Abstract
Due to the public health importance of flagellar genes for typing, it is important to understand mechanisms that could alter their expression or presence. Phenotypic novelty in flagellar genes arise predominately through accumulation of mutations but horizontal transfer is known to occur. A linear plasmid termed pBSSB1 previously identified in Salmonella Typhi, was found to encode a flagellar operon that can mediate phase variation, which results in the rare z66 flagella phenotype. The identification and tracking of homologs of pBSSB1 is limited because it falls outside the normal replicon typing schemes for plasmids. Here we report the generation of nine new pBSSB1-family sequences using Illumina and Nanopore sequence data. Homologs of pBSSB1 were identified in 154 genomes representing 25 distinct serotypes from 67,758 Salmonella public genomes. Pangenome analysis of pBSSB1-family contigs was performed using roary and we identified three core genes amenable to a minimal pMLST scheme. Population structure analysis based on the newly developed pMLST scheme identified three major lineages representing 35 sequence types, and the distribution of these sequence types was found to span multiple serovars across the globe. This in silico pMLST scheme has shown utility in tracking and subtyping pBSSB1-family plasmids and it has been incorporated into the plasmid MLST database under the name “pBSSB1-family”.
Introduction
Serotyping is the current standard for classification of Salmonella isolates according to the reaction of antisera against the surface lipopolysaccharide layer (LPS) (O antigen) and flagellar (H antigens) [1–3]. Based on the combination of antigens and biochemical characteristics an isolate is categorized into a serotype according to the White-Kauffman Le Minor (WKL) scheme [1–3]. The rfb locus is important in determining the LPS layer phenotype but there is a complex genetic basis for O antigen phenotypes [4,5]. The majority of Salmonella serovars possess two chromosomally encoded flagellar genes termed fliC and fljB that encode the H antigens. These flagellar proteins are alternately expressed as cells undergoing phase changes switch between transcription of the two genes [6]. Phenotypic novelty in these important cellular components arise predominately through accumulation of mutations but horizontal gene transfer (HGT) is known to occur [4,7–9]. An example of HGT affecting serologically important phenotypes is the plasmid mediated O antigen changes in the rare Salmonella serotypes Crossness and Borreze [10,11]. Flagellar antigens have also been documented as being affected by HGT such as the case of Salmonella Typhi which normally expresses either the d or j flagella antigen [12,13] but a rare plasmid-borne variant expressing the z66 antigen exists [14]. The novel z66 flagellar gene was localized to a linear plasmid termed pBSSB1, which was able to mediate phase variation despite not being localized in the chromosome through silencing of the chromosomal fliC by expression of a plasmid encoded fljA[15].
Whole genome sequencing (WGS) is revolutionizing the field of public health and it is replacing traditional serological testing as the primary diagnostic test for Salmonella and other pathogens [16]. WGS provides an extraordinary level of discrimination of isolates, allows multiple tests to be run on the same data and provides a rich resource for the research community to answer novel questions which are not within the scope of traditional surveillance [17–19]. However, the existing surveillance systems and historical data are dependent on serotype information and in order to maintain a connection to this important data, multiple tools have been developed for the purposes of predicting serotype based on sequence data [1,20]. The Salmonella in silico Typing Resource (SISTR) identifies the genetic determinants for the O and H antigens from draft genome assemblies and uses 330 core gene to predict serotype with a high degree of accuracy [1,16]. Presence of plasmid-encoded alleles of flagellar or O-antigen genes can confound WGS-based prediction of serotypes as these schemes currently do not account for the presence of multiple alleles of these genes.
Linear plasmids are extremely rare in Enterobacteraceae [15] and pBSSB1 is the only case described in Salmonella. Typing of plasmids is traditionally based on replicon incompatibility where plasmids are grouped based on the ability to be stably maintained in a cell [21]. The identification and tracking of this linear plasmid in bacterial populations is limited since pBSSB1 replicates through a different mechanism from the circular plasmids normally occurring in Enterobacteraceae and due to its linear nature does not possess a relaxase; so, it falls outside the existing typing schemes for plasmids currently in use. Plasmid Multi-locus sequence typing (pMLST) is a technique for categorizing genetic diversity through assigning unique numeric identifiers for alleles of a set of genes which define the scheme [22]. Traditional MLST schemes are based on a small subset of genes but the approach can be extended to any number of genes [1,23–25]. pMLST schemes have been developed for IncA/C, IncH, IncI and IncN replicon families, which facilitates the tracking of these plasmids through populations [26–29].
To date pBSSB1 had only been reported in Salmonella Typhi isolates from Indonesia presenting a z66 phenotype [14,15,30]. Here we present an in sillico pMLST typing scheme for the pBSSB1 plasmid backbone and information on the broad distribution of this plasmid in Salmonella. Based on phylogenetic analyses of the flagella and plasmid sequences, we have found evidence to support potential interspecies transfer of an intact flagellar operon from Citrobacter to Salmonella, which has implications for serology-based identification of Salmonella.
Materials and methods
DNA preparation and sequencing
The OIE Reference Laboratory for Salmonellosis performed phenotypic serotyping according to accredited procedures. Genomic DNA was extracted using the Qiagen EZ1 robotic extraction system according to manufacturer’s instructions. DNA concentration was measured using the Invitrogen Qubit™ system, and quality of the DNA template was evaluated using the Agilent TapeStation™. Illumina MiSeq sequencing libraries were prepared using the NexteraXT kit according to the manufacturer’s protocol for 600-cycle sequencing. Nanopore sequencing was performed using the RAD002 or RBK004 rapid library preparation kit according to the manufacturer’s instructions on a R9.4 flow cell. Raw sequence data generated from this study was deposited into NCBI and the accession numbers are listed in S1 Table.
Genome assembly
Hybrid assembly using MiSeq and Nanopore reads was performed using Unicycler v. 0.4.5 with the default parameters [31]. Each assembly was examined to confirm that every component was closed and circularized with the exception of the pBSSB1 plasmid. The terminal inverted repeats flanking pBSSB1-family plasmids were found to be difficult to assemble due to low sequencing coverage of the ends and the collapsing of repeats and assignment to either the 5’ or 3’ end of the plasmid (data not shown). This issue was not resolved by using Canu v. 1.8 [32], so the ends of the plasmids are likely incomplete. Each assembly was iteratively polished with Racon v 1.3.2 (https://github.com/isovic/racon) and Pilon v. 1.23 (https://github.com/broadinstitute/pilon) until no changes were made to the assembly. Unicycler with the default parameters was used to assemble publicly available MiSeq data for other isolates where long reads were unavailable in order to minimize variability due to differences in assembly procedure.
In silico analysis of pBSSB1
Previously, we assembled 67,758 Salmonella genomes from the SRA [33] and each of these assemblies was checked for the presence of plasmids homologous to pBSSB1 (referred to hereafter as “pBSSB1-family plasmids”) using MOB-recon which can detect plasmid contigs through an ensemble approach which utilizes the presence of defined replicon and relaxase biomarkers as well as contig coverage of reference plasmid sequences [34]. The Salmonella in silico typing resource SISTR [1] was used to predict the serotype of each Salmonella assembly found to contain a pBSSB1 homolog. Serotypes for E. coli genomes were predicted using ECTyper v. 0.81 (https://github.com/phac-nml/ecoli_serotyping). MOB-recon reconstructed plasmids were annotated using Prokka v. 1.19 [35] and pangenome analyses were performed using Roary v. 3.12.0 with the identity threshold relaxed to 90% for core genes [36]. A multiple sequence alignment for each gene was constructed using MAFFT v. 7.221 with the auto flag enabled [37]. Tajima’s D statistic was calculated for each multiple sequence alignment using MEGA 7 with all three codon positions used [38]. A maximum likelihood tree was generated for the concatenated multiple sequence alignments for each ST using MEGA 7 with the following parameters (100 bootstraps, Kimura 2-parameter model, gamma distributed rate, all coding positions). Population structure of the Salmonella isolates was visualized using GrapeTree with the Enterobase cgMLST scheme [25,39]. pMLST allele calls were extracted using the MLST tool (https://github.com/tseemann/mlst) using the S. enterica or pBSSB1 schema based on the three genes soj, higB and mqsA.
In silico flagellar gene analyses
Prokka 1.19 [35] was run on the sequences of pBSSB1-family plasmids which had been reconstructed using MOB-recon v. 1.4.8 [34] and genes annotated as “Flagellin” were selected for further analyses. Identical and truncated subsequences were identified using cd-hit-est [40] using an identity threshold of 1. The resulting unique set of sequences was subject to clustering in a second round with cd-hit-est using a threshold of 0.9 to identify any similar flagella alleles.
Results
Closed pBSSB1-family plasmid analysis
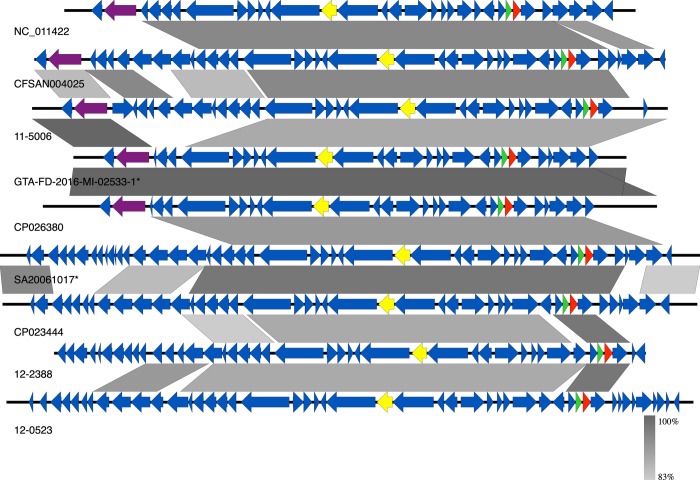
Long read sequencing using Nanopore was performed on nine Salmonella isolates found to contain a pBSSB1-family plasmid based on their Illumina sequence data. These newly closed plasmid genomes were analyzed along with three pBSSB1-like sequences from NCBI (NC_011422: Salmonella Typhi, CP026380: Salmonella Senftenberg, CP023444: Klebsiella pneumoniae) which were the only hits obtained by using the newly generated sequences as queries to BLASTn. The accessions for all newly generated sequences are available in S1 Table. The closed pBSSB1-family plasmids ranged in size from 26kb to 33Kb with an average GC% of 36%. Pangenome analysis using Roary estimated a core genome of 14 genes (Table 1). Gene synteny was visualized for the closed plasmids using EasyFig with the following blast parameters (evalue < = 1e-8, length> = 1500bp, identity > = 75%) [41] (Fig 1). Overall, there is a conserved central core region of the plasmid but the ends of the plasmids carry significantly different sequence content. Only six out of the 12 plasmids contained a flagella gene (Fig 1). The plasmids from isolates SA20061017 and SA20130280 are nearly identical across their length. The sequence CP026380 clusters tightly with our newly generated sequences 11–5006 and GTA-FD-2016-MI-02533-1 to GTA-FD-2016-MI-02533-3.
Table 1. Core genes from closed pBSSB1-family plasmid sequences were tested for selection using Tajima’s D statistic using MEGA 7.
| Gene | Annotation | Average Length (bp) | Number of Alleles | m | S | ps | Θ | π | D |
|---|---|---|---|---|---|---|---|---|---|
| group_13 | hypothetical protein | 410 | 6 | 12 | 47 | 0.11 | 0.04 | 0.05 | 0.91 |
| group_7 | hypothetical protein | 742 | 6 | 12 | 68 | 0.09 | 0.03 | 0.03 | 0.57 |
| soj | Chromosome-partitioning ATPase Soj | 626 | 6 | 12 | 126 | 0.2 | 0.07 | 0.08 | 1.29 |
| group_14 | hypothetical protein | 332 | 7 | 12 | 33 | 0.11 | 0.04 | 0.04 | 0.09 |
| mqsA | Antitoxin MqsA | 290 | 7 | 12 | 15 | 0.05 | 0.02 | 0.02 | -0.36 |
| group_1 | hypothetical protein | 695 | 8 | 12 | 85 | 0.13 | 0.04 | 0.04 | 0.17 |
| group_10 | hypothetical protein | 2333 | 8 | 12 | 362 | 0.16 | 0.05 | 0.06 | 0.7 |
| group_2 | hypothetical protein | 1121 | 8 | 12 | 143 | 0.13 | 0.04 | 0.05 | 0.5 |
| group_32 | hypothetical protein | 305 | 8 | 12 | 29 | 0.09 | 0.03 | 0.03 | 0.27 |
| group_33 | hypothetical protein | 344 | 8 | 12 | 29 | 0.09 | 0.03 | 0.03 | 0.27 |
| group_44 | hypothetical protein | 374 | 8 | 12 | 18 | 0.05 | 0.02 | 0.02 | 0.06 |
| group_8 | hypothetical protein | 254 | 8 | 12 | 32 | 0.13 | 0.04 | 0.04 | 0 |
| higB-2 | Toxin HigB-2 | 353 | 8 | 12 | 14 | 0.04 | 0.01 | 0.01 | 0.06 |
| traC | DNA primase TraC | 1099 | 8 | 12 | 57 | 0.08 | 0.03 | 0.03 | 0.82 |
m = number of sequences, n = total number of sites, S = Number of segregating sites, ps = S/n, Θ = ps/a1, π = nucleotide diversity, and D is the Tajima test statistic.
Fig 1. The sequence conservation for closed pBSSB1-family plasmids was visualized using EasyFig.
Boxed arrows represent the position and transcriptional direction of ORFs. Shaded grey areas indicate conserved blocks with an evalue > = 1e-8. The locations of flagella genes are highlighted in purple. Genes associated selected for the three pMLST scheme are highlighted in yellow (soj), green (higB), red (mqsA). Sequences with an asterisk indicate multiple samples with nearly identical sequences with a representative for that group: (SA20061017, SA20130280) and (GTA-FD-2016-MI-02533-1 to GTA-FD-2016-MI-02533-3).
Development of a pBSSB1-family plasmid pMLST scheme
In order to facilitate tracking of different lineages of the pBSSB1-family plasmid backbone, we developed a minimal pMLST scheme based on its plasmid sequences. The distinct number of alleles for each of the core genes was determined and is listed in Table 1. Nine of the genes had 8 alleles with the remaining genes having either 6 or 7 alleles. Each of 14 core genes was tested for neutral evolution using Tajima’s D test in MEGA v. 7 (Table 1). None of the genes showed strong evidence for selection with soj showing the highest deviation from neutral with a Tajima’s D of 1.2 (Table 1). Since no significant selective pressure was observed for the core genes, all of them were considered viable pMLST candidates. We identified three genes, which were good candidates for use as typing markers. We selected the sporulation inhibition homolog soj, along with the bacterial toxin/antitoxin (TA) genes higB and mqsA. The gene set resulted in 8 pMLST profiles for the 12 closed plasmid sequences. Genes that contained multiple indels were excluded as candidates for pMLST marker genes. The developed scheme has been deposited into pubMLST (https://pubmlst.org/plasmid/) under the name “pBSSB1-family” using the BIGSdb platform [42,43]. The selected genes for the MLST scheme are highly specific to pBSSB1 since BLASTn (September 2019) against the NCBI nucleotide database only obtained hits from pBSSB1 sequences associated with Salmonella isolates.
Distribution of pBSSB1-family plasmids
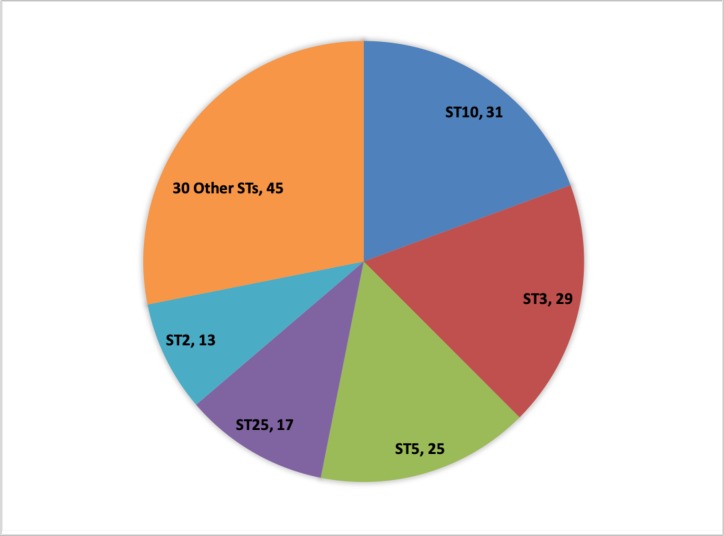
A total of 154 Salmonella genomes out of the 67,758 SRA genomes were found to contain pBSSB1-family plasmids based on the results of MOB-recon. Each of these positive isolates was typed according to the S. enterica MLST scheme and then with the newly developed scheme for pBSSB1-family plasmids (S2 Table). A total of 35 pBSSB1-family sequence types were identified in the dataset with five sequence types accounting for 75% of the pBSSB1-family plasmids (Fig 2). A minimum spanning tree based on the Enterobase cgMLST scheme was constructed using GrapeTree and overlaid with the pBSSB1-family sequence type to determine if the predominant sequence types were due to repeated samples from genetically similar members of a serovar (Fig 3).
Fig 2. Pie chart indicating the pMLST sequence type composition of identified pBSSB1-family STs in Salmonella.
Counts of each sequence type are listed in each slice.
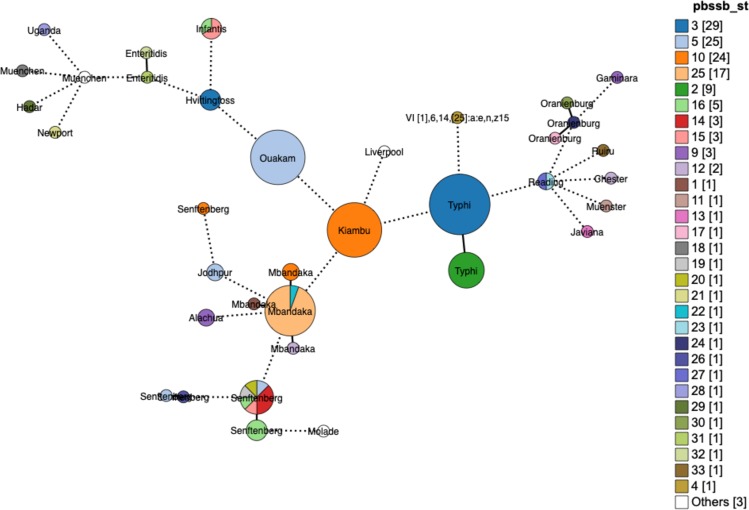
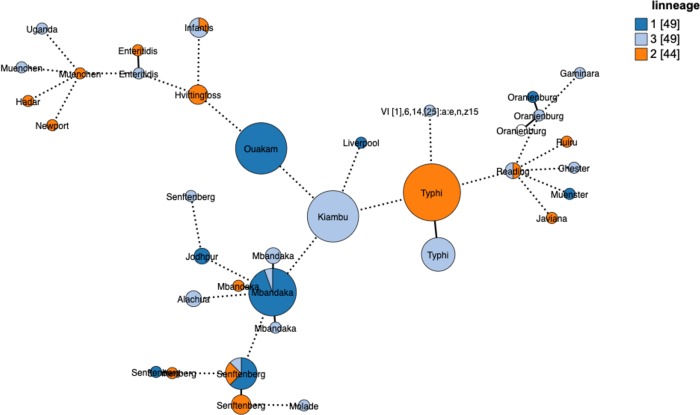
Fig 3. GrapeTree minimum-spanning tree based on the Enterobase cgMLST and colored based on the pBSSB1 sequence type present in the genome.
Nodes differing by fewer than 50 alleles were collapsed together and branches longer than 500 alleles different were shortened and are indicated with a hashed line. Size of the nodes indicates the number of samples contained in them.
The pBSSB1-family pMLST Sequence Type 10 (ST 10) primarily consists of serovar Kiambu isolates belonging to a single cluster (Fig 3), which is indicative of repeated sampling of closely related isolates. This pattern is consistent for the remaining isolates of ST 10 within different serotypes Mbandaka and Senftenberg (Fig 3). A single cluster of Typhi isolates account for the majority of ST 3 isolates with a small cluster of Hvittingfoss accounting for the remaining three isolates (Fig 3). A separate cluster of Typhi contains z66-positive ST 2, which indicates that not all pBSSB1 homologues in Typhi carry the z66 flagella (Fig 3). A cluster of Ouakam contains the majority of ST 5, with isolates of Jodhpur and Senftenberg containing the others (Fig 3). Infantis, Reading and Senftenberg are interesting cases because single clusters contain multiple pBSSB1-family sequence types (Fig 3).
Population structure of pBSSB1-family plasmids
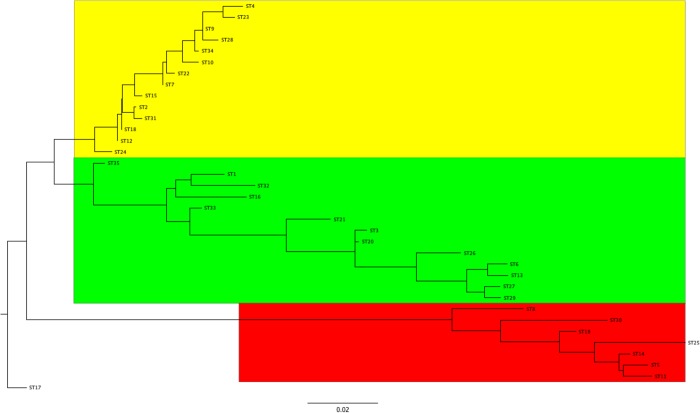
A maximum likelihood tree based on the concatenated pMLST gene sequences for each of the pBSSB1-family sequence types identified three major clades (Fig 4). Both clades 1 and 2 contain considerable sequence divergence, which is in contrast to clade 3 where the sequences form a tighter association. When the lineage information of pBSSB1-family plasmids is overlaid on the Salmonella population structure, there is evidence for both clonal expansion and horizontal transfer of lineages (Fig 5). Each of the three different lineages are distributed across diverse serotypes (Fig 5). The two clusters of Typhi contain either lineage 1 or 2 exclusively (Fig 5). This is in contrast to Mbandaka, Senftenberg, Infantis and Reading where there are multi-lineage clusters occurring (Fig 5). These results are consistent with repeated introductions of divergent plasmids into these serovars rather than spread and diversification of a single plasmid.
Fig 4. Maximum likelihood phylogenetic analysis of pBSSB1-family plasmids using concatenated sequences of the MLST genes soj, mqsA, higB.
The sequence types have been divided into three major clades coloured in red (1), green (2) and yellow (3).
Fig 5. GrapeTree minimum-spanning tree based on the Enterobase cgMLST and coloured based on the pBSSB1-family lineages present in the genome.
Nodes differing by fewer than 50 alleles were collapsed together and branches longer than 500 alleles different were shortened and are indicated with a hashed line. Size of the nodes indicates the number of samples contained in them.
Plasmid mediated flagellar genes
Due to the presence of an intact fliC operon in some members of the pBSSB1-family, we examined the flagella sequences in detail to ascertain their similarity to other known Enterobacteracea flagella sequences. Flagellar genes were found in 104 of the 154 pBSSB1-family plasmids, which are distributed in 15 pBSSB1 STs and in all three lineages (S2 Table). There are total of 13 distinct flagella alleles including z66 from Typhi, which forms four clusters using cd-hit-est with a 0.9 threshold for identity. Web-based BLASTn searches were performed using each of the allele sequences against the NCBI nucleotide database to identify possible sources of the flagellar genes (Table 2). Flagella cluster 1 and 2 both had their top hit as Citrobacter portucalensis (CP012554) but cluster 1 had much higher identity with 99.37% compared to 78.76% for cluster 2 (Table 2). The downstream fljA sequence was also present in CP012554 at 100% coverage and 97% identity for both cluster 1 and 2. Our samples 11–5006 and GTA-FD-2016-MI-02533-1 to GTA-FD-2016-MI-02533-3 belong to the flagella cluster 1 and our phenotypic serotyping results identified the z35 antigen but were unable to detect the normal g,[s],t flagella expression. This indicates that the genes encoding flagella on the identified pBSSB1-family plasmids are functional and these plasmid-encoded alleles are dominant relative to chromosomally-encoded flagellar genes and their presence masks the detection of the endogenous flagella. Sequences from cluster 1 share very little similarity with other z35 flagella in Salmonella, which is suggestive that there is cross-reactivity within the z35 antisera. Cluster 3 matched to the pBSSB1 plasmid NC_011422 from Salmonella Typhi and so represents the z66 flagella (Table 2). The fourth cluster matches with a chromosomal C. freundii flagella but overall had only 61% coverage and 84% identity (Table 2).
Table 2. Blast result summary from NCBI web-blast using a single representative per flagella sequence cluster.
| Allele | Representative | Length | Closest NCBI Hit | Hit Species | Total Score | Query Coverage (%) |
E-value | Percent Identity (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | SRR3606556 | 1578 | CP012554 | C. portucalensis | 3337 | 100 | 0 | 99.37 |
| 2 | SRR3372244 | 1572 | CP012554 | C. portucalensis | 1803 | 100 | 0 | 78.76 |
| 3 | ERR1764822 | 1527 | NC_011422 | S. Typhi | 2809 | 100 | 0 | 100 |
| 4 | SRR3210535 | 1341 | CP037734 | C. freundii | 873 | 61 | 1e-150 | 84.57 |
Discussion
Given the importance of classification of Salmonella into serotypes, it is critical to characterize and understand the mechanisms, which generate novel antigenic combinations. The presence of variants of Salmonella Typhi containing a novel flagellar gene has been known since the 1980s [44], and in 2007 the linear plasmid pBSSB1 containing the z66 fliC was described [15]. The plasmid pBSSB1 represents the only known vector for transferring an intact flagella operon in Salmonella and, based on the available data, it was only known to occur in Typhi isolates originating from some parts of Indonesia [15]. This work represents the first description of pBSSB1 in diverse serovars and geographic locations. Analysis of 67,758 publicly available genomes from a previous study [33] shows that the plasmid is in fact globally distributed and present in a variety of serotypes (Fig 2). The wide distribution of pBSSB1-family in a variety of serotypes and species indicates that this plasmid backbone could contribute to the generation of novel flagellar phenotypes through inter-species transfer. The transfer of this plasmid is known to be dominantly expressed over the endogenous fliC, which can result in incomplete typing of isolates by phenotypic methods [15]. This is of concern to public health since serotype information is a critical piece of outbreak detection and response.
The circulating pBSSB1-family plasmids identified in this study represent diverse lineages rather than clonal spread of a single plasmid backbone (Fig 2). The analysis using GrapeTree based on the Enterobase [25] cgMLST scheme overlaid with pBSSB1-family ST information, highlights that there has been repeated sampling of closely related isolates within serotypes (Fig 3). Senftenberg is notable since within cgMLST clusters there exist multiple pBSSB1-family sequence types (Fig 3). These results support the hypotheses that there were multiple independent acquisitions of the plasmid within this serotype. Estimates of the frequency of pBSSB1 homologues in Salmonella as a whole based on the SRA data should be undertaken with caution since the dataset is heavily biased towards repeated sampling of outbreaks and human clinical cases. However, given that pBSSB1 homologues were found in less than 0.3% of samples it is suggestive that it is not common within Salmonella of clinical relevance.
Conclusion
This is the first documentation of plasmids similar to pBSSB1 outside of Indonesian Salmonella Typhi and provides evidence for global distribution. These results are of consequence to public health since serological classification of Salmonella is still the global standard and plasmids belonging to the pBSSB1-family can be vectors that can alter the flagellar phenotype of an isolate. These classification issues will still be present even after the public health reference laboratory community switches to WGS since serotype information remains critically important for investigations and reporting. The development of a pBSSB1-family pMLST will aid in the tracking of these plasmids through different bacterial populations.
Supporting information
(XLSX)
(XLSX)
Acknowledgments
We thank our colleagues within the National Microbiology Laboratory’s Reference Services Laboratory and the OIE Salmonella Reference Laboratory within the Division of Enteric Diseases for their assistance with phenotypic testing of the isolates. In addition, we would like to thank Paul Manninger for preforming WGS of some of the samples, Andrew Low for bioinformatics support, as well as Adam Koziol and Moe Elmufti for their comments and critiques during the review process. We also would like to thank the Food and Drug Administration, Center For Food Safety And Applied Nutrition (CFSAN) for providing the isolate of CFSAN004025. Finally, we would like to thank Marc Stevens and Dr. Roger Stephan from Institute of Food Safety, University of Zurich who provided the raw PacBio data for CP026380.
Abbreviations
- cgMLST
core gene multi-locus sequence typing
- MLST
multi-locus sequence typing
- ST
sequence type
- WGS
whole genome sequencing
- WKL
White-Kauffman Le Minor serotyping scheme
Data Availability
Raw sequence data and assemblies for newly sequenced isolates have been deposited into NCBI accession numbers: SAMN06030139; SAMN06030177; SAMN06030204; SAMN11029488; SAMN11029489; SAMN10501886; SAMN10501887; SAMN10501888; SAMN02352702
Funding Statement
This work was funded by the Public Health Agency of Canada and the Government of Canada’s Genomics R&D Initiative Phase VI Shared Priority Project Management Plan on Antimicrobial Resistance.
References
- 1.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, et al. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLOS ONE. 2016. January 22;11(1):e0147101 10.1371/journal.pone.0147101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franklin K, Lingohr EJ, Yoshida C, Anjum M, Bodrossy L, Clark CG, et al. Rapid Genoserotyping Tool for Classification of Salmonella Serovars▿. J Clin Microbiol. 2011. August;49(8):2954–65. 10.1128/JCM.02347-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida C, Gurnik S, Ahmad A, Blimkie T, Murphy SA, Kropinski AM, et al. Evaluation of molecular methods for the identification of Salmonella serovars. J Clin Microbiol. 2016. May 18;JCM.00262-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broadbent SE, Davies MR, van der Woude MW. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol Microbiol. 2010. July;77(2):337–53. 10.1111/j.1365-2958.2010.07203.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnaitman CA, Klena JD. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993. September;57(3):655–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman M, Zieg J, Hilmen M, Simon M. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc Natl Acad Sci U S A. 1979. January;76(1):391–5. 10.1073/pnas.76.1.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltran P, Musser JM, Helmuth R, Farmer JJ, Frerichs WM, Wachsmuth IK, et al. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci. 1988. October 1;85(20):7753–7. 10.1073/pnas.85.20.7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kropinski AM, Kovalyova IV, Billington SJ, Patrick AN, Butts BD, Guichard JA, et al. The Genome of ε15, a Serotype-Converting, Group E1 Salmonella enterica-Specific Bacteriophage. Virology. 2007. December 20;369(2):234–44. 10.1016/j.virol.2007.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright A. Mechanism of Conversion of the Salmonella O Antigen by Bacteriophage ε34. J Bacteriol. 1971. March;105(3):927–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keenleyside WJ, Whitfield C. A Novel Pathway for O-Polysaccharide Biosynthesis in Salmonella enterica Serovar Borreze. J Biol Chem. 1996. November 8;271(45):28581–92. 10.1074/jbc.271.45.28581 [DOI] [PubMed] [Google Scholar]
- 11.Rowe B, Hall ML, McCoy JH. Salmonella crossness—a new serotype containing a new comatic (O) antigen, 67. J Hyg (Lond). 1976. December;77(3):355–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everest P, Wain J, Roberts M, Rook G, Dougan G. The molecular mechanisms of severe typhoid fever. Trends Microbiol. 2001. July;9(7):316–20. 10.1016/s0966-842x(01)02067-4 [DOI] [PubMed] [Google Scholar]
- 13.Kidgell C, Reichard U, Wain J, Linz B, Torpdahl M, Dougan G, et al. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect Genet Evol. 2002. October;2(1):39–45. 10.1016/s1567-1348(02)00089-8 [DOI] [PubMed] [Google Scholar]
- 14.Pa G, Wh J, Hm M, L LM, R B. An unusual H antigen (Z66) in strains of Salmonella typhi. Ann Microbiol (Paris). 1980 1981;132(3):331–4. [PubMed] [Google Scholar]
- 15.Baker S, Hardy J, Sanderson KE, Quail M, Goodhead I, Kingsley RA, et al. A Novel Linear Plasmid Mediates Flagellar Variation in Salmonella Typhi. PLOS Pathog. 2007. May 11;3(5):e59 10.1371/journal.ppat.0030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yachison CA, Yoshida C, Robertson J, Nash JHE, Kruczkiewicz P, Taboada EN, et al. The Validation and Implications of Using Whole Genome Sequencing as a Replacement for Traditional Serotyping for a National Salmonella Reference Laboratory. Front Microbiol [Internet]. 2017. [cited 2017 Jul 17];8 Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01044/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair S, Ashton P, Doumith M, Connell S, Painset A, Mwaigwisya S, et al. WGS for surveillance of antimicrobial resistance: a pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J Antimicrob Chemother. 2016. September 1;dkw318. [DOI] [PubMed] [Google Scholar]
- 18.Nutrition C for FS and A. Whole Genome Sequencing (WGS) Program—GenomeTrakr Fast Facts [Internet]. [cited 2016 Nov 25]. Available from: http://www.fda.gov/Food/FoodScienceResearch/WholeGenomeSequencingProgramWGS/ucm403550.htm
- 19.Wyres KL, Conway TC, Garg S, Queiroz C, Reumann M, Holt K, et al. WGS Analysis and Interpretation in Clinical and Public Health Microbiology Laboratories: What Are the Requirements and How Do Existing Tools Compare? Pathogens. 2014. June 11;3(2):437–58. 10.3390/pathogens3020437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, Yin Y, Jones MB, Zhang Z, Kaiser BLD, Dinsmore BA, et al. Salmonella Serotype Determination Utilizing High-Throughput Genome Sequencing Data. J Clin Microbiol. 2015. May 1;53(5):1685–92. 10.1128/JCM.00323-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014. July;58(7):3895–903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiden MCJ, van Rensburg MJJ, Bray JE, Earle SG, Ford SA, Jolley KA, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013. October;11(10):728–36. 10.1038/nrmicro3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, Sangal V, et al. Multilocus Sequence Typing as a Replacement for Serotyping in Salmonella enterica. PLOS Pathog. 2012. June 21;8(6):e1002776 10.1371/journal.ppat.1002776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Been M de, Pinholt M, Top J, Bletz S, Mellmann A, Schaik W van, et al. Core Genome Multilocus Sequence Typing Scheme for High-Resolution Typing of Enterococcus faecium. J Clin Microbiol. 2015. December 1;53(12):3788–97. 10.1128/JCM.01946-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alikhan N-F, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella. PLOS Genet. 2018. April 5;14(4):e1007261 10.1371/journal.pgen.1007261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancock SJ, Phan M-D, Peters KM, Forde BM, Chong TM, Yin W-F, et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother. 2017;61(2):e01740–16. 10.1128/AAC.01740-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.García-Fernández A, Carattoli A. Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J Antimicrob Chemother. 2010. June;65(6):1155–61. 10.1093/jac/dkq101 [DOI] [PubMed] [Google Scholar]
- 28.García-Fernández A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, et al. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008. June;61(6):1229–33. 10.1093/jac/dkn131 [DOI] [PubMed] [Google Scholar]
- 29.García-Fernández A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, et al. Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother. 2011. September;66(9):1987–91. 10.1093/jac/dkr225 [DOI] [PubMed] [Google Scholar]
- 30.Zhang H, Zhu Y, Xie X, Wang M, Du H, Xu S, et al. Identification and Characterization of a Gene stp17 Located on the Linear Plasmid pBSSB1 as an Enhanced Gene of Growth and Motility in Salmonella enterica Serovar Typhi. Front Cell Infect Microbiol [Internet]. 2016. October 5;6 Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5050219/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017. June 8;13(6):e1005595 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017. March 15;gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robertson J, Yoshida C, Kruczkiewicz P, Nadon C, Nichani A, Taboada EN, et al. Comprehensive assessment of the quality of Salmonella whole genome sequence data available in public sequence databases using the Salmonella in silico Typing Resource (SISTR). Microb Genomics [Internet]. 2018. [cited 2018 Apr 3];4(2). Available from: http://mgen.microbiologyresearch.org/content/journal/mgen/10.1099/mgen.0.000151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genomics. 2018;4(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014. July 15;30(14):2068–9. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 36.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015. November 15;31(22):3691–3. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002. July 15;30(14):3059–66. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018. July 26;gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006. July 1;22(13):1658–9. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 41.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011. April 1;27(7):1009–10. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010. December 10;11(1):595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res [Internet]. 2018. September 24 [cited 2019 Apr 9];3 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6192448/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guinée PA, Jansen WH, Maas HM, Le Minor L, Beaud R. An unusual H antigen (Z66) in strains of Salmonella typhi. Ann Microbiol (Paris). 1981. June;132(3):331–4. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
Raw sequence data and assemblies for newly sequenced isolates have been deposited into NCBI accession numbers: SAMN06030139; SAMN06030177; SAMN06030204; SAMN11029488; SAMN11029489; SAMN10501886; SAMN10501887; SAMN10501888; SAMN02352702