Abstract
Objective
Single-nucleotide polymorphism (SNP) rs143383 (T to C) in the 5′-untranslated region (5′-UTR) of GDF5 has recently been reported to be associated with osteoarthritis (OA) susceptibility, with lower expression of the risk-associated T allele observed in vitro and in vivo. The in vivo studies were performed on cartilage tissue from OA patients. The present study was undertaken to expand the analysis of the effect of this SNP on GDF5 allelic expression to more joint tissue types, to investigate for cis and trans factors that interact with the SNP, and to examine novel cis-acting GDF5 regulatory polymorphisms.
Methods
Tissue samples were collected from OA patients undergoing joint replacement of the hip or knee. Nucleic acid was extracted, and, using rs143383 and an assay that discriminates and quantifies allelic expression, the relative amount of GDF5 expression from the T and C alleles was measured. Additional common variants in the GDF5 transcript sequence were interrogated as potential regulatory elements using allelic expression and luciferase reporter assays, and electrophoretic mobility shift assays were used to search for trans factors binding to rs143383.
Results
We observed a consistent allelic expression imbalance of GDF5 in all tissues tested, implying that the functional effect mediated by rs143383 on GDF5 expression is joint-wide. We identified a second polymorphism, located in the 3′-UTR of GDF5, that influenced allelic expression of the gene independent of rs143383. Finally, we observed differential binding of deformed epidermal autoregulatory factor 1 (DEAF-1) to the 2 alleles of rs143383.
Conclusion
These findings show that the OA susceptibility mediated by polymorphism in GDF5 is not restricted to cartilage, emphasizing the need to consider the disease as involving the whole joint. The existence of an additional cis-acting regulatory polymorphism highlights the complexity of the regulation of expression of this important OA susceptibility locus. DEAF-1 is a trans-acting factor that merits further investigation as a potential tool for modulating GDF5 expression.
Genetic association of osteoarthritis (OA) with polymorphisms from within a number of human genes has now been reported (1). One of the most compelling and robust is with the single-nucleotide polymorphism (SNP) rs143383, a T-to-C transition located within the 5′-untranslated region (5′-UTR) of the growth differentiation factor 5 gene GDF5 (2).
Growth differentiation factor 5, also known as cartilage-derived morphogenetic protein 1, is a member of the transforming growth factor β superfamily and participates in the development, maintenance, and repair of bone, cartilage, and other tissues of the synovial joint (3–8). The SNP rs143383 was reported as being associated with OA initially in Asian, and subsequently in European, populations (2,9,10). A luciferase reporter assay in the Asian study demonstrated a direct functional effect of the SNP on the expression of a reporter construct in a chondrogenic cell line, with the OA-associated T allele showing reduced expression relative to the C allele (2). Additional evidence of a functional effect of the SNP came from our differential allelic expression (DAE) analysis, which revealed that the transcript containing the T allele had up to a 27% lower expression level than the C allele in RNA extracted from the articular cartilage of OA patients who had undergone elective joint replacement surgery (9). These findings suggest that a slight reduction in the expression of GDF5 in cartilage increases an individual’s risk of developing OA.
In addition to the degeneration of articular cartilage, OA is characterized by structural and histologic changes in other joint tissues, including bone, synovial tissue, ligaments, and intraarticular fat pad, some of which may precede the cartilage loss (11,12). Since growth differentiation factor 5 is involved in the functional regulation of these tissues, we hypothesized that the effect of rs143383 on GDF5 expression, and therefore on OA susceptibility, could be joint-wide rather than restricted to cartilage. To test this hypothesis we performed GDF5 allelic expression analysis, using SNP rs143383, on a variety of tissues collected from the hip and knee joints of OA patients. We also sought to identify trans-acting factors that demonstrated differential binding to this polymorphism. Finally, we sought to identify additional cis polymorphisms in GDF5 that could influence the allelic expression of the gene independent of rs143383.
PATIENTS AND METHODS
Patients
Joint tissue was obtained from individuals undergoing elective joint replacement for OA of the hip (total hip replacement [THR]) or the knee (total knee replacement [TKR]), as described in detail previously (13). In patients undergoing TKR, tissue samples were obtained from cartilage, infrapatellar fat pad, meniscus, anterior cruciate ligament, and synovium. In THR patients, the tissue sources were cartilage, synovium, and ligamentum teres femoris. Ethics approval for tissue collection was granted by the local ethics committees, and informed consent was obtained from each donor.
Case–control cohort
The cases (n = 1,944; 1,184 women and 760 men) were ascertained through the Nuffield Orthopaedic Centre, Oxford, UK. All had primary OA and had undergone THR (n = 1,325; 813 women and 512 men), TKR (n = 519; 309 women and 210 men), or both THR and TKR (n = 100; 62 women and 38 men). In our study, case status was determined according to the presence of signs and symptoms that were sufficiently severe to necessitate joint replacement surgery. The control group (n = 850; 382 women and 468 men) comprised individuals who had no signs or symptoms of arthritis or joint disease (pain, swelling, tenderness, or restriction of movement). All patients and all controls were UK Caucasians. A full description of the cases and controls has been published previously (9). Allele and genotype distributions between cases and controls were compared using standard chi-square analysis of contingency tables.
DAE
Using a protocol described by us previously (13), nucleic acid was extracted from the joint tissue of OA patients who had undergone THR or TKR. The genomic DNA was used to genotype the patients for rs143383 and for SNP 2250ct, using polymerase chain reaction (PCR) restriction enzyme assays. The RNA from patients heterozygous for one or both SNPs was then taken forward for allelic expression analysis, using a single-base extension (SBE) assay that we have also described in detail previously (13). In summary, at least 800 ng of RNA was used for each complementary DNA (cDNA) synthesis, using random hexamers and the Superscript kit (Invitrogen, Paisley, UK). Two reactions were performed for each assay: with reverse transcriptase (RT) and without RT. From each reaction with RT, 20 individual PCR amplifications were carried out using forward primer 5′-CTTCAAGCCCTCAGTCAGTTG-3′ and reverse primer 5′-CGGGTGTGTGTTTGTATCCAG-3′ for rs143383, and forward primer 5′-TAAGCACCTCTCAGGAGAGC-3′ and reverse primer 5′-ACAGTCTAACAGCCTCACAC-3′ for 2250ct. Controls performed without RT did not yield detectable PCR product. The primer 5′-CTCGTTCTTGAAAGGAGAAAGCC-3′, which is located immediately adjacent to rs143383, and the primer 5′-CAGTCAGCTTCTCAACTGTCCC-3′, which is located immediately adjacent to 2250ct, were then used for the respective SBE assays. To ascertain the peak pattern for an assumed 1:1 ratio between alleles, we performed, for each SNP, 5 individual PCR and SBE reactions on the cartilage genomic DNA of each patient tested.
The same PCR primers and SBE primer were used for the cDNA and genomic template. This use of the same analytic conditions for the cDNA and genomic DNA measurements allowed us to use the average of the genomic DNA allelic ratio measurements (representing the assumed 1:1 ratio between alleles) to correct the allelic ratios obtained from the cDNA measurements, and thus to account for differences in fluorescent yield and terminator dye incorporation specific to an assay. With this correction, we were able to obtain exact values of the relative allelic expression of each cDNA measurement. To determine if there was a significant difference in allelic expression in a particular tissue from an individual patient, the cDNA allelic ratio in that patient’s tissue was compared with the pooled genomic allelic ratios, using a 2-tailed Mann-Whitney exact test. To determine if there was an overall difference in expression between alleles for a particular tissue type, the mean allelic ratios for patient cDNA were compared with the mean allelic ratios for patient genomic cDNA, using one-way analysis of variance followed by Tukey’s honest significant difference test or Dunnett’s T3 test. P values were uncorrected.
Qualitative gene expression
The expression of GDF5 (determined using forward primer 5′-CTGTCCGATGCTGACAGAAAG-3′ and reverse primer 5′-AACACGTACCTCTGCTTCCTG-3′) and DEAF1 (using forward primer 5′-GCTGCTGCAGACAATGTCTTC-3′ and reverse primer 5′-GTCTCCACGATGCTCCCATC-3′) in different joint tissues and human cell lines was analyzed by RT-PCR with HPRT1 (using forward primer 5′-CTGAACGTCTTGCTCGAGATG-3′ and reverse primer 5′-TGCGACCTTGACCATCTTTGG-3′) as a housekeeping control.
Genotyping, sequencing of GDF5, and molecular haplotyping
SNPs were genotyped by PCR restriction enzyme assay. (Information on the primer sequences and enzymes used is available upon request from the corresponding author.) Sequencing of the transcript sequence, promoter region, and 2 alternative exons upstream of GDF5 was performed on overlapping PCR products generated from genomic DNA. PCR products were prepared for sequencing using a PCR product pre-sequencing kit (USB, Staufen, Germany) and sequenced using the BigDye Terminator v3.1 sequencing kit, an ABI 3100 Genetic Analyzer, and SeqScape software (Applied Biosystems, Warrington, UK). Molecular haplotyping was used to determine the haplotype structure between rs143383 and 2250ct, 2 SNPs that are not in high linkage disequilibrium (LD). The 2 SNPs were amplified in a single PCR fragment using genomic DNA as template, and the product was then subcloned into Escherichia coli using the TOPO XL PCR cloning kit (Invitrogen). Transformed colonies were then genotyped.
Construction of luciferase reporter plasmids
The promoter/5′-UTR of GDF5 (−34 to +367 with respect to the transcriptional start site described previously [2]), encompassing and containing specific alleles of rs143383 and rs143384, was PCR amplified using primers flanked by either a Kpn I (5′-GAAGGTAC/CGGATTCAAAACTAGGGGG-3′) or a Hind III (5′-GAAA/AGCTTCCGCTGAATGACACCAAAG-3′) restriction enzyme site. To generate the haplotype CT for rs143383 and rs143384, a reverse primer was used to force a T at rs143384 onto a CC haplotype (5′-GAAA/AGCTTCCGCTGAATGACACCAAAGAGAACAGCGGCAGCAGCAAA-GG-3′ [underlined nucleotide represents the forced change]). The PCR products were digested with Kpn I and Hind III (New England Biolabs, Beverly, MA), dephosphorylated with shrimp alkaline phosphatase (GE Healthcare, Piscataway, NJ), and purified using the QIAquick PCR purification kit (Qiagen, Crawley, UK). The Kpn I and Hind III restriction sites of pGL3 basic luciferase reporter vector (Promega, Southampton, UK) were used to generate the promoter/5′-UTR–luciferase constructs using T4 ligase (Promega). The 3′-UTR of GDF5 (1895–2460 with respect to the transcriptional start site described previously [2]), encompassing and containing the appropriate alleles of SNPs 12164tg and 2250ct, was PCR amplified using primers flanked by either an Xba I (forward 5′-AATT/CTAGACAGCACTGGCCCTCTGTCTTC-3′) or a Bam HI (reverse 5′-AGAG/GATCCCAGTTTTAGGCACA-GTTTTGC-3′) restriction enzyme site. The PCR products were digested with Xba I and Bam HI, dephosphorylated, and purified. Subsequently, the products were cloned into the Xba I and Bam HI restriction sites of the GDF5 promoter/5′-UTR–luciferase reporter vector (haplotype TC for rs143383 and 143384).
To obtain all 4 possible haplotypes for the combination of 12164tg and 2250ct, site-directed mutagenesis was performed using a QuikChange II Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). In all cloning experiments, positive clones were isolated using a GenElute HP Plasmid MidiPrep Kit (Sigma-Aldrich, Poole, UK) and sequenced to ensure the correct sequence of the constructs.
Cell cultures and luciferase assays
Three human cell lines were used for transfection experiments: MG63 (human osteosarcoma cell line), SW872 (human liposarcoma cell line), and CH8 (human articular chondrocyte cell line) (14). Twenty-four hours before transfection, cells were seeded at a density of ~2.0 × 104 cells/cm2 per well of 6-well tissue culture plates. Using GeneJuice transfection reagent (Novagen, Madison, WI), cells were transfected with 500 ng of the experimental vectors and with 500 ng of pSV-galactosidase control vector or 10 ng of pRL-TK (Promega). For overexpression experiments, 7.5 × 103 MG63 cells in 96-well tissue culture plates were cotransfected with 50 ng of a deformed epidermal autoregulatory factor 1 (DEAF-1)–related transcriptional regulator 1 expression vector or its control pCMV6-XL5 (OriGene, Rockville, MD), and 50 ng of the appropriate GDF5 promoter/5′-UTR–luciferase constructs. After 48 hours, luciferase activity was measured on a GloMax 20/20 Luminometer, using the Luciferase Assay System or Dual Luciferase Assay System (Promega). Activity of β-galactosidase was measured, by spectrophotometric assay, in 150 μl 2.9 mM 2-nitrophenyl β-D-galactopyranoside/1 mM MgCl2/14.1 mM β-mercaptoethanol/82 mM Na2HPO4⋅2H2O/18 mM NaH2PO4⋅2H2O, and absorbance was measured at 405 nm; β-galactosidase (Sigma-Aldrich) was used as a standard. In overexpression studies of the promoter/5′-UTR constructs, we were not able to obtain constant expression of the control vectors. Therefore, we normalized the luciferase activity to the total protein content of the cell lysate using a bicinchoninic acid–based assay (Pierce, Chester, UK), as described previously (15,16). Statistical comparisons were performed with Student’s 2-tailed t-test. All P values were uncorrected.
Preparation of nuclear extracts
Nuclear extracts from MG63, CH8, and SW872 cells were prepared as described previously (17), with minor modifications. Briefly, 5 × 106 cells were washed twice with 3 ml of ice-cold phosphate buffered saline and scrapped in 1 ml ice-cold buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol [DTT], with 1 tablet Complete Mini protease inhibitor cocktail [Roche, Lewes, UK] added per 10 ml of solution just before use). Following incubation on ice for 15 minutes, 66 μl of 10% Nonidet P40 was added to 1 ml of cell suspension, followed by vortexing and incubation on ice for 2 minutes. The cell suspension was centrifuged at 12,000 revolutions per minute for 30 seconds at 4°C, and following removal of the supernatant, the pellet was washed with 500 μl of ice-cold buffer A, vortexed, and centrifuged at 12,000 rpm for 30 seconds at 4°C. The pelleted nuclei were resuspended in 3 volumes of ice-cold buffer B (20 mM HEPES [pH 7.9], 0.4M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 tablet Complete Mini protease inhibitor cocktail [Roche] added per 10 ml of solution just before use). After incubation on ice for 1 hour with intermittent strong vortexing, the extracts were centrifuged at 12,000 rpm for 5 minutes at 4°C, and the supernatant containing the nuclear proteins was collected and stored at −80°C in small aliquots until use. Protein concentration was measured using an ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Electrophoretic mobility shift assay (EMSA)
To create double-stranded probes, oligonucleotides were annealed at 100°C for 5 minutes in a solution containing 1× restriction enzyme buffer H (Roche) and allowed to cool to room temperature. The double-stranded probes (3 pmoles) were then radiolabeled by incubation with 0.74 MBq (6.7 pmoles) of α32P-dCTP (110 TBq/mmole; GE Healthcare) and 5 units of Klenow enzyme (Promega) for 40 minutes at room temperature. Radiolabeled probes were purified on Mini Quick Spin DNA columns (Roche). Binding was performed for 15 minutes at room temperature in 16-μl reaction mixtures containing 60 fmoles (40,000 counts per minute) of labeled probe, 4 μg of nuclear proteins, 20 mM HEPES (pH 7.9), 2 mM EDTA, 2 mM EGTA, 25% glycerol, and 1 μg poly(dI-dC) (GE Healthcare). For competition experiments, unlabeled double-stranded oligonucleotides were added in 100-fold molar excess 5 minutes before the addition of the radiolabled probe. Gels were transferred to 3MM paper (Whatman, Kent, UK), dried, exposed to a Phosphor Storage Screen (Molecular Dynamics, Chesham, UK) for 3 days, and scanned with a STORM 860 scanner (Molecular Dynamics). The DEAF-1 oligonucleotides were 5′-AGCTTTCGGACTGATTCGGCTTCCCACTTCGGGGAACTTCG-3′ and 5′-AGCTCGAAGTTCCCCGAAGTGGGAAGCCGAATCAGTCCGAA-3′ (18), and the activator protein 1 oligonucleotides were 5′-AGCTCGCTTGATGAGTCAGCCGGAA-3′ and 5′-AGCTTTCCGGCTGACTCATCAAGCG-3′ (Promega).
RESULTS
Evidence that differential allelic expression of GDF5 is joint-wide
Using qualitative PCR, we demonstrated expression of GDF5 in all joint tissues tested, i.e., cartilage, synovium, meniscus, ligament, tendon, and fat pad (results not shown). We subsequently identified 10 patients (5 women and 5 men; ages 50–88 years at the time of surgery) who were heterozygous at rs143383 and from whom we had collected samples of at least 2 joint tissue types. Nine of the 10 patients had undergone TKR and 1 (female) patient had undergone THR. Joint tissue RNA from the 10 patients was subjected to DAE analysis.
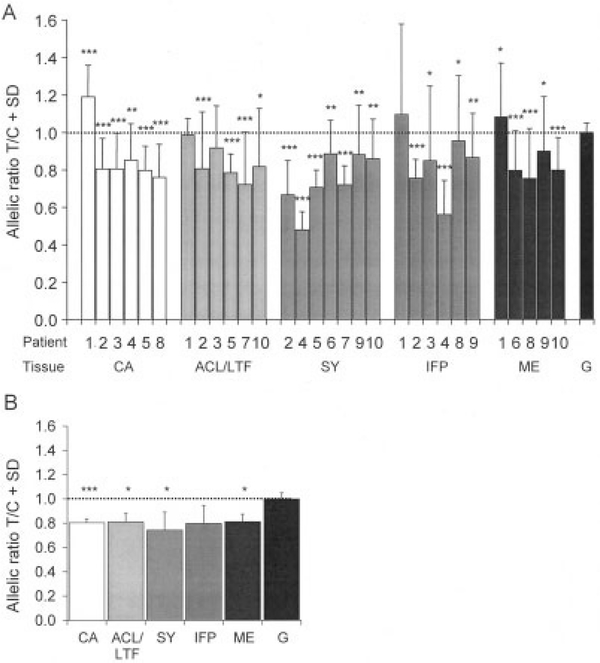
Eight of the 10 patients (patient 2 and patients 4–10 [Table 1 and Figure 1A]) exhibited significantly decreased expression of the T allele compared with the C allele in all of the tissues tested (nominal P ≤ 0.05). In patient 3, 2 of the 3 tissue types examined (cartilage and fat pad) showed significantly decreased expression of the T allele, and the decrease in T allele expression in the third tissue (ligament) approached significance (P = 0.08). In contrast, in patient 1, there was no significant difference in allelic expression in the ligament and fat pad, and in the cartilage and meniscus, the expression of the C allele was significantly decreased relative to the T allele. We have so far examined GDF5 allelic expression at rs143383 in the cartilage of 15 unrelated patients (6 in this study and 9 described in a previous report [9]), and this is the first time we have observed a relative decrease in expression of the C allele. It implies the existence of at least 1 other cis-acting polymorphism for GDF5.
Table 1.
Mean allelic ratios from the differential allelic expression analysis of GDF5 using single-nucleotide polymorphism rs143383 and RNA extracted from the joint tissues of 10 osteoarthritis patients
| Patient, tissue | Mean allelic ratio, T/C | n* | Nominal P† | Bonferroni-corrected P‡ |
|---|---|---|---|---|
| Patient 1 | ||||
| Cartilage | 1.19 | 19 | 2.73 × 10−8 | 8.21 × 10−7 |
| Ligament | 0.986 | 19 | 0.782 | 1 |
| Fat pad | 1.095 | 18 | 0.243 | 1 |
| Meniscus | 1.082 | 20 | 0.020 | 0.606 |
| Patient 2 | ||||
| Cartilage | 0.805 | 20 | 2.94 × 10−9 | 8.83 × 10−8 |
| Synovium | 0.667 | 20 | 3.17 × 10−11 | 9.50 × 10−10 |
| Fat pad | 0.755 | 20 | 3.84 × 10−14 | 1.15 × 10−12 |
| Ligament | 0.806 | 20 | 0.0002 | 0.0059 |
| Patient 3 | ||||
| Cartilage | 0.805 | 19 | 1.18 × 10−5 | 0.00035 |
| Ligament | 0.918 | 19 | 0.081 | 1 |
| Fat pad | 0.849 | 20 | 0.011 | 0.328 |
| Patient 4 | ||||
| Cartilage | 0.852 | 19 | 0.00082 | 0.025 |
| Synovium | 0.479 | 18 | 4.13 × 10−19 | 1.24 × 10−17 |
| Fat pad | 0.561 | 20 | 1.43 × 10−14 | 4.28 × 10−13 |
| Patient 5 | ||||
| Cartilage | 0.797 | 20 | 7.37 × 10−10 | 2.21 × 10−8 |
| Ligament | 0.784 | 20 | 2.39 × 10−11 | 7.17 × 10−10 |
| Synovium | 0.707 | 20 | 1.40 × 10−17 | 4.20 × 10−16 |
| Patient 6 | ||||
| Meniscus | 0.797 | 20 | 7.76 × 10−6 | 0.00023 |
| Synovium | 0.886 | 20 | 0.001 | 0.0305 |
| Patient 7 | ||||
| Ligament | 0.722 | 19 | 1.87 × 10−5 | 0.00056 |
| Synovium | 0.720 | 20 | 2.18 × 10−16 | 6.53 × 10−15 |
| Patient 8 | ||||
| Cartilage | 0.760 | 20 | 1.99 × 10−9 | 5.97 × 10−8 |
| Fat pad | 0.954 | 20 | 0.012 | 0.363 |
| Meniscus | 0.754 | 20 | 3.07 × 10−5 | 0.00092 |
| Patient 9 | ||||
| Fat pad | 0.866 | 20 | 0.0024 | 0.0725 |
| Meniscus | 0.900 | 20 | 0.0089 | 0.267 |
| Synovium | 0.882 | 20 | 0.0036 | 0.109 |
| Patient 10 | ||||
| Ligament | 0.817 | 20 | 0.0058 | 0.174 |
| Meniscus | 0.799 | 20 | 9.49 × 10−8 | 2.85 × 10−6 |
| Synovium | 0.859 | 20 | 0.0007 | 0.021 |
| Genomic DNA (patients 1–10) | 1.000 | 70 | – | – |
Number of replicates performed in order to obtain the mean allelic ratio.
By 2-tailed Mann-Whitney exact test.
A conservative correction in which the nominal P values were multiplied by 30 to account for the 10 patients studied and the average of 3 tissues per patient.
Figure 1.
Allelic expression analysis of GDF5 using single-nucleotide polymorphism rs143383 and RNA extracted from various types of joint tissue from 10 osteoarthritis patients. A, Data on each tissue type obtained from each patient. The cDNA allelic ratio in each specimen was compared with the pooled genomic allelic ratio (G) (1:1; dotted line) by 2-tailed Mann-Whitney exact test. B, Combined patient data on patients 2–10 for each tissue type. The mean allelic ratios in the patient cDNA were compared with the mean allelic ratio in the patient genomic cDNA (1:1; dotted line) by one-way analysis of variance. Values are the mean and SD of at least 18 replicates per patient per tissue. * = P < 0.05; ** = P < 0.01; *** = P < 0.001. CA = cartilage; ACL = anterior cruciate ligament; LTF = ligamentum teres femoris; SY = synovium; IFP = infrapatellar fat pad; ME = meniscus.
We combined the data from the 9 patients who exhibited a relative reduction in expression of the T allele. A significant difference was obtained for each tissue type examined except for fat pad (Figure 1B). The results for all tissue types were strikingly comparable, and the mean T-to-C allelic ratio was 0.79 for the tissues combined. Our data imply that the rs143383 polymorphism has a similar effect on the expression of GDF5 across all of the joint tissue types tested.
Evidence that rs143383 is functional and its effect on gene expression is influenced by a second 5′-UTR SNP
As described above, our DAE data were consistent in 9 of the 10 patients, with patient 1 being the exception. Findings in this patient implied the existence of at least 1 other cis-acting GDF5 regulatory polymorphism, and we hypothesized that this would become apparent by the demonstration of a unique genotype at one or more polymorphisms in patient 1 relative to the other 9 patients. Using public databases, we identified 14 polymorphisms from within a 5.5-kb interval encompassing the GDF5 transcript sequence, and then genotyped these polymorphisms in the 10 patients.
The only polymorphism for which patient 1 had a unique genotype was SNP rs143384, which, like rs143383, is located in the 5′-UTR of GDF5, but closer to the translational start codon of the gene. Patient 1 was CC homozygous at rs143384, whereas the other 9 patients were all CT heterozygous. Sequence analysis of the transcript region of GDF5 in patient 1 did not reveal any further polymorphisms, and we therefore focused our attention on rs143384. From the HapMap project, the reported pairwise r2 between rs143383 and rs143384 is 0.75 in the CEPH (Centre d’Etude du Polymorphisme Humain) population, demonstrating relatively high LD between the 2 SNPs. In patient 1, the OA-risk T allele of rs143383 was found to be on a haplotype with a C allele of rs143384, since this patient was heterozygous at rs143383 but homozygous (CC) at rs143384. Patient 1 therefore carried a TC haplotype (rs143383–rs143384) and a CC haplotype. The remaining 9 patients were all compound heterozygotes at the 2 SNPs and, based on the high LD between the SNPs, the T allele of rs143383 is on the same chromosome as the T allele of rs143384, such that these individuals carried a CC haplotype and a TT haplotype.
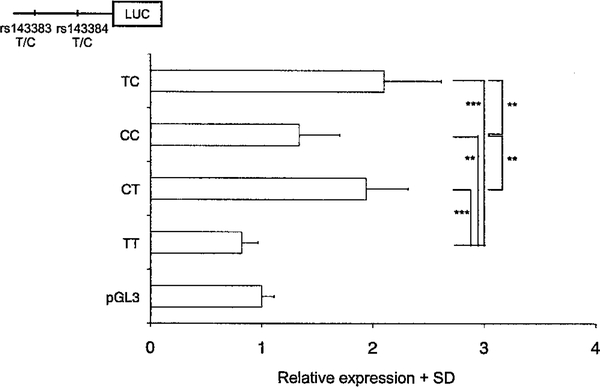
The involvement in transcriptional regulation of the 4 possible haplotypes of the 2 SNPs (TC, CC, CT, and TT) was assessed by luciferase reporter assay in 3 different cell lines: CH8 cells (chondrogenic), SW872 cells (adipogenic), and MG63 cells (osteogenic). Figure 2 presents data obtained with CH8 cells; the same expression patterns were found with SW872 and MG63 cells (data not shown). The OA-risk T allele of rs143383 was found to mediate a reduction in luciferase activity relative to the C allele only when it was on the background of a T allele at rs143384 (the TT haplotype in Figure 2). These luciferase assay data therefore support the observations from our DAE studies: patient 1 carried the TC and CC haplotypes, and in this combination the T allele of rs143383 will show greater expression relative to the C allele, as was observed in the cartilage and meniscus of this patient (Figure 1A). Patients 2–10 were carriers of CC and TT haplotypes, and in this combination the T allele of rs143383 will show less expression relative to the C allele, which is again what was observed in the tissues from these patients. Our data emphasize the complex nature by which cis-acting polymorphisms interact to modulate gene expression. They also demonstrate how an in vitro expression assay can be used to analyze in more detail an expression observation made in vivo.
Figure 2.
Results of luciferase (LUC) reporter assays of GDF5 promoter/5′-untranslated region (5′-UTR) constructs in the chondrogenic cell line CH8, for the 4 possible haplotypes for rs143383 and rs143384. A schematic drawing of the promoter/5′-UTR constructs is shown at the top left. Data are the fold expression in relation to control pGL3 vector, and are shown as the mean and SD from 3 independent experiments performed in triplicate. ** = P < 0.01; *** = P < 0.001.
DAE independent of rs143383
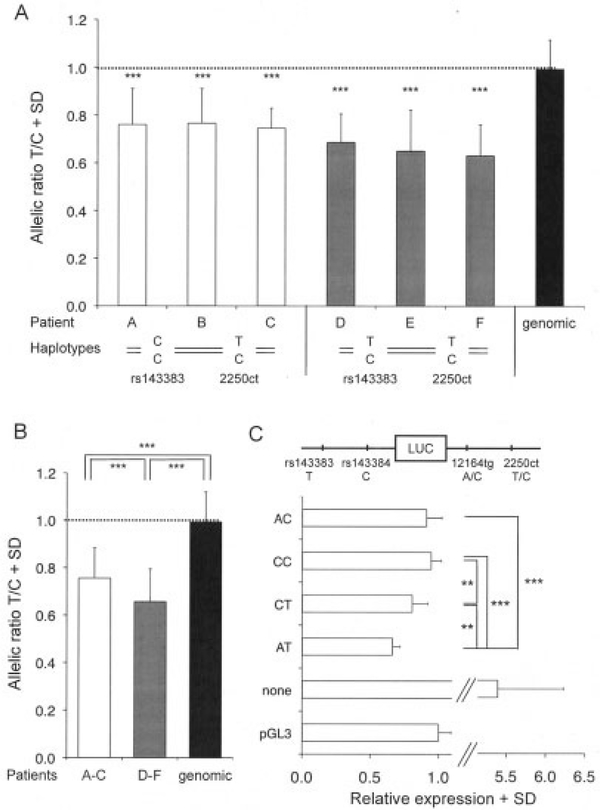
Our investigation for additional polymorphisms in GDF5 highlighted a SNP in the 3′-UTR of the gene that had been reported previously and had been given the name 2250ct (2). This SNP does not show strong LD with rs143383, and therefore it provided us with the opportunity to assess whether DAE at GDF5 occurred independent of rs143383. To assess this, we screened >100 patients, and we were able to identify 3 new patients (patients A–C in Figure 3A) from whom cartilage tissue was available and who were homozygous (CC) at rs143383 but heterozygous (TC) at 2250ct. All 3 patients exhibited a strikingly similar imbalance of expression at 2250ct, with significantly lower expression of the T allele relative to the C allele (ranging from 23% to 25%). This finding demonstrates the existence of a regulatory polymorphism of GDF5 that acts independent of rs143383.
Figure 3.
Functional analysis of the GDF5 3′-UTR single-nucleotide polymorphisms 12164tg and 2250ct. A, Results of allelic expression analysis using 2250ct and RNA extracted from articular cartilage of 6 osteoarthritis patients who were either homozygous (patients A–C) or heterozygous (patients D–F) at rs143383. Patients A, C, and F were women who had undergone total hip replacement (THR), patients B and E were men who had undergone total knee replacement, and patient D was a man who had undergone THR. Values are the mean and SD of at least 19 replicates per patient. B, Results of combined allelic expression analysis in the 3 homozygous patients and in the 3 heterozygous patients. Values are the mean and SD. Dotted lines in A and B indicate equal allelic expression (1:1 ratio), as depicted in the genomic DNA column. C, Findings of luciferase reporter assays with the GDF5 3′-UTR inserted into a promoter/5′-UTR–luciferase construct and transfected into CH8 cells. The 4 possible haplotypes for 12164tg and 2250ct (AC, CC, CT, and AT) were examined. “None” represents a construct containing the promoter/5′-UTR but no 3′-UTR. Data are the fold expression in relation to control pGL3 vector, and are shown as the mean and SD from 3 independent experiments performed in triplicate. ** = P < 0.01; *** = P < 0.001. A schematic drawing of the constructs is shown at the top. See Figure 2 for other definitions.
We subsequently assessed whether the independent effects of rs143383 and 2250ct are additive. We identified 3 further patients (patients D–F in Figure 3A) who were compound heterozygous for both SNPs and from whom cartilage tissue was available. Using molecular haplotyping, we demonstrated that the patients carried a TT haplotype (rs143383–2250ct) and a CC haplotype (i.e., the T alleles that are associated with reduced expression of each SNP were present together on the same chromosome). In all 3 patients, an enhanced imbalance was observed, with lower expression of the TT haplotype relative to the CC haplotype (ranging from 30% to 35%). A comparison of pooled data on patients A–C and pooled data on patients D–F (Figure 3B) revealed that the relative reduction in GDF5 expression associated with the T allele at 2250ct (haplotype CT) was reduced by a further 13% with the inclusion of the T allele at rs143383 (haplotype TT). The independent effects therefore appeared to be partly, but not completely, additive.
The SNP 2250ct is in complete LD (r2 = 1.0) with a second 3′-UTR SNP, 12164tg (2), with the haplotype AT (12164tg–2250ct) being the most common in our population (frequency 96%). To assess whether either SNP was itself functional, the full-length 3′-UTR of GDF5 was cloned into a GDF5 promoter/5′-UTR–luciferase construct. Using site-directed mutagenesis, the 4 possible haplotypes of the 2 SNPs (12164tg–2250ct; AC, CC, CT and AT) were created, and their effects on luciferase expression were investigated in CH8, SW872, and MG63 cells. Figure 3C presents data obtained in studies of CH8 cells, with SW872 and MG63 cells exhibiting the same expression patterns (data not shown). In comparison with a construct lacking the 3′-UTR, the 3′-UTR constructs all led to a reduction in luciferase activity. This general effect of 3′-UTRs on luciferase activities has been reported previously (19).
Taking into account this consistent effect across 3′-UTR constructs, we subsequently examined each allele and haplotype. For 2250ct, the T allele (haplotypes CT and AT) mediated reduced luciferase activity compared with the C allele (haplotypes AC and CC), whereas no consistent difference was observed between the 2 alleles of 12164tg, implying that 2250ct is functional whereas 12164tg is not. However, the T allele at 2250ct did appear to be influenced to a small degree by the allele at 12164tg, with the AT haplotype showing a significant reduction relative to the CT haplotype (P < 0.01). The effect, though, was not as dramatic as that seen for SNPs rs143383 and rs143384.
The finding of reduced luciferase activity mediated by the T allele of 2250ct supports our DAE observations and implies that 2250ct is itself functional. Many 3′-UTRs are known to harbor regulatory elements (20). However, using the public databases SRS and RegRNA, we were not able to identify a stability-affecting sequence such as an AU-rich element or a microRNA target site at or adjacent to 2250ct. Thus, the role of this polymorphism in the regulation of GDF5 expression requires additional investigation.
When we genotyped 2250ct in our UK case–control cohort we found that the T allele had a frequency of 95.5% in cases and 95.1% in controls (P = 0.47). Stratification by sex or by joint replaced did not reveal any significant associations (data not shown).
Differential interaction of DEAF-1 with the 2 alleles of rs143383
To further characterize the function of rs143383, we attempted to identify trans-acting factors that bind differentially to the 2 alleles of the SNP. EMSAs were performed using nuclear extracts from MG63, CH8, and SW872 cells, and 2 radioactively labeled GDF5 promoter/5′-UTR sequence probes containing either the T or the C allele of rs143383. The probes were found to have differing mobilities in each cell line, suggesting some differences in the protein complexes binding to the 2 alleles (Figure 4A, lanes 2 and 8). The specificity of the assay was confirmed by allele-specific competition of protein binding using excessive unlabeled T and C allele probes (Figure 4A, lanes 3 and 10).
Figure 4.
Differential interaction of deformed epidermal autoregulatory factor 1 (DEAF-1) with the alleles of rs143383. A, Results of electrophoretic mobility shift assays (EMSAs) performed on probes containing the T allele (lanes 1–6) or the C allele (lanes 7–12) and nuclear extract from MG63 cells. The arrow on the left indicates the faster mobility band observed with the T allele; the arrow on the right indicates the slower mobility band observed with the C allele. B, Results of EMSAs performed using a probe containing a consensus sequence of DEAF-1 and nuclear extract from MG63 cells. C, Results of luciferase reporter assay of GDF5 promoter/5′-UTR constructs in MG63 cells with cotransfection of DEAF-1 expression vector. Data are the fold expression in DEAF-1–transfected cells in relation to control pCMV vector after normalization to expression of the pGL3 control vector, and are shown as the mean and SD from 3 independent experiments performed in triplicate. A schematic drawing of the constructs is shown at the top. * = P < 0.05. AP-1 = activator protein 1 (see Figure 2 for other definitions).
Since the binding of the protein(s) to the C allele probe was competed by an excess of the T allele probe (Figure 4A, lane 9) but not vice versa (Figure 4A, lane 4), it is probable that some of the same proteins bind to the 2 probes, but that the T allele binds a unique protein that makes the T allele complex unresponsive to the C allele probe. Using the public databases TESS, Tfsearch, and MatInspector, we identified several transcription factors that were predicted to bind in an allele-specific manner to the GDF5 promoter region, and we subsequently focused on 3: the Myb protooncogene protein c-Myb, early growth response protein 1 (EGR-1), and DEAF-1. Using 3 different c-Myb consensus sequences, we were not able to compete either the T allele or the C allele in EMSAs (results not shown), implying that c-Myb is not involved in the allele-specific regulation of GDF5. We obtained similar negative results with EGR-1 (results not shown). In contrast, with the DEAF-1 consensus probe, we were able to compete the binding of the T allele protein complex, but not the C allele protein complex (Figure 4A, lanes 5 and 11). This was confirmed by the greater competition of unlabeled T allele competitor relative to unlabeled C allele competitor for protein binding to the DEAF-1–labeled probe (Figure 4B, lanes 3 and 4).
An independent functional role of DEAF-1 in the allele-specific regulation of rs143383 was explored by performing cotransfection experiments with GDF5 promoter/5′-UTR constructs and a DEAF-1 expression vector. Regardless of the allele at rs143383, both GDF5 promoter/5′-UTR constructs were repressed in the presence of DEAF-1, when compared with its empty control vector pCMV6-XL5 (Figure 4C). However, in accordance with the stronger binding of DEAF-1 observed in the EMSA, the T allele was significantly more repressed, by ~40%, than the C allele (P < 0.05). Finally, using qualitative gene expression, we confirmed the expression of DEAF1 in patient joint tissues, including cartilage, and in all 3 of the cell lines used in our study (results not shown).
DISCUSSION
The genetic association of the GDF5 5′-UTR SNP rs143383 with OA is one of the most robust reported to date for this common disease, with association confirmed in different ethnic groups and by meta-analysis (10). We previously reported that the OA-risk T allele at the SNP correlated with reduced expression of the gene in OA patient cartilage (9), a result that supported in vitro data from the original association report (2). We have now extended our analysis to many other soft tissues of the synovial joint and have demonstrated that the differential allelic effect seen in cartilage is also present in all other tissues tested. This observation is very significant, since it demonstrates that OA risk mediated by this locus is not restricted to cartilage, but rather is joint-wide. This highlights the need to ensure that in studies investigating potential diagnostic, prognostic, or therapeutic uses of the genetic data, the whole joint must be considered, rather than focusing on just one tissue.
We subsequently identified, using luciferase reporter assays, an additional polymorphism in the 5′-UTR of the gene, rs143384, which can modulate the effect of rs143383 in vitro, emphasizing the complex and subtle nature by which cis-acting polymorphism can regulate gene expression. This work involved an analysis of the 4 possible haplotype combinations of rs143383 and rs143384. These haplotypes were also investigated in the originally reported study of the association of rs143383 with OA, but, in contrast with the present findings, a significant effect of rs143384 on rs143383 was not observed in that study (2). The chondrogenic cell line HCS-2/8 was used in the earlier investigation, and this difference in cell lines used in the 2 studies may account for the disparate findings regarding the functional effect of rs143384.
We also identified a polymorphism, 2250ct, that influenced GDF5 allelic expression independent of rs143383. The effect of 2250ct on GDF5 allelic expression was comparable with that seen for rs143383, i.e., a consistent but moderate relative reduction in expression, on the order of 20–25%. Genotyping of 2250ct in 2,794 UK OA cases and controls did not reveal significant association. This is not particularly surprising. The minor allele of the SNP had a frequency of only 4.9% in our controls, which is considerably less than the minor allele frequency (MAF) of rs143383 in our controls (i.e., 38.0%) (9). When studying polymorphisms with low MAFs, large sample sizes are needed in order to detect robust genetic association, particularly when the genetic effect is relatively modest: for rs143383, a meta-analysis of >11,000 individuals was needed to confirm the association with OA, despite its relatively high MAF (10). SNP 2250ct would therefore need to be genotyped in considerably larger sample sizes than those currently available to the OA research community, to detect and replicate a robust association. It would also be beneficial to genotype all other variants within the GDF5 locus in such large cohorts once they are assembled, to scrutinize haplotypes harboring multiple independent effects that are each modest in nature, and to identify possible rare variants that have large singular effects on GDF5 expression.
Using EMSAs, we scrutinized 3 potential trans-acting factors that may interact differentially with rs143383, and we were able to demonstrate that one of these, DEAF-1, does indeed do so. DEAF-1 is expressed in a number of tissue types, and it has been previously reported that it can act as a transcriptional enhancer or repressor and is involved in skeletal patterning, with malformations in the cartilaginous rib cage of Deaf-1–mutant mice (18,21–23). However, its role in cartilage formation and homeostasis is currently unknown, although we were able to demonstrate expression of the gene in patient joint tissues. DEAF-1 represents a valid candidate for an allele-specific trans-regulator at SNP rs143383 and now merits more intensive investigation.
Overall, we have extended the functional analysis of regulatory polymorphisms of the GDF5 gene. We have demonstrated the joint-wide effect of rs143383 on GDF5 expression, identified novel cis and trans factors that interact with this SNP, and identified a novel cis-acting polymorphism in the 3′-UTR of the gene. The OA susceptibility mediated by regulatory polymorphism at this gene is subtle but consistent within the different joint tissues in individuals, and between the joint tissues of different individuals. Recently it was reported that rs143383 was also associated with variation in human height, with the OA-risk T allele associated with reduced height (24). This study demonstrates the pleiotropic potential of DNA polymorphism on common traits and implies that the functional effect of rs143383 on GDF5 allelic expression is also active during skeletal development. In conclusion, GDF5 is a fascinating OA susceptibility gene, with small but consistent effects on allelic expression imbalance across joint tissues acting to modulate OA risk.
ACKNOWLEDGMENTS
We thank Andrew Price, Nick Athanasou, and Bridget Watkins for helping to organize the collection of samples from the subjects evaluated in this study, Irina Udalova for technical advice regarding EMSAs, and Tim McCaffrey and Yang Zhaoqing for supplying the EGR-1 expression plasmids.
Supported by Research into Aging and the Arthritis Research Campaign. Dr. Egli was recipient of a postdoctoral fellowship from the Swiss National Science Foundation and the Hans-Neuenschwander Foundation. Ms Southam’s work was supported by a grant from the European Community Seventh Framework Programme collaborative project Treat-OA. Dr. Wilkins was recipient of a Marshall Scholarship. Dr. Pombo-Suarez was recipient of a Fundacion Espanola de Reumatologia bursary. Dr. Gonzalez’ work was supported by the Instituto de Salud Carlos III, Spain, with participation from the European Union FEDER Fund, and by the Fundacion MMA, Spain.
REFERENCES
- 1.Bos SD, Slagboom PE, Meulenbelt I. New insights into osteoarthritis: early developmental features of an ageing-related disease. Curr Opin Rheumatol 2008;20:553–9. [DOI] [PubMed] [Google Scholar]
- 2.Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet 2007;39:529–33. [DOI] [PubMed] [Google Scholar]
- 3.Edwards CJ, Francis-West PH. Bone morphogenetic proteins in the development and healing of synovial joints. Semin Arthritis Rheum 2001;31:33–42. [DOI] [PubMed] [Google Scholar]
- 4.Luyten FP. Cartilage-derived morphogenetic protein-1. Int J Biochem Cell Biol 1997;29:1241–4. [DOI] [PubMed] [Google Scholar]
- 5.Mikic B Multiple effects of GDF-5 deficiency on skeletal tissues: implications for therapeutic bioengineering. Ann Biomed Eng 2004;32:466–76. [DOI] [PubMed] [Google Scholar]
- 6.Francis-West PH, Abdelfattah A, Chen P, Allen C, Parish J, Ladher R, et al. Mechanisms of GDF-5 action during skeletal development. Development 1999;126:1305–15. [DOI] [PubMed] [Google Scholar]
- 7.Harada M, Takahara M, Zhe P, Otsuji M, Iuchi Y, Takagi M, et al. Developmental failure of the intra-articular ligaments in mice with absence of growth differentiation factor 5. Osteoarthritis Cartilage 2007;15:468–74. [DOI] [PubMed] [Google Scholar]
- 8.Hatakeyama Y, Tuan RS, Shum L. Distinct functions of BMP4 and GDF5 in the regulation of chondrogenesis. J Cell Biochem 2004;91:1204–17. [DOI] [PubMed] [Google Scholar]
- 9.Southam L, Rodriguez-Lopez J, Wilkins JM, Pombo-Suarez M,Snelling S, Gomez-Reino JJ, et al. An SNP in the 5′-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum Mol Genet 2007;16:2226–32. [DOI] [PubMed] [Google Scholar]
- 10.Chapman K, Takahashi A, Meulenbelt I, Watson C, Rodriguez-Lopez J, Egli R, et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5′ UTR of GDF5 with osteoarthritis susceptibility. Hum Mol Genet 2008;17: 1497–504. [DOI] [PubMed] [Google Scholar]
- 11.Mansell JP, Collins C, Bailey AJ. Bone, not cartilage, should be the major focus in osteoarthritis. Nat Clin Pract Rheumatol 2007;3:306–7. [DOI] [PubMed] [Google Scholar]
- 12.Quasnichka HL, Anderson-MacKenzie JM, Bailey AJ. Subchondral bone and ligament changes precede cartilage degradation in guinea pig osteoarthritis. Biorheology 2006;43:389–97. [PubMed] [Google Scholar]
- 13.Wilkins JM, Southam L, Price AJ, Mustafa Z, Carr A, Loughlin J. Extreme context specificity in differential allelic expression. Hum Mol Genet 2007;16:537–46. [DOI] [PubMed] [Google Scholar]
- 14.Yoshimatsu T, Saitoh A, Ryu JN, Shima D, Handa H, Hiramoto M, et al. Characterization of immortalized human chondrocytes originated from osteoarthritis cartilage. Int J Mol Med 2001;8: 345–51. [DOI] [PubMed] [Google Scholar]
- 15.Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, Lightowlers RN, et al. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum Mol Genet 2004;13:2519–34. [DOI] [PubMed] [Google Scholar]
- 16.Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci U S A 2002;99:5400–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res 1989;17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelson RJ, Collard MW, Ziemba AJ, Persinger J, Bartholomew B, Huggenvik JI. Nuclear DEAF-1-related (NUDR) protein contains a novel DNA binding domain and represses transcription of the heterogeneous nuclear ribonucleoprotein A2/B1 promoter. J Biol Chem 1999;274:30510–9. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Zhang J, Zhang R, Xue F, Sun Y, Han X. Limitation in use of luciferase reporter genes for 3′-untranslated region analysis. Biotechnol Lett 2007;29:1691–6. [DOI] [PubMed] [Google Scholar]
- 20.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell 2008;132:9–14. [DOI] [PubMed] [Google Scholar]
- 21.Czesak M, Lemonde S, Peterson EA, Rogaeva A, Albert PR. Cell-specific repressor or enhancer activities of Deaf-1 at a serotonin 1A receptor gene polymorphism. J Neurosci 2006;26: 1864–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahm K, Sum EY, Fujiwara Y, Lindeman GJ, Visvader JE, Orkin SH. Defective neural tube closure and anteroposterior patterning in mice lacking the LIM protein LMO4 or its interacting partner Deaf-1. Mol Cell Biol 2004;24:2074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barker HE, Smyth GK, Wettenhall J, Ward TA, Bath ML, Lindeman GJ, et al. Deaf-1 regulates epithelial cell proliferation and side-branching in the mammary gland. BMC Dev Biol 2008; 8:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, Bonnycastle LL, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat Genet 2008;40: 198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]