Abstract
Background
Epithelial ovarian cancer (EOC) is the fifth most common cause of cancer death in women. Due to a lacking of early detection method, its five-year survival rate is only 30%. Nevertheless, novel biomarkers for diagnosis remain to be discovered. The potential of microRNA signatures in the diagnosis of EOC has been especially described in recent years. In our previous experiments, we identified that circBNC2 was downregulated in EOC specimens, and was associated with advanced tumor stage and lymph node metastasis (LNM) by performing circRNA-sequencing analysis. The aim of this study was to explore the diagnostic value of circBNC2 in patients with epithelial ovarian cancer (EOC).
Methods
Plasma from 249 age and menopause-matched women (83 with EOC; 83 with benign ovarian cyst; 83 were healthy volunteers) was collected prior to surgery. CircBNC2 was analyzed using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Cancer antigen 125 (CA125) and human epididymis protein 4 (HE4) were analyzed using enzyme-linked immunosorbent assay (ELISA). Receiver operating curve (ROC), the area under the curve (AUC), sensitivity and specificity were estimated.
Results
CircBNC2 was downregulated in EOC and had higher ROC AUC in comparing EOC to benign (ROC AUC 0.879, sensitivity 96.4%, specificity 80.7%) or healthy (ROC AUC 0.923, sensitivity 95.2%, specificity 85.5%) cohorts than HE4 (ROC AUC: 0.742, benign cohort; 0.779, healthy cohort) and CA125 (ROC AUC: 0.373, benign cohort; 0.713, healthy cohort). Early stage EOC vs benign (ROC AUC 0.864, sensitivity 92.0%, specificity 80.7%) and healthy (ROC AUC 0.908, sensitivity 92.0%, specificity 85.5%) cohorts could be significantly separated by circBNC2. CircBNC2 performed alike in pre- and postmenopausal women, within EOC compared to the benign or healthy cohort.
Conclusion
CircBNC2 is downregulated in EOC (both in tissue and plasma samples) and might present promising novel biomarker for EOC. Further studies are needed to verify our results.
Impact
CircBNC2 is downregulated in EOC and warrants investigation in a screening study in females at risk for EOC.
Keywords: epithelial ovarian cancer, circular RNA, noncoding RNA, diagnosis, biomarker
Introduction
Ovarian cancer (OC) is the most lethal gynecologic malignancy, accounting for 5% of all female cancer deaths.1,3 The number of new cases of ovarian cancer worldwide was estimated at about 295,400 and the number of deaths was about 184,700 in 2018.2 Epithelial ovarian cancer (EOC) is the most common subtype, accounting for 90% of all OC cases.4 More than 70% of EOC patients are diagnosed with advanced diseases, largely due to lacking of effective detection method.6,7 As a result, the 5-year survival rate for EOC cases is merely 30%.2 Since patients with stage I disease have a cure rate of 93%,3 improved early detection method is a research priority in order to improve the survival outcome of EOC patients.
CA125 has been used in OC diagnosis for 30 years and was the only biomarker for OC to be approved by the US Food and Drug Administration (FDA) before the year 2008.7 Although CA125 is elevated in approximately 80% of EOC patients with advanced diseases,8 it increased in only 50% of patients with stage I disease.9 Additionally, CA125 is also elevated in other gynecological cancers,10 benign gynecological diseases and non-gynecological malignancies.11 He4 is the second FDA-approved biomarker for EOC at the year 2008.7 Unlike CA125, HE4 does not elevate in benign gynecological diseases (including endometriosis).12 However, conflicts also arise on the relatively low sensitivity of HE4 (approximately 70%).7 Giving these, instead of early detection, both CA125 and HE4 are used as aids in monitoring tumor progression or recurrence in EOC cases.7 Therefore, considerable efforts are underway to identify novel biomarkers to improve early detection of EOC.
Circular RNAs (circRNAs) were first identified as a viroid in RNA (ribonucleic acid) as early as 197613 and were thought to be the results of splicing errors for several decades.14 Until the twenty-first century, with the development of high-throughput sequencing and novel computational approaches, circRNAs from back-spliced exons have been identified as a naturally occurring family of noncoding RNAs.15 They are characterized by a covalently closed loop structure without 5’ caps or 3’ tails.16 Increasingly more evidence showed that circRNAs were closely linked to the occurrence and development of various diseases (including cancer) by regulating gene expression via multiple mechanisms.17,18 Unlike linear RNAs, circRNAs are relatively stable due to their tolerance to RNA exonucleases.19 Additionally, some researchers have found that circRNAs are abundant in eukaryotic cells.20 Due to their abundance, conservation, and tissue specificity, circRNAs might be promising biomarkers for detection of human diseases.21 Many clinical trials have been conducted to identify the role of circRNAs in various diseases using clinical blood samples. During these clinical trials, it was identified that patients who suffered from type 2 diabetes mellitus had upregulated hsa_circ_0054633 in the peripheral blood samples.22 In case of gastric cancer, downregulated hsa_circ_0001649 was observed in serum samples.21 However, the diagnostic value of circRNAs in EOC remains largely unknown.
CircBNC2 (hsa_circ_0008732) is spliced from the zinc-finger protein BNC2, a possible transcription factor, which is highly expressed in reproductive tissue and may be involved in the differentiation of spermatozoa and oocytes.23 Besides, BNC2 has been described as a susceptibility gene for epithelial ovarian cancer, significantly associated with glioblastoma multiforme and esophageal squamous cell carcinoma, and it is a candidate gene mediating continuous skin coloration.24,25 Nevertheless, the characterization of circBNC2 in epithelial ovarian cancer (EOC) remains largely unknown. In this study, we aimed to evaluate the diagnostic value of circBNC2 in EOC by comparing with that of CA125 and HE4. We especially wanted to investigate levels of circBNC2, CA125, and HE4 with regard to tumor stage.
Materials And Methods
Study Population
A total of 254 women were enrolled in this study. Five malignant tumors were excluded because of non-epithelial ovarian cancer (n = 4; 2 granulosa cell tumor and 2 dysgerminoma) and metastasis from gastric cancer (n = 1). The eligible study population (n = 249) comprised age and menopause-matched women with EOC (n = 83), benign ovarian cysts (n = 83), and healthy individuals (n = 83). The study protocol was approved by the local ethics committee at Peking Union Medical College Hospital (PUMCH) and was conducted in accordance with the Declaration of Helsinki. Written informed consent for taking the venous blood was obtained from all of the patients and healthy volunteers. The main clinicopathologic data, including age, menopause, histological grade, histological subtype, FIGO stage, LNM, and distant metastasis are shown in Table 1. The inclusion criteria in this study were: histological diagnosis of EOC (Stage I–IV, Grade 1–3, histology including serous, mucinous, endometrioid, clear cell, and mixed); patients did not undergo preoperative chemotherapy, radial therapy or target therapy; no history of other invasive cancer; and follow-up information. Patients with borderline tumors of the ovary were excluded. Histological subtype, grade, and tumor stage were determined according to the FIGO classification.26 Patients in the benign ovarian cyst cohort had mature teratoma, serous cystadenoma, mucinous cystadenoma, and endometriosis (Table 1). The healthy individuals had no concomitant illnesses.
Table 1.
The Main Clinicopathologic Characteristics Of Enrolledwomen (N = 249)
| N (%) | |
|---|---|
| Age, average, rangea | 54 (32–73) |
| Menopauseb | |
| Pre-M | 132 (53) |
| Post-M | 117 (47) |
| Epithelial ovarian cancerc | |
| Histological grade | |
| G3 | 62 (75) |
| G1 | 21 (25) |
| Histological subtype | |
| Serous | 56 (67) |
| Others | 27 (33) |
| FIGO stage | |
| I, II | 25 (30) |
| III, IV | 58 (70) |
| LNM | |
| No | 69 (83) |
| Yes | 14 (17) |
| Distant metastasis | |
| Yes | 33 (40) |
| No | 50 (60) |
| Benign ovarian cystd | |
| Mature teratoma | 9 (11) |
| Serous cystadenoma | 11 (13) |
| Mucinous cystadenoma | 15 (18) |
| Endometriosis | 48 (58) |
Notes: aAverage age for 249 age-matched enrolled women. bMenopause for 249 menopause-matched enrolled women. cClinicopathologic characteristics of patients with epithelial ovarian cancer (n = 83). dHistological subtype of patients with benign ovarian cyst (n = 83).
Abbreviations: M, menopause; G, grade; FIGO, International Federation of Gynecology and Obstetrics; LNM, lymph node metastasis; N, number.
Sample Collection
Patients were prospectively and consecutively included when admitted for surgery for a benign or clinically suspicious malignant ovarian cyst at the Department of Gynecology and Obstetrics, PUMCH, Beijing, China, from 2015 to 2018. Peripheral venous blood samples were collected on the surgery day in sterile plastic tubes containing 3.8% tri-sodium citrate dehydrate, final volume 1/10, and immediately centrifuged at 3000 rotations per minute for 5 mins. Plasma was aliquoted and stored at −80°C until use.
RNA Preparation, Quality Assessment And RT-qPCR
Total RNA was extracted from 200 μL of the plasma using TRIzol reagent (Takara Bio, Nojihigashi, Kusatsu, Japan) according to the manufacturer’s instructions. RNA concentration was measured using NanoDrop 1000 spectrophotometers. RNA was set at an OD A260/280 ratio between 1.8 and 2.1 and an OD A260/230 ratio >1.8. RT-qPCR was performed using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara Bio, Nojihigashi, Kusatsu, Japan) and SYBR® Premix Ex Taq™ II (Tli RNaseHPlus) (Takara Bio, Nojihigashi, Kusatsu, Japan) according to the manufacturer’s instructions. GAPDH was used as an internal reference gene. The RT-qPCR protocol included an initial denaturation step (95°C for 30 s) and 40 cycles of denaturation (95°C for 5 s) and annealing (60°C for 40 s). The relative expression levels were calculated using 2−ΔΔCq method.27 The primer sequences were as follows: circBNC2 (forward, 5ʹ-GCAGTTCGGAACCAGAACGAC-3ʹ and reverse, 5ʹ-ATGCTGGCCAGTCTTGCTCAC-3ʹ), and GAPDH (forward, 5ʹ-AACGTGTCAGTGGTGGACCTG-3ʹ and reverse, 5ʹ-GAGACCACCTGGTGCTCAGTG-3ʹ).
ELISA
ELISA analyses were performed on plasma according to the manufacturer’s instructions to measure CA125 plasma concentrations (Quantikine Human CA125 Immunoassay; R&D Systems, Minneapolis, USA) and plasma HE4 levels (Quantikine Human HE4 Immunoassay; R&D Systems, Minneapolis, USA). The assays were performed on coded samples.
Statistical Analysis
Statistical differences between two groups were evaluated using the unpaired test (normal distribution data) or Mann–Whitney test (abnormal distribution data). The predicted probabilities for each marker were used to construct receiver operating characteristic (ROC) curves, and the area under the curve (AUC) values were calculated. Sensitivity and specificity were calculated for individual marker. Considering the upper limit of the normal reference range for HE4 is 55.86 pmoL/L and for CA125 is 35U/mL clinically, cut-off for CA125 < 35 U/mL and cut-off for HE4 <55.86 pmol/L was used. For circBNC2, cut-off value was calculated using Youden index (specificity + sensitivity-1). Cases with marker levels above (CA125 and HE4) or below (circBNC2) threshold levels were considered to have a positive result. P values < 0.05 were considered significant for all statistical comparisons. Statistical analyses were performed using SPSS 22.0 (SPSS Inc., Chicago, IL, USA).
Results
Patient Material
Of 249 women eligible for analysis, 83 had EOC, 83 had benign ovarian cyst, and 83 were healthy volunteers. The average age was 54 (range, 32 to 73) years old. Nearly half of the women (47%) were postmenopausal (Table 1).
The Diagnostic Value Of circBNC2, HE4 And CA125 In EOC Patients
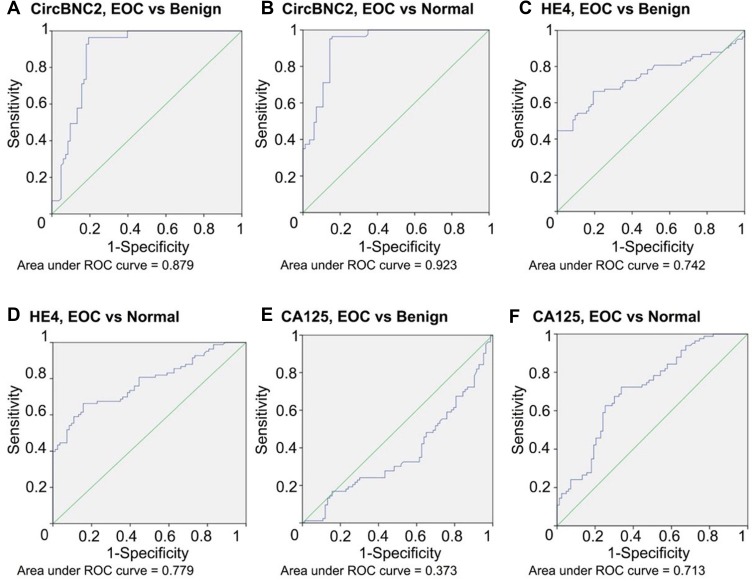
CircBNC2, HE4 and CA125 significantly separated the EOC cohort from the benign (p< 0.05) and healthy cohorts (p< 0.05) (Table 2; Figure 1). The median value in healthy cohort for CA125 was 4.0 U/mL and increased to 16.4 U/mL and 9.0 U/mL in benign and EOC cohorts. The median value in healthy cohort for HE4 was 53.1 pmol/L and increased to 56.5 and 175.1 pmol/L in benign and EOC cohorts. The difference was even more notable for circBNC2 where median value of circBNC2 ranged from 1.008 in healthy cohort to 0.680 in benign cohort and 0.230 in EOC cohort (Table 3). The ROC AUC was, according to these findings, highest for circBNC2 in discrimination between EOC and benign (AUC = 0.879) or healthy cohort (AUC = 0.923). The ROC AUC was relatively low for HE4 (AUC: 0.742, benign cohort; 0.779, healthy cohort). And the ROC AUC was lowest for CA125 (AUC: 0.373, benign cohort; 0.713, healthy cohort). In discrimination between EOC and benign cohorts, circBNC2 carried the highest sensitivity and specificity (96.4%; 80.7%), followed by HE4 (80.7%; 47.0%) and CA125 (24.1%; 66.3%). In discrimination between EOC and healthy cohorts, the sensitivity was highest for circBNC2 (95.2%), followed by HE4 (80.7%) and CA125 (24.1%); and the specificity for circBNC2 (85.5%) and CA125 (91.6%) was better than HE4 (55.4%) (Table 2; Figure 1).
Table 2.
CircBNC2, HE4 And CA125levels According To Histology, Menopause Status And Tumor Stage; ROC AUC, Sensitivity, Specificity And Significant Difference in EOC Vs Benign Ovarian Cyst And Healthy Individual Cohorts
| EOC vs Benign Ovarian Cyst | EOC vs Healthy Individuals | |||||||
|---|---|---|---|---|---|---|---|---|
| ROC AUC (95% CI) | Sen | Spe | P-Value | ROC AUC (95% CI) | Sen | Spe | P-Value | |
| circBNC2 | 0.879 (0.822–0.937) | 96.4% | 80.7% | 0.00 | 0.923 (0.882–0.965) | 95.2% | 85.5% | 0.00 |
| Pre-M | 0.854 (0.759–0.949) | 100.0% | 81.8% | 0.00 | 0.913 (0.852–0.975) | 100.0% | 81.8% | 0.00 |
| Post-M | 0.893 (0.821–0.966) | 92.3% | 79.5% | 0.00 | 0.928 (0.871–0.985) | 92.3% | 89.7% | 0.00 |
| Early stagea | 0.864 (0.795–0.933) | 92.0% | 80.7% | 0.00 | 0.908 (0.852–0.965) | 92.0% | 85.5% | 0.00 |
| Late stagea | 0.886 (0.828–0.944) | 98.3% | 80.7% | 0.00 | 0.930 (0.888–0.972) | 98.3% | 84.3% | 0.00 |
| HE4 | 0.742 (0.664–0.821) | 80.7% | 47.0% | 0.00 | 0.779 (0.709–0.850) | 80.7% | 55.4% | 0.00 |
| Pre-M | 0.654 (0.535–0.773) | 63.6% | 70.5% | 0.01 | 0.721 (0.614–0.827) | 63.6% | 81.8% | 0.00 |
| Post-M | 0.913 (0.852–0.974) | 100.0% | 20.5% | 0.00 | 0.929 (0.873–0.985) | 100.0% | 25.6% | 0.00 |
| Early stagea | 0.560 (0.398–0.723) | 60.0% | 50.0% | 0.36 | 0.633 (0.491–0.774) | 60.0% | 55.4% | 0.05 |
| Late stagea | 0.821 (0.746–0.895) | 90.0% | 50.0% | 0.00 | 0.843 (0.776–0.909) | 90.0% | 55.4% | 0.00 |
| CA125 | 0.373 (0.288–0.459) | 24.1% | 66.3% | 0.01 | 0.713 (0.635–0.791) | 24.1% | 91.6% | 0.00 |
| Pre-M | 0.344 (0.230–0.459) | 15.9% | 65.9% | 0.01 | 0.702 (0.591–0.813) | 15.9% | 90.9% | 0.00 |
| Post-M | 0.431 (0.300–0.562) | 33.3% | 66.7% | 0.30 | 0.717 (0.603–0.832) | 33.3% | 92.3% | 0.00 |
| Early stagea | 0.204 (0.095–0.312) | 12.5% | 66.3% | 0.00 | 0.546 (0.424–0.668) | 12.5% | 91.6% | 0.49 |
| Late stagea | 0.442 (0.345–0.539) | 28.8% | 66.3% | 0.24 | 0.781 (0.706–0.856) | 28.8% | 91.6% | 0.00 |
Notes: Early stage = I + II, late stage = III + IV; aAccording to FIGO staging.
Abbreviations: pre-M, premenopausal, post-M, postmenopausal.
Figure 1.
ROC AUC for circBNC2, HE4 and CA125 in epithelial ovarian cancer (EOC). It contains (A) ROC AUC for circBNC2 by EOC and benign ovarian cyst and (B) by EOC and normal controls; (C) ROC AUC for HE4 by EOC and benign ovarian cyst and (D) by EOC and normal controls; (E) ROC AUC for CA125 by EOC and benign ovarian cyst and (F) by EOC and normal controls.
Table 3.
The Expression Levels Of circBNC2, HE4, And CA125 With Regard To Histology, Menopause Status, And Tumor Stage
| Group | Total n = 249 | circBNC2, Median (Range) | HE4, pmol/L, Median (Range) | CA125, U/mL, Median (Range) |
|---|---|---|---|---|
| Epithelial ovarian cancer | n = 83 | 0.230 (0.081–0.511) | 175.1 (29.3–246.8) | 9.0 (2.1–52.9) |
| Pre-M | n = 44 | 0.250 (0.116–0.355) | 60.3 (29.3–215.8) | 8.1 (2.8–45.5) |
| Post-M | n = 39 | 0.204 (0.081–0.511) | 210.4 (57.2–246.8) | 12.5 (2.1–52.9) |
| Early stagea | n = 25 | 0.137 (0.120–0.511) | 60.3 (29.3–214.1) | 3.4 (2.3–45.5) |
| Late stagea | n = 58 | 0.230 (0.081–0.490) | 190 (39.4–246.8) | 11.2 (2.1–52.9) |
| Benign ovarian cyst | n = 83 | 0.680 (0.091–1.555) | 56.5 (38.1–189.3) | 16.4 (2.1–53.1) |
| Pre-M | n = 44 | 0.950 (0.092–1.555) | 49.2 (38.1–162.8) | 13.3 (3.2–52.1) |
| Post-M | n = 39 | 0.443 (0.091–1.450) | 64.6 (39.4–189.3) | 18.7 (2.1–53.1) |
| Normal cohort | n = 83 | 1.008 (0.155–1.932) | 53.1 (17.8–201.2) | 4.0 (1.0–43.1) |
| Pre-M | n = 44 | 1.040 (0.164–1.932) | 49.8 (22.4–184.4) | 4.1 (1.0–43.1) |
| Post-M | n = 39 | 0.928 (0.155–1.328) | 64.9 (17.8–201.2) | 3.9 (1.8–38.6) |
Note: aAccording to FIGO staging.
Abbreviations: M, menopause; n, number.
CircBNC2, CA125, And HE4 Evaluation In Early And Late Stage EOC
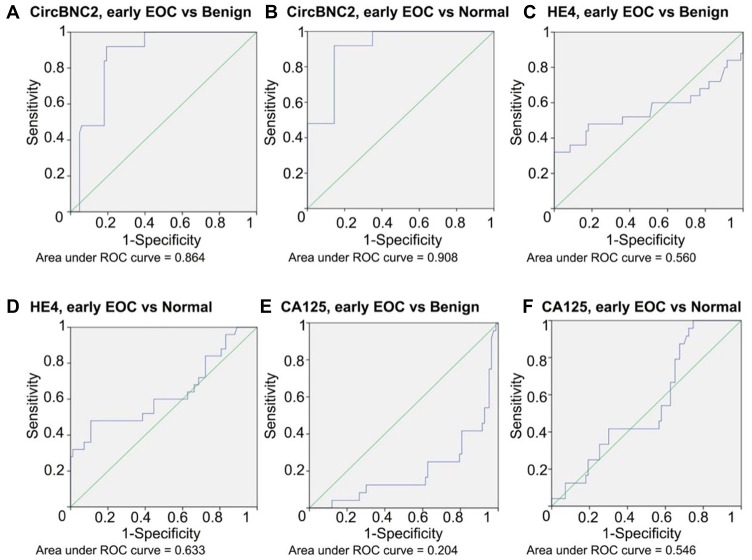
In the next step, we wanted to evaluate the diagnostic value of circBNC2, HE4, and CA125 with regard to tumor stage. EOC was divided into early stage (FIGO I + II; n = 25; 30%) and late stage (FIGO III + IV; n = 58; 70%) (Table 1), and compared to the benign and healthy cohorts. In discrimination between early stage EOC and benign or healthy cohort, statistically significant difference was noted in comparison of circBNC2 and CA125 (Table 2; Figure 2). The median values for circBNC2 in early and late stage EOC were 0.137 and 0.230, respectively (Table 3). When comparing early stage EOC with benign or healthy cohort, circBNC2 had the highest ROC AUC (AUC: 0.864, benign cohort; 0.908, healthy cohort), followed by HE4 (AUC: 0.560, benign cohort; 0.633, healthy cohort) and CA125 (AUC: 0.204, benign cohort; 0.546, healthy cohort). Individually used, circBNC2 carried the highest sensitivity in early stage EOC cohort compared to benign (92.0%) or healthy (92.0%) cohort. In discrimination between early stage EOC cohort and benign cohort, circBNC2 carried higher specificity (80.7%) than HE4 (50.0%) and CA125 (66.3%). In discrimination between early stage EOC cohort and healthy cohort, the specificity was higher for circBNC2 (85.5%) and CA125 (91.6%) than HE4 (55.4%) (Table 2; Figure 2). When comparing late stage EOC with benign or healthy cohort, circBNC2 also had the highest ROC AUC (AUC: 0.886, benign cohort; 0.930, healthy cohort), followed by HE4 (AUC: 0.821, benign cohort; 0.843, healthy cohort) and CA125 (AUC: 0.442, benign cohort; 0.781, healthy cohort). Individually used, circBNC2 carried the highest sensitivity in late stage EOC cohort compared to benign (98.3%) or healthy (98.3%) cohort. In discrimination between late stage EOC cohort and benign cohort, circBNC2 carried higher specificity (80.7%) than HE4 (50.0%) and CA125 (66.3%). In discrimination between late stage EOC cohort and healthy cohort, the specificity was higher for circBNC2 (84.3%) and CA125 (91.6%) than HE4 (55.4%) (Table 2).
Figure 2.
ROC AUC for circBNC2, HE4 and CA125 in early stage epithelial ovarian cancer (EOC). Itcontains (A) ROC AUC for circBNC2 by early stage EOC and benign ovarian cyst and (B) by early stage EOC and normal controls; (C) ROC AUC for HE4 by early stage EOC and benign ovarian cyst and (D) by early stage EOC and normal controls; (E) ROC AUC for CA125 by early EOC and benign ovarian cyst and (F) by early stage EOC and normal controls.
CircBNC2, CA125, And HE4 Evaluation In Pre- And Postmenopausal EOC
We then wanted to investigate the diagnostic value of circBNC2, HE4 and CA125 in pre- and postmenopausal women, within EOC compared to the benign and healthy cohorts. Statistically significant differences were found between all groups except for the comparisons of CA125 within EOC compared to benign cohort in postmenopausal women (Table 2). The median values for circBNC2 in EOC in pre- and postmenopausal women were 0.250 and 0.204, respectively (Table 3). In premenopausal women, circBNC2 had the highest AUC ROC in comparing EOC with benign (0.854) and healthy (0.913) cohorts, followed by HE4 (AUC: 0.654, benign cohort; 0.721, healthy cohort) and CA125 (AUC: 0.344, benign cohort; 0.702, healthy cohort). Individually used, circBNC2 carried the highest sensitivity in EOC cohort compared to benign (100.0%) or healthy (100.0%) cohort in premenopausal women. In discrimination between EOC cohort and benign cohort in premenopausal women, circBNC2 carried higher specificity (81.8%) than HE4 (70.5%) and CA125 (65.9%). In discrimination between EOC cohort and healthy cohort in premenopausal women, the specificity was alike for circBNC2 (81.8%), HE4 (81.8%), and CA125 (90.0%) (Table 2). In postmenopausal women, both circBNC2 and HE4 had higher AUC ROC when comparing EOC with benign (0.893, circBNC2; 0.913, HE4) and healthy (0.928, circBNC2; 0.929, HE4) cohorts than CA125 (AUC: 0.431, benign cohort; 0.717, healthy cohort). Individually used, circBNC2 and HE4 carried higher sensitivity in EOC cohort compared to benign (circBNC2, 92.3%; HE4, 100.0%) or healthy (circBNC2, 92.3%; HE4, 100.0%) cohort than CA125 (benign cohort, 33.3%; healthy cohort, 33.3%) in postmenopausal women. In discrimination between EOC cohort and benign cohort in postmenopausal women, circBNC2 carried higher specificity (79.5%) than HE4 (20.5%) and CA125 (66.7%). In discrimination between EOC cohort and healthy cohort in postmenopausal women, the specificity was higher for circBNC2 (89.7%) and CA125 (92.3%) than HE4 (25.6%) (Table 2).
The Association Between circBNC2 And The Clinicopathologic Variables Of EOC
The association between circBNC2 expression level and various clinicopathologic features of EOC was evaluated in 83 patients with EOC. Our data showed that downregulation of circBNC2 was significantly associated with higher histological grade (p < 0.01), serous subtype (p < 0.01), LNM (p < 0.01), and distant metastasis (p = 0.03) (Table 4).
Table 4.
The Association Between circBNC2 Expression Level And Clinicopathologic Parameters Of Epithelial Ovarian Cancer (N = 83)
| N (%) | circBNC2, Mean ± SD | P | |
|---|---|---|---|
| Age | 0.37 | ||
| ≤50 | 32 (39) | 0.24 ±0.08 | |
| >50 | 51 (61) | 0.24 ±0.11 | |
| Histological grade | <0.01* | ||
| G3 | 62 (75) | 0.20 ±0.09 | |
| G1 | 21 (25) | 0.35±0.05 | |
| Histological type | <0.01* | ||
| Serous | 56 (67) | 0.20±0.09 | |
| Others | 27 (33) | 0.30± 0.10 | |
| FIGO stage | 0.28 | ||
| I, II | 25 (30) | 0.26 ±0.13 | |
| III, IV | 58 (70) | 0.23 ±0.09 | |
| LNM | <0.01* | ||
| No | 69 (83) | 0.25 ±0.10 | |
| Yes | 14 (17) | 0.16 ±0.07 | |
| Distant metastasis | 0.03* | ||
| Yes | 33 (40) | 0.20±0.09 | |
| No | 50 (60) | 0.26 ±0.10 |
Note: *P-value less than 0.05.
Abbreviations: N, number; SD, Standard Deviation; G, grade; FIGO, International Federation of Gynecology and Obstetrics; LNM, lymph node metastasis.
Discussion
Recently, a body of studies has suggested that circRNAs might play an important role in the initiation and development of cancer and might act as potential diagnostic biomarkers in carcinoma.28,29 It was found by Chen et al30 that hsa_circ_0000190 was downregulated in both gastric cancer (GC) tissues and plasma from patients with GC might be a potential biomarker for the diagnosis of GC. Additionally, Qin et al31 identified hsa_circ_0001649 as a potential diagnosis biomarker in hepatocellular carcinoma (HCC) due to its dramatic discrepancy in the expression in HCC cells and adjacent liver tissues. Several studies have also reported the potential diagnostic value of circRNAs in non-small-cell lung cancer,32 colon cancer,33 laryngeal squamous cell cancer,34 and bladder carcinoma.35
Screening strategies have failed to effectively identify EOC cases, especially those with early stage. In this study, we aimed to evaluate the diagnostic value of the biomarker circBNC2 by comparing with that of CA125 and HE4 in EOC patients. Our results found that circBNC2 could distinguish EOC from benign and healthy cohorts (AUC: 0.879, benign cohort; 0.923, healthy cohort). CircBNC2 could also discriminate early stage EOC from benign or healthy cohort. CircBNC2 performed alike in distinguishing EOC cohort from benign or healthy cohort in both pre- and postmenopausal women. Our data suggested that circBNC2 might be a promising novel diagnostic biomarker for EOC. To the best of our knowledge, we are the first to report the diagnostic value of circBNC2 in EOC.
The high morbidity and mortality of EOC severely threaten female health.3 Detecting early stage EOC is the ultimate goal to increase the survival of EOC patients.36 The current study revealed significant differences between the expression level of circBNC2 in the plasma of EOC patients and those in benign and healthy cohorts. Specifically, our data showed that circBNC2 could help distinguishing early stage EOC from benign and healthy cohorts. These results suggested that circBNC2 might have the potential to be used as a novel diagnostic biomarker for early stage EOC in clinical practice.
In accordance with our previous studies,25 we also found a strong association between tumor characteristics and plasma concentrations of circBNC2. Low plasma circBNC2 was linked to serous cancer subtype, higher histological grade, LNM and distant metastasis. The field of circRNAs is quite new, and thus, to the best of our knowledge, no definite evidence demonstrating the functions of circBNC2 is available. The results of gene oncology (GO) analysis in our previous studies25 revealed that circBNC2 was highly involved in regulating gene expression. It has been revealed in recent studies that circRNAs might participate in the occurrence and development of various diseases (including cancer) via various mechanisms, such as competing endogenous RNAs or miRNA sponges,37 interaction with RNA binding proteins,38 modulating the stability of mRNAs,39 regulating gene transcription,40 and translating proteins.41 Therefore, future mechanism research needs to be carried out to investigate the role of circBNC2 in the development of EOC.
It should be noted that the expression level of CA125 in benign cohort (Median,16.4 U/mL) was higher than that in EOC cohort (Median, 9.0 U/mL) in this present study. This might be due to that more than half of the patients had endometriosis (n = 48, 58%) in our cohort of benign ovarian cyst.42 This further validated the limitation of CA125 in discriminating EOC from benign ovarian cysts.43
Our study had some limitations. Firstly, the sample size in our cohort was small. Future large-scale study needs to be performed to validate the diagnostic value of circBNC2 in EOC cases. Furthermore, the evaluation of circBNC2 as a true detection biomarker was limited in this study of patients with preoperative benign ovarian cysts or suspicious malignancies, formal investigation in a screening cohort of women at risk for EOC is warranted.
In conclusion, this study suggested that circBNC2 might present promising novel biomarker for EOC cases. Further well-designed large-scale studies are needed to verify our results.
Acknowledgments
We would like to thank our dedicated research and clinical support staff for making this project possible. This study was supported by the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (grant no. CAMS-2017-I2M-1-002).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.National Cancer Institute. Surveillance, Epidemiology, and End Results (SEER) program. Available from: http://seer.cancer.gov
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [PubMed: 30207593]. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 3.Previs RA, Secord AA. Ovarian cancer: Clinical trial breakthroughs and impact on management. Obstet Gynecol Clin North Am. 2019;46(1):67–88. [PubMed: 30683267]. doi: 10.1016/j.ogc.2018.09.005 [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. [PubMed: 29809280]. doi: 10.3322/caac.21456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Page C, Provencher D, Maugard CM, Ouellet V, Mes-Masson AM. Signature of a silent killer: expression profiling in epithelial ovarian cancer. Expert Rev Mol Diagn. 2004;4(2):157–167. [PubMed: 14995903]. doi: 10.1586/14737159.4.2.157 [DOI] [PubMed] [Google Scholar]
- 6.Rauh-Hain JA, Krivak TC, Del Carmen MG, Olawaiye AB. Ovarian cancer screening and early detection in the general population. Rev Obstet Gynecol. 2011;4(1):15–21. [PubMed: 21629494]. [PMC free article] [PubMed] [Google Scholar]
- 7.Li F, Tie R, Chang K, et al. Does risk for ovarian malignancy algorithm excel human epididymis protein 4 and CA125 in predicting epithelial ovarian cancer: a meta-analysis. BMC Cancer. 2012;12:258 [PubMed: 22712526]. doi: 10.1186/1471-2407-12-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis HM, Zurawski VR Jr, Bast RC Jr, Klug TL. Characterization of the CA125 antigen associated with human epithelial ovarian carcinomas. Cancer Res. 1986;46(12 Pt 1):6143–6148. [PubMed: 2430690]. [PubMed] [Google Scholar]
- 9.Bast RC Jr, Badgwell D, Lu Z, et al. New tumor markers: CA125 and beyond. Int J Gynecol Cancer. 2005;15(Suppl 3):274–281. [PubMed: 16343244]. doi: 10.1111/j.1525-1438.2005.00441.x [DOI] [PubMed] [Google Scholar]
- 10.Niloff JM, Klug TL, Schaetzl E, Zurawski VR Jr, Knapp RC, Bast RC Jr. Elevation of serum CA125 in carcinomas of the fallopian tube, endometrium, and endocervix. Am J Obstet Gynecol. 1984;148(8):1057–1058. [PubMed: 6201072]. doi: 10.1016/S0002-9378(84)90444-7 [DOI] [PubMed] [Google Scholar]
- 11.Park Y, Lee JH, Hong DJ, Lee EY, Kim HS. Diagnostic performances of HE4 and CA125 for the detection of ovarian cancer from patients with various gynecologic and non-gynecologic diseases. Clin Biochem. 2011;44(10–11):884–888. [PubMed: 21549107]. doi: 10.1016/j.clinbiochem.2011.04.011 [DOI] [PubMed] [Google Scholar]
- 12.Hellström I, Raycraft J, Hayden-Ledbetter M, et al. The HE4 (WFDC2) proteins a biomarker for ovarian carcinoma. Cancer Res. 2003;63(13):3695–3700. [PubMed: 12839961]. [PubMed] [Google Scholar]
- 13.Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215 [PubMed: 27050392]. doi: 10.1038/ncomms11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. [PubMed: 24811520]. doi: 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [PubMed: 23446348]. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 16.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNAsponges. Nature. 2013;495(7441):384–388. [PubMed: 23446346]. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 17.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. [PubMed: 25242144]. doi: 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 18.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. [PubMed: 25768908]. doi: 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 19.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2015;160(6):1125–1134. [PubMed: 25768908]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885. [PubMed: 25921068]. doi: 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 21.Ojha R, Nandani R, Chatterjee N, Prajapati VK. Emerging role of circular RNAs as potential biomarkers for the diagnosis of human diseases. Adv Exp Med Biol. 2018;1087:141–157. [PubMed: 30259364]. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54(3):237–245. [PubMed: 27878383]. doi: 10.1007/s00592-016-0943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romano RA, Li H, Tummala R, Maul R, Sinha S. Identification of Basonuclin2, a DNA-binding zinc-finger protein expressed in germ tissues and skin keratinocytes. Genomics. 2004;83(5):821–833. [PubMed: 15081112]. doi: 10.1016/j.ygeno.2003.11.009 [DOI] [PubMed] [Google Scholar]
- 24.Winham SJ, Armasu SM, Cicek MS, et al. Genome-wide investigation of regional blood-based DNA methylation adjusted for complete blood counts implicates BNC2 in ovarian cancer. Genet Epidemiol. 2014;38(5):457–466. [PubMed: 24853948]. doi: 10.1002/gepi.2014.38.issue-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shpak M, Hall AW, Goldberg MM, et al. An eQTL analysis of the human glioblastoma multiforme genome. Genomics. 2014;103(4):252–263. [PubMed: 24607568]. doi: 10.1016/j.ygeno.2014.02.005 [DOI] [PubMed] [Google Scholar]
- 26.Javadi S, Ganeshan DM, Qayyum A, Iyer RB, Bhosale P. Ovarian cancer, the revised FIGO staging system, and the role of imaging. AJR Am J Roentgenol. 2016;206(6):1351–1360. [PubMed: 27042752]. doi: 10.2214/AJR.15.15199 [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. [PubMed: 11846609]. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 28.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94 [PubMed: 28535767]. doi: 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Jiang L, Sun D, Hou J, Zhenling J. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. [PubMed: 28721656]. doi: 10.1007/s12282-017-0793-9 [DOI] [PubMed] [Google Scholar]
- 30.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta.2017;466:167–171. [PubMed: 28130019]. doi: 10.1016/j.cca.2017.01.025 [DOI] [PubMed] [Google Scholar]
- 31.Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. [PubMed: 26600397]. doi: 10.3233/CBM-150552 [DOI] [PubMed] [Google Scholar]
- 32.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-Catenin pathway. Biomed Res Int.2016;2016:1579490 [PubMed: 27642589]. doi: 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou Y, Cha DJ, Franklin JL, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982 [PubMed: 27892494]. doi: 10.1038/srep37982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xuan L, Qu L, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8(2):932–939. [PubMed: 27158380]. [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep.2016;6:30919 [PubMed: 27484176]. doi: 10.1038/srep30919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manenti L, Paganoni P, Floriani I, et al. Expression levels of vascular endothelial growth factor, matrix metalloproteinases 2 and 9 and tissue inhibitor of metalloproteinases 1 and 2 in the plasma of patients with ovarian carcinoma. Eur J Cancer. 2003;39(13):1948–1956. [PubMed: 12932675]. doi: 10.1016/S0959-8049(03)00427-1 [DOI] [PubMed] [Google Scholar]
- 37.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–5612. [PubMed: 24014594]. doi: 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- 38.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. [PubMed: 26861625]. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2011;30(21):4414–4422. [PubMed: 21964070]. doi: 10.1038/emboj.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. [PubMed: 24035497]. doi: 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–179. [PubMed: 25449546]. doi: 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vercellini P, Viganò P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10(5):261–275. [PubMed: 24366116]. [DOI] [PubMed] [Google Scholar]
- 43.Mckinnon B, Mueller MD, Nirgianakis K, Bersinger NA. Comparison of ovarian cancer markers in endometriosis favours HE4 over CA125. Mol Med Rep. 2015;12(4):5179–5184. [PubMed: 26165164]. doi: 10.3892/mmr.2015.4062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- National Cancer Institute. Surveillance, Epidemiology, and End Results (SEER) program. Available from: http://seer.cancer.gov