While FCF off targets effects have been reported in mammalian cells93, it is possible that they are an indirect consequence of alteration of septins dynamics given the larger number of organelles and functions they partake. Further research is needed to clarify whether or not SEPT9, and other septins, also affects the transport of MT1-MMP to FAs and to elucidate the mechanism by which it promotes MMP secretion at FA sites. Elucidating the role of SEPT9 in matrix metalloproteinases dynamics is significant since these proteins are required for the initial metastatic steps and have been proposed as pharmacological targets for anti-metastatic therapy94, 95.
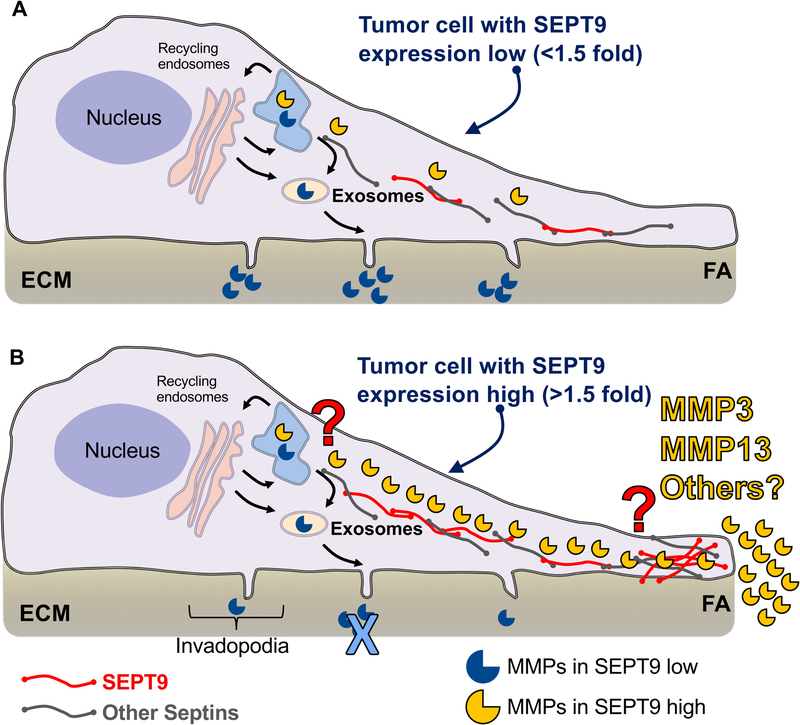
Collectively, previous studies linking SEPT9, FAs, and microtubules, together with the data presented here, imply that the interaction of SEPT9 with the cytoskeleton promotes both trafficking and secretion of MMPs near FAs, thus enhancing migration and invasion of breast cancer epithelial cells. In our model, tumor cells without SEPT9 overexpression (Figure 8A) (i.e. lower than 1.5-fold the mean expression as suggested by the results shown in Figure 3D) degrade the ECM mainly through mature ventral invadopodia projections which have been shown to function as regulatory structures for the secretion of MMP2 and MMP935, 37–42. SEPT9 overexpression in cancer cells (Figure 8B) promotes the assembly of a septin network that includes SEPT9 and likely other septin members, which promotes the delivery of MMP3, MMP13, and possibly other proteases to the plasma membrane at FAs. As a result, ECM degradation is highly enhanced at FAs. The molecular mechanisms by which this process is regulated, the key proteins responsible for the delivery of proteases to FAs, and whether proteolytic enzymes and MMPs other than MMP3 and MMP13 are involved in this process remain to be elucidated.
Figure 8: General model of functional consequences of SEPT9 overexpression in breast cancer cells.
A. In the absence of SEPT9 upregulation, degradation of the ECM in cancer cells occurs at invadopodia (protrusions that assemble at the ventral side of cells). After synthesis, MMPs - mainly MMP2 and MMP9 (open mouth blue circles) - are directed to the endoplasmic reticulum (ER, light blue structure at the top of the cell) and after Golgi processing (pink large vesicular structures), MMPs are delivered to the plasma membrane primarily via exosome secretion. B. In SEPT9-overexpressing cells, septin filaments (red = SEPT9 and gray = other septins) assemble into a network directed to FAs. The septin network promotes the delivery of MMP3 and MMP13 (open mouth yellow circles) to FAs, which represents the main ECM degradation pathway in these cells. The mechanisms by which SEPT9 and possibly other septins promote MMP3 and MMP13 secretion at FAs remains to be elucidated (red question mark). Likewise, whether SEPT9 promotes the secretion of MMPs or ECM-degrading enzymes other than MMP3 and MMP13 at FAs requires further investigation.
Protease secretion at invadopodia is regulated by cortactin, which was proposed to link vesicular trafficking and dynamic branched actin assembly37. Phosphorylation of cortactin was shown to lead to activation of the actin binding protein, cofilin, which induces F-actin polymerization in the invadopodium core and consequently MMP recruitment70, 96. F-actin polymerization at FAs is directly driven by SEPT919, which was shown to bind F-actin at sites that overlap with regions involved in the binding of cofilin75. It is noteworthy that invadopodia assemble at the proximal tip of FAs69, and the later maturation of invadopodia involves transition of FA proteins to invadopodia97–99. We therefore propose that SEPT9-mediated actin polymerization and phosphoinositol mobilization at nascent FAs enhances the formation of invadopodium precursor assembly leading to MMP secretion and activation at FAs. Thus, FAs serve as sites for enhanced vesicular trafficking, protease secretion, and ECM degradation in SEPT9 overexpressing cells.
Materials and Methods
Cell lines
MCF7 clones overexpressing GFP chimera SEPT9_v1, SEPT9_v2, and SEPT9_v3 isoforms were previously described14. MDA-MB-231 and T47D cells (DTP/DCTD/NCI Tumor Repository) expressing the same GFP-fused SEPT9 isoform constructs were generated using the approach previously described for MCF7 cells5. All cell lines were maintained at low, post-G418 selection passage, and cultured only for up to one month; all lines were grown in low-glucose DMEM supplemented with 8% fetal bovine serum and incubated in 5% CO2/37°C. FACS for the removal of GFP negative cells was performed using live MCF7 clones. About 5×106 cells were trypsinized, washed in 2ml of 0.5%BSA in PBS, re-suspended in 0.5ml of 0.5%BSA in PBS, and then filtered using a 40μm cell strainer (Falcon Corning, Corning NY). GFP negative cells were used as a control and to set the threshold of positive intensities. FACS was performed using the BD FACSAria II (BD Biosciences, San Jose, CA) equipped with a 100μm nozzle and a pressure of 20psi at 4°C. Cells were sorted in 5ml of complete media and immediately plated overnight to allow recovery.
RNA isolation and cDNA synthesis
Low passage MCF7_SEPT9_v1, MCF7_SEPT9_v2, and MCF7_SEPT9_v3-overexpressing cell lines and the MCF7 control cells (MCF7_c) were placed in culture simultaneously and split to establish three biological replicates; the plates were taken from culture when they reached 70% confluence. The cells were then lysed directly on the plate with Qiazol lysis reagent (Qiagen, Valencia, CA) and placed at −80°C until all samples were ready for RNA extraction. Total RNA was isolated using the miRNeasy kit (Qiagen, Valencia, CA) and assessed using an Agilent Bioanalyzer. All RNA had an RNA integrity number (RIN) score of 10.
Library Preparation and RNA-sequencing
A total of 12 samples were used for RNA-Seq analysis (three biological replicates from each of the MCF-7 isoforms and control cells). Sequencing libraries for the Illumina 2500 platform were created from the polyadenylated fraction of RNA from each cell line. mRNA was then isolated with Dyna1 oligo-dT beads (Invitrogen, Carlsbad CA) from 10μg of total RNA. The mRNA was randomly fragmented using the RNA fragmentation kit from Ambion. First-strand cDNA synthesis was performed using random primers and SuperScriptII reverse-transcriptase (Invitrogen, Carlsbad CA). This was followed by second-strand cDNA synthesis using DNA Polymerase I and RNase H (Invitrogen, Carlsbad CA). The Illumina adaptor was ligated to the ends of the double-stranded cDNA fragments and a 200bp size-selection of the final product was performed by gel-excision, following the Illumina-recommended protocol. Template molecules of 200bp cDNA with the adaptor attached were enriched by PCR to create the final library. Libraries were pooled and sequenced in two lanes on the Illumina HiSeq (2500) Sequencing System (Illumina, San Diego, CA) using 150bp reads at the New York Genome Center (New York, NY). Sequences in fastq format as well as normalized data files have been deposited into Gene Expression Omnibus (GEO)100 (Project ID# GSE119449 entitled: ”Septin 9 over expression in MCF7”).
RNA-sequencing analysis and identification of candidate genes
We generated a total of 4.3×107 reads, an average of ~36 million reads per library (Supplementary Table 1 and Supplementary Figure 1B), of which 92.3–93.8% could be uniquely mapped to the NCBI Build37/hg19 of the human reference genome. To rule out 3’ bias of poly A+ libraries, we plotted the coverage across the gene body (5’ -> 3’) in all cell lines analyzed by RNA-Seq. We determined that the highest percentage of reads was observed between the 25th and 85th percentile of the gene bodies, therefore excluding bias towards the 5’ end of the mRNA sequences and thus excluding technical confounding factors (Supplementary Figure 1C). On average 88.38% of the reads mapped to coding sequences (CDS) (87–90%) (Supplementary Figure 1D). We found, across all samples analyzed, that 16,743 genes had five or more reads corresponding to 80.93% of the MCF7 genome being transcribed.
Pass filter sequences were aligned to the NCBI Build 37hg19 reference genome. Alignments were then referenced against an annotation database that combines ENSEMBL (release 71), GENBANK/NCBI (release 196), and REFSEQ (release 60). All statistical methods, and data analysis were conducted using the R statistical software101. For QC of sequencing results we used the RSeQC package102, uniquely aligned sequences were analyzed using the DESeq package, and to illustrate the expression differences for each gene and to plot the average of the normalized counts per million (CPM) across biological replicates and the standard error for each condition, we used the ggplot2 package103. Heatmaps were created using the heatmap.3 function in the GMD package104.
For all analyses, a P-value <0.05 was considered significant and all P-values were adjusted for multiple testing using Benjamini-Hochberg correction where a cutoff FDR value <0.1 was considered significant.
Gene ontology was determined using g:profiler including, as a background, all genes across the three MCF7 clones with >5 reads and FDR<0.5, adjusted for multiple testing using the Benjamini & Hochberg method. Enrichment maps were visualized using The Enrichment Map Cytoscape Plugin. Gene sets comprising hallmarks of apical surface (M5916), hallmarks of protein secretion (M5910), EGD1 signature (Phospholipids as signaling intermediaries, M10579), and members of the TGFβ pathway (M18933) were obtained from the Molecular Signatures Database (MSigDB)105. Pathway analysis was performed using the web interphase Reactome.org106.
siRNA transfections
For silencing of SEPT9, we used the siRNA SMARTpool (Dharmacon Cat #L-006373, Lafayette USA). ON-TARGET SEPT9 oligos were used for gene specific downregulation and same MCF7 or MDA-MB-231 cells transfected with the ON-TARGET Non-targeting (scramble) siRNA Control Pools were used as a reference. siRNA pools were resuspended using manufacturer procedures in RNase-free 1x siRNA Buffer at a final concentration of 20 mM. Cells were transfected using DharmaFECT Transfection Reagent according to manufacturer’s instructions. After transfections, cells were allowed to grow for 48 hours before analysis of specific endpoints.
Real-Time qRT-PCR
Total RNA was isolated from cells as reported previously8. cDNA was reverse-transcribed from 5μg of total RNA using random primers and SuperScript II Reverse Transcriptase (Invitrogen). Primer3 software107 was used to design SEPT9 primers (F- 5’- CCCCAGAAGGAATTTGATGA −3’; R- 5’- TGGTACCCCACTTGGTCTTC −3’) along with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers (F- 5’-CCACATCGCTCAGACACCAT-3’; R- 5’-CCAGGCGCCCAATACG-3’). Primers for MMP1, MMP2, MMP3, MMP9, and MMP13 were selected from KiCqStart Primers (MilliporeSigma, Burlington, MA) by choosing those mapping to exons spanning all known isoforms for each gene: H_MMP1_2, H_MMP2_3, H_MMP3_2, H_MMP9_3, H_MMP13_2. H_GAPDH_1 was also obtained from Sigma and tested together with the custom-designed GAPDH primers described above. Due to the low expression levels of MMP2 and MMP9, a TaqMan assay was also performed for these two genes using MMP2 - Hs00234422_m1, MMP9 - Hs00234579_m1, GAPDH - Hs99999905_m1 (Roche LifeScience) as we previously described8. Predesigned TaqMan probes and primers for VCAN (Hs00171642_m1), TBX15 (Hs00537087_m1), SULF1 (Hs00290918_m1), GPC6 (Hs00170677_m1), KRT20 (Hs00300643_m1), FGF21 (Hs00173927_m1) and GAPDH (Hs99999905_m1) were obtained from ThermoFisher.
Real-Time qRT-PCR was performed using Applied Biosystems Fast SYBR Green Master Mix and TaqMan Fast Advanced Master Mix with the StepOnePlus Real-Time PCR System (Life Technologies Corp., Carlsbad, CA, USA). Correlation analyses were performed by computing the normalized read counts for each analyzed MCF7 line with the levels obtained by TaqMan assay. The Pearson correlation coefficient (r) and the p value were determined in each case.
The Cancer Genome Atlas (TCGA) mining and statistical analysis of MMP expression in SEPT9 overexpressing human breast cancer samples
We mined the provisional dataset of TCGA using the cBIOportal interface108 to download the normalized expression level of SEPT9 and all known MMPs. MMP2, MMP9, MMP11, MMP14, MMP15, MMP19, MMP24, and MMP25 had expression data which were detected in all 1093 breast cancer patients for whom global transcriptomic profiling was available; these expression data were therefore used for further analysis. Next, the cases were assigned to group A or group B based on SEPT9 mRNA expression level. Group A included cases (n=210) in which SEPT9 expression level was 1.5 standard deviations higher than the mean based on the z-Score (RNA Seq V2 RSEM) compared to the expression distribution of SEPT9 in tumors that are diploid for this gene; group B (n=883) included all the remaining cases. Group A and group B were compared for the expression of the MMPs listed above by multiple testing analysis (ANOVA).
Western Blotting
Cell lysates were prepared by washing cells in cold PBS followed by addition of SDS-PAGE sample buffer containing Protease Inhibitor Cocktail (SIGMA Cat#P2714–1BTL). Cells were transferred to a 1.5ml tube, sonicated, and heated at 95°C for 5 min. Proteins were separated by electrophoresis in NuPAGE 10% Bis-Tris gels and transferred to PVDF membranes (Milipore- Immobilon, FL, Cat #IPFL00010- pore size: 0.45μm) by electroblotting. Membranes were blocked in PBS+ 5% milk powder and incubated with primary antibodies and anti-SEPT9 rabbit antibody (Proteintech Cat#10769–1-AP) and α-TUBULIN mouse antibody (Sigma Cat #T9026) overnight at 4°C. The other antibodies used were MMP3 rabbit antibody (Arigo Biolaboratories Cat#ARG55262), CD9 mouse antibody (Proteintech Cat#60232–1-Ig), or Actin mouse antibody (Proteintech Cat#66009–1-Ig). After three washes in PBS-T, membranes were incubated with secondary antibodies (Mouse 680 and Rabbit 800 [LI-COR Biosciences]) for one hour at room temperature, washed three times in PBS-T, and scanned using a high-sensitivity Odyssey Infrared Imaging System (Li-COR Biosciences). α-TUBULIN was used as a loading control. ImageJ was used for quantification of bands intensities.
Migration assay and colony size assessment
Transwell inserts (8μM; Millipore, Billerica, MA, USA) were coated with 25μg/mL type I rat tail collagen (BD Biosciences Franklin Lakes, NJ, USA). Cells (5 × 104 in DMEM 0.3% BSA) were added in triplicate for each sample to the transwell inserts and allowed to migrate for 10 hours at 37°C. Nonmigratory cells were removed and filters were fixed in 3.7% formaldehyde/PBS for 15 minutes and stained with 0.2% crystal violet dye for 10 minutes. Migrated cells were counted and averaged from 10 fields of view per filter at 20 × magnification using an Axio Observer.A1 inverted microscope (Carl Zeiss MicroImaging, Inc.). Two independent biological replicate assays were analyzed.
Gelatin coating and degradation assay
The ability of MDA-MB-231 cells to degrade gelatin was assessed by plating cells onto fluorescently-labeled gelatin. Gelatin MatTek dishes were prepared as described previously109. Briefly, acid-washed MatTek dishes were coated with 50μg/ml poly-L-lysine followed by 0.2% gelatin labeled with either Alexa Fluor 405 dye (Life Technologies, Cat #A30000) or with Oregon Green® 488 (Molecular probes, Cat #G-13186) and sequentially treated with 0.5% glutaraldehyde for 15 minutes at room temperature and then with 5mg/ml NaBH4. Cells (2×105) were seeded in complete medium and allowed to attach and degrade for 4–6 hours at 37°C in 5% CO2. Cells were then fixed with 200μl of 4% PFA for 15 minutes, permeabilized with 0.1% Triton-X for 10 minutes, and counterstained with Alexa Fluor® 647 Phalloidin (ThermoFisher Scientific Cat #A22287).
Fixed cells were imaged in 1x PBS at room temperature. Images were acquired on a DeltaVision Core Microscope (Applied Precision, Issaquah, WA) using a CoolSnap HQ2 camera (Photometrics, Tucson, AZ), 60X/numerical aperture (NA) 1.4 oil objective, standard four-channel filter set, and softWoRx software. Custom ImageJ macros were used for the following image processing: to split and save the individual channels of the DeltaVision .dv files as .tif files; to find and save the best focus slice from the individual z-stacks; and to correct the uneven focus in the degradation channel. Because the number of cells in each field varied and because MDA-MB-231 cells tend to move, three panels of 5×4 fields (each field 110×110 μm2) were imaged for each clone. Matrix degradation activity was measured by the local loss of gelatin fluorescence. Gelatin degradation areas (μm2) were measured as the total area covered by degradation holes/field in thresholded images using the Analyze Particles tool in Fiji (National Institutes of Health, Bethesda, MD), and cell area (μm2) was measured as the area covered by cells/field in thresholded images. For each panel, total gelatin degradation area (sum of degradation areas in all 20 fields) was normalized by total cell area (sum of cell areas in all 20 fields) to give degradation area as % of cell area.
Immunofluorescence microscopy
Cells were seeded on MatTek dishes coated with 0.2% gelatin and then fixed and permeabilized as described above. Fixed cells were counterstained with Alexa Fluor® 647 Phalloidin (ThermoFisher Scientific Cat#A22287) or with Alexa Fluor® 633 Phalloidin (ThermoFisher Scientific Cat #A22284), for TIRF microscopy. Antibodies used for immunofluorescence were: anti-VINCULIN (Sigma Cat# V9131) 1:400, anti-SEPT9 (Proteintech Cat#10769–1-AP) 1:200, and anti-MMP3 (Arigo Biolaboratories Cat#ARG55262) 1:200. All Alexa Fluor secondary antibodies were from Molecular Probes (Life Technologies, Carlsbad, CA). All primary and secondary antibodies were diluted in blocking buffer (1% goat serum + 1% BSA in PBS). Cells were imaged on a DeltaVision Core Microscope (Applied Precision, Issaquah, WA) using a CoolSnap HQ2 camera (Photometrics, Tucson, AZ), 60X/numerical aperture (NA) 1.4 oil objective, standard four-channel filter set, and softWoRx software.
Focal adhesions and MMP3 analysis
Images (tif) were used for analysis of FAs and the MMP3 signal. FAs were analyzed from the vinculin images, and MMP3 signal from the MMP3 images, using the Focal Adhesion Analysis server (http://faas.bme.unc.edu/) [Berginski ME, Gomez SM. (2013) The Focal Adhesion Analysis Server: a web tool for analyzing focal adhesion dynamics [v1; ref status: indexed, http://f1000r.es/yc] F1000Research 2013, 2:68 (doi: 10.3410/f1000research.2-68.v1)]. Phalloidin images were used as reference for the cell boundaries.
Protease Array assay
Secreted protease profiling was performed with Human Proteome Profiler Protease Array from R & D Systems (Minneapolis, Minnesota, Cat#ARY021B), according to the manufacturer’s instructions. To generate conditioned cell culture medium for the protease arrays, 4×105 MDA-MB-231_SEPT9_v3 (overexpressing SEPT9_v3) cells and MDA-MB-231 cells transfected with SEPT9 siRNA, were seeded in 60 cm plates (4×105 cells) and grown in 3ml DMEM supplemented with 8% FBS. After 24 hours the media was changed to DMEM supplemented with 0.5% FBS and cells continued to grow for an additional 24 hours. The conditioned media was collected, filtered through a 22μm syringe filter unit (Millex-GP), and kept at −80°C until use. The protease array membranes were incubated for one hour in Assay Buffer 6. One mL of conditioned medium was combined with 500μL of Buffer 6 and 15μL of Detection Antibody Cocktail and incubated for one hour. Assay Buffer 6 was aspirated from the wells containing the membranes. The prepared sample/antibody mixtures were added to the membranes and incubated overnight. Membranes were then washed with 1× Wash Buffer 3 times and incubated with diluted streptavidin-HRP for 30 minutes. Membranes were washed and treated with Chemi Reagent Mix for 1 minute. The chemiluminescent signals of proteases were measured with a LI-COR Odyssey Fc scanner. ImageJ was used to determine the intensities of the signals. Background levels were subtracted from the protease signal levels and the signals were normalized by the intensities of the reference signals in each blot.
Isolation of Exosomes from cell culture medium
To generate conditioned cell culture medium, 2.5×105 cells were seeded on 10cm plates coated with 0.2% gelatin and grown in 10ml DMEM supplemented with 10% FBS. After 24 hours cells were washed and media changed to DMEM supplemented with 0.5% exosome-free fetal bovine serum (prepared by centrifugation at 120,000 × g for 24 hours). Cells continued to grow for an additional 72 hours. The conditioned media was collected and cells were counted for normalization.
For EV Analysis:
Normalized volumes of conditioned media (~6ml) were centrifuged at 2000 × g for 20 minutes to eliminate cells, and apoptotic bodies. The supernatants were then mixed with 3ml of Total Exosome Isolation Reagent (ThermoFisher Scientific #4478359) and mixed well by vortexing. The mixtures were incubated at 4°C overnight and then centrifuged at 10,000 × g for one hour at 4°C. The exosomes were resuspended in 1ml of 1x PBS and were analyzed by laser scattering microscopy or by dynamic light scattering48, 49.
For Western Blot analysis:
Cell lysates were prepared for WB as described above.
Normalized volumes of conditioned media (~20ml) were centrifuged at 2,000 × g for 20 minutes to eliminate cells, and apoptotic bodies. The supernatants were then centrifuged at 100,000 × g for 70 min at 4°C. Next, supernatants were concentrated ~80 times using Millipore Amicon Ultra-4 30k (Cat #UFC803024) and pellets, which contain the EVs, were resuspended in 50μl of RIPA buffer (made from 10x SIGMA #20–188) containing Protease Inhibitor Cocktail (SIGMA #P2714–1BTL). SDS-PAGE sample buffer was added to the samples and they were heated at 95°C for 5 min.
Supplementary Material
Acknowledgements
We would like to thank the following shared resources at the Albert Einstein College of Medicine: Molecular Cytogenetic Core, in particular Dr. Jidong Shan; and the Analytical Imaging Facility in particular Dr. Vera DesMarais. Research reported in this publication was supported by the Albert Einstein Cancer Center support grant of the National Institutes of Health under award number P30CA013330 and CA150344.
Footnotes
Conflict of interest statement
The authors have no conflict of interest to declare.
References
- 1.Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nature reviews Molecular cell biology 2012; 13: 183–194. [DOI] [PubMed] [Google Scholar]
- 2.Osaka M, Rowley JD, Zeleznik-Le NJ. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25). Proceedings of the National Academy of Sciences of the United States of America 1999; 96: 6428–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerveira N, Bizarro S, Teixeira MR. MLL-SEPTIN gene fusions in hematological malignancies. Biological chemistry 2011; 392: 713–724. [DOI] [PubMed] [Google Scholar]
- 4.McDade SS, Hall PA, Russell SE. Translational control of SEPT9 isoforms is perturbed in disease. Human molecular genetics 2007; 16: 742–752. [DOI] [PubMed] [Google Scholar]
- 5.Connolly D, Hoang HG, Adler E, Tazearslan C, Simmons N, Bernard VV et al. Septin 9 amplification and isoform-specific expression in peritumoral and tumor breast tissue. Biological chemistry 2014; 395: 157–167. [DOI] [PubMed] [Google Scholar]
- 6.Russell SE, McIlhatton MA, Burrows JF, Donaghy PG, Chanduloy S, Petty EM et al. Isolation and mapping of a human septin gene to a region on chromosome 17q, commonly deleted in sporadic epithelial ovarian tumors. Cancer research 2000; 60: 4729–4734. [PubMed] [Google Scholar]
- 7.Sorensen AB, Warming S, Fuchtbauer EM, Pedersen FS. Alternative splicing, expression, and gene structure of the septin-like putative proto-oncogene Sint1. Gene 2002; 285: 79–89. [DOI] [PubMed] [Google Scholar]
- 8.Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T et al. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer research 2003; 63: 2179–2187. [PubMed] [Google Scholar]
- 9.Montagna C, Bejerano-Sagie M, Zechmeister JR. Mammalian septins in health and disease. Journal of Research and Reports in Biochemistry 2015; 2015:5: 59–72. [Google Scholar]
- 10.Angelis D, Spiliotis ET. Septin Mutations in Human Cancers. Front Cell Dev Biol 2016; 4: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015; 163: 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez ME, Peterson EA, Privette LM, Loffreda-Wren JL, Kalikin LM, Petty EM. High SEPT9_v1 expression in human breast cancer cells is associated with oncogenic phenotypes. Cancer research 2007; 67: 8554–8564. [DOI] [PubMed] [Google Scholar]
- 13.Verdier-Pinard P, Salaun D, Bouguenina H, Shimada S, Pophillat M, Audebert S et al. Septin 9_i2 is downregulated in tumors, impairs cancer cell migration and alters subnuclear actin filaments. Sci Rep 2017; 7: 44976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly D, Yang Z, Castaldi M, Simmons N, Oktay MH, Coniglio S et al. Septin 9 isoform expression, localization and epigenetic changes during human and mouse breast cancer progression. Breast cancer research : BCR 2011; 13: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amir S, Golan M, Mabjeesh NJ. Targeted knockdown of SEPT9_v1 inhibits tumor growth and angiogenesis of human prostate cancer cells concomitant with disruption of hypoxia-inducible factor-1 pathway. Molecular cancer research : MCR 2010; 8: 643–652. [DOI] [PubMed] [Google Scholar]
- 16.Chacko AD, McDade SS, Chanduloy S, Church SW, Kennedy R, Price J et al. Expression of the SEPT9_i4 isoform confers resistance to microtubule-interacting drugs. Cellular oncology 2012; 35: 85–93. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez ME, Makarova O, Peterson EA, Privette LM, Petty EM. Up-regulation of SEPT9_v1 stabilizes c-Jun-N-terminal kinase and contributes to its pro-proliferative activity in mammary epithelial cells. Cellular signalling 2009; 21: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chacko AD, Hyland PL, McDade SS, Hamilton PW, Russell SH, Hall PA. SEPT9_v4 expression induces morphological change, increased motility and disturbed polarity. The Journal of pathology 2005; 206: 458–465. [DOI] [PubMed] [Google Scholar]
- 19.Dolat L, Hunyara JL, Bowen JR, Karasmanis EP, Elgawly M, Galkin VE et al. Septins promote stress fiber-mediated maturation of focal adhesions and renal epithelial motility. The Journal of cell biology 2014; 207: 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.TCGA. (TCGA) The Cancer Genome Atlas, 2014. [Google Scholar]
- 21.Golan M, Mabjeesh NJ. SEPT9_i1 is required for the association between HIF-1alpha and importin-alpha to promote efficient nuclear translocation. Cell cycle 2013; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vardi-Oknin D, Golan M, Mabjeesh NJ. Forchlorfenuron disrupts SEPT9_i1 filaments and inhibits HIF-1. PloS one 2013; 8: e73179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amir S, Wang R, Simons JW, Mabjeesh NJ. SEPT9_v1 up-regulates hypoxia-inducible factor 1 by preventing its RACK1-mediated degradation. The Journal of biological chemistry 2009; 284: 11142–11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall PA, Jung K, Hillan KJ, Russell SE. Expression profiling the human septin gene family. The Journal of pathology 2005; 206: 269–278. [DOI] [PubMed] [Google Scholar]
- 25.Wolfgang CD, Essand M, Vincent JJ, Lee B, Pastan I. TARP: a nuclear protein expressed in prostate and breast cancer cells derived from an alternate reading frame of the T cell receptor gamma chain locus. Proceedings of the National Academy of Sciences of the United States of America 2000; 97: 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda K, Nakanishi N, Rogers BL, Haser WG, Shitara K, Yoshida H et al. Expression of the T-cell receptor gamma-chain gene products on the surface of peripheral T cells and T-cell blasts generated by allogeneic mixed lymphocyte reaction. Proceedings of the National Academy of Sciences of the United States of America 1987; 84: 6536–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfgang CD, Essand M, Lee B, Pastan I. T-cell receptor gamma chain alternate reading frame protein (TARP) expression in prostate cancer cells leads to an increased growth rate and induction of caveolins and amphiregulin. Cancer research 2001; 61: 8122–8126. [PubMed] [Google Scholar]
- 28.Wu XR, Kong XP, Pellicer A, Kreibich G, Sun TT. Uroplakins in urothelial biology, function, and disease. Kidney Int 2009; 75: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee G Uroplakins in the lower urinary tract. Int Neurourol J 2011; 15: 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nile AH, Tripathi A, Yuan P, Mousley CJ, Suresh S, Wallace IM et al. PITPs as targets for selectively interfering with phosphoinositide signaling in cells. Nat Chem Biol 2014; 10: 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagano T, Takehara S, Takahashi M, Aizawa S, Yamamoto A. Shisa2 promotes the maturation of somitic precursors and transition to the segmental fate in Xenopus embryos. Development 2006; 133: 4643–4654. [DOI] [PubMed] [Google Scholar]
- 32.Tai KY, Shieh YS, Lee CS, Shiah SG, Wu CW. Axl promotes cell invasion by inducing MMP-9 activity through activation of NF-kappaB and Brg-1. Oncogene 2008; 27: 4044–4055. [DOI] [PubMed] [Google Scholar]
- 33.Byers JR. Metamorphosis of the perirectal malpighian tubules in the mealworm Tenebrio molitor L. (Coleoptera, Tenebrionidae). II. Ultrastructure and role of autophagic vacuoles. Can J Zool 1971; 49: 1185–1191. [DOI] [PubMed] [Google Scholar]
- 34.Weidmann MD, Surve CR, Eddy RJ, Chen X, Gertler FB, Sharma VP et al. Mena(INV) dysregulates cortactin phosphorylation to promote invadopodium maturation. Sci Rep 2016; 6: 36142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou ZN, Sharma VP, Beaty BT, Roh-Johnson M, Peterson EA, Van Rooijen N et al. Autocrine HBEGF expression promotes breast cancer intravasation, metastasis and macrophage-independent invasion in vivo. Oncogene 2014; 33: 3784–3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacob A, Jing J, Lee J, Schedin P, Gilbert SM, Peden AA et al. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. Journal of cell science 2013; 126: 4647–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark ES, Weaver AM. A new role for cortactin in invadopodia: regulation of protease secretion. European journal of cell biology 2008; 87: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monsky WL, Kelly T, Lin CY, Yeh Y, Stetler-Stevenson WG, Mueller SC et al. Binding and localization of M(r) 72,000 matrix metalloproteinase at cell surface invadopodia. Cancer research 1993; 53: 3159–3164. [PubMed] [Google Scholar]
- 39.Nakahara H, Howard L, Thompson EW, Sato H, Seiki M, Yeh Y et al. Transmembrane/cytoplasmic domain-mediated membrane type 1-matrix metalloprotease docking to invadopodia is required for cell invasion. Proceedings of the National Academy of Sciences of the United States of America 1997; 94: 7959–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bourguignon LY, Gunja-Smith Z, Iida N, Zhu HB, Young LJ, Muller WJ et al. CD44v(3,8–10) is involved in cytoskeleton-mediated tumor cell migration and matrix metalloproteinase (MMP-9) association in metastatic breast cancer cells. Journal of cellular physiology 1998; 176: 206–215. [DOI] [PubMed] [Google Scholar]
- 41.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer research 2006; 66: 3034–3043. [DOI] [PubMed] [Google Scholar]
- 42.Poincloux R, Lizarraga F, Chavrier P. Matrix invasion by tumour cells: a focus on MT1-MMP trafficking to invadopodia. Journal of cell science 2009; 122: 3015–3024. [DOI] [PubMed] [Google Scholar]
- 43.Montagna C, Bejerano-Sagie M, Zechmeister J. Mammalian septins in health and disease. Research and Reports in Biochemistry 2015; 2015:5: 59–72. [Google Scholar]
- 44.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 2010; 141: 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016; 164: 1226–1232. [DOI] [PubMed] [Google Scholar]
- 46.Shimoda M, Khokha R. Metalloproteinases in extracellular vesicles. Biochimica et biophysica acta 2017; 1864: 1989–2000. [DOI] [PubMed] [Google Scholar]
- 47.Filipe V, Hawe A, Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res 2010; 27: 796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehdiani A, Maier A, Pinto A, Barth M, Akhyari P, Lichtenberg A. An innovative method for exosome quantification and size measurement. J Vis Exp 2015: 50974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PloS one 2017; 12: e0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int 2012; 82: 1024–1032. [DOI] [PubMed] [Google Scholar]
- 51.Nabeshima K, Inoue T, Shimao Y, Sameshima T. Matrix metalloproteinases in tumor invasion: role for cell migration. Pathol Int 2002; 52: 255–264. [DOI] [PubMed] [Google Scholar]
- 52.Stehbens SJ, Paszek M, Pemble H, Ettinger A, Gierke S, Wittmann T. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nature cell biology 2014; 16: 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt K, Nichols BJ. Functional interdependence between septin and actin cytoskeleton. BMC Cell Biol 2004; 5: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagata K, Kawajiri A, Matsui S, Takagishi M, Shiromizu T, Saitoh N et al. Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules. The Journal of biological chemistry 2003; 278: 18538–18543. [DOI] [PubMed] [Google Scholar]
- 55.Hu J, Bai X, Bowen JR, Dolat L, Korobova F, Yu W et al. Septin-driven coordination of actin and microtubule remodeling regulates the collateral branching of axons. Current biology : CB 2012; 22: 1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sellin ME, Stenmark S, Gullberg M. Mammalian SEPT9 isoforms direct microtubule-dependent arrangements of septin core heteromers. Molecular biology of the cell 2012; 23: 4242–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spiliotis ET, Hunt SJ, Hu Q, Kinoshita M, Nelson WJ. Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. The Journal of cell biology 2008; 180: 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bai X, Karasmanis EP, Spiliotis ET. Septin 9 interacts with kinesin KIF17 and interferes with the mechanism of NMDA receptor cargo binding and transport. Molecular biology of the cell 2016; 27: 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, Nagata K. SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling. Human mutation 2007; 28: 1005–1013. [DOI] [PubMed] [Google Scholar]
- 60.Tokhtaeva E, Capri J, Marcus EA, Whitelegge JP, Khuzakhmetova V, Bukharaeva E et al. Septin dynamics are essential for exocytosis. The Journal of biological chemistry 2015; 290: 5280–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott M, Hyland PL, McGregor G, Hillan KJ, Russell SE, Hall PA. Multimodality expression profiling shows SEPT9 to be overexpressed in a wide range of human tumours. Oncogene 2005; 24: 4688–4700. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Wang H, Li J, Entenberg D, Xue A, Wang W et al. Direct visualization of the phenotype of hypoxic tumor cells at single cell resolution in vivo using a new hypoxia probe. Intravital 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hecht M, Rosler R, Wiese S, Johnsson N, Gronemeyer T. An Interaction Network of the Human SEPT9 Established by Quantitative Mass Spectrometry. G3 (Bethesda) 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang J, Kong C, Xie H, McPherson PS, Grinstein S, Trimble WS. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Current biology : CB 1999; 9: 1458–1467. [DOI] [PubMed] [Google Scholar]
- 65.Barral Y, Kinoshita M. Structural insights shed light onto septin assemblies and function. Current opinion in cell biology 2008; 20: 12–18. [DOI] [PubMed] [Google Scholar]
- 66.Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Buvelot Frei S, Snapp EL et al. A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. Elife 2014; 3: e01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spiliotis ET, Nelson WJ. Here come the septins: novel polymers that coordinate intracellular functions and organization. Journal of cell science 2006; 119: 4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akil A, Peng J, Omrane M, Gondeau C, Desterke C, Marin M et al. Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV. Nature communications 2016; 7: 12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma VP, Eddy R, Entenberg D, Kai M, Gertler FB, Condeelis J. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Current biology : CB 2013; 23: 2079–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eddy RJ, Weidmann MD, Sharma VP, Condeelis JS. Tumor Cell Invadopodia: Invasive Protrusions that Orchestrate Metastasis. Trends in cell biology 2017; 27: 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valenzuela-Iglesias A, Sharma VP, Beaty BT, Ding Z, Gutierrez-Millan LE, Roy P et al. Profilin1 regulates invadopodium maturation in human breast cancer cells. European journal of cell biology 2015; 94: 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beaty BT, Sharma VP, Bravo-Cordero JJ, Simpson MA, Eddy RJ, Koleske AJ et al. beta1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Molecular biology of the cell 2013; 24: 1661–1675, S1661–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, McNiven MA. Invasive matrix degradation at focal adhesions occurs via protease recruitment by a FAK-p130Cas complex. The Journal of cell biology 2012; 196: 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Noordstra I, Akhmanova A. Linking cortical microtubule attachment and exocytosis. F1000Res 2017; 6: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith C, Dolat L, Angelis D, Forgacs E, Spiliotis ET, Galkin VE. Septin 9 Exhibits Polymorphic Binding to F-Actin and Inhibits Myosin and Cofilin Activity. J Mol Biol 2015; 427: 3273–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bai X, Bowen JR, Knox TK, Zhou K, Pendziwiat M, Kuhlenbaumer G et al. Novel septin 9 repeat motifs altered in neuralgic amyotrophy bind and bundle microtubules. The Journal of cell biology 2013; 203: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nistico P, Bissell MJ, Radisky DC. Epithelial-mesenchymal transition: general principles and pathological relevance with special emphasis on the role of matrix metalloproteinases. Cold Spring Harbor perspectives in biology 2012; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen QK, Lee K, Radisky DC, Nelson CM. Extracellular matrix proteins regulate epithelial-mesenchymal transition in mammary epithelial cells. Differentiation 2013; 86: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell 1999; 98: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McGowan PM, Duffy MJ. Matrix metalloproteinase expression and outcome in patients with breast cancer: analysis of a published database. Ann Oncol 2008; 19: 1566–1572. [DOI] [PubMed] [Google Scholar]
- 81.Cichon MA, Nelson CM, Radisky DC. Regulation of epithelial-mesenchymal transition in breast cancer cells by cell contact and adhesion. Cancer Inform 2015; 14: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Flores-Pliego A, Espejel-Nunez A, Castillo-Castrejon M, Meraz-Cruz N, Beltran-Montoya J, Zaga-Clavellina V et al. Matrix Metalloproteinase-3 (MMP-3) Is an Endogenous Activator of the MMP-9 Secreted by Placental Leukocytes: Implication in Human Labor. PloS one 2015; 10: e0145366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. The Journal of biological chemistry 1999; 274: 13066–13076. [DOI] [PubMed] [Google Scholar]
- 84.Simian M, Hirai Y, Navre M, Werb Z, Lochter A, Bissell MJ. The interplay of matrix metalloproteinases, morphogens and growth factors is necessary for branching of mammary epithelial cells. Development 2001; 128: 3117–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wiseman BS, Sternlicht MD, Lund LR, Alexander CM, Mott J, Bissell MJ et al. Site-specific inductive and inhibitory activities of MMP-2 and MMP-3 orchestrate mammary gland branching morphogenesis. The Journal of cell biology 2003; 162: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fata JE, Werb Z, Bissell MJ. Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast cancer research : BCR 2004; 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khokha R, Werb Z. Mammary gland reprogramming: metalloproteinases couple form with function. Cold Spring Harbor perspectives in biology 2011; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dickson SR, Warburton MJ. Enhanced synthesis of gelatinase and stromelysin by myoepithelial cells during involution of the rat mammary gland. J Histochem Cytochem 1992; 40: 697–703. [DOI] [PubMed] [Google Scholar]
- 89.Knauper V, Bailey L, Worley JR, Soloway P, Patterson ML, Murphy G. Cellular activation of proMMP-13 by MT1-MMP depends on the C-terminal domain of MMP-13. FEBS letters 2002; 532: 127–130. [DOI] [PubMed] [Google Scholar]
- 90.Cazorla M, Hernandez L, Nadal A, Balbin M, Lopez JM, Vizoso F et al. Collagenase-3 expression is associated with advanced local invasion in human squamous cell carcinomas of the larynx. The Journal of pathology 1998; 186: 144–150. [DOI] [PubMed] [Google Scholar]
- 91.Kotepui M, Punsawad C, Chupeerach C, Songsri A, Charoenkijkajorn L, Petmitr S. Differential expression of matrix metalloproteinase-13 in association with invasion of breast cancer. Contemp Oncol (Pozn) 2016; 20: 225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Neubauer K, Zieger B. The Mammalian Septin Interactome. Front Cell Dev Biol 2017; 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heasley LR, Garcia G 3rd, McMurray MA. Off-target effects of the septin drug forchlorfenuron on nonplant eukaryotes. Eukaryot Cell 2014; 13: 1411–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Winer A, Adams S, Mignatti P. Matrix Metalloproteinase Inhibitors in Cancer Therapy: Turning Past Failures Into Future Successes. Mol Cancer Ther 2018; 17: 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Winer A, Janosky M, Harrison B, Zhong J, Moussai D, Siyah P et al. Inhibition of Breast Cancer Metastasis by Presurgical Treatment with an Oral Matrix Metalloproteinase Inhibitor: A Preclinical Proof-of-Principle Study. Mol Cancer Ther 2016; 15: 2370–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Magalhaes MA, Larson DR, Mader CC, Bravo-Cordero JJ, Gil-Henn H, Oser M et al. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. The Journal of cell biology 2011; 195: 903–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoshino D, Jourquin J, Emmons SW, Miller T, Goldgof M, Costello K et al. Network analysis of the focal adhesion to invadopodia transition identifies a PI3K-PKCalpha invasive signaling axis. Sci Signal 2012; 5: ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan KT, Cortesio CL, Huttenlocher A. FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion. The Journal of cell biology 2009; 185: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Genna A, Lapetina S, Lukic N, Twafra S, Meirson T, Sharma VP et al. Pyk2 and FAK differentially regulate invadopodia formation and function in breast cancer cells. The Journal of cell biology 2018; 217: 375–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research 2002; 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing., 2016. [Google Scholar]
- 102.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 2012; 28: 2184–2185. [DOI] [PubMed] [Google Scholar]
- 103.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics 2015; 31: 2912–2914. [DOI] [PubMed] [Google Scholar]
- 104.Zhao X, Valen E, Parker BJ, Sandelin A. Systematic clustering of transcription start site landscapes. PloS one 2011; 6: e23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015; 1: 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R et al. The Reactome pathway Knowledgebase. Nucleic acids research 2016; 44: D481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in molecular biology 2000; 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 108.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sharma VP, Entenberg D, Condeelis J. High-resolution live-cell imaging and time-lapse microscopy of invadopodium dynamics and tracking analysis. Methods in molecular biology 2013; 1046: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.