Abstract
The study of cellular processes occurring inside intact organisms requires methods to visualize cellular functions such as gene expression in deep tissues. Ultrasound is a widely used biomedical technology enabling non-invasive imaging with high spatial and temporal resolution. However, no genetically encoded molecular reporters are available to connect ultrasound contrast to gene expression in mammalian cells. To address this limitation, we introduce mammalian acoustic reporter genes. Starting with a gene cluster derived from bacteria, we engineered a eukaryotic genetic program whose introduction into mammalian cells results in the expression of intracellular air-filled protein nanostructures called gas vesicles, which produce ultrasound contrast. Mammalian acoustic reporter genes allow cells to be visualized at volumetric densities below 0.5% and permit high-resolution imaging of gene expression in living animals.
One Sentence Summary:
Acoustic reporter genes enable non-invasive ultrasound imaging of gene expression in mammalian cells.
The study of cellular function within the context of intact living organisms is a grand challenge in biological research and synthetic biology (1). Addressing this challenge requires imaging tools to visualize specific cells in tissues ranging from the developing brain to tumors, and to monitor gene- and cell-based therapeutic agents in vivo (2). However, most common methods for imaging cellular processes such as gene expression rely on fluorescent or luminescent proteins, which have limited performance in intact animals due to the poor penetration of light in biological tissue (3, 4). On the other hand, ultrasound easily penetrates most tissues, enabling deep non-invasive imaging with excellent spatial and temporal resolution (~100 µm and ~1 ms, respectively) (2, 5). These capabilities, along with its safety, portability and low cost, have made ultrasound a widely used technology in biomedicine. Despite these advantages, to date ultrasound has played a relatively small role in cellular imaging due to the lack of appropriate genetically encoded reporters.
Recently, biomolecular contrast agents for ultrasound were introduced based on gas vesicles, air-filled protein nanostructures which evolved in certain waterborne bacteria and archaea to provide cellular buoyancy (6, 7). Gas vesicles comprise a 2 nm-thick protein shell enclosing a gas compartment with dimensions on the order of 100 nm. The acoustic impedance mismatch between their gas interior and surrounding aqueous media allows gas vesicles to strongly scatter sound waves and thereby serve as ultrasound contrast agents (8–12). In their native organisms, gas vesicles are encoded by clusters of 8–14 genes, including one or two primary structural proteins, and several other essential genes encoding putative assembly factors or minor shell constituents.
The use of gas vesicles as reporter genes requires the heterologous expression of their cognate multi-gene operon in a new cellular host, ensuring proper transcription and translation of each gene, functional folding of each corresponding protein and appropriate stoichiometry and co-localization of the constituents for gas vesicle assembly. Recently, a genetic engineering effort succeeded in expressing gas vesicles as acoustic reporter genes (ARGs) in commensal bacteria, allowing their imaging in the mouse gastrointestinal tract (13). If ARGs could be developed for mammalian cells, this would enable the study of how such cells develop, function and malfunction within the context of model organisms and enable the in vivo imaging of mammalian cells engineered to perform diagnostic or therapeutic functions (14–16). However, developing ARGs for mammalian cells represents an even greater synthetic biology challenge due to the differences in transcription, translation, co-localization and protein folding between prokaryotes and eukaryotes (17–19). To our knowledge, no genetic operon larger than 6 genes has been moved between these domains of life (20).
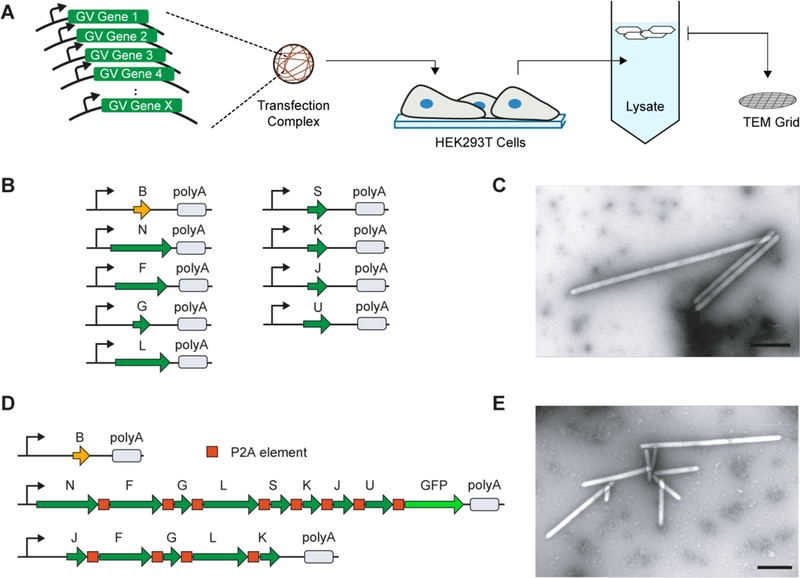
Here, we describe the expression of ARGs in mammalian cells to enable ultrasound imaging of mammalian gene expression. To identify a set of genes capable of encoding gas vesicle assembly in mammalian cells, we synthesized individual gas vesicle genes from three different microbial species using codons optimized for human expression, cloned each gene into a separate monocistronic plasmid and transiently co-transfected mixtures of the genes from each species into HEK293T cells (Fig. 1A). After allowing 72 hours for protein expression, we gently lysed the cells (~2×106 cells per sample), and centrifugated the lysate to enrich for buoyant particles, which would include any gas vesicles. The top fraction of the centrifugated lysate was then screened for gas vesicles using transmission electron microscopy (TEM). These experiments took advantage of the intrinsic stochasticity of transient co-transfection, in terms of the ratios of genes and the overall DNA quantity delivered to each cell, to simultaneously sample a broad range of gene stoichiometries and expression levels without prior knowledge of parameters leading to gas vesicle formation.
Fig. 1. Engineering of mammalian acoustic reporter genes.
(A) Schematic of the transient co-transfection assay used to identify combinations of genes capable of producing gas vesicles in mammalian cells. (B) Schematic of nine genes from B. megaterium capable of encoding gas vesicle expression in mammalian cells. Thin arrow denotes CMV promoter. polyA denotes SV40 polyadenylation element. (C) Representative TEM image of purified gas vesicles expressed in HEK293T cells. (D) Gene cassettes comprising the mammalian acoustic reporter gene construct, mARG. (E) Representative TEM image of gas vesicles purified from HEK293T cells transiently transfected with mARGs for 72 hours. All scale bars represent 500 nm.
The co-transfection of the gas vesicle genes from Halobacterium salinarum and Anabaena flos-aquae did not lead to the formation of detectable gas vesicles. However, the co-transfection of 9 gas vesicle-forming genes from Bacillus megaterium (Fig. 1B) resulted in the production of unmistakable gas vesicles as evidenced by their appearance in TEM images (Fig. 1C). The 9 genes originate from an eleven-gene B. megaterium gene cluster previously used to express gas vesicles in E. coli (13, 21), with the exception of GvpR and GvpT, which were found to be unnecessary for gas vesicle formation (Fig. S1).
Using the 9 genes identified in our stochastic screen, we set out to construct a polycistronic mammalian operon for consistent gas vesicle expression by joining these genes using the viral co-translational self-cleavage peptide P2A (22). Having determined that all genes except GvpB could tolerate P2A peptide additions (Fig. S2 and Table S1), we constructed a polycistronic plasmid containing the 8 P2A-tolerant gas vesicle genes connected by P2A sequences, and co-transfected it into HEK293T cells together with a plasmid encoding GvpB. Unfortunately, this did not result in the production of gas vesicles. We hypothesized that one or more of the genes in our polycistronic plasmid was expressed at an insufficient level, and used a complementation assay to identify GvpJ, GvpF, GvpG, GvpL and Gvpft as bottleneck genes (Fig. S3). This led us to construct a polycistronic “booster” plasmid containing these five genes, ordered to minimize P2A modifications to GvpJ and Gvpft, which were found to be most limiting. The co-transfection of the booster plasmid together with the two plasmids above (Fig. 1D) enabled robust expression of gas vesicles in cells (Fig. 1E). We named this set of three genetic constructs mammalian acoustic reporter genes, or mARGs.
After establishing polycistronic constructs for mammalian gas vesicle assembly, we used an integrase (23, 24) to incorporate them into the cellular genome for stable expression under a doxycycline-inducible TRE3G promoter, with fluorescent proteins added to each construct as transfection indicators (Fig. 2A). We transfected these plasmids into HEK293-tetON cells and used flow cytometry to sort cells according to their expression level of each fluorescent reporter. We found that the cell population combining the strongest expression of each construct produced the largest quantity of gas vesicles (Fig. 2B, Fig. S4, A–D). To ensure that mARG expression was not limited to HEK293 cells, we also transfected Chinese hamster ovary cells (CHO-K1), and obtained similar results (Fig. S4, E–G).
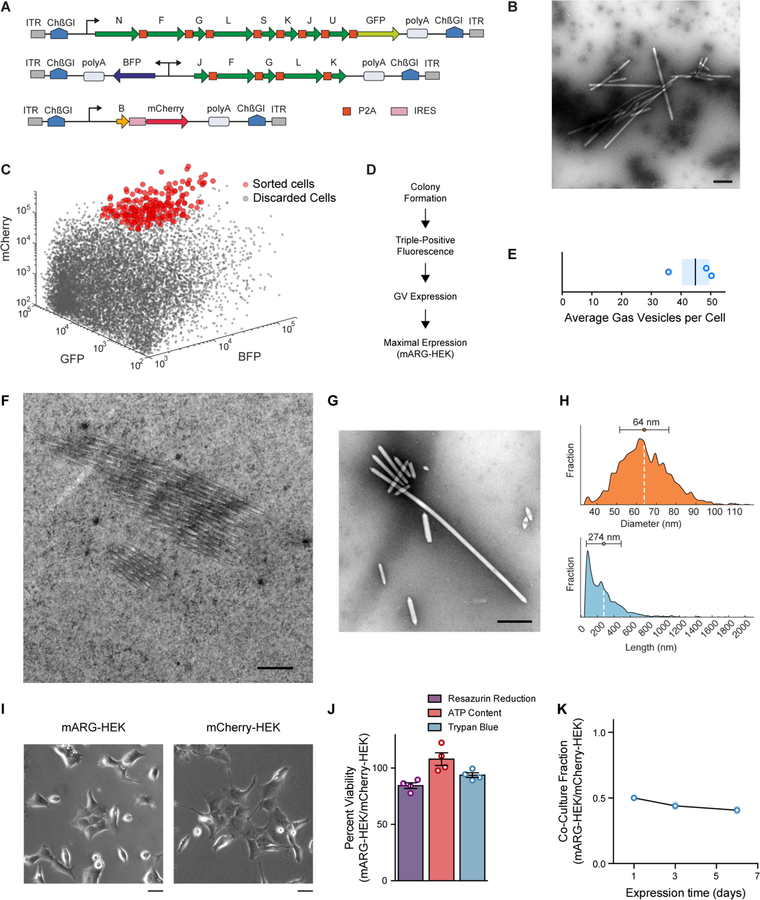
Fig. 2. Formation, properties and non-toxicity of gas vesicles in cells with genome-integrated mammalian acoustic reporter genes.
(A) Schematic of mARG constructs used for genomic integration into cells with the piggyBac transposase system. ITR, inverted terminal repeat; ChβGI, Chicken beta-globin insulator; GFP, Emerald green fluorescent protein; BFP, enhanced blue fluorescent protein 2. (B) Representative TEM image of buoyancy-enriched lysate from HEK293-tetON cells transfected with the constructs in (A) and sorted for high expression of all three operons. (C) Fluorescence-activated cell sorting of HEK293-tetON cells transfected with the constructs in (A). Red circles denote individual cells selected by sorting to form monoclonal cell lines. (D) Selection process for monoclonal cell lines, including assays for viability, fluorescence intensity and gas vesicle yield. (E) Number of gas vesicles expressed by monoclonal HEK293-tetON cells after 72 hours of induced expression, as counted in lysates using TEM. Bar represents the mean and the shaded area represents SEM (n=3, each from two technical replicates). (F) Representative TEM image of a 60-nm section through an mARG-HEK cell showing an angled slice through two bundles of gas vesicles in the cytosol. (G) Representative TEM image of gas vesicles purified from mARG-HEK cells. (H) Size distribution of gas vesicles expressed in mARG-HEK cells. The mean and standard deviation of both distributions is illustrated as a circle and with error bars. (n=1828) (I) Phase contrast images of mARG-HEK and mCherry-HEK cells 72 hours after induction with 1 µg/mL doxycycline and 5 mM sodium butyrate. (J) Cell viability of mARG-HEK cells relative to mCherry-HEK cells after 72 hours of gene expression. Error bars indicate SEM. (K) Fraction of mARG-HEK cells in co-culture with mARG-mCherry cells seeded in equal numbers over 6 days of gene expression (n=3 biological replicates, each from 4 technical replicates, with darker symbols showing the mean). Scale bars in B, F, G represent 500 nm. Scale bar in I represents 20 µm.
To generate a stable monoclonal cell line expressing mARGs for detailed analysis, we sorted individual high-expression HEK293-tetON cells for monoclonal growth (Fig. 2C), producing 30 cell lines, which we screened for viability, fluorescence and gas vesicle formation (Fig. 2D, Table S2). The number of gas vesicles per cell was then estimated from TEM images, and a cell line yielding the largest quantity of gas vesicles was selected and named mARG-HEK. When induced for 72 hours with 1 µg/mL of doxycycline and 5 mM sodium butyrate (to reduce epigenetic silencing), this cell line produced on average 45 gas vesicles per cell (Fig. 2E). Using thin-section TEM, gas vesicles could clearly be seen in the cytosol of individual mARG-HEK cells (Fig. 2F). From TEM images of cell lysates, we measured the average dimensions of gas vesicles produced in this cell line to be 64 ± 12 nm wide (standard deviation, n=1828) and 274 ± 212 nm long (standard deviation, n=1828), with some reaching lengths greater than 1 micron (aspect ratios greater than 30) (Fig. 2, G–H). This corresponds to an average gas vesicle volume of 0.605 attoliters. Together, the 45 gas vesicles expressed in an average mARG-HEK cell are expected to occupy just 0.0027% of the cell’s cytosolic volume.
The expression of gas vesicles did not change the gross morphology of mARG-HEK cells (Fig. 2I), and was non-toxic as determined by three different assays (Fig. 2J), as compared to a similarly prepared control cell line (mCherry-HEK) (Fig. S5 A–B). During a 6-day co-culture, mARG-HEK cells showed only a minor growth disadvantage compared to mCherry-HEK cells (Fig. 2K). As expected, both engineered cell lines grew more slowly than wild-type HEK293T cells (Fig. S6).
Having engineered mARG-HEK cells, we sought to image their expression of acoustic reporters with ultrasound. Gas vesicles encoded by the B. megaterium gene cluster are expected to produce linear ultrasound scattering (21). However, since mammalian cells themselves also produce significant linear contrast, detecting gas vesicles expressed in such cells using linear methods is challenging. To enable more selective imaging of mARG-expression, we took advantage of the ability of gas vesicles to collapse irreversibly above specific ultrasound pressure thresholds (8, 9, 13, 21). A switch in the incident ultrasound pressure from below to above such a threshold results in a strong transient signal from the gas vesicles, which decays to a lower level in the next ultrasound frame due to immediate dissolution of their gas contents and the elimination of ultrasound scattering (Fig. 3, A–B). Meanwhile, background tissue scattering rises with the increase in incident pressure and remains constant at the new level. Thus, images formed by taking the difference in signal between the collapsing and post-collapse frames reveal specifically the presence of gas vesicles.
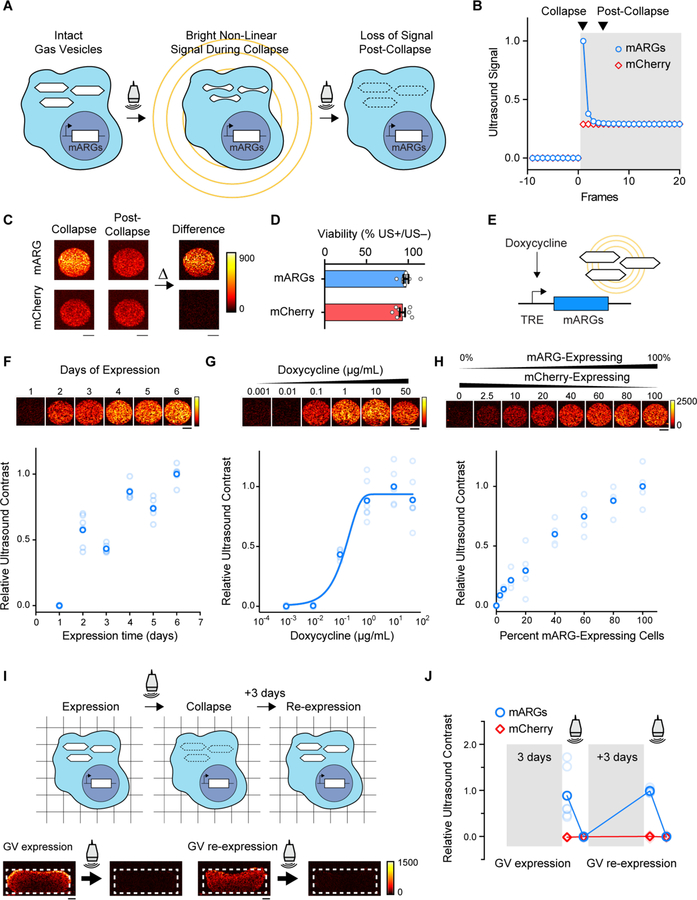
Fig. 3. Ultrasound imaging of mammalian gene expression in vitro.
(A) Illustration of the collapse-based ultrasound imaging paradigm used to generate gas vesicle-specific ultrasound contrast from mARG-expressing cells. (B) Representative non-linear signal recorded during a step change in the incident acoustic pressure, from 0.27 MPa in the white-shaded region to 1.57 MPa in the grey-shaded region. (C) Representative collapse and post-collapse ultrasound images of mARG-HEK and mCherry-HEK cells acquired during this ultrasound imaging paradigm and their difference, indicating gas vesicle-specific contrast. (D) Cellular viability after being insonated under 3.2 MPa acoustic pressures, as measured using the MTT assay. (E) Schematic of a chemically inducible gene circuit with mARG expression as its output. All three mARG cassettes in mARG-HEK cells are under the control of the doxycycline-inducible TRE3G promoter (TRE), with expression triggered by incubation with doxycycline. (F) Representative ultrasound images and contrast measurements in mARG-HEK cells as a function of time following induction with 1 µg/mL of doxycycline and 5 mM sodium butyrate (n=6, with the darker dots showing the mean). (G) Representative ultrasound images and contrast measurements in mARG-HEK cells as a function of doxycycline induction concentrations. Cells were allowed to express gas vesicles for 72 hours in the presence of 5 mM sodium butyrate. (n=6, with the darker dots showing the mean). A sigmoidal function is fitted as a visual guide. (H) Representative ultrasound images and contrast measurements in mARG-HEK cells mixed with mCherry-HEK cells in varying proportions. Cells were induced with 1 µg/mL of doxycycline and 5 mM sodium butyrate for 72 hours prior to imaging. (n=4, with the darker dots showing the mean) (I) Schematic and representative ultrasound images from mARG-HEK cells in Matrigel re-expressing gas vesicles after acoustic collapse. Cells were induced with 1 µg/mL of doxycycline and 5 mM sodium butyrate for 72 hours before and after 3.2 MPa acoustic insonation. Ultrasound images were acquired after an additional 72 hours in culture following collapse. (J) Ultrasound contrast in mARG-HEK and mCherry-HEK cells after initial expression, after collapse, after re-expression and after second collapse. (n=7, with the darker dots showing the mean). GV, gas vesicles. All scale bars represent 1 mm.
We implemented this collapse-based imaging approach using an amplitude modulation pulse sequence (10), which we found to provide the best cancellation of non-gas vesicle signals. When hydrogels containing mARG-HEK cells were imaged using this technique at 18 MHz, they were easily distinguishable from mCherry-HEK controls based on their contrast dynamics (Fig. 3C). Critically, while this imaging paradigm requires the collapse of gas vesicles inside cells, this does not affect cell viability (Fig. 3D).
To test if mARGs can faithfully monitor circuit-driven gene expression (25, 26), we measured the dynamic ultrasound response of mARG-HEK cells under the control of a doxycycline-inducible promoter (Fig. 3E). After induction with 1 µg/mL doxycycline, the cells showed a gradual buildup of ultrasound signal, with clear contrast appearing on day two and increasing over the next 4 days (Fig. 3F). These kinetics are similar to those observed with fluorescent indicators (Fig. S7A). When the gene circuit was driven using a range of inducer concentrations, the ultrasound contrast followed the expected transfer function of the promoter (Fig. 3G, Fig. S7B).
To determine how sensitively mARG-expressing cells could be detected in a mixed cell population, we combined mARG-HEK cells with mCherry-HEK cells at varying ratios. We were able to detect the presence of mARG-expressing cells in these mixtures down to 2.5% of total cells (Fig. 3H), corresponding to less than 0.5% volumetric density, or approximately 3 cells or 135 gas vesicles per voxel with dimensions of 100 µm. A similar voxel-averaged concentration of gas vesicles was detectable in a monoculture of mARG-HEK cells induced to express 1.4 ± 0.6 gas vesicles per cell (Fig. S8).
In many imaging experiments, the output of a gene circuit is read out only once. However, in some cases it may be desirable to track gene expression over time. We therefore tested whether mARG-expressing cells in which the gas vesicles have been collapsed during imaging could re-express these reporters to allow additional imaging. mARG-HEK cells cultured a nutrient-supported hydrogel produced clear ultrasound contrast 3 days after induction, and were able to re-express their acoustic reporters over 3 additional days (Fig. 3, I–J).
Having engineered mammalian cells to stably express gas vesicles and characterized their ability to produce ultrasound contrast in vitro, we next tested the ability of mARG expression to be visualized in vivo with high spatial resolution. We formed model tumor xenografts in immunocompromised mice by inoculating mARG-HEK cells in Matrigel subcutaneously in their left flanks (Fig. 4A). In the same mice, the right flanks were inoculated with mCherry-HEK control cells. We induced reporter gene expression in both tumors for 4 days through systemic injections of doxycycline and sodium butyrate (Fig. 4B). We expected these nascent tumors to be mostly vascularized at their perimeter, resulting in the strongest inducible gene expression at the tumor periphery (Fig. 4A). Ultrasound, with its sub-100-µm spatial resolution (at 18 MHz), should be able to discern this gene expression pattern, whereas attaining such resolution would be challenging with optical techniques.
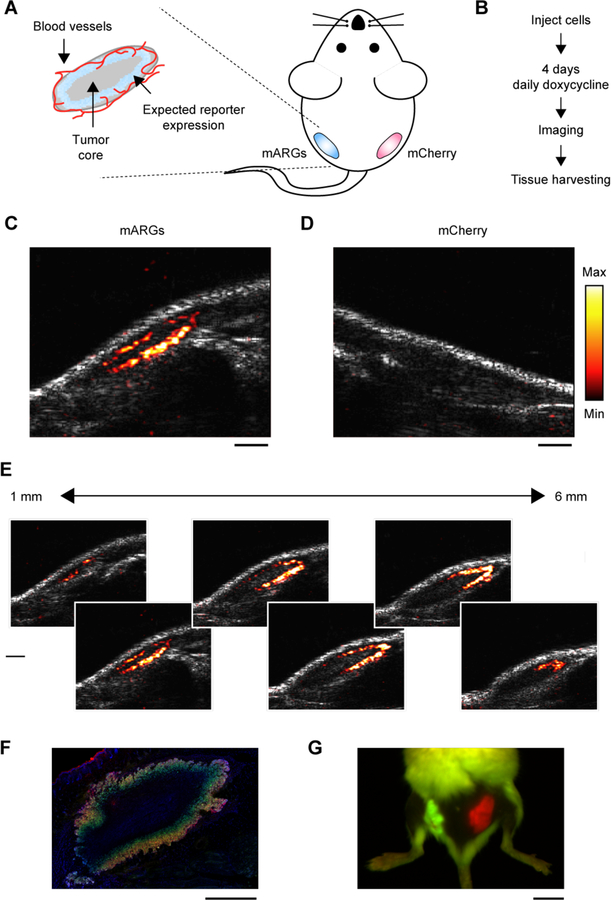
Fig. 4. Ultrasound imaging of mammalian gene expression in vivo.
(A) Diagram of a mouse implanted with a subcutaneous tumor model, and the expected spatial pattern of vascularization and doxycycline-induced reporter gene expression. (B) Experimental timeline. (C) Representative ultrasound image of tumors containing mARG-HEK cells after 4 days of doxycycline administration. mARG-specific contrast shown in the hot colormap is overlaid on an anatomical B-mode image showing the background anatomy. (D) Representative ultrasound image of tumors containing mCherry-HEK cells after 4 days of doxycycline administration. (E) Ultrasound images of adjacent planes in the mARG-HEK tumor acquired at 1 mm intervals. The minimum and maximum values of color bars in C-E are 4000 and 40000 au, respectively. (F) Representative fluorescence image of a histological tissue section of an mARG-HEK tumor. Blue color shows the TO-PRO3 nucleus stain, green color shows GFP fluorescence and red color shows mCherry fluorescence. (G) Fluorescence image of a mouse implanted with mARG-HEK and mCherry-HEK tumors on the left and right flanks, respectively, after 4 days of expression. Scale bars for are 1 mm for C–F and 1 cm for G.
After 4 days of induction, we observed clear ultrasound contrast in the flank inoculated with mARG-HEK cells, which was absent from the contralateral side (Fig. 4, C–D). As expected, the pattern observed with ultrasound revealed mARG expression at the perimeter of the tumor, while the core remained dark, and the imaging of adjacent ultrasound planes revealed this pattern of gene expression to persist across the tumor mass (Fig. 4E, Fig. S9).
The ultrasound-observed spatial distribution of gene expression was consistent with the low vascularity in the tumor core, as observed with Doppler ultrasound (Fig. S10). The peripheral gene expression pattern was confirmed with subsequent histological examination of the tissue (Fig. 4F, Fig. S11). In comparison, our in vivo fluorescence images just showed the presence of signal somewhere in the tissue and not its precise distribution (Fig. 4G). These results, which were consistent across 5 animals (Fig. S12A), demonstrate that mARGs enable gene expression imaging in vivo and highlight the ability of ultrasound to visualize intricate patterns of gene expression non-invasively. We imaged 3 of the animals again after an additional 4 days to look for re-expression of the collapsed gas vesicles, and observed ultrasound contrast in each case (Fig. S12B).
Our results establish the ability of an engineered genetic construct encoding prokaryote-derived gas vesicles to serve as a mammalian reporter gene for ultrasound, providing the ability to monitor cellular location and function inside living organisms. mARGs provide many of the capabilities associated with established genetically encoded optical reporters, including imaging cellular dynamics via promoter-driven expression and mapping cellular populations in complex samples. While optical reporter genes mainly provide these capabilities in culture and surgically accessed tissues, mARGs enable gene expression to be resolved non-invasively in vivo.
While the genetic constructs described in this work should be immediately useful in a variety of contexts, significant scope exists for further optimization to make acoustic reporter genes as widely useful as GFP (5, 12). For example, accelerating mARG expression beyond the day-scale kinetics shown in this study and developing sensitive imaging paradigms that do not require gas vesicle collapse would enable the imaging of more dynamic cellular processes. In addition, while this study demonstrated essential mARG functionality with clonally selected cell lines, the expression of mARGs in primary cells, their delivery to endogenous cells via viral vectors, and their expression in transgenic animals would greatly expand the utility of this technology. To facilitate such uses, it would be helpful to further condense the mARG constructs. For example, genes could be consolidated into fewer clusters, and preliminary experiments show that gvpB can be combined with the 8-gene polycistron encoding gvpN-gvpU via an internal ribosome entry sequence (IRES) (Fig. S13). In addition, the total length of the coding sequence contained in mARG could be reduced from 7.6 kb to 4.8 kb by eliminating the need for redundant booster genes, relying instead on non-coding elements such as different-strength promoters to tune expression stoichiometry. Further optimization of mARG genetic constructs is also needed to reduce epigenetic silencing and metabolic burden (27–29). Just as the engineering of GFP over many years yielded brighter and more colorful reporters enabling new uses of fluorescence microscopy, further engineering of the genetic constructs comprising mARGs would help cellular ultrasound penetrate and enable new areas of mammalian biology and biomedicine.
Supplementary Material
Acknowledgments:
The authors thank David Maresca, Bill Ling and Avinoam Bar-Zion for help with ultrasound imaging, Noushin Koulena for assistance with tissue histology, Mohamad Abedi, Justin Lee and Mei Yi You for assistance with Flow Cytometer experiments, Andres Collazo for confocal microscopy, Chris Buser and Oak Crest Institute of Science for cell sectioning and staining, Erik Jue and William Chour for assistance with initial experiments. Electron microscopy was performed at the Beckman Institute Resource Center for Transmission Electron Microscopy at Caltech. Fluorescence imaging of tissues was performed in the Biological Imaging Facility of the Caltech Beckman Institute with support from the Arnold and Mabel Beckman Foundation. We appreciate the help of Audrey Lee-Gosselin and Caltech’s Office of Laboratory Animal Research with animal protocols and husbandry.
Funding: Arash Farhadi was supported by the NSERC graduate fellowship. Daniel Sawyer was supported by the NSF graduate research fellowship (Grant No. 1745301). This research was supported by the National Institutes of Health (grant numbers R01EB018975 and U54CA199090 to M.G.S.), the Heritage Medical Research Institute (M.G.S.), the Packard Fellowship for Science and Engineering (M.G.S.), and the Burroughs Welcome Fund Career Award at the Scientific Interface (M.G.S.).
Footnotes
Competing interests: The authors declare no competing financial interests. The authors are inventors on patent applications related to acoustic reporter genes filed by the California Institute of Technology.
Data and materials availability: The mARG genetic construct will be deposited on Addgene at the time of manuscript publication. Raw data is available from the authors upon reasonable request.
References and Notes:
- 1.Tsien RY, Imagining imaging’s future. Nature Reviews Molecular Cell Biology 4, SS16–SS21 (2003). [PubMed] [Google Scholar]
- 2.Piraner DI et al. , Going Deeper: Biomolecular Tools for Acoustic and Magnetic Imaging and Control of Cellular Function. Biochemistry 56, 5202–5209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu J et al. , A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat Biotech 34, 760–767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos EB et al. , Sensitive in vivo imaging of T cells using a membrane-bound Gaussia princeps luciferase. Nat Med 15, 338–344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maresca D et al. , Biomolecular Ultrasound and Sonogenetics. Annu Rev Chem Biomol Eng 9, 229–252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeifer F, Distribution, formation and regulation of gas vesicles. Nat Rev Microbiol 10, 705–715 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Walsby AE, Gas vesicles. Microbiol Rev 58, 94–144 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro MG et al. , Biogenic gas nanostructures as ultrasonic molecular reporters. Nature Nanotechnology 9, 311–316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lakshmanan A et al. , Molecular Engineering of Acoustic Protein Nanostructures. ACS Nano 10, 7314–7322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maresca D et al. , Nonlinear ultrasound imaging of nanoscale acoustic biomolecules. Appl Phys Lett 110, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maresca D, Sawyer DP, Renaud G, Lee-Gosselin A, Shapiro MG, Nonlinear X-Wave Ultrasound Imaging of Acoustic Biomolecules. Phys Rev X 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu GJ, Farhadi A, Mukherjee A, Shapiro MG, Proteins, air and water: reporter genes for ultrasound and magnetic resonance imaging. Curr Opin Chem Biol 45, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourdeau RW et al. , Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature 553, 86–90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis MM, Tato CM, Furman D, Systems immunology: just getting started. Nat Immunol 18, 725–732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marblestone AH et al. , Physical principles for scalable neural recording. Front Comput Neurosci 7, 137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeder T, Imaging stem-cell-driven regeneration in mammals. Nature 453, 345–351 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Gradinaru V et al. , Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shieh YW et al. , Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350, 678–680 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Natan E, Wells JN, Teichmann SA, Marsh JA, Regulation, evolution and consequences of cotranslational protein complex assembly. Curr Opin Struct Biol 42, 90–97 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Close DM et al. , Autonomous bioluminescent expression of the bacterial luciferase gene cassette (lux) in a mammalian cell line. PLoS One 5, e12441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farhadi A et al. , Recombinantly Expressed Gas Vesicles as Nanoscale Contrast Agents for Ultrasound and Hyperpolarized MRI. AIChE J 64, 2927–2933 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szymczak AL, Vignali DAA, Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opin Biol Th 5, 627–638 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Ding S et al. , Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Wilson MH, Coates CJ, George AL Jr., PiggyBac transposon-mediated gene transfer in human cells. Mol Ther 15, 139–145 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Elowitz MB, Leibler S, A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Gardner TS, Cantor CR, Collins JJ, Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Gaidukov L et al. , A multi-landing pad DNA integration platform for mammalian cell engineering. Nucleic Acids Res 46, 4072–4086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jusiak B et al. , Comparison of Integrases Identifies Bxb1-GA Mutant as the Most Efficient Site-Specific Integrase System in Mammalian Cells. ACS Synth Biol 8, 16–24 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Neville JJ, Orlando J, Mann K, McCloskey B, Antoniou MN, Ubiquitous Chromatin-opening Elements (UCOEs): Applications in biomanufacturing and gene therapy. Biotechnol Adv 35, 557–564 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Schindelin J et al. , Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demene C et al. , Spatiotemporal Clutter Filtering of Ultrafast Ultrasound Data Highly Increases Doppler and fUltrasound Sensitivity. IEEE Trans Med Imaging 34, 2271–2285 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.